Significance
The inactivation of bacteria by UV light involves sequential dynamic processes that occur from femtoseconds to minutes and span from simple molecules to proteins and organisms. Detailed understanding of the inactivation mechanism is important and the key to efficient inactivation of harmful pathogens whose antibiotic resistance is critically increasing. In this study, we show that, for Escherichia coli and Bacillus subtilis bacteria, their tryptophan fluorescence intensity increased within the first minute of UV irradiation and subsequently decreased continuously. These data suggest that, shortly after exposure to UV light, tryptophan may be released from the bacterial membrane and protein unfolding which contributes to the increased tryptophan fluorescence intensity. This study provides a further means for monitoring the rate of bacteria inactivation.
Keywords: bacteria inactivation, synchronous fluorescence, photodissociation kinetics
Abstract
The UV photodissociation kinetics of tryptophan amino acid, Trp, attached to the membrane of bacteria, Escherichia coli and Bacillus subtilis, have been studied by means of normal and synchronous fluorescence. Our experimental data suggest that the fluorescence intensity of Trp increases during the first minute of irradiation with 250 nm to ∼ 280 nm, 7 mW/cm2 UV light, and subsequently decreases with continuous irradiation. During this short, less than a minute, period of time, 70% of the 107 cell per milliliter bacteria are inactivated. This increase in fluorescence intensity is not observed when tryptophan is in the free state, namely, not attached to a protein, but dissolved in water or saline solution. This increase in fluorescence is attributed to the additional fluorescence of tryptophan molecules formed by protein unfolding, the breakage of the bond that attaches Trp to the bacterial protein membrane, or possibly caused by the irradiation of 2 types of tryptophan residues that photolyze with different quantum yields.
UV irradiation is critically important in many situations where inactivation of bacteria is required to prevent contamination or even infection. This is especially true where environmental decontamination is necessary (1, 2). Yet, despite the fact that UV light has been widely used for many years (3), features of the inactivation process by UV light remain unknown, and in-depth knowledge of the UV light interaction with biological species may lead to a more effective use. In this regard, we report on important phases of the lethal effect of UV light on 2 bacterial species of medical relevance.
Tryptophan (Trp), an aromatic amino acid, is a constituent of peptides/proteins and also a precursor of serotonin and other derivatives such as adenine dinucleotides. Trp is known to be the most intensely fluorescing component of Escherichia coli and Bacillus subtilis bacteria, and the photooxidation of proteins after absorption of UV radiation is known to proceed by either direct photooxidation that generates excited singlet and triplets states or formation of singlet oxygen, ROS, by energy transfer to ground-state molecular oxygen, process I and process II, respectively (4–7). Although the photostability of Trp-containing proteins has been investigated, the detailed mechanism of this photochemical process is not completely known. To that effect, the data presented in this article provide information that advances knowledge concerning the detail mechanism of the initial bacterial photoinactivation process.
While peptide bonds exhibit a weak absorption band, n → π* transition at 210 nm to ∼220 nm, ε = 100 dm3⋅mol−1⋅cm−1, the absorption spectrum of Trp, a major chromophoric amino acid, exhibits a strong band at 220 nm and a weaker one at 280 nm with molar extinction coefficients of 22,900 and 3,800, respectively. This suggests that the indole moiety of the Trp molecule is the Trp component that absorbs in the near UV and generates the ROS that reacts efficiently with and photodegrade proteins (8–10). Experimental data indicate that singlet oxygen and possibly other radical species are mostly responsible for the photodissociation of Trp. The dissociation of Trp and tyrosine (Tyr) induced by UV irradiation was determined by the change in their fluorescence intensity as a function of UV dose by us (11) and others (12–16). Previous experimental data show that there is a bend in the linear decay as a function of UV dose (17), which may be assigned to irradiation of undamaged residues or to the presence of 2 types of Trp residues that are photolyzed with varying UV quantum yields. Our study shows that the quantum yield of Trp photolysis in proteins is several times larger than that in free amino acids, and may vary with protein type. To that effect, our experimental fluorescence data regarding bacteria inactivation with UV light are interpreted using a mechanism which indicates that the initial photodissociation process is the detachment of some Trp molecules from the membrane protein(s) followed by dissociation and dimerization of the DNA thymine bases.
