Significance
The structural dynamics of the dendritic synapse, arising from the remodeling of actin cytoskeletons, has been widely associated with memory and cognition. The remodeling is regulated by intracellular Ca2+ levels. Under low Ca2+ concentration, actin filaments are bundled by a calcium signaling protein, CaMKII. When the Ca2+ concentration is raised, CaMKII dissociates from actin and opens the window for actin remodeling. At present, the molecular details of the actin bundling and regulation are elusive. Herein we use experimental tools along with molecular simulations to construct a model of how CaMKII bundles actin and how the CaMKII–actin architecture is regulated by Ca2+ signals. In this way, our results explain how Ca2+ signals ultimately change the structure of the dendritic synapse.
Keywords: CaMKII, actin remodeling, calmodulin
Abstract
Calcium/calmodulin-dependent kinase II (CaMKII) plays a key role in the plasticity of dendritic spines. Calcium signals cause calcium−calmodulin to activate CaMKII, which leads to remodeling of the actin filament (F-actin) network in the spine. We elucidate the mechanism of the remodeling by combining computer simulations with protein array experiments and electron microscopic imaging, to arrive at a structural model for the dodecameric complex of CaMKII with F-actin. The binding interface involves multiple domains of CaMKII. This structure explains the architecture of the micrometer-scale CaMKII/F-actin bundles arising from the multivalence of CaMKII. We also show that the regulatory domain of CaMKII may bind either calmodulin or F-actin, but not both. This frustration, along with the multipartite nature of the binding interface, allows calmodulin transiently to strip CaMKII from actin assemblies so that they can reorganize. This observation therefore provides a simple mechanism by which the structural dynamics of CaMKII establishes the link between calcium signaling and the morphological plasticity of dendritic spines.
Hebbian learning in the brain requires somehow transforming ephemeral electrical and chemical signals at the synapse into long-lasting changes in the strength of synaptic connections (1). One form of this strengthening, called long-term potentiation (LTP), has been directly observed accompanying the changes in synaptic plasticity. It originates from patterns of neuronal activity that elevate intracellular calcium (2) and subsequently modulate the architecture of the actin cytoskeleton (3). A key actin-binding protein in the calcium signaling cascade is Ca2+/calmodulin-dependent kinase II (CaMKII) (4, 5). The high abundance of CaMKII in dendritic synapses (1 to 2% of the total brain protein) suggests that, in addition to its catalytic function, CaMKII might participate in establishing the structure of the cytoskeleton in the dendritic spine. The dual role of CaMKII allows it directly to transmit a calcium signal to the morphology of a dendritic spine and thus change synaptic strength in response to electrochemical inputs (6). Supporting this attractive hypothesis for the physical basis of synaptic plasticity is the fact that, at low Ca2+concentration, CaMKII binds to actin filaments and bundles them together, stabilizing a rigid dendritic spine structure (7, 8). Yet, when the Ca2+ concentration is raised, the Ca2+ activates another signaling protein, calmodulin, which, in turn, triggers the dissociation of CaMKII from F-actin bundles. Once unbundled, during a brief time interval, the actin cytoskeleton can be remodeled by a panoply of other proteins that includes cofilin and Arp 2/3 (9). Herein, we employ a combined approach of computer simulations, kinetic models, protein array experiments, and electron microscopy, to reveal the regulatory role of CaMKII in connecting signal transduction and actin reorganization through integrative modeling from the nanoscale to the microscale. Together with characterizing the structural basis of CaMKII/F-actin architectures, we suggest a mechanism by which calcium-activated calmodulin directly catalyzes the disassembly of CaMKII/F-actin complexes. Moreover, this minimal set of molecules can form the core of a signal decoding network to support dendrite growth, branching, and structural changes at synapses.
Owing to both the complexity of the CaMKII molecule itself and the variety of structures it can form with F-actin at higher levels of organization (4), no high-resolution structures of CaMKII/F-actin assemblies have been obtained directly from experiment to date. CaMKII is a unique dodecameric holoenzyme (Fig. 1A) with an interesting geometry of doubly decked hexamers. Each CaMKII monomer consists of a catalytic domain, a regulatory domain, a linker, and an association domain that forms the “hub” of a dodecamer. The crystal structure of CaMKII holoenzymes has, so far, only been obtained by amputating the flexible linker domain (10) (Fig. 1A; the position of the missing linker is indicated by the dashed orange arrow). In the full-length CaMKII holoenzyme, electron microscopy confirms that the linker adopts an extended, highly flexible structure (11, 12). Despite its flexibility, the linker plays an important role in CaMKII binding to F-actin. The CaMKIIβ isoform that has a long linker of 93 amino acids (aa), binds more strongly to F-actin than does the CaMKIIα isoform that has only a short linker of 30 aa (8, 13). The association domain of CaMKII is also important for actin binding (14, 15). Therefore, it is highly probable that all of these domains are important in forming CaMKII/F-actin complexes.
