Significance
Constant exposure of aquatic plants to freely exchangeable nutrients and pathogenic microbes requires regulation of gene expression different to land plants. However, short-read sequencing platforms fail to provide vital information that comprises genes involved in the response to such a challenge. Here, we applied long-read sequencing to retrieve missing sequences to the duckweed species of Spirodela polyrhiza. Evolution of the genetic network and root morphology show that roots play a function as sea anchors rather than nutrient uptake. Moreover, disease-resistance gene clusters are constitutively active whereas they are silenced by phasiRNA in land plants.
Keywords: long reads, root evolution, disease resistance, tandem duplication, aquatic adaptation
Abstract
Aquatic plants have to adapt to the environments distinct from where land plants grow. A critical aspect of adaptation is the dynamics of sequence repeats, not resolved in older sequencing platforms due to incomplete and fragmented genome assemblies from short reads. Therefore, we used PacBio long-read sequencing of the Spirodela polyrhiza genome, reaching a 44-fold increase of contiguity with an N50 (a median of contig lengths) of 831 kb and filling 95.4% of gaps left from the previous version. Reconstruction of repeat regions indicates that sequentially nested long terminal repeat (LTR) retrotranspositions occur early in monocot evolution, featured with both prokaryote-like gene-rich regions and eukaryotic repeat islands. Protein-coding genes are reduced to 18,708 gene models supported by 492,435 high-quality full-length PacBio complementary DNA (cDNA) sequences. Different from land plants, the primitive architecture of Spirodela’s adventitious roots and lack of lateral roots and root hairs are consistent with dispensable functions of nutrient absorption. Disease-resistant genes encoding antimicrobial peptides and dirigent proteins are expanded by tandem duplications. Remarkably, disease-resistant genes are not only amplified, but also highly expressed, consistent with low levels of 24-nucleotide (nt) small interfering RNA (siRNA) that silence the immune system of land plants, thereby protecting Spirodela against a wide spectrum of pathogens and pests. The long-read sequence information not only sheds light on plant evolution and adaptation to the environment, but also facilitates applications in bioenergy and phytoremediation.
Greater duckweeds, i.e., Spirodela polyrhiza, are small and fast-growing aquatic plants, belonging to the early-diverging monocot order of the Alismatales (1) (SI Appendix, Fig. S1). As one of the most yielding biomass species, they can be found from temperate to tropical regions, surviving in freshwater ponds and animal waste lagoons. The plant is composed of a tiny frond and a few adventitious roots (ARs), providing unique material to study root evolution at early angiosperm lineages. Two other genera of Lemnaceae (Wolffiella and Wolffia) appear to lack roots, based on close morphological and microscopic inspections (2). Because of larger fronds of the genus Spirodela, sticky roots perhaps help maintain their upright position in the water and promote vegetative dispersal with the attachment to animals, rather than act primarily as an organ for the uptake of water and nutrients (3). On the other hand, the presence of water brings the fronds into immediate contact with a diverse population of microbes. Interestingly, the duckweed crude extract exhibited antimicrobial resistance toward waterborne fungi and bacteria (4, 5), which could be a potential source for medicinal herbs and new antimicrobial therapeutics. To understand the molecular nature of these properties, a comprehensive repertoire of genes is essential, not available with older sequencing platforms. Due to larger gaps from short-read (6) sequence assembly, missing information could be critical for understanding the molecular mechanism of root formation and defenses against diseases. The previous assembly of the Spirodela genome was based on a read length of ∼350 base pairs (bp) (7), too short to assess critical properties of plant aquatic adaptation. Indeed, the assembly included more than 16,055 small contigs and 11.8% of missing sequences, limiting access to total gene content. Here, we were able to extend read length to more than 10 kilobases (kb), using PacBio long-read sequencing. We filled 95.4% of preexisting gaps, and, with PacBio sequencing of full-length complementary DNA (cDNA), we could identify 18,708 protein-coding genes, many of which were larger than previously thought. In particular, the expansion of disease-resistant gene families by tandem duplications can explain the innate immunity and worldwide distribution of the species. Furthermore, new sequence information gives us new insights into the minimal morphological requirements of a higher plant and its environmental adaptation.
Results and Discussion
Genome Sequencing, Assembly, and Annotation.
A clone of S. polyrhiza 7498 from North Carolina, United States, had been subjected to physical mapping and whole-genome shotgun sequencing using the “454” platform (7). Because of the short reads (∼350 bp) of the “454” system, the genome of Spirodela (Sp7498V2) was highly fragmented and could not resolve complex regions, leading to genome misassembly or misannotations. Although shotgun DNA sequencing (8) facilitated the assembly of a complete genome (9), the presence of tandemly repeated sequences, like gene copies, transposable elements, or satellite repeats of large genomes, prevents earlier developed sequencing platforms from delivering contiguous sequence information. PacBio with long reads (>10 kb) can cover sequence junctions between single repeats, such as retrotransposons, and can overcome assembly problems in terms of gap filling and repeat reconstruction in many plants, such as maize (10, 11), Oropetium (12), quinoa (13), and broomcorn millet (14). To this end, we resequenced S. polyrhiza 7498 with ∼126-fold coverage of PacBio long reads, resulting in 411 contigs and an N50 length (a median of contig lengths) of 831 kb (15) (SI Appendix, Figs. S2 and S3 and Tables S1 and S2), which improved contiguous sequence information 44 times compared with the previous version (Table 1) (7). The new genome assembly version, named Sp7498V3, presents the highest contiguity and the fewest gaps compared to another ecotype, S. polyrhiza 9509 (16), and to Lemna minor (17) assembled from Illumina short reads. The older version, Sp7498V2, had 13,459 gaps, with 11.8% unknown sequences, whereas Sp7498V3 had only 270 gaps, leaving only 4.6% of missing sequences (Fig. 1 and SI Appendix, Fig. S4 and Table S3).
Table 1.
