Significance
The morphology of male genitalia evolves rapidly, probably driven by sexual selection. However, little is known about the genes underlying genitalia differences between species. Identifying these genes is key to understanding how sexual selection acts to produce rapid phenotypic change. We have found that the gene tartan underlies differences between male Drosophila mauritiana and Drosophila simulans in the size and bristle number of the claspers—genital projections that grasp the female during copulation. Moreover, since tartan encodes a protein that is involved in cell interactions, this may represent an alternative developmental mechanism for morphological change. Therefore, our study provides insights into the genetic and developmental bases for the rapid evolution of male genitalia and organ size more generally.
Keywords: evolution, development, genetics, Drosophila, genitalia
Abstract
Male genital structures are among the most rapidly evolving morphological traits and are often the only features that can distinguish closely related species. This process is thought to be driven by sexual selection and may reinforce species separation. However, while the genetic bases of many phenotypic differences have been identified, we still lack knowledge about the genes underlying evolutionary differences in male genital organs and organ size more generally. The claspers (surstyli) are periphallic structures that play an important role in copulation in insects. Here, we show that divergence in clasper size and bristle number between Drosophila mauritiana and Drosophila simulans is caused by evolutionary changes in tartan (trn), which encodes a transmembrane leucine-rich repeat domain protein that mediates cell–cell interactions and affinity. There are no fixed amino acid differences in trn between D. mauritiana and D. simulans, but differences in the expression of this gene in developing genitalia suggest that cis-regulatory changes in trn underlie the evolution of clasper morphology in these species. Finally, analyses of reciprocal hemizygotes that are genetically identical, except for the species from which the functional allele of trn originates, determined that the trn allele of D. mauritiana specifies larger claspers with more bristles than the allele of D. simulans. Therefore, we have identified a gene underlying evolutionary change in the size of a male genital organ, which will help to better understand not only the rapid diversification of these structures, but also the regulation and evolution of organ size more broadly.
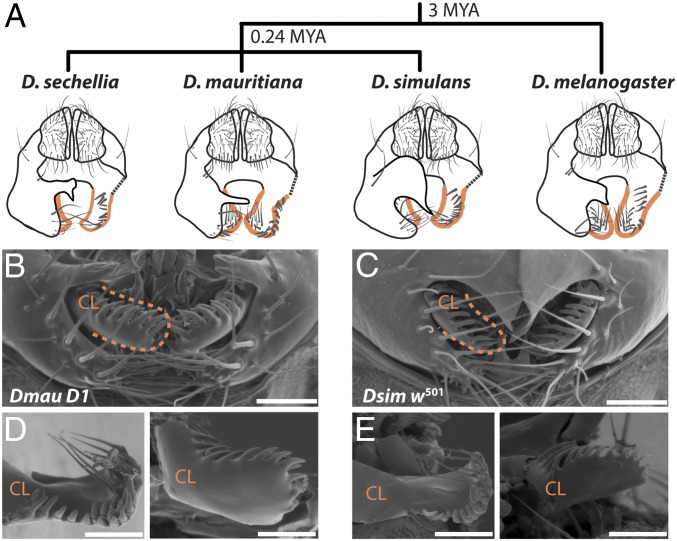
The morphology of male genitalia can differ dramatically even between very closely related animal species (1). In Drosophila mauritiana males, for example, the size, shape, and bristle morphology of the claspers (surstyli), posterior lobes (epandrial posterior lobes), and anal plates (cerci) are strikingly different from those of its sister species Drosophila simulans and Drosophila sechellia (Fig. 1). Moreover, these differences have evolved in only the last 240,000 y since these species last shared a common ancestor (2) (Fig. 1A).
Fig. 1.
Divergence in periphallic structures in the D. simulans clade and its relationship to the outgroup D. melanogaster (2). (A) Schematic representation of the male analia and external genitalia (posterior view). Posterior lobes are illustrated as dissected away on the right-hand side, in order to facilitate visualization of the claspers (outlined in orange), which are typically covered by the posterior lobes. While the shape and size of the posterior lobes is species-specific, the claspers and anal plates are very similar between D. simulans and D. sechellia, which are smaller and have less bristles than those of D. mauritiana and D. melanogaster. In addition, the clasper bristles of D. mauritiana are shorter and thicker than those of the other three species (19, 20, 48). (B–E) Scanning electron micrographs of external male genitalia (B and C) and dissected claspers (D and E) of Dmau D1 and Dsim w501, respectively. (Scale bars, 50 μm.)
As in other animal groups (1, 3–5), interspecific differences in the morphology of genital structures are thought to have been driven by sexual selection (6). However, the mechanisms [female choice, sperm competition, or sexual antagonism (5)], and their contribution to reproductive isolation between populations and species, have been difficult to address and resolve theoretically (7–9) and experimentally (10, 11). Genetic manipulation of the evolved loci would allow us to test directly the effect of male genital divergence on mating behavior and reproductive fitness and, therefore, facilitate the empirical study of these questions (12, 13). Although quantitative mapping studies of morphological differences in male genitalia between species of the D. simulans clade were first carried out more than three decades ago (14–21), the genetic bases of male genital divergence between these species has remained elusive. This is due, at least in part, to the large number of loci found to contribute to variation in size and shape of these structures (18, 19, 21).
