Significance
Our comprehensive genomic comparison of matched tumor, brain tumor-initiating cells (BTICs), and xenografts will help inform the observations made when using these model systems and guide the experimental design for drug screening and hypothesis testing. In addition, paired genomic and transcriptomic data we generated for tumors and their matched in vitro and in vivo models provide valuable resources for the future study of glioblastoma at the genetic and molecular levels.
Keywords: glioblastoma, genome, therapy, transcriptome, BTICs
Abstract
Glioblastoma multiforme (GBM) is the most deadly brain tumor, and currently lacks effective treatment options. Brain tumor-initiating cells (BTICs) and orthotopic xenografts are widely used in investigating GBM biology and new therapies for this aggressive disease. However, the genomic characteristics and molecular resemblance of these models to GBM tumors remain undetermined. We used massively parallel sequencing technology to decode the genomes and transcriptomes of BTICs and xenografts and their matched tumors in order to delineate the potential impacts of the distinct growth environments. Using data generated from whole-genome sequencing of 201 samples and RNA sequencing of 118 samples, we show that BTICs and xenografts resemble their parental tumor at the genomic level but differ at the mRNA expression and epigenomic levels, likely due to the different growth environment for each sample type. These findings suggest that a comprehensive genomic understanding of in vitro and in vivo GBM model systems is crucial for interpreting data from drug screens, and can help control for biases introduced by cell-culture conditions and the microenvironment in mouse models. We also found that lack of MGMT expression in pretreated GBM is linked to hypermutation, which in turn contributes to increased genomic heterogeneity and requires new strategies for GBM treatment.
Glioblastoma multiforme (GBM) is the most deadly form of brain cancer, with almost all patients dying of the disease within 2 y of initial diagnosis. Treatment regimens combining surgical resection with radiotherapy and concurrent adjuvant temozolomide (TMZ) chemotherapy have led to modest gains in survival, with 2- and 5-y survival rates of 25 and 10%, respectively (1). The main barriers to successful treatment include the invasive nature and low proliferative activity of GBM that impede complete surgical removal of the invasive cells while also promoting resistance to chemotherapy and radiotherapy.
Cell lines and xenograft models are frequently used for investigating cancer biology and therapeutics. Several reports have described GBM cell lines and xenograft models (2–5). In particular, the identification of brain tumor-initiating cells (BTICs) more than a decade ago led to the hypothesis that a subpopulation of treatment-resistant cells may possess “stem cell-like” properties and spawn recurrences (6). By adapting neural stem cell tissue-culture techniques (7), a library of BTICs was generated by culturing patient-derived GBM tumor cells as spheres in neural stem cell media. These BTICs mimic the phenotypic aspects of human GBM when implanted orthotopically in mice and capture the genetic diversity of GBM more faithfully than traditional immortalized cell lines (2, 8). However, it is unclear how closely BTICs in culture or grown orthotopically in mice resemble the genetic, epigenetic, and transcriptomic landscape of tumors from which they were derived.
The Cancer Genome Atlas (TCGA) has generated a comprehensive catalog of molecular alterations and deregulated signaling pathways in GBM patient samples (9–11). Such surveys illustrate the power of systematic sequencing approaches to identify molecular genetic alterations of biological significance and, in due course, candidate therapeutic targets. TCGA resource also provides an important resource against which to compare data from other GBMs and materials derived from them. Such a data resource and in-depth analysis are needed for cell lines and xenografts that are heavily used in the study of disease biology and treatment. Here, we report comprehensive whole-genome, transcriptome, and methylome datasets derived from matched GBM tumor, BTIC, and xenograft samples, comparing these with each other and GBM samples from TCGA. The comprehensive molecular profiling of BTICs and xenografts provides insights into GBM biology and reveals how in vitro and in vivo growth environments affect the genomes and transcriptomes of these model systems. Our findings will help to interpret drug-screen results from these models, refine future strategies for the development of novel therapeutic approaches, and avoid biases introduced by culture conditions.
Results
To investigate how the GBM genomic landscape is shaped during the production of BTICs from human tumor tissues and their subsequent growth in xenografts, we sequenced and analyzed the genomes of 201 samples, including 46 matched parent tumor–BTIC pairs and 15 matched trios of tumor–BTIC–xenograft, together with their matched blood samples (Table 1). In addition, we performed RNA sequencing on 108 samples, including 37 pairs and 10 trios, to determine changes in gene expression. To assess differences in DNA methylation, 35 pairs and 4 trios were profiled using Illumina’s Infinium Human Methylation 450K array. In addition to newly diagnosed tumors, our analyses also included posttreatment, recurrent GBM (GBM-R) cases (including a secondary GBM sample) to provide insights into the mechanisms of treatment resistance.
Table 1.
