Significance
In the classic view, telomerase is silenced in terminally differentiated cells and its reactivation supports immortalization and unlimited growth of most cancers. Here, we determine the involvement of telomerase during replicative senescence in primary fibroblasts from mouse and human. In both cases, we find that cells that are unable to produce telomerase approach cellular senescence earlier and exhibit a significantly higher rate of malignant transformation than the control cells. Furthermore, an evident upregulation of telomerase expression is detected in wild-type control cells at the presenescence stage, which is accountable for the protection. In conclusion, this study suggests that telomerase has a previously underappreciated, protective role in buffering senescence stresses due to short, dysfunctional telomeres, thereby preventing malignant transformation.
Keywords: telomerase, senescence, tumorigenesis, ATM
Abstract
Telomerase is an enzymatic ribonucleoprotein complex that acts as a reverse transcriptase in the elongation of telomeres. Telomerase activity is well documented in embryonic stem cells and the vast majority of tumor cells, but its role in somatic cells remains to be understood. Here, we report an unexpected function of telomerase during cellular senescence and tumorigenesis. We crossed Tert heterozygous knockout mice (mTert+/−) for 26 generations, during which time there was progressive shortening of telomeres, and obtained primary skin fibroblasts from mTert+/+ and mTert−/− progeny of the 26th cross. As a consequence of insufficient telomerase activities in prior generations, both mTert+/+ and mTert−/− fibroblasts showed comparable and extremely short telomere length. However, mTert−/− cells approached cellular senescence faster and exhibited a significantly higher rate of malignant transformation than mTert+/+ cells. Furthermore, an evident up-regulation of telomerase reverse-transcriptase (TERT) expression was detected in mTert+/+ cells at the presenescence stage. Moreover, removal or down-regulation of TERT expression in mTert+/+ and human primary fibroblast cells via CRISPR/Cas9 or shRNA recapitulated mTert−/− phenotypes of accelerated senescence and transformation, and overexpression of TERT in mTert−/− cells rescued these phenotypes. Taking these data together, this study suggests that TERT has a previously underappreciated, protective role in buffering senescence stresses due to short, dysfunctional telomeres, and preventing malignant transformation.
Telomerase is a ribonucleoprotein complex that protects and extends the telomeres of the chromosome (1–3). It consists of 2 essential subunits, the template RNA (TR; telomerase RNA) and the reverse-transcriptase catalytic subunit (TERT; telomerase reverse transcriptase) (1, 4). Telomerase activity is required for the maintenance of stemness in stem cells (5), and its expression is precisely regulated in stem and progenitor cells and generally suppressed in differentiated somatic cells (6–9). Somatic cells without telomerase activity exhibit a limited replicative capacity and after a finite number of cell divisions reach a state known as replicative senescence that can be abrogated by ectopic telomerase expression (10–12).
Replicative senescence is triggered by critically shortened telomeres and serves as a natural barrier to tumorigenesis (13, 14). While senescent cells undergo up-regulation of tumor-suppressor genes and cell cycle inhibitors to arrest cell cycle, they also gradually develop a senescence-associated secretory phenotype (SASP), which can transform senescent cells into proinflammatory cells (15, 16). Those cells can escape senescence arrest and undergo continuous proliferation (17), eventually via either an alternative mechanism of telomere elongation (alternative lengthening of telomere, ALT) or reactivation of TERT expression to promote malignant transformation (18, 19). In this case, telomerase reactivation or an ALT supports immortalization and the unlimited growth of most cancers.
In the past 2 decades, independent groups have constructed telomerase-deficient (mTR−/− or mTert−/−) mouse models (20–23). While those telomerase-deficient mice did not show any noticeable defects during the early generations of intercross between deficient mice, late-generation animals show phenotypes, including short telomere length (TL), shortened lifespan, progressive tissue atrophy, reduced ability for stress response, and notably, spontaneous malignancies (22, 24–26). Since laboratory mouse models usually carry very long telomeres in comparison with those of human cells (27), in the absence of sufficient telomerase activity, it takes many generations for the telomerase-deficient models to reach a critically short telomere stage. It appears that the lack of TERT expression plays a crucial role in causing the above phenotypes in telomerase-deficient mice, as reactivation of TERT expression via an inducible promotor effectively ameliorates the phenotypes in late-generation mTert−/− mice (26). Based on this observation, we speculate that telomerase activity in senescent somatic cells with short telomeres might have additional roles beyond promoting tumor formation.
To test this idea, we utilized an experimental system that allows us to conduct direct studies of the function of TERT in mouse somatic cells with extremely short telomeres. Gender- and age-matched mouse primary skin fibroblasts were obtained from mTert+/+ and mTert−/− siblings, which were the progenies from late-generation (26th) mTert+/− parents with C57BL/6 (B6) genotype (23, 28). mTert+/+ and mTert−/− are not only genotypically identical except for the mTert gene, but also carry similarly short telomeres. By comparing their senescence and tumorigenesis behavior in cell culture, we can determine the potential involvement of the mTert gene in genotypically mTert+/+ fibroblasts during replicative senescence. If mTert is completely silenced or carries no function during presenescence and senescence stages (18, 29), we expect to observe no differences between the mTert−/− and mTert+/+ cells. However, the results of this experiment demonstrate that mTert−/− cells approached cellular senescence earlier and exhibited a significantly higher rate of malignant transformation than mTert+/+ cells. Furthermore, an evident up-regulation of TERT expression was detected in mTert+/+ cells at the presenescence stage. Moreover, removal or down-regulation of TERT expression via CRISPR/Cas9 or short-hairpin RNA (shRNA) in wild-type mouse and human cells recapitulated mTert−/− phenotypes, and overexpression of TERT in mTert−/− cells rescued the phenotypes. Taking these data together, this study suggests that TERT has a previously underappreciated, protective role in buffering senescence stresses due to short, dysfunctional telomeres, thereby preventing malignant transformation.
Result
Fibroblasts from Tert+/+ and Tert−/− Offspring of Late-Generation Tert+/− Breeding Carry Extremely Short Telomeres in Comparison to Normal Control B6 Fibroblasts.
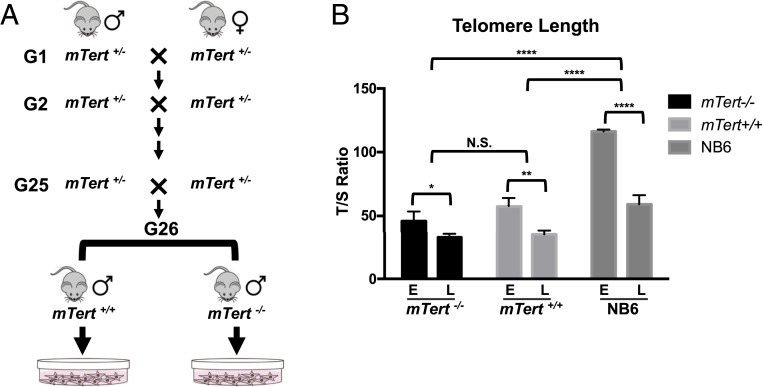
Mice heterozygous for the mTert deletion (mTert+/−) were bred for 25 consecutive generations, with progressive telomere shortening occurring this time, as previously reported (28). Skin fibroblasts were isolated from ear punches of mTert+/+ and mTert−/− juvenile offspring of the 26th intercross and cultured in vitro for 8 mo (Fig. 1A). Fibroblasts from age- and gender-matched normal B6 (NB6) mice were used as controls (30). During the culturing process, cell pellets were collected, and TL was determined by the multiplexed qPCR and telomere Q-FISH analyses (23, 28, 31). Based on the relative cell proliferation rate (SI Appendix, Fig. S1A) and p16 expression levels (SI Appendix, Fig. S1B) in mTert+/+ and mTert−/− cells, the in vitro cell-culturing process was grouped into 4 distinct stages: Early stage (E), middle stage (M), late stage (L), and final stage (F) (SI Appendix, Fig. S1C). Each stage lasted at least 10 passages.