The claspers are periphallic structures with an essential role in grasping and proprioception of the female and in securing genital coupling (12, 22–27). Previously, we found that multiple loci contribute to divergence in clasper size and bristle number between D. simulans and D. mauritiana (19). Here, we report the identification of one of these loci, tartan (trn). In addition, mapping and functional experiments strongly suggest that cis-regulatory changes in this gene underlie differences in clasper morphology between these two species.
Results and Discussion
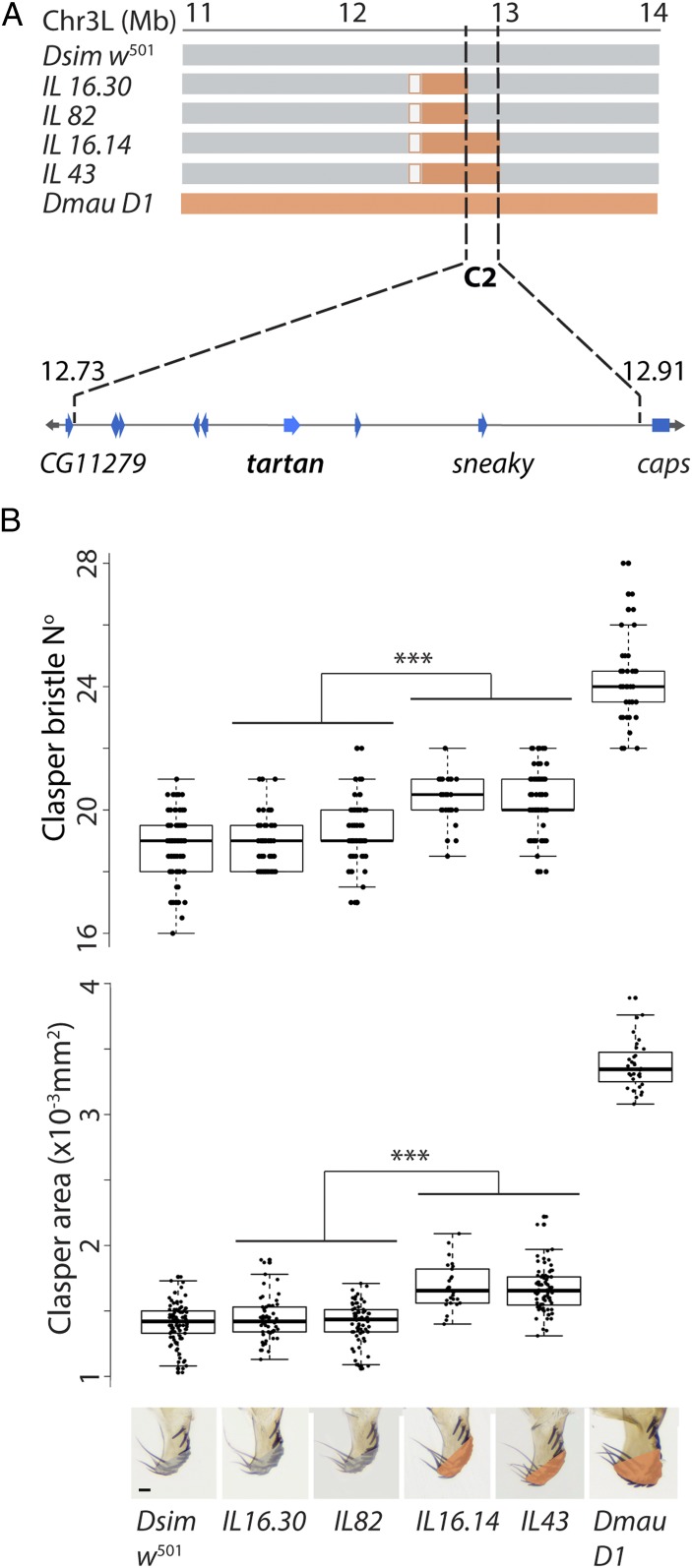
Previously, we identified two regions on the left arm of chromosome 3 that contribute to differences in clasper size and bristle number between D. mauritiana and D. simulans (19). Here, we have generated recombinant introgression lines (ILs) between the D. mauritiana D1 (Dmau D1) and D. simulans w501 (Dsim w501) strains (Dataset S1) to increase the resolution of one of these regions, C2, from ∼3.5 Mb (24) to 177 kb. This interval explains about 16.3% of the difference in clasper size (and 37.9% of clasper bristle number) between the two parental strains (Fig. 2 and Datasets S2 a and b). The claspers of lines that are homozygous for introgressed D. mauritiana DNA in C2 are significantly larger than those of natural strains of D. simulans (P < 0.001, SI Appendix, Fig. S1). The change in clasper size caused by differences in C2 is therefore outside the range of variation in clasper size in D. simulans, suggesting that C2 underlies interspecific divergence between D. mauritiana and D. simulans and not merely intraspecific polymorphism in clasper size in either or both of these species.