Matched tumor–BTIC–xenograft blood samples with whole-genome sequencing, transcriptome sequencing, and methylation array data
| Samples and sequencing data | Clinical data | |||||||||
| Patient ID | BTIC ID | Tumor* | BTIC | Xenograft | Blood | Tumor MGMT methylation status† | New or recurrent‡ | Treatment§ | Age, y | Gender |
| PT-VO7089 | BT-100 | W, T, M | W, T, M | W | U | N | No treatment | 63 | M | |
| PT-AB0029 | BT-106 | W, T | W | W | N | RT alone | 83 | M | ||
| PT-GC1519 | BT-108 | W, T | W, T | W | N | RT+TMZ + 2 cycles TMZ only | 46 | M | ||
| PT-AB6372 | BT-119 | W, T, M | W, T, M | W | M | R | RT+TMZ, TMZ | 69 | F | |
| PT-HN6692 | BT-126 | W, M | W, T, M | W | U | Unknown | Unknown | 67 | M | |
| PT-BM772 | BT-127 | W, M | W, T, M | W | U | N | RT+TMZ, TMZ | 55 | M | |
| PT-BK0248 | BT-134 | W | W, T | W | W | N | CRT | 50 | F | |
| PT-RL7940 | BT-140 | W, T, M | W, T, M | W | U | N | RT+TMZ, TMZ | 63 | M | |
| PT-EV3071 | BT-142 | W, T | W, T | W, T | W | N | RT+TMZ, TMZ | 38 | M | |
| PT-MB9777 | BT-143 | W, T, M | W, T, M | W, T, M | W | M | R | RT+TMZ, TMZ | 39 | F |
| PT-MB9777 | BT-143-Y | W, T | W, T | W | W | R | RT+TMZ, TMZ | 39 | F | |
| PT-MB9777 | BT-143-Z | W | W, T | W | R | RT+TMZ, TMZ | 39 | F | ||
| PT-DS9789 | BT-147 | W, T, M | W, T, M | W, T, M | W | U | R | RT+TMZ, TMZ | 55 | M |
| PT-MD9088 | BT-156 | W, T, M | W, T, M | W | U | R | RT+TMZ, TMZ | 51 | M | |
| PT-SK0976 | BT-161 | W, T, M | W, T, M | W | W | M | N | RT+TMZ+placebo or bevacizumab followed by TMZ | 55 | F |
| PT-JW6420 | BT-166 | W, T | W, T | W | N | RT+TMZ, TMZ | 69 | M | ||
| PT-LR9369 | BT-167 | W, T, M | W, T, M | W | U | R | RT+TMZ, TMZ | 63 | M | |
| PT-RD1291 | BT-169 | W, T | W, T | W | N | RT+TMZ, TMZ | 41 | M | ||
| PT-KM5291 | BT-172 | W, T, M | W, T, M | W | W | U | N | RT+TMZ, TMZ | 51 | M |
| PT-JP2405 | BT-181 | W, T, M | W, T, M | W | M | R | RT and TMZ before diagnosis of GBM | 47 | M | |
| PT-SJ5453 | BT-191 | W, T, M | W, T, M | W, T | U | N | RT+TMZ | 44 | F | |
| PT-AK7565 | BT-194 | W, T, M | W, T, M | W | M | N | CRT (Stupp), then RESCUE after progression/recurrence | 59 | M | |
| PT-LS4891 | BT-198 | W, T, M | W, T, M | W | W | U | N | No treatment | 52 | F |
| PT-PV2594 | BT-206 | W, T, M | W, T, M | W, T | M | N | RT+TMZ, TMZ | 68 | M | |
| PT-PV2594 | BT-206-R | W, T | M | N | RT+TMZ, TMZ | 68 | M | |||
| PT-AH1410 | BT-208 | W, M | W, T, M | W | M | N | RT+TMZ, TMZ | 69 | M | |
| PT-SS3647 | BT-220 | W, T, M | W, T, M | U | N | RT alone | 84 | M | ||
| PT-GJ3716 | BT-238-X | W | W, T | W | W | N | RT+TMZ | 61 | M | |
| PT-GJ3716 | BT-238-Z | W, T | W | W | N | RT+TMZ | 61 | M | ||
| PT-JE6375 | BT-241 | W, T | W, T | T | W | N | RT followed by TMZ | 79 | F | |
| PT-CA2271 | BT-245 | W, T, M | W, T, M | W, T | W | U | N | RT followed by TMZ | 70 | M |
| PT-GR2309 | BT-246 | W, M | W, T, M | W | M | N | CRT (Stupp) | 60 | F | |
| PT-CM1209 | BT-248-XY | W, T | W, T | W, T | W | R | RT+TMZ, TMZ | 55 | F | |
| PT-CM1209 | BT-248-Z | W, T | W, T | W | W | R | RT+TMZ, TMZ | 55 | F | |
| PT-HS9105 | BT-266 | W | W | W | R | CRT (Stupp), then RESCUE after progression/recurrence | 54 | F | ||
| PT-TM5196 | BT-280 | W, T, M | W, T, M | W | U | R | RT alone (CEC1 trial), then RESCUE after recurrence | 35 | M | |
| PT-WP9124 | BT-284 | W, T, M | W, T, M | W | M | R | CRT (Stupp), then bevacizumab after recurrence | 50 | M | |
| PT-GB9483 | BT-287 | W | W | W | R | CRT (Stupp) | 52 | M | ||
| PT-PD6881 | BT-41 | W, T, M | W, T, M | W | U | R | RT+TMZ | 49 | M | |
| PT-DF5919 | BT-53 | W, T, M | W, T, M | M | W | M | N | Unknown | 59 | M |
| PT-JB1730 | BT-63 | W, T | W, T | W | N | TMZ followed by TMZ+RAD001C (clinical trial) | 60 | F | ||
| PT-FB6711 | BT-67 | W, T, M | W, T, M | W | M | N | RT+TMZ | 44 | M | |
| PT-HO0394 | BT-69 | W, M | W, T, M | W, T | W | U | N | No treatment | 51 | M |
| PT-RW9277 | BT-73 | W, T, M | W, T, M | W, T, M | W | U | Unknown | Unknown | 52 | M |
| PT-RL5404 | BT-75 | W, T, M | W, T, M | W | U | N | RT alone | 74 | M | |
| PT-AL4257 | BT-84 | W, T, M | W, T, M | W | M | N | Unknown | Unknown | M | |
| PT-GB9186 | BT-85 | W, T, M | W, T, M | W | M | N | RT followed by TMZ | 74 | M | |
| PT-SO0258 | BT-89 | W, T, M | W, T, M | W | M | N | RT followed by TMZ | 60 | F | |
| PT-AR5365 | BT-92 | W, T | W, T | W | R | RT+TMZ, TMZ | 23 | M | ||
| PT-LC3356 | BT-94 | W, T, M | W, T, M | W, T | W | M | N | RT+TMZ | 60 | F |
M, methylation profiling; T, transcriptome sequencing by RNA-seq; W, whole-genome sequencing.
M, methylated; U, unmethylated.
N, newly diagnosed; R, recurrent after treatment.
CRT, concurrent chemoradiotherapy; RESCUE, a phase II TMZ trial; RT, radiation therapy; RT+TMZ, concurrent radiation therapy and temozolomide; Stupp, the Stupp protocol for glioblastoma; RT+TMZ, TMZ, concurrent radiation therapy and temozolomide followed by temozolomide.
Conservation of Genomic Features among Parent Tumors, Matched Cell Lines, and Xenografts.
