Significance
Metal clusters play important roles in a wide variety of proteins. In cyanobacteria, algae, and plants, photosystem II uses light energy to oxidize water and release O2 at an active site that contains 1 calcium and 4 manganese atoms. This cluster must be built within the protein environment through a process known as photoassembly. Through experiments and simulations, we found that the efficiency of photoassembly was highly dependent on protons and chloride. Surprisingly, when the solvent was switched from H2O to deuterated water, D2O, the yield of photoassembly was higher. These results provide insights into the stepwise mechanism of photoassembly that can inform synthesis and repair strategies being developed for artificial photosynthesis technologies.
Keywords: photosystem II, assembly, inverse isotope effect, proton transfer, electrostatics
Abstract
Photosystem II (PSII) performs the solar-driven oxidation of water used to fuel oxygenic photosynthesis. The active site of water oxidation is the oxygen-evolving complex (OEC), a Mn4CaO5 cluster. PSII requires degradation of key subunits and reassembly of the OEC as frequently as every 20 to 40 min. The metals for the OEC are assembled within the PSII protein environment via a series of binding events and photochemically induced oxidation events, but the full mechanism is unknown. A role of proton release in this mechanism is suggested here by the observation that the yield of in vitro OEC photoassembly is higher in deuterated water, D2O, compared with H2O when chloride is limiting. In kinetic studies, OEC photoassembly shows a significant lag phase in H2O at limiting chloride concentrations with an apparent H/D solvent isotope effect of 0.14 ± 0.05. The growth phase of OEC photoassembly shows an H/D solvent isotope effect of 1.5 ± 0.2. We analyzed the protonation states of the OEC protein environment using classical Multiconformer Continuum Electrostatics. Combining experiments and simulations leads to a model in which protons are lost from amino acid that will serve as OEC ligands as metals are bound. Chloride and D2O increase the proton affinities of key amino acid residues. These residues tune the binding affinity of Mn2+/3+ and facilitate the deprotonation of water to form a proposed μ-hydroxo bridged Mn2+Mn3+ intermediate.
Inorganic cofactors facilitate chemical reactions in a wide variety of enzymes. For multinuclear cofactors, individual metal ions must be assembled into clusters with specific geometries and ligands. These clusters can be assembled externally in specialized scaffold proteins, such as those commonly used for iron/sulfur clusters (1), or can be assembled in situ directly in the apo enzyme as found for the Mn4CaO5 oxygen-evolving complex (OEC) in photosystem II (PSII) (2–4). The OEC has a complex geometry that includes a Mn3CaO4 heterocubane linked to a fourth Mn via an additional μ-oxo bridge (5, 6). Most of the Mn and Ca ligands are amino acid side chains from PSII protein subunits (6). This specialized ligand environment results in a cluster that cannot be extracted without destruction (3).
In its functional form, the PSII OEC has high-valent Mn3+ and Mn4+ ions (7). During cluster assembly, light excitation of the primary chlorophyll-a donor, P680, leads to oxidation of labile Mn2+ via a redox-active tyrosine residue. This light-dependent process does not require metallochaperones and has been termed photoactivation or photoassembly (reviewed in refs. 2, 3).
The oxidation of water to O2 by PSII requires highly oxidizing reduction potentials for the OEC (standard reduction potential (Em) = 0.9 V) and the primary chlorophyll-a donor, P680 (Em = 1.2 V) (4). These values are among the most positive found in biology. In vivo, PSII is subjected to oxidative damage and must be frequently repaired (8, 9). The cell includes repair machinery for complex turnover so that a functional population is maintained.
Instead of replacing the entire complex, only the protein subunit that is most likely to be damaged, D1 (10), is replaced (11, 12). This subunit contains most of the OEC ligating residues (6). A new D1 subunit is then translated and inserted into the PSII complex. Following the processing of a C-terminal tail, a new OEC must be photoassembled before PSII can be functional (9). The lifetime of PSII is ∼20 to 40 min in the chloroplast (13), and it is repaired at a high bioenergetic cost (14, 15).
The complex yet efficient process of PSII repair makes this reaction center a model catalytic system. Nature builds a scaffold in which a water-oxidation catalyst is self-assembled from earth-abundant, nontoxic Mn, Ca, and H2O. The same protein scaffold that facilitates catalytic activity also facilitates assembly. If understood at a mechanistic level, these remarkable features will inform practical inorganic catalytic systems, including those used in artificial photosynthesis technologies.
The net reaction of OEC photoassembly to form the first catalytically active intermediate, the S0 state (16, 17), is shown in Eq. 1:
| [1] |
This process includes 5 sequential Mn2+ and Ca2+ bindings, 5 one-electron Mn oxidations, and proton releases from 5 waters to form multiple μ-oxo or μ-hydroxo bridges. In addition to these 5 water molecules, several other waters must be removed from [Mn(H2O)6]2+ and [Ca(H2O)]2+ as these ligands are replaced by amino acid residues. The functional OEC retains 4 terminal water ligands (6).
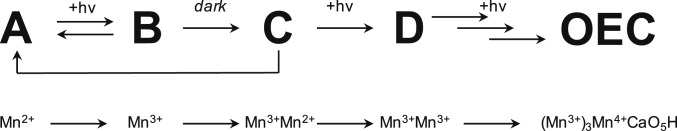
An early study by Radmer and Cheniae (18) established that OEC photoassembly is a “2-quantum” process. In this model shown in Fig. 1, which includes later insights from other groups (reviewed in ref. 2), an initiating light-dependent step is followed by a slow light-independent step and another light-dependent step. First, 1 Mn2+ binds to a single high-affinity site (HAS). The dissociation constant (Kd; at equilibrium) of the first Mn2+ at the HAS is 40 to 50 μM (19). This intermediate is notated as A in Fig. 1. Next, Mn2+ in the HAS is oxidized to Mn3+ through photochemical charge separation (20) to form intermediate B. B is slowly converted to C with a half-life of 100 to 150 ms. This conversion does not require light and involves a second metal binding event. Contrasting reports suggest that the second metal that binds is Ca2+ or Mn2+ (19, 21–23). The net conversion of A to C has a low quantum yield because both B and C can spontaneously decay back to A (24). Therefore, the production of C is controlled by equilibrium processes and is rate-determining for photoassembly. C is converted to D through a second light-driven Mn oxidation event. D is stable in the dark, and following the last light-induced oxidation, it is converted to a functional OEC rapidly with a high quantum yield (18, 24–26).