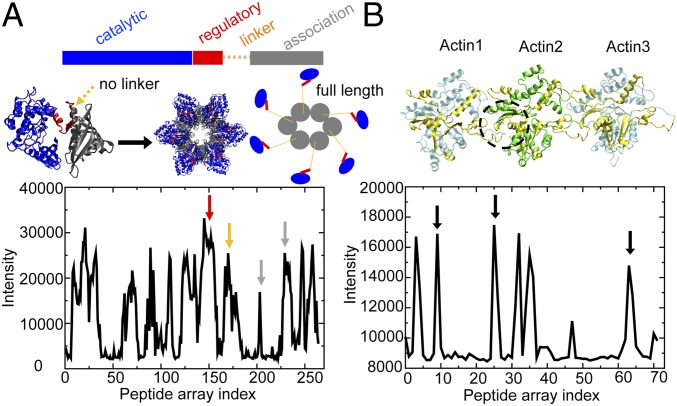
Fig. 1.
Structure and experimental peptide array data for CaMKIIβ and actin. (A) The crystal structure of a CaMKII construct without linker (PDB ID code 3SOA). (Left) CaMKII monomer; (Middle) CaMKII dodecamer; (Right) in comparison, a full-length CaMKII construct with linker displays an extended structure. The intensity profile of CaMKIIβ peptides binding to actin in the peptide arrays is shown below. Multiple CaMKII domains show binding with actin (red arrow, regulatory domain; orange arrow, linker; gray arrow, association domain). (B) The structure of 3 consecutive actins (Actin1 to Actin3) in a near-atomic model for an actin filament (PDB ID code 3J8I). The intensity profile of actin peptides binding to CaMKIIβ in the peptide arrays is shown below. Actin regions showing a high degree of binding with CaMKIIβ are colored in yellow on the ribbon diagram. The dashed black circle represents the hydrophobic cleft of F-actin, a conserved binding pocket for many actin-binding proteins (shown in SI Appendix, Fig. S1). Corresponding peptide signals are identified by black arrows.
In this study, we propose a nanoscale structural model for the binding complex of the CaMKIIβ isoform with F-actin, which combines peptide array experiments to guide the selection of possible binding regions with computer modeling that uses a coarse-grained predictive energy landscape, Associative Memory, Water Mediated, Structure and Energy Model (AWSEM) (16). By extracting geometric constraints from this structural model of the CaMKII−actin complex and acknowledging that the multivalency of the CaMKII holoenzyme originates from its unique 6-fold geometry, we derive simple rules to construct micrometer-scaled models for the bundled CaMKII−actin networks. The predicted large-scale architectures and critical stoichiometries in the CaMKIIβ−actin networks match the electron microcopy images. Using the same computational tools, we also study the catalysis of disassembly of a CaMKIIβ−actin complex by Ca2+-bound calmodulin. Together, these simulations suggest a simple mechanism for how CaMKIIβ participates in actin reorganization, establishing the link between calcium signaling and the morphological plasticity of dendritic spines.
Results
The Binding Complex of CaMKII and F-Actin.
Peptide arrays using overlapping sequential fragments of CaMKIIβ allow us to determine the regions of CaMKIIβ most critical to actin binding. Fluorescently labeled F-actin is washed over the array to identify potential binding regions. Peptides from the linker domain of CaMKIIβ strongly bind F-actin (Fig. 1A, solid orange arrow), suggesting that the linker may serve as an important binding interface as hypothesized previously (7). In the peptide arrays, the regulatory domain (solid red arrow) and the association domain (solid gray arrow) of CaMKIIβ also display interactions with strong binding signals. This observation supports the notion that the binding between CaMKIIβ and actin involves several of the domains in the CaMKIIβ monomer. Although the catalytic domain in this array assay displays significant F-actin binding, previous experiments (7) did not find that the deletion of the catalytic domain greatly influences the interaction between CaMKIIβ and F-actin. To identify the peptide fragments of F-actin that form the binding interface, we constructed an array of actin peptide fragments and probed their binding to CaMKIIβ (Fig. 1B). The regions of F-actin with strong CaMKIIβ binding are highlighted in the structure of 3 consecutive actin regions (labeled Actin1 to Actin3), which were derived from a near-atomic modeling for the actin filament (Protein Data Bank [PDB] ID code 3J8I). Together, these binding regions form a hydrophobic binding pocket at the interface between adjacent actin regions (Fig. 1B, black circle between Actin1 and Actin2, black arrows in the peptide array data). This same pocket serves as the binding region for at least 18 other actin-binding proteins whose structures have been determined by X-ray crystallography (SI Appendix, Fig. S1). Our simulations below confirm that this binding pocket also plays an important role in the CaMKII binding to actin.
