Significance
Vitamin A is an essential nutrient for immune system development and function. After absorption from the diet, vitamin A is converted to retinol, which is delivered to cells and tissues by retinol-binding proteins. Serum amyloid A (SAA) proteins are retinol-binding proteins that transport retinol specifically during an infection. In this study, we illuminate the molecular details of how SAA proteins bind to retinol. We present the cocrystal structure of mouse SAA3 bound to retinol, which reveals that 3 molecules of SAA3 assemble to form a deep binding pocket that protects retinol, a fat-soluble molecule, from the aqueous environment. Our findings thus provide structural insight into how retinol is transported throughout the body during infection.
Keywords: vitamin A, retinol, crystal structure, infection, immunity
Abstract
Serum amyloid A (SAA) proteins are strongly induced in the liver by systemic infection and in the intestine by bacterial colonization. In infected mice, SAA proteins circulate in association with the vitamin A derivative retinol, suggesting that SAAs transport retinol during infection. Here we illuminate a structural basis for the retinol–SAA interaction. In the bloodstream of infected mice, most SAA is complexed with high-density lipoprotein (HDL). However, we found that the majority of the circulating retinol was associated with the small fraction of SAA proteins that circulate without binding to HDL, thus identifying free SAA as the predominant retinol-binding form in vivo. We then determined the crystal structure of retinol-bound mouse SAA3 at a resolution of 2.2 Å. Retinol-bound SAA3 formed a novel asymmetric trimeric assembly that was generated by the hydrophobic packing of the conserved amphipathic helices α1 and α3. This hydrophobic packing created a retinol-binding pocket in the center of the trimer, which was confirmed by mutagenesis studies. Together, these findings illuminate the molecular basis for retinol transport by SAA proteins during infection.
Vitamin A and its derivative retinol are essential for immunity to infection (1). Retinol is necessary for the development of adaptive immune cells, particularly at barrier sites such as the intestine (2, 3). It is also required for the expression of certain antimicrobial proteins in skin epithelial cells (4). A vitamin A-deficient diet can lead to increased risk of infection, highlighting the importance of vitamin A and its derivatives for host immunity (5).
Vitamin A is absorbed by intestinal epithelial cells, converted to retinol, and stored in the liver as retinyl esters (6). Following a microbial exposure, retinol must be mobilized from the intestine and liver and delivered to epithelial cells and developing immune cells. Because retinol is lipid-soluble, it must circulate bound to proteins with deep hydrophobic binding pockets that protect the retinol from the aqueous environment. Plasma retinol-binding proteins (RBPs) and cellular retinol-binding proteins (CRBPs) are 2 well-studied protein families that bind retinol and play important roles in retinol transport within cells and among tissues (7, 8). However, during infection, when retinol transport is especially important, circulating RBP concentrations drop dramatically (9), raising the question of how retinol is transported during infection.
We previously identified serum amyloid A (SAA) proteins as retinol-binding proteins that transport retinol specifically during bacterial infection (10). The SAA family encompasses a group of secreted proteins whose members are encoded in the genomes of almost all vertebrates and are highly conserved (11, 12). SAA proteins are expressed predominantly in the intestine and the liver and circulate in the serum (10, 12–15). SAA expression in both intestine and liver requires dietary vitamin A and the transcription factor retinoic acid receptor β (10, 16).
SAA expression is also strongly stimulated by exposure to microbes. SAA expression is induced in intestinal epithelial cells by the microbiota (14–17), and liver and serum SAAs are markedly elevated by systemic bacterial or viral infection (18, 19). Serum concentrations of SAAs can increase by approximately 1,000-fold during an infection, reaching concentrations of >1 mg/mL (20). SAA proteins bind tightly to retinol (with a nanomolar binding affinity) and circulate in association with retinol in the bloodstream of infected mice (10). However, the molecular details of how SAA proteins bind retinol remain unclear.
In this study, we illuminate the structural basis for retinol binding by SAAs. Although most circulating SAAs are complexed with high-density lipoprotein (HDL), we show that in infected mice, retinol circulates primarily with SAA proteins that are not complexed with HDL. This identifies free SAA as the predominant retinol-binding form in vivo. We present the cocrystal structure of mouse SAA3 complexed with retinol, which provides structural insight into the retinol–SAA3 binding interaction. These findings provide mechanistic insight into how retinol is transported during infection.
Results
Serum Retinol Is Bound to Freely Circulating SAAs in the Bloodstream of Infected Mice.
Humans and mice each encode 4 SAA family members in their genomes. On acute infection, SAA proteins are produced by hepatocytes and other cells and are secreted into the circulation. Although the majority of circulating SAAs are associated with HDL (13, 21), a fraction of SAAs circulate freely in the mouse bloodstream after acute infection (22).