Fig. 1.
mTert+/+ and mTert−/− mouse skin fibroblasts have extremely short telomeres in comparison to NB6 control fibroblasts. (A) Tert-heterozygous mice (mTert+/−) were intercrossed for 26 generations. Primary skin fibroblasts were cultured from ear punches from mTert+/+ and mTert−/− progenies of the 26th intercross. (B) Telomere length at E and L stages measured by the multiplexed qPCR analysis. Relative telomere length was shown as the T/S ratio. mTert+/+ and mTert−/− fibroblast cells showed significantly shorter telomere length than NB6 at the E stage. There was a slight decrease in mTert+/+ and mTert−/− TL from E to L. NB6 telomeres dramatically shortened from E to L stages. T/S ratio: The ratio of telomere repeat copy number to single-copy gene copy number. Error bars represent SD between 3 independent biological repeats (n = 3). Statistical significance was determined using a 2-way ANOVA with Bonferroni’s posttest. N.S., not significant; *P < 0.05; **P < 0.01; ****P < 0.0001.
In comparison to NB6, both mTert+/+ and mTert−/− showed significantly shorter TL at the beginning of cell culture (Fig. 1B and SI Appendix, Fig. S2 A–C). Although mTert+/+ has the wild-type genotype, its telomeres were at a similar length as those in mTert−/− cells and significantly shorter than NB6 due to the haploinsufficient telomerase activity in their ancestors (28). During the cell culture, the TL in mTert+/+ and mTert−/− only decreased slightly by 10 to 20% (Fig. 1B and SI Appendix, Fig. S2 D and E). In contrast, NB6 showed a drastic reduction in TL during the same process (SI Appendix, Fig. S2F). Its TL was shortened by almost 50% from E to L stage and became comparable with the E-stage TL in mTert+/+ and mTert−/− cells (Fig. 1B).
mTert−/− Fibroblasts Prematurely Commit to Replicative Senescence and Cancer Transformation at the M Stage.
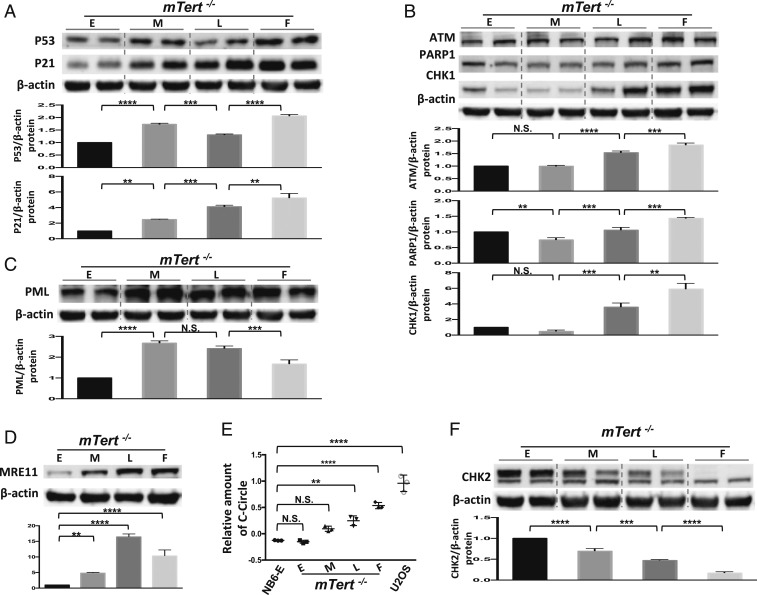
To monitor the cell senescence and transformation behavior of mTert−/− and mTert+/+ cells, we analyzed 3 groups of molecular biomarkers, including tumor suppressors/senescence biomarkers, DNA damage response (DDR) factors, and checkpoint proteins (32–35). In mTert−/− cells, the levels of the senescence biomarkers p53 and p21 were increased sharply at the M stage (Fig. 2A), in response to critically shortened telomeres at the same stage (SI Appendix, Fig. S2D). DDR factors ataxia telangiectasia mutated (ATM) and poly(ADP-ribose) polymerase (PARP)1 and checkpoint protein CHK1 are typically triggered when senescence biomarkers P53/p21 become activated (36–43). Unexpectedly, in mTert−/− cells, we observed a significant delay for over 10 passages in the up-regulation of the expression of ATM, PARP1, and CHK1; their expressions were kept at the basal level at the M stage and became up-regulated at the L stage (Fig. 2B). In contrast, ALT cancer biomarkers promyelocytic leukemia (PML) and MRE11, which are generally at basal expression in normal somatic cells (44–47), were drastically increased at the M stage, suggesting that some mTert−/− cells have adopted a telomerase-independent mechanism to immortalize and transform at the M stage (Fig. 2 C and D). This ALT-mediated mechanism was further confirmed by the detection of significant up-regulation of the C-circle, the only known ALT-specific molecule (Fig. 2E) (48). Consistently, at the M stage, mTert−/− cells showed a gradual loss of the checkpoint protein CHK2, a dominant tumor suppressor at the DNA damage checkpoint (49–51), further supporting the notion that some mTert−/− cells have surpassed replicative senescence and initiated transformation at the M stage (Fig. 2F).
Fig. 2.
mTert−/− fibroblasts prematurely committed to replicative senescence and transformation at the M stage. (A) Western blotting analysis to detect the protein level of p53 and p21 in mTert−/− cells. Normalized relative p53 and p21 protein levels from E to F stages are shown in the lower graph. Two technical repeats are shown at each stage and quantified by the mean value. Error bars represent the SD of 3 biological repeats (n = 3). (B) Western blotting analysis to detect the protein level for ATM, PARP1, and CHK1 in mTert−/− cells. Normalized relative ATM, PARP1, and CHK1 protein levels from E to F stages are shown in the lower graph. Two technical repeats are shown at each stage and quantified by the mean value. Error bars represent the SD of 3 biological repeats (n = 3). (C) Western blotting analysis to detect the protein level of PML in mTert−/− cells. Normalized relative PML protein levels from E to F stages are shown in the lower graph. Two technical repeats are shown at each stage and quantified by the mean value. Error bars represent the SD of 3 biological repeats (n = 3). (D) Western blotting analysis to detect the protein level of MRE11 in mTert−/− cells from E to F stage. Normalized relative MRE11 protein levels from E to F stages are shown in the lower graph. Error bars represent the SD of 3 biological repeats (n = 3). (E) The C-circle assay was conducted to determine ALT transformation in mTert−/− cells. The relative C-circle level was detected by qPCR in mTert−/− cells from E to F stage. NB6-E is used as a negative control, and U2OS as a positive control. Error bars represent the SD of 3 biological repeats (n = 3). (F) Western blotting analysis to detect the protein level for CHK2 in mTert−/− cells. Normalized relative CHK2 protein levels from E to F stages are shown in the lower graph. Two technical repeats are shown at each stage and quantified by the mean value. Error bars represent the SD of 3 biological repeats (n = 3). Statistical significance in A to F was determined using a 1-way ANOVA with Tukey’s posttest. N.S., not significant; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Taken together, these results suggested that once mTert−/− cells reached critically shortened TL at the M stage, some quickly transformed into ALT cancer cells. This process is accompanied by the loss of CHK2 that allows cells to escape senescence arrest. Moreover, the delayed up-regulation of DDR factors (especially PARP1) until the L stage implies activation of ALT-nonhomologous end-joining repair in mTert−/− cells, which may further induce genome instability and tumorigenesis in the absence of CHK2.
mTert+/+ Fibroblasts Showed Delayed Replicative Senescence and Reduced Transformation in Comparison with mTert−/− Cells.