Comparison of genome assembly of Spirodela from short reads and long reads
| Parameters | Sp7498V2 | Sp7498V3 |
| Platform | ABI3730 and 454 | PacBio |
| Sequencing depth | 21x | 126x |
| Genome size, Mb | 145 | 138 |
| Contig number | 16,055 | 411 |
| Contig N50, kb | 18.9 | 831 |
| Scaffold number | 1,071 | 227 |
| Scaffold N50, Mb | 3.8 | 3.3 |
| Gap, % | 11.8 | 4.6 |
| Repeat, % | 17.3 | 30.91 |
| Protein coding genes | 19,623 | 18,708 |
| Mean gene length, bp | 3,458 | 4,342 |
| Mean CDS length, bp | 1,108 | 1,217 |
| Mean exon per gene | 4 | 5.7 |
| Mean exon length, bp | 213 | 281 |
| Mean intron length, bp | 560 | 585 |
The Sp7498V2 genome was assembled and annotated from short reads while Sp7498V3 was generated by PacBio long-read sequencing.
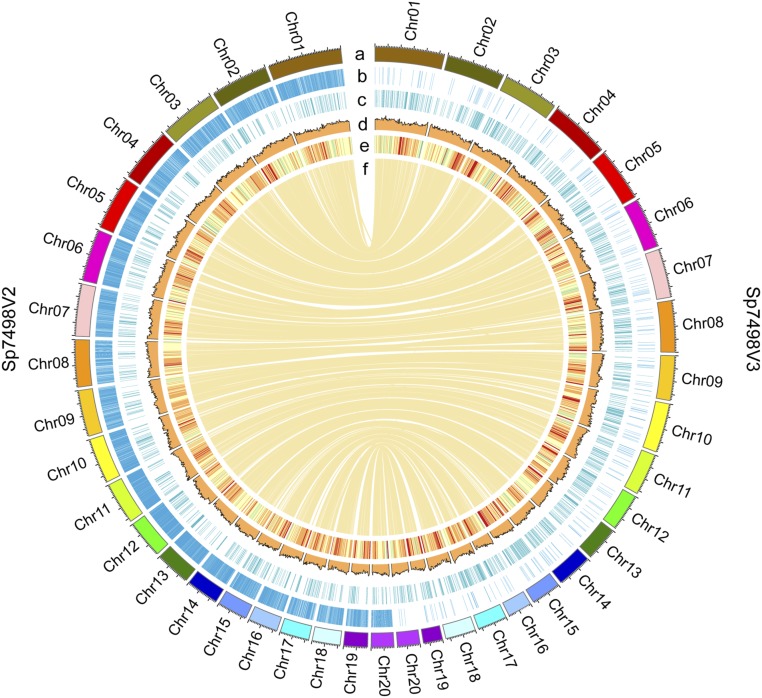
Fig. 1.
Comparison of genome assembly from short reads and long reads. From outside to inside, the circles represent karyotype (a), sequence gaps (b), GC content (c), full-length LTRs (d), gene density (e), and syntenous connections (f). The metrics are calculated in 1-Mb sliding windows. The right half circle represents genome assembly from long reads (Sp7498V3). The left half circle represents genome assembly from short reads (Sp7498V2). Every blue vertical bar indicates one gap in layer b. There are 270 gaps in Sp7498V3 and 13,459 gaps in Sp7498V2. The inner lines denote the synteny of two versions of genomes. Chr, chromosome.
Combining bacterial artificial chromosome (BAC) end sequences (BESs), 411 contigs were placed into 227 scaffolds with an N50 of 3.3 Mb (Table 1), merged into 20 chromosomes with the aid of FISH (18) (Fig. 1 and SI Appendix, Table S3). Still, there were certain contigs equal to 7.1 Mb unassigned to any chromosome. The quality and integrity of chromosomes were confirmed with the physical map and 20 fully sequenced 40-kb fosmids (Fig. 1) (7). The fosmids matched the Spirodela genome assembly (Sp7498V3) with more than 98.6% sequence identity, all within a single contig from the long-read assembly (SI Appendix, Table S4).
A total of 545,753 full-length transcript sequences were generated by PacBio isoform sequencing (19) (SI Appendix, Table S5), which resulted in 492,435 high-quality consensus sequences. The annotation of the Spirodela genome was improved in combination with de novo prediction and cDNA data, leading to a level of 74.6% of 18,708 protein-coding genes supported by full-length transcripts. The number for predicted gene models is slightly lower (4.66%) than those from Sp7498V2 (19,623), mainly due to the merger of gene fragments. For example, the genes of Spo013477, Spo003046, and Spo000617 in Sp7498V3 were concatenated from 4, 3, and 2 genes in Sp7498V2, respectively (SI Appendix, Fig. S5). The average gene length was significantly increased by 24.5% from 3,458 to 4,342 bp, with longer and higher exon numbers for each gene (Table 1). BUSCO (Benchmarking Universal Single-Copy Orthologs) evaluation revealed that Sp7498V3 had a higher rate (86%) of complete homologous proteins, compared with 79% of Sp7498V2 (SI Appendix, Table S6), indicating a high-quality assembly and annotation of Sp7498V3.
Repetitive elements play important roles in shaping genome architecture and gene regulation. Most repeats were incomplete, unassembled, or highly collapsed in Sp7498V2 due to short-read assembly. The improved genome of Sp7498V3 allowed us to accurately survey its completeness of repetitive features. We found that repetitive sequences account for 30.91% of the genome (SI Appendix, Fig. S6 and Table S7), 18.61% of which were long terminal repeat (LTR) retrotransposons that arose before 2 million years ago (MYA) and were species-specific (SI Appendix, Fig. S7 and Table S8). A total of 156 retrotranspositions occurred on top of each other (nested versions) whereas 1,544 intact LTR retrotransposons were present in Sp7498V3, which was comparable to that of only 722 in Sp7498V2. A close view of scaffold 15 illustrated that genes and transposable elements preferentially were located in individual islands, resembling both prokaryotic and eukaryotic features in Spirodela chromosomes (Fig. 2).