Fig. 2.
High-resolution mapping of differences in clasper morphology between Dsim w501 and Dmau D1. (A) Introgression line breakpoints on chromosome arm 3L define the 177-kb region C2 (gray, orange, and white boxes indicate DNA regions from Dsim w501, Dmau D1, or not verified, respectively). Coordinates are given in megabases with respect to the D. simulans genome (Flybase R2.02). This region contains eight protein coding genes including trn and is flanked by CG11279 and caps. (B) Introgression lines containing region C2 from Dmau D1 (IL43 and IL16.14) contribute 37.9% of the difference in bristles (Upper graph) and 16.3% of the clasper size difference (Lower graph) of this strain compared to Dsim w501 (Dataset S2a). IL43 and IL16.14 differed significantly from IL16.30 and IL82 in clasper bristle number and in clasper area (P < 0.001). Asterisks indicate significance comparisons where P < 0.001 (Dataset S2c). Shading in the pictures underneath the Lower graph indicates the area measured at the distal end of the claspers in lines containing Dsim w501 (gray) or Dmau D1 (orange) regions for C2. Boxes indicate the range, upper and lower quartiles, and median for each sample. (Scale bar, 20 μm.)
C2 contains eight protein-coding genes with orthologs in Drosophila melanogaster. RNA-Sequencing (RNA-Seq) data suggests that only one of these genes, trn, is expressed in the terminalia of D. simulans and D. mauritiana when the difference in clasper morphology develops between these two species (Dataset S3 and SI Appendix, Fig. S3). However, if the causative gene has a spatially restricted pattern of expression it may not have been detected in the RNA-Seq. Therefore, we knocked down the expression of all genes in the candidate region (with the exception of CG34429, for which there was no available UAS line) using RNA interference (RNAi) in D. melanogaster to test if these positional candidates are involved in clasper development (Dataset S4). In addition, we knocked down CG11279 and capricious (caps), a gene closely related to trn and that functionally overlaps with trn in some contexts (28–34). CG11279 and caps flank C2, but their cis-regulatory sequences may still be within this region (Fig. 2A). We found that while knockdown of trn significantly reduced the size of the claspers (Dataset S4 and SI Appendix, Fig. S2), RNAi against the other nine genes tested, including caps, had no effect on clasper morphology in D. melanogaster (Dataset S4). Note that trn RNAi had no effect on the posterior lobes consistent with region C2 only affecting the claspers (Dataset S4).
trn encodes a leucine-rich repeat transmembrane protein (28, 30, 32, 33, 35, 36), and it is thought that its main function is to confer differences in affinity between cells and mediate their correct allocation to compartments in developing tissues such as the nervous system, trachea, eyes, wings, and legs (28, 30, 32, 35, 37, 38). Intriguingly, changes in trn expression can affect the allocation of cells between compartments, cause misspecification of compartmental boundaries, and even result in invasive movements of cells across such boundaries (33, 35, 38).
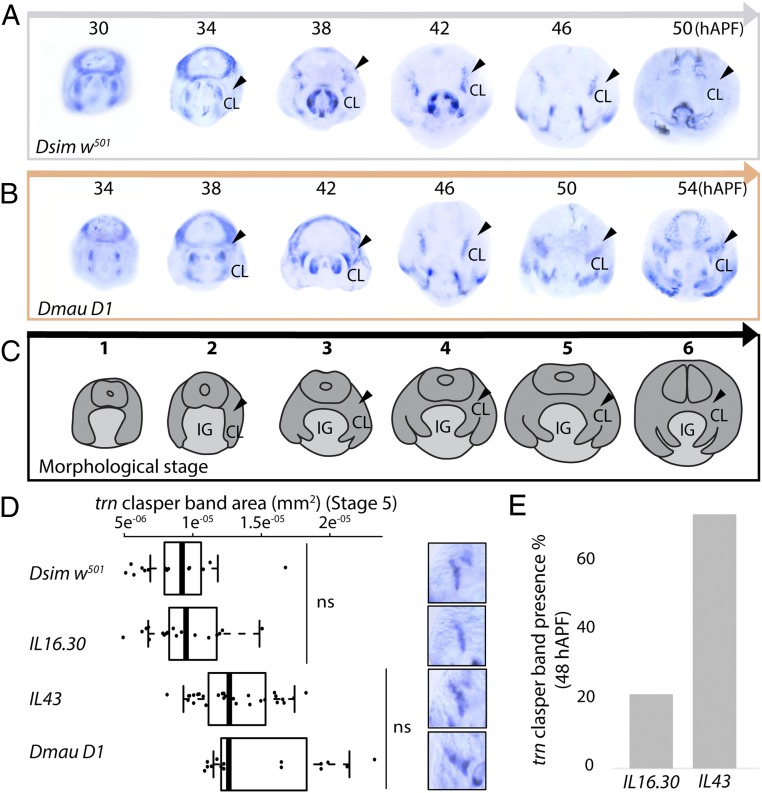
Our RNA-Seq data indicates that trn is more highly expressed in D. simulans during early terminalia development but is subsequently up-regulated in D. mauritiana at a later stage (Dataset S3). However, these data correspond to the sum of all of the expression domains of trn throughout the terminalia at each of these stages and may conceal more subtle localized expression differences between these species in specific tissues like the developing claspers. Therefore, we investigated the spatial pattern of trn expression throughout terminalia development using mRNA in situ hybridization (ISH) in Dmau D1 and Dsim w501 (Fig. 3 A–C and SI Appendix, Fig. S3). Concomitantly, we observed a 4-h difference in the timing of terminalia development between the two strains used (Fig. 3 A–C and SI