Significance
The identification of cancer drivers is essential for realizing the goal of precision medicine in cancer. By integrating 3D protein structures and dynamics, we describe a framework to identify cancer driver genes using a sensitive search of mutational hotspot communities in 3D structures. Our workflow identifies previously identified driver genes as well as unidentified putative drivers.
Keywords: cancer driver, hotspot communities, protein dynamics, TCGA, PanCancer
Abstract
Large-scale exome sequencing of tumors has enabled the identification of cancer drivers using recurrence-based approaches. Some of these methods also employ 3D protein structures to identify mutational hotspots in cancer-associated genes. In determining such mutational clusters in structures, existing approaches overlook protein dynamics, despite its essential role in protein function. We present a framework to identify cancer driver genes using a dynamics-based search of mutational hotspot communities. Mutations are mapped to protein structures, which are partitioned into distinct residue communities. These communities are identified in a framework where residue–residue contact edges are weighted by correlated motions (as inferred by dynamics-based models). We then search for signals of positive selection among these residue communities to identify putative driver genes, while applying our method to the TCGA (The Cancer Genome Atlas) PanCancer Atlas missense mutation catalog. Overall, we predict 1 or more mutational hotspots within the resolved structures of proteins encoded by 434 genes. These genes were enriched among biological processes associated with tumor progression. Additionally, a comparison between our approach and existing cancer hotspot detection methods using structural data suggests that including protein dynamics significantly increases the sensitivity of driver detection.
Large-scale cancer genome studies, such as The Cancer Genome Atlas (TCGA) project (1, 2) and the International Cancer Genome Consortium (ICGC) (3, 4), have generated comprehensive catalogs of somatic alterations for various cancer cohorts. The majority of these somatic variants incur little or no functional consequence on tumor progression and are thus often termed neutral “passengers.” In contrast, a handful of “driver” mutations are considered to provide a selective advantage to cancer cells. One of the critical goals of TCGA and ICGC projects has been to distinguish between these positively selected “driver mutations” (5–7) from a large number of neutral passenger mutations.
A majority of the cancer-driver detection algorithms quantify the recurrence of mutations to identify significantly mutated genes and noncoding genomic elements (8–11). However, the somatic mutational landscapes of cancer genomes are highly heterogeneous (12–14) and exhibit a long tail of low-frequency mutations (11, 13, 15–17). The presence of this long tail of rare somatic mutations, along with limited cohort sizes, makes recurrence-based driver identification very challenging. This long tail often contains many latent drivers (18, 19): That is, variants which may not individually confer selective advantages to tumor cells, but which can potentially drive tumor growth in the presence of other mutations. Thus, canonical recurrence-based approaches are likely to overlook such latent drivers.
An alternative is to employ algorithms that aggregate mutation recurrence on gene/element-levels (11, 20) or to predict the molecular functional impact of mutations (21) to distinguish drivers from passengers. Compared to protein-truncating mutations and large structural variants, missense mutations induce subtle changes, which are often difficult to interpret on the phenotypic level. Thus, identifying missense driver mutations based on their molecular functional impact (22) is also challenging. However, the signal of positive selection aggregated on functional elements or subregions of the coding genome [such as protein domains (23–25), posttranslational modification sites (26–28), protein interaction interfaces (29, 30), and mutation cluster/hotspots (31–33)] has been shown to be effective. We note that these approaches are inherently limited by the fact that only a subset of mutations might occupy these functional elements or subregions.
Prior studies have identified driver mutations based on their presence in mutational clusters (31–33), which are often called “hotspot” regions. These mutational clusters are defined based on the proximity of somatic mutations within the primary sequence (31, 33) or 3D structure of a given protein (34–38). Linear sequence-based mutational cluster identification algorithms (31, 33, 39) discover significantly mutated genes while considering an appropriate background mutation model, trinucleotide context, and distribution of silent mutations. However, sequence-based approaches miss many hotspot regions, as they ignore spatial proximity between residues that may be far apart in sequence but very close in 3D space (40, 41). In contrast, despite being inherently limited due to incomplete structural coverage of the proteome, 3D structure-based mutational cluster definitions often provide physical intuition or mechanistic insights into the roles of such clusters in cancer progression (29, 35–38, 40, 42). These structure-based methods compute residue distances or generate residue–residue contact networks in the 3D structures of proteins to identify a group of spatially proximal residues. Furthermore, mutation shuffling is performed to identify significantly mutated residue clusters or hotspots on protein structures. However, current approaches under this framework have failed to consider protein dynamics.
Proteins are inherently dynamic and sample large ensembles of conformations (43–46). The energy landscape underlying the distribution of structures in these ensembles are often altered based on external (thermodynamic) (45, 47) or internal (allosteric) signals (46, 48–50). Previous biophysical studies have clearly shown the crucial role of protein motions in conferring protein functionality (51–55). Thus, prior structure-based driver-detection methods that employ only static structures of proteins are generally less sensitive when attempting to identify functional residues under the mutation clustering framework.
In particular, a static crystal structure provides only 1 limited snapshot of the protein, most likely close to (or at) the bottom of the free-energy landscape. In contrast, motion-weighted community detection approaches more accurately reflect the physical reality in which proteins undergo 2 general types of dynamics. First, a protein can dynamically oscillate around the bottom of its energetic well, and second, dynamics may arise when the underlying free-energy landscape itself changes in distinct ways, thereby shifting the protein conformation to an alternative functional state. In each of these scenarios, communication between different communities plays a pivotal role in the proper functioning of the protein. We posit that hotspot communities exist in large part because certain select communities either play essential roles in these functional dynamics or because their contributions to such dynamics are especially sensitive to mutations. Static representations of protein structures can fail to sensitively define communities in light of the essential role of dynamics in function. Furthermore, such static models potentially miss many critical mutational clusters with a potential role in cancer progression.
In the present work, we address this issue by explicitly incorporating protein dynamics into our framework to identify mutational hotspot communities in protein structures. We applied this framework to the TCGA PanCancer Atlas catalog of missense mutations to identify genes with significantly mutated residue communities in protein structures. Our pan-cancer analysis identifies 434 unique genes with at least 1 hotspot community in the corresponding protein structure. The majority of these genes are involved in critical biological processes and pathways that play a vital role in cancer progression, including DNA repair, signal transduction, apoptosis, and posttranslational modifications. As expected, we observed higher cross-species conservation scores and greater functional impact scores for mutations within these hotspot communities. Furthermore, our prediction includes previously characterized driver genes with hotspot communities in corresponding protein structures. Additionally, we also identified genes with at least 1 hotspot community that were not detected by other mutation cluster algorithms lacking information on protein dynamics. Finally, we highlight some examples of driver genes containing hotspot communities that are predicted to play vital roles in cancer progression.
Materials and Methods
SNV Dataset and Mapping to Protein Structures.