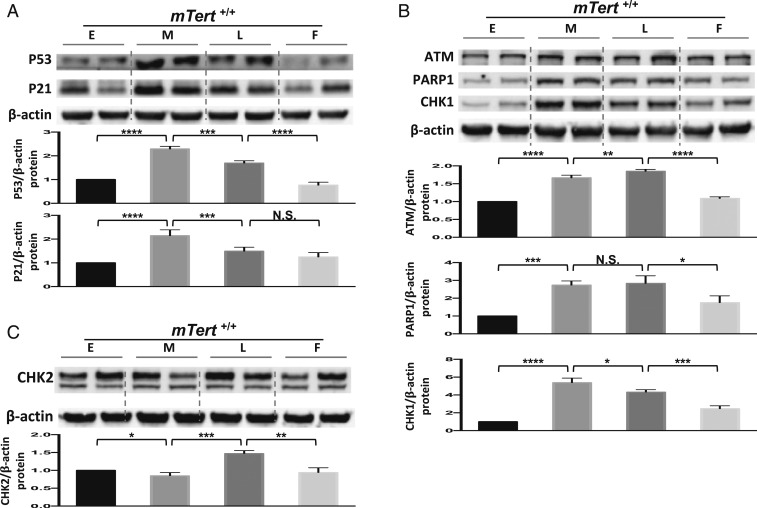
Similarly, we monitored the behavior of mTert+/+ cells using the 3 groups of molecular biomarkers. At the M stage, consistent with mTert−/− cells, senescence markers p53/p21 were activated in mTert+/+ cells (Fig. 3A), due to their critically short telomeres (SI Appendix, Fig. S2E). However, unlike mTert−/− cells, the activation of the p53/p21 pathway in mTert+/+ cells immediately triggered the DDR response and up-regulated the expression of ATM, PARP1, and CHK1 at the M stage, and their elevated expressions were maintained through the L stage (Fig. 3B). In addition, CHK2 levels were relatively stable (Fig. 3C), suggesting that the vast majority of mTert+/+ cells did not transform at the M stage. In support of this notion, SASP marker IL6 was maintained at the basal level at the M stage and became activated at the L stage (SI Appendix, Fig. S3A) (16). Side-by-side Western blotting analysis confirmed that DDR factors ATM, PARP1, and CHK1 and CHK2 were much lower in mTert−/− cells than in mTert+/+ cells at the M stage (SI Appendix, Fig. S3B). Furthermore, when M stage mTert+/+ cells were treated with ATM or PARP1 inhibitors for 72 h, senescence rapidly occurred, as indicated by a significant up-regulation of p16 (SI Appendix, Fig. S3 C and D).
Fig. 3.
mTert+/+ fibroblasts showed delayed replicative senescence and reduced transformation in comparison with mTert−/− cells. (A) Western blotting analysis to detect the protein level of p53 and p21 in mTert+/+ cells. Normalized relative p53 and p21 protein levels from E to F stages are shown in the lower graph. Two technical repeats are shown at each stage and quantified by the mean value. Error bars represent the SD of 3 biological repeats (n = 3). (B) Western blotting analysis to detect the protein level of ATM, PARP1, and CHK1 in mTert+/+ cells. Normalized relative ATM, PARP1, and CHK1 protein levels from E to F stages are shown in the lower graph. Two technical repeats are shown at each stage and quantified by the mean value. Error bars represent the SD of 3 biological repeats (n = 3). (C) Western blotting analysis to detect the protein level for CHK2 in mTert+/+ cells. Normalized relative CHK2 protein levels from E to F stages are shown in the lower graph. Two technical repeats are shown at each stage and quantified by the mean value. Error bars represent the SD of 3 biological repeats (n = 3). Statistical significance in A to C was determined using a 1-way ANOVA with Tukey’s posttest. N.S., not significant; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Interestingly, this observation indicates that mTert+/+ cells behaved differently from mTert−/− cells in response to short telomere-induced DNA damage signals. Both cells can detect telomere-induced DNA damage efficiently by activating the p53/p21 pathway. mTert−/− cells committed to replicative senescence immediately upon critically short telomeres and a detectable fraction of mTert−/− cells transformed subsequently. In contrast, mTert+/+ cells were able to mediate the telomere stress by up-regulating DDR factors and postponing replicative senescence and transformation.
Cell Cycle Analysis Supported Premature Senescence and Transformation in mTert−/− Cells.
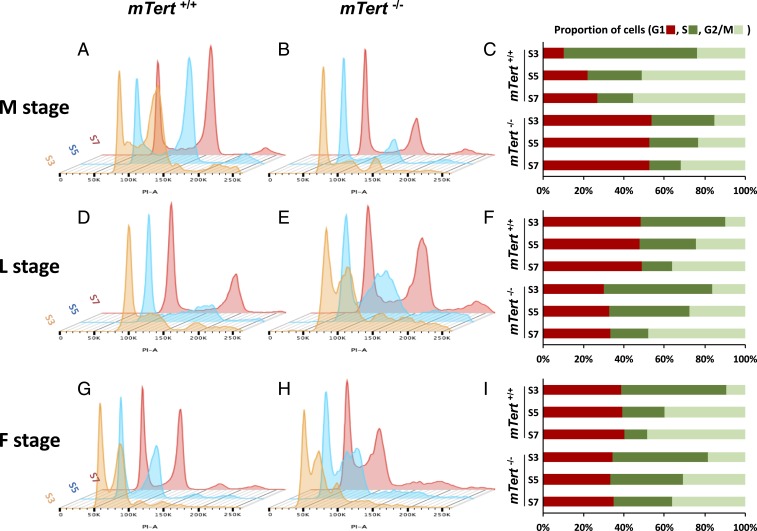
To further elucidate the difference between mTert+/+ and mTert−/− cells in handling telomere stress, we performed cell cycle analysis at the M, L, and F stages. mTert+/+ and mTert−/− cells at different stages were blocked at the entrance of S phase by aphidicolin treatment for 24 h, then released by changing into the fresh medium. We then harvested cells after release at different time intervals (S3, 3 h; S5, 5 h; S7, 7 h). The harvested cells were stained with propidium iodide (PI) to reveal DNA content and sorted with flow cytometry. We expect that the cells in active cell cycle will be arrested with aphidicolin at the G1/S junction, which will be reflected by accumulation at S3, and move gradually to G2/M and G1 at the S5 and S7 time points.
Indeed, in M-stage mTert+/+ cells, we found 66% cells at the S phase, 24% cells at the G2/M phase, and 10% at the G1 phase at S3 (Fig. 4 A and C). At S5 and S7, a decrease in S phase population and a corresponding increase in G2/M was observed; G2/M phase cells accumulated from 24% in S3 to 55% in S7, and G1 phase cells were also increased from 10% in S3 to 27% in S7. Taking these data together, this experiment suggests that at the M stage, the majority of mTert+/+ cells were in the active cell cycle and responded to the drug arrest, despite the presence of short telomeres and activated p53/p21. In contrast, in the M-stage mTert−/− cells, only about 31% cells were in S phase at S3, and more than 50% of mTert−/− cells remained in G1 during S3, S5, and S7 (Fig. 4 B and C). Thus, at any time points during the 7-h release, only about less than 50% of mTert−/− cells were in the cell cycle, which could also include the population of transformed ALT cancer cells. Thus, this cell cycle study supported the molecular biomarker analysis (Figs. 2 and 3) and indicated that significantly more mTert−/− cells were at replicative senescence arrest than mTert+/+ cells at the M stage.
Fig. 4.
Cell cycle analysis supported delayed senescence and transformation in mTert+/+ cells. (A, B, D, E, G, and H) Cell cycle analysis in mTert+/+ and mTert−/− cells from M to F stage. The released cells from 12-h aphidicolin treatment were harvested at 3 h (S3), 5 h (S5), and 7 h (S7) time points. The x axis and y axis represent cell number and DNA content, respectively. (C, F, and I) The relative percentage of cells at G1, S, and G2/M phase of the cell cycle at given time points in mTert+/+ and mTert−/− cells.
Next, we conducted the same analysis for L-stage mTert+/+ and mTert−/− cells. Differing from the results at the M stage, nearly half of mTert+/+ cells stayed in G1 phase at every time point (S3, S5, and S7) (Fig. 4 D and F), suggesting cell cycle arrest and replicative senescence. In contrast, the majority of mTert−/− cells (over 65%), which likely had escaped replicative arrest at this point, reentered the “normal” cell cycle (Fig. 4 E and F). At the F stage, both mTert+/+ and mTert−/− cells showed a similar pattern as the L-stage mTert −/− cells (Fig. 4 G–I).