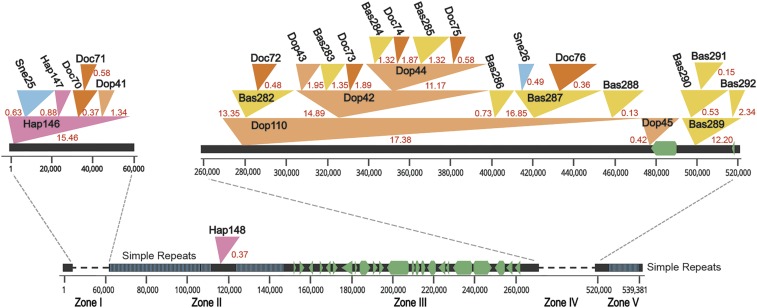
Fig. 2.
A benchmark of the nested LTRs in Spirodela. Scaffold 15 with contiguous sequence length of 539 kb exhibits regions of TE islands and a gene cluster. Zone I and IV are nested with LTR regions. Red numbers next to LTRs represent insertion ages in million years ago (MYA). Retrotransposons are indicated by filled triangles of different colors. Zone II and Zone V are simple repeat regions. Simple repeats are represented by blue vertical lines. Zone III contains 23 genes without any transposon interruption. The genes are shown as green pentagons or triangles. Genes and LTRs have been drawn to scale. Hap, Doc, Sne, Sle, Dop, and Bas indicate different types of LTR retrotransposons.
Comparative Genomics.
The identification of syntenic regions between the Spirodela genome itself revealed two rounds of whole genome duplications (WGDs), as expected (SI Appendix, Fig. S8) (7). The dot plot map showed that more syntenic segmental blocks were retrieved in Sp7498V3 than Sp7498V2 (SI Appendix, Fig. S8), indicating that the improved genome assembly uncovered more intragenomic collinearity. Conserved single copy protein sequences were aligned to form a divergence tree for 9 species [Zostera marina (20), Phoenix dactylifera (21), Ananas comosus (22), Oryza sativa (23), Sorghum bicolor (24), Zea mays (10), Arabidopsis thaliana (25), Solanum lycopersicum (26), and S. polyrhiza]. Spirodela and Zostera were clustered together as sister clades compared with all other monocots. They diverged between 118.5 and 129.0 million years ago (MYA). Spirodela and other terrestrial monocots were separated somewhere between 130.4 and 140.4 MYA (SI Appendix, Fig. S9). The tree could help us to understand how plants adapt to different living media, including sea zone, freshwater area, and terrestrial land with a spectrum of sequenced genomes.
One answer to such a question comes from the comparison of gene families of Spirodela, Zostera, Zea, Oryza, and Arabidopsis. There were 7,647 gene families shared by all 5 species whereas 674 families were specifically lost in Zostera and present in the other 4 species (SI Appendix, Fig. S10). Gene ontology (GO) analysis showed significant enrichment for positive regulation of stomatal complex development, response to ultraviolet (UV), starch catabolic process, regulation of DNA repair, glyoxylate cycle, tryptophan biosynthetic process, gamma-tubulin complex localization, and regulation of proteasomal ubiquitin-dependent protein catabolic process. The lost functions were consistent with that Zostera need not to cope with intense UV radiation and not to exchange gas with stomata under marine conditions (20). In contrast, there were 420 gene families that were lost in Spirodela and Zostera, in comparison with rice, maize, and Arabidopsis. GO enrichment analysis indicated that the biosynthetic and metabolic processes of secondary metabolites, including sesquiterpene, terpene, terpenoid, and isoprenoid, were overrepresented in these species. These secondary metabolites are a large and diverse class of naturally occurring organic chemicals, playing a key role as precursors to overcome gravity and dry land (27). We hypothesize that Spirodela and Zostera do not require complex metabolites to cope with variable temperature, light, and nutrients under relatively stable aquatic environments.
Evolution of Root Development.
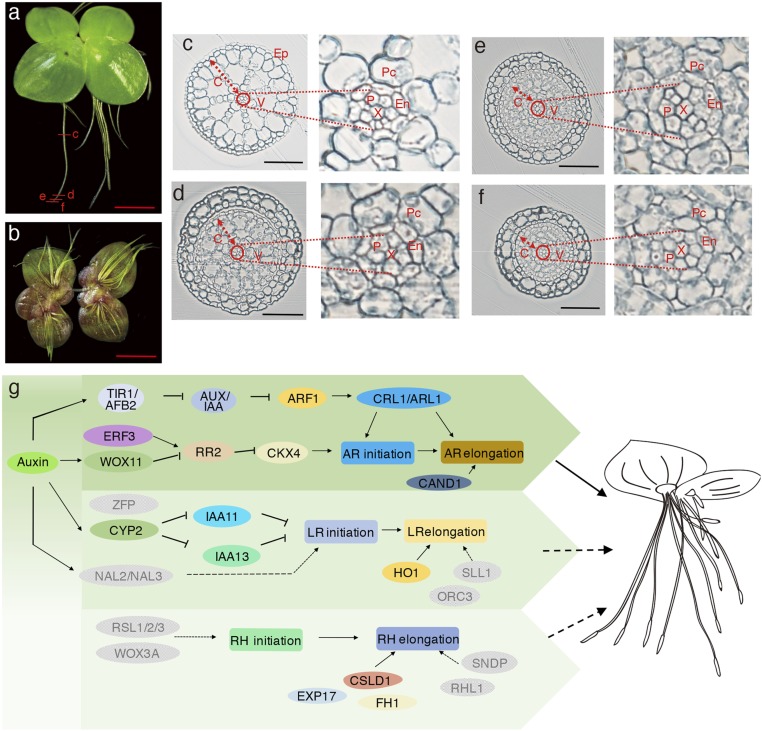
A major difference between land and aquatic plants is the intake pathway of water and nutrients. The Spirodela sticky root system appears to be more critical for securing an anchoring position and promoting a wide distribution rather than absorbing nutrients. It was reported that the lower surface of the frond served as the main organ for nutrient uptake (2). The attachment of roots to animals and birds allows duckweeds to be transported to a distant location, thus aiding geographic dispersal without human intervention (28). When the lower surface was painted with waterproof lanolin, duckweeds grew more slowly than control plants (http://www.missouribotanicalgarden.org). As we cut the roots from mother fronds, Spirodela continued to grow by producing daughter fronds after 3 d, keeping pace with the control plants bearing intact roots (SI Appendix, Fig. S11). Spirodela has as many as 12 adventitious roots (ARs) per frond but lacks secondary lateral roots (LRs) and root hairs (RHs) (Fig. 3 A and B). In addition, the genera of Wolffiella and Wolffia do not even have any roots (2). ARs, usually derived from shoots, stems, or leaves, are prevalent in monocots, which is different from eudicots containing lateral roots (LRs) (29). We examined the root structures and observed epidermis, cortex, and vascular tissue, but no root hairs (RHs) in the cross-section of ARs (Fig. 3 C–F). The epidermis is the outermost boundary. The cortex is loosely packed in parenchymatous cells with large intercellular air spaces, allowing gaseous exchange and providing buoyancy. The vascular tissue is highly primitive in Spirodela, with a tracheary element in the middle surrounded by a ring of phloem tissue (Fig. 3 C–F). The simple architecture is consistent with its function of maintaining the stability of the plant body.