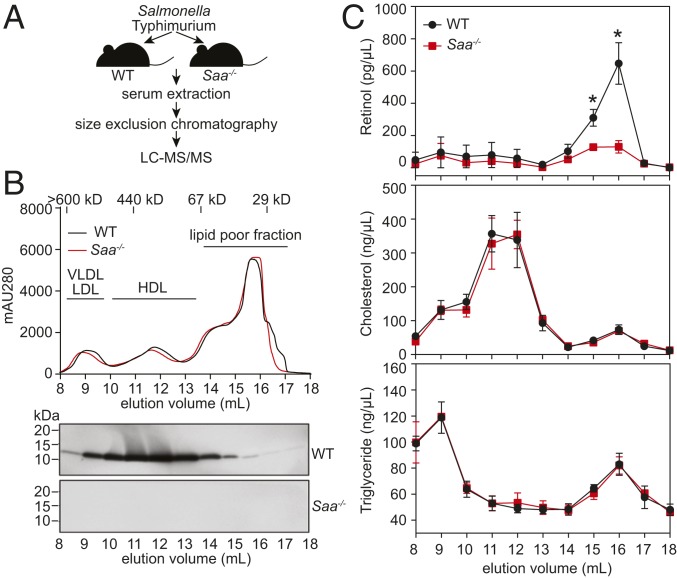
To establish a biological basis for our structural analysis, we first sought to identify the predominant form of retinol-bound SAA in the circulation. We therefore asked whether serum retinol is associated with HDL-bound SAAs or with freely circulating SAAs during infection. We used CRISPR/Cas9-mediated gene targeting to delete the entire mouse Saa locus (encompassing Saa1 to Saa4) (SI Appendix, Fig. S1A), and verified that SAA transcript and protein expression was lost in the Saa−/− mice (SI Appendix, Fig. S1 B and C). We then fractionated serum from Salmonella typhimurium-infected wild-type and Saa−/− mice by size-exclusion chromatography and analyzed the fractions for SAA content (Fig. 1 A and B). In the infected wild-type mice, the majority of serum SAA was associated with the HDL peak, as expected (Fig. 1B) (21, 22). However, SAAs also eluted beyond the HDL peak, suggesting that a fraction of SAA circulates freely and is not associated with HDL. As expected, no SAAs were detected in the sera of infected Saa−/− mice (Fig. 1B).
Fig. 1.
Serum retinol is bound to freely circulating SAAs in the bloodstream of infected mice. (A) Schematic representation of the experiment. (B) Wild-type mice or mice lacking the entire Saa locus (Saa−/−) were infected i.p. with S. typhimurium, and serum was collected 5 h later. The serum was fractionated by size-exclusion chromatography (Top), and SAA proteins in each fraction were detected by immunoblotting with a pan-SAA antibody (Bottom). (C) Fractions from wild-type or Saa−/− mice were hexane-extracted and analyzed by LC-MS/MS for retinol, cholesterol, and triglyceride. Data were derived from 3 independent experiments and plotted as mean ± SEM, *P < 0.05 as determined by 2-tailed Student's t test. WT, wild-type.
Liquid chromatography tandem mass spectrometry (LC-MS/MS) analysis of the serum fractions revealed that the majority of serum retinol recovered from the infected wild-type mice eluted at 14 to 18 mL with a peak at 16 mL, corresponding to the elution volume that includes low molecular weight proteins (30 to 40 kDa) (Fig. 1C). In contrast, retinol concentrations in the sera of infected Saa−/− mice were much lower (Fig. 1C), similar to those seen in uninfected wild-type or Saa−/− mice (SI Appendix, Fig. S2). There were no differences in serum retinol concentrations between uninfected wild-type and Saa−/− mice (SI Appendix, Fig. S2). In uninfected wild-type and Saa−/− mice, the majority of the serum retinol eluted at 13 to 17 mL with the peak at 15 mL (SI Appendix, Fig. S2), corresponding to the elution volume of the RBP4-transthyretin complex (∼77 kDa), which circulates retinol to support normal tissue functions in the absence of infection (23). As a comparison, there was no difference in serum cholesterol and triglyceride concentrations between the infected wild-type and Saa−/− mice (Fig. 1C).
These results suggested that SAA proteins account for the difference in serum retinol levels between wild-type and Saa−/− mice. However, it remained formally possible that differential production of other serum retinol-binding proteins, such as RBP4, serum albumin, and apolipoproteins, might be differentially expressed in the infected wild-type and Saa−/− mice, accounting for the lower serum retinol concentrations. To rule out this possibility, we analyzed the proteome of the retinol-containing fractions and found that the concentrations of other retinol-binding proteins were comparable between the wild-type and Saa−/− mice (SI Appendix, Table S1). Thus, freely circulating SAAs, rather than HDL-associated SAAs, are the predominant retinol-binding SAA form in the bloodstream of infected mice.
Crystallization and Overall Structure of the Mouse SAA3-Retinol Complex.
Structural studies have shown that the 2 known families of retinol-binding proteins, RBP and CRBP, bind retinoid ligands in highly hydrophobic β-barrel pockets (SI Appendix, Fig. S3 A and B) (24–26). However, SAA family proteins share a conserved 4-helix bundle structure (SI Appendix, Fig. S4), and thus lack a similar hydrophobic pocket that can accommodate retinoid ligands in their monomeric forms (10, 27). This raises the question of how SAAs bind their retinoid ligands.
To answer this question, we sought to determine the 3D structure of a mouse SAA3-retinol complex by X-ray crystallography. We were unable to obtain crystals through direct crystallization of the SAA3-retinol complex, likely due to the lability of retinol and the long time scales required for crystal growth. However, size-exclusion chromatography revealed that retinol binding did not change the oligomerization state of SAA3 in solution (SI Appendix, Fig. S5 A and B). Therefore, we used crystal soaking to determine the 3D structure of the SAA3-retinol complex (SI Appendix, Fig. S6A).
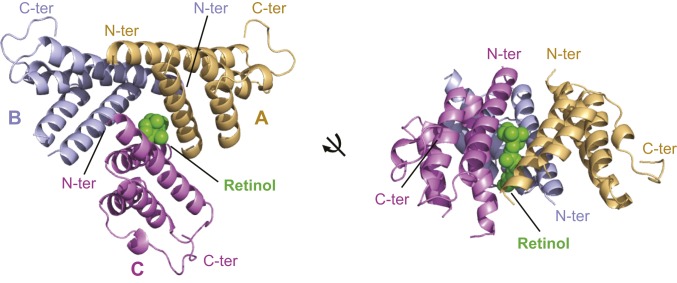
The structures of retinol-free and retinol-bound SAA3 were determined to a resolution of 1.7 Å and 2.2 Å, respectively [Fig. 2 and SI Appendix, Fig. S6B and Table S2 (28, 29)]. Both structures show trimeric packing of SAA3, which is consistent with the molecular size of SAA3 in solution (SI Appendix, Fig. S5A). Consistent with our size-exclusion chromatography results (SI Appendix, Fig. S5A), there were no marked structural changes between retinol-free and retinol-bound SAA3, with the exception of an α-helix that contacts retinol (SI Appendix, Fig. S6C).
