Significance
More than 160 rhinovirus (RV) types cause about a billion respiratory infections annually in the United States alone, contributing to influenza-like illness. This diversity makes vaccination impractical. Existing small-molecule inhibitors target RVs by binding to a hydrophobic pocket in the capsid but exhibit side effects, resistance, and/or mutational escape, impeding registration as drugs. The pyrazolopyrimidine OBR-5-340 acts like other capsid binders by preventing conformational changes required for genome release. However, by using cryo-EM, we show that OBR-5-340 inhibits the naturally pleconaril-resistant RV-B5 by attaching close to the pocket entrance in a binding geometry different from that of most capsid binders. Combinations of inhibitors with disparate binding modes might thus effectively combat RVs while reducing the risk of resistance development.
Keywords: rhinovirus, capsid binder, inhibitor, 3D structure, cryo-EM
Abstract
Viral inhibitors, such as pleconaril and vapendavir, target conserved regions in the capsids of rhinoviruses (RVs) and enteroviruses (EVs) by binding to a hydrophobic pocket in viral capsid protein 1 (VP1). In resistant RVs and EVs, bulky residues in this pocket prevent their binding. However, recently developed pyrazolopyrimidines inhibit pleconaril-resistant RVs and EVs, and computational modeling has suggested that they also bind to the hydrophobic pocket in VP1. We studied the mechanism of inhibition of pleconaril-resistant RVs using RV-B5 (1 of the 7 naturally pleconaril-resistant rhinoviruses) and OBR-5-340, a bioavailable pyrazolopyrimidine with proven in vivo activity, and determined the 3D-structure of the protein-ligand complex to 3.6 Å with cryoelectron microscopy. Our data indicate that, similar to other capsid binders, OBR-5-340 induces thermostability and inhibits viral adsorption and uncoating. However, we found that OBR-5-340 attaches closer to the entrance of the pocket than most other capsid binders, whose viral complexes have been studied so far, showing only marginal overlaps of the attachment sites. Comparing the experimentally determined 3D structure with the control, RV-B5 incubated with solvent only and determined to 3.2 Å, revealed no gross conformational changes upon OBR-5-340 binding. The pocket of the naturally OBR-5-340-resistant RV-A89 likewise incubated with OBR-5-340 and solved to 2.9 Å was empty. Pyrazolopyrimidines have a rigid molecular scaffold and may thus be less affected by a loss of entropy upon binding. They interact with less-conserved regions than known capsid binders. Overall, pyrazolopyrimidines could be more suitable for the development of new, broadly active inhibitors.
Rhinoviruses (RVs), including the species A, B, and C, are the most common respiratory pathogens (1) within the genus Enterovirus, family Picornaviridae (2, 3). RVs are the main cause of the common cold and are adapted to optimally grow at 33 to 34 °C, the mean temperature of the upper respiratory tract (4). However, some RVs can also efficiently replicate at 37 °C and cause pulmonary infections (5).
Biology and pathogenesis of RVs are active fields of research due to their high medical and economic impact and the lack of vaccines and efficient medication (6, 7). RVs are icosahedrons, with a protein shell of about 30 nm in diameter containing a ss(+)RNA genome of about 7,200 bases. The capsid is composed of 60 copies each of the four proteins VP1, VP2, VP3, and VP4 arranged in a pseudo T = 3 lattice (8). VP4 is ∼7 kDa and located at the inner side, close to the RNA. VP1, VP2, and VP3 (∼25 to 35 kDa) adopt antiparallel 8-stranded β-barrel folds connected by loops. The surface-exposed loops are recognized by serotype-specific neutralizing antibodies. VP1, VP2, and VP3 interact with cell-surface molecules (receptors) for viral attachment. Most RV-A and all RV-B exploit the intercellular adhesion molecule-1 (ICAM-1) as receptor (9). Twelve RV-A bind at least two members of the low-density lipoprotein receptor (LDLR) family (10). RV-C use the cadherin-related family member 3 (CDHR3) for cell entry (11). Whereas ICAM-1 binds into the canyon, a deep cleft encircling the star-shaped dome at the 5-fold axes of symmetry (12), members of the LDLR family instead bind to the dome itself, closer to the symmetry axis (13). The binding site of CDHR3 on RV-C has not yet been disclosed.
Among the most conserved regions in the structural and nonstructural proteins are a hydrophobic pocket in the capsid of RV-A and RV-B [RV-C lack a similar pocket (14)] and the active sites of the viral enzymes. Therefore, these regions are obvious targets of inhibitors acting against a broad spectrum of RVs. However, due to the insufficient efficacy, mutational escape, and/or side effects observed in clinical studies, none of these antivirals has been approved (15–17). Recently, it was discovered that the small cellular phospholipase pla2g16 aids the transfer of the viral RNA genome into the cytosol (18, 19). It may be a potential host target for the treatment of RVs and enteroviruses (EVs). Nevertheless, a full understanding of pla2g16’s function in the uninfected cell will be necessary before inhibitors can be brought to the clinic.
Severe disease outbreaks caused by RV-related EVs (20) restimulated the search for antivirals (21). In addition to the ongoing quest for compounds targeting nonstructural viral proteins and cellular factors, capsid binders like pleconaril (SI Appendix, Fig. S1) and vapendavir (22) have remained in the focus. They bind into the hydrophobic pocket lined by amino acid residues from VP1, with some contribution of VP2 and VP3 (23). Most of the residues are highly conserved within the genus Enterovirus (23, 24), enabling the inhibition of a broad spectrum of RVs and EVs with capsid binders (25). Capsid binders can exert virucidal activity against RVs and EVs (24, 26), impede viral adsorption by modifying the conformation of the receptor binding site (24, 27, 28), and/or prevent structural changes required for RNA uncoating (29). As suggested by the example of pleconaril, occupancy of the hydrophobic pocket can be higher when the capsid binder is introduced during viral assembly (25).