Fig. 3.
Anatomy of the Spirodela root. (A). A dorsal overview of a Spirodela plant, showing the location of cross-sections. (Scale bar: 500 μm.) The cross-sections were sampled at 8 (c), 1.5 (d), 0.5 (e), and 0.2 mm (f) from the root tip, corresponding to Fig. 3 C–F, respectively. (B). A ventral overview of a Spirodela plant exhibits as many as 12 adventitious roots. (Scale bar: 500 μm.) (C–F) Cross-sections of Spirodela roots, illustrating the structures of the epidermis (Ep), cortex (C), endodermis (En), and vascular tissue (V). A close investigation shows vascular tissues. The central cell (X), located in the middle, is a tracheary element, which is surrounded by a ring of phloem tissue (P). The endodermis (En) is between the vascular tissue and the cortex parenchyma cells (Pc). (Scale bars: 50 μm.) (G) The genetic pathways involved in adventitious root (AR), lateral root (LR), and root hair (RH) development. The homologous genes in Spirodela are defined by using the rice protein sequences.
The layers of pith, conjunctive tissue, xylem (protoxylem and metaxylem), and phloem (protophloem and metaphloem) have been investigated in roots of rice, which are required to provide mechanical support and to improve water and nutrient transportation over a short or long distance (30). Therefore, we used rice protein sequences involved in AR root development to search for homologous genes in Spirodela. Spirodela shared all of the genes with rice for AR’s initiation and elongation (Fig. 3G), supporting a conserved mechanism of AR evolution as early as in early-diverging monocots. But, comparably, Spirodela had lost gene members associated with LR initiation (ZFP, NAL2, and NAL3), as well as with LR elongation (ORC3 and SLL1). Genes responsible for RH development (RSL, WOX3A, SNDP, and RHL1) were also absent, resulting in an incomplete RH pathway. The loss of lateral roots and root hairs is consistent with the reduced functions of nutrient uptake in duckweeds but would not significantly affect their function as a sea anchor, as well as helping vegetative dispersal.
Disease-Resistant Genes in Tandem Duplications.
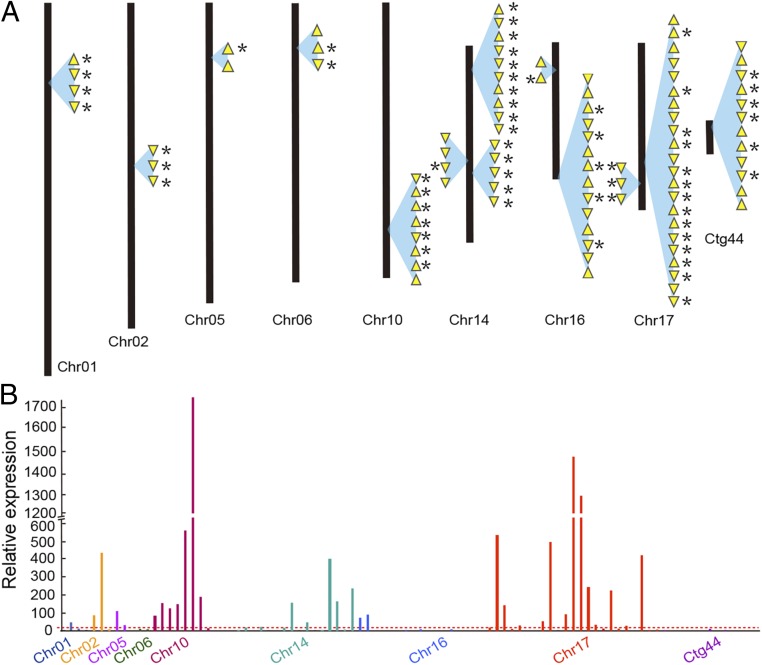
Whole-genome shotgun DNA sequencing based on long reads is critical for assemblies of tandemly repeated gene copies (31). In Spirodela, this became evident because disease-resistant genes are mostly enriched in tandem duplications, including gene families encoding antimicrobial peptides and dirigent proteins (SI Appendix, Fig. S12). Plant antimicrobial peptides (AMPs) are small defense proteins that constitute a first-line protection against pathogens (32). Compared with 46 antimicrobial peptides (AMPs) in maize (33), a total of 108 AMP members were found in Spirodela, which included many families, such as defensin, snaking, lipid transfer protein, hevein, and cyclotide, among which the cyclotide family was the most dominant (SI Appendix, Fig. S13). It is worth noting that 92 out of 108 AMP genes were tandemly clustered with up to 22 copies in Spirodela (Fig. 4A), compared to the array of two copies at most in maize (33). Dirigent proteins are also reported to be involved in defense response against fungi and insects (34). They were also induced by abiotic stress, such as drought, low temperature, and abscisic acid (35). There were 23 gene copies with the largest cluster of 12 tandem duplications in the assembled genome (SI Appendix, Fig. S12). The tandemly duplicated genes generally maintain a similar function due to their adjacent location and the sharing of the same regulatory elements (36), which is different from dispersed gene copies that tend to evolve novel functions. Thus, gene amplification affects gene dosage and correlates with a higher level of gene expression (37). Here, 62 out of 92 AMP genes were supported by full-length cDNA sequences (Fig. 4A), and 36 ones were expressed higher than the medium level (Fig. 4B), compared with all expressed genes from RNA-Seq analysis (38), indicating their constitutive expression to maintaining their active pathogen-resistant ability. Different from land plants, where expression of disease resistance genes is inducible by exposure to pathogens, aquatic plants are in constant contact with a diverse population of microorganisms. This adaptation is also consistent with the low level of the 24-nt siRNAs, possibly due to reduced expression of DCL3 and PolIV in Spirodela fronds (Table 2), which are known to guide DNA methylation and be involved in the silencing of repeat sequences like retrotransposons and tandemly repeated disease-resistant genes in angiosperms (39, 40). Because of the neotenous growth of Spirodela, there is less of a need to guard against retrotransposition during meiosis than in land plants, but, as we can see here, also for the repression of its immune system.