Taking these data together, the cell cycle analysis supported that mTert−/− cells showed a premature onset of replicative senescence and transformation, suggesting that the presence of the mTert gene alters the DDR and checkpoint responses to short telomere-induced DNA damages.
mTert−/− Cells Showed a Higher Transformation Rate than mTert+/+ Cells.
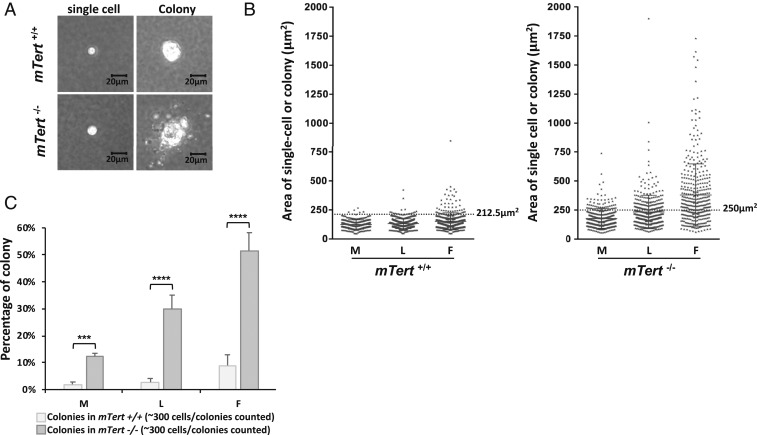
To study the rates of transformation, we conducted the soft agar colony formation assay (52). After 4-wk growth in soft agar, only immortalized cells with the capability of proliferation can form large cell aggregates (Fig. 5A). To define the immortalized cells, we measure the area of a normal single cell in soft agar for mTert+/+ and mTert−/− at the E stage (SI Appendix, Fig. S4A). Based on these data, an mTert+/+ colony is defined as an aggregate with an area over 212.5 μm2 and an mTert−/− colony with an area over 250 μm2 (SI Appendix, Fig. S4A). Here, we applied a more stringent criterion for mTert−/− colony definition due to the slightly larger cell size of mTert−/− cells.
Fig. 5.
mTert−/− cells showed a higher transformation rate than mTert+/+ cells. (A) Cell suspension containing mTert+/+ and mTert−/− cells were cultured in soft agar medium for 30 d. Representative single-cell and colony images of mTert+/+ and mTert−/− samples are shown; 3 biological experiments (n = 3). (B) Quantification of the areas of cell/colony size in mTert+/+ and mTert−/− cells. n > 300 for each sample; t3 biological experiments (n = 3). Statistical significance was determined using a 1-way ANOVA with Tukey’s posttest. ***P < 0.001, ****P < 0.0001. (C) Comparison of percentage of colonies identified in B in mTert+/+ and mTert−/− samples. Error bars represent the SD among 3 biological experiments (n = 3). Statistical significance was determined using a 2-way ANOVA with Bonferroni’s posttest. ***P < 0.001, ****P < 0.0001.
Next, at the beginning of M, L, and F stages, we seeded the cells in soft agar and measured the area of every cell aggregate in mTert+/+ and mTert−/− samples 4 wk after seeding (Fig. 5B). Consistent with previous observations (Figs. 2–4), the soft agar assay revealed that mTert+/+ cells had few, sporadic transformation events during the M and L stages, and the transformation rate significantly increased to about 10% at the F stage (Fig. 5C). In contrast, the transformation rate of mTert−/− cells was much higher: More than 10% at the M stage and 30% at the L stage. At the F stage, more than half of the mTert−/− cells showed the transformation capability (Fig. 5C). Moreover, when the size of the colonies is compared, mTert−/− showed significantly larger colonies than mTert+/+, implying their more aggressive or invasive characteristics (Fig. 5B). This experiment further implies that the presence of the mTert gene protects mTert+/+ cells from tumorigenesis.
mTert+/+ Cells Up-Regulated TERT Expression at the M Stage.
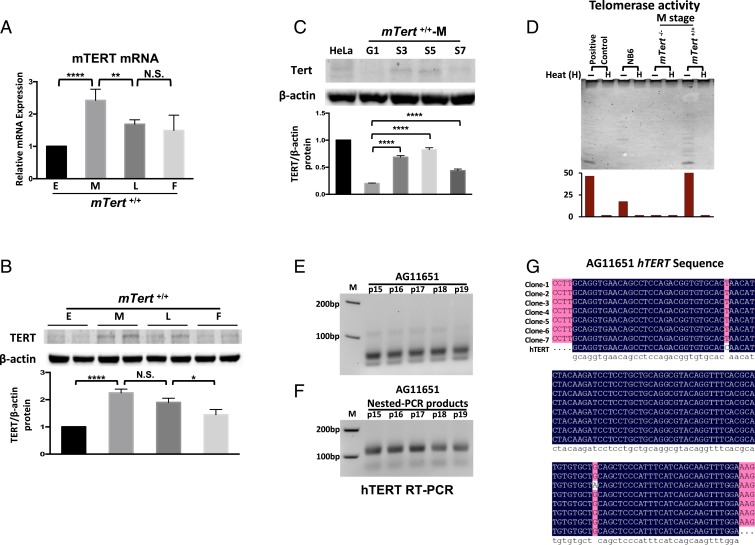
Thus far, the results suggest that mTert+/+ cells have a delayed onset of replicative senescence and a reduced rate of transformation. To understand how the presence of the mTert gene affects those responses in mTert+/+ cells, particularly at the M stage, we determined the expression of TERT by qRT-PCR in mTert+/+ cells. mTert−/− was included as a negative control. Interestingly, up-regulation of mTert mRNA was observed around the M stage when shortened telomeres trigger DDR responses in mTert+/+ cells (Fig. 6A), and NB6 showed a lower, basal level of mTert mRNA expression relative to the E stage mTert+/+ (SI Appendix, Fig. S4B). Consistent with the genotype, mTert−/− cells showed no mTert mRNA expression (Fig. 6A). Furthermore, Western blotting revealed that TERT protein exhibited a weak signal at the E stage, was elevated at the M stage, and became reduced during the L and F stages (Fig. 6B).
Fig. 6.
Detection of telomerase expression in mTert +/+ and human primary fibroblast cells. (A) qRT-PCR analysis reflecting the significantly up-regulated expression of mTERT mRNA in mTert+/+ cells from the M to L stage. Consistent with the genotype, mTert−/− fibroblasts showed no mTERT expression. Two technical repeats are shown at each stage and quantified by the mean value. Error bars represent the SD of 3 biological repeats (n = 3). (B) Western blotting analysis to detect the protein level for mTERT in mTert+/+ cells. Normalized relative mTERT protein levels from E to F stage are shown in the lower graph. Two technical repeats are shown at each stage and quantified by the mean value. Error bars represent the SD of 3 biological repeats (n = 3). (C) Western blotting analysis to detect the protein level for TERT at G1, S3, S5, and S7 time points in mTert+/+ cells during the M stage. HeLa, a telomerase-dependent cancer line, was included as a positive control. Normalized relative TERT levels at M stage are shown in the lower graph. Error bars represent the SD among 3 independent experiments (n = 3). Statistical significance in A tIo C was determined using a 1-way ANOVA with Tukey’s posttest. N.S., not significant; *P < 0.05; **P < 0.01; ****P < 0.0001. (D) Estimation of telomerase activity by TRAP. Representative profiles of TRAP assay performed using cell lysates from NB6, mTert+/+ and mTert−/− cells at the M stage. Each lysate was tested for unheated (-) activation and heated (H) inactivation. (E) Human TERT mRNA expression was detected by RT-PCR analysis in a human primary fibroblast line AG11651 from passage 15 (P15) to passage 19 (P19). PCR products amplified by hTERT-F/R primers were loaded 50 μL and separated by 1.5% agarose gel. The size of the amplified fragments was 113 bp. (F) Nested-PCR was conducted to enhance the signals of hTert; 200 μL of hTERT-F/R PCR products was separated by 1.5% agarose gel, purified, and used as the template for PCR with hTERT-nested-F/R primer. The nested-PCR products were loaded 10 μL and detected by 1.5% agarose gel. The size of the amplified fragments was 106 bp. (G) The sequencing alignment analysis for hTERT-nested PCR products. The nested-PCR products of AG11651-P19 were cloned into TA vector and transformed into the DH5α competent cells. The plasmids were extracted from clone-1 to clone-7 to sequence them. The alignment between hTERT–Exon11-13 and clone sequences showed that the nested-PCR products were the hTERT sequence.