Naturally occurring as well as experimentally selected resistance of RVs and EVs to capsid binders is based mainly on a polymorphism of amino acid residues in the center of the hydrophobic binding pocket (23, 28, 30–33). For example, in the pleconaril-resistant RV-B4, -B5, -B42, -B84, -B93, -B97, and -B99 (30) the amino acid residues F1152 and L1191 (first digit refers to the VP; i.e., “1” denotes VP1) were detected. In pleconaril-susceptible serotypes these positions are occupied by Y1152 and V/I/T1191, respectively (34). It has been shown that the amino acid substitutions Y1152F and V1191L in VP1 of the naturally pleconaril-susceptible RV-B14 confer full resistance to pleconaril (34). The 3D structures of the pleconaril-susceptible RV-B14 (8) and RV-A16 (25) have been determined to atomic resolution by X-ray crystallography. The results suggest that the bulkier L1191 narrows the hydrophobic pocket and sterically hinders pleconaril binding, in particular by interfering with its central ring (34). A similar resistance mechanism was proposed for vapendavir in EV-68 and EV-71 (22, 35). Accordingly, chemical modifications of the central ring of pleconaril led to a regain of its anti-enteroviral activity (36). In addition, full pleconaril resistance was detected in coxsackievirus B3 (CVB3), another EV, following I1207R/K substitutions near the hydrophobic binding pocket (37). In the absence of atomic structures of pleconaril-resistant RVs, the differences in the capsid structures of pleconaril-susceptible and -resistant RVs remain to be determined.
Pyrazolopyrimidines are relatively new capsid-binding inhibitors. They show strong activity against pleconaril-resistant EVs and RVs (24). Like pleconaril and vapendavir, they are three-ring compounds. The structural similarity and inhibition profile, together with results of computational modeling of CVB3 in complex with 3-(4-trifluoromethylphenyl)amino-6-phenylpyrazolo[3,4-d]pyrimidine-4-amine (OBR-5-340) (SI Appendix, Fig. S1), taken as an example, suggested that pyrazolopyrimidines would also bind to the hydrophobic pocket (24). However, the differences in the spectrum of pleconaril- and OBR-5-340-susceptible RVs suggest differences in the binding.
Here, we aimed to decipher the mechanism of inhibition of fully pleconaril-resistant RVs (30) by pyrazolopyrimidines as well as the atomic structure of a RV–pyrazolopyrimidine complex. These studies were performed with the pleconaril-resistant RV-B5 and OBR-5-340, the most promising, orally bioavailable pyrazolopyrimidine. For comparison, the pleconaril-sensitive RV-A2 and RV-A89 and pleconaril were included in some of the experiments. The anti–RV-B5 activity of OBR-5-340 under single growth cycle conditions was confirmed. Steps of the replication cycle inhibited by OBR-5-340 were identified. Uncoating inhibition was confirmed by stabilization of RV-B5 by OBR-5-340 against temperature-dependent conversion into the permeable subviral A-particle, an uncoating intermediate. Since RV-B5 was found to grow poorly in spinner cultures, as employed for mass production of numerous other serotypes (38), we solved the 3D structure to 3.6 Å with the available small amounts of RV-B5 complexed to OBR-5-340 by using cryogenic electron microscopy (cryo-EM). For comparison, 3D structures of RV-B5 (in the absence of OBR-5-340) and of naturally drug-resistant RV-A89 (in the presence of OBR-5-340) were determined at 3.2 and 2.9 Å, respectively. Unexpectedly, OBR-5-340 was found to bind closer to the entrance of the pocket than most capsid binders studied so far, with only partially overlapping binding sites. This suggests that suitable combinations of antivirals might alleviate resistance to more efficiently fight the common cold.
Results
Effect of OBR-5-340 on Single-Step Growth of RV-B5.
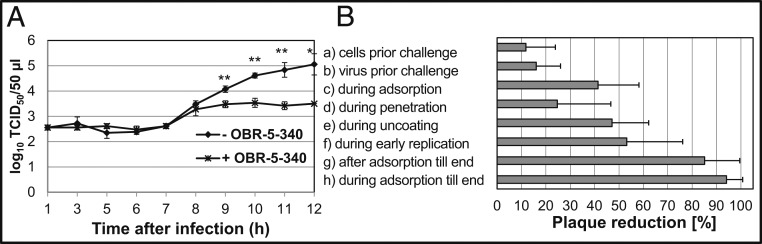
Because RV-B5 is highly susceptible to OBR-5-340 in cytopathic effect (CPE) inhibition assays (50% inhibitory concentration, IC50 = 0.08 µM; compound 36 in the Supporting Information of ref. 24) and fully resistant to pleconaril (IC50 > 12.5 µM; higher concentrations were cytotoxic) (24, 30), we selected it for the present study. Single-step growth curves with RV-B5 were recorded in the absence and presence of the highly effective OBR-5-340 concentration of 1 µM in HeLa cells. The time-dependent increase in infectious units (measured in a separate assay as 50% tissue culture infectious dose; TCID50) in the presence of OBR-5-340 was marginal, whereas in the control without OBR-5-340 the virus titer increased by about 2.5 log units within 10 h, the time needed to complete a single growth cycle. The reduction in virus titer was thus about 1.5 log units (Fig. 1A).
Fig. 1.
(A) OBR-5-340 strongly reduces the single-step growth of RV-B5. HeLa cells were challenged with RV-B5 at a multiplicity of infection of 1 TCID50/cell ± OBR-5-340 at 1 µM, and the virus titer was determined at the times indicated. Mean and SD of 3 parallels per time point are shown. Significant differences were determined with Welch’s t test: *P < 0.05 and **P < 0.01. (B) Impact of OBR-5-340 on different steps of the replication cycle of RV-B5. Plaque reduction assays with 15 to 50 plaque-forming units of RV-B5 in HeLa cells were performed. OBR-5-340 was added to HeLa cells and/or RV-B5 at 0.5 µM at different times. The number of plaques, recorded without addition of OBR-5-340 but otherwise conducted identically, was set to 100%. The bars indicate the percentage of plaque reduction by OBR-5-340. From top to bottom: (a) Cells prior to challenge. HeLa cells were treated with OBR-5-340 for 0.5 h at 33 °C before RV-B5 infection. (b) Virus prior to challenge. RV-B5 was incubated with OBR-5-340 for 1 h at 33 °C and subsequently diluted to noneffective inhibitor concentrations (1:5,000) prior to challenge to HeLa cells. (c) During adsorption. RV-B5 and OBR-5-340 were incubated with HeLa cells at 4 °C for 2 h, a temperature and time allowing for viral adsorption but not replication. (d) During penetration. RV-B5 was allowed to attach to the cells as in c but without OBR-5-340. After washing away nonadsorbed RV-B5, OBR-5-340 was added to the infected HeLa cells for 1 h at room temperature, allowing for RV-B5 penetration but no further replication. (e and f) During uncoating and early replication. OBR-5-340 was added for 2 or 6 h at 33 °C, respectively, after RV-B5 adsorption and penetration as in c and d but without OBR-5-340. In a–f, plaques were allowed to develop under agar without OBR-5-340 for 72 h at 33 °C. (g) After adsorption. RV-B5 was allowed to attach to the cells as in c but without OBR-5-340. After washing away nonadsorbed RV-B5, OBR-5-340 was added with the agar overlay to the RV-B5–infected HeLa cells for 72 h. (h) During adsorption until the end. RV-B5 was allowed to attach to the cells as in c. After washing away nonadsorbed RV-B5, OBR-5-340 was added with the agar overlay to the RV-B5–infected HeLa cells for 72 h. After crystal violet staining, plaques were counted.