We leveraged the MC3 (multiple-center mutation calling in multiple cancer) (56) somatic mutation dataset generated as part of the TCGA PanCancer Atlas project. Briefly, the MC3 call set was generated using ∼10,000 tumor/normal whole-exome sequences belonging to 33 different cancer types. Multiple callers, including MuTect (57), RADIA (58), SomaticSniper (59), and VarScan (60) were applied to obtain high-confidence variant calls. Subsequent filtering removed mutations due to lack of coverage, potential germline contamination, and other artifacts. We utilized v2.8 of the publicly accessible MC3 variant call set (5). Furthermore, we only analyzed missense mutations that were designated as “PASS” based on the filtering criterion. Moreover, we only analyzed variants from samples that were included in the whitelist samples and were not hypermutated. This subset comprises 2.85 million mutations from 8,937 samples in the PanCancer Atlas project. Approximately 2.75 million mutations in this subset occupy the coding regions of the genome that consists of 1.5 million missense mutations, 0.6 million silent mutations, 1.18 million nonsense mutations, and 3.7K splice mutations.
We applied the Variant Annotation Tool (VAT) (61) to map TCGA missense mutations to protein structures. For each missense mutation, VAT provides an annotation that includes the gene name, transcript name, and the position of the affected residue in the translated protein sequence. Additionally, it also provides the residue identities of both the wild-type and variant residues. Subsequently, we integrated VAT annotations with a BioMart-derived identifier map (62), which consists of the gene identifier, transcript identifier, and the corresponding PDB ID code, if available. We restricted our analyses to mutations that map to crystal structures having resolutions that are better than 3.0 Å. Overall, we mapped 0.329 million missense mutations on ∼17,300 crystal structures in the present study.
Workflow to Identify 3D Hotspot Communities in Cancer.
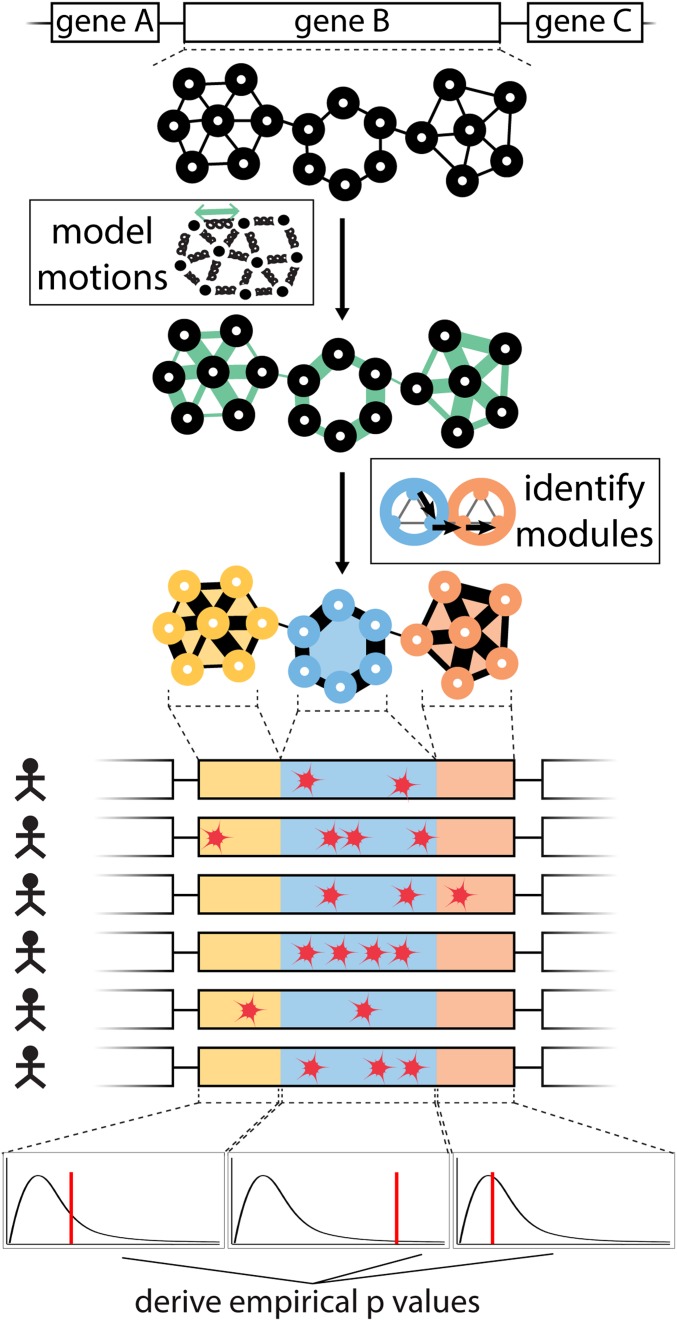
As discussed above, our framework to predict driver genes by identifying hotspot communities is distinct from previous methods in that we explicitly included protein dynamics in our workflow (Fig. 1). Briefly, we modeled large-scale conformational changes of each protein to identify nonoverlapping subregions (or “communities”). The large-scale conformational changes are modeled using anisotropic network models (48, 63). Subsequently, we modeled each protein structure as a residue-interaction network, wherein each residue constitutes a node in the network, and edges (or connections between these nodes, where connections are defined by close physical proximity) form the physical interactions between these nodes. Furthermore, edges in a network can be “weighted” using the extent to which contacting residues exhibit correlated motions within the dynamic structure of the protein. Highly correlated motions between 2 residues that are physically in contact (though not necessarily covalently linked) suggest that knowledge of the motions for one residue can provide a great deal of information regarding the motions of the other residue. This mutual knowledge, in a sense, suggests a strong degree of informational flow between residues. The weight for each edge in the network corresponds to the “effective distance” of this edge, in which a strong degree of correlated motion results in a short distance, and a weak correlation in the motions results in a long distance. With this motion-weighted protein network, communities of resides are defined with the Girvan–Newman algorithm (64). A community constitutes a group of residues in which each residue is connected to other residues of the same community, and only weakly connected (if at all) to residues outside the immediate community. These network-weighted communities thus form densely interconnected neighborhoods.
Fig. 1.
Workflow of HotCommics to identify putative driver genes: This integrative approach utilizes protein community information along with mapped mutations to identify significantly mutated communities in protein structures. Fisher’s method is employed to quantify the significance of variant enrichment in each community with mapped mutations (thereby defining the hotspot communities).
To identify mutational hotspot communities in a given structure, we first mapped missense mutations from TCGA cohorts to 3D protein structures. We then computed the frequency of mapped mutations for each community on the pan-cancer level as well as in specific cancer cohorts. Furthermore, for each community with mapped mutations, we performed Fisher’s exact test to determine whether a given community is more frequently mutated than what would be expected by chance. Fisher’s exact test assigns an empirical P value to each community, which is corrected for multiple hypothesis testing using the Benjamini–Hochberg method. Finally, these multiple hypothesis-corrected P values are used to identify significantly mutated hotspot communities encoded by a particular gene. We note that, for a substantial number of genes, there are multiple PDB structures available. We removed this structural redundancy using structural coverage (highest fraction of residues covered in the structure) as a filter to provide 1-to-1 mapping between each PDB structure and its corresponding gene. The source code for the workflow is available on the project’s Github page (https://github.com/gersteinlab/HotCommics) (65).
Downstream Analyses.