Appendix, Fig. S3). We found that during early pupal stages trn is more highly expressed in Dsim w501 compared to Dmau D1 at the center of the terminalia, from where the internal genital structures will develop, which may explain the overall higher expression of trn in D. simulans at 30 h after puparium formation (hAPF) according to the RNA-Seq data (Fig. 3 A and B and Dataset S3). However, during later stages, the expression of trn is detected in a wider domain and persists for longer at the base of the developing claspers of Dmau D1 compared to Dsim w501 (Fig. 3 A and B). This is consistent with higher expression of trn in D. mauritiana detected in the RNA-Seq data at ∼50 hAPF (Dataset S3). These results are also consistent with the RNAi results in D. melanogaster where knockdown of trn results in the loss of trn expression at the base of the claspers (SI Appendix, Fig. S2B) and the development of smaller claspers (SI Appendix, Fig. S2A). Together, these results suggest that the higher and/or more persistent expression of the trnmau allele relative to the trnsim allele in the developing claspers is at least partially responsible for the larger claspers in D. mauritiana.
Fig. 3.
The spatial and temporal expression of trn differs in the developing claspers of D. simulans and D. mauritiana. Expression shown at 4-h intervals hAPF in Dsim w501 (A) and Dmau D1 (B). (C) Illustration of the developing structures at each morphological stage (SI Appendix, Fig. S3). Black arrowheads indicate expression at the base of the developing claspers. CL, clasper; IG, internal genitalia. (D) Analysis of trn expression domain at the base of the developing clasper at stage 5. trnsim males, Dsim w501 and IL 16.30, exhibit significantly smaller expression domains than trnmau males, IL 43 and Dmau D1 (all comparisons in trn expression domain between lines are significant [P < 0.001], except for those indicated by ns (nonsignificant), see also Dataset S5b). Boxes show the range, upper and lower quartiles, and the median for each sample. Representative trn expression at the base of the claspers is shown on the right-hand side. (E) The proportion of males with trn expression at the base of the clasper at 48 hAPF (between stages 5 and 6) in IL 16.30 and IL 43. IL43 males exhibit on average 51.9% more trn expression at the base of the claspers than IL 16.30 males (Dataset S5c).
Quantitative analysis of trn ISH confirmed that males containing trnmau (Dmau D1 and IL43) exhibit a larger expression domain at the base of the developing claspers at stage 5 (50 hAPF for Dmau D1 and 46 hAPF for Dsim w501 and ILs) than those containing trnsim (Dsim w501 and IL16.30) (Fig. 3D and Dataset S5a). Moreover, although at stage 6 IL43 and IL16.30 seem to recapitulate the pattern observed in Dsim w501 (i.e., trn expression no longer detected), we found that just before this, between stages 5 and 6 (48 hAPF in these ILs and D. simulans, SI Appendix, Fig. S3), there was variability in the presence of trn expression at the base of the developing claspers: expression was observed in 21% of IL16.30 males (i.e., males with trnsim) but in 74% of IL43 males (i.e., males with trnmau) (Fig. 3E and Dataset S5b). These data further support the hypothesis that spatial and/or temporal divergence in the expression of trn underlies differences in clasper size between D. simulans and D. mauritiana.
We also carried out ISH for CG11279 and caps (which are both also expressed in the terminalia, Dataset S3) and CG34429 (which we were unable to knock down in D. melanogaster). This showed that, unlike trn, these genes are either not expressed in the developing genitalia or at least not in a pattern consistent with a role in clasper development and evolution (SI Appendix, Fig. S4). For example, although caps expression in the male genitalia is generally similar to that of trn, caps transcripts were never detected at the base of the developing claspers (Fig. 3 A and B and SI Appendix, Fig. S4A).