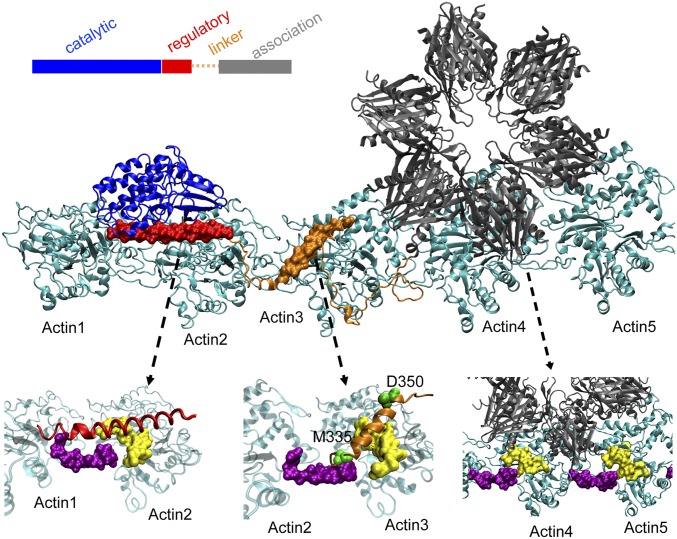
Guided by these peptide array experiments, we carried out molecular dynamics simulations with AWSEM (16) to develop a near-atomic structural model (see details in SI Appendix) of the binding complex of CaMKIIβ isoform with F-actin (Fig. 2). In the resulting model, CaMKII utilizes its multiple domains to bind 5 consecutive actin regions (Actin1 to Actin5) along an F-actin filament. Our simulations suggest that the regulatory domain in a full-length CaMKIIβ monomer forms a helical structure and docks to the binding pocket between Actin1 and Actin2 (Fig. 2, left arrow). The central linker region (Fig. 2, middle arrow) also forms a helical structure which docks to the binding pocket between Actin2 and Actin3. The central linker region is followed by a long coil that spreads over the surfaces of Actin3 and Actin4. This coil connects to the association domain (Fig. 2, right arrow) which docks to the surface of Actin4 and Actin5. As mentioned above, the association domains in CaMKIIβ assemble to form a dodecameric structure (called the “hub” below). As shown in Fig. 2, the 3 consecutive monomers in the hub that interact with Actin4 and Actin5 define a unique orientation of CaMKIIβ with respect to the F-actin fiber. In the next section, we show that this geometric arrangement provides the structural basis for CaMKIIβ to both bundle and arborize actin fibers.
Fig. 2.
Computational model for the binding complex of CaMKII–actin filament. Five consecutive actins (Actin1 to Actin5) are colored in cyan. The binding interfaces of CaMKII in its regulatory domain (red), linker domain (orange), and association domain (gray) are zoomed in, pointed by the dashed black lines. The full regulatory domain binds to the hydrophobic cleft of F-actin whose surface is colored in purple and yellow. For the linker domain, its central part ranging from M335 to D350 forms a helix and serves as the main binding interface. For the association domain, its first 30 residues serve as the main binding interface.
The Structural Basis of Actin Bundling by CaMKIIβ.
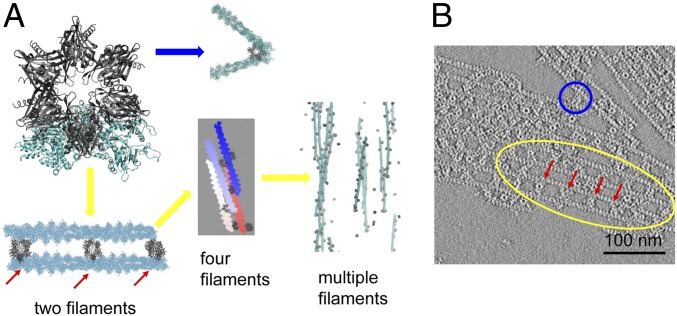
Actin fibers often bundle in parallel in the presence of CaMKIIβ (8, 17). This structural feature arises from the multivalence of CaMKIIβ which allows CaMKIIβ to bind multiple actin fibers simultaneously. To explore the structural basis of this parallel alignment, we first created models of 2 bundled actin fibers based on the predicted structure of the complex of CaMKIIβ with a single F-actin (Fig. 2), and then mounted the second F-actin onto the opposing subunits of the CaMKIIβ hub (Fig. 3A). Because the orientation between CaMKIIβ and F-actin is uniquely determined, the resulting offset angle between the long axes of the 2 actin filaments that both dock on CaMKIIβ is small but nonzero, approximately 2°. Thus, the hub serves as a structural anchor to restrict the orientations of neighboring actin filaments. Because of the small offset angle, CaMKIIβ allows the formation of nearly parallel aligned F-actin bundles as we increase the number of filaments in the computational model (Fig. 3A, yellow arrow). This predicted near-parallel alignment of actin filaments agrees with what is seen in images from electron microscopy (Fig. 3B) and other studies (7, 8). The multivalence of CaMKIIβ also permits 2 filaments to bind on the neighboring subunits in the dodecameric association domain at an acute angle of roughly 60°. This structure allows the formation of a branched actin network (Fig. 3A, blue arrow). As depolarization-mediated calcium entry and CaMKII activation subsides, this arborization of the F-actin network mediated by CaMKIIβ bundling of F-actin may support the mushroom shape of the dendritic spine during the maintenance phase of LTP (7).