We performed a number of downstream analyses to further validate our predictions. We extracted PhyloP (66) and CADD (67) scores for each mutation mapped to a structure. Furthermore, we classified mutations into hotspot and nonhotspot variants based on whether mutations are mapped to residues belonging to hotspot communities or otherwise. We then compared the phyloP score and CADD score distributions for hotspot and nonhotspot mutations. We performed two-sided Kolmogorov–Smirnov (KS) test to assess the significance of conservation score differences between hotspot and nonhotspot mutations. We applied the same method to quantify such disparities for the molecular functional impact (CADD) score for hotspot and nonhotspot mutations. Here, our null hypothesis is that the conservation or impact score for hotspot and nonhotspot mutations are not (on average) different as they would be drawn from the same distribution.
We also performed gene ontology (GO) enrichment and pathway enrichment analyses to further validate the role of our putative driver genes in tumor progression. For the GO analysis, we calculated the enrichment based on biological processes available from the GO database (68), and we performed pathway enrichment analysis using the Reactome (69) as well as the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (70). We visualized the enrichment analysis result using the clusterProfiler (71) package available in Bioconductor.
Additionally, we also compared our predicted driver gene list derived from our hotspot community analysis with other cluster-based approaches (at the level of both sequences and structures). One of the key differences between our approach and other approaches is that we employ information on protein dynamics (along with structural data) to determine hotspot communities. For structure-based methods, we obtained the lists of predicted genes derived from HotSpot3D (37), 3DHotSpot (36), and HotMap (38) algorithms. All 3 of these algorithms were previously applied to TCGA PanCancer Atlas data (5), which allows us to make direct comparisons with our work. However, we also note small differences in our workflow compared to other structure-based approaches. In contrast to many other methods that rely only of experimentally determined structures, HotMap also employs homology models in order to expand structural coverage. Moreover, our method was applied only on crystal structures of poorer resolution (in contrast to other methods that included NMR as well as crystal structures of higher resolution). As part of our comparisons, we also included predicted driver genes from a sequence-based cluster analysis tool [OncodriverClust (33)] as well as previously curated driver genes in the Cancer Gene Census (CGC) database (72, 73). We note that we excluded driver genes in CGC that play roles in cancer through INDELs, copy number aberrations, or other structural variations. We used the UpsetR (74) package in R to visualize the multiway comparisons among predicted driver genes from various tools and CGC database. In addition to these, we modified our original framework to identify putative driver, where we don’t include motion-weighted edges to define communities on protein structures. We performed comparisons between the lists of putative drivers for our weighted and unweighted approach.
Finally, we also performed gene-expression analysis to validate the role of our putative driver genes in cancer at the transcriptome level. For this analysis, we obtained the TCGA RNA-sequencing quantification available for samples in the PanCancer Atlas Project (2). For each gene in our putative driver gene list (based on hotspot community information), we compared the gene-expression distribution for samples that harbor missense mutations to those that are not mutated. We performed a 2-sided KS test to evaluate the significance value for each gene in our putative gene list. These significance tests were carried out separately for each cancer type. However, we combined the significance level (P value) for each gene across multiple cancer types using the Fisher method. We visualized significantly differentially expressed genes using a standard QQ plot.
Results
Pan-Cancer Analysis of Genes Containing Mutation Clusters.
We applied our workflow to identify significantly mutated hotspot communities for each cancer cohort as well as on the pan-cancer level. As expected, we observed a relatively higher number of genes with at least 1 hotspot community on the pan-cancer level compared to cancer-specific analysis. Our pan-cancer analysis identifies hotspot communities in protein structures of 434 unique genes (Fig. 2A and Dataset S1). In contrast, a cancer-specific analysis revealed 56 potential driver genes with 186 significantly mutated hotspot communities in the corresponding protein structure (Dataset S2). Some of these genes (including TP53, PIK3CA, BRAF, SPOP, KRAS, HRAS, and PTEN) have previously been shown to be drivers for different cancer types. However, we also identified numerous genes containing hotspot communities that might drive cancer progression. Previous studies suggest that newly identified driver genes, including RHOC, NCOA1, and KLHL12, are involved in various signaling pathways. Similarly, PSPC1, FOXO3, and XRCC5 are known to be pivotal for immune response, apoptosis, and DNA repair, respectively. Furthermore, among these 434 genes, 12 had 5 or more hotspot communities, whereas 352 genes had just 1 hotspot community. These results highlight the efficacy of our approach in identifying novel and low-frequency putative driver genes with hotspot communities.
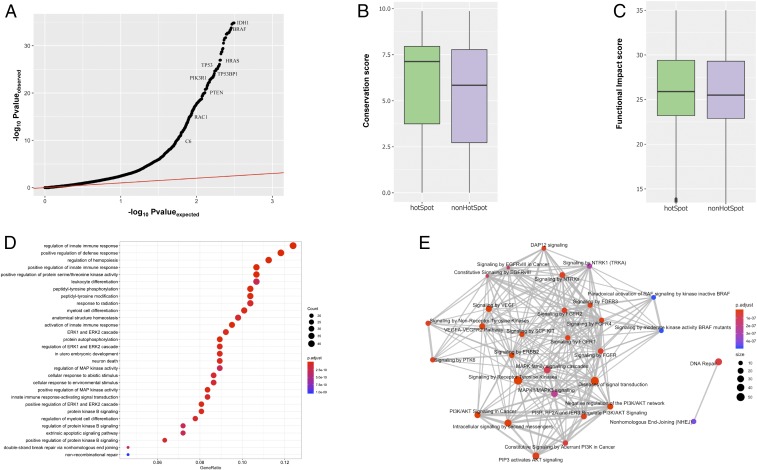
Fig. 2.
Pan-cancer analysis of putative driver genes with hotspot communities. (A) Pan-cancer QQ plot for genes with hotspot communities. (B) PhyloP conservation score comparisons between mutations occupying hotspot communities against nonhotspot communities on protein structures. (C) CADD score correlation between mutations occupying hotspot communities and nonhotspot communities on protein structures. (D) Biological process enrichment analysis for putative driver genes with at least 1 hotspot. The x axis corresponds to the gene ratio quantifying the fraction of putative driver genes belonging to a particular biological process. The color code and size correspond to corrected P value and number of genes involved in the biological process, respectively. (E) Reactome-based pathway enrichment analysis. The color code and size quantify to corrected P value and number of genes involved in the biological process, respectively.
Mutational cluster-based approaches assume that residues constituting such clusters are essential for protein function. Thus, a majority of cancer missense mutations occupying these hotspot communities are very likely to disrupt the protein functionality. In order to validate this assumption, we quantified the cross-species conservation measure [PhyloP score (66)] for mutations in hotspot as well as nonhotspot communities. As expected, we observe higher average conservation scores for mutations associated with residues in hotspot communities compared to those outside of hotspots. Furthermore, the observed difference in conservation was statically significant (2-sided KS test, P < 2e-5) (Fig. 2B). Similarly, the putative molecular functional impact [CADD score (67)] of mutations occupying hotspot communities was significantly higher compared to those mapping to nonhotspot communities (2-sided KS test, P < 2e-5) (Fig. 2C).