Mechanism of Viral Inhibition.
We investigated which stage of the infection cycle was most affected by OBR-5-340 by using a previously established plaque-reduction assay (24). OBR-5-340 (0.5 µM) was added at different time points before and after challenging the HeLa cells with RV-B5. As seen from the plaque-reduction measurements reported in Fig. 1B, incubation of HeLa cells with OBR-5-340 before viral challenge (Fig. 1 B, a), of RV-B5 prior to challenge of HeLa cells (Fig. 1 B, b), and during RV-B5 penetration (Fig. 1 B, d) was ineffective. Formation of RV-B5–induced plaques was reduced by 40 to 50% when OBR-5-340 treatment was performed only during RV-B5 adsorption (2 h at 4 °C; Fig. 1 B, c), during uncoating (2 h at 33 °C after RV-B5 adsorption and penetration Fig. 1 B, e), and during early replication (6 h at 33 °C after RV-B5 adsorption and penetration; Fig. 1 B, f). The strongest reduction of plaque formation was detected when OBR-5-340 was present for 72 h in the agar overlay and could act during multiple rounds of RV replication. This is shown in Fig. 1 B, g and h, where OBR-5-340 was added immediately after RV-B5 adsorption and kept until the end of the experiment or was present throughout the entire experiment, respectively.
OBR-5-340 Stabilizes RV-B5 Against Heat-Triggered RNA Exposure but RV-A2 and RV-A89 Are Unaffected.
Several capsid binders have been shown to stabilize the virion against the conformational alteration associated with uncoating of the genomic RNA (27, 28, 39–41). Conversion of the native virion into the expanded A-particle with holes for the RNA exit is facilitated by expulsion of the natural pocket factor, probably a fatty acid (42). Capsid binders stabilize the virion against the conversion into the permeable A-particle and thus block the release of the RNA (43). Heating to ≥50 °C also triggers this conversion and/or the release of the RNA, depending on the buffer composition (44, 45). This process is inhibited by capsid binders.
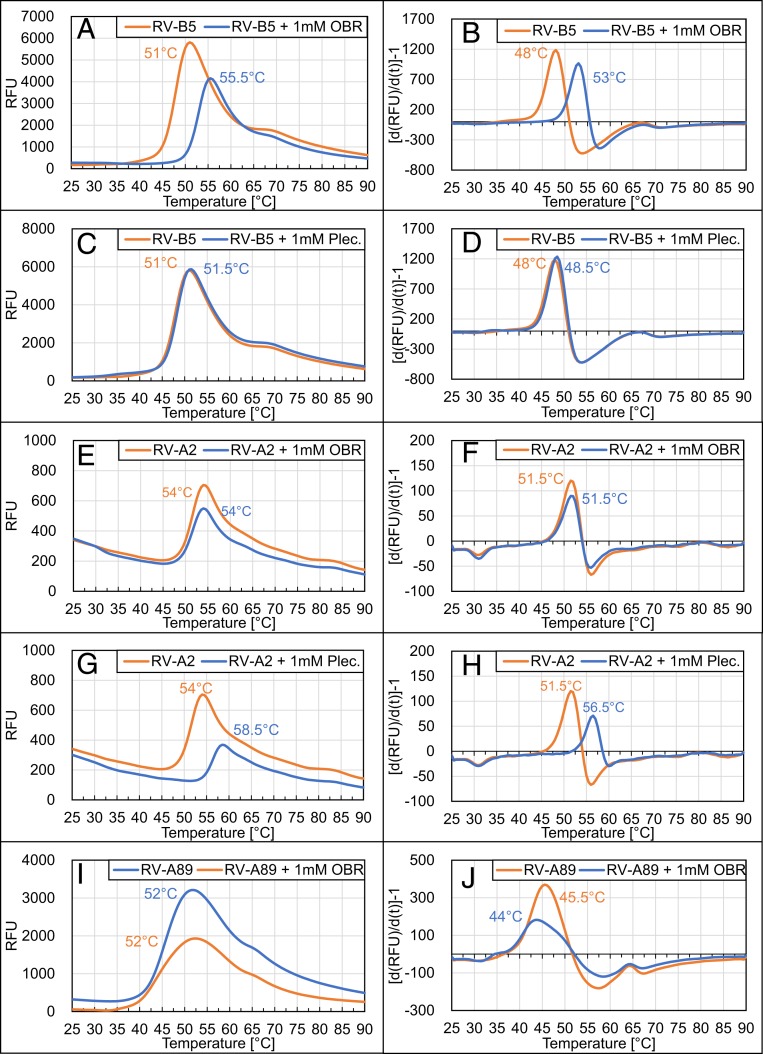
Results from plaque-reduction assays showed inhibition of RV-B5 uncoating (Fig. 1 B, e). To further confirm this, RV-B5 was preincubated for 1 h at 37 °C with 1 mM of the respective inhibitor to allow for saturation of binding sites and displacement of natural factors. Then, Syto-82, a dye, which becomes fluorescent upon binding to RNA, was added and the temperature was increased (46). Syto-82 cannot penetrate into the native virion. However, upon conversion into subviral particles, the RNA is accessible for the dye either through the holes in the shell or upon its release into solution; both result in fluorescence. As seen in Fig. 2A, a fluorescence peak appeared at 51 °C when the virus was heated in the absence of OBR-5-340. In its presence, this peak was shifted toward 55.5 °C. Decline of the signal upon further rise of the temperature is probably due to RNA unfolding. When the RNA had been released prior to the experiment, fluorescence at the start of temperature ramping was similar to the maximum attained with virus at around 51 °C and similarly diminished upon further temperature increase. Using bovine serum albumin as a control, no peaks were observed, excluding interactions between the dye and OBR-5-340 and/or the solvent dimethylsulfoxide (DMSO) (SI Appendix, Fig. S2).