Fig. 1.
Proposed sequence of intermediates in OEC photoassembly. hν, light energy.
Given the relatively slow rate of conversion of B to C, Cheniae and coworkers (27) and Dismukes and coworkers (28–30) proposed that this step involves a protein conformation change. However, a recent structural study by Zouni and coworkers (31) showed that the OEC protein environment is essentially unchanged when the OEC is partially removed, fully removed, or partially reassembled. This result is consistent with an earlier mutagenesis study that indicated little change in protein conformation (32). This structural insight motivated us to reexamine the rate-determining step of photoassembly to more fully explain its chemical mechanism. We present data here to support an updated photoassembly mechanism in which the migration of Mn3+ within the apo site and the extraction of proton(s) to form a μ-hydroxo bridged Mn2+Mn3+ dimer control the yield of photoassembly.
Results
The apo-OEC PSII membranes from spinach used here have been treated with 1 M CaCl2 to remove the extrinsic subunits PsbO, PsbP, and PsbQ, and with 5 mM NH2OH to remove the OEC. PsbO is known to alter Mn2+ access to the apo site and is not essential for photoassembly (33, 34). The samples used have no detectable O2 evolution activity (SI Appendix, Table S1), >85% of total Mn content has been removed (SI Appendix, Fig. S1), and >95% of extrinsic subunits have been removed (SI Appendix, Fig. S2).
In vitro photoassembly conditions of apo-OEC PSII membranes were optimized under continuous light. Given our sample sizes and chlorophyll concentrations, optimal photoassembly was observed after 20 min at 50 μE⋅m−2⋅s−1 (SI Appendix, Fig. S3). Consistent with earlier results from Ananyev and Dismukes (29), we observe that the ratio of Ca2+ to Mn2+ is a strong determinant of photoassembly yield (SI Appendix, Fig. S4). For all subsequent experiments, we used the optimized concentrations of 40 mM Ca2+ and 0.16 mM Mn2+ ([Ca2+]/[Mn2+] = 250).
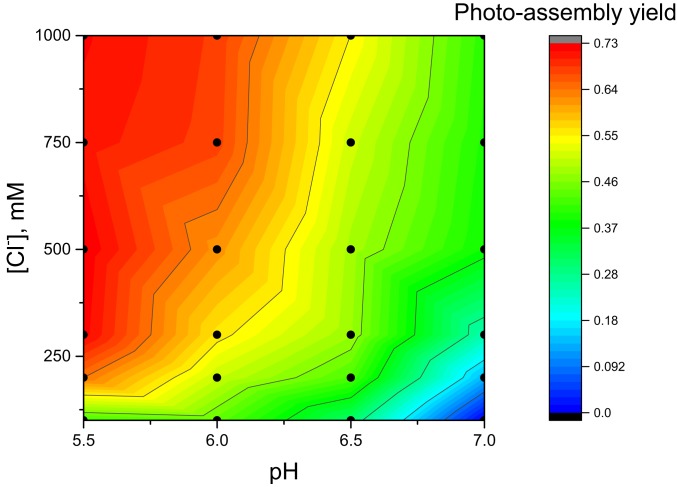
As previously observed for apo-OEC PSII membranes with PsbO removed, maximum photoassembly yields require high chloride concentrations (>150 mM) (35). Chloride dependence varies with the negative logarithm of hydrogen ion concentration (pH) (Fig. 2). At all pH values tested, photoassembly yield increases as chloride concentration increases. Highest yields of photoassembly are observed at pH 5.5 with 250 to 1,000 mM chloride.
Fig. 2.
Two-dimensional analysis of [Cl−] and pH dependence of photoassembly. All samples contained 0.25 mM 2-(N-morpholino)ethanesulfonic acid (pH 5.5 to 7.0), 40 mM CaCl2, 0.16 mM MnCl2, 8 mM NaHCO3, 400 mM sucrose, 10 μM dichlorophenolindophenol (DCIP), and 0.25 mg/mL chlorophyll as apo-OEC PSII membranes. Variable concentrations of NaCl were added to control [Cl−]. Photoassembly was performed at 50 μE⋅m−2⋅s−1 for 20 min. Dots represent the 24 buffer combinations used to generate the plot (pH values of 5.5, 6.0, 6.5, and 7.0, and chloride concentrations of 100, 200, 300, 500, 750, and 1,000 mM). Yields were normalized to the rate of extrinsics-depleted PSII membranes under identical measurement conditions [44.1 μmol of O2 (mg Chl)−1⋅h−1; SI Appendix, Table S1].
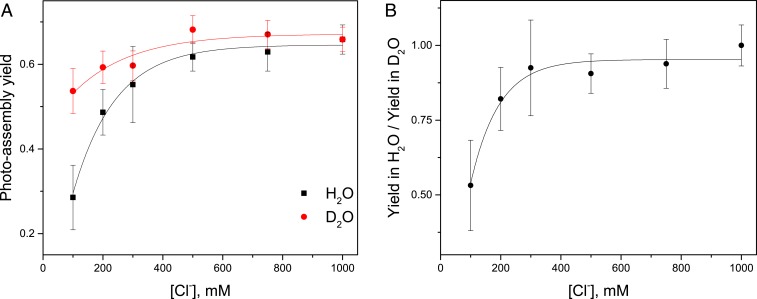
When the photoassembly solvent was replaced with deuterated water, D2O, chloride dependence decreased (Fig. 3A). At pL 6.05, where L represents either H+ or D+, photoassembly yield in D2O is approximately 2-fold higher at 100 mM chloride than in H2O.