In all 3 types of samples, tumor, BTIC, and xenograft, we generally observed conserved patterns of driver events in GBM that were described previously, such as PTEN mutation, EGFR amplification and point mutation, as well as homozygous loss of CDKN2A and CDKN2B (Fig. 1) (9, 10). In particular, from the analysis of copy-number alterations (CNAs), we found that the conserved pattern of CNAs among the 3 types of samples was not limited to alterations known to be characteristic of GBM, such as gains of chromosome 7 and losses of chromosome 10, but across the whole genome (SI Appendix, Fig. S1).
Fig. 1.
Distribution of somatic aberrations in genes found significantly altered in previous GBM studies. Aberration types are encoded by different colors. Letters are used when the aberration is not shared in a pair or trio. C, private in a BTIC; D, private in a tumor and xenograft in a trio; F, private in a BTIC and xenograft in a trio; P, private in a tumor and BTIC in a trio; T, private in a tumor; X, private in a xenograft. MGMT and EGFR vIII status tiles are split when a tumor (Upper) and BTIC (Lower) differ. Loss of tumor suppresser and gain of oncogene are well-conserved in pairs and trios. The private aberrations often have low functional impact, such as shallow copy changes or a variant of unknown significance, or passenger mutations in hypermutated samples. (A) Aberrations in paired tumor and BTIC samples. (B) Aberrations in trios of tumor–BTIC–xenograft samples.
Using the whole-genome sequencing data, we also investigated the mutation landscape of tumors, BTICs, and xenografts. As our collection included both pre- and posttreatment tumor samples, we observed 2 levels of mutation burden: 39 samples showed an average of 3.9 somatic mutations per million bp in the genome (range, 0.95 to 16.09), including pre- and posttreatment samples. Additionally, 10 samples displayed a hypermutation phenotype, exhibiting a much higher rate, with an average of 93 mutations per million bases (range, 41 to 140) (SI Appendix, Fig. S2A). Hypermutation is a genomic feature found in many posttreatment GBMs (12), and all hypermutated samples in our collection were derived from recurrent GBM treated with TMZ in the primary setting; these hypermutated samples have mutations in mismatch repair (MMR) genes (SI Appendix, Table S1). However, not all TMZ-treated recurrent GBM demonstrated a hypermutation phenotype, suggesting alternative mechanisms of resistance that are discussed later.
In contrast to CNAs, mutational patterns showed a larger diversity between matched pairs, especially in hypermutated samples. The proportion of shared single-nucleotide variants (SNVs) between tumors and matched BTICs varied across samples, ranging from 12 to 64% in samples without hypermutation, with a median of 34% (SI Appendix, Figs. S2B and S3). A generally lower percentage of mutations was shared between hypermutated tumors and their matched BTICs (0.7 to 52%, median 21%; SI Appendix, Fig. S2B); the level of SNV overlap appeared to be related to the timing of mutations in MMR genes, especially MSH6. In samples without shared MMR gene mutations (mutations acquired independently after diverging from an ancestral clone), only ∼0.7% of mutations were common between pairs. In samples that had 1 shared MMR mutation, ∼10% of mutations were shared between the matched tumor and BTIC, and in pairs with 2 shared MMR mutations >20% mutations were in common (SI Appendix, Fig. S2C).
Using the 15 trios of tumor, BTIC, and xenograft samples, we further investigated the genomic similarities and differences among tumor, BTIC, and xenograft samples. Our analysis showed that in most trios, the majority of mutations are shared among all 3 sample types from the same patient (34 to 80%, median 51%; SI Appendix, Fig. S2D). As expected, the commonly mutated genes are enriched in glioma-related pathways, such as the PI3K pathway, MAPK pathway, and p53 signaling, suggesting that cancer-driving mutations are well-conserved in the in vitro and in vivo models.
Disparate Genomic Features Revealed in Comparisons among Tumors, Cell Lines, and Xenografts.
Comparison between tumors and their matched BTICs revealed aberrations that were private to each sample type. BTIC-specific mutations ranged from 8 to 59% of all mutations in a given pair (SI Appendix, Fig. S3). Few genes were mutated privately in more than 2 BTIC lines. There were ∼700 genes that were uniquely mutated in only 1 of the BTIC lines. These did not reveal significant enrichment in any particular biological pathway or process. This may indicate that the majority of new aberrations gained in the cell lines were likely due to tumor heterogeneity or random and ongoing intrinsic mutational processes associated with proliferation in vitro.
We also identified variants in the tumors but not in the matched BTICs. Out of the total mutations in each pair, ∼2 to 60% of mutations are found in tumors only. This is expected, since BTICs are derived from only a few cells of the original tumor and therefore tumor-specific mutations may reflect tumor heterogeneity. Alternately, selective pressures of in vitro growth conditions may lead to preferential loss of some clones, especially when the same observation was made in several tumor–BTIC pairs. Among such variants, of particular interest were PDGFRA amplification and PDGFRA p. P343L/T mutations. In 2 patients (PT-LS4891 and PT-RL5404), PDGFRA was within high-level focal amplifications in the tumor sample, and most of the amplified copies had the same mutation in proline 343 (35 out of 40 copies in PT-LS4891, and 71 out of 77 copies in PT-RL5404). In contrast, neither the amplification nor the mutation was found in BTICs derived from these 2 tumor samples, suggesting a different selection preference for clones with PDGFRA aberrations in BTICs.
Another difference between tumors and BTICs was observed in 2 samples with the IDH1 mutation. BT-142 from patient PT-EV3071 and BT-92 from patient PT-AR5365 each had an IDH1 R132H mutation in the tumor, but the BT-142 cell line lost the wild-type allele while the BT-92 cell line lost the mutated allele, suggesting that this allele has to be homozygous in order to establish cell lines. This finding was confirmed with more samples and these results have been previously published (13).
Among the 15 trios of tumor, BTIC, and xenograft samples, private mutations in each sample type were sparse: 0 to 4% in tumors, 0 to 27% in BTICs, and 6 to 37% in xenografts. We did not observe any functional enrichment of statistical significance. However, among mutations private to BTICs, several were found in genes involved in cell-matrix interaction including cell-matrix adhesion, cell-substrate adhesion, and extracellular matrix organization, such as ITGA2, ITGA8, and NID1. Private mutations observed in xenografts are found in several genes involved in brain functions, including ciliary or flagellar motility, sensory organ development, and regulation of synaptic transmission. Larger sample sizes are needed to determine if these private mutations are a consequence of adaptation to the distinct growth environments of each model system.