It is known that telomerase activity and cell cycle progression are linked, and telomerase is preferentially up-regulated and recruited to the telomeres at the middle to late S phase (53). Thus, we synchronized the M stage mTert+/+ cells with aphidicolin and released the cells at S3, S5, and S7 and analyzed telomerase protein expression. Indeed, Western blotting revealed an evident, cell cycle-dependent up-regulation of TERT protein at S3 and S5 (Fig. 6C), suggesting TERT is up-regulated at the M stage when the critically short telomeres trigger the senescence and DDR responses (Fig. 3A). To determine the telomerase activity, we applied the TRAP assay (54) in NB6, mTert−/−, and mTert+/+ cells at the M stage, together with a positive control provided by the kit. Consistent with the genotype, mTert−/− cells were devoid of telomerase activity. mTert+/+ cells showed a robust telomerase activity, similar to the level of the positive control. NB6 also showed a basal level activity (Fig. 6D). While much less abundant in comparison to that in mouse cells, hTERT mRNA was also detected in primary human fibroblast cells at the presenescence stage, using nested-PCR (Fig. 6 E–G). Together, these data suggested that wild-type cells induced hTERT expression and activity at the presenescence, which may provide a protective role in buffering senescence stress and reducing transformation.
CRISPR/Cas9 and shRNA of mTert Confirmed the Protective Role of Telomerase in Mediating Telomere Stress in mTert+/+ Cells.
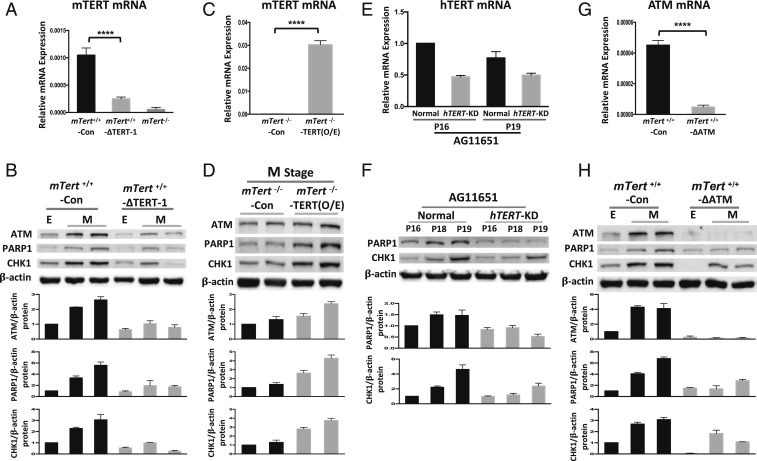
To determine directly whether expression of TERT is a primary cause of the difference in response to short telomere stress between mTert+/+ and mTert−/− cells, we removed the mTert gene in mTert+/+ cells using CRISPR/Cas9 technology with an mTert-specific single-guide RNA (sgRNA) at the E stage. After puromycin selection, we identified 2 single-cell clones, mTert+/+–ΔTERT-1 and mTert+/+–ΔTERT-2, which showed internal deletions in both mTert alleles and a subsequent premature translational termination (SI Appendix, Figs. S5 and S6 A and B). A control sgRNA was used to generate an mTert+/+-Con cell line that underwent the same experimental procedure as mTert+/+-ΔTERT. We found that the mTert mRNA was significantly decreased in mTert+/+-ΔTERT, compared with mTert+/+-Con (Fig. 7A). Moreover, similar to mTert−/− cells, mTert+/+-ΔTERT showed reduced expression of DDR factors (ATM, PARP1, and CHK1) at the presenescence stage (Fig. 7B and SI Appendix, Fig. S6C).
Fig. 7.
CRISPR/Cas9 knockout/knockdown and overexpression (O/E) of TERT confirmed the protective role of the TERT expression in mediating telomere stress. (A) CRISPR/Cas9/sgRNA lentivirus was introduced in mTert+/+ cells. The single-cell cloning lines of mTert+/+-Con, mTert+/+-ΔTERT, and mTert−/− were used for qRT-PCR analysis to check mTERT mRNA expression. mTert+/+-Con: Control sgRNA introduced into mTert+/+ cells; mTert+/+-ΔTERT: TERT sgRNA introduced into mTert +/+ cells. Error bars represent the SD among 3 independent experiments (n = 3). Statistical significance was determined using a 1-way ANOVA with Tukey’s posttest. ****P < 0.0001. (B) Western blotting analysis to detect the protein level for DDR factors (ATM, PARP1, and CHK1) in mTert+/+-Con and mTert+/+-ΔTERT at E and M stages. Normalized relative ATM, PARP1, and CHK1 levels from E to M stages are shown in the lower graph. Error bars represent the SD among 3 independent experiments (n = 3). (C) Overexpression of mTERT by retroviruses in mTert−/− cells at the M stage. The expression of mTERT mRNA in mTert−/−-Con and mTert−/−-TERT(O/E) was examined by qRT-PCR. mTert−/−-Con: Control retrovirus; mTert−/−-TERT(O/E): The retrovirus carrying the mTert gene. Error bars represent the SD among 3 independent experiments (n = 3). Statistical significance was determined using a 2-tailed t test. ****P < 0.0001. (D) Western blotting analysis to detect the protein level of DDR factors (ATM, PARP1, and CHK1) in mTert−/−-Con and mTert−/−-TERT(O/E) at the M stage. Normalized relative ATM, PARP1, and CHK1 levels from E to F stages are shown in the lower graph. Error bars represent the SD among 3 independent experiments (n = 3). (E) Nested qRT-PCR analysis showing the down-regulated expression of hTERT mRNA in hTERT-KD cells in comparison with normal cells. CRISPR/Cas9/sgRNA lentivirus was introduced in AG11651. hTERT mRNA was detected by nested qRT-PCR in the puromycin-selected cell population of normal and hTERT-KD at passage 16 (P16) and passage 19 (P19). Normal: Control sgRNA (Ctr2) introduced into AG11651; hTERT-KD: hTERT-Ex2 sgRNA introduced into AG11651. (F) Western blotting analysis to directly compare the protein level of PARP1, and CHK1 between normal and hTERT-KD. Normalized relative PARP1and CHK1 levels are shown in the lower graph. Error bars represent the SD between 3 independent experiments (n = 3). (G) CRISPR/Cas9/sgRNA lentivirus was introduced in mTert+/+ cells. The single-cell cloning population from mTert+/+-Con and mTert+/+-ΔATM were selected for qRT-PCR analysis to check ATM mRNA expression. mTert+/+-Con: Control sgRNA introduced into mTert+/+ cells; mTert+/+-ΔATM: Mouse ATM sgRNA introduced into mTert+/+ cells. Error bars represent the SD among 3 independent experiments (n = 3). Statistical significance was determined using a 2-tailed t test. ****P < 0.0001. (H) Western blotting analysis to detect the protein level for DDRs (ATM, PARP1, and CHK1) in mTert+/+-Con and mTert+/+-ΔATM at the E and M stages. Normalized relative ATM, PARP1, and CHK1 levels from E to F stages are shown in the lower graph. Error bars represent the SD among 3 independent experiments (n = 3).
We also knocked down TERT expression by an shRNA method. After mTERT-shRNA retrovirus transfection, mTERT-shRNA reduced mTert mRNA (SI Appendix, Fig. S7A). We then performed single-colony selection and found that the N4 colony showed significantly down-regulated mTert mRNA in comparison to control shRNA (N2) (SI Appendix, Fig. S7B). Again, consistent with the CRISPR study (Fig. 7 A and B), the expression of ATM, PARP1, and CHK1 were dramatically reduced at the M stage in N4 cells (SI Appendix, Fig. S7C). Furthermore, overexpression of mTert in the M-stage mTert−/− cells with retroviruses rescued the DDR phenotype, leading to an increase in the expression of ATM, PARP1, and CHK1(Fig. 7 C and D).