Fig. 2.
Structural overview of the mouse SAA3-retinol complex. The overall structure of the SAA3-retinol complex is shown in 2 different orientations. The 3 SAA3 chains are labeled and shown in different colors as indicated. The bound retinol is shown in green.
In the structure of retinol-free SAA3, 1 molecule of the detergent decyl β-d-maltopyranoside (DM), which was used to prevent protein aggregation during purification, is bound at the center of the SAA3 trimer (SI Appendix, Fig. S6B). In the structure of retinol-bound SAA3, 1 molecule of retinol also binds at the center of the SAA3 trimer but with a different orientation (Fig. 2 and SI Appendix, Fig. S6C). To validate the retinol density observed in the retinol-bound SAA3 structure, we aligned the 2 SAA3 structures and compared the electron densities of retinol and DM (SI Appendix, Fig. S6 C and D). Although there is overlap, the density observed in the retinol-bound SAA3 cannot be derived from DM (or another compound) for several reasons. First, DM and retinol show different orientations, and most of their electron densities do not overlap. Second, the electron density of the maltose group of DM, observed in the retinol-free SAA3 structure, is completely absent in the retinol-bound SAA3 structure. Third, the SAA3 crystals changed from clear to yellow after retinol soaking (SI Appendix, Fig. S6A), consistent with the presence of retinol in the structure. Fourth, the SAA3-retinol complex has a 3:1 SAA3:retinol molar ratio in solution (SI Appendix, Fig. S5C), which is consistent with the SAA3-retinol structure. Finally, as detailed below, our mutagenesis analysis supports our structural findings.
In the SAA3 trimeric structure, each of the 3 SAA3 molecules in an asymmetric unit shares a conserved architecture consisting of a 4-helix bundle (designated α1–4 from the N to the C termini). The monomeric structure of retinol-bound mouse SAA3 is highly similar to the structures of ligand-free human SAA1 and mouse SAA3 (10, 27), suggesting that retinol binding does not cause marked conformational changes in mouse SAA3 (Fig. 2 and SI Appendix, Fig. S7 A–C).
Helix-wheel diagrams of the SAA monomer identified a highly conserved hydrophobic surface on 1 side of the 4-helix bundle (SI Appendix, Fig. S8 A–C). This structural feature suggests that SAA proteins may be unstable as free monomers in solution, and that other hydrophobic interactions, such as with HDL or other SAA monomers, may be needed to bury their exposed hydrophobic surfaces. In support of the latter possibility, the 3 SAA3 molecules from an asymmetric unit pack against each other tightly, with their hydrophobic surfaces buried in the center of the trimer. The trimeric packing is also supported by Protein Interfaces, Surfaces and Assemblies (PDBePISA) analysis (SI Appendix, Table S3) (30). Packing of the SAA3 molecules results in formation of a hydrophobic pocket in which retinol binds (Fig. 2).
Asymmetric Assembly of the SAA3 Trimer.
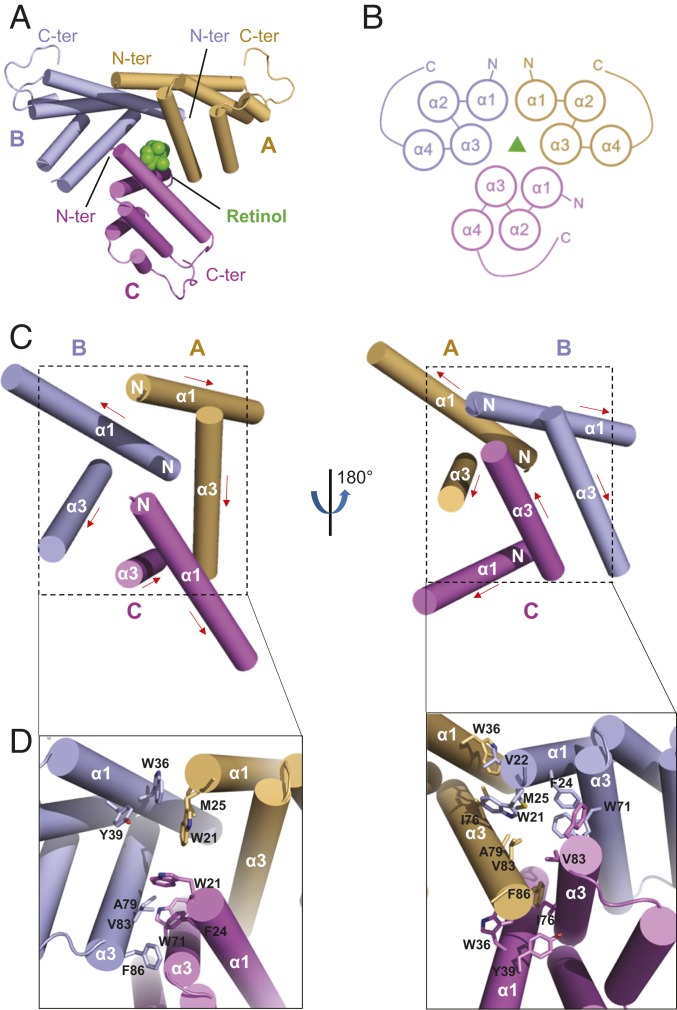
The amphipathic helices α1 and α3 of SAA3 form a V-shaped motif (i.e., V motif), with its apolar side forming an extended hydrophobic surface and a hydrophobic cavity between the 2 helices (i.e., V-motif cavity) (SI Appendix, Fig. S4). The 3 chains of the SAA3 trimer associate with each other around this hydrophobic surface in an asymmetric manner. In the SAA3 trimer, chains A and C are oriented similarly with only a 37° rotation relative to one another, whereas chain B is in the opposite orientation and is rotated 172° relative to chain A and 167° relative to chain C (Fig. 3 A and B).