Fig. 3.
Chloride dependence of photoassembly yield varies in H2O and D2O. (A) Photoassembly was performed in buffers containing H2O or D2O for 20 min at 50 μE⋅m−2⋅s−1 at variable chloride concentrations. Buffer pL was 6.05. Other conditions were similar to Fig. 2. Photoassembly yield was measured as O2 evolution rates in H2O. (B) Ratio of the observed photoassembly yield in H2O vs. D2O varies with chloride concentration. Data with error bars represent average values and SE (n = 3 to 6).
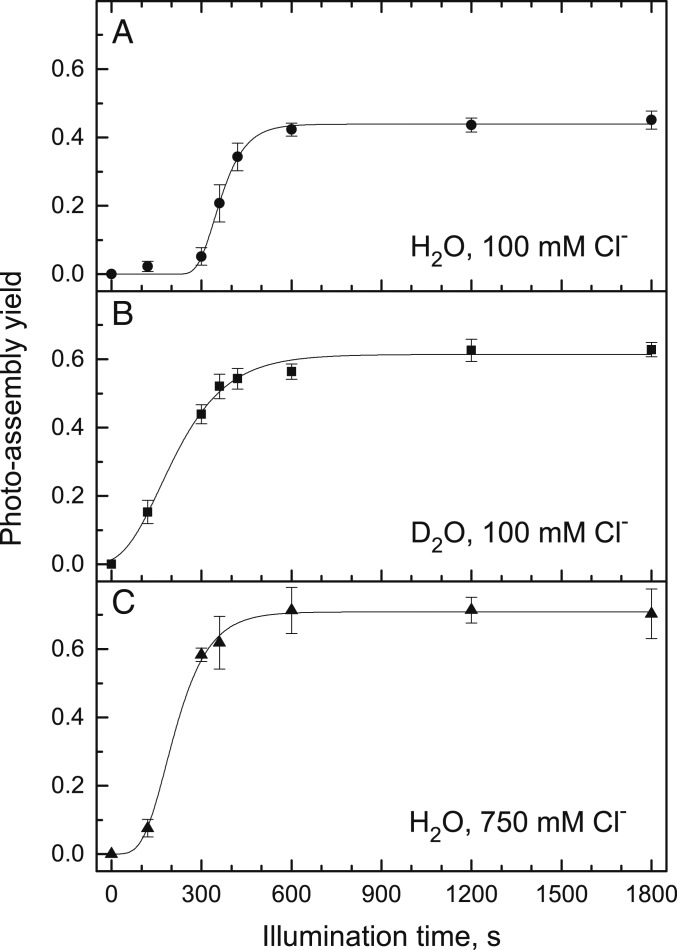
To kinetically resolve phases of photoassembly, we shifted from steady-state experiments (Fig. 3) to kinetic experiments (Fig. 4). The yield of photoassembly with respect to illumination time is sigmoidal in H2O with a subsaturating chloride concentration of 100 mM at pH 6.05 (Fig. 4A). The lag phase is significantly shorter when H2O is replaced with D2O at 100 mM chloride (Fig. 4B). A similar trend is observed in H2O with a saturating chloride concentration of 750 mM (Fig. 4C).
Fig. 4.
Rates of photoassembly are dependent on solvent and chloride concentration. Samples were photoassembled at 50 μE⋅m−2⋅s−1 for variable times. Sample conditions are identical to Fig. 3. Photoassembly was performed in H2O, pH 6.05, with 100 mM total Cl− (A); D2O, pL 6.05, with 100 mM total Cl− (B); or H2O, pH 6.05, with 750 mM total Cl− (C). Data with error bars represent average values and SE (n = 3 to 6). Traces represent kinetic analyses described in Table 1.
The kinetic data in Fig. 4 were fit to a sigmoidal function with a lag phase and growth phase that reach a steady-state value (Table 1). Based on the resulting rate constants, the photoassembly lag phase at 100 mM chloride has an inverse apparent H/D solvent isotope effect of 0.14 ± 0.05, with the rate being faster in D2O than in H2O at pL 6.05. The growth phase at 100 mM chloride has a normal apparent H/D solvent isotope effect of 1.5 ± 0.2.
Table 1.
Rates of photoassembly determined by fitting data from Fig. 4
| Solvent | [Cl−], mM | Lag phase duration, Tlag, s | Average lag phase rate, klag, s−1 | Growth phase rate, kgrowth, s−1 |
| H2O | 100 | 287 ± 7 | 0.0036 ± 0.0001 | 0.0029 ± 0.0003 |
| D2O | 100 | 41 ± 15 | 0.024 ± 0.009 | 0.0019 ± 0.0001 |
| H2O | 750 | 104 ± 7 | 0.0096 ± 0.0007 | 0.0033 ± 0.0002 |
Errors represent uncertainty of the fit.
Photoassembly kinetics are altered by the presence of the extrinsic subunit PsbO. As shown in SI Appendix, Fig. S5 and Table S2, the extent of the lag phase is decreased when PsbO is bound compared with when it is absent (Fig. 4 and Table 1). Nonetheless, at low chloride concentrations, the lag phase duration decreases when H2O is replaced with D2O and an inverse H/D solvent isotope effect is observed. The same decrease in the lag phase duration occurs when a saturating concentration of chloride is added.
The Mn4CaO5 core complex has a charge of +6 in the initial S0 or stable S1 state. Anionic ligands of the OEC include 2 aspartates (D1-D170 and D1-D342), 3 glutamates (D1-E189, D1-E333, and CP43-E354), and the C terminus of the D1 peptide (D1-A344), in addition to 1 neutral histidine (D1-H332). D1-H337 and CP43-R357 make hydrogen bonds with μ-oxo ligands, while D1-D61 makes a hydrogen bond with a terminal water ligand (6). The nearby D2-K317 is involved in binding chloride (36). Comparison of the crystal structure of the apo-OEC PSII complex and earlier structures of the intact PSII complex show that the positions of these residues are remarkably similar (31). However, structural studies provide little insight into how the protonation of these residues in the apo structure responds to the removal of the positively charged cluster.