Although our expression analyses of developing claspers suggest that cis-regulatory changes in trn are likely to contribute to differences in clasper morphology between D. mauritiana and D. simulans, we also found a total of 22 nucleotide differences in the coding sequence of trn between our mapped strains, Dmau D1 and Dsim w501. Only three of these differences are nonsynonymous and none is fixed between the two species (Dataset S6). In addition, a comparison of clasper size between strains of D. simulans and D. mauritiana with different combinations of amino acids at these three sites suggests that none of these substitutions is sufficient to explain the contribution of trn to the difference in clasper size between the species (SI Appendix, Fig. S5 and Dataset S2e). However, although the clasper size of the two mapped strains is also well within the range of their species (SI Appendix, Fig. S1 and Dataset S2d), we cannot rule out that one or more of these three amino acid substitutions may contribute to the difference in clasper size between the two strains used in this study.
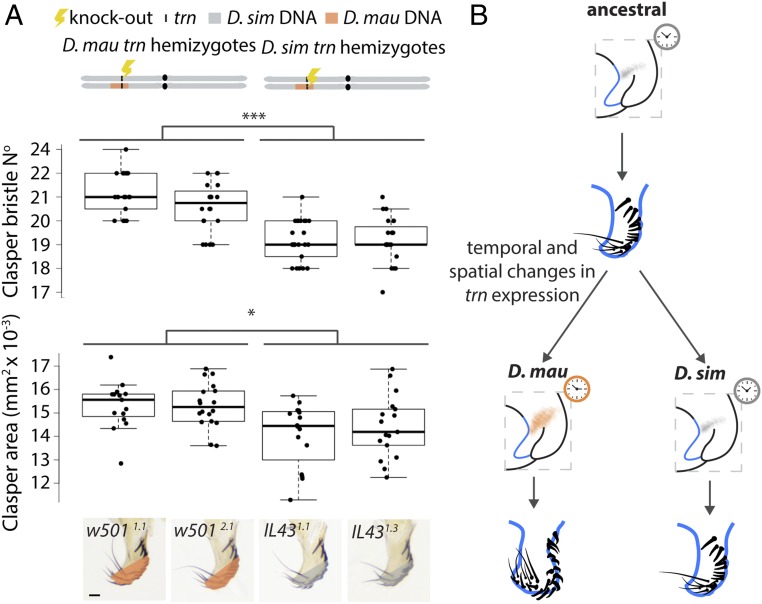
To confirm that sequence divergence in trn contributes to the difference in clasper morphology between Dmau D1 and Dsim w501, we used CRISPR/Cas9 to make null alleles of D. simulans trn (in Dsim w501) and D. mauritiana trn (in IL43, see Fig. 2 and SI Appendix, Fig. S6). We then generated reciprocal hemizygotes for trn i.e., genetically identical male flies that differ only in whether they have a functional copy of trn from D. mauritiana or D. simulans (Fig. 4A) (39). Comparison of the claspers between male reciprocal hemizygotes of trn shows that flies with a functional D. mauritiana trn allele have significantly larger claspers (P < 0.05) with more bristles (P < 0.001) than those with a functional D. simulans trn allele (Fig. 4B and Dataset S2c). This confirms that, consistent with the effects of the introgressions containing trn (Fig. 2), D. mauritiana trn has evolved to confer larger claspers than D. simulans trn.
Fig. 4.
Reciprocal hemizygotes of trn show that this locus contributes to evolutionary differences in male clasper morphology. (A) Schematic at the top illustrates the third chromosome of the reciprocal hemizygotes carrying a functional allele of trn from only Dmau D1 (Left) and Dsim w501. We found a significant difference in their clasper area (F(3, 61) = 7.012, P < 0.001) and clasper bristle number (F(3, 83) = 26.29, P < 0.001), shown in the boxplots underneath. Flies with a functional trn allele from D. mauritiana (IL431.1 and IL431.3) have significantly larger claspers (*P < 0.05) with more bristles (***P < 0.001) than those with a functional D. simulans trn allele, w5011.1 and w5012.1 (Dataset S2d). Boxes show the range, upper and lower quartiles, and the median for each sample. (B) Evolutionary changes increased the spatial domain and temporal expression of trn during clasper development in D. mauritiana have led to larger claspers with more bristles in this species compared to D. simulans. Orange and gray shading indicate broad and narrow expression of trn at the base of the developing claspers in D. mauritiana and D. simulans, respectively. The correspondingly colored clocks indicate differences in the persistence of this expression domain. (Scale bar, 20 μm.)
We have found that trn is a gene that underlies the rapid evolution in the size of a male genital organ and more generally a gene that contributes to differences in animal organ size (e.g., refs. 40–42). Many examples of phenotypic evolution, including differences in genital bristles between other Drosophila species (43), have been found to be caused by changes in the expression of transcription factors (44). However, trn is a transmembrane protein that appears to mediate differences in cell-cell contact directly through its extracellular domain, directing cells toward their correct positions via cues that are currently unknown (33, 35). Our results suggest that differences in trn expression in Drosophila are able to alter clasper size. Therefore, changes in cell affinity caused by variation in the temporal and/or spatial expression of transmembrane proteins that mediate cell interactions may represent another mechanism for the evolution of organ size. There is also some evidence, however, that trn could act as a ligand and may transduce signals, although its intracellular domain appears to be dispensable for most of its functions (28, 30, 33, 36). Therefore, further study of the function of trn and characterization of its role in organ size regulation and evolution is required.