Fig. 2.
Pleconaril and OBR-5-340 differently stabilize RVs against heat-triggered uncoating. The specified virus types were preincubated with or without 1 mM of the indicated inhibitors for 1 h at 37 °C. Syto-82 was then added, and the temperature was continuously increased while monitoring fluorescence as detailed in Materials and Methods. (Left) Relative fluorescence units measured at the temperatures given on the abscissa. The temperature at which the peak of the respective fluorescence intensity was observed is indicated. (Right) The corresponding first derivative of the curve. Here, the peaks denote the highest change in fluorescence, which was taken as the onset of RNA accessibility. Note that RV-B5 is stabilized by OBR-5-340 (A and B) but not by pleconaril (C and D) and that RV-A2 is stabilized by pleconaril (G and H) but neither RV-A2 (E and F) nor RV-A89 (I and J) is stabilized by OBR-5-340. Control experiments with free RNA obtained by prior heating of RV-B5 to 60 °C followed by cooling demonstrated a fluorescence plateau that started to decline at about 40 °C, presumably because of dissociation of the dye from unfolding RNA (SI Appendix, Fig. S2). Note that the 2 effects occur in parallel with the first one overwhelming the second one. For lower concentrations of OBR-5-340 inhibiting RV-B5, see SI Appendix, Fig. S3.
For better identification of the temperature of fluorescence onset, we plotted the first derivative against the temperature (Fig. 2, Right). Accordingly, viral conversion was initiated at 48 °C in the absence of OBR-5-340 and at 53 °C in its presence (Fig. 2B). It is thus obvious that OBR-5-340 stabilizes RV-B5 against conversion into (permeable) subviral particles. As control, we also studied the effect of pleconaril (Fig. 2, C and D). In agreement with RV-B5 being resistant against pleconaril (34), no shift of the fluorescence signal toward higher temperature was observed. As additional controls we used the pleconaril-sensitive RV-A2 and RV-A89 (30, 47). Several compounds have been shown to inhibit RV-A2 conversion into the A-particle/empty capsid by heat or low pH using other means of detection (40, 43). Fig. 2, E and F, shows that heat-triggered uncoating of RV-A2 is not affected by OBR-5-340 [IC50 = 13.77 µM; compound 36, in the Supplementary Material of ref. 24), as compared to IC50 ∼ 0.08 µM for RV-B5]. However, as expected (48), RV-A2 was stabilized by pleconaril (Fig. 2, G and H). Additionally, we studied the pleconaril-sensitive RV-A89 (47). As seen in Fig. 2, I and J, OBR-5-340 also had no impact on the temperature sensitivity of RV-A89, which is consistent with its natural resistance against this compound, as was also confirmed in our CPE inhibition assays; no inhibitory activity of OBR-5-340 at 100 µM was recorded for RV-A89. Conversely, RV-A89 was inhibited by pleconaril with IC50 0.02 ± 0.01 µM in the CPE inhibitory assay. Of note, OBR-5-340 stabilized RV-B5 somewhat at 1 µM and strongly at 10 and 100 µM (SI Appendix, Fig. S3).
Cryo-EM 3D Structures of RV-B5 and RV-A89.
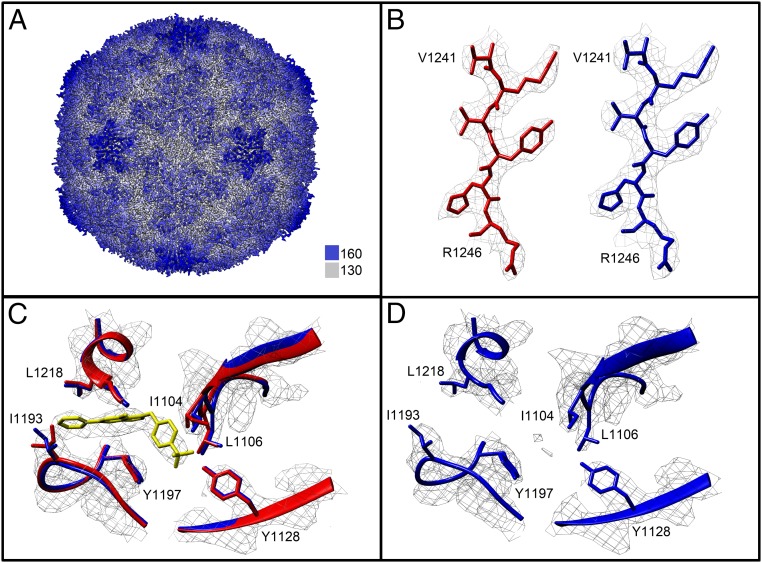
Poor growth of RV-B5 in suspension culture did not yield quantities necessary for crystallization trials as required for X-ray analysis (49). We thus collected cryo-EM data of the available small amounts of virus incubated with 1 mM OBR-5-340, the highest concentration achievable without exceeding 10% DMSO used as solvent. For direct comparability, incubation conditions were kept identical to those of the stabilization assays described above, except that the virus concentration was 0.6 mg/mL. Imposing icosahedral symmetry, we determined the 3D structure of RV-B5 to 3.6 Å based upon the Fourier shell correlation resolution criteria 0.143 (Fig. 3A, SI Appendix, Table S1 and Fig. S4, Left and refs. 50 and 51). The quality of the maps was high (Fig. 3 B, Left). Residues included in the atomic model are Q1016-Y1288 for VP1 (not visible: G1001-Q1015); G2008-I2259 for VP2 (not visible: S2001-C2007 and Q2230); G3001-Q3231 for VP3 (not visible: T3232); and I4030-N4069 for VP4 (not visible G1001-V4029 and S4070). Additional density not attributable to amino acid residues was identified in the hydrophobic pocket (Fig. 3C); this density accommodates OBR-5-340 well. In the absence of OBR-5-340, RV-B5 shows very weak density of dissimilar shape in the deeper binding position (Fig. 3D, SI Appendix, Table S1 and Fig. S4 and refs. 52 and 53). It might stem from a natural pocket factor present at low occupancy. The pocket of the OBR-5-340-resistant RV-A89, identically incubated with OBR-5-340 and analyzed as an additional negative control (SI Appendix, Figs. S4 and S6 and Table S1), was empty. Notably, radial average curves of all calibrated volumes showed the same diameter of 302 Å. This excludes that OBR-5-340 binding modifies the size of the virion (SI Appendix, Fig. S5).