The classical Multiconformer Continuum Electrostatics program MCCE (37) was used to determine amino acid protonation states as a function of pH and chloride in the apo structure from Zouni and coworkers (Protein Data Bank [PDB] ID code 5MX2) (31) and in a structure optimized around the S1 state with the cluster removed (38, 39). The protonation states of all residues in the full PSII structure are allowed to change. As shown in SI Appendix, Fig. S6, with the exception of D1-D170, which is predicted to be ionized under all conditions tested, and the D1 C-terminal A344, which is predicted to be neutral under all conditions tested, the residues closely associated with the OEC (D1-E189, D1-E333, D1-D342, CP43-E354, and D1-H332) are found in a distribution of protonation states. If the considered residues (7 carboxylates, 1 arginine, 1 lysine, and 2 histidines) had standard solution charges, the total charge would be −3 to −5 depending on the histidine protonation states. However, the average net charge ranges from −0.26 (PDB ID code 5MX2 with chloride at pH 5.5) to −0.49 (S1 without chloride at pH 7.0). Thus, several of the acidic ligands bind protons to compensate for the loss of the OEC cluster. This allows them to maintain their positions without encountering major electrostatic repulsion. The protonation of OEC ligands D1-D170, D1-E189, the D1 C-terminal A344, and D1-H332, as well as the nearby residues CP43-R357 and D2-K317, change very little with pH or the presence of chloride (SI Appendix, Fig. S6). The proton affinities of D1-D61, D1-E333, and CP43-E354 decrease when chloride is removed, while the proton affinity of D1-D342 increases when chloride is removed (SI Appendix, Fig. S6).
The predicted protonation states are similar when the calculations are carried out on the true apo structure (31) compared with the results when the Mn4CaO5 cluster and bound waters are removed from the holo PSII (SI Appendix, Fig. S6). Only D1-H337 is especially sensitive to the starting structure. As shown in SI Appendix, Fig. S6, it has lower proton affinity and is more chloride-sensitive in the true apo-PSII structure (PDB ID code 5MX2) compared with the OEC-deleted S1 structure. These results confirm that the function and structure of the OEC binding site change little when the metals are removed.
Discussion
PSII is a system where a binding site is prearranged to facilitate assembly of its active site as seen in the comparison of the crystal structures with and without the OEC (31). The protein prebuilds the binding site and constructs the OEC within this scaffold. The current measurements of pH and chloride dependence, H/D solvent isotope effects, and MCCE simulations of the pH and chloride dependence show the important role protons and chloride play in stabilizing the highly acidic region in a rigid structure prepared for assembly.
As illustrated in Fig. 5, we identify 2 key components and roles of the apo-OEC protein environment in the early stages of photoassembly: Protons must be shuttled away from the active site during μ-hydroxo or μ-oxo bridge formation, as well as removing the protons on the acidic ligands. The Kd for binding the first Mn to the HAS must be tuned to facilitate Mn2+ binding, Mn2+ oxidation, and Mn3+ translocation. As discussed below, we propose that D1-D61 facilitates proton transfer and D1-E333 tunes the relative affinity of the HAS for Mn2+ or Mn3+.
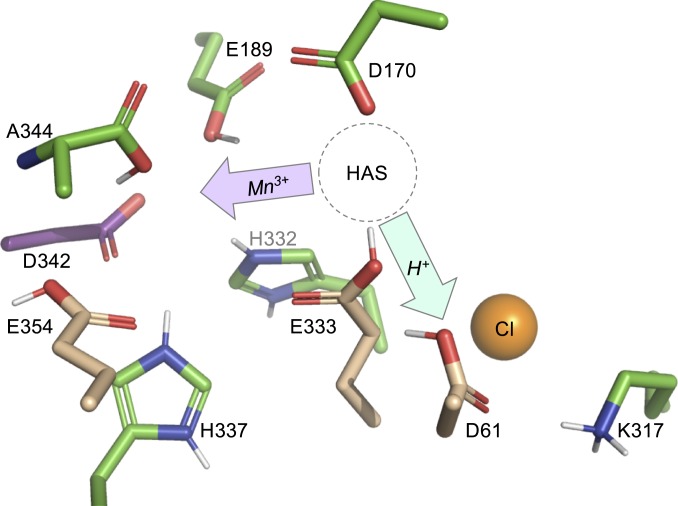
Fig. 5.
Protein environment of the apo-OEC binding site from Zouni and coworkers (PDB ID code 5MX2) (31). Protons have been added to titratable groups based on the predominant species observed in MCCE calculations (SI Appendix, Fig. S6). The HAS represents the location where Mn2+ binds during photoassembly. To form intermediate C (Fig. 1), oxidized Mn3+ must migrate deeper into the apo site (purple arrow). Water must be deprotonated for μ-hydroxo bridge formation, and the proton(s) must be shuttled out of the site (cyan arrow). Residues D1-E333 and D1-61 facilitate these 2 processes. When chloride is removed, the pKa values of CP43-E354, D1-E333, and D1-D61 (tan sticks) decrease and the pKa value of D1-D342 (purple sticks) increases.
D1-D61 and Chloride Facilitate Proton Release.
Residue D1-D61 is located 2.9 Å from D1-E333 in the apo-OEC structure (31), at the entrance of both the narrow and broad hydrogen-bonded channels (40). D1-D61 is ionized when the OEC is present (38) and acts as a hydrogen-bond acceptor to terminal water W1 (6). Earlier experiments and simulations have shown the coupled effects of protonation and chloride occupancy on proton release from the OEC to the thylakoid lumen during oxygen evolution (40–43). In intact PSII, chloride binds to D2-K317 (36, 44, 45). Upon chloride removal, D2-K317 forms a salt bridge with D1-D61, decreasing the negative logarithm of the acid dissociation constant (pKa) of D1-D61 (44) and slowing the release of protons (36).