Fig. 3.
Structural basis for CaMKIIβ in bundling actin filaments. (A) Actin network produced in simulations by docking multiple actin filaments onto the structure in Fig. 2. Multiple actin strands can either align in parallel (yellow arrow) or form branched structures (blue arrow). (B) EM of negative stained CaMKII/actin rafts formed on a lipid monolayer. Actin filaments are arranged either in parallel (yellow circle) or branched (blue circle). For both simulations and experiments, the spacing between 2 adjacent CaMKII particles in the complexes is nearly fixed at 36 nm (red arrow).
Actin Twisting Controls the Structural Pattern of CaMKIIβ−Actin Bundling.
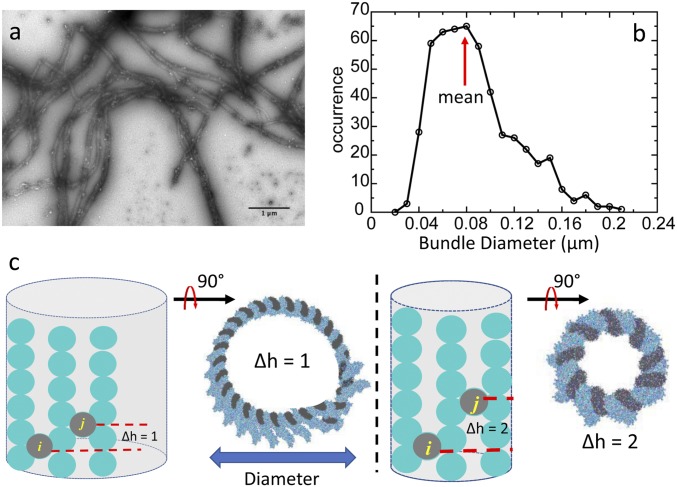
A single actin filament exhibits a helical twist along its long axis whereby the rise is 2.76 nm and the twist is −166.6 ± 0.6° per molecule (18). The innate twist of an actin filament plays 2 roles in the architecture of the CaMKIIβ−actin system. First, the twist implies that the sites for CaMKIIβ interaction are not uniformly distributed along a bundle. Instead, there is a periodic binding interface along F-actin for CaMKIIβ. This periodicity enforces an even spacing of around 36 nm between 2 adjacent CaMKIIβ molecules along the F-actin as observed in electron microscopy images, and in computational simulations (red arrows in Fig. 3). Second, the helical twist provides a small mismatch between the pitches of individual actin filaments, leading to an intrinsic limit to bundle growth shown in electron microcopy (Fig. 4 A and B). Our micrometer-scaled structural model shows that, when CaMKII bundles F-actins, aligned filaments form a curved surface (Fig. 4C). The curvature depends on the packing pattern of CaMKIIβ along the actin filaments. When CaMKIIβ binds to different grooves in an actin filament, the curvature of the filament will vary. The difference in the number of registers where CaMKIIβ binds, called “∆h,” dictates the curvature of actin filaments that can assemble into a barrel. The diameter of the barrel decreases with ∆h. When ∆h = 1, the predicted diameter of the barrel achieves a maximum of 93 nm. This prediction is close to the mean value of 83 nm obtained from the experimental measurements of the diameter of CaMKIIβ/actin bundles (red arrow in Fig. 4B). The idea that the diameter of a bundle formed by actin-binding protein and actin filaments is limited by the helical twist of an actin filament was proposed by Claessens et al. (19) in their α-actinin/F-actin bundles, and can be observed in our detailed structural model of CaMKIIβ/F-actin bundles.
Fig. 4.