The response of bacteria and other microorganisms to UV radiation may be influenced by several factors including 1) the growth medium, 2) the stage of the culture, and 3) the strain of the microorganisms (18, 19). Photoreactivation is another important factor which determines bacterial response to UV radiation. Most, if not all studies, attribute the “death” of a bacterium by UV irradiation to the damage induced by lesions of its nucleic acids which, subsequently, form pyrimidine dimers and other photoproducts (20). The dominant product of the UV-induced DNA lesion is cyclobutane pyrimidine dimers generated by the adjacent thymine bases. The consequence of these photoproducts is the prohibition of the normal DNA cellular replication process. Such prohibition process leads to cell death and immune suppression. Topically, DNA is located, practically, at the center of the bacterium, and therefore it is shielded from the UV irradiation by the enveloping membranes that contain several amino acids, such as Trp and Tyr, which are characterized by high absorption cross-sections in the 200- to ∼300-nm UV region, where the DNA maximum absorption peak is also located. Therefore, it is expected that Trp and Tyr absorb the UV radiation, and, after partial photolysis, sufficient UV light is allowed to penetrate through the membrane and enter the inner bacterium, causing DNA lysis followed by bacteria inactivation and/or death. It is also known that the thymine dimer induced by the UV radiation may undergo further photolysis that regenerates DNA. To that effect, the thymine dimers distort the DNA double helix and thus reduce or forbid normal DNA replication. Subsequent exposure of the UV-damaged cells to visible light often repairs the damage to the DNA by restoring the dimers, caused by the UV irradiation, a process known as photoreactivation.
In this study, we found that the Trp fluorescence intensity of E. coli and B. subtilis bacteria increased within the first minute of UV irradiation and then decreased continuously. The recorded spectroscopic data indicate that, during the first 30 s of E. coli and B. subtilis bacteria under UV irradiation, the Trp, 350-nm, fluorescence band intensity maximum increased by 7 to 10%. However, no fluorescence intensity increase was detected in the free state of Trp dissolved in solvents such as saline and subjected to UV irradiation. The observed fluorescence intensity increase in bacterial samples is attributed to the formation of additional Trp molecules by protein unfolding, the breakage of the bond that attaches Trp to the bacterial protein membrane, or possibly the irradiation of 2 types of Trp residues that photolyze with different quantum yields. Our experimental data also suggest that irradiation of E. coli and B. subtilis bacteria in water with 7 mW/cm2, 250- to ∼270-nm UV light, for 10 min is sufficient to induce inactivation of practically all E. coli and B. subtilis bacteria; this is based on the fact that, during this time period, the number of live bacteria decreased from 107 or 108 cells per milliliter to 103 cells per milliliter.