In the ensemble of states found in MCCE calculations on the apo-OEC structure, D1-D61 is likely to be a protonated carboxylic acid (SI Appendix, Fig. S6). In the absence of chloride, more anionic D1-D61 is present in the ensemble at a given pH, indicating a lower proton affinity (i.e., a lower apparent pKa). This result was also found for PSII containing the OEC (38). Therefore, D1-D61 is a better Brønsted base when chloride is present in apo-OEC PSII. The formation of a metastable Mn dimer intermediate (discussed below) requires the formation of a μ-oxo or μ-hydroxo bridge. Thus, to form this ligand, water must be deprotonated and the proton efficiently shuttled away. We propose that D1-D61 serves this role during photoassembly.
Location of the HAS.
The specific location of the HAS is disputed. Both experimental and computational studies have suggested that D1-H332 is a ligand to the first Mn2+ during photoassembly, which would place the HAS at the Mn1 site in the intact OEC (46, 47). However, other evidence suggests that D1-D170 is a component of the HAS, which would place the HAS at the dangler Mn4 site in the intact OEC (48–52). The findings of a pulsed electron paramagnetic resonance (EPR) experiment by Asada and Mino (53) that measured the distance between Mn2+ in the HAS and the stable tyrosine-D radical are consistent with the latter HAS location (Fig. 5).
As reviewed by Bao and Burnap (2), mutagenesis studies also inform our understanding of the structure of the HAS. Mutants of D1-D170 strongly perturb the ability of PSII to bind Mn2+ and assemble a functional OEC (48–52). Only substitution with glutamate maintains high activity (48, 50, 52). On the other hand, mutagenesis of D1-E333 has a smaller effect on OEC assembly, with substitution of D1-E333 with glutamine supporting moderate PSII activity (32). This glutamate is calculated to be protonated in the apo structure. As described below, our data support a HAS at the Mn4 position ligated by D1-D170 and D1-E333.
Tuning the Kd of the HAS Facilitates Mn3+ Translocation.
At the HAS, D1-D170 is calculated to be always fully ionized, while D1-E333 is in a mixture of protonated and deprotonated states. These results support earlier mutagenesis studies: D1-D170 can only be replaced by a residue that provides a strong Mn2+/3+ ligand, while D1-E333 is a weak Mn2+/3+ ligand and can be replaced by other residues. In the presence of chloride, the free energy of D1-E333 favors the neutral state by ∼1 kcal⋅mol−1 at pH 5.5 (average charge of −0.2). The propensity to be ionized increases as the pH is increased to 7.0 (average charge of −0.4; SI Appendix, Fig. S6). The small change in protonation with pH shows the strong thermodynamic coupling of proton affinity among the OEC ligands. We predict that the HAS will have the highest affinity for Mn2+ when both D1-D170 and D1-E333 are ionized and that the Kd for Mn2+ will increase as D1-E333 is protonated. Using a nonequilibrium assay, Ono and Mino (20) measured the effective Kd for Mn2+ as a function of pH. They found that the Kd decreased as pH was increased from 5.0 to 7.0 and remained constant at pH ≥ 7.0. In their discussion of these data, they proposed that “a residue responsible for Mn2+ binding has been protonated below pH 7 . . .” (20). Based on their study, the likely location of the HAS (53), and our current MCCE calculations, we propose that D1-E333 has an apparent pKa value of ∼7 and tunes the Kd of Mn2+ binding in a pH- and chloride-dependent manner.
Zouni and coworkers (31) recently proposed a mechanism in which Mn2+ first binds to the HAS and is later translocated to a different site deeper in the OEC binding pocket. Next, a second Mn2+ binds to the now vacant HAS and is oxidized. We note that observed Kd values for Mn2+ binding of ∼50 µM for both the first (19) and second (24) binding events are consistent with this mechanism.
Potential Identity of Intermediate C.
In this mechanism, the first metal dimer intermediate (C in Fig. 1) is Mn2+Mn3+, which is consistent with previous spectroscopic (21–23) and structural (31) studies. However, Dismukes and coworkers (19) presented compelling evidence that a µ-hydroxy or µ-oxo bridged Mn3+Ca2+ dimer formed first. In their model, Ca2+ binds early in the photoassembly process. In other models, Ca2+ binds as late as the very last step (26). Using parallel mode EPR detection of Mn3+, they observed that the ligand field of the first Mn3+ is highly symmetrical at pH ≥ 7.5 and becomes distorted as the pH decreases to 6.0. All of these experiments were performed at temperatures below −20 °C, so that diffusion of ions is limited (19). In light of the discussion above, these results have 2 implications. First, a more symmetrical ligand field for Mn3+ locked in the HAS is consistent with D1-E333 being deprotonated as we predict at high pH. At lower pH values, the Mn3+ will have less favorable interaction with a protonated carboxylic acid group in an axial position. Second, the use of low temperatures to prevent diffusion does not facilitate Mn3+ translocation in subsequent steps. Therefore, it is possible that the observed Mn3+Ca2+ species is only present when Mn3+ is prevented from vacating the HAS. We propose that at higher temperatures, Mn3+ migrates and a second Mn2+ binds to the HAS. Ca2+ is clearly an essential component of photoassembly (27), but the precise step in which it is incorporated is not fully understood and motivates future studies.
Protons and Chloride Gate OEC Photoassembly.
In Fig. 2, both pH and chloride concentration affect photoassembly yield. At pH 5.5, when chloride is not saturating (lower left corner of Fig. 2), our simulations suggest that D1-E333 is mostly ionized and that D1-D61 is mostly protonated. In this regime, Mn3+ translocation will be hindered because the HAS has a low Kd. While proton transfer is favored, the Mn2+Mn3+ dimer intermediate is unable to efficiently form. At pH 5.5, when chloride is saturating (upper left corner of Fig. 2), our simulations suggest that both D1-E333 and D1-D61 are mostly protonated. Here, both Mn3+ translocation and proton transfer are favored, and photoassembly yield is high. At pH 7.0, the proton affinity of D1-E333 is not strongly affected by the presence of chloride. However, the proton affinity of D1-D61 is at least 2-fold greater when chloride is present. At this pH value, D1-E333 remains partially protonated, Mn3+ translocation is favored, and the observed enhancement of photoassembly yield by the addition of chloride is only affected by proton release.