Limited diameter of actin bundles. (A) Electron micrograph of negative-stained CaMKIIβ/actin bundles. (B) Diameter distribution of actin bundles in the experiment. (C) Actin forms a curved surface in simulations due to the helical twist of actin filament. The curvature depends on the difference in the number of registers where adjacent CaMKII holoenzymes bind, called “∆h.” If all CaMKII holoenzymes pack with the same pattern, the curved surface will form a barrel. The outer boundary of the barrel dictates the diameter of actin bundles. CaMKII particles are represented by gray circles. Actin subunits are represented by cyan circles.
Ca2+/Calmodulin Regulates the Binding Between CaMKIIβ and Actin through a ter-Molecular Reaction.
While the association domain of CaMKIIβ serves as the structural anchor for actin bundling, the regulatory domain is critically important for Ca2+/calmodulin regulation. Our structure shows that the regulatory domain directly binds to F-actin (Fig. 5A, red surfaces); thus it enhances the stability of the CaMKIIβ−actin complex. The same regulatory domain, however, is also the binding target of Ca2+/calmodulin. Aligning the crystal structure of Ca2+/calmodulin–CaMKII peptide binding complex (PDB ID code 1CDM) that includes the same sequence of the regulatory domain against our model leads to serious steric clashes between Ca2+/calmodulin and F-actin (red arrow in Fig. 5A). Therefore, the formation of a binary complex between the regulatory domain and F-actin is mutually exclusive with the formation of a binary complex between the regulatory domain and calmodulin. This frustration suggests that calmodulin might catalyze disassembly of CaMKIIβ−actin networks (20).
Fig. 5.
The structural basis for Ca2+/calmodulin regulating the binding between CaMKII and actin. (A) The binding complex between the regulatory domain (colored in red), part of the linker (colored in yellow), and F-actin (colored in cyan). The structure of Ca2+/calmodulin (colored in purple) is docked to the regulatory domain by utilizing the crystal structure of Ca2+/calmodulin–CaMKII peptide binding complex (PDB ID code 1CDM). There are atom clashes between Ca2+/calmodulin and F-actin (shown by the red arrow). (B) Kinetic model for CaMKII binding to actin regulated by Ca2+/calmodulin. The letter “R” represents the regulatory domain, “L” represents the linker, “A” represents the association domain, and “C” represents Ca2+/calmodulin. The cyan line represents F-actin. Arrows represent the association or dissociation events; k0 represents the association rate of Ca2+/calmodulin to CaMKII, while [C] represents the concentration of Ca2+/calmodulin. Once Ca2+/calmodulin binds to the regulatory domain (dashed arrow), the regulatory domain cannot bind to F-actin (red cross). (C) Free-energy profiles of individual CaMKII domains binding to F-actin measured in simulations, as a function of the distance between the center of mass of each individual CaMKII domain and the center of mass of the conserved binding pocket of actin. The free-energy barriers, ∆G, are used to estimate the relative rates in the kinetic model shown in B. (Left) The regulatory domain. (Middle) The linker. (Right) The association domain. (D) The overall dissociation rate of CaMKII from F-actin with increasing concentrations of Ca2+/calmodulin divided by the dissociation rate without Ca2+/calmodulin.
We developed a kinetic scheme to explain the disassembly of CaMKIIβ−actin networks (Fig. 5B and SI Appendix, Fig. S6). Owing to the multipartite nature of the binding interface, a ter-molecular reaction can occur where Ca2+/calmodulin binds to the regulatory domain destabilizing the CaMKIIβ/actin interaction due to the atom clashes between Ca2+/calmodulin and actin, initiating disassembly. In addition, once Ca2+/calmodulin binds to the regulatory domain, CaMKII may become autophosphorylated, increasing the binding with Ca2+/calmodulin (21, 22). This step would significantly block the regulatory domain from participating in actin assembly.
We computed the individual dissociation rate coefficients in the kinetic scheme for each domain using their corresponding binding free-energy profiles to approach the actin fiber computed via the AWSEM simulations with the weighted histogram method (Fig. 5C). Then the rate coefficients were estimated using a 1-dimensional Kramers model (see detailed calculations in SI Appendix). Fig. 5D shows that the effective dissociation rate of CaMKIIβ from F-actin is increased roughly 10-fold over its uncatalyzed value once the concentration of Ca2+/calmodulin reaches 10 μM. Our result from the kinetic modeling agrees reasonably well with an experimental measurement of disassembly kinetics using fluorescence video microscopy and image analysis (20) (A detailed discussion is provided in SI Appendix). These results provide a plausible mechanism for how short-term, robust, changes in Ca2+ concentration are decoded by CaMKII and lead directly to reorganization of the spine actin cytoskeleton.