We also preformed GO (71) and pathway enrichment analysis to decipher the biological functions of genes with predicted hotspot communities. The biological process-based GO enrichment analysis implicates putative driver genes in diverse biological functions, including a role in the immune response, cell differentiation, kinase activities, posttranslational modifications, apoptosis, and DNA repair (Fig. 2D and Dataset S3). Similarly, reactome (69) pathway-based enrichment analysis suggests that putative driver genes with hotspot communities play roles in various signaling pathways (Dataset S4), including NTRK signaling, DAP12 signaling, EGFR signaling, and MAP kinase-associated signaling. Additionally, these genes are also enriched among DNA repair and nonhomologous end-joining–associated pathways (Fig. 2E). Furthermore, KEGG (75) pathway-based enrichment analysis indicates that our identified putative driver genes play roles in various cancer subtypes (bladder, pancreatic, breast, chronic myeloid leukemia, melanoma, acute myeloid leukemia, glioma) (SI Appendix, Fig. S1 and Dataset S5).
Comparisons of 3D Structure-Based Clustering Methods.
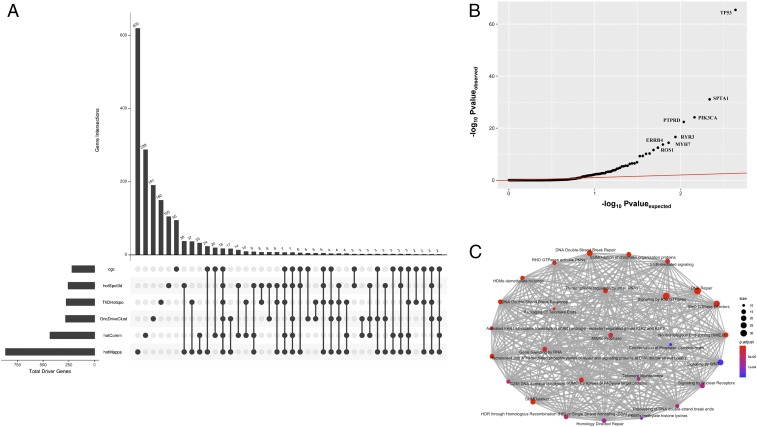
We compared our set of predicted drivers to the predicted drivers from other methods, including the set of curated genes in the COSMIC (72) database (Fig. 3A). Furthermore, we also performed a comparison between putative driver genes identified using our workflow and genes identified as drivers by other mutation cluster detection algorithms that do not take protein dynamics into account. The majority of these additional algorithms employ the 3D structure of a protein to identify mutational clusters, with the exception of OncoDriveClust (33), which searches for hotspot mutations at the sequence level. Overall, our workflow identified many additional genes (288 genes) with hotspot communities compared to other mutation hotspot analysis tools (Fig. 3A). One exception was the HOTMAP (38) algorithm, which utilizes protein homology models in addition to protein structure. Thus, it identifies a significantly higher number of unique genes (620 genes) with mutation clusters compared to any other tool. Furthermore, our approach identified 146 genes (34% of our gene list) with hotspot communities that are either curated as driver genes in COSMIC or predicted to contain a mutation cluster by another tool (Fig. 3A). Among these 146 genes, 89 genes overlapped with putative driver genes identified by the HOTMAP algorithm, whereas 63 genes overlapped with drivers in COSMIC. As expected, we observed the lowest overlap (33 genes, 7% of our putative driver gene list) with the sequence-based method (OncoDriveClust) (Fig. 3A).
Fig. 3.
Comparison with other hotspot detection tools. (A) Comparison of multiple driver detection algorithms represented using the upset plot. We used the most recent version of the CGC database for this analysis. All algorithms were run on the TCGA-MC3 variant call set. Numbers of identified driver genes common to different sets of methods are shown in the bar chart (Upper), and those unique to specific methods in each set are indicated with solid points below the bar chart. (B) QQ plot highlighting differentially expressed putative driver genes across multiple cancer types. (C) Pathway-level enrichment analysis of those singleton genes identified by HotCommics that were novel (with respect to putative driver genes identified by other algorithms and/or the CGC database).
To evaluate the added predictive contribution of protein dynamics, we performed a controlled, comparative study in which we identify driver genes under 2 schemes: first in which the edges are weighted using the models of protein motions, and second in which the edges are left unweighted (i.e., wherein all edges are weighted the same, as in a static structure). We applied our workflow on the same set of protein structures using these 2 approaches. Overall, we observed that, relative to the unweighted static networks, we identified 49% more genes with 1 or more hotspot communities using the motion-weighted networks compared to the unweighted approach (SI Appendix, Fig. S3). This observation highlights the advantage of employing protein dynamics-based community definitions. The motion-weighted network definition tends to result in larger sized (and thus fewer) communities (SI Appendix, Fig. S4) relative to unweighted networks. The larger community definitions provide higher statistical power to detect low-frequency drivers. Additionally, we found that communities identified using motion-weighted network edges performed better at capturing biological annotations relative to unweighted networks (SI Appendix, Figs. S5 and S6).
Additionally, we analyzed TCGA expression data to obtain further evidence corroborating the biological validity of putative driver genes identified through our workflow. For each candidate gene, we quantified the statistical significance in expression distribution differences using a 2-sided KS test. We performed this test for individual cancer type, and the corresponding P values were combined across cancer types using Fisher’s method to provide a pan-cancer significance measure. Overall, our analysis identified 60 genes, including TP53, SPTA1, PIK3CA, KRAS, and EGFR that were differentially expressed across cancer types (Fig. 3B and Dataset S6). A subset of these differentially expressed genes, such as MYH7, ROS1, TIAM1, PTPRD, and HUWE1 are potentially novel driver genes with predicted hotspot communities (Fig. 3B and Dataset S6). Moreover, we note that 76% of our putative driver gene list with significantly mutated hotspot communities were differentially expressed in at least 1 TCGA cancer cohort.
Finally, we also performed GO and pathway enrichment analysis on genes that have not been previously reported to be cancer driver but for which we identified mutational hotspot communities. These genes are defined to be those that were neither present in the COSMIC driver database nor were predicted to encompass mutation clusters using other hotspot identification tools. We observed significant enrichment of these genes in crucial biological processes (Dataset S7), including DNA conformation change, regulation of immune response, regulation of stem cell differentiation, nucleosome organization, and endothelial cell apoptotic processes (SI Appendix, Fig. S2). Similarly, pathway enrichment analysis implicates their role in DNA repair, SUMOylation, RHO GTPase activity, telomere maintenance, and various signaling pathways (Fig. 3C and Dataset S8).
Case Studies Highlighting the Roles of Hotspot Communities in Deciphering Driver Mechanisms.
Integrating knowledge of 3D structures and protein dynamics to identify driver genes has a clear advantage over other methods that do not leverage protein structure or dynamics. Our method allows us to investigate disruption in protein structure and function induced by missense mutations within predicted hotspot communities. We also note that the majority of our hotspot communities encompass residues that are pivotal for important protein functions, including allostery, bimolecular signaling, protein binding, and posttranslation modifications. The sensitive detection of functional sites on protein structure helps to decipher the underlying biophysical mechanism that plays a crucial role in cancer growth. Here, we highlight 3 examples to showcase the utility of our framework in gaining biophysical insights into cancer progression through disruption of predicted hotspot communities. These examples include an oncogene (BRAF), a tumor suppressor (PIK3R1), and a previously unreported putative driver (PTPRD), all of which are predicted to contain multiple hotspot communities on their respective structures. PTPRD is a transmembrane protein containing a cytoplasmic tyrosine phosphatase domain. PTPRD is absent in the COSMIC driver gene database, and existing methods which ignore protein dynamics do not identify this gene as a cancer driver. Besides, the “static version” of our framework (i.e., wherein network communities are identified without weighing the edges using dynamics) failed to identify PTPRD as a driver. Thus, through this example, we demonstrate that including dynamics constitutes an essential feature in the search for novel drivers.