Fig. 3.
(A) Cryo-EM structure of RV-B5 complexed to OBR-5-340 colored radially as indicated by the color bar. Distance from the viral center is 130 Å (white) to 160 Å (dark blue). (B) Example of the quality of the maps of RV-B5 with OBR-5-340 (Left) and without OBR-5-340 (Right). (C) View centered on OBR-5-340 (yellow) in complex with RV-B5 (red). For comparison, the control, i.e., RV-B5 solved in the absence of inhibitor (blue), is overlaid. Residues nearby and contributed by VP1 are labeled. (D) RV-B5 solved in the absence of OBR-5-340. Note the absence of density at the position where the inhibitor is seen in the complex.
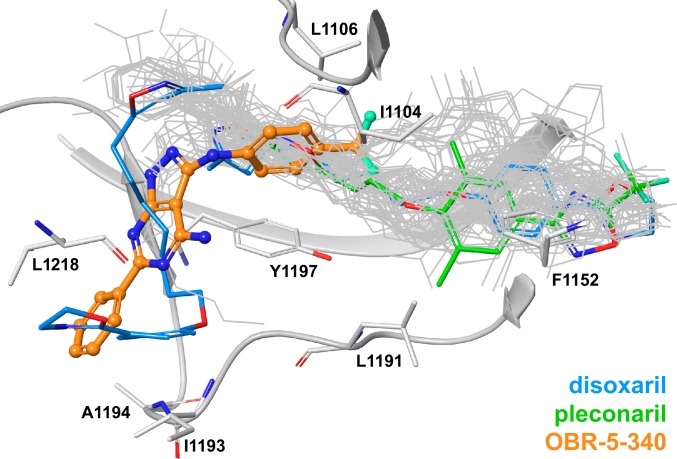
Strikingly, OBR-5-340 was observed to bind closer to the entrance of the hydrophobic pocket than most capsid binders, including pleconaril (Fig. 4 and SI Appendix, Table S2). The only instance in which weak density was also observed at the entrance (or “mouth”) of the hydrophobic pocket is the structure of disoxaril (WIN51711) bound to CoxA9. This density could stem from a second disoxaril molecule, with the first one sitting in the deep parts of the pocket (54) [Protein Data Bank (PDB) 1D4M]. Few structures of Enterovirus capsids have been deposited in the PDB where pocket factors bind to deep parts of the pocket and protrude to the entry/mouth area of the binding site. The best examples are PDB 1MQT, where C8 ceramide is located mainly in the β-barrel region but elongates into the entry area, and PDB 1AYM, where lauric acid is straddled between the two regions. The small molecules observed or postulated to bind to the mouth of the hydrophobic pocket exhibit enormous conformational flexibility that is based on their long, aliphatic moieties. This makes their binding entropically unfavorable. In contrast, OBR-5-340 is a rigid molecule with few rotatable bonds. Therefore, it is expected to be less affected by loss of entropy upon binding and to be more suitable as a template for the development of new drugs.
Fig. 4.
Superposition of 82 crystal structures of capsid binders (most depicted in gray wires) in complex with Enterovirus coat proteins (PDB IDs listed in SI Appendix, Table S2). OBR-5-340 is shown in ball-and-stick representation (with orange carbon atoms) in complex with RV-B5. The 2 binding poses of disoxaril from PDB 1D4M are shown in stick representation with blue carbon atoms; pleconaril from PDB 1NCQ is shown in stick representation with green carbon atoms. Parts of the protein backbone and selected amino acids of VP1 are depicted in gray.
OBR-5-340 forms a tight network of interactions with the VPs. Its pyrazolopyrimidine core forms two hydrogen bonds with N1198, one with the amide NH and one with the amide carbonyl oxygen. The trifluorophenyl moiety forms an H-bond with Y1128. The ligand core and the trifluorophenyl moiety are engaged in pi-stacking interactions with Y1197. OBR-5-340 also forms hydrophobic interactions with I1104, L1106, I1193, A1194, and L1218.
In the absence of OBR-5-340, RV-B5 shows very weak density of dissimilar shape in the deeper binding position (Fig. 3D and SI Appendix, Table S1 and Fig. S4). This density might originate from a natural pocket factor, presumably myristate, being present at low occupancy. The pocket of the OBR-5-340-resistant RV-A89 identically incubated with OBR-5-340 and analyzed as additional negative control (SI Appendix, Fig. S6, Table S1, and Fig. S4, Right and refs. 55 and 56) was empty.
Discussion
We demonstrate that infection of HeLa cells by RV-B5, a naturally pleconaril-resistant rhinovirus, is strongly inhibited by OBR-5-340. This compound represents a class of pyrazolopyrimidines active against various picornaviruses of the genus Enterovirus (24). In addition to RV-B5, OBR-5-340 also strongly inhibits other pleconaril-resistant RVs at low micromolar concentrations (24). Whether the compound is similarly active against the remaining pleconaril-resistant RV-B4, -B84, -B93, -B97, and -B99 (34) remains to be determined.