Transcriptome and Methylome Reveal Differences among Tumor, BTIC, and Xenograft Samples.
We generated and compared RNA-sequencing data from 108 samples, including 37 matched tumor–BTIC pairs and 10 matched tumor–BTIC–xenograft trios, to observe how gene expression differs between tumors and the in vitro and in vivo model systems. From principal-component analysis (PCA) of the expression data, we found the clear separation of tumor samples from BTIC and xenograft samples along PC1, while BTIC and xenograft clusters were completely overlapping on PC1 and partially overlapping along PC2 (Fig. 2). A heuristic clustering analysis also showed that samples from the same patient did not group together. Thus, higher similarity at the expression level was observed in samples of the same type (tumor, BTIC, xenograft) rather than those of samples from the same patient.
Fig. 2.
Principal-component analysis plot of tumors, BTICs, and xenografts based on RNA expression. Color indicates sample type, while shape represents subtype. Samples show higher similarity within that same sample type, suggesting the growth condition has a higher impact on the gene expression than the individual genetic makeup.
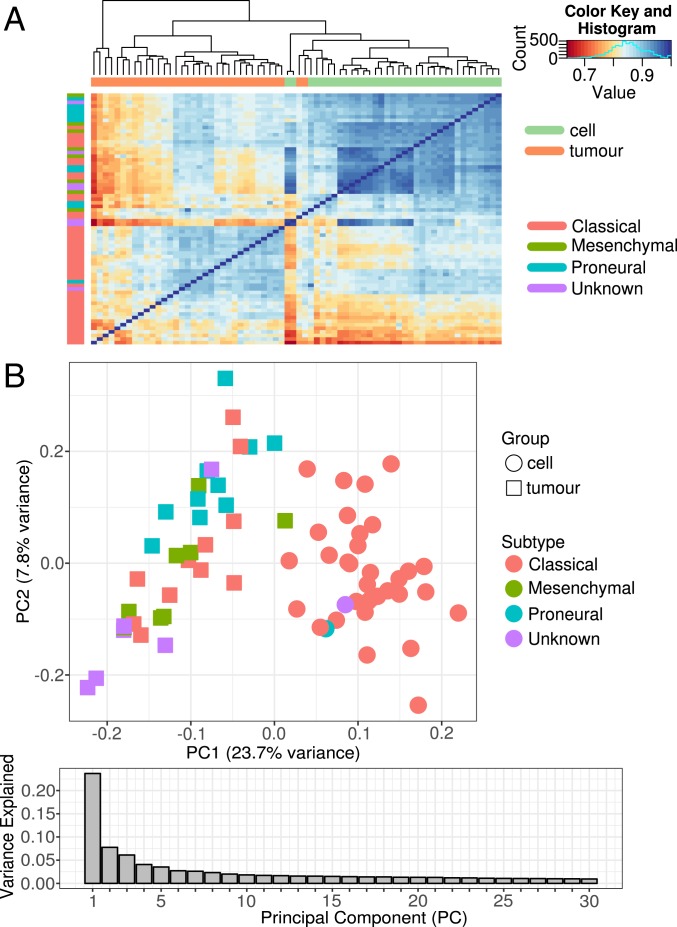
Similar differences were observed between GBM tumors and matched BTIC samples when DNA methylation (DNAm) profiles were interrogated across ∼420,000 CpG sites in the genome. Based on unsupervised hierarchical clustering of DNAm profiles, GBM tumors clustered distinctly away from BTIC samples, indicating global differences in DNAm signatures between the paired tissue sources (Fig. 3A). In addition, no discernible clustering in DNAm profiles was observed based on sample subtype classification (either classical, mesenchymal, or proneural) (Fig. 3A). These results were corroborated by PCA, which showed that tissue source and subtype were associated with PC1, comprising 23.7% of the variance in DNAm (Fig. 3B). Moreover, PC1 was largely driven by differences in tissue source (i.e., tumor versus BTIC) followed by subtype class (Fig. 3B), similar to what was observed in the clustering results of RNA expression data.
Fig. 3.
Global differences in DNA methylation profiles between BTICs and matched GBM tumors. (A) Heatmap of sample-to-sample correlations of global DNA methylation profiles from matched GBM tumors and BTICs shows that samples largely cluster by tissue source. There was no apparent clustering by sample subtype. (B) Scatterplot of PC1 versus PC2 scores shows that the first 2 PCs are largely associated with tissue source (BTIC vs. GBM tumor) followed by sample subtype classification.
As well as transcriptionally defined subtypes, we also calculated subgroups based on methylation patterns as described previously (14). All 33 tumor methylation samples with suitable sample quality were classified as GBM, with 14 classified as RTK I subtype, 10 as RTK II, and 9 as mesenchymal. Compared with tumors, cell lines in general have lower scores, which may result from lack of stromal cells. These methylation-defined subtypes are not entirely equivalent to the proneural, classical, and mesenchymal subtypes but show overlap as described previously (15) (SI Appendix, Table S2).
Differential Expression Analysis across Matched Pairs and Trios.
We further analyzed differentially expressed genes (DEGs) across tumor, BTIC, and xenograft samples in order to determine which genes and biological pathways had altered expression in the comparison between parent tumor samples and tumor-derived model systems. We found 3,172 genes up-regulated and 321 down-regulated in tumor samples relative to BTIC samples, while 3,032 were up-regulated and 1,158 were down-regulated in tumor samples relative to xenograft samples (Datasets S1 and S2). Many of the significantly enriched biological process gene ontology terms for the DEGs that separate tumor from either BTIC or xenograft samples are associated with the immune response, and 8 of the top 10 terms are shared between the 2 comparisons (Fig. 4 and Dataset S3). In both comparisons of tumor versus tumor-derived model, tumor samples form immune-high and immune-low clusters, supporting the notion that immune microenvironment is a major contributor to the observed expression patterns (SI Appendix, Fig. S4). The immune-high clusters have an overrepresentation of mesenchymal vs. nonmesenchymal subtype samples (P = 0.0034 and P = 0.0010, Fisher’s exact test), and the immune-low clusters are overrepresented with proneural samples (P = 0.0021 and P = 0.0157, Fisher’s exact test). In addition to the immune association, we find many DEGs with increased expression in tumor relative to BTIC or xenograft samples that belong to gene families associated with oncogenic signaling pathways, such as colony-stimulating factors (CSFs) and CSF receptors, fibroblast growth factors (FGFs) and FGF receptors, phosphatidylinositol 3-kinases, and vascular endothelial growth factor receptors (Datasets S1 and S2). This has implications for future drug-targeting studies that utilize these model systems, as targeting these components or their pathways in an in vitro or in vivo model is less likely to provide a response representative of the original tumor environment.