Maintaining D1-E333 and D1-D61 in environments where their pKa values are relatively high facilitates Mn3+ translocation and proton transfer, respectively, thus increasing the yield of photoassembly. The MCCE simulations show that adding chloride to the system has this desired effect. The pKa values of carboxylate groups can also be increased by replacing H2O with D2O (54). In general, this replacement increases pKa values by ∼0.5 pH units (55, 56). In Fig. 3, we observed that photoassembly yields are higher in D2O compared with H2O only when chloride is limiting. As described below, we interpret these data as the result of an inverse H/D solvent equilibrium isotope effect. At saturating chloride concentrations at pL 6, the proton affinities of D1-E333 and D1-D61 are already optimized for photoassembly, and the addition of D2O has little effect.
Kinetic resolution of these data show that photoassembly at pH 6 in H2O with 100 mM chloride has a pronounced lag phase (Fig. 4A). We attribute this lag phase to the decay of intermediates B and C in the early stages of photoassembly. Dramatically, this lag phase is effectively eliminated when H2O is replaced with D2O (Fig. 4B), resulting in an apparent H/D solvent isotope effect of 0.14 ± 0.05 (Table 1). Because the pKa values of D1-D61 and D1-E333 increase in D2O, both μ-hydroxo bridge formation and Mn3+ translocation will be favored even though chloride is limiting. Therefore, intermediate C accumulates at earlier time points. The reversible nature of intermediates B and C makes this process dependent on equilibrium. The shortened lag phase of photoassembly in H2O with saturating chloride concentrations mimics that of D2O at lower chloride concentrations (Fig. 4C).
The growth phase of photoassembly represents the irreversible and non–rate-determining steps following the formation of intermediate C. Here, the rate is insignificantly different in H2O at low and high chloride concentrations (Fig. 4 A and C). In D2O, the rate is slower, giving an apparent H/D solvent isotope effect of 1.5 ± 0.2 (Table 1). These later steps of photoassembly are kinetically controlled.
Conclusion
In our current mechanism of OEC photoassembly based on our present data and a critical examination of the literature, Mn2+ first binds to the HAS in the apo-OEC protein environment. This environment is already in a “preorganized shell,” (31) and no protein conformational changes are required, but protons may need to be removed. The proton affinities of carboxylic acid groups in the pocket are relatively high, and electrostatic repulsion is minimized. The first Mn2+ is oxidized to Mn3+ and is in a sufficiently labile ligand environment to facilitate translocation to a site deeper in the apo-OEC pocket. A second Mn2+ binds to the now vacated HAS. This second Mn2+ is trapped in place by the formation of a μ-hydroxo bridge with the Mn3+ ion. Formation of this ligand requires proton release and an efficient Brønsted base located nearby. These early steps are controlled by equilibrium and show strong chloride dependence and an inverse H/D solvent equilibrium isotope effect. Next, the second Mn2+ is oxidized to Mn3+, forming a Mn3+Mn3+ dimer. This intermediate is highly stable and templates the remaining photoassembly process. All remaining steps are kinetically rapid. These latter steps do not show chloride dependence and have a moderate H/D solvent kinetic isotope effect greater than 1.
Materials and Methods
Detailed methods on sample preparation, characterization, in vitro photoassembly, analysis, and MCCE simulations are provided in SI Appendix, Supplemental Materials and Methods.
Supplementary Material
Acknowledgments
We thank Prof. Gary Brudvig for helpful discussions and early suggestions about the deprotonation requirements of μ-oxo bridge formation. We also thank Prof. Terry Bricker for helpful discussions, frequent troubleshooting, and the gift of the α-PsbO antibody. The experimental work was funded by Louisiana State University and the Herman Frasch Fund for Chemical Research Grant 822-HF17 (to D.J.V.). The computation work was funded by the Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences, US Department of Energy, Photosynthesis Systems Grant DE-SC0001423 (to M.R.G.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1910231116/-/DCSupplemental.
References
- 1.Liu J., et al. , Metalloproteins containing cytochrome, iron-sulfur, or copper redox centers. Chem. Rev. 114, 4366–4469 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bao H., Burnap R. L., Photoactivation: The light-driven assembly of the water oxidation complex of photosystem II. Front. Plant Sci. 7, 578 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dasgupta J., Ananyev G. M., Dismukes G. C., Photoassembly of the water-oxidizing complex in photosystem II. Coord. Chem. Rev. 252, 347–360 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vinyard D. J., Ananyev G. M., Dismukes G. C., Photosystem II: The reaction center of oxygenic photosynthesis. Annu. Rev. Biochem. 82, 577–606 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Ferreira K. N., Iverson T. M., Maghlaoui K., Barber J., Iwata S., Architecture of the photosynthetic oxygen-evolving center. Science 303, 1831–1838 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Umena Y., Kawakami K., Shen J.-R., Kamiya N., Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 Å. Nature 473, 55–60 (2011). [DOI] [PubMed] [Google Scholar]