Fig. 3.
Structural details of the SAA3 trimer assembly. (A) Top view of the assembly of SAA3 trimers. Three SAA3 chains are labeled and shown in different colors as indicated. The α-helices are depicted as cylinders, and the bound retinol is shown in green. (B) Topology diagrams of the SAA3 trimer assembly. (C) Helices involved in the assembly of SAA3 trimer shown in 2 different orientations. Three SAA3 molecules are shown in different colors, and helices α1 and α3 from each molecule are labeled. The red arrow indicates the direction from the N terminus to the C terminus. “N” represents the N terminus of each protomer. (D) Amino acids involved in assembly of the SAA3 trimer. Side chains are shown as sticks.
For the interaction between chain A and chain B, the N-terminal hydrophobic residues of the α1 helix of chain B (Trp21, Val22, and Met25) pack against the V-motif cavity of chain A (Trp36, Tyr39, Ile76, and Ala79) (Fig. 3 C and D). However, the N-terminal side of the α1 helix of chain A does not pack against the V-motif cavity of chain B but instead forms weak interactions with the residues at the edge of the α1 helix of chain B. The C-terminal hydrophobic residues of the α3 helix of chain A (Ala82 and Phe86) pack against the V-motif cavity of chain C (Trp36, Tyr39, Ile76, Ala79, and Val83), forming the chain A–chain C interface. The interaction between chain B and chain C is mainly mediated by the α3 helices from the 2 chains, and residues involved in this interaction include Trp71, Val75, Ala79, and Ala82 from chain B and Trp71 and Val75 from chain C (Fig. 3 C and D). In addition, the 3 interchain interfaces are close to one another and form cross-over interactions. Thus, the cooperative and extensive packing of the 3 V motifs gives rise to the asymmetric assembly of the SAA3 trimer, with a buried surface area of 3,090 Å2.
Retinol-Binding Pocket of SAA3.
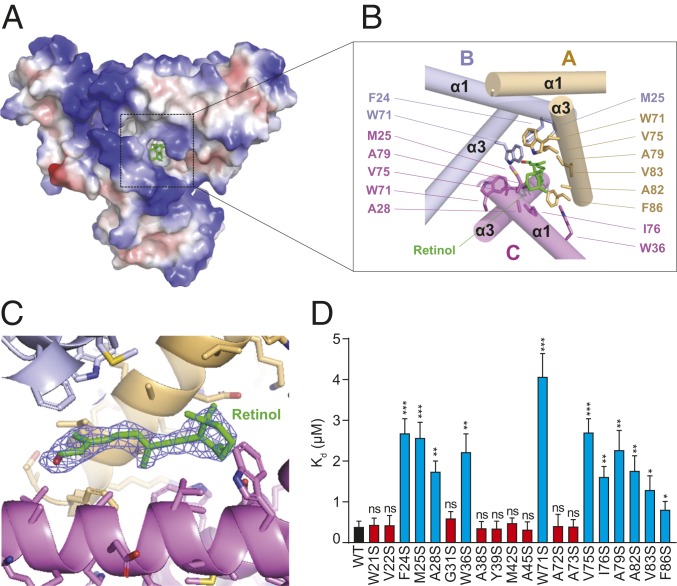
Retinol is caged in a corn-shaped hydrophobic pocket located at the center of the trimeric SAA3 assembly measuring ∼17.4 Å × 8.6 Å × 8.9 Å (Fig. 4 A–C), which is comparable to the dimensions of the RBP retinol-binding pocket (24). Although the 3D structures of SAAs and RBPs are completely distinct, their retinol-binding pockets are remarkably similar, indicating a conserved molecular mechanism of retinol binding among different retinol-binding proteins (SI Appendix, Fig. S9 A–D).
Fig. 4.
Retinol-binding pocket of SAA3. (A) Electrostatic surface representations of the SAA3 trimer with the bound retinol shown as green sticks. White, blue, and red indicate neutral, positive, and negative surfaces, respectively. (B) Detailed interactions between SAA3 and retinol from the area highlighted in A. Residues from each chain are labeled and shown in different colors. (C) Electron density, 2Fo − Fc (blue, contoured at 0.8 σ), of the bound retinol under Coot. The retinol molecule was omitted for calculation of the molecule electron density. (D) Mutagenesis of SAA3 identifying key residues that mediate the interaction between SAA3 and retinol. Dissociation constant (Kd) values between retinol and each SAA3 mutant were determined by a fluorometric retinol-binding assay (SI Appendix, Fig. S11) and are shown as mean ± SEM. ns, not significant; *P < 0.05; **P < 0.01; ***P < 0.001 as determined by 2-tailed Student’s t test.
The retinol-binding pocket is composed of several apolar residues located on the V motifs of each of the 3 SAA3 protomers (Fig. 4B). These residues include Trp71, Val75, Ala79, Ala82, Val83, and Phe86 from chain A; Phe24, Met25, and Trp71 from chain B; and Met25, Ala28, Trp36, Trp71, Val75, Ile76, and Ala79 from chain C. All these residues interact with retinol through hydrophobic packing. Structure-based sequence alignment showed that most of the retinol-interacting residues are highly conserved in different SAA members and in SAAs from different species (SI Appendix, Fig. S8C).