Missense hot spot communities in PIK3R1.
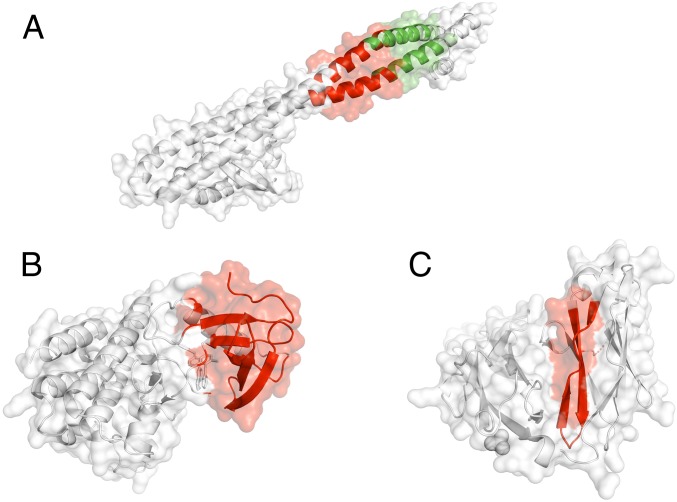
The PI3KR1 gene encodes the α-subunit of the enzyme Phosphatidylinositol 3-kinase, which plays a crucial role in a variety of cellular processes, including cell survival, regulation of gene expression, cell metabolism, and cytoskeletal rearrangement (76). Mutations in PIK3KR1 (a tumor suppressor gene) have previously been implicated in breast cancer. Recent therapeutic studies have targeted PI3K inhibition resulting in a decrease in cellular proliferation and reduced metastasis in the mouse model. PI3Ks are obligate heterodimers composed of a p110 subunit and a regulatory subunit. Previous studies have identified 4 distinct domains belonging to the catalytic P110 α-subunit that harbor somatic mutations leading to an increase in PI3K activity. We observed 2 distinct hotspot communities (Fig. 4A) on the cocrystal structure (PDB ID code 2V1Y) of the protein complex that compromises the adaptor-binding domain (ABD) of the P110 α-subunit and the iSH2 domain of the p85 α-regulatory subunit (76). The 2 hotspot communities are composed of 28 (community 5) and 26 (community 7) residues, respectively (Fig. 4A). On the pan-cancer level, we observed 24 and 16 mutations that map to community 5 and community 7 on the cocrystal structure, respectively. These distinct hotspot communities are adjacent to each other in the same helical structure. However, we observed a small kink in this helical structure, which presumably leads to distinct protein motions associated with these 2 different hotspot communities. Additionally, both these communities occupy the iSH2 domain that plays an essential role in proper binding to the ABD domain (76). Thus, the presence of these mutational hotspot communities in the iSH2 domain is likely to influence the ABD–iSH2 interaction in tumor samples. Furthermore, modification in this interaction might affect the binding between ABD and the catalytic region of the p110 subunit. The altered interaction may trigger hyperactivation of the PI3K pathways (77), which are often implicated in various types of cancer.
Fig. 4.
Examples of a tumor-suppressor gene, an oncogene, and a putative driver with hotspot communities. (A) Hotspot communities (shown in red) in PIK3R1, as identified by our workflow. Previous studies have also identified the PIK3R1 gene as a tumor-suppressor gene. (B) Hotspot communities in BRAF, as identified by our workflow. Previous studies have identified BRAF1 gene as an oncogene. (C) Hotspot communities in PTPRD, as identified by our workflow. PTPRD is an example of a novel putative driver gene.
Missense hotspot communities in BRAF.
The BRAF gene encodes a protein belonging to the serine/threonine protein kinase family that regulates MAP kinase and ERK signaling pathway (78). This pathway is considered to be essential for a number of biological functions, including cell differentiation, cellular growth, senescence, and apoptosis. Somatic mutations in the BRAF gene are often implicated in various cancer subtypes, including melanoma, colorectal cancer, prostate cancer, nonsmall-cell lung cancer, and papillary thyroid tumors (79). The BRAF protein comprises 3 distinct conserved regions: CR1, CR2, and CR3. The CR1 region constitutes the RAS-binding domain and functions as an autoinhibitor. The BRAF kinase domain is encoded by the CR3 region of the BRAF protein. The N terminus of the CR3 region contains the P-loop region that stabilizes ATP binding. Additionally, the CR3 region also comprises an αC-helix and the dimerization interface, which maintains the inactive state of BRAF. Finally, the C-terminal end of the CR3 region consists of a catalytic loop, the DFG motif, and the activation loop. These elements in the CR3 region facilitate binding of substrate proteins to BRAF and maintains the protein in the inactive state. It has been proposed that mutations in BRAF induce dysregulation in the binding of Ras to Raf and MEK proteins within the Ras/RAF/MEK/ERK signaling cascade, thereby leading to overactivation of the signaling pathway and subsequent oncogenesis (79). Multiple enzyme inhibitors have been designed to target BRAF kinase. One such inhibitor (aminoisoquinoline) has been cocrystallized with the BRafV600E kinase domain at a resolution of 2.7 Å (PDB ID code 3IDP) (80). In our study, we identified 1 hotspot community in this cocrystal structure (Fig. 4B). This hotspot community is composed of 52 residues that adopt a β-sheet of residues at the dimerization interface, catalytic loop, and the DFG motifs in the CR3 region of the BRAF protein. All of these elements of the CR3 region play vital roles in maintaining the inactive state of the native BRAF protein. Thus, recurrent cancer mutations can facilitate changes in the conformation of BRAF from its inactive state to an active state, thereby potentially driving tumor progression.
Missense hotspot community in PTPRD.
The PTPRD gene encodes a protein that belongs to the protein tyrosine phosphatase (PTP) family. PTP proteins are considered to be essential for regulating cellular proliferation, differentiation, and oncogenic transformation. The PTPRD gene encodes a transmembrane protein containing a cytoplasmic tyrosine phosphatase domain. Previous studies have shown that PTPRD genes are frequently deleted in various cancer types, including glioma, neuroblastoma, and lung cancer (81). However, we note that PTPRD is not identified as missense driver in COSMIC (82). Moreover, previous studies did not identify mutational hotspot communities in the PTPRD gene. In contrast, our analysis identifies 1 hotspot community in the crystal structure (PDB ID code 2YD7) of the receptor protein tyrosine phosphatase (RPTP) σ-subunit.