- 7.Krewald V., et al. , Metal oxidation states in biological water splitting. Chem. Sci. 6, 1676–1695 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Järvi S., Suorsa M., Aro E.-M., Photosystem II repair in plant chloroplasts—Regulation, assisting proteins and shared components with photosystem II biogenesis. Biochim. Biophys. Acta 1847, 900–909 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Komenda J., Sobotka R., Nixon P. J., Assembling and maintaining the photosystem II complex in chloroplasts and cyanobacteria. Curr. Opin. Plant Biol. 15, 245–251 (2012). [DOI] [PubMed] [Google Scholar]
- 10.Frankel L. K., et al. , Radiolytic mapping of solvent-contact surfaces in photosystem II of higher plants: Experimental identification of putative water channels within the photosystem. J. Biol. Chem. 288, 23565–23572 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lindahl M., et al. , The thylakoid FtsH protease plays a role in the light-induced turnover of the photosystem II D1 protein. Plant Cell 12, 419–431 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sirpiö S., et al. , AtCYP38 ensures early biogenesis, correct assembly and sustenance of photosystem II. Plant J. 55, 639–651 (2008). [DOI] [PubMed] [Google Scholar]
- 13.Reisman S., Ohad I., Light-dependent degradation of the thylakoid 32 kDa QB protein in isolated chloroplast membranes of Chlamydomonas reinhardtii. Biochim. Biophys. Acta 849, 51–61 (1986). [Google Scholar]
- 14.Miyata K., Noguchi K., Terashima I., Cost and benefit of the repair of photodamaged photosystem II in spinach leaves: Roles of acclimation to growth light. Photosynth. Res. 113, 165–180 (2012). [DOI] [PubMed] [Google Scholar]
- 15.Noguchi K., et al. , Costs of protein turnover and carbohydrate export in leaves of sun and shade species. Aust. J. Plant Physiol. 28, 37–47 (2001). [Google Scholar]
- 16.Kok B., Forbush B., McGloin M., Cooperation of charges in photosynthetic O2 evolution-I. A linear four step mechanism. Photochem. Photobiol. 11, 457–475 (1970). [DOI] [PubMed] [Google Scholar]
- 17.Pal R., et al. , S0-State model of the oxygen-evolving complex of photosystem II. Biochemistry 52, 7703–7706 (2013). [DOI] [PubMed] [Google Scholar]
- 18.Radmer R., Cheniae G. M., Photoactivation of the manganese catalyst of O2 evolution. II. A two-quantum mechanism. Biochim. Biophys. Acta 253, 182–186 (1971). [DOI] [PubMed] [Google Scholar]
- 19.Tyryshkin A. M., et al. , Spectroscopic evidence for Ca2+ involvement in the assembly of the Mn4Ca cluster in the photosynthetic water-oxidizing complex. Biochemistry 45, 12876–12889 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Ono T. A., Mino H., Unique binding site for Mn2+ ion responsible for reducing an oxidized YZ tyrosine in manganese-depleted photosystem II membranes. Biochemistry 38, 8778–8785 (1999). [DOI] [PubMed] [Google Scholar]
- 21.Barra M., et al. , Intermediates in assembly by photoactivation after thermally accelerated disassembly of the manganese complex of photosynthetic water oxidation. Biochemistry 45, 14523–14532 (2006). [DOI] [PubMed] [Google Scholar]
- 22.Cole J. L., et al. , Structure of the manganese complex of photosystem II upon removal of the 33-kilodalton extrinsic protein: An X-ray absorption spectroscopy study. Biochemistry 26, 5967–5973 (1987). [DOI] [PubMed] [Google Scholar]
- 23.Pospísil P., Michael H., Dittmer J., Solé V. A., Dau H., Stepwise transition of the tetra-manganese complex of photosystem II to a binuclear Mn2(micro-O)2 complex in response to a temperature jump: A time-resolved structural investigation employing x-ray absorption spectroscopy. Biophys. J. 84, 1370–1386 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller A. F., Brudvig G. W., Manganese and calcium requirements for reconstitution of oxygen-evolution activity in manganese-depleted photosystem II membranes. Biochemistry 28, 8181–8190 (1989). [DOI] [PubMed] [Google Scholar]
- 25.Ono T.-A., Inoue Y., Reductant-sensitive intermediates involved in multi-quantum process of photoactivation of latent O2-evolving system. Plant Cell Physiol. 28, 1293–1299 (1987). [Google Scholar]
- 26.Tamura N., Cheniae G., Photoactivation of the water-oxidizing complex in photosystem II membranes depleted of Mn and extrinsic proteins. I. Biochemical and kinetic characterization. Biochim. Biophys. Acta 890, 179–194 (1987). [Google Scholar]
- 27.Chen C., Kazimir J., Cheniae G. M., Calcium modulates the photoassembly of photosystem II (Mn)4-clusters by preventing ligation of nonfunctional high-valency states of manganese. Biochemistry 34, 13511–13526 (1995). [DOI] [PubMed] [Google Scholar]
- 28.Ananyev G. M., Dismukes G. C., High-resolution kinetic studies of the reassembly of the tetra-manganese cluster of photosynthetic water oxidation: Proton equilibrium, cations, and electrostatics. Biochemistry 35, 14608–14617 (1996). [DOI] [PubMed] [Google Scholar]
- 29.Ananyev G. M., Dismukes G. C., Assembly of the tetra-Mn site of photosynthetic water oxidation by photoactivation: Mn stoichiometry and detection of a new intermediate. Biochemistry 35, 4102–4109 (1996). [DOI] [PubMed] [Google Scholar]
- 30.Zaltsman L., Ananyev G. M., Bruntrager E., Dismukes G. C., Quantitative kinetic model for photoassembly of the photosynthetic water oxidase from its inorganic constituents: Requirements for manganese and calcium in the kinetically resolved steps. Biochemistry 36, 8914–8922 (1997). [DOI] [PubMed] [Google Scholar]
- 31.Zhang M., et al. , Structural insights into the light-driven auto-assembly process of the water-oxidizing Mn4CaO5-cluster in photosystem II. eLife 6, e26933 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cohen R. O., Nixon P. J., Diner B. A., Participation of the C-terminal region of the D1-polypeptide in the first steps in the assembly of the Mn4Ca cluster of photosystem II. J. Biol. Chem. 282, 7209–7218 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Burnap R. L., Qian M., Pierce C., The manganese stabilizing protein of photosystem II modifies the in vivo deactivation and photoactivation kinetics of the H2O oxidation complex in Synechocystis sp. PCC6803. Biochemistry 35, 874–882 (1996). [DOI] [PubMed] [Google Scholar]