Fig. 4.
Gene ontology biological process terms enriched in the top 10% of DEGs up-regulated in each sample type relative to one another. Terms were ranked by false discovery rate (FDR) and the top 10 are shown for each comparison. The vertical black line represents the threshold for a significant P-value cutoff (FDR < 0.05). (A) DEGs up-regulated in tumors relative to BTICs. (B) DEGs up-regulated in tumors relative to xenografts. (C) DEGs up-regulated in BTICs relative to xenografts. (D) DEGs up-regulated in xenografts relative to BTICs. (E) DEGs up-regulated in BTICs relative to tumors. A plot for DEGs up-regulated in xenografts relative to tumors is absent, as the DEG set from this comparison produced no enriched terms.
In contrast to the imbalance of up-regulated DEGs in the tumor versus BTIC and xenograft sample comparisons, a similar number of up- and down-regulated DEGs were found when comparing the in vitro and in vivo models, with 1,491 up-regulated and 1,828 down-regulated in BTIC samples relative to xenograft samples (Dataset S4). The DEGs up-regulated in BTIC samples were significantly enriched for cell adhesion, extracellular matrix organization, and the collagen catabolic process, in line with their growth environment. Interestingly, the DEGs most strongly up-regulated in xenograft samples were not significantly enriched for any biological processes, suggesting that fewer pathway-specific expression differences exist between tumor and xenograft samples than either sample type compared with BTICs. Overall, it appears that immune-related gene expression is a major contributor to the expression differences between tumors and tumor-derived models. Such differences may be contributed by stromal cells or other noncancer cell types in the tumor sample, which are absent in the BTIC and xenograft samples with mouse sequences removed. Additionally, this may indicate that substantial differences in drug-target genes can occur within in vitro and in vivo model systems depending on the growth environment, and it is important to know which genes are sensitive to such environmental changes and which are more tolerant.
To identify clinically relevant expression signatures well-modeled in vitro and in vivo relative to parent tumors, we performed enrichment analysis of a set of genes with high expression that are not differentially expressed between sample types. Several KEGG pathways, including glioma, cell cycle, Erbb signaling, mTOR signaling, and pathways in cancer, were found to be significantly enriched, and include genes frequently altered in GBM such as EGFR, PTEN, CDK6, RB1, and TP53. Genes highly expressed in vivo and in vitro involved in GBM-related pathways provide a reasonable starting point for drug screens.
Of particular interest is the expression and methylation status of MGMT in matched tumors, BTICs, and xenografts, as this gene is an important marker for TMZ response in the clinic. Our data suggest that the MGMT expression level is stable across the matched samples (SI Appendix, Table S3). Methylation array data showed that 27 out of 35 pairs had the same MGMT status in tumors and BTICs, while the remaining 8 pairs had MGMT unmethylated in tumors and methylated in BTICs (Fig. 1). Six of the 8 pairs had matched genomic and transcriptomic data to infer tumor content and MGMT expression levels. Tumor content in 3 of these 6 pairs was low (<30% in the matched tumors of BT-69, BT-126, and BT-238), so the status change may come from enrichment of tumor cells in the BTIC samples. Two BTICs (BT-147 and BT-245) still showed a low level of MGMT expression, which suggested that the sample may contain a mixed population of methylated and unmethylated cells.
Microenvironment of Tumor Samples Contributes to the Differential Expression across Models and Subtypes.
The DEGs most strongly up-regulated in tumor samples appear to be associated with tumor microenvironment. To evaluate how the growth environment affects the differential expression between tumors and their in vitro and in vivo models, we used Cibersort (16) and xCell (17), 2 methods of determining cell composition via different algorithms. The Cibersort absolute score was significantly higher for tumor samples than either BTIC (P < 2.2e-16) or xenograft (P = 5.6e-8) samples, reflecting the presence of nontumor cells in the original tumor samples (SI Appendix, Fig. S5). A similar pattern was found for the xCell microenvironment score, which is the sum of stromal and immune cell scores, with higher scores in tumor samples than BTIC (P = 8.5e-14) and xenograft (P = 1.1e-3; nonhuman sequences were removed) samples (SI Appendix, Fig. S5). As expected, both methods found significantly higher nonmalignant cell-type association scores in tumor samples, which may partly contribute to the differential expression among tumors and the in vitro and in vivo models.