Mutagenesis Analysis of SAA3.
To critically test our structural model, we performed serine-scanning mutagenesis of the hydrophobic surface of SAA3. We mutated each hydrophobic residue on the amphipathic helices α1 and α3 to serine, a hydrophilic residue (SI Appendix, Fig. S10A). We then expressed and purified the mutant proteins (SI Appendix, Fig. S10B) and determined dissociation constants (Kd) values between retinol and each SAA3 mutant using a fluorometric binding assay (SI Appendix, Fig. S11) (10, 31). Approximately one-half of the SAA3 mutants, having mutations in both α1 and α3 helices, showed a 4- to 10-fold reduced binding affinity for retinol compared with wild-type SAA3 (Fig. 4D). The other half of the mutant SAA3 proteins had retinol-binding affinities similar to those of wild-type SAA3 (Fig. 4D). Consistent with our crystal structure, all mutations that reduced binding affinity mapped to amino acids that are either directly involved with retinol binding or are close to the retinol-binding pocket. In contrast, the mutations with no impact on binding affinity mapped to amino acids located far from the retinol-binding pocket (Fig. 4B and SI Appendix, Fig. S12). Among the mutations that reduced retinol-binding affinity, W71S had the greatest impact, reducing binding affinity by ∼10-fold (Fig. 4D). Consistent with this finding, the structure of the SAA3-retinol complex reveals that Trp71 is the only residue that all 3 SAA3 protomers contribute to the retinol-binding pocket (Fig. 4B).
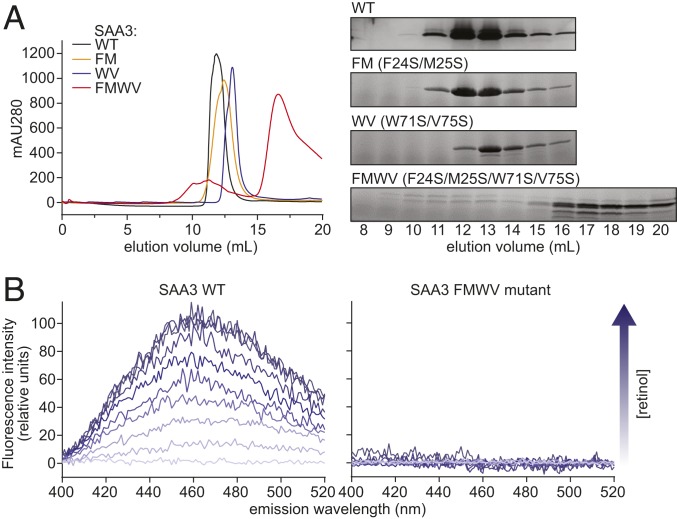
Each of the mutants with reduced retinol-binding affinity nevertheless retained some residual retinol-binding activity (Fig. 4D). Therefore, we tested whether multiple mutations might have an additive effect on retinol binding. We generated double mutations on either the α1 (F24S/M25S; FM) or the α3 helix (W71S/V75S; WV), and a quadruple mutation on both the α1 and α3 helices (F24S/M25S/W71S/V75S; FMWV). Size-exclusion chromatography showed that the FM and WV mutants maintained an oligomeric state similar to that of wild-type SAA3. In contrast, the quadruple mutation FMWV abrogated oligomerization of SAA3 (Fig. 5A). The fluorometric retinol-binding assay showed that the quadruple mutant FMWV completely lost its retinol-binding ability, as no fluorescence enhancement was detected during retinol titration (Fig. 5B). This indicates that SAA3 oligomerization is essential for retinol binding. Collectively, the results of our mutagenesis analyses support our structural model for retinol binding by SAA3.
Fig. 5.
Mutations in helices α1, α2, and α3 disrupt SAA3 packing and retinol binding. (A) Wild-type or mutant SAA3 proteins were expressed, purified, and analyzed by size-exclusion chromatography. (Left) Size-exclusion chromatography profiles of wild-type and mutated SAA3. FM, WV, and FMWV denote the F24S/M25S, W71S/V75S, and F24S/M25S/W71S/V75S SAA3 mutants, respectively. (Right) Samples of peak fractions shown at the left were visualized by Coomassie blue staining after SDS/PAGE. (B) Retinol-binding assay of wild-type SAA3 (Left) or FMWV mutant (Right). 0.5 μM SAA3 protein (wild-type or FMWV mutant) was incubated with different retinol concentrations, and the fluorescence intensity was monitored following excitation at 348 nm. The arrow indicates increasing retinol concentrations (0 to 5 μM).
Discussion
Vitamin A and its derivative retinol are essential for the development of immunity to infection. Despite the immune system’s increased demand for retinol, concentrations of serum RBP decline precipitously during acute infection (9). We previously identified SAAs as a retinol-binding protein family that transports retinol specifically during infection and is distinct from other known retinol-binding proteins (10). Accordingly, SAA-deficient mice show enhanced susceptibility to bacterial infection and inflammatory disease (10, 14). We speculate that SAAs support immune function by selectively delivering retinol to cells of the immune system, in contrast to RBPs, which circulate retinol to support normal tissue functions. This idea is currently under investigation.