It should be noted that, in the kinetic scheme (SI Appendix, Fig. S6), we considered an intermediate state where Ca2+/calmodulin only partially binds to CaMKII as proposed by previous theoretical studies (23). In this intermediate state, the atom clashes between Ca2+/calmodulin and actin might be resolved by partially unfolding Ca2+/calmodulin and by partially breaking up the interface between the regulatory domain of CaMKII with actin. However, once the regulatory domain dissociates from actin, Ca2+/calmodulin will quickly adjust its binding interface to fully bind to the regulatory domain (23). Therefore, the specific form of this intermediate state will not qualitatively influence the regulation role of Ca2+/calmodulin, as demonstrated by our calculations shown in SI Appendix, Fig. S6.
Discussion
The Binding Interface of a Full-Length CaMKII with F-Actin Involves Multiple Domains in CaMKII.
The predicted structural model of the CaMKII/F-actin assemblies indicates that the complex binding interface involves not only the linker but also the regulatory domain and the association domain of CaMKII. The 3 domains each bind to the conserved hydrophobic pockets within clefts between consecutive monomers in the actin fiber (Figs. 1B and 2). Two previous in situ experiments are consistent with this prediction and show that the association domain can colocalize with F-actin (14, 15). In addition, both our experiments and simulations show that the central part of the linker directly binds to actin (Fig. 2) and contributes 40% of the overall binding free energy of CaMKII to actin. The critical role of linker binding explains the difference in the actin-binding affinity between the 2 CaMKII isoforms: CaMKIIα and CaMKIIβ. CaMKIIα, which lacks the linker, has a much weaker actin binding interaction than CaMKIIβ (13, 17).
Our investigation shows that a smaller contribution, roughly 20% of the binding free energy between CaMKII and F-actin, comes from the regulatory domain. We calculate that the linker and the association domain contribute nearly equally (Fig. 5C). The 3 domains of CaMKII monomers spread apart and bind to the interfaces between 5 consecutive actin monomers in the actin filament (Fig. 2). This design has 3 major advantages. First, the binding affinity of each domain is relatively weak. This allows a rapid response to Ca2+/calmodulin binding by dissociating the regulatory domain from F-actin, initiating unzippering of the CaMKII monomer from the actin filament. Second, CaMKII is a dodecamer that multivalently binds several F-actins. So CaMKII/F-actin bundles are quite stable. Third, the hydrophobic pocket where CaMKII binds to F-actins is also utilized by many other actin-remodeling proteins such as Arp2/3 and cofilin (24) which must compete with CaMKII for this site. Therefore, the binding between CaMKII and F-actin should inhibit the ability of cofilin to bind to and then sever F-actin (9). Our model of CaMKII and F-actin assemblies thus provides a molecular basis for the Ca2+-induced plasticity of the actin cytoskeleton while providing the needed long-term stability to maintain the actin cytoskeleton of the dendritic spine.
CaMKII/F-Actin Bundles Are Determined by Simple Geometric Rules Implied by the Predicted CaMKII/F-Actin Complex Structure.
Our structural model of the nanoscale complex provides insight into how CaMKII can influence cytoskeletal architectures in synapses by bundling F-actins at the microscale. The association domain serves as the structural anchor that bundles actin filaments. The formation of ordered, near-parallel F-actin bundles restrains both the distances between neighboring CaMKII-bound actin filaments and their orientations. The linker domain, although contributing to the thermodynamics of binding, by itself does not provide strong geometrical constraints because of its conformational flexibility (11). In contrast, the well-structured association domain with higher rigidity is able to restrain the angle between neighboring CaMKII-bound actin filaments. The strict geometry of the binding interface between the association domain and actin guarantees that, if 2 actin filaments bind to the opposite sides of a dodecameric CaMKII holoenzyme, they will be aligned in parallel. This property is important for the formation of an ordered actin network at large scales (Fig. 3A, yellow arrow) and thus distinguishes CaMKII as one of the few macromolecules that can bundle actin filaments. We also note that multiple positively charged residues of CaMKII distribute in or near the binding interface between CaMKII and F-actin (SI Appendix, Fig. S2) and therefore neutralize the negative charges of F-actin. This effect also favors the bundling of actin filaments by reducing their mutual electrostatic repulsion.
The Interplay between Ca2+ Signaling Transduction and Actin Reorganization Lies in the Regulatory Domain of CaMKII.
Electron microscopy images of native CaMKII holoenzymes containing the linker region demonstrate that the regulatory and catalytic domains extend away from the core (11, 12), making them available for interactions with actin and other proteins. Our studies indicate that the regulatory domain of CaMKII binds to F-actin directly. This binding provides a plausible mechanism for Ca2+/CaM regulation by competing for this binding interface.