OBR-5-340 treatment for 2 h at 4 °C inhibited RV-B5 adsorption to HeLa by 40%. This agrees with the inhibition of RV-B14 adsorption by 8 other capsid binders (27). However, the strongest plaque reduction was observed when OBR-5-340 was present for 72 h (overall incubation time for the plaque assay), i.e., during multiple rounds of replication. In each round, inhibition of RV-B5 adsorption and uncoating can occur. Furthermore, it is possible that OBR-5-340, similar to pleconaril (25), incorporates better into progeny viruses during assembly, thereby increasing the antiviral effect. Zhang et al. observed a lower degree of incorporation of pleconaril into RV-B14 and RV-A16 crystals upon soaking than in cases where pleconaril was present during viral assembly (25). The authors concluded that pleconaril acts more strongly on the infectivity of RV-A16 progeny virions than on the parent input virus. This was not studied here but might help to further explain our data.
The moderate inhibition of RV-B5 uncoating is in line with the low but detectable in vitro stabilization of RV-B5 by 1 µM OBR-5-340 (SI Appendix, Fig. S3). The binding site of OBR-5-340 overlaps only marginally with that of other capsid binders as it localizes more closely to the entryway of the hydrophobic pocket (Fig. 4). Similar to other capsid binders, OBR-5-340 prevents conformational changes of the native virion that are essential for RNA release. Incubation of RV-B5 with OBR-5-340 at concentrations between 1 µM and 1 mM stabilized the virion against heat-triggered exposure of the RNA as indicated by in vitro uncoating occurring at an up to 5 °C higher temperature as compared to virus without OBR-5-340 (Fig. 2A and SI Appendix, Fig. S3). During viral replication OBR-5-340 might become incorporated into the capsid, rendering progeny virus noninfectious and thus unable to spread.
Of note, the conformation of the pocket was virtually identical, regardless of the presence or absence of OBR-5-340 in RV-B5 (Fig. 3 C and D). Hence, filling the pocket does not seem to result in major structural rearrangements as were observed upon binding of WIN 52035–2 to RV-B14 (28). Furthermore, the diameter of the virion was the same regardless of the presence or absence of OBR-5-340 (SI Appendix, Fig. S5). The pocket of RV-B5 is somewhat more open when compared to the one of RV-A89 (SI Appendix, Fig. S6D); several residues in close proximity to OBR-5-340 in the complex with RV-B5 are different in RV-A89 (SI Appendix, Fig. S6 and Table S3). This might explain why OBR-5-340 is unable to bind to RV-A89, even at a concentration as high as 1 mM.
OBR-5-340 is a drug-like compound with a rigid molecular scaffold that binds to the entrance of the hydrophobic pocket. This adds another dimension to the quest for capsid-binding drugs and suggests that selected drug combinations might become useful in combating RV infections.
Materials and Methods
Cells and Viruses.
HeLa Ohio cells were used throughout. Virus stocks were grown from seed obtained from the American Type Culture Collection (ATCC). Viral identity was verified via partial RNA and tandem mass spectrometry protein sequencing as well as by neutralization with ATCC type-specific antisera.
Effect of OBR-5-340 on Single-Step Growth of RV-B5 in HeLa Cells.
The time-dependent production of viral progeny without (control) and with the addition of 1 µM OBR-5-340 was recorded. At specified times, infected cells were subjected to 3 rounds of freeze–thawing to release intracellular virus, and the titer was determined as TCID50.
Mechanism of Action Studies with RV-B5 and OBR-5-340 Using a Modified Plaque Reduction Assay.
Cells or virus were preincubated with OBR-5-340, or OBR-5-340 was added and kept present for the different time periods as indicated and described previously for CVB3 (24). Three days post infection cells were fixed and stained with a crystal violet-formalin solution, and plaques were counted.
Temperature-Dependent In Vitro Uncoating of Native Virus and Its Inhibition by Drugs.
Temperature-stability assays were carried out similarly as for foot-and-mouth-disease virus (46). Virus ± inhibitor was heated in the presence of Syto-82. The onset of fluorescence was taken as the temperature at which the virus was converted into the permeable A- and/or B-particle with the RNA becoming accessible for the dye.
Cryo-EM Analysis of RV-B5 OBR-5-340 Complexes and of RV-A89 in the Presence of OBR-5-340.
The respective virus was either incubated with OBR-5-340 or with solvent, applied onto carbon-coated grids, and flash-frozen with a Vitrobot. Movies were collected and submitted to drift correction, and the 3D structure was determined with routine procedures by using Relion-3 (57). Fitting the coordinates was carried out with Phenix (58) and Coot (59). The models were visualized with Chimera (60).
Comparison of Enterovirus Ligand Complexes in the PDB Database with OBR-5-340 Complexed to RV-B5.
The PDB (https://www.rcsb.org, accessed February 22, 2019) was searched for any structures of enteroviral capsid proteins that have at least 1 ligand present. This resulted in 82 structures that we aligned with Maestro (release 2018–3, Schrödinger).
Supplementary Material
Acknowledgments
This work was funded by the Austrian Science Fund project #27444 (D.B.). M.P. and I.Z. were supported by Erasmus fellowships. N.M. and J. K. were supported by the Bergen Research Foundation (BFS2017TMT01). T.C.M. was supported by funds through the Behörde für Wissenschaft, Forschung, und Gleichstellung of the City of Hamburg. Data for RV-B5 and RV-A89, both incubated with OBR-5-340, were collected at the Vienna Biocenter Electron Microscopy Facility. Data collection of the control sample (RV-B5 without OBR-5-340) was funded by iNEXT Grant 5950. iNEXT (project no. 653706) was funded by the Horizon 2020 program of the European Union. This article reflects only the author's view and the European Commission is not responsible for any use that may be made of the information it contains. Czech Infrastructure for Integrative Structural Biology research infrastructure project LM2015043, funded by Ministry of Education, Youth and Sports of the Czech Republic is gratefully acknowledged for the financial support of the measurements at the Central Facility Cryo-electron Microscopy and Tomography Central European Institute of Technology, Masaryk University. We thank Pavel Plevka for freezing and Jirka Novacek for collecting the corresponding data, David Haselbach for advice and help with Phenix and Coot, and Irene Goesler for virus production and purification.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The 3D structures reported in this paper have been deposited in the Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank, http://www.rcsb.org/ and the Electron Microscopy Data Bank, https://www.ebi.ac.uk/pdbe/emdb/ (accession nos.: RV-B5 complexed to OBR-5-340, PDB 6SK5 and EMD-10220; RV-B5, PDB 6SK6 and EMD-10221; RV-A89, PDB 6SK7 and EMD-10222).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1904732116/-/DCSupplemental.