Significantly, in primary human fibroblasts, when hTERT gene is knockdown via CRSPR/Cas9 (Fig. 7E and SI Appendix, Fig. S7D), we observed a similar down-regulation of PARP1 and CHK1 as in mTert+/+-ΔTERTcells (Fig. 7F). We also found a similar up-regulation of PML and p16 as in mTert−/− cells (SI Appendix, Fig. S7 E and F, respectively), suggesting that human cells might use the same mechanism and need the function of TERT at the presenescence stage.
It is known that ATM is required to assemble at the telomeres to recruit telomerase in telomerase+ cells (55, 56). Here, we found that in the absence of telomerase, the ATM-mediated DDR pathway in response to dysfunctional telomere is also impaired. We further tested the involvement of ATM as a downstream effector of TERT induction at presenescence. We knocked out both alleles of the ATM gene with the CRISPR/Cas9 method (SI Appendix, Fig. S8 A–C) and conducted single-cell colony selection. In the representative colony mTert+/+-ΔATM, the level of ATM mRNA and protein appeared significantly lower than mTert+/+-Con (Fig. 7 G and H). Correspondingly, PARP1 could not be promptly activated at the M stage in mTert+/+-ΔATM cells (Fig. 7H). Similar to the results in SI Appendix, Fig. S3C using ATM inhibitor, premature senescence is observed in in mTert+/+-ΔATM cells (SI Appendix, Fig. S8D).
Discussion
Replicative Senescence Response in Murine and Human Cells.
Human somatic cells that can be grown in culture undergo cellular senescence in vitro. In most cases, telomerase activity is missing and its reactivation may result in indefinite cell proliferation (7, 12, 57–59). In contrast, the view of telomere shortening-mediated senescence response in mouse cells has been evolving. In the first study to describe the removal of telomerase in mice, Blasco et al. (20) demonstrated that mTR−/− fibroblasts remained capable of bypassing crisis and of exhibiting cellular transformation in vivo. These results led to the notion that murine cells do not exhibit the same senescence response to telomere dysfunction as human cells. Along this line, Smogorewska and de Lange (32) showed a fundamental difference in telomere damage signaling in human and mouse cells, mostly involving the p16/RB response to telomere dysfunction. Interestingly, more recent work reported that mouse telomeres shorten ∼100 times faster than human telomeres, and demonstrated that short telomeres have a direct and similar impact on longevity in both human and mouse cells (60). Here, we report that telomerase is transiently up-regulated at the presenescence stage in both murine and human cells (Fig. 6), and the cells that are unable to induce telomerase expression approach senescence earlier and exhibit a significantly higher rate of transformation (Figs. 2, 5, and 7). In light of the previous findings, this study suggests that, at the cellular level, there may be more commonalities between murine and human senescence than previously appreciated.
The Basal Level of Telomerase Expression in Mouse and Human Cells.
In mouse fibroblast cells, there is a low but detectable basal expression of mTERT (Fig. 6 and SI Appendix, Fig. S4B). In SI Appendix, Fig. S4B we directly compared the mRNA levels of mTERT in NB6 with that in the E-stage mTert+/+ cells. This experiment indicated that in mouse wild-type primary fibroblast NB6, basal expression of telomerase is detectable. Consistently, in Fig. 6D a weak telomerase activity was detected in NB6 cells. However, this basal level of telomerase appears not able to prevent telomere from shortening, as shown in Fig. 1B and SI Appendix, Fig. S2 E and F, and progressive telomere erosion occurred in both NB6 and the E-stage mTert+/+ cells. Moreover, a comparable rate of TL reduction is observed in mTert−/− and mTert+/+ cells at the E stage, further supporting that the basal dosage of TERT is not sufficient to prevent telomere shortening.
There have been precedents for insufficient expression of telomerase in primary human cells/tissues (5, 61). For example, human T lymphocytes express telomerase upon activation but its expression is insufficient to grant immortality, resulting in a limited replicative life span of T cells (61). Interestingly, blockage of endogenous hTERT expression by ectopic expression of dominant-negative hTERT led to precocious senescence and cytogenetic abnormalities in T lymphocytes. These results are in agreement with our findings and suggest that while transient and limited expression of TERT cannot prevent overall telomere shortening, it has a major influence on cell longevity (61).
The Acute Up-Regulation of Telomerase at the Presenescence Stage.
Besides the basal level of TERT expression, in mTert+/+ cells, we report an acute up-regulation of TERT at the M stage followed by significant down-regulation at the L stage (Fig. 6 A and B). The quick down-regulation of mTERT at the L stage implies that the induction of TERT expression at the M stage is a transient event, not a result of the gradual expansion of transformed cells. Otherwise, we would expect a progressive increase in TERT at the L stage. Furthermore, IL-6, the major biomarker of SASP, was absent at the M stage and drastically activated at the L stage in mTert+/+ cells, supporting the induction of telomerase indeed happened at presenescence.
When this up-regulation occurred in mTert+/+ cells, concomitantly, mTert−/− cells started behaving differently in terms of delayed DDR, premature senescence, and increased transformation (Fig. 2). Moreover, when TERT is knockdown in human fibroblast cells using CRISPR/Cas9, an increase in senescence and transformation markers, p16 and PML respectively, was observed at the presenescence stage (SI Appendix, Fig. S7 D–F). Together, these results support that the transient induction of telomerase at presenescence is of importance in cellular senescence and transformation.
The Protective Role of Telomerase in Cellular Senescence.
A number of previous studies have explored the transient and limited expression of telomerase in mammalian systems. In 2000, Steinert et al. (62) showed that the transient induction of hTERT in normal human BJ cells led to a significant extension in the subsequent number of replicative passages prior to senescence. In an mTert+/− murine model, despite haploinsufficiency for maintaining long telomeres, the heterozygous progenies were protected from germline exhaustion and bone marrow failure compared to their homozygous mTert−/− littermates (63, 64). These results have led to the speculations that low levels of telomerase may offset the effects of tissue aging in vivo. Indeed, adeno-associated viral vectors carrying the telomerase gene (AAV9-Tert) has been shown promising efficacy in mouse models of cardiac infarct, aplastic anemia, and pulmonary fibrosis (65–67).
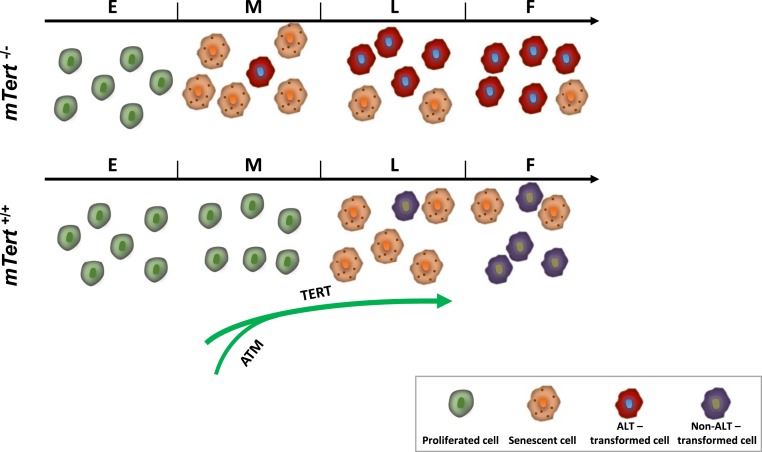
In this study, we demonstrate an acute up-regulation of TERT expression at the presenescence stage in Tert+/+ cells. In comparison with their littermate Tert−/−cells, it is clear that TERT expression is required in properly activating the short-telomere–induced DDR pathway, buffering short telomere stress and reducing transformation (Fig. 8). Furthermore, in Q-FISH analysis (SI Appendix, Fig. S2), about 3 to 4% of telomere signal-free ends were detected in mTert+/+ and mTert−/− cells at the E stage. Previous works have shown unequivocally that is the presence of critically short telomeres that dictate the cellular phenotypes in telomerase-deficient models (23, 63, 68). We speculate that the transiently up-regulated TERT in mTert+/+ cells may function to extend the telomere signal-free ends and direct proper DDR on those ends. Future work on how TERT expression is triggered, where the induced TERT locates relative to the telomere signal-free ends, and DDR pathway factors will be required to fully appreciate the role of telomerase in cellular senescence.