Our model suggests that, in the basal state of the synapse, CaMKII binds to F-actin tightly. When the concentration of calcium in the synapse rises, calcium turns a significant pool of apo-calmodulin in the synapse into its active holo form. Once Ca2+/calmodulin binds to the regulatory domain, the regulatory domain displaces F-actin due to steric effects. The physiological concentration of CaM in the brain varies from 17 μM to 26 μM (25). At this range of Ca2+/calmodulin concentration, our calculations show that the overall dissociation rate of CaMKII is increased 10-fold (Fig. 5D). Therefore, the interaction of Ca2+/calmodulin with the regulatory domain is the lynchpin that provides a direct connection between calmodulin-dependent calcium signal transduction and actin reorganization. Autophosphorylation and CaM trapping can dynamically adjust the propensity of individual CaMKII subunits within a holoenzyme to participate in binding to F-actin. Future experiments will be required to address the kinetics and geometrical consequences of subunit specific alterations in the context of CaMKII holoenzyme/F-actin interactions. The kinetic model for this reorganization resembles the molecular stripping mechanism observed in other kinetically controlled biological systems (26). In conclusion, our integrative structure modeling provides insight into the molecular basis of the structural plasticity in dendritic spines mediated by CaMKII and F-actins.
Methods
Experiments.
Protein binding to blots or slide arrays.
Fluorescently labeled kinase or actin (2 nM to 10 nM; see detailed methodology in SI Appendix) diluted in blocking buffer, was applied to the membrane for 20 min to 30 min at room temperature with constant shaking. Blots or slides were then washed 3 times with binding buffer and immediately imaged. Dylight800-deltaCaMKII binding was quantified using a Licor Imaging Station (Licor Biotechnology), and binding of Alexa647-actin and Alexa647-CaMKII was quantified using a Typhoon Trio+ Imaging Station (Molecular Dynamics). Immediately before adding labeled protein, each blot/array was imaged under the same conditions (laser and gain settings) to establish background counts. For quantification, regions of interest were created with the MicroArray Profile in ImageJ, and spot intensity was quantified in blots imaged under nonsaturating conditions for the detector. The identical region-of-interest pattern was placed over the imaged blots/arrays before addition of fluorescently labeled binding protein to serve as background subtraction.
Actin/CaMKII binding assessed by electron microscopy.
The morphology of CaMKII/F-actin complexes was assessed by negative stain electron microscopy basically, as previously described (17), and detailed methodology is provided in SI Appendix. CaMKIIβ was added at various ratios to a fixed actin concentration, and the reactions were incubated at room temperature for 1 h. Aliquots were added to freshly glow-discharged carbon-coated grids for 1 min, washed twice with water, negative-stained with 0.75% freshly prepared uranyl formate for 30 s, and air dried. For higher-resolution imaging, F-actin/CaMKIIβ complexes were prepared on lipid monolayers. The monolayer was then negative-stained as above. Negative-stained samples were imaged either on a JEOL1400 operated at 120 kV using an Orius (Gatan) side-mounted charge-coupled device camera at 18 Å/pixel at the image plane or on a Polara G2 (FEI) cryoelectron microscope operated at 300 kV in low-dose mode with images collected on a K2 summit direct electron detector (Gatan) at 2.6 Å/pixel at the image plane as described previously (8, 17). Bundle diameter was determined using the measure tool in ImageJ.
Computational modeling of the CaMKII−actin complex.
Due to the large size of CaMKII holoenzyme, we first divided the full-length CaMKII into several overlapped segments and predicted the binding between each individual segment and F-actin. Next, we combined all of the segments together to construct a model for the binding complex of the full-length CaMKII and F-actin. Due to space considerations of Methods, the detailed protocol is included in SI Appendix.
Supplementary Material
Acknowledgments
This work was supported by the Center for Theoretical Biological Physics sponsored by NSF Grant PHY-1427654. This work was also supported by NSF Grants CHE-1614101 and CHE-1743392. Additional support was provided by the D.R. Bullard-Welch Chair at Rice University, Grant C-0016, and an endowment from the William Wheless III Professorship at University of Texas Health Science Center at Houston.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1911452116/-/DCSupplemental.