Materials and Methods
Introgression Mapping and Phenotyping.
We increased the resolution of the previously predicted C2 region by generating recombinants between introgression line D11.01 and Dsim w501 (19) (SI Appendix, SI Materials and Methods). Flies were phenotyped and genotyped as described previously (19) using molecular markers (Dataset S7). All stocks and crosses were maintained on a standard cornmeal diet at 25 °C under a 12-h:12-h dark/light cycle unless otherwise stated. The periphalic structures and T1 legs were dissected, imaged, and measured as described in ref. 19 (SI Appendix, SI Materials and Methods and Dataset S2). All statistical analyses were conducted in R Studio.
RNA Sequencing and Differential Expression Analysis.
Three independent biological replicates of RNA-Seq libraries were generated from abdominal tip tissue dissected from 20 to 30 males for Dsim w501 and Dmau w− males at 30 and 50 hAPF (SI Appendix, Fig. S3). Indexed libraries were sent to Macrogen Japan for sequencing in a single lane of HiSeq4000 (Illumina), producing 100-bp paired-end reads. Raw fastq files are deposited at DDBJ under the accession no’s. DRA006755 and DRA006758 for D. mauritiana and D. simulans, respectively. Genes were considered not expressed if reads per kilobase million (RPKM) was below 1.5. RNA-Seq analysis of genes in C2 as well as CG11279 and caps is summarized in Dataset S3.
RNAi Knockdown of C2 Candidate Genes.
We conducted RNAi knockdown of all of the genes within region C2 (with the exception CG34429 for which there was no available UAS line) in D. melanogaster using UAS-RNAi lines from Vienna Drosophila Resource Center (VDRC, www.vdrc.at) and from Bloomington Drosophila Stock Center (Dataset S8). UAS males of our candidate genes were crossed to NP6333–GAL4 driver female virgins (P(GawB)PenNP6333) (45) carrying the transgene UAS-Dicer-2 P(UAS-Dcr-2.D). Crosses for the RNAi were carried out at 25 °C. The genital morphology of the male knockdowns was compared to NP6333-GAL4, UAS-Dicer, and UAS-RNAi controls. Clasper bristle number and tibia length were measured for 16 individuals of each genotype. Differences in clasper bristle number and tibia size were assessed using a one-way ANOVA followed by a Tukey’s test (Dataset S4). For raw phenotypic data, see Dataset S2a.
trn Sequence Analysis.
To assess the number of interspecific nucleotide substitutions in the coding sequence of trn, we used polymorphism data from Pool-seq data from 107 strains of D. mauritiana and from 50 strains of sub-Saharan D. simulans (46, 47) available at http://www.popoolation.at/pgt/ as well as from whole genome data for 10 strains of each species submitted to the SRA database by the University of Rochester (D. mauritiana lines: SRX135546, SRX688576, SRX688581, SRX688583, SRX688588, SRX688609, SRX688610, SRX688612, SRX688710, SRX688712; D. simulans lines: SRX497551, SRX497574, SRX497553, SRX497563, SRX497558, SRX497564, SRX497559, SRX495510, SRX495507, SRX497557). The sequence analysis is summarized in Dataset S6.
In Situ Hybridization.
We performed in situ hybridization to detect trn, CG11279, CG34429, and caps expression in the male terminalia of Dmau D1, Dsim w501, and D. melanogaster w1118 at a range of developmental time-points (SI Appendix, Fig. S3).
Quantification of Temporal and Spatial trn Expression.
To investigate potential differences in the trn expression domain between introgression lines used to map the C2 interval, in situ hybridizations were carried out at 46 hAPF in Dsim w501, IL 16.30, IL 43, and 50 hAPF in Dmau D1. It is at these time points that the largest differences in trn expression can be detected between the two parental species (Fig. 3 A and B). Dsim w501, IL 16.30, and IL 43 are morphologically equivalent at these stages (SI Appendix, Fig. S3).
Generation of Reciprocal Hemizygotes and Statistical Analysis.
We inserted 3xP3-DsRed to disrupt the reading frame of trn in Dsim w501 and IL43 using CRISPR/Cas9 (SI Appendix, Fig. S6). Injections were carried by The University of Cambridge Department of Genetics Fly Facility. Transgenic Dsim w501 and IL43 males heterozygous for the mutation were then crossed to noninjected IL43 and D. simulans w501 virgin females, respectively, to generate F1 males carrying the mutation (i.e., hemizygous for trn allele).