RPTPs are cell surface proteins with intracellular PTP activity and extracellular domains that are sequentially homologous to cell adhesion molecules. Moreover, the RPTP σ-subunit is considered necessary for nervous system development and function. In our analysis, somatic mutations occur in 2 communities (communities 2 and 4) on the crystal structure of the RPTP σ-subunit. Our workflow predicts 1 hotspot community that comprises 47 residues in the crystal structure of PTPRD (Fig. 4C) and constitutes a β-sheet conformation (83). This hotspot community comprises residues primarily belonging to the Ig1 and Ig2 domains of the RPTP σ-subunit, which facilitate binding to heparan-sulfate glycosaminoglycans (HSGAGs) polysaccharides. HSGAGs modulate cell signaling and tumorigenesis by regulating autocrine signaling loops (84). The presence of predicted hotspots in the Ig1-2 domain of the RPTP σ-subunit is likely to alter its binding to HSGAGs and may play role in tumor progression.
Discussion
The underlying heterogeneous nature of cancer (14) makes interpretability of genomic alterations in a given cancer genome very challenging. In particular, genomic heterogeneity poses a major challenge in identifying key cancer-driver mutations. Large-scale genome sequencing efforts have helped us to generate comprehensive catalogs of driver mutations (5) in various cancer types. However, the canonical recurrence-based driver-detection algorithms have failed to identify low-frequency or rare drivers. The limited cohort size (11) and heterogeneity (14) in cancer genomes provides limited power to identify low-frequency drivers using the canonical position-level recurrence algorithms. A simplistic approach to address the issue of missing rare drivers would be to sequence more patients for a given cancer type. However, this will be particularly challenging for highly heterogeneous cancer cohorts with multiple subtypes (85).
Moreover, this approach will not be practical for certain rare cancers, such as neuroblastoma, angiosarcoma, Hodgkin’s lymphoma, and various pediatric cancers. One potential remedy is to quantify recurrence over functional elements, such as posttranslational modification sites (27, 28) and protein interaction interfaces (30). However, many rare and latent drivers (19) may not fall within well-defined functional annotation sites. Thus, a suitable alternative is to measure recurrence of variants within entire subregions of genes (86), thereby identifying mutational clusters or neighborhoods (35–38, 40). Aggregating multiple variants into such clusters can mitigate the issues posed by the limited statistical power of quantifying position-level recurrence of individual variants.
In particular, many driver-detection algorithms search for the presence of mutational hotspots in 3D-protein structures to identify putative driver genes. Compared to sequence-based driver-detection methods, using protein structural data can help to decipher the underlying molecular mechanisms that influence cancer progression. However, current approaches to identify such hotspots and their corresponding host driver genes completely ignore the role of protein dynamics, which are essential for protein function. Thus, here we propose a framework that integrates protein dynamics and 3D-structures to identify missense hotspot communities and their associated putative driver genes.
Overall, our workflow identified 802 hotspot communities on crystal structures of proteins corresponding to 434 unique genes on the pan-cancer level. We also compared our putative driver-gene list with derived driver-gene lists generated in previous experimental and prediction-based studies. Among our putative driver-gene list, we found 36% of genes are either known or predicted to be driver genes based on previous studies. We term the remaining 64% of genes “novel drivers.” We performed many downstream analyses on our putative driver genes to highlight their roles in cancer progression.
Our framework assumes that a residue community on a protein structure represents a putative functional subunit of a protein. Thus, high mutation densities in such communities (compared to a random expectation) is very likely to alter protein function. One would expect that mutations influencing residues in these communities will have a high functional impact as they can drive cancer progression. Our observation is consistent with this hypothesis, as we find that missense mutations occupying hotspot communities in protein structures are highly conserved across species and have a higher molecular functional impact compared to those outside such hotspot communities.
Furthermore, we also observed significantly higher enrichment of our putative driver genes with predicted hotspot communities in vital biological processes and pathways that are relevant for oncogenesis. For example, our ontological analysis indicates enrichment of our putative driver genes in biological processes associated with regulation and activation of the innate immune response. This observation is consistent with the current notion that dysfunction in the immune response (as a result of genomic alterations) may allow tumor cells to evade immune detection.
Additionally, we also observed a significant enrichment of putative driver genes in cell differentiation and cell growth processes, such as the regulation of hematopoiesis and myeloid cell differentiation, which were previously implicated in tumor growth. Moreover, we observed a high enrichment of our putative driver genes in the regulation of kinase activities, including protein serine/threonine and MAP kinase activities. Additionally, these genes are also enriched among ERK1/ERK2 signaling cascade, protein kinase B signaling, PI3K/AKT signaling, FGFR1 signaling, NTRK1 signaling, apoptosis signaling, and various other signaling pathways. Aberrant signaling pathways constitute an essential hallmark of cancer. Thus, the enrichment of our putative driver genes in critical signaling pathways provides clear biological evidence for their role in cancer. Moreover, these genes are enriched for DNA repair function via nonhomologous end joining and other nonrecombination-based repair mechanisms. Finally, we note that we observed the same enrichment for the subset of novel genes that have not been identified as driver genes in previous studies.
Genomic alterations that are consequential for tumor growth are often manifested on the transcriptome level such that mutated driver genes are often differentially expressed compared to a healthy population or patients without any mutation in driver genes. We leveraged the transcriptome data from TCGA to identify genes among our list of putative driver genes that are also differentially expressed. We identified 60 genes among our predicted driver genes that were differentially expressed in tumor samples. These differentially expressed putative driver genes include novel as well as previously established driver genes. As with genomic data, the amount of transcriptomic data for each individual cohort is not sufficiently large to provide enough statistical power for identifying differentially expressed genes. However, we note that 76% of our putative driver genes were differentially expressed in at least 1 TCGA cancer cohort. These analyses further validate our hotspot community-based driver-detection approach. Finally, we note that our current framework identifies the hotspot communities in putative driver genes without specifying putative driver mutations. However, a close inspection of molecular functional impact score and residue-level annotation of mutations in our putative hotspot communities can be utilized to identify the putative driver mutations.
In the context of investigating the molecular mechanism underlying tumor growth, protein structure-based driver-detection methods offer significant advantages over approaches that are limited to sequence space. However, structure-based methods suffer from limited coverage of the human proteome. Thus, the applicability of structure-based methods is, of course, limited only to mutations that can be mapped to protein structures. A prior study (38) has applied homology model-derived structures to circumvent the issue of limited structural coverage. However, the accuracy of homology-based models has shown to be limited for various protein complexes and transmembrane proteins. Moreover, modeling protein motions for homology-model–derived protein structures would most likely be less accurate, thereby affecting sensitivity. Nevertheless, significant technical improvements in crystallographic and cryo-EM techniques (87) are expected to expand the current structurally resolved proteome. In particular, cryo-EM technologies (87) now allow us to obtain a high-resolution structure of large proteins and biomolecular complexes that were previously elusive. Thus, we anticipate an essential role of our approach in future studies aimed at discovering low-frequency drivers in various cancer cohorts. Additionally, knowledge of protein motions (along with structures) can potentially help uncover druggable hotspot communities. Such studies are likely to open new therapeutic avenues for various cancers and will help in realizing the goal of precision medicine in cancer.