References
- 1.Nicoll R. A., A brief history of long-term potentiation. Neuron 93, 281–290 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Okamoto K., Bosch M., Hayashi Y., The roles of CaMKII and F-actin in the structural plasticity of dendritic spines: A potential molecular identity of a synaptic tag? Physiology (Bethesda) 24, 357–366 (2009). [DOI] [PubMed] [Google Scholar]
- 3.Nakahata Y., Yasuda R., Plasticity of spine structure: Local signaling, translation and cytoskeletal reorganization. Front. Synaptic Neurosci. 10, 29 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lisman J., Schulman H., Cline H., The molecular basis of CaMKII function in synaptic and behavioural memory. Nat. Rev. Neurosci. 3, 175–190 (2002). [DOI] [PubMed] [Google Scholar]
- 5.Zalcman G., Federman N., Romano A., CaMKII isoforms in learning and memory: Localization and function. Front. Mol. Neurosci. 11, 445 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim K., Saneyoshi T., Hosokawa T., Okamoto K., Hayashi Y., Interplay of enzymatic and structural functions of CaMKII in long-term potentiation. J. Neurochem. 139, 959–972 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Okamoto K., Narayanan R., Lee S. H., Murata K., Hayashi Y., The role of CaMKII as an F-actin-bundling protein crucial for maintenance of dendritic spine structure. Proc. Natl. Acad. Sci. U.S.A. 104, 6418–6423 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanabria H., Swulius M. T., Kolodziej S. J., Liu J., Waxham M. N., betaCaMKII regulates actin assembly and structure. J. Biol. Chem. 284, 9770–9780 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim K., et al. , A temporary gating of actin remodeling during synaptic plasticity consists of the interplay between the kinase and structural functions of CaMKII. Neuron 87, 813–826 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chao L. H., et al. , A mechanism for tunable autoinhibition in the structure of a human Ca2+/calmodulin- dependent kinase II holoenzyme. Cell 146, 732–745 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Myers J. B., et al. , The CaMKII holoenzyme structure in activation-competent conformations. Nat. Commun. 8, 15742 (2017). Erratum in Nat. Commun. 9, 16180 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaertner T. R., et al. , Comparative analyses of the three-dimensional structures and enzymatic properties of alpha, beta, gamma and delta isoforms of Ca2+-calmodulin-dependent protein kinase II. J. Biol. Chem. 279, 12484–12494 (2004). [DOI] [PubMed] [Google Scholar]
- 13.Shen K., Teruel M. N., Subramanian K., Meyer T., CaMKIIbeta functions as an F-actin targeting module that localizes CaMKIIalpha/beta heterooligomers to dendritic spines. Neuron 21, 593–606 (1998). [DOI] [PubMed] [Google Scholar]
- 14.Caran N., Johnson L. D., Jenkins K. J., Tombes R. M., Cytosolic targeting domains of gamma and delta calmodulin-dependent protein kinase II. J. Biol. Chem. 276, 42514–42519 (2001). [DOI] [PubMed] [Google Scholar]
- 15.Lin Y. C., Redmond L., CaMKIIβ binding to stable F-actin in vivo regulates F-actin filament stability. Proc. Natl. Acad. Sci. U.S.A. 105, 15791–15796 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davtyan A., et al. , AWSEM-MD: Protein structure prediction using coarse-grained physical potentials and bioinformatically based local structure biasing. J. Phys. Chem. B 116, 8494–8503 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoffman L., Farley M. M., Waxham M. N., Calcium-calmodulin-dependent protein kinase II isoforms differentially impact the dynamics and structure of the actin cytoskeleton. Biochemistry 52, 1198–1207 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujii T., Iwane A. H., Yanagida T., Namba K., Direct visualization of secondary structures of F-actin by electron cryomicroscopy. Nature 467, 724–728 (2010). [DOI] [PubMed] [Google Scholar]
- 19.Claessens M. M., Semmrich C., Ramos L., Bausch A. R., Helical twist controls the thickness of F-actin bundles. Proc. Natl. Acad. Sci. U.S.A. 105, 8819–8822 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khan S., Downing K. H., Molloy J. E., Architectural dynamics of CaMKII-actin networks. Biophys. J. 116, 104–119 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanson P. I., Meyer T., Stryer L., Schulman H., Dual role of calmodulin in autophosphorylation of multifunctional CaM kinase may underlie decoding of calcium signals. Neuron 12, 943–956 (1994). [DOI] [PubMed] [Google Scholar]
- 22.Putkey J. A., Waxham M. N., A peptide model for calmodulin trapping by calcium/calmodulin-dependent protein kinase II. J. Biol. Chem. 271, 29619–29623 (1996). [DOI] [PubMed] [Google Scholar]
- 23.Wang Q., et al. , Protein recognition and selection through conformational and mutually induced fit. Proc. Natl. Acad. Sci. U.S.A. 110, 20545–20550 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Galkin V. E., et al. , Remodeling of actin filaments by ADF/cofilin proteins. Proc. Natl. Acad. Sci. U.S.A. 108, 20568–20572 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kakiuchi S., et al. , Quantitative determinations of calmodulin in the supernatant and particulate fractions of mammalian tissues. J. Biochem. 92, 1041–1048 (1982). [DOI] [PubMed] [Google Scholar]
- 26.Potoyan D. A., Zheng W., Komives E. A., Wolynes P. G., Molecular stripping in the NF-κB/IκB/DNA genetic regulatory network. Proc. Natl. Acad. Sci. U.S.A. 113, 110–115 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.