Fig. 4.
Genomic distribution and transcriptomic expression of tandemly duplicated disease-resistant genes. (A) Disease-resistant gene copies at each locus in the genome are illustrated as yellow arrowheads. The yellow arrowheads indicate the gene coding direction. The sign of * next to the yellow arrowhead indicates that the gene is supported by full-length cDNA evidence. Contig 44 (Ctg44) is one of contigs that could not be incorporated into chromosomes with current sequence data. (B) Gene expression level is represented in the y axis with the value of fragments per kilobase of transcript per million mapped reads (FPKM) analyzed from RNA-Seq. The horizontal dotted line shows the average gene expression of total expressed genes. The bars are labeled in different colors based on chromosomes.
Table 2.
Gene expression in the pathway of RNA-directed DNA methylation (RdDM)
| Description/gene | Arabidopsis | Leaf | Spirodela | Frond |
| RdDM | ||||
| AGO4* | AT2G27040 | 9.81 | Spo000559 | 0.00 |
| AGO6* | AT2G32940 | 0.77 | Spo000559 | 0.00 |
| AGO9 | AT5G21150 | 0.00 | Spo000559 | 0.00 |
| CLSY1* | AT3G42670 | 1.31 | Spo009900 | 0.00 |
| DCL3* | AT3G43920 | 1.37 | Spo005963 | 0.42 |
| DMS1* | AT2G16390 | 2.54 | Spo009900 | 0.00 |
| DMS4* | AT2G30280 | 6.52 | Spo000389 | 1.91 |
| DRM2* | AT5G14620 | 4.90 | Spo007386 | 2.30 |
| HEN1 | AT4G20910 | 2.79 | Spo016688 | 2.99 |
| KTF1* | AT5G04290 | 7.51 | Spo001256 | 0.27 |
| RDM1* | AT3G22680 | 1.91 | Spo004351 | 0.14 |
| RDR2* | AT4G11130 | 0.94 | Spo003647 | 0.24 |
| RNAP IV subunit 1* | AT1G63020 | 1.60 | Spo003863 | 0.33 |
| RNAP IV subunit 7 | AT3G22900 | 1.39 | Spo009872 | 1.65 |
| RNAP V subunit 1* | AT2G40030 | 1.02 | Spo004499 | 0.08 |
| RNAP V subunit 5A* | AT3G57080 | 7.62 | Spo008450 | 0.20 |
| RNAP V subunit 5C | AT3G54490 | 0.20 | Spo008450 | 0.20 |
| RNAP V subunit 7 | AT4G14660 | 2.59 | Spo009872 | 1.65 |
| SHH1 | AT1G15215 | 1.77 | Spo018440 | 10.68 |
| SUVH2* | AT2G33290 | 2.29 | Spo017635 | 0.01 |
| SUVH4* | AT5G13960 | 2.64 | Spo001865 | 0.70 |
| SUVH5 | AT2G35160 | 0.40 | Spo006758 | 1.27 |
| SUVH6* | AT2G22740 | 9.35 | Spo006758 | 1.27 |
| SUVH9* | AT4G13460 | 14.40 | Spo017635 | 0.01 |
| Control | ||||
| Actin2 | AT3G18780 | 460.25 | Spo017354 | 241.30 |
| Ubiquitin | AT5G25760 | 25.11 | Spo014772 | 18.86 |
The gene expression value was normalized in FPKM. AGO, Argonaute; CLSY, CLASSY; DCL, Dicer-like; DMS, defective in meristem silencing; HEN, HUA enhancer; KTF, KOW domain-containing transcription factor; RDM, RNA-directed DNA methylation; RDR, RNA-dependent RNA polymerase; RNAP, RNA polymerase; SHH, SAWADEE homeodomain homolog; SUVH, SUVAR homolog. RNA-Seq for Arabidopsis (accession no. GSM3120107) (61) and for Spirodela (accession no. PRJNA557001).
Indicates that gene expression was significantly reduced in Spirodela compared with Arabidopsis.
Our study illustrates how sequencing with long-read technology can resolve complex repeat regions and gene loci with tandem duplications. Such regions contain critical genomic information that is relevant for the evolution of species and their adaptation to their growth environment, thus helping improve crops through genomic breeding approaches (41). Structural and physiological adaptations to fresh waters learned from this research effort might accelerate potential applications of duckweed in bioreactors, bioremediation, and biofuel.
Methods
Whole-Genome Shotgun PacBio Sequencing.
S. polyrhiza 7498 was grown in SH medium under 16 h light/8 h dark at 28 °C for 7 d. More than 50 μg of genomic DNA was prepared from S. polyrhiza 7498. The SMRTbell library with 20-kb insert was constructed with BluePippin size selection (Sage Science). The resulting SMRTbell templates were sequenced with four SMRT Cells of the PacBio Sequel platform (Pacific Biosciences, Frasergen, Wuhan, China).
PacBio Isoform Sequencing.