In this study, we have illuminated the structural basis for retinol binding by SAA proteins. We first showed that freely circulating SAA proteins, rather than HDL-associated SAA proteins, bind the majority of retinol in the bloodstream of Salmonella-infected mice. Based on our identification of the predominant retinol-binding form of SAA, we determined the crystal structure of free SAA3 complexed with retinol. The SAA3-retinol cocrystal structure has a unique protein architecture consisting of an unconventional asymmetric trimeric packing of SAA3. This structure is distinct from previous crystal structures of ligand-free mouse SAA3 or human SAA1, which exhibit a symmetric packing of tetramer or hexamer (10, 27). It remains possible that retinol may also be bound by other oligomeric forms of SAA, as SAAs can adopt different oligomeric states in solution (27, 32).
The asymmetric packing of SAA3 forms an asymmetric hydrophobic pocket for retinol binding in the center of the SAA3 trimer. Interestingly, a similar asymmetric hydrophobic pocket is also present in the structures of RBPs and CRBPs (24–26), suggesting a conserved molecular mechanism of retinol binding among different retinol-binding proteins.
In conclusion, our findings provide structural insight into how SAAs transport retinol during infection. These findings lay the groundwork for altering SAA function to enhance resistance to infection and control inflammation during disease.
Materials and Methods
Mice.
Saa−/− mice lacking all 4 mouse Saa genes (Saa1 to Saa4) were generated using CRISPR/Cas9 genome editing with guide RNAs that target regions upstream and downstream of the Saa locus (SI Appendix, Fig. S1). Guide RNAs were injected into fertilized C57BL/6J embryos along with in vitro transcribed Cas9 mRNA at the University of Texas (UT) Southwestern Transgenic Core Facility. Healthy blastocysts were implanted into pseudopregnant mice. The resulting litters were screened by genomic sequencing to detect the deletion of the Saa locus, and mice harboring the deleted allele were bred to homozygosity. To minimize off-target effects, offspring of 2 independent founders were analyzed (SI Appendix, Fig. S1). All mice were housed and bred in the specific pathogen-free facility at the UT Southwestern Transgenic Core Facility according to protocols approved by the Institutional Animal Care and Use Committee of UT Southwestern Medical Center.
S. typhimurium Infection.
Salmonella enterica serovar Typhimurium (SL1344) was grown overnight in Luria-Bertani broth at 37 °C. Ten- to 16-wk old mice were infected i.p. with 104 CFU per mouse. Mice were killed after 5 h, and serum was collected.
Serum Analysis by Size-Exclusion Chromatography.
Sera were pooled from 4 mice, and 1 mL of the pooled serum was separated by size-exclusion chromatography on a Superdex 200 Increase 10/300 GL column (GE Healthcare Life Sciences). Samples were processed at a flow rate of 0.2 mL/min in PBS with 1.5 mM EDTA. The eluate was collected as 1-mL fractions, snap-frozen in liquid nitrogen, and stored at −80 °C until analysis.
LC-MS/MS Analysis of Retinol.
Retinol was quantified at the UT Southwestern Metabolic Phenotyping Core as described previously (10), with minor modifications. For retinol extraction, 5 mL of hexane and 20 µL of internal standard stock solution were added to 1 mL of sample. Samples were vortexed and then centrifuged at 2,000 × g for 10 min at 4 °C. The upper organic layer was transferred to a new glass tube, and the lower aqueous phase was re-extracted with 5 mL of hexane. Both organic layers were combined and dried under a nitrogen stream with no heat. Then 220 µL of acetonitrile was added to the dried residue, and the samples were sonicated in a chilled sonic bath for 5 min. The reconstituted samples were centrifuged at maximum speed for 10 min at 4 °C, and the supernatants were transferred to autosampler glass GC vials with 300-µL deactivated glass inserts.
Retinol was quantified by LC-MS/MS technology using a Nexera X2 UHPLC system coupled with a Shimadzu LCMS-8060 triple-quadrupole mass spectrometer (Shimadzu Scientific Instruments). Metabolites were detected using atmosphere pressure chemical ionization (APCI) in the positive mode with the mass spectrometer in the selective reaction monitoring scan mode. Metabolites were separated by reverse-phase liquid chromatography on a 2.1 × 100-mm Titan C18 1.9-µm column heated to 30 °C (Supelco) under a gradient elution over 12 min (mobile phase A: water, 0.1% formic acid; mobile phase B: acetonitrile, 0.1% formic acid). Then 10 µL of sample was injected into the LC-MS/MS system.
Quantification of Cholesterol and Triglyceride.
Total cholesterol and triglyceride concentrations in each serum fraction were measured by an enzymatic assay using the Infinity Triglyceride and Cholesterol Kit (Thermo Fisher Scientific).
Immunoblotting.
Fractionated serum samples were separated using a 4 to 20% gradient SDS/PAGE gel and then transferred to a PVDF membrane. Membranes were blocked with 5% nonfat milk in TBS-T buffer (0.1% Tween-20 in Tris-buffered saline), then sequentially incubated with pan-anti-SAA antiserum (10) and HRP-conjugated secondary antibody. Membranes were visualized using a Bio-Rad ChemiDoc Touch system.
Protein Expression and Purification.