Supplementary Material
Acknowledgments
We thank Jonathan Warrell and Timur Galeev for providing valuable feedback for improving the manuscript. We acknowledge support from the NIH Grant (R01 HG 008126) and from A. L. Williams Professorship funds.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The scripts and code reported in this article have been deposited in GitHub (https://github.com/gersteinlab/HotCommics).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1901156116/-/DCSupplemental.
References
- 1.Weinstein J. N., et al. ; Cancer Genome Atlas Research Network , The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 45, 1113–1120 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ding L., et al. ; Cancer Genome Atlas Research Network , Perspective on oncogenic processes at the end of the beginning of cancer genomics. Cell 173, 305–320.e10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hudson T. J., et al. ; International Cancer Genome Consortium , International network of cancer genome projects. Nature 464, 993–998 (2010). Erratum in: Nature465, 966 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campbell P. J., et al. , Pan-cancer analysis of whole genomes. bioRxiv:10.1101/162784 (12 July 2017).
- 5.Matthew Bailey A. H., et al. , Comprehensive characterization of cancer driver genes and mutations article comprehensive characterization of cancer driver genes and mutations. Cell 173, 371–376.e18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rheinbay E., et al. , Discovery and characterization of coding and non-coding driver mutations in more than 2,500 whole cancer genomes. bioRxiv:10.1101/237313 (23 December 2017).
- 7.Sabarinathan R., et al. , The whole-genome panorama of cancer drivers. bioRxiv:10.1101/190330 (20 September 2017).
- 8.Ding L., Wendl M. C., McMichael J. F., Raphael B. J., Expanding the computational toolbox for mining cancer genomes. Nat. Rev. Genet. 15, 556–570 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raphael B. J., Dobson J. R., Oesper L., Vandin F., Identifying driver mutations in sequenced cancer genomes: Computational approaches to enable precision medicine. Genome Med. 6, 5 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tamborero D., et al. , Comprehensive identification of mutational cancer driver genes across 12 tumor types. Sci. Rep. 3, 2650 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lawrence M. S., et al. , Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 505, 495–501 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garraway L. A., Lander E. S., Lessons from the cancer genome. Cell 153, 17–37 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Stratton M. R., Exploring the genomes of cancer cells: Progress and promise. Science 331, 1553–1558 (2011). [DOI] [PubMed] [Google Scholar]
- 14.Lawrence M. S., et al. , Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499, 214–218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Armenia J., et al. ; PCF/SU2C International Prostate Cancer Dream Team , The long tail of oncogenic drivers in prostate cancer. Nat. Genet. 50, 645–651 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greenman C., et al. , Patterns of somatic mutation in human cancer genomes. Nature 446, 153–158 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beerenwinkel N., et al. , Genetic progression and the waiting time to cancer. PLoS Comput. Biol. 3, e225 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nussinov R., Jang H., Tsai C.-J., Cheng F., Precision medicine review: Rare driver mutations and their biophysical classification. Biophys. Rev. 11, 5–19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nussinov R., Tsai C. J., ‘Latent drivers’ expand the cancer mutational landscape. Curr. Opin. Struct. Biol. 32, 25–32 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Dees N. D., et al. , MuSiC: Identifying mutational significance in cancer genomes. Genome Res. 22, 1589–1598 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gonzalez-Perez A., Lopez-Bigas N., Functional impact bias reveals cancer drivers. Nucleic Acids Res. 40, e169 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar S., Clarke D., Gerstein M., Localized structural frustration for evaluating the impact of sequence variants. Nucleic Acids Res. 44, 10062–10073 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nehrt N. L., Peterson T. A., Park D., Kann M. G., Domain landscapes of somatic mutations in cancer. BMC Genomics 13 (suppl. 4), S9 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peterson T. A., Gauran I. I. M., Park J., Park D., Kann M. G., Oncodomains: A protein domain-centric framework for analyzing rare variants in tumor samples. PLoS Comput. Biol. 13, e1005428 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang F., et al. , Protein domain-level landscape of cancer-type-specific somatic mutations. PLoS Comput. Biol. 11, e1004147 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reimand J., Bader G. D., Systematic analysis of somatic mutations in phosphorylation signaling predicts novel cancer drivers. Mol. Syst. Biol. 9, 637 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Narayan S., Bader G. D., Reimand J., Frequent mutations in acetylation and ubiquitination sites suggest novel driver mechanisms of cancer. Genome Med. 8, 55 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reimand J., Wagih O., Bader G. D., The mutational landscape of phosphorylation signaling in cancer. Sci. Rep. 3, 2651 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Porta-Pardo E., Godzik A., e-Driver: A novel method to identify protein regions driving cancer. Bioinformatics 30, 3109–3114 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Porta-Pardo E., Garcia-Alonso L., Hrabe T., Dopazo J., Godzik A., A pan-cancer catalogue of cancer driver protein interaction interfaces. PLoS Comput. Biol. 11, e1004518 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller M. L., et al. , Pan-cancer analysis of mutation hotspots in protein domains. Cell Syst. 1, 197–209 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van den Eynden J., Fierro A. C., Verbeke L. P. C., Marchal K., SomInaClust: Detection of cancer genes based on somatic mutation patterns of inactivation and clustering. BMC Bioinformatics 16, 125 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tamborero D., Gonzalez-Perez A., Lopez-Bigas N., OncodriveCLUST: Exploiting the positional clustering of somatic mutations to identify cancer genes. Bioinformatics 29, 2238–2244 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Ryslik G. A., et al. , A spatial simulation approach to account for protein structure when identifying non-random somatic mutations. BMC Bioinformatics 15, 231 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kamburov A., et al. , Comprehensive assessment of cancer missense mutation clustering in protein structures. Proc. Natl. Acad. Sci. U.S.A. 112, E5486–E5495 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao J., et al. , 3D clusters of somatic mutations in cancer reveal numerous rare mutations as functional targets. Genome Med. 9, 4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Niu B., et al. , Protein-structure-guided discovery of functional mutations across 19 cancer types. Nat. Genet. 48, 827–837 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tokheim C., et al. , Exome-scale discovery of hotspot mutation regions in human cancer using 3D protein structure. Cancer Res. 76, 3719–3731 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ye J., Pavlicek A., Lunney E. A., Rejto P. A., Teng C. H., Statistical method on nonrandom clustering with application to somatic mutations in cancer. BMC Bioinformatics 11, 11 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ryslik G. A., Cheng Y., Cheung K. H., Modis Y., Zhao H., A graph theoretic approach to utilizing protein structure to identify non-random somatic mutations. BMC Bioinformatics 15, 86 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ryslik G. A., Cheng Y., Modis Y., Zhao H., Leveraging protein quaternary structure to identify oncogenic driver mutations. BMC Bioinformatics 17, 137 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meyer M. J., et al. , mutation3D: Cancer gene prediction through atomic clustering of coding variants in the structural proteome. Hum. Mutat. 37, 447–456 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frauenfelder H., Sligar S., Wolynes P., The energy landscapes and motions of proteins. Science 254, 1598–1603 (1991). [DOI] [PubMed] [Google Scholar]