Results and Discussion
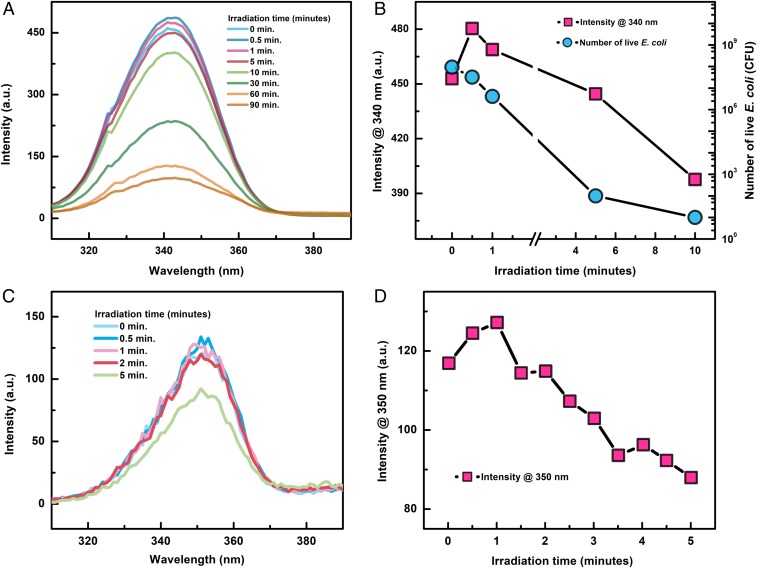
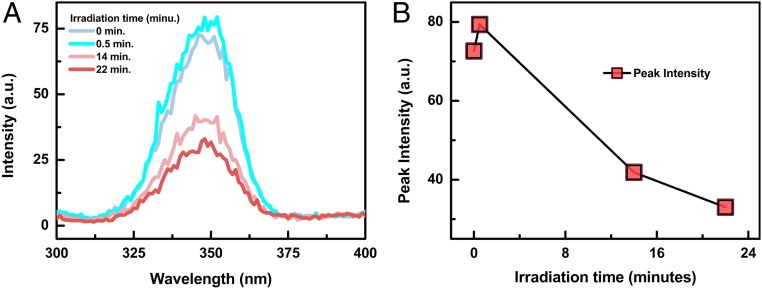
The fluorescence spectra and their peak intensity change are shown in Fig. 1 for E. coli and B. subtilis irradiated with varying UV doses. Fig. 1 B and D show that the peak fluorescence intensity increases during the first minute of irradiation and then decreases continuously with further irradiation. For the B. subtilis bacteria, its maximum fluorescence intensity at 350 nm initially increased by 8.4% within the first minute of UV irradiation and then decreased continuously within the subsequent 4 min of irradiation. For the E. coli bacteria, its maximum fluorescence intensity at 340 nm initially increased by 6.6% within the first minute of UV irradiation and then decreased continuously. Incubation of the E. coli bacteria sample after each UV irradiation indicates that, within the first minute of UV irradiation, 70% of the 107 cell per milliliter E. coli bacteria are inactivated, which is also shown in Fig. 1B.
Fig. 1.
Fluorescence intensity change for (A) E. coli and (C) B. subtilis as a function of UV irradiation time. The change of the peak fluorescence intensity at 340 nm and 350 nm is shown in B and D for E. coli and B. subtilis, respectively. The number of live E. coli as a function of UV irradiation time is also shown in B and is labeled by the right axis.
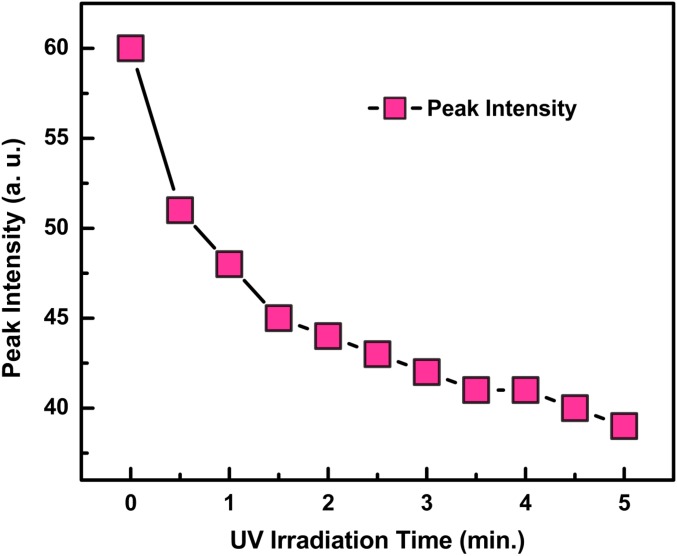
We observed that this phenomenon occurred only when Trp was attached to bacteria such as E. coli and B. subtilis and nonbacterial proteins that we studied. As shown in Fig. 2, repeating the same experiment with pure Trp, alone, in water and saline solutions, we observed only a continuous decrease in the fluorescence intensity, with no apparent intensity increase during the first minute or any time during UV irradiation.