Mouse Saa3 (minus the encoded signal sequence) was amplified from a reverse-transcribed intestinal cDNA and cloned into the pET28(a)+ expression vector. The amplicon was placed between the NdeI and XhoI restriction endonuclease sites with an N-terminal hexa-histidine tag, followed by a thrombin cleavage site and a C-terminal stop codon. Expression and purification were carried out as described previously (10), with some modifications. Proteins were expressed in Escherichia coli BL21-CodonPlus (DE3)-RILP cells (Stratagene) by induction with 0.4 mM isopropyl-β-d-galactoside for 12 h at 18 °C. Cells were harvested, resuspended in buffer containing 25 mM Tris⋅HCl pH 8.0 and 150 mM NaCl, and lysed by sonication. After sonication, decyl maltopyranoside (DM) (Avanti Polar Lipids) was added to a final concentration of 10 mM, followed by incubation for 3 h at 4 °C. The mixture was pelleted by centrifugation at 10,000 × g for 30 min, after which the supernatant was loaded onto a Ni2+ metal affinity column (Qiagen) preequilibrated with 4 mM DM in lysis buffer. Nonspecific contaminants were washed away with 40 mM imidazole in DM buffer, and the protein was eluted in DM buffer containing 300 mM imidazole. Subsequent buffers were detergent-free. The eluate was desalted on a PD-10 desalting column (GE Life Sciences) into resuspension buffer. Thrombin (Roche) was added (1 unit/1.5 mg protein), followed by incubation overnight at 4 °C. Undigested protein was removed by passing the digest over a Ni2+ affinity matrix and collecting the flow-through. The eluate was concentrated in a 3K cutoff Amicon Ultra centrifugal device (Millipore) and further purified by size-exclusion chromatography on a Superdex 75 10/300 GL column (GE Life Sciences) in 10 mM Tris pH 8.0 and 100 mM NaCl. For crystallization, SAA3 was concentrated to ∼5 mg/mL.
Crystallization.
Protein crystals were generated by mixing the protein with an equal amount of well solution (1.5 μL) by the sitting-drop vapor-diffusion method at 18 °C. Crystals appeared overnight under multiple conditions, with the best being 0.2 M trisodium citrate, 0.1 M Hepes pH 7.5, and 10% isopropanol, which was further refined by the addition of 6 to 8% PEG 6000. To obtain the SAA3-retinol cocrystal, SAA3 crystals were soaked with 5 mM retinol for 2 d in the crystallization buffer at 18 °C protected from light. Crystals were directly flash-frozen in liquid nitrogen with 30% glycerol as the cryoprotectant.
Data Collection and Structure Determination.
Data were collected at 100 K under a nitrogen gas stream at the Advanced Photon Source beamlines 19ID or 23ID of the Argonne National Laboratory. All datasets were processed with the HKL3000 package (33). The crystal structure of the SAA3-retinol complex was determined by molecular replacement with PHASER (34) using the structure of SAA3 monomer (PDB ID code 4Q5G) as a search model. The model from molecular replacement was built using Coot (35) and subsequently subjected to refinement by PHENIX (36). The structure of the SAA3-retinol complex was determined to a resolution of 2.2 Å, while that of retinol-free SAA3 was determined to 1.7 Å. Data collection and refinement statistics are summarized in SI Appendix, Table S2. The structure figures were prepared using PyMOL (www.pymol.org).
Mutagenesis.
Mouse SAA3 mutants were generated by a standard PCR-based cloning strategy, and their identities were confirmed by sequencing. Protein expression and purification were done as for wild-type SAA3.
Retinol-Binding Assay.
Retinol-binding assays were done as reported previously (10, 31). In brief, for retinol titrations, 0.5 μM wild-type or mutant SAA3 protein was incubated with varying concentrations of retinol, and the fluorescence intensity was monitored at 460 nm following excitation at 348 nm using a QuantaMaster 40 spectrofluorometer (Photon Technology International) and FelixGX software. All assays were done in 10 mM Tris pH 8.0 and 100 mM NaCl at 20 °C.
Statistics.
Statistical differences were calculated using the unpaired 2-tailed Student’s t test with GraphPad Prism software. Results are expressed as mean ± SEM.
Data Deposition.
Crystallography, atomic coordinates, and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID codes 6PY0 and 6PXZ).
Supplementary Material
Acknowledgments
We thank Dr. Ruth Gordillo, Preethi Csudae, and Duyen Do (UT Southwestern Metabolic Phenotyping Core) for mass spectrometry analysis of retinoids in biological samples and Shimadzu Scientific Instruments for the collaborative effort. Structures shown in this report were derived from work performed at Argonne National Laboratory, Structural Biology Center at the Advanced Photon Source. Argonne is operated by UChicago Argonne, LLC, for the US Department of Energy, Office of Biological and Environmental Research under Contract DE-AC02-06CH11357. This work was supported by NIH Grant R01 DK070855 (to L.V.H.), Welch Foundation Grant I-1874 (to L.V.H.), the Walter M. and Helen D. Bader Center for Research on Arthritis and Autoimmune Diseases (L.V.H.), and the Howard Hughes Medical Institute (L.V.H.). Y.-J.B. was supported by a Crohn’s and Colitis Foundation of America Research Fellowship Award.
Footnotes
The authors declare no conflict of interest.
Data deposition: Crystallography, atomic coordinates, and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID codes 6PY0 and 6PXZ).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1910713116/-/DCSupplemental.