Fig. 8.
A working model. The results from this study support a model that in comparison to mTert+/+ cells, mTert−/− cells approached cellular senescence earlier and exhibited a significantly higher rate of transformation. This phenotype is tightly associated with the expression and activity of telomerase, possibly through a TERT-mediated activation of ATM.
Materials and Methods
Mouse.
Mice were bred and maintained in our animal facility in accordance with US National Institutes of Health guidelines. All animal experiments were approved by the National Cancer Institute and Animal Care and Use Committees and carried out according to the National Institutes of Health Guide for Care and Use of Laboratory Animals (69).
Cell Lines and Cell Culture.
Fig. 1A shows the strategy used in the breeding of Tert+/− mice. Ear skin explants (1- to ∼2-mm2 pieces) from control normal C57 BL/6 (NB6) or from the 26th generation of juvenile Tert+/− crossed mice were excised, placed in a 100-mm dish, and cultured in DMEM (#12-614Q; Lonza) supplemented with 2 mM glutamine (#25-005-CI; Corning) and 10% FBS (#97068-085; VWR) at 37 °C in a humidified atmosphere of 5% CO2. The normal human skin fibroblast (AG11651) was bought from the Coriell Institute and cultured as the same condition as mouse fibroblast. When fibroblasts had reached confluence in the dish, the explant was removed, and the cells were trypsinized and passaged. For passaging, cells were split at a ratio of 1:4 at 90% to ∼5% confluency.
Statistical Analysis.
Results are presented as mean ± SD. Data were analyzed by 1- or 2-way ANOVA followed by Bonferroni’s/Tukey’s post hoc test, as well as a 2-tailed Student t test. All data and statistical analysis were performed by GraphPad Prism 7 or Excel software. P < 0.05 was considered significant. Asterisks indicate statistical difference as follows: N.S., not significant; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Supplementary Material
Acknowledgments
We thank members in the F.S.C., R.J.H., and and K.C. laboratories for helpful discussion; and Ken Class and Amy Beaven (flow cytometry and imaging cores at the University of Maryland College Park) for technical support. This work was supported by NIH Grant R01HL126784 (to K.C.) and the NIH intramural research fund (to F.S.C. and R.J.H.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1907199116/-/DCSupplemental.
References
- 1.Greider C. W., Blackburn E. H., Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 43, 405–413 (1985). [DOI] [PubMed] [Google Scholar]
- 2.Blackburn E. H., et al. , Recognition and elongation of telomeres by telomerase. Genome 31, 553–560 (1989). [DOI] [PubMed] [Google Scholar]
- 3.Masutomi K., et al. , Telomerase maintains telomere structure in normal human cells. Cell 114, 241–253 (2003). [DOI] [PubMed] [Google Scholar]
- 4.Greider C. W., Blackburn E. H., A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature 337, 331–337 (1989). [DOI] [PubMed] [Google Scholar]
- 5.Wright W. E., Piatyszek M. A., Rainey W. E., Byrd W., Shay J. W., Telomerase activity in human germline and embryonic tissues and cells. Dev. Genet. 18, 173–179 (1996). [DOI] [PubMed] [Google Scholar]
- 6.Hoffmeyer K., et al. , Wnt/β-catenin signaling regulates telomerase in stem cells and cancer cells. Science 336, 1549–1554 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Lin S.-Y., Elledge S. J., Multiple tumor suppressor pathways negatively regulate telomerase. Cell 113, 881–889 (2003). [DOI] [PubMed] [Google Scholar]
- 8.Yao Y., Bellon M., Shelton S. N., Nicot C., Tumor suppressors p53, p63TAα, p63TAy, p73α, and p73β use distinct pathways to repress telomerase expression. J. Biol. Chem. 287, 20737–20747 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weng N. P., Levine B. L., June C. H., Hodes R. J., Human naive and memory T lymphocytes differ in telomeric length and replicative potential. Proc. Natl. Acad. Sci. U.S.A. 92, 11091–11094 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hayflick L., Moorhead P. S., The serial cultivation of human diploid cell strains. Exp. Cell Res. 25, 585–621 (1961). [DOI] [PubMed] [Google Scholar]
- 11.Rossiello F., Herbig U., Longhese M. P., Fumagalli M., d’Adda di Fagagna F., Irreparable telomeric DNA damage and persistent DDR signalling as a shared causative mechanism of cellular senescence and ageing. Curr. Opin. Genet. Dev. 26, 89–95 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bodnar A. G., et al. , Extension of life-span by introduction of telomerase into normal human cells. Science 279, 349–352 (1998). [DOI] [PubMed] [Google Scholar]
- 13.Xu Z., Duc K. D., Holcman D., Teixeira M. T., The length of the shortest telomere as the major determinant of the onset of replicative senescence. Genetics 194, 847–857 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feldser D. M., Greider C. W., Short telomeres limit tumor progression in vivo by inducing senescence. Cancer Cell 11, 461–469 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodier F., et al. , Persistent DNA damage signalling triggers senescence-associated inflammatory cytokine secretion. Nat. Cell Biol. 11, 973–979 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kang C., et al. , The DNA damage response induces inflammation and senescence by inhibiting autophagy of GATA4. Science 349, aaa5612 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Romanov S. R., et al. , Normal human mammary epithelial cells spontaneously escape senescence and acquire genomic changes. Nature 409, 633–637 (2001). [DOI] [PubMed] [Google Scholar]
- 18.Bednarek A., Budunova I., Slaga T. J., Aldaz C. M., Increased telomerase activity in mouse skin premalignant progression. Cancer Res. 55, 4566–4569 (1995). [PubMed] [Google Scholar]
- 19.Bryan T. M., Englezou A., Gupta J., Bacchetti S., Reddel R. R., Telomere elongation in immortal human cells without detectable telomerase activity. EMBO J. 14, 4240–4248 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blasco M. A., et al. , Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell 91, 25–34 (1997). [DOI] [PubMed] [Google Scholar]
- 21.Yuan X., et al. , Presence of telomeric G-strand tails in the telomerase catalytic subunit TERT knockout mice. Genes Cells 4, 563–572 (1999). [DOI] [PubMed] [Google Scholar]
- 22.Liu Y., et al. , The telomerase reverse transcriptase is limiting and necessary for telomerase function in vivo. Curr. Biol. 10, 1459–1462 (2000). [DOI] [PubMed] [Google Scholar]
- 23.Chiang Y. J., et al. , Expression of telomerase RNA template, but not telomerase reverse transcriptase, is limiting for telomere length maintenance in vivo. Mol. Cell. Biol. 24, 7024–7031 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rudolph K. L., et al. , Longevity, stress response, and cancer in aging telomerase-deficient mice. Cell 96, 701–712 (1999). [DOI] [PubMed] [Google Scholar]
- 25.Hathcock K. S., et al. , Haploinsufficiency of mTR results in defects in telomere elongation. Proc. Natl. Acad. Sci. U.S.A. 99, 3591–3596 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jaskelioff M., et al. , Telomerase reactivation reverses tissue degeneration in aged telomerase-deficient mice. Nature 469, 102–106 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kipling D., Cooke H. J., Hypervariable ultra-long telomeres in mice. Nature 347, 400–402 (1990). [DOI] [PubMed] [Google Scholar]