Fig. 2.
Fluorescence intensity change of pure Trp in saline solution as a function of UV irradiation time. No fluorescence intensity increase was observed in the first few minutes after UV irradiation.
We define bacterial inactivation, in our experiments, as the decrease in the number of replicable bacteria during the period of time that the live bacteria were exposed to UV light. The dynamics of inactivation are essentially the same for the bacterial Trp and Tyr studied, where their fluorescence decay may be represented by the first-order expression
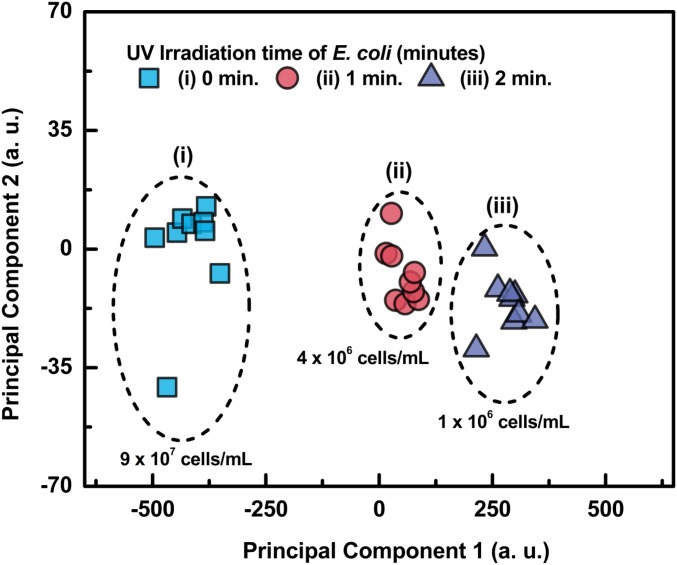
where the bacterial fluorescence intensity decreases with fluence, and the inactivation rate, , is a function of the UV dose; is the inactivation rate constant in units of square centimeters per millijoule; represents the number of live bacteria. The rate of inactivation of the same concentration of E. coli and B. subtilis bacteria was also determined by culture and incubation on agar plates for 24 h, at 37 °C of the bacteria and subsequently counting the number of colony-forming units before and after UV irradiation. We find that the rate of inactivation, number of bacteria killed by the UV, determined by these 2 methods, fluorescence decay and incubation, was practically the same. In addition, we measured the live/dead bacteria ratio by subjecting the bacterial fluorescence, before and after irradiation, to principal component analysis (PCA) (21). The plot of the PCA data Fig. 3 shows a clear separation of the live and dead bacteria as a function of UV irradiation dose from which their ratio can also be determined. The fluorescence spectra were also processed in order to generate excitation−emission matrix (EEM) plots which reveal the most appropriate scanning Δλ to be used for recording their synchronous fluorescence spectra.
Fig. 3.
PCA plot of nonirradiated and irradiated E. coli bacteria. Note the separation as a function of irradiation dose.