S. polyrhiza 7498 was treated by multiple conditions, as follows: 37 °C, 0 °C, desiccation, pH value of 9, UV exposure, 20 mg/L CuCl2, 300 mg/L KNO3, 250 nM ABA, 10 mM kinetin, and 300 mM mannitol. Total RNA was isolated using TRIzol reagent (Invitrogen) and purified with the RNeasy Mini kit after DNase I digestion (Qiagen). RNA was pooled with identical quantities and then subjected to PacBio isoform sequencing (Iso-Seq) using a Clontech SMARTer PCR cDNA synthesis Kit (Clontech). cDNA was size-selected for library construction of 1 to 2 kb, 2 to 3 kb, 3 to 6 kb, and 5 to 10 kb. The templates were sequenced by using P6 polymerase and C4 chemistry. The downstream Iso-Seq analysis was processed by using SMRT-Analysis software v2.3.0 (Pacific Biosciences), including full-length cDNA identification, isoform-level clustering, and final consensus corrections.
De Novo Genome Assembly.
The PacBio raw reads were corrected to Preads by Falcon, and then Preads longer than 8 kb were assembled to contigs by using two assemblers, Falcon (42) and Canu (43), with optimized parameters. The short Preads smaller than 8 kb were applied to fill gaps. The scaffolds were created by mapping BAC-end sequencing (BES) data to the contigs. The scaffolds were ordered into 20-chromosome-level pseudomolecules after the integration of the last Spirodela genome version (Sp7498V2) (7), resulting in the final version of Sp7498V3.
Transposable Elements Prediction.
De novo repeat identification was conducted by Repeat Modeler (http://www.RepeatMasker.org) and Repeat Masker (44). Long terminal repeat (LTR) retrotransposons were predicted by LTRdigest (45) and LTRharvest (46). Helitrons were predicted by default parameters in HelitronScanner (47). Short interspersed nuclear elements (SINEs) were defined by SINE-finder (48). Terminal inverted repeats were predicted from the pipeline TARGeT (49). The LTR integration time was estimated based on the sequence similarity of 3′ LTR and 5′ LTR aligned with MAFFT (50). Kimura parameter distances (K) between 5′ LTR and 3′ LTR repeats of each LTR element were calculated with the EMBOSS program (51). The divergence time (T) of intact LTR retrotransposons was calculated using the formula T = K/2r (r is the neutral substitution rate of 1.3 × 10−8 substitutions per site per million years) (52).
Annotation.
The structural annotation for protein-coding genes was based on de novo prediction, homologous alignment, and full-length cDNA sequences from PacBio isoform sequencing. The programs of Augustus and GlimmerHMM were utilized to ab initio predict gene loci and structures on the repeat-masked genome, with parameters trained from a set of high-quality proteins and full-length cDNAs, which were manually curated. The protein sets were collected and chosen as homology-based evidence from species of: Z. marina (ORCAE v. 2.1) (20), O. sativa (Phytozome v. 9.0) (53), Z. mays (Phytozome v. 9.0) (10), and A. thaliana (TAIR10) (54). A number of 492,435 high-quality full-length cDNAs generated by isoform sequencing from Spirodela multiple tissues were used as transcript-based evidence, as well as transcripts assembled from RNA-Seq reads. The pipeline of MAKER was used to combine all evidence to generate nonredundant gene models (55). The functional annotation was assigned by using a sequence homology search. The derived Gene Ontology terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway were assigned with the alignment against the databases of InterPro, GO, KEGG, Swiss-Prot, TrEMBL, and NR (56). The completeness of genome assembly and annotation was assessed by using the analysis of Benchmarking Universal Single-Copy Orthologs (BUSCO). The syntenic dot plot was generated by using the CoGe platform to perform comparative genomics. The web-based tool of SynMap was used to compare the Spirodela genome to itself to identify syntenic regions with the default blast parameters and an e-value of 0.001 (57). Following an “all-versus-all” BlastP alignment for the selected species with the e-value of 1e−5, identity of 70%, and coverage of 50%, OrthoMCL was used to identify orthologous groups between and within species applying a Markov Cluster algorithm (58).
Phylogenetic Tree.
All single-copy orthologs with a minimal length of 100 amino acids across the selected genomes were extracted and aligned using MAFFT (v7.221). The alignment was transformed into a supergene alignment with phy format by a custom perl script. A phylogenetic tree was constructed using RAxML (v8.0.26) Maximum Likelihood analysis with a high bootstrap of 100. The divergence time was estimated by r8s based on the topology of the phylogenic tree with the combination of known divergence time (http://www.timetree.org). To further study gene family expansion or contraction, the sizes of all gene families (excluding orphans and species-specific families) were calculated using CAFÉ (v4.2).
Tandem Duplicated Genes and Homologous Gene Search.
Tandem duplicated genes were detected by MCscanX with the similarity search of BlastP (e-value < 1e−10) (59). The dedicated searches of homologous genes involved in the initiation and elongation of adventitious roots, lateral roots, and root hairs were performed. Putative orthologs were defined using BlastP, with the query chosen from documented pathways in Oryza (29, 60) against the database of Spirodela proteins.
Histological Analysis of Spirodela Roots.
Whole roots were fixed with acetic acid:ethanol (1:3 vol:vol) for 24 h, followed with a serial ethanol dehydration (70, 80, 90, 95, and 100%) at room temperature for 0.5 to 1 h at each step. Samples were embedded in Technovit 7100 resin and cut at a thickness of 8 µm. The slides were dried at 65 °C and observed under a microscope (Nikon).
Data Availability.
Genome assembly and consensus sequences were deposited into NCBI GenBank with an accession ID of SWLF00000000. Full-length cDNA sequences were derived by error correction of multiple reads, resulting in 492,435 high-quality isoforms that were uploaded with the ID of SRX5321175. RNA-Seq was deposited at GenBank BioProject of PRJNA557001.