- 44.Tsai C.-J., Nussinov R., The free energy landscape in translational science: How can somatic mutations result in constitutive oncogenic activation? Phys. Chem. Chem. Phys. 16, 6332–6341 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boehr D. D., Nussinov R., Wright P. E., The role of dynamic conformational ensembles in biomolecular recognition. Nat. Chem. Biol. 5, 789–796 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nussinov R., Tsai C.-J., Allostery in disease and in drug discovery. Cell 153, 293–305 (2013). [DOI] [PubMed] [Google Scholar]
- 47.Onuchic J. N., Luthey-Schulten Z., Wolynes P. G., Theory of protein folding: The energy landscape perspective. Annu. Rev. Phys. Chem. 48, 545–600 (1997). [DOI] [PubMed] [Google Scholar]
- 48.Clarke D., et al. , Identifying allosteric hotspots with dynamics: Application to inter- and intra-species conservation. Structure 24, 826–837 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ming D., Wall M. E., Quantifying allosteric effects in proteins. Proteins 59, 697–707 (2005). [DOI] [PubMed] [Google Scholar]
- 50.del Sol A., Fujihashi H., Amoros D., Nussinov R., Residues crucial for maintaining short paths in network communication mediate signaling in proteins. Mol. Syst. Biol. 2, 2006.0019 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Henzler-Wildman K., Kern D., Dynamic personalities of proteins. Nature 450, 964–972 (2007). [DOI] [PubMed] [Google Scholar]
- 52.Ghosh A., Vishveshwara S., Variations in clique and community patterns in protein structures during allosteric communication: Investigation of dynamically equilibrated structures of methionyl tRNA synthetase complexes. Biochemistry 47, 11398–11407 (2008). [DOI] [PubMed] [Google Scholar]
- 53.Mitternacht S., Berezovsky I. N., Binding leverage as a molecular basis for allosteric regulation. PLoS Comput. Biol. 7, e1002148 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rousseau F., Schymkowitz J., A systems biology perspective on protein structural dynamics and signal transduction. Curr. Opin. Struct. Biol. 15, 23–30 (2005). [DOI] [PubMed] [Google Scholar]
- 55.Agajanian S., Odeyemi O., Bischoff N., Ratra S., Verkhivker G. M., Machine learning classification and structure-functional analysis of cancer mutations reveal unique dynamic and network signatures of driver sites in oncogenes and tumor suppressor genes. J. Chem. Inf. Model. 58, 2131–2150 (2018). [DOI] [PubMed] [Google Scholar]
- 56.Ellrott K., et al. ; MC3 Working Group; Cancer Genome Atlas Research Network , Scalable open science approach for mutation calling of tumor exomes using multiple genomic pipelines. Cell Syst. 6, 271–281.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cibulskis K., et al. , Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 31, 213–219 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Radenbaugh A. J., et al. , RADIA: RNA and DNA integrated analysis for somatic mutation detection. PLoS One 9, e111516 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Larson D. E., et al. , SomaticSniper: Identification of somatic point mutations in whole genome sequencing data. Bioinformatics 28, 311–317 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Koboldt D. C., et al. , VarScan: Variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics 25, 2283–2285 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Habegger L., et al. , VAT: A computational framework to functionally annotate variants in personal genomes within a cloud-computing environment. Bioinformatics 28, 2267–2269 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smedley D., et al. , The BioMart community portal: An innovative alternative to large, centralized data repositories. Nucleic Acids Res. 43, W589–W598 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sethi A., Eargle J., Black A. A., Luthey-Schulten Z., Dynamical networks in tRNA:protein complexes. Proc. Natl. Acad. Sci. U.S.A. 106, 6620–6625 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Girvan M., Newman M. E. J., Community structure in social and biological networks. Proc. Natl. Acad. Sci. U.S.A. 99, 7821–7826 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kumar S., Clarke D., Gerstein M., HotCommics. GitHub. https://github.com/gersteinlab/HotCommics. Deposited 29 December 2018. [Google Scholar]
- 66.Pollard K. S., Hubisz M. J., Rosenbloom K. R., Siepel A., Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 20, 110–121 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kircher M., et al. , A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 46, 310–315 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.The Gene Ontology Consortium , Expansion of the gene ontology knowledgebase and resources. Nucleic Acids Res. 45, D331–D338 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fabregat A., et al. , The reactome pathway knowledgebase. Nucleic Acids Res. 46, D649–D655 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Marisa L., et al. , KEGG: Kyoto encyclopedia of genes and genomes. Nature 10, 1350–1356 (2013). [Google Scholar]
- 71.Yu G., Wang L.-G., Han Y., He Q.-Y., clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Futreal P. A., et al. , A census of human cancer genes. Nat. Rev. Cancer 4, 177–183 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Forbes S. A., et al. , COSMIC: Exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res. 43, D805–D811 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Conway J. R., Lex A., Gehlenborg N., UpSetR: An R package for the visualization of intersecting sets and their properties. Bioinformatics 33, 2938–2940 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kanehisa M., Furumichi M., Tanabe M., Sato Y., Morishima K., KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 45, D353–D361 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cantley L. C., The phosphoinositide 3-kinase pathway. Science 296, 1655–1657 (2001). [DOI] [PubMed] [Google Scholar]
- 77.Cheung L. W., Mills G. B., Targeting therapeutic liabilities engendered by PIK3R1 mutations for cancer treatment. Pharmacogenomics 17, 297–307 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Davies H., et al. , Mutations of the BRAF gene in human cancer. Nature 417, 949–954 (2002). [DOI] [PubMed] [Google Scholar]
- 79.Dankner M., Rose A. A. N., Rajkumar S., Siegel P. M., Watson I. R., Classifying BRAF alterations in cancer: New rational therapeutic strategies for actionable mutations. Oncogene 37, 3183–3199 (2018). [DOI] [PubMed] [Google Scholar]
- 80.King A. J., et al. , Demonstration of a genetic therapeutic index for tumors expressing oncogenic BRAF by the kinase inhibitor SB-590885. Cancer Res. 66, 11100–11105 (2006). [DOI] [PubMed] [Google Scholar]
- 81.Veeriah S., et al. , The tyrosine phosphatase PTPRD is a tumor suppressor that is frequently inactivated and mutated in glioblastoma and other human cancers. Proc. Natl. Acad. Sci. U.S.A. 106, 9435–9440 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Forbes S. A., et al. , COSMIC: Somatic cancer genetics at high-resolution. Nucleic Acids Res. 45, D777–D783 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Coles C. H., et al. , Proteoglycan-specific molecular switch for RPTP clustering and neuronal extension. Science 332, 484–488 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sasisekharan R., Shriver Z., Venkataraman G., Narayanasami U., Roles of heparan-sulphate glycosaminoglycans in cancer. Nat. Rev. Cancer 2, 521–528 (2002). [DOI] [PubMed] [Google Scholar]
- 85.Hoadley K. A., et al. ; Cancer Genome Atlas Network , Cell-of-origin patterns dominate the molecular classification of 10,000 tumors from 33 types of cancer. Cell 173, 291–304.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Porta-Pardo E., et al. , Comparison of algorithms for the detection of cancer drivers at subgene resolution. Nat. Methods 14, 782–788 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nogales E., The development of cryo-EM into a mainstream structural biology technique. Nat. Methods 13, 24–27 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.