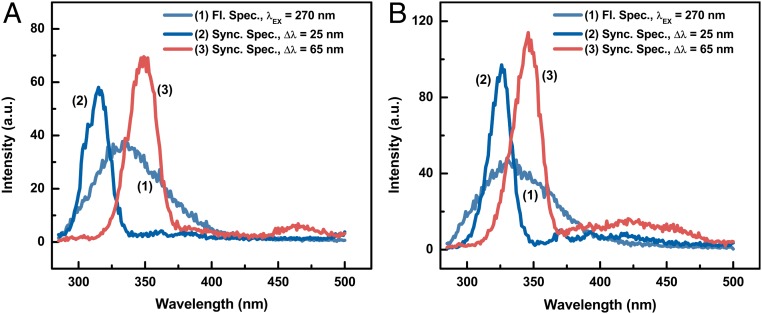
The synchronous fluorescence spectra made it possible to display, clearly, the spectra of the bacterial major molecular components such as Trp, Tyr, and DNA and their decay as a function of irradiation dose. The synchronous fluorescence spectra of bacteria were recorded using Δλ = 25 nm and Δλ = 65 nm for Tyr and Trp, respectively. The normal and synchronous fluorescence spectra of E. coli and B. subtilis are shown in Fig. 4A and Fig. 4B, respectively. Most bacteria, including E. coli and B. subtilis, are characterized by a broad, rather structureless, fluorescence band spanning the wavelength region from 300 nm to ∼600 nm. The fluorescence spectrum of E. coli, shown in Fig. 4A, does not show any band(s). In contrast, the synchronous fluorescence spectra shown Fig. 4A depict 2 rather distinct intense bands, with maxima at 310 nm and 350 nm, which correspond to the synchronous fluorescence of Tyr and Try components, respectively. We have recorded similar spectra for B. subtilis as shown in Fig. 4B.
Fig. 4.
Fluorescence and synchronous fluorescence spectra of (A) E. coli and (B) B. subtilis bacteria. The Δλ used in the synchronous fluorescence measurements are 25 nm and 65 nm for the Tyr and Trp components, respectively.
It is rather interesting to note that several amino acids, including Tyr and Trp, which are components of the inner and outer membranes of bacteria, absorb quite intensely in the 200- to ∼270-nm region where DNA also has its strongest absorption cross-section. However, the absorption cross-sections for Trp and Tyr are much larger than the DNA absorption cross-section. Therefore, the DNA absorption band at 260 nm is not observed, because it is masked by the intense absorption of the Trp and Tyr bands. The ratio of the normal fluorescence peak intensity over the Trp peak intensity is 1.8 for the Gram-negative bacteria E. coli, while the ratio for Gram-positive bacteria B. subtilis is 2.7. This ratio may be a potential means for the distinguishing of Gram-positive and Gram-negative bacteria. The normal and synchronous fluorescence spectra of pure Tyr and Trp, dissolved in saline and water solutions, are practically identical to the synchronous fluorescence spectra exhibited by E. coli and B. subtilis bacteria dispersed in the same solvents.
Our data show that the doses of UV light necessary for the inactivation of E. coli bacteria were several times smaller than the UV doses required to inactivate an equal number, in cells per milliliter, of viruses and bacterial spores. These results are in agreement with the data presented in ref. 18. The higher resistance of spores than bacteria to UV is most probably due to the many additional, protective layers that surround the bacteria when they are in the form of spores, which suggests that the DNA is further shielded from UV radiation. In the present study, we are concerned, mostly, with bacteria in their natural state, and, in the case of B. subtilis spores, the bacteria were removed from their spore environment before absorption, emission, and UV irradiation measurements were performed.
Sensitivity to UV disinfection can vary for a certain species of a microorganism species with strain type, growth medium, and stage of the culture (19). The E. coli bacterial samples in our experiments were irradiated with a nearly collimated beam whose UV power at the sample was measured using a calibrated power meter. And the absorption spectra of bacterial samples were recorded by a Shimadzu UV 160 spectrophotometer.
The average intensity, , in the optical cell that contain the bacteria was calculated by ref. 18,
where is the incident light intensity, is the absorbance per centimeter, and is the path length. As shown previously in Fig. 2, such increase in the fluorescence intensity was unobservable, when Trp was alone, not attached to a protein, and irradiated with UV light. In contrast, similar experiments performed with Human Serum Albumin (HSA) (Fig. 5) shows the same fluorescence intensity changes as a function of UV irradiation dose as the E. coli bacteria.
Fig. 5.
HSA fluorescence spectra vs. UV irradiation time (A) and the correspondent fluorescence peak intensity vs. UV irradiation time (B). Fluorescence intensity increase was observed in the first minute after UV irradiation. HAS concentration in water solution, 100 μg/mL; UV intensity, 1.3 mW/cm2.