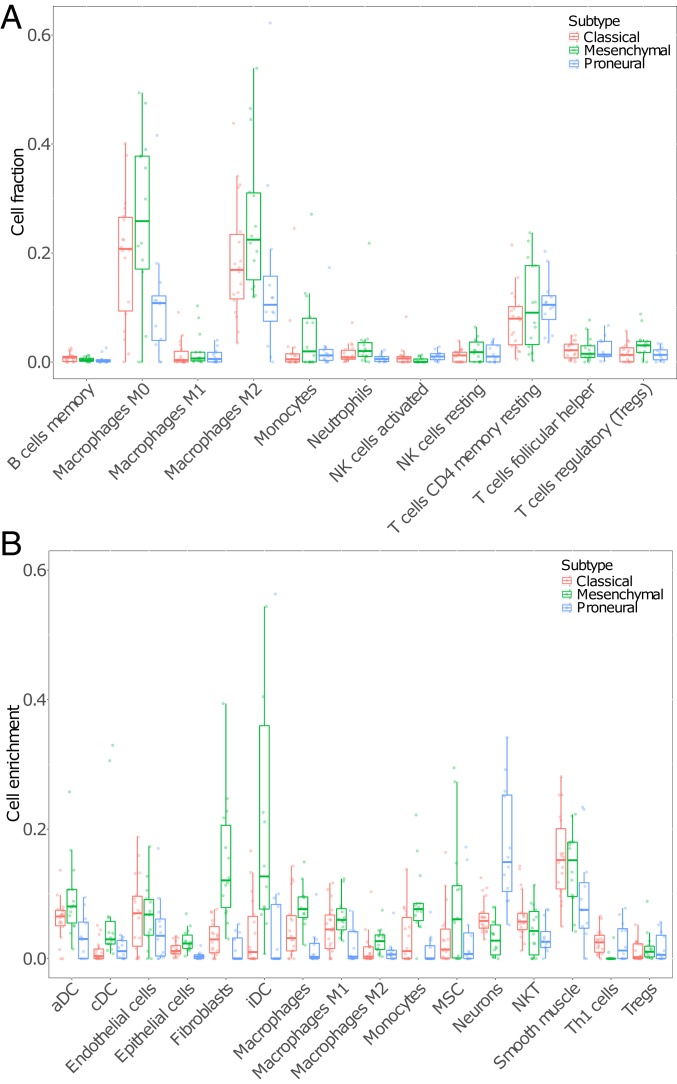
We also observed differences in nonmalignant cells among tumor subtypes. Our collection of tumor samples included 3 GBM subtypes based on the expression of the subtype signature genes defined in previous studies (11). We found that 18 out of the 44 tumor samples (41%) displayed features of the classical subtype, 12 (25%) were classified as proneural, and another 14 (32%) were classified as mesenchymal. Both Cibersort and xCell results showed that tumor samples classified as mesenchymal contained a larger variety of cell types (Fig. 5). Specifically, we found a significant positive association between several immune-suppressing cell types and the mesenchymal subtype (SI Appendix, Figs. S6 and S7). Of particular note is the higher proportion of M2 macrophages in mesenchymal tumors relative to classical and proneural tumors. Monocyte, fibroblast, and dendritic cell enrichment values are also higher in mesenchymal samples than nonmesenchymal subtype samples (SI Appendix, Fig. S7B). Overall, our result suggested that GBM transcriptional subtype is associated with immune microenvironment, especially in the mesenchymal subtype.
Fig. 5.
Cibersort cell fractions (A) and xCell enrichment scores (B) across multiple cell types with classical, mesenchymal, and proneural subtypes shown as red, green, and blue boxes and points, respectively. Only selected cell types of interest and with a median value greater than 0.001 from xCell are shown in B; for all cell types, see SI Appendix, Fig. S7. Two outlier values were excluded from B.
Integrated Genomic and Transcriptomic Analysis Reveals Two Mutually Exclusive Mechanisms of TMZ Resistance.
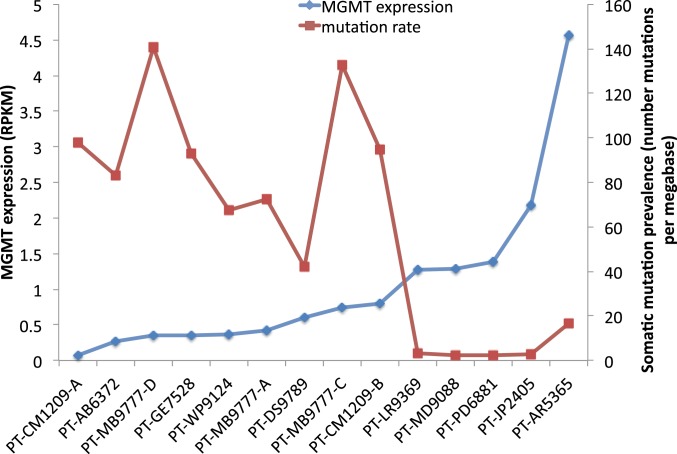
As observed before, many recurrent GBM tumors derived after TMZ treatment of the primary tumor showed a hypermutated phenotype, likely caused by mutation in MMR genes and therefore MMR deficiency (12). This is a known mechanism of TMZ resistance. However, not all posttreatment samples were hypermutated. In our collection, only 9 out of the 14 GBM-R samples showed a hypermutated phenotype and had mutations in MMR genes (Fig. 6). Another 5 GBM-R samples showed a normal mutational load, without loss-of-function mutation in MMR genes or TMZ-related mutation signature. Therefore, we examined the genome and transcriptome data of these posttreated samples to determine an alternative mechanism of resistance. One major difference between the 2 groups was the expression of the MGMT gene. All recurrent samples without hypermutation showed MGMT expression >1 RPKM (reads per kilobase of transcript per million mapped reads), while MGMT expression was not detected or at a very low level (<1 RPKM) in recurrent samples with hypermutation. This observation led to the hypothesis that MMR deficiency and MGMT expression are 2 mutually exclusive mechanisms of TMZ resistance.
Fig. 6.
MGMT expression and MMR deficiency as 2 mutually exclusive mechanisms of TMZ resistance. MGMT expression levels are indicated by RPKM value, and mutation load is shown by the number of somatic mutations per megabase from posttreatment samples. Samples with >20 mutations per Mb have a very low level of MGMT RNA expression (<1 RPKM), and they all contain a somatic MSH loss-of-function mutation and have a hypermutation phenotype. Samples with MGMT expression over 1 RPKM have a low mutation load (<20 mutations per Mb).
We tested this hypothesis of the exclusive relationship between MGMT expression and MMR deficiency in BTICs and xenografts from 2 newly diagnosed GBM samples, BT-206 and BT-73. BTICs from these pretreatment samples showed sensitivity to TMZ, and were implanted into mouse brains to establish xenografts. Both mice were treated with TMZ, and developed TMZ resistance. Cell lines were then explanted from these xenografts (BT-206-R and BT-73-R), sequenced, and compared with the original TMZ-sensitive lines. Both TMZ-sensitive BTICs (pretreatment) had no somatic mutation in the MMR gene and no detectable MGMT expression. After TMZ treatment, 1 of the TMZ-resistant cell lines, BT-206-R, acquired a mutation in MSH6 (p.T1219I) and displayed a hypermutation phenotype while MGMT expression remained undetectable. The other cell line, BT-73-R, did not have hypermutation or mutation in any MMR genes, but displayed a significantly increased level of MGMT expression (RPKM values from 0 in the pretreatment sample to 5.5 in the posttreatment sample, which is the highest level among all of the sequenced samples in this collection). Such MGMT expression change was also observed at the protein level (18). Thus, the 2 experiments showed each of the mutually exclusive mechanisms of resistance to TMZ treatment.