Supplementary Material
Acknowledgments
We thank Zehua Liu, Lu Wang, Shijun Xiao, and Xiaofei Zeng (Frasergen) for bioinformatic analysis. The project was supported by National Natural Science Foundation of China Grant 31670366 (to W.W.) and the Waksman Chair in Molecular Genetics (J.M.).
Footnotes
The authors declare no conflict of interest.
Data deposition: Genome assembly and consensus sequences have been deposited in the National Center for Biotechnology Information (NCBI) GenBank database, https://www.ncbi.nlm.nih.gov (accession no. SWLF00000000). Full-length cDNA sequences have been uploaded to the NCBI Genbank database (accession no. SRX5321175). RNA-Seq data have been deposited in the NCBI GenBank BioProject database (accession no. PRJNA557001).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1910401116/-/DCSupplemental.
References
- 1.Les D., Crawford D., Landolt E., Gabel J., Kemball R., Phylogeny and systematics of Lemnaceae, the duckweed family. Syst. Bot. 27, 221–240 (2002). [Google Scholar]
- 2.Sree K. S., et al. , The duckweed Wolffia microscopica: A unique aquatic monocot. Flora Morphol. Distrib. Funct. Ecol. Plants 210, 31–39 (2015). [Google Scholar]
- 3.Landolt E., The Family of Lemnaceae–A Monographic Study (Veroffentlichungen des Geobotanischen Institutes der Eidgenossischen Technischen Hochschule, Stiftung Rubel, 1986), vol. 1. [Google Scholar]
- 4.Almahy H., Antibacterial activity of methanol extracts of the leaves of Lemna minor against eight different bacterial species. Int. J. Pharm. 5, 46–50 (2015). [Google Scholar]
- 5.Tan L. P., et al. , Antibacterial activity and toxicity of Duckweed, Lemna minor L. (Arales: Lemnaceae) from Malaysia. Malaysian J. Microbiol. 14, 387–392 (2018). [Google Scholar]
- 6.Zhao Q., et al. , Pan-genome analysis highlights the extent of genomic variation in cultivated and wild rice. Nat. Genet. 50, 278–284 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Wang W., et al. , The genome of the primordial monocotyledonous Spirodela polyrhiza: Neotenous reduction, fast growth, and aquatic lifestyle. Nat. Commun. 5, 3311 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Messing J., Crea R., Seeburg P. H., A system for shotgun DNA sequencing. Nucleic Acids Res. 9, 309–321 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gardner R. C., et al. , The complete nucleotide sequence of an infectious clone of cauliflower mosaic virus by M13mp7 shotgun sequencing. Nucleic Acids Res. 9, 2871–2888 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiao Y., et al. , Improved maize reference genome with single-molecule technologies. Nature 546, 524–527 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun S., et al. , Extensive intraspecific gene order and gene structural variations between Mo17 and other maize genomes. Nat. Genet. 50, 1289–1295 (2018). [DOI] [PubMed] [Google Scholar]
- 12.VanBuren R., et al. , Single-molecule sequencing of the desiccation-tolerant grass Oropetium thomaeum. Nature 527, 508–511 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Zou C., et al. , A high-quality genome assembly of quinoa provides insights into the molecular basis of salt bladder-based salinity tolerance and the exceptional nutritional value. Cell Res. 27, 1327–1340 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi J., et al. , Chromosome conformation capture resolved near complete genome assembly of broomcorn millet. Nat. Commun. 10, 464 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.An D., Zhou Y., Wang W., Assembled genome sequences, Spirodela polyrhiza genome sequencing and assembly. NCBI GenBank. https://www.ncbi.nlm.nih.gov/bioproject/520740 (accession no. SWLF00000000). Deposited 2 February 2019.
- 16.Michael T. P., et al. , Comprehensive definition of genome features in Spirodela polyrhiza by high-depth physical mapping and short-read DNA sequencing strategies. Plant J. 89, 617–635 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Van Hoeck A., et al. , The first draft genome of the aquatic model plant Lemna minor opens the route for future stress physiology research and biotechnological applications. Biotechnol. Biofuels 8, 188 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cao H. X., et al. , The map-based genome sequence of Spirodela polyrhiza aligned with its chromosomes, a reference for karyotype evolution. New Phytol. 209, 354–363 (2016). [DOI] [PubMed] [Google Scholar]
- 19.An D., Zhou Y., Wang W., Full-length cDNA sequences, Isoform sequencing of Spirodela polyrhiza. NCBI GenBank database. https://www.ncbi.nlm.nih.gov/sra/SRX5321175). Deposited 30 January 2019. [Google Scholar]
- 20.Olsen J. L., et al. , The genome of the seagrass Zostera marina reveals angiosperm adaptation to the sea. Nature 530, 331–335 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Al-Dous E. K., et al. , De novo genome sequencing and comparative genomics of date palm (Phoenix dactylifera). Nat. Biotechnol. 29, 521–527 (2011). [DOI] [PubMed] [Google Scholar]
- 22.Ming R., et al. , The pineapple genome and the evolution of CAM photosynthesis. Nat. Genet. 47, 1435–1442 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.International_Rice_Genome_Sequencing , The map-based sequence of the rice genome. Nature 436, 793–800 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Paterson A. H., et al. , The Sorghum bicolor genome and the diversification of grasses. Nature 457, 551–556 (2009). [DOI] [PubMed] [Google Scholar]
- 25.Lamesch P., et al. , The Arabidopsis information resource (TAIR): Improved gene annotation and new tools. Nucleic Acids Res. 40, D1202–D1210 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sato S., et al. ; Tomato Genome Consortium , The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485, 635–641 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mizutani M., Ohta D., Diversification of P450 genes during land plant evolution. Annu. Rev. Plant Biol. 61, 291–315 (2010). [DOI] [PubMed] [Google Scholar]
- 28.Cross J. W., Duckweed roots: Their role in vegetative dispersal. ISCDRA 5, 58–59 (2017). [Google Scholar]
- 29.Bellini C., Pacurar D. I., Perrone I., Adventitious roots and lateral roots: Similarities and differences. Annu. Rev. Plant Biol. 65, 639–666 (2014). [DOI] [PubMed] [Google Scholar]