We also observed an initial increase in fluorescence intensity with pure proteins, egg ovalbumin, as a function of UV light irradiation dose. Such initial increase in the fluorescence of pure proteins as a function of UV irradiation has been observed earlier with bovine growth hormone (5) and tentatively attributed to protein(s) unfolding (22, 23), which is a form of UV-induced damage to the proteins that compromise their functions in the cell and may also induce toxicity. However, as proteins unfold, a number of Trp amino acids become exposed to direct UV radiation, which results in initial fluorescence increase, followed by subsequent intensity decrease as Trp is denatured by the continuous UV radiation. This phenomenon resembles, closely, the initial increase observed for E. coli and B. subtilis described earlier.
The mechanism for this initial increase in fluorescence may be different in pure proteins and bacteria. The initial increase in fluorescence upon UV radiation represents damage to the proteins which is correlated with the death of bacteria cells. It is known that bacterial inactivation does not take place at very low UV fluencies, most probably because the low rate of inactivation is balanced by the high replication rate of bacteria (24–27). However, as discussed previously, exposure to higher UV fluence, even for very short periods of time, does result in observable bacterial inactivation.
It is known that the inactivation of bacteria irradiated with UV of wavelengths lower than 255 nm, such as 214 nm to ∼220 nm, is more effective than irradiation with the 253.7-nm mercury emission line. This may be due to the higher absorption cross-section of the bacteria at the shorter wavelengths and the fact that the higher photon energy of the shorter wavelengths is absorbed by the single C−C and C−H bonds of the bacterial membranes and nucleic acids, which do not absorb at the longer wavelengths. The experimental data also suggest that, when the bacteria were irradiated with 270-nm UV light, the fluorescence spectrum displayed a similar fluorescence intensity behavior. Namely, the bacterial fluorescence intensity decreases, after an initial increase, which scales with bacteria inactivation, and, at longer irradiation time periods, a continuous photodegradation of Trp and Tyr is observed which correlates with further bacterial inactivation.
The inactivation rate constant of E. coli and B. subtilis bacteria in water has been reported by several investigators (17, 28–32) and is summarized in ref. 33. Pathogenic bacterial cells have been found to be more susceptible to UV irradiation than viruses, about which we are not concerned in this study. The inactivation rate constant K (square centimeters per millijoule) for the bacteria irradiated in the present studies varied from 0.065 for B. subtilis to 1.37 for Vibrio cholera (33). The rate constant for E. coli found in our studies varies from 0.63 to 0.52, which is also in very good agreement with the literature value of 0.54 (30, 34). It might be interesting to note that the sensitivity to monochromatic and polychromatic UV radiation in the same narrow wavelength region and same dose was found to be very similar when applied with the same UV flux. These experimental data are not surprising when one considers that the absorption band of the bacteria is broad and of similar optical density in this wavelength range. Our data indicate that B. subtilis and especially spores are less sensitive to UV than the E. coli bacteria. The B. subtilis rate constant was found to be 0.62 in our study, which is similar to the reported literature values (18, 29, 33).
One advantage of UV irradiation over other bacterial inactivation bacterial processes is the fact that the quantum yield of UV inactivation is not affected by temperature, pH, and most non−UV-absorbing impurities. However, several factors may play a role in the UV inactivation rate, including 1) the physiological state of the organism such as particle associations and repair mechanisms, including DNA double helix restoration after dimer photolysis, and 2) fluence distribution in the sample: absorption, reflection, and scattering.