References
- 1.Jacobs S. E., Lamson D. M., St George K., Walsh T. J., Human rhinoviruses. Clin. Microbiol. Rev. 26, 135–162 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Palmenberg A. C., et al. , Sequencing and analyses of all known human rhinovirus genomes reveal structure and evolution. Science 324, 55–59 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Royston L., Tapparel C., Rhinoviruses and respiratory enteroviruses: Not as simple as ABC. Viruses 8, E16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tyrrell D. A., Parsons R., Some virus isolations from common colds. III. Cytopathic effects in tissue cultures. Lancet 1, 239–242 (1960). [DOI] [PubMed] [Google Scholar]
- 5.Papadopoulos N. G., Sanderson G., Hunter J., Johnston S. L., Rhinoviruses replicate effectively at lower airway temperatures. J. Med. Virol. 58, 100–104 (1999). [DOI] [PubMed] [Google Scholar]
- 6.Papadopoulos N. G., et al. , Promising approaches for the treatment and prevention of viral respiratory illnesses. J. Allergy Clin. Immunol. 140, 921–932 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rollinger J. M., Schmidtke M., The human rhinovirus: Human-pathological impact, mechanisms of antirhinoviral agents, and strategies for their discovery. Med. Res. Rev. 31, 42–92 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rossmann M. G., et al. , Structure of a human common cold virus and functional relationship to other picornaviruses. Nature 317, 145–153 (1985). [DOI] [PubMed] [Google Scholar]
- 9.Greve J. M., et al. , The major human rhinovirus receptor is ICAM-1. Cell 56, 839–847 (1989). [DOI] [PubMed] [Google Scholar]
- 10.Hofer F., et al. , Members of the low density lipoprotein receptor family mediate cell entry of a minor-group common cold virus. Proc. Natl. Acad. Sci. U.S.A. 91, 1839–1842 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bochkov Y. A., et al. , Cadherin-related family member 3, a childhood asthma susceptibility gene product, mediates rhinovirus C binding and replication. Proc. Natl. Acad. Sci. U.S.A. 112, 5485–5490 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Olson N. H., et al. , Structure of a human rhinovirus complexed with its receptor molecule. Proc. Natl. Acad. Sci. U.S.A. 90, 507–511 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Verdaguer N., Fita I., Reithmayer M., Moser R., Blaas D., X-ray structure of a minor group human rhinovirus bound to a fragment of its cellular receptor protein. Nat. Struct. Mol. Biol. 11, 429–434 (2004). [DOI] [PubMed] [Google Scholar]
- 14.Liu Y., et al. , Atomic structure of a rhinovirus C, a virus species linked to severe childhood asthma. Proc. Natl. Acad. Sci. U.S.A. 113, 8997–9002 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bauer L., Lyoo H., van der Schaar H. M., Strating J. R., van Kuppeveld F. J., Direct-acting antivirals and host-targeting strategies to combat enterovirus infections. Curr. Opin. Virol. 24, 1–8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Senior K., FDA panel rejects common cold treatment. Lancet Infect. Dis. 2, 264 (2002). [DOI] [PubMed] [Google Scholar]
- 17.Ulferts R., et al. , Screening of a library of FDA-approved drugs identifies several enterovirus replication inhibitors that target viral protein 2C. Antimicrob. Agents Chemother. 60, 2627–2638 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elling U., et al. , A reversible haploid mouse embryonic stem cell biobank resource for functional genomics. Nature 550, 114–118 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Staring J., et al. , PLA2G16 represents a switch between entry and clearance of Picornaviridae. Nature 541, 412–416 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Willyard C., An unknown enemy: Drugs sought against EV-68 as paralysis link is explored. Nat. Med. 21, 419–421 (2015). [DOI] [PubMed] [Google Scholar]
- 21.van der Linden L., Wolthers K. C., van Kuppeveld F. J., Replication and inhibitors of enteroviruses and parechoviruses. Viruses 7, 4529–4562 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tijsma A., et al. , The capsid binder Vapendavir and the novel protease inhibitor SG85 inhibit enterovirus 71 replication. Antimicrob. Agents Chemother. 58, 6990–6992 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Badger J., et al. , Structural analysis of a series of antiviral agents complexed with human rhinovirus 14. Proc. Natl. Acad. Sci. U.S.A. 85, 3304–3308 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Makarov V. A., et al. , Pyrazolopyrimidines: Potent inhibitors targeting the capsid of rhino- and enteroviruses. ChemMedChem 10, 1629–1634 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y., et al. , Structural and virological studies of the stages of virus replication that are affected by antirhinovirus compounds. J. Virol. 78, 11061–11069 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andries K., et al. , In vitro activity of pirodavir (R 77975), a substituted phenoxy-pyridazinamine with broad-spectrum antipicornaviral activity. Antimicrob. Agents Chemother. 36, 100–107 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pevear D. C., et al. , Conformational change in the floor of the human rhinovirus canyon blocks adsorption to HeLa cell receptors. J. Virol. 63, 2002–2007 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shepard D. A., Heinz B. A., Rueckert R. R., WIN 52035-2 inhibits both attachment and eclipse of human rhinovirus 14. J. Virol. 67, 2245–2254 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muckelbauer J. K., et al. , The structure of coxsackievirus B3 at 3.5 A resolution. Structure 3, 653–667 (1995). [DOI] [PubMed] [Google Scholar]