See extended methodological details in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Christian Schlötterer, Christina Muirhead, and Daven Presgraves for facilitating access to population genetic data. This work was funded by Natural Environment Research Council (NERC) Grant NE/M001040/1 and Biotechnology and Biological Sciences Research Council (BBSRC) Grant BB/M020967/1 (to A.P.M.), Japan Society for the Promotion of Science (JSPS) Grants-in-Aid for Scientific Research - KAKENHI Grant 15J05233 (to K.M.T.), and a Genetics Society Summer Studentship grant (to A.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Raw fastq files for the RNA sequencing data reported in this paper have been deposited at DNA Data Bank of Japan under accession nos. DRA006755 and DRA006758 for D. mauritiana and D. simulans, respectively.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1909829116/-/DCSupplemental.
References
- 1.Eberhard W. G., Sexual Selection and Animal Genitalia (Harvard University Press, Cambridge, MA, 1985), p. x, 244 pp.
- 2.Garrigan D., et al. , Genome sequencing reveals complex speciation in the Drosophila simulans clade. Genome Res. 22, 1499–1511 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eberhard W. G., Evolution of genitalia: Theories, evidence, and new directions. Genetica 138, 5–18 (2010). [DOI] [PubMed] [Google Scholar]
- 4.Hosken D. J., Stockley P., Sexual selection and genital evolution. Trends Ecol. Evol. (Amst.) 19, 87–93 (2004). [DOI] [PubMed] [Google Scholar]
- 5.Simmons L. W., Sexual selection and genital evolution. Austral Entomol. 53, 1–17 (2014). [Google Scholar]
- 6.House C. M., et al. , Sexual and natural selection both influence male genital evolution. PLoS One 8, e63807 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Panhuis T. M., Butlin R., Zuk M., Tregenza T., Sexual selection and speciation. Trends Ecol. Evol. (Amst.) 16, 364–371 (2001). [DOI] [PubMed] [Google Scholar]
- 8.Ritchie M. G., Sexual selection and speciation. Annu. Rev. Ecol. Evol. Syst. 38, 79–102 (2007). [Google Scholar]
- 9.Servedio M. R., Bürger R., The counterintuitive role of sexual selection in species maintenance and speciation. Proc. Natl. Acad. Sci. U.S.A. 111, 8113–8118 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frazee S. R., Masly J. P., Multiple sexual selection pressures drive the rapid evolution of complex morphology in a male secondary genital structure. Ecol. Evol. 5, 4437–4450 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.LeVasseur-Viens H., Polak M., Moehring A. J., No evidence for external genital morphology affecting cryptic female choice and reproductive isolation in Drosophila. Evolution 69, 1797–1807 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Masly J. P., Kamimura Y., Asymmetric mismatch in strain-specific genital morphology causes increased harm to Drosophila females. Evolution 68, 2401–2411 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Tanaka K. M., Kamimura Y., Takahashi A., Mechanical incompatibility caused by modifications of multiple male genital structures using genomic introgression in Drosophila. Evolution 72, 2406–2418 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Coyne J. A., Genetic basis of differences in genital morphology among three sibling species of Drosophila. Evolution 37, 1101–1118 (1983). [DOI] [PubMed] [Google Scholar]
- 15.Laurie C. C., True J. R., Liu J., Mercer J. M., An introgression analysis of quantitative trait loci that contribute to a morphological difference between Drosophila simulans and D. mauritiana. Genetics 145, 339–348 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.LeVasseur-Viens H., Moehring A. J., Individual genetic contributions to genital shape variation between Drosophila simulans and D. mauritiana. Int. J. Evol. Biol. 2014, 808247 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu J., et al. , Genetic analysis of a morphological shape difference in the male genitalia of Drosophila simulans and D. mauritiana. Genetics 142, 1129–1145 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Masly J. P., Dalton J. E., Srivastava S., Chen L., Arbeitman M. N., The genetic basis of rapidly evolving male genital morphology in Drosophila. Genetics 189, 357–374 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tanaka K. M., et al. , Genetic architecture and functional characterization of genes underlying the rapid diversification of male external genitalia between Drosophila simulans and Drosophila mauritiana. Genetics 200, 357–369 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.True J. R., Liu J., Stam L. F., Zeng Z.-B., Laurie C. C., Quantitative genetic analysis of divergence in male secondary sexual traits between Drosophila simulans and Drosophila mauritiana. Evolution 51, 816–832 (1997). [DOI] [PubMed] [Google Scholar]
- 21.Zeng Z. B., et al. , Genetic architecture of a morphological shape difference between two Drosophila species. Genetics 154, 299–310 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Acebes A., Cobb M., Ferveur J. F., Species-specific effects of single sensillum ablation on mating position in Drosophila. J. Exp. Biol. 206, 3095–3100 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Jagadeeshan S., Singh R. S., A time-sequence functional analysis of mating behaviour and genital coupling in Drosophila: Role of cryptic female choice and male sex-drive in the evolution of male genitalia. J. Evol. Biol. 19, 1058–1070 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Kamimura Y., Mitsumoto H., Comparative copulation anatomy of the Drosophila melanogaster species complex (Diptera: Drosophilidae). Entomol. Sci. 14, 399–410 (2011). [Google Scholar]