Current Treatment Regimen Potentially Causes a Higher Level of Heterogeneity.
As mentioned above, our posttreatment samples showed higher mutational divergence between tumors and their matched cell lines, suggesting higher levels of heterogeneity in posttreatment GBM. To further investigate the increased treatment-associated heterogeneity, we collected and sequenced multiple spatially separated samples from posttreatment tumors from 2 patients, PT-MB9777 and PT-CM1209. BTICs were also generated from at least 2 regions of the tumor. In addition, a BTIC from the pretreatment tumor of PT-CM1209 was included in this analysis.
Patient PT-MB9777 (BT-143) presented with recurrent glioblastoma after treatment with TMZ and concurrent radiation therapy followed by adjuvant TMZ. Four tumor sections were taken from distinct regions of the debulked recurrent tumor. Sequencing data showed that each section had many private SNVs, ranging from ∼2,000 to 5,000, while only 61 mutations were shared among all 4 sections (<0.4% of total mutations from all sections combined; SI Appendix, Fig. S8A). These 61 core mutations may derive from the original founding clone, and include PIK3CA p.Q546H, which may be the initial driver mutation. All 4 sections showed hypermutation phenotypes and strong TMZ mutation signatures, suggesting that the high mutation burden resulted from TMZ-induced mutagenesis. Interestingly, each of the 4 sections contained 2 distinct MSH6 mutations, and there was no shared MSH6 mutation between any pair of sections, suggesting that these MSH6 mutations each arose independently from distinct clones. All these MSH6 mutations are G>A or C>T, consistent with the pattern of TMZ-introduced mutations. Some of these mutations have also been reported in other cancers, such as p.T1219I, p.D575Y, and p.T757I, and likely cause MSH6 loss of function. These observations indicate that the exposure to TMZ led to MSH6 mutations rapidly arising in different cells, exemplifying how an often-used anticancer DNA-damaging agent can generate mutations leading to resistance.
Patient PT-CM1209 (BT-248) had 2 sections taken from a posttreatment tumor, and a BTIC derived from a pretreatment tumor. BTICs derived from each section of the posttreatment tumor were also sequenced. Comparison among the genomes of the 5 samples showed that each of the posttreatment samples accumulated thousands of mutations, but only 41 of them are shared with the pretreatment sample (SI Appendix, Fig. S8B). Among the shared ones are a PTEN homozygous p.R130* nonsense mutation and TP53 p.R273C missense mutation, which were likely present as initial drivers. The 2 posttreatment samples and their derived BTICs shared 1,159 mutations (including 2 MSH6 mutations: p.G864E and p.E1193L). As described earlier, the shared MSH6 mutations may explain the higher level of overlap among the sections.
Comparison between pre- and posttreatment samples showed 48 mutations in the pretreatment samples that are absent in the posttreatment ones, including TP53 p.V157fs and JAK1 p.R219*. The loss of a TP53 mutation in a recurrent tumor was also observed in another study, in which 2 out of 7 cases seem to have lost TP53 mutations in the recurrent tumor (19). In contrast, another TP53 mutation, R273C, is still present in both pre- and posttreatment samples. R273C is described as oncomorphic, by increasing expression of activated progrowth pathways including phosphorylation of EGFR, Her2, and retinoblastoma protein (20). The switch of function of TP53 due to R273C mutation may confer a growth advantage and lead to retention of the variant in different clones.
Discussion
To investigate how faithfully GBM cell lines and xenografts capture the molecular features of their parental tumors, we compared the genomic and transcriptomic features of matched tumor, BTIC, and xenograft samples. Multiple genome features are well-conserved across the 3 types of samples, including somatic driver mutations and genome-wide CNAs.
As expected, differences are also observed at the genome level. Most of the nonconserved mutations are from genes without known significance to cancer, and are not shared in multiple samples, suggesting they are likely from intratumor heterogeneity. Yet, in rare cases, a difference in mutations from key GBM genes was also observed, as the PDGFRA amplification and mutation described above. The PDGFRA proline 343 mutation together with high-level amplification may provide a growth advantage to tumors in the presence of PDGF-family mitogens. Such an advantage is likely not maintained within the BTIC culture conditions that lack PDGF in the media. The loss of amplification of PDGFRA proline 343 in BTICs suggests that in cell-culture conditions lacking PDGF, they may be selected in favor of cells expressing growth factor receptors that respond to those factors present in the neural stem cell growth medium.
Unlike genomic changes that take generations to accumulate, gene expression has the potential to more easily and quickly adapt to changes in the environment. Therefore, more transcriptome-level changes are expected than at the genome level. Such differences are well-captured by our analysis of cell-type composition. We used Cibersort and xCell to assess the association of GBM tumor subtype with immune cell enrichment and microenvironment. Cibersort results show that M0 and M2 macrophages represent the dominant immune cell types in our 44 tumor samples, with CD4 memory resting T cells being the next-largest fraction (Fig. 5A and SI Appendix, Fig. S7A). Since xCell has a much broader set of cell-type signatures, these results showed enrichment of several cell types other than macrophages. These include dendritic cell (DC) types, most notably immature DCs, fibroblasts, mesenchymal stem cells, endothelial cells, neurons, and smooth muscle cells, among others (Fig. 5B and SI Appendix, Fig. S7B).
The mesenchymal subtype of GBM is associated with higher cellular heterogeneity and shorter median survival than other expression subtypes (21). This GBM subtype has also been shown to be enriched for macrophage M1 and M2 polarized cells and neutrophils, while having a reduced NK cell signature (21). M2 macrophages are known to play an immunosuppressive role by inhibiting phagocytosis and T cell proliferation (22), and this immune inactivation may play a part in the shorter median survival seen in mesenchymal GBM. Monocyte, fibroblast, and dendritic cell enrichment values are also higher in mesenchymal than nonmesenchymal subtype samples (SI Appendix, Fig. S7B). Cancer-associated fibroblasts have been shown to play a role in the growth and migration of glioma cells (23), while dendritic cells are typically associated with coordination and activation of an immune response. It is possible that the monocyte signature is enriched due to the presence of myeloid-derived suppressor cells (MDSCs), as they are derived from monocytes and there is no specific signature for MDSCs in either Cibersort or xCell. This is supported by the significantly higher expression of the MDSC markers CD33 and S100A9 in mesenchymal subtype samples relative to nonmesenchymal samples (P = 0.0028 and P = 0.021). MDSCs have strong immunosuppressive activity through inhibition of CD8+ T cells and increasing production of tumor-specific regulatory T cells (24). We also find differences in immune-promoting cells. There was an elevated Th1 cell signature in classical and proneural tumors compared with mesenchymal tumors (SI Appendix, Fig. S7B). Th1 cells are thought to be tumor-suppressing by playing a role in activating cytotoxic cells (25). The effect of the immune microenvironment on glioma progression has been known for some time and has implications for treatment improvement and survival (26–31), and therefore the use of expression-based cell deconvolution and enrichment analyses of the tumor microenvironment may help to reveal novel pathways for targeted treatment via immune modulation.