- 30.Chhun T., Taketa S., Tsurumi S., Ichii M., Interaction between two auxin-resistant mutants and their effects on lateral root formation in rice (Oryza sativa L.). J. Exp. Bot. 54, 2701–2708 (2003). [DOI] [PubMed] [Google Scholar]
- 31.Dong J., et al. , Analysis of tandem gene copies in maize chromosomal regions reconstructed from long sequence reads. Proc. Natl. Acad. Sci. U.S.A. 113, 7949–7956 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hammami R., Ben Hamida J., Vergoten G., Fliss I., PhytAMP: A database dedicated to antimicrobial plant peptides. Nucleic Acids Res. 37, D963–D968 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Noonan J., Williams W. P., Shan X., Investigation of antimicrobial peptide genes associated with fungus and insect resistance in maize. Int. J. Mol. Sci. 18, E1938 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li N., et al. , A novel soybean dirigent gene GmDIR22 contributes to promotion of lignan biosynthesis and enhances resistance to Phytophthora sojae. Front. Plant Sci. 8, 1185 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ralph S., Park J. Y., Bohlmann J., Mansfield S. D., Dirigent proteins in conifer defense: Gene discovery, phylogeny, and differential wound- and insect-induced expression of a family of DIR and DIR-like genes in spruce (Picea spp.). Plant Mol. Biol. 60, 21–40 (2006). [DOI] [PubMed] [Google Scholar]
- 36.Miclaus M., Xu J. H., Messing J., Differential gene expression and epiregulation of alpha zein gene copies in maize haplotypes. PLoS Genet. 7, e1002131 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kondrashov F. A., Gene duplication as a mechanism of genomic adaptation to a changing environment. Proc. Biol. Sci. 279, 5048–5057 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.An D., Zhou Y., Wang W., RNA-Seq, Transcriptome profiling of different tissues of Spirodela polyrhiza 7498. NCBI GenBank. https://www.ncbi.nlm.nih.gov/sra/PRJNA557001 (accession no.PRJNA557001). Deposited 26 July 2019. [Google Scholar]
- 39.Fourounjian P., et al. , Post-transcriptional adaptation of the aquatic plant Spirodela polyrhiza under stress and hormonal stimuli. Plant J. 98, 1120–1133 (2019). [DOI] [PubMed] [Google Scholar]
- 40.Cai Q., et al. , The disease resistance protein SNC1 represses the biogenesis of microRNAs and phased siRNAs. Nat. Commun. 9, 5080 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wing R. A., Purugganan M. D., Zhang Q., The rice genome revolution: From an ancient grain to green super rice. Nat. Rev. Genet. 19, 505–517 (2018). [DOI] [PubMed] [Google Scholar]
- 42.Chin C. S., et al. , Phased diploid genome assembly with single-molecule real-time sequencing. Nat. Methods 13, 1050–1054 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koren S., et al. , Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27, 722–736 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smit A., Hubley R., Green P., RepeatMasker Open-3.0. Version: RepeatMasker 4.0.9. http://www.repeatmasker.org/. 1996. Accessed 9 April 2019.
- 45.Steinbiss S., Willhoeft U., Gremme G., Kurtz S., Fine-grained annotation and classification of de novo predicted LTR retrotransposons. Nucleic Acids Res. 37, 7002–7013 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ellinghaus D., Kurtz S., Willhoeft U., LTRharvest, an efficient and flexible software for de novo detection of LTR retrotransposons. BMC Bioinformatics 9, 18 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiong W., He L., Lai J., Dooner H. K., Du C., HelitronScanner uncovers a large overlooked cache of Helitron transposons in many plant genomes. Proc. Natl. Acad. Sci. U.S.A. 111, 10263–10268 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wenke T., et al. , Targeted identification of short interspersed nuclear element families shows their widespread existence and extreme heterogeneity in plant genomes. Plant Cell 23, 3117–3128 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Han Y., Burnette J. M. 3rd, Wessler S. R., TARGeT: A web-based pipeline for retrieving and characterizing gene and transposable element families from genomic sequences. Nucleic Acids Res. 37, e78 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Katoh K., Standley D. M., MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rice P., Longden I., Bleasby A., EMBOSS: The European molecular biology open software suite. Trends Genet. 16, 276–277 (2000). [DOI] [PubMed] [Google Scholar]
- 52.Ma J., Bennetzen J. L., Rapid recent growth and divergence of rice nuclear genomes. Proc. Natl. Acad. Sci. U.S.A. 101, 12404–12410 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu J., et al. , A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296, 79–92 (2002). [DOI] [PubMed] [Google Scholar]
- 54.Arabidopsis_Genome_Initiative , Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815 (2000). [DOI] [PubMed] [Google Scholar]
- 55.Campbell M. S., Holt C., Moore B., Yandell M., Genome annotation and curation using MAKER and MAKER-P. Curr. Protoc. Bioinformatics 48, 1–40 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mitchell A., et al. , The InterPro protein families database: The classification resource after 15 years. Nucleic Acids Res. 43, D213–D221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Haug-Baltzell A., Stephens S. A., Davey S., Scheidegger C. E., Lyons E., SynMap2 and SynMap3D: Web-based whole-genome synteny browsers. Bioinformatics 33, 2197–2198 (2017). [DOI] [PubMed] [Google Scholar]
- 58.Li L., Stoeckert C. J. Jr, Roos D. S., OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome Res. 13, 2178–2189 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang Y., et al. , MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, e49 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mai C. D., et al. , Genes controlling root development in rice. Rice (N. Y.) 7, 30 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ferreira-Saab M., et al. Compounds released by the biocontrol yeast Hanseniaspora opuntiae protect plants against Corynespora cassiicola and Botrytis cinerea. Front Microbiol. 9:1596 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genome assembly and consensus sequences were deposited into NCBI GenBank with an accession ID of SWLF00000000. Full-length cDNA sequences were derived by error correction of multiple reads, resulting in 492,435 high-quality isoforms that were uploaded with the ID of SRX5321175. RNA-Seq was deposited at GenBank BioProject of PRJNA557001.