References
- 1.Stephensen C. B., Vitamin A, infection, and immune function. Annu. Rev. Nutr. 21, 167–192 (2001). [DOI] [PubMed] [Google Scholar]
- 2.Mucida D., et al. , Reciprocal TH17 and regulatory T cell differentiation mediated by retinoic acid. Science 317, 256–260 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Hall J. A., et al. , Essential role for retinoic acid in the promotion of CD4(+) T cell effector responses via retinoic acid receptor alpha. Immunity 34, 435–447 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harris T. A., et al. , Resistin-like molecule α provides vitamin-A-dependent antimicrobial protection in the skin. Cell Host Microbe 25, 777–788.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sommer A., Vitamin a deficiency and clinical disease: A historical overview. J. Nutr. 138, 1835–1839 (2008). [DOI] [PubMed] [Google Scholar]
- 6.Harrison E. H., Mechanisms of digestion and absorption of dietary vitamin A. Annu. Rev. Nutr. 25, 87–103 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Noy N., Retinoid-binding proteins: Mediators of retinoid action. Biochem. J. 348, 481–495 (2000). [PMC free article] [PubMed] [Google Scholar]
- 8.Blaner W. S., Retinol-binding protein: The serum transport protein for vitamin A. Endocr. Rev. 10, 308–316 (1989). [DOI] [PubMed] [Google Scholar]
- 9.Rosales F. J., Ritter S. J., Zolfaghari R., Smith J. E., Ross A. C., Effects of acute inflammation on plasma retinol, retinol-binding protein, and its mRNA in the liver and kidneys of vitamin A-sufficient rats. J. Lipid Res. 37, 962–971 (1996). [PubMed] [Google Scholar]
- 10.Derebe M. G., et al. , Serum amyloid A is a retinol binding protein that transports retinol during bacterial infection. Elife 3, e03206 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uhlar C. M., Burgess C. J., Sharp P. M., Whitehead A. S., Evolution of the serum amyloid A (SAA) protein superfamily. Genomics 19, 228–235 (1994). [DOI] [PubMed] [Google Scholar]
- 12.Uhlar C. M., Whitehead A. S., Serum amyloid A, the major vertebrate acute-phase reactant. Eur. J. Biochem. 265, 501–523 (1999). [DOI] [PubMed] [Google Scholar]
- 13.Whitehead A. S., et al. , Identification of novel members of the serum amyloid A protein superfamily as constitutive apolipoproteins of high-density lipoprotein. J. Biol. Chem. 267, 3862–3867 (1992). [PubMed] [Google Scholar]
- 14.Eckhardt E. R., et al. , Intestinal epithelial serum amyloid A modulates bacterial growth in vitro and pro-inflammatory responses in mouse experimental colitis. BMC Gastroenterol. 10, 133 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reigstad C. S., Lundén G. O., Felin J., Bäckhed F., Regulation of serum amyloid A3 (SAA3) in mouse colonic epithelium and adipose tissue by the intestinal microbiota. PLoS One 4, e5842 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gattu S., et al. , Epithelial retinoic acid receptor β regulates serum amyloid A expression and vitamin A-dependent intestinal immunity. Proc. Natl. Acad. Sci. U.S.A. 116, 10911–10916 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ivanov I. I., et al. , Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139, 485–498 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meek R. L., Benditt E. P., Amyloid A gene family expression in different mouse tissues. J. Exp. Med. 164, 2006–2017 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miwata H., et al. , Serum amyloid A protein in acute viral infections. Arch. Dis. Child. 68, 210–214 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shainkin-Kestenbaum R., Winikoff Y., Cristal N., Serum amyloid A concentrations during the course of acute ischaemic heart disease. J. Clin. Pathol. 39, 635–637 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benditt E. P., Eriksen N., Hanson R. H., Amyloid protein SAA is an apoprotein of mouse plasma high-density lipoprotein. Proc. Natl. Acad. Sci. U.S.A. 76, 4092–4096 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tannock L. R., et al. , Serum amyloid A3 is a high-density lipoprotein-associated acute-phase protein. J. Lipid Res. 59, 339–347 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mody N., Graham T. E., Tsuji Y., Yang Q., Kahn B. B., Decreased clearance of serum retinol-binding protein and elevated levels of transthyretin in insulin-resistant ob/ob mice. Am. J. Physiol. Endocrinol. Metab. 294, E785–E793 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zanotti G., Ottonello S., Berni R., Monaco H. L., Crystal structure of the trigonal form of human plasma retinol-binding protein at 2.5 A resolution. J. Mol. Biol. 230, 613–624 (1993). [DOI] [PubMed] [Google Scholar]
- 25.Kleywegt G. J., et al. , Crystal structures of cellular retinoic acid binding proteins I and II in complex with all-trans-retinoic acid and a synthetic retinoid. Structure 2, 1241–1258 (1994). [DOI] [PubMed] [Google Scholar]
- 26.Menozzi I., et al. , Structural and molecular determinants affecting the interaction of retinol with human CRBP1. J. Struct. Biol. 197, 330–339 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Lu J., Yu Y., Zhu I., Cheng Y., Sun P. D., Structural mechanism of serum amyloid A-mediated inflammatory amyloidosis. Proc. Natl. Acad. Sci. U.S.A. 111, 5189–5194 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hooper L., Hu Z., Bang Y.-J.. (2019) Crystal structure of mouse serum amyloid A3 (SAA3) in the trimeric form. Worldwide Protein Data Bank. https://www.rcsb.org/structure/6PXZ. Deposited 28 July, 2019.
- 29.Hooper L., Hu Z., Bang Y.-J.. (2019) Crystal structure of mouse serum amyloid A3 (SAA3) bound with retinol. Worldwide Protein Data Bank. https://www.rcsb.org/structure/6PY0. Deposited 28 July, 2019.
- 30.Krissinel E., Henrick K., Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Cogan U., Kopelman M., Mokady S., Shinitzky M., Binding affinities of retinol and related compounds to retinol binding proteins. Eur. J. Biochem. 65, 71–78 (1976). [DOI] [PubMed] [Google Scholar]
- 32.Wang Y., et al. , Serum amyloid A 2.2 refolds into a octameric oligomer that slowly converts to a more stable hexamer. Biochem. Biophys. Res. Commun. 407, 725–729 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Minor W., Cymborowski M., Otwinowski Z., Chruszcz M., HKL-3000: The integration of data reduction and structure solution—From diffraction images to an initial model in minutes. Acta Crystallogr. D Biol. Crystallogr. 62, 859–866 (2006). [DOI] [PubMed] [Google Scholar]
- 34.McCoy A. J., et al. , Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adams P. D., et al. , PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 (2002). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.