- 25.Mattei A. L., Riccio M. L., Avila F. W., Wolfner M. F., Integrated 3D view of postmating responses by the Drosophila melanogaster female reproductive tract, obtained by micro-computed tomography scanning. Proc. Natl. Acad. Sci. U.S.A. 112, 8475–8480 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robertson H. M., Mating asymmetries and phylogeny in the Drosophila melanogaster species complex. Pac. Sci. 42, 72–80 (1988). [Google Scholar]
- 27.Yassin A., Orgogozo V., Coevolution between male and female genitalia in the Drosophila melanogaster species subgroup. PLoS One 8, e57158 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang Z., et al. , Molecular and genetic characterization of the Drosophila tartan gene. Dev. Biol. 160, 315–332 (1993). [DOI] [PubMed] [Google Scholar]
- 29.Hong W., et al. , Leucine-rich repeat transmembrane proteins instruct discrete dendrite targeting in an olfactory map. Nat. Neurosci. 12, 1542–1550 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krause C., Wolf C., Hemphälä J., Samakovlis C., Schuh R., Distinct functions of the leucine-rich repeat transmembrane proteins capricious and tartan in the Drosophila tracheal morphogenesis. Dev. Biol. 296, 253–264 (2006). [DOI] [PubMed] [Google Scholar]
- 31.Kurusu M., et al. , A screen of cell-surface molecules identifies leucine-rich repeat proteins as key mediators of synaptic target selection. Neuron 59, 972–985 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mao Y., Kerr M., Freeman M., Modulation of Drosophila retinal epithelial integrity by the adhesion proteins capricious and tartan. PLoS One 3, e1827 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Milán M., Pérez L., Cohen S. M., Boundary formation in the Drosophila wing: Functional dissection of Capricious and Tartan. Dev. Dyn. 233, 804–810 (2005). [DOI] [PubMed] [Google Scholar]
- 34.Shishido E., Takeichi M., Nose A., Drosophila synapse formation: Regulation by transmembrane protein with Leu-rich repeats, CAPRICIOUS. Science 280, 2118–2121 (1998). [DOI] [PubMed] [Google Scholar]
- 35.Milán M., Weihe U., Pérez L., Cohen S. M., The LRR proteins capricious and Tartan mediate cell interactions during DV boundary formation in the Drosophila wing. Cell 106, 785–794 (2001). [DOI] [PubMed] [Google Scholar]
- 36.Bugga L., Ratnaparkhi A., Zinn K., The cell surface receptor Tartan is a potential in vivo substrate for the receptor tyrosine phosphatase Ptp52F. Mol. Cell. Biol. 29, 3390–3400 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Milán M., Pérez L., Cohen S. M., Short-range cell interactions and cell survival in the Drosophila wing. Dev. Cell 2, 797–805 (2002). [DOI] [PubMed] [Google Scholar]
- 38.Sakurai K. T., Kojima T., Aigaki T., Hayashi S., Differential control of cell affinity required for progression and refinement of cell boundary during Drosophila leg segmentation. Dev. Biol. 309, 126–136 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Stern D. L., Identification of loci that cause phenotypic variation in diverse species with the reciprocal hemizygosity test. Trends Genet. 30, 547–554 (2014). [DOI] [PubMed] [Google Scholar]
- 40.Indjeian V. B., et al. , Evolving new skeletal traits by cis-regulatory changes in bone morphogenetic proteins. Cell 164, 45–56 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Loehlin D. W., Werren J. H., Evolution of shape by multiple regulatory changes to a growth gene. Science 335, 943–947 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lamichhaney S., et al. , A beak size locus in Darwin’s finches facilitated character displacement during a drought. Science 352, 470–474 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Nagy O., et al. , Correlated evolution of two copulatory organs via a single cis-regulatory nucleotide change. Curr. Biol. 28, 3450–3457.e13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martin A., Orgogozo V., The Loci of repeated evolution: A catalog of genetic hotspots of phenotypic variation. Evolution 67, 1235–1250 (2013). [DOI] [PubMed] [Google Scholar]
- 45.Chatterjee S. S., Uppendahl L. D., Chowdhury M. A., Ip P. L., Siegal M. L., The female-specific doublesex isoform regulates pleiotropic transcription factors to pattern genital development in Drosophila. Development 138, 1099–1109 (2011). [DOI] [PubMed] [Google Scholar]
- 46.Nolte V., Pandey R. V., Kofler R., Schlötterer C., Genome-wide patterns of natural variation reveal strong selective sweeps and ongoing genomic conflict in Drosophila mauritiana. Genome Res. 23, 99–110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pandey R. V., Kofler R., Orozco-terWengel P., Nolte V., Schlötterer C., PoPoolation DB: A user-friendly web-based database for the retrieval of natural polymorphisms in Drosophila. BMC Genet. 12, 27 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsacas L., David J., Drosophila mauritiana n. sp. du groupe melanogaster de I’lle Maurice. Bull. Soc. Entomol. Fr. 79, 42–46 (1974). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.