- 30.Ledford R. M., et al. , VP1 sequencing of all human rhinovirus serotypes: Insights into genus phylogeny and susceptibility to antiviral capsid-binding compounds. J. Virol. 78, 3663–3674 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pevear D. C., et al. , Relationship of pleconaril susceptibility and clinical outcomes in treatment of common colds caused by rhinoviruses. Antimicrob. Agents Chemother. 49, 4492–4499 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmidtke M., et al. , Susceptibility of coxsackievirus B3 laboratory strains and clinical isolates to the capsid function inhibitor pleconaril: Antiviral studies with virus chimeras demonstrate the crucial role of amino acid 1092 in treatment. J. Antimicrob. Chemother. 56, 648–656 (2005). [DOI] [PubMed] [Google Scholar]
- 33.Heinz B. A., et al. , Genetic and molecular analyses of spontaneous mutants of human rhinovirus 14 that are resistant to an antiviral compound. J. Virol. 63, 2476–2485 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ledford R. M., Collett M. S., Pevear D. C., Insights into the genetic basis for natural phenotypic resistance of human rhinoviruses to pleconaril. Antiviral Res. 68, 135–138 (2005). [DOI] [PubMed] [Google Scholar]
- 35.Sun L., et al. , Antiviral activity of broad-spectrum and enterovirus-specific inhibitors against clinical isolates of enterovirus D68. Antimicrob. Agents Chemother. 59, 7782–7785 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmidtke M., Wutzler P., Zieger R., Riabova O. B., Makarov V. A., New pleconaril and [(biphenyloxy)propyl]isoxazole derivatives with substitutions in the central ring exhibit antiviral activity against pleconaril-resistant coxsackievirus B3. Antiviral Res. 81, 56–63 (2009). [DOI] [PubMed] [Google Scholar]
- 37.Braun H., et al. , Molecular mechanism of a specific capsid binder resistance caused by mutations outside the binding pocket. Antiviral Res. 123, 138–145 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Weiss V. U., et al. , Capillary electrophoresis, gas-phase electrophoretic mobility molecular analysis, and electron microscopy: Effective tools for quality assessment and basic rhinovirus research. Methods Mol. Biol. 1221, 101–128 (2015). [DOI] [PubMed] [Google Scholar]
- 39.Reisdorph N., et al. , Human rhinovirus capsid dynamics is controlled by canyon flexibility. Virology 314, 34–44 (2003). [DOI] [PubMed] [Google Scholar]
- 40.Gruenberger M., Pevear D., Diana G. D., Kuechler E., Blaas D., Stabilization of human rhinovirus serotype 2 against pH-induced conformational change by antiviral compounds. J. Gen. Virol. 72, 431–433 (1991). [DOI] [PubMed] [Google Scholar]
- 41.Dewindt B., van Eemeren K., Andries K., Antiviral capsid-binding compounds can inhibit the adsorption of minor receptor rhinoviruses. Antiviral Res. 25, 67–72 (1994). [DOI] [PubMed] [Google Scholar]
- 42.Garriga D., et al. , Insights into minor group rhinovirus uncoating: The X-ray structure of the HRV2 empty capsid. PLoS Pathog. 8, e1002473 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Okun V. M., Nizet S., Blaas D., Kenndler E., Kinetics of thermal denaturation of human rhinoviruses in the presence of anti-viral capsid binders analyzed by capillary electrophoresis. Electrophoresis 23, 896–902 (2002). [DOI] [PubMed] [Google Scholar]
- 44.Korant B. D., Lonberg-Holm K., Noble J., Stasny J. T., Naturally occurring and artificially produced components of three rhinoviruses. Virology 48, 71–86 (1972). [DOI] [PubMed] [Google Scholar]
- 45.Lonberg-Holm K., Yin F. H., Antigenic determinants of infective and inactivated human rhinovirus type 2. J. Virol. 12, 114–123 (1973). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kotecha A., et al. , Application of the thermofluor PaSTRy technique for improving foot-and-mouth disease virus vaccine formulation. J. Gen. Virol. 97, 1557–1565 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lacroix C., et al. , In vitro characterisation of a pleconaril/pirodavir-like compound with potent activity against rhinoviruses. Virol. J. 12, 106 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kaiser L., Crump C. E., Hayden F. G., In vitro activity of pleconaril and AG7088 against selected serotypes and clinical isolates of human rhinoviruses. Antiviral Res. 47, 215–220 (2000). [DOI] [PubMed] [Google Scholar]
- 49.Verdaguer N., Blaas D., Fita I., Structure of human rhinovirus serotype 2 (HRV2). J. Mol. Biol. 300, 1179–1194 (2000). [DOI] [PubMed] [Google Scholar]
- 50.Wald J., Goessweiner-Mohr N., Blaas D., Pasin M., Cryo-EM structure of rhinovirus-B5 complexed to antiviral OBR-5-340. Protein Data Bank. https://www.rcsb.org/structure/6SK5. Deposited 14 August 2019. [DOI] [PMC free article] [PubMed]
- 51.Wald J., Goessweiner-Mohr N., Blaas D., Pasin M., Cryo-EM structure of rhinovirus-B5 complexed to antiviral OBR-5-340. Electron Microscopy Data Bank. http://www.ebi.ac.uk/pdbe/entry/emdb/EMD-10220. Deposited 14 August 2019.
- 52.Wald J., Goessweiner-Mohr N., Blaas D., Pasin M., Cryo-EM structure of rhinovirus-B5. Protein Data Bank. https://www.rcsb.org/structure/6SK6. Deposited 14 August 2019.
- 53.Wald J., Goessweiner-Mohr N., Blaas D., Pasin M., Cryo-EM structure of rhinovirus-B5. Electron Microscopy Data Bank. http://www.ebi.ac.uk/pdbe/entry/emdb/EMD-10221. Deposited 14 August 2019.
- 54.Hendry E., et al. , The crystal structure of coxsackievirus A9: New insights into the uncoating mechanisms of enteroviruses. Structure 7, 1527–1538 (1999). [DOI] [PubMed] [Google Scholar]
- 55.Wald J., Goessweiner-Mohr N., Blaas D., Pasin M., Cryo-EM structure of rhinovirus-A89. Protein Data Bank. https://www.rcsb.org/structure/6SK7. Deposited 14 August 2019.
- 56.Wald J., Goessweiner-Mohr N., Blaas D., Pasin M., Cryo-EM structure of rhinovirus-A89. Electron Microscopy Data Bank. http://www.ebi.ac.uk/pdbe/entry/emdb/EMD-10222. Deposited 14 August 2019.
- 57.Scheres S. H., RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Adams P. D., et al. , PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pettersen E. F., et al. , UCSF Chimera: A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.