- 34.Qian M., Al-Khaldi S. F., Putnam-Evans C., Bricker T. M., Burnap R. L., Photoassembly of the photosystem II (Mn)4 cluster in site-directed mutants impaired in the binding of the manganese-stabilizing protein. Biochemistry 36, 15244–15252 (1997). [DOI] [PubMed] [Google Scholar]
- 35.Miyao M., Inoue Y., Enhancement by chloride ions of photoactivation of oxygen evolution in manganese-depleted photosystem II membranes. Biochemistry 30, 5379–5387 (1991). [DOI] [PubMed] [Google Scholar]
- 36.Pokhrel R., Service R. J., Debus R. J., Brudvig G. W., Mutation of lysine 317 in the D2 subunit of photosystem II alters chloride binding and proton transport. Biochemistry 52, 4758–4773 (2013). [DOI] [PubMed] [Google Scholar]
- 37.Song Y., Mao J., Gunner M. R., MCCE2: Improving protein pKa calculations with extensive side chain rotamer sampling. J. Comput. Chem. 30, 2231–2247 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaur D., et al. , Relative stability of the S2 isomers of the oxygen evolving complex of photosystem II. Photosyn. Res, 10.1007/s11120-019-00637-6 (2019). [DOI] [PubMed] [Google Scholar]
- 39.Luber S., et al. , S1-state model of the O2-evolving complex of photosystem II. Biochemistry 50, 6308–6311 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vogt L., Vinyard D. J., Khan S., Brudvig G. W., Oxygen-evolving complex of photosystem II: An analysis of second-shell residues and hydrogen-bonding networks. Curr. Opin. Chem. Biol. 25, 152–158 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Debus R. J., Evidence from FTIR difference spectroscopy that D1-Asp61 influences the water reactions of the oxygen-evolving Mn4CaO5 cluster of photosystem II. Biochemistry 53, 2941–2955 (2014). [DOI] [PubMed] [Google Scholar]
- 42.Hundelt M., Hays A.-M. A., Debus R. J., Junge W., Oxygenic photosystem II: The mutation D1-D61N in synechocystis sp. PCC 6803 retards S-state transitions without affecting electron transfer from YZ to P680+. Biochemistry 37, 14450–14456 (1998). [DOI] [PubMed] [Google Scholar]
- 43.Dilbeck P. L., et al. , The D1-D61N mutation in Synechocystis sp. PCC 6803 allows the observation of pH-sensitive intermediates in the formation and release of O2 from photosystem II. Biochemistry 51, 1079–1091 (2012). [DOI] [PubMed] [Google Scholar]
- 44.Pokhrel R., McConnell I. L., Brudvig G. W., Chloride regulation of enzyme turnover: Application to the role of chloride in photosystem II. Biochemistry 50, 2725–2734 (2011). [DOI] [PubMed] [Google Scholar]
- 45.Rivalta I., et al. , Structural-functional role of chloride in photosystem II. Biochemistry 50, 6312–6315 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dasgupta J., Tyryshkin A. M., Dismukes G. C., ESEEM spectroscopy reveals carbonate and an N-donor protein-ligand binding to Mn2+ in the photoassembly reaction of the Mn4Ca cluster in photosystem II. Angew. Chem. Int. Ed. Engl. 46, 8028–8031 (2007). [DOI] [PubMed] [Google Scholar]
- 47.Nakamura S., Noguchi T., Initial Mn2+ binding site in photoassembly of the water-oxidizing Mn4CaO5 cluster in photosystem II as studied by quantum mechanics/molecular mechanics calculations. Chem. Phys. Lett. 721, 62–67 (2019). [Google Scholar]
- 48.Chu H.-A., Nguyen A. P., Debus R. J., Site-directed photosystem II mutants with perturbed oxygen-evolving properties. 1. Instability or inefficient assembly of the manganese cluster in vivo. Biochemistry 33, 6137–6149 (1994). [DOI] [PubMed] [Google Scholar]
- 49.Chu H.-A., Nguyen A. P., Debus R. J., Amino acid residues that influence the binding of manganese or calcium to photosystem II. 1. The lumenal interhelical domains of the D1 polypeptide. Biochemistry 34, 5839–5858 (1995). [DOI] [PubMed] [Google Scholar]
- 50.Nixon P. J., Diner B. A., Aspartate 170 of the photosystem II reaction center polypeptide D1 is involved in the assembly of the oxygen-evolving manganese cluster. Biochemistry 31, 942–948 (1992). [DOI] [PubMed] [Google Scholar]
- 51.Diner B. A., Nixon P. J., The rate of reduction of oxidized redox-active tyrosine, Z+, by exogenous Mn2+ is slowed in a site-directed mutant, at aspartate 170 of polypeptide D1 of photosystem II, inactive for photosynthetic oxygen evolution. Biochim. Biophys. Acta 1101, 134–138 (1992). [Google Scholar]
- 52.Boerner R. J., Nguyen A. P., Barry B. A., Debus R. J., Evidence from directed mutagenesis that aspartate 170 of the D1 polypeptide influences the assembly and/or stability of the manganese cluster in the photosynthetic water-splitting complex. Biochemistry 31, 6660–6672 (1992). [DOI] [PubMed] [Google Scholar]
- 53.Asada M., Mino H., Location of the high-affinity Mn2+ site in photosystem II detected by PELDOR. J. Phys. Chem. B 119, 10139–10144 (2015). [DOI] [PubMed] [Google Scholar]
- 54.Mora-Diez N., Egorova Y., Plommer H., Tremaine P. R., Theoretical study of deuterium isotope effects on acid–base equilibria under ambient and hydrothermal conditions. RSC Adv. 5, 9097–9109 (2015). [Google Scholar]
- 55.Delgado R., et al. , Dissociation constants of Brønsted acids in D2O and H2O: Studies on polyaza and polyoxa-polyaza macrocycles and a general correlation. Anal. Chim. Acta 245, 271–282 (1991). [Google Scholar]
- 56.Krȩżel A., Bal W., A formula for correlating pKa values determined in D2O and H2O. J. Inorg. Biochem. 98, 161–166 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.