Conclusion
UV inactivation experiments with E. coli, B. subtilis, and other bacteria show that, during the first 30 s to 1 min of irradiation (∼7 mW/cm2, 220- to ∼270-nm UV light), a number of the Trp amino acids attached to the membrane proteins are exposed to the radiation, which causes an initial increase in the number of fluorescing Trp molecules, and, consequently, the fluorescence intensity increases temporarily. The cause of Trp molecules exposure is attributed to protein unfolding and release of Trp and Tyr amino acid molecules which may fluoresce with varying quantum yields. During this initial irradiation time period, ∼40% of the 107 to 108 bacteria irradiated were found to be inactivated. Further UV irradiation of the bacteria results in the continuous decrease in the fluorescence intensity. The UV light absorbed by the Trp and Tyr amino acid components of the bacterium induces their photodissociation, generating “a hole” in the membrane through which the UV radiation enters the interior of the bacterium where it interacts and dimerizes DNAs, which, in turn, inhibit DNA replication and result in bacterial inactivation.
Acknowledgments
We thank Arjun Khrishnamoorthi for technical assistance. This work was supported, in part, by the Welch Foundation Grant 1501928, the Air Force Office of Scientific Research Grant FA9550-18-1-0100, and the Texas A&M Engineering Experiment Station funds.
Footnotes
The authors declare no conflict of interest.
References
- 1.Weber D. J., et al. , Effectiveness of ultraviolet devices and hydrogen peroxide systems for terminal room decontamination: Focus on clinical trials. Am. J. Infect. Control 44 (suppl. 5), e77–e84 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ali S., Yui S., Muzslay M., Wilson A. P. R., Comparison of two whole-room ultraviolet irradiation systems for enhanced disinfection of contaminated hospital patient rooms. J. Hosp. Infect. 97, 180–184 (2017). [DOI] [PubMed] [Google Scholar]
- 3.AMA Council on Physical Medicine , Acceptance of ultraviolet lamps for disinfecting purposes. JAMA 137, 1600–1603 (1948). [Google Scholar]
- 4.Pattison D. I., Rahmanto A. S., Davies M. J., Photo-oxidation of proteins. Photochem. Photobiol. Sci. 11, 38–53 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Miller B. L., Hageman M. J., Thamann T. J., Barròn L. B., Schöneich C., Solid-state photodegradation of bovine somatotropin (bovine growth hormone): Evidence for tryptophan-mediated photooxidation of disulfide bonds. J. Pharm. Sci. 92, 1698–1709 (2003). [DOI] [PubMed] [Google Scholar]
- 6.Lippitz M., Erker W., Decker H., van Holde K. E., Basché T., Two-photon excitation microscopy of tryptophan-containing proteins. Proc. Natl. Acad. Sci. U.S.A. 99, 2772–2777 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caldwell C. R., Ultraviolet-induced photodegradation of cucumber (Cucumis sativus L.) microsomal and soluble protein tryptophanyl residues in vitro. Plant Physiol. 101, 947–953 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davies M. J., Fu S., Dean R. T., Protein hydroperoxides can give rise to reactive free radicals. Biochem. J. 305, 643–649 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Igarashi N., Onoue S., Tsuda Y., Photoreactivity of amino acids: Tryptophan-induced photochemical events via reactive oxygen species generation. Anal. Sci. 23, 943–948 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Krieger-Liszkay A., Singlet oxygen production in photosynthesis. J. Exp. Bot. 56, 337–346 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Li R., et al. , In situ detection of live-to-dead bacteria ratio after inactivation by means of synchronous fluorescence and PCA. Proc. Natl. Acad. Sci. U.S.A. 115, 668–673 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Piras R., Vallee B. L., Effect of ultraviolet irradiation on composition and function of carboxypeptidase A. Biochemistry 5, 849–854 (1966). [DOI] [PubMed] [Google Scholar]