- 28.Chiang Y. J., et al. , Telomere length is inherited with resetting of the telomere set-point. Proc. Natl. Acad. Sci. U.S.A. 107, 10148–10153 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prowse K. R., Greider C. W., Developmental and tissue-specific regulation of mouse telomerase and telomere length. Proc. Natl. Acad. Sci. U.S.A. 92, 4818–4822 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Varga R., et al. , Progressive vascular smooth muscle cell defects in a mouse model of Hutchinson-Gilford progeria syndrome. Proc. Natl. Acad. Sci. U.S.A. 103, 3250–3255 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cawthon R. M., Telomere length measurement by a novel monochrome multiplex quantitative PCR method. Nucleic Acids Res. 37, e21 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smogorzewska A., de Lange T., Different telomere damage signaling pathways in human and mouse cells. EMBO J. 21, 4338–4348 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Deursen J. M., The role of senescent cells in ageing. Nature 509, 439–446 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.d’Adda di Fagagna F., et al. , A DNA damage checkpoint response in telomere-initiated senescence. Nature 426, 194–198 (2003). [DOI] [PubMed] [Google Scholar]
- 35.Feng F. Y., de Bono J. S., Rubin M. A., Knudsen K. E., Chromatin to clinic: The molecular rationale for PARP1 inhibitor function. Mol. Cell 58, 925–934 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Herbig U., Jobling W. A., Chen B. P. C., Chen D. J., Sedivy J. M., Telomere shortening triggers senescence of human cells through a pathway involving ATM, p53, and p21(CIP1), but not p16(INK4a). Mol. Cell 14, 501–513 (2004). [DOI] [PubMed] [Google Scholar]
- 37.Wieler S., Gagné J. P., Vaziri H., Poirier G. G., Benchimol S., Poly(ADP-ribose) polymerase-1 is a positive regulator of the p53-mediated G1 arrest response following ionizing radiation. J. Biol. Chem. 278, 18914–18921 (2003). [DOI] [PubMed] [Google Scholar]
- 38.Doksani Y., de Lange T., Telomere-internal double-strand breaks are repaired by homologous recombination and PARP1/Lig3-dependent end-joining. Cell Rep. 17, 1646–1656 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao H., et al. , PARP1- and CTCF-mediated interactions between active and repressed chromatin at the lamina promote oscillating transcription. Mol. Cell 59, 984–997 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Banin S., et al. , Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science 281, 1674–1677 (1998). [DOI] [PubMed] [Google Scholar]
- 41.Artandi S. E., Attardi L. D., Pathways connecting telomeres and p53 in senescence, apoptosis, and cancer. Biochem. Biophys. Res. Commun. 331, 881–890 (2005). [DOI] [PubMed] [Google Scholar]
- 42.Denchi E. L., de Lange T., Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature 448, 1068–1071 (2007). [DOI] [PubMed] [Google Scholar]
- 43.Erdmann N., Harrington L. A., No attenuation of the ATM-dependent DNA damage response in murine telomerase-deficient cells. DNA Repair (Amst.) 8, 347–353 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yeager T. R., et al. , Telomerase-negative immortalized human cells contain a novel type of promyelocytic leukemia (PML) body. Cancer Res. 59, 4175–4179 (1999). [PubMed] [Google Scholar]
- 45.Draskovic I., et al. , Probing PML body function in ALT cells reveals spatiotemporal requirements for telomere recombination. Proc. Natl. Acad. Sci. U.S.A. 106, 15726–15731 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhong Z. H., et al. , Disruption of telomere maintenance by depletion of the MRE11/RAD50/NBS1 complex in cells that use alternative lengthening of telomeres. J. Biol. Chem. 282, 29314–29322 (2007). [DOI] [PubMed] [Google Scholar]
- 47.Hu J., et al. , Antitelomerase therapy provokes ALT and mitochondrial adaptive mechanisms in cancer. Cell 148, 651–663 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Henson J. D., et al. , DNA C-circles are specific and quantifiable markers of alternative-lengthening-of-telomeres activity. Nat. Biotechnol. 27, 1181–1185 (2009). [DOI] [PubMed] [Google Scholar]
- 49.Hirao A., et al. , Chk2 is a tumor suppressor that regulates apoptosis in both an ataxia telangiectasia mutated (ATM)-dependent and an ATM-independent manner. Mol. Cell. Biol. 22, 6521–6532 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Squatrito M., et al. , Loss of ATM/Chk2/p53 pathway components accelerates tumor development and contributes to radiation resistance in gliomas. Cancer Cell 18, 619–629 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Matsuoka S., et al. , Reduced expression and impaired kinase activity of a Chk2 mutant identified in human lung cancer. Cancer Res. 61, 5362–5365 (2001). [PubMed] [Google Scholar]
- 52.Kusakawa S., Yasuda S., Kuroda T., Kawamata S., Sato Y., Ultra-sensitive detection of tumorigenic cellular impurities in human cell-processed therapeutic products by digital analysis of soft agar colony formation. Sci. Rep. 5, 17892 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Londoño-Vallejo J. A., Wellinger R. J., Telomeres and telomerase dance to the rhythm of the cell cycle. Trends Biochem. Sci. 37, 391–399 (2012). [DOI] [PubMed] [Google Scholar]
- 54.Yang G., et al. , Knockdown of p53 combined with expression of the catalytic subunit of telomerase is sufficient to immortalize primary human ovarian surface epithelial cells. Carcinogenesis 28, 174–182 (2007). [DOI] [PubMed] [Google Scholar]
- 55.Lee S. S., Bohrson C., Pike A. M., Wheelan S. J., Greider C. W., ATM kinase is required for telomere elongation in mouse and human cells. Cell Rep. 13, 1623–1632 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tong A. S., et al. , ATM and ATR signaling regulate the recruitment of human telomerase to telomeres. Cell Rep. 13, 1633–1646 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang S., Zhao Y., Hu C., Zhu J., Differential repression of human and mouse TERT genes during cell differentiation. Nucleic Acids Res. 37, 2618–2629 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.de Lange T., Activation of telomerase in a human tumor. Proc. Natl. Acad. Sci. U.S.A. 91, 2882–2885 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Akincilar S. C., Unal B., Tergaonkar V., Reactivation of telomerase in cancer. Cell. Mol. Life Sci. 73, 1659–1670 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vera E., Bernardes de Jesus B., Foronda M., Flores J. M., Blasco M. A., The rate of increase of short telomeres predicts longevity in mammals. Cell Rep. 2, 732–737 (2012). [DOI] [PubMed] [Google Scholar]
- 61.Roth A., et al. , Telomerase levels control the lifespan of human T lymphocytes. Blood 102, 849–857 (2003). [DOI] [PubMed] [Google Scholar]
- 62.Steinert S., Shay J. W., Wright W. E., Transient expression of human telomerase extends the life span of normal human fibroblasts. Biochem. Biophys. Res. Commun. 273, 1095–1098 (2000). [DOI] [PubMed] [Google Scholar]
- 63.Erdmann N., Liu Y., Harrington L., Distinct dosage requirements for the maintenance of long and short telomeres in mTert heterozygous mice. Proc. Natl. Acad. Sci. U.S.A. 101, 6080–6085 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Meznikova M., Erdmann N., Allsopp R., Harrington L. A., Telomerase reverse transcriptase-dependent telomere equilibration mitigates tissue dysfunction in mTert heterozygotes. Dis. Model. Mech. 2, 620–626 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Le Saux C. J., et al. , A novel telomerase activator suppresses lung damage in a murine model of idiopathic pulmonary fibrosis. PLoS One 8, e58423 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Muñoz-Lorente M. A., et al. , AAV9-mediated telomerase activation does not accelerate tumorigenesis in the context of oncogenic K-Ras-induced lung cancer. PLoS Genet. 14, e1007562 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Povedano J. M., et al. , Therapeutic effects of telomerase in mice with pulmonary fibrosis induced by damage to the lungs and short telomeres. eLife 7, e31299 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hemann M. T., Strong M. A., Hao L.-Y., Greider C. W., The shortest telomere, not average telomere length, is critical for cell viability and chromosome stability. Cell 107, 67–77 (2001). [DOI] [PubMed] [Google Scholar]
- 69.National Research Council , Guide for the Care and Use of Laboratory Animals (National Academies Press, Washington, DC, ed. 8, 2011). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.