- 13.Vladimirov I. U. A., Rashchupkin D. I., Studies on primary photochemical processes in proteins. I. 2 photochemical reactions with aromatic amino acids [in Russian]. Biofizika 9, 282–292 (1964). [PubMed] [Google Scholar]
- 14.Imahori K., The ultraviolet inactivation of pepsin. Biochim. Biophys. Acta 18, 216–220 (1955). [DOI] [PubMed] [Google Scholar]
- 15.Fujimori E., Ultraviolet light irradiated collagen macromolecules. Biochemistry 5, 1034–1040 (1966). [DOI] [PubMed] [Google Scholar]
- 16.Cooper D. R., Davidson R. J., The effect of ultraviolet irradiation on soluble collagen. Biochem. J. 97, 139–147 (1965). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harris G. D., et al. , Ultraviolet inactivation of selected bacteria and viruses with photoreactivation of the bacteria. Water Res. 21, 687–692 (1987). [Google Scholar]
- 18.Chang J. C., et al. , UV inactivation of pathogenic and indicator microorganisms. Appl. Environ. Microbiol. 49, 1361–1365 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morton R. A., Haynes R. H., Changes in the ultraviolet sensitivity of Escherichia coli during growth in batch cultures. J. Bacteriol. 97, 1379–1385 (1969). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rastogi R. P., et al. , Molecular mechanisms of ultraviolet radiation-induced DNA damage and repair. J. Nucleic Acids 2010, 592980 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wold S., Esbensen K., Geladi P., Principal component analysis. Chemom. Intell. Lab. Syst. 2, 37–52 (1987). [Google Scholar]
- 22.Prompers J. J., Hilbers C. W., Pepermans H. A., Tryptophan mediated photoreduction of disulfide bond causes unusual fluorescence behaviour of Fusarium solani pisi cutinase. FEBS Lett. 456, 409–416 (1999). [DOI] [PubMed] [Google Scholar]
- 23.Havel H. A., Kauffman E. W., Elzinga P. A., Fluorescence quenching studies of bovine growth hormone in several conformational states. Biochim. Biophys. Acta 955, 154–163 (1988). [DOI] [PubMed] [Google Scholar]
- 24.Hoyer O., Testing performance and monitoring of UV systems for drinking water disinfection. Water Supply 16, 424–429 (1998). [Google Scholar]
- 25.Sommer R., et al. , Time dose reciprocity in UV disinfection of water. Water Sci. Technol. 38, 145–150 (1998). [Google Scholar]
- 26.Mamane-Gravetz H., Linden K. G., Relationship between physiochemical properties, aggregation and u.v. inactivation of isolated indigenous spores in water. J. Appl. Microbiol. 98, 351–363 (2005). [DOI] [PubMed] [Google Scholar]
- 27.Knudson G. B., Photoreactivation of UV-irradiated Legionella pneumophila and other Legionella species. Appl. Environ. Microbiol. 49, 975–980 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Butler R. C., Lund V., Carlson D. A., Susceptibility of Campylobacter jejuni and Yersinia enterocolitica to UV radiation. Appl. Environ. Microbiol. 53, 375–378 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sommer R., et al. , UV-inactivation of microorganisms in water [in German]. Zentralbl. Hyg. Umweltmed. 189, 214–224 (1989). [PubMed] [Google Scholar]
- 30.Oguma K., Katayama H., Ohgaki S., Photoreactivation of Escherichia coli after low- or medium-pressure UV disinfection determined by an endonuclease sensitive site assay. Appl. Environ. Microbiol. 68, 6029–6035 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zimmer J. L., Slawson R. M., Potential repair of Escherichia coli DNA following exposure to UV radiation from both medium- and low-pressure UV sources used in drinking water treatment. Appl. Environ. Microbiol. 68, 3293–3299 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hijnen W. A., et al. , Increased resistance of environmental anaerobic spores to inactivation by UV. Water Sci. Technol. Water Supply 4, 55–61 (2004). [Google Scholar]
- 33.Hijnen W. A., Beerendonk E. F., Medema G. J., Inactivation credit of UV radiation for viruses, bacteria and protozoan (oo)cysts in water: A review. Water Res. 40, 3–22 (2006). [DOI] [PubMed] [Google Scholar]
- 34.Sommer R., Cabaj A., Sandu T., Lhotsky M., Measurement of UV radiation using suspensions of microorganisms. J. Photochem. Photobiol. B 53, 1–6 (1999). [DOI] [PubMed] [Google Scholar]