Whole-genome and RNA sequencing of both pre- and posttreatment samples in our collection enabled us to compare their genomes and transcriptomes and infer potential mechanisms of treatment resistance. From our data, we hypothesize that MMR deficiency and MGMT expression are 2 mutually exclusive mechanisms of TMZ resistance. Conceptually, this can be explained by the hypothetical mechanism of how MGMT expression repairs TMZ-induced damage. MGMT encodes O6-alkylguanine DNA alkyltransferase that can remove the damage caused by TMZ, and therefore the tumor cells are not under the selection pressure of accumulated mutations. When MGMT expression is absent, C>T (or G>A) mutations introduced by TMZ accumulate in the genome. When these mutations occur in MMR genes causing a loss of function, clones deficient in MMR cannot trigger apoptosis, leading to accumulation of cells that survive with higher mutation loads. This phenomenon has been alluded to in previous literature (32, 33), but we now demonstrate a clear mutually exclusive relationship between MGMT expression and MMR deficiency.
It is well-established that GBM displays high levels of inter- and intratumoral heterogeneity and several studies have revealed spatial heterogeneity in multiregional biopsies (34, 35). In posttreatment tumors, TMZ may induce hypermutation, which can lead to potentially new mutations that drive tumor progression. Indeed, previous studies have demonstrated that specific alterations and evolutionary patterns are associated with treatment (36), and therefore increase heterogeneity and challenge the targeted posttreatment of GBM. A study using 127 GBM samples showed that genomic characterization of spatial and temporal heterogeneity in multiple sections of GBM can inform targeted treatment (37), yet clinically it could be challenging to administer multiple drugs simultaneously. Although hypermutation can impose treatment challenges, it has been linked to positive outcomes in immune checkpoint inhibitor treatment (38), and several clinical trials have been initiated in GBM patients (39). A comprehensive study of hypermutation in human cancers showed hypermutation as a consequence of treatment by chemotherapies such as alkylators, as commonly observed in GBM treated with TMZ (40). However, we show here that not all tumors developing resistance to TMZ become hypermutated. Understanding whether TMZ induces hypermutation in an individual patient and whether hypermutation creates a vulnerability to immune therapy may shed light on this promising new type of treatment.
Conclusion
Through genomic characterization and comparison of GBM tumors, cell lines, and xenografts, we found that genomic aberrations are well-conserved across matched tumors, cell lines, and xenografts. In contrast, gene expression and methylation showed higher levels of divergence, likely due to the different in vivo and in vitro growth environments among sample types. These findings suggest that an understanding of genomic information is crucial to the interpretation of drug-test results from in vitro and in vivo models, as well as the understanding of drug resistance observed in current treatment. Also, this knowledge can guide the design of drug-screening experiments to avoid bias introduced by culture conditions.
Materials and Methods
Tumor and Nucleic Acid Samples.
Formalin-fixed paraffin-embedded tumor specimens and fresh-frozen tumor specimens were collected with consent per protocols approved by the Conjoint Health Research Ethics Board of the University of Calgary. Only tumors classified histologically as GBM according to 2007 WHO CNS tumor classification criteria were included. DNA and RNA were extracted from frozen tumor material using DNeasy (Qiagen) and TRIzol (Invitrogen), respectively, per the manufacturers’ protocols.
Establishment of BTIC Lines and Xenografts.
Surgical samples from patients with newly diagnosed and recurrent glioblastoma were obtained from the Tumor Tissue Bank within the Arnie Charbonneau Cancer Institute, transported to the BTIC Core Facility, and established as described previously (2). Patient-derived BTICs were implanted into the brains of SCID mice as described previously (18, 41). Following sacrifice, the brains were removed, and tumors were excised and flash-frozen in liquid nitrogen. All procedures were reviewed and approved by the University of Calgary Animal Care Committee (Animal Protocol #AC17-0230). A more detailed description is included in SI Appendix.
Whole-Genome Sequencing and Analysis.
We implemented an automated version of the TruSeq DNA PCR-Free Kit (FC-121-1002; Illumina). Sheared DNA was end-repaired and size-selected using AMPure XP beads targeting a 300- to 400-bp fraction. After 3′ A tailing, full-length TruSeq adapters were ligated. The PCR-free library concentration was quantified using a qPCR Library Quantification Kit (KK4824; KAPA). After passing QC, the library was sequenced using the Illumina HiSeq 2000 and 2500. The sequencing depth is ∼30× for each tumor genome.
Reads were aligned to the human genome (GRCh37-lite) using BWA (v0.5.7) (42). Reads from multiple lanes were merged and duplicate-marked using Picard (v1.71) (43). Variants were called using mpileup (SAMtools v0.1.17) (44) and subsequently filtered with the built-in varFilter function. Each tumor sample (metastasis and primary) was compared with the normal sample to identify somatic copy-number variants (CNAseq v0.0.6) (45), loss-of-heterozygosity (LOH) events (APOLLOH v0.1.1) (46), single-nucleotide variants [SAMtools v0.1.17; MutationSeq v1.0.2 (47); Strelka v0.4.6.2 (48)], and small insertions and deletions (Strelka v0.4.6.2). Variants were annotated to genes with SnpEff (v3.2a) (49) using the Ensembl database (v69) (50). For tumor and xenograft samples, we evaluated tumor purity based on copy number, LOH, and variant allele frequency, and removed samples with tumor purity less than 20%.
Transcriptome Sequencing and Analysis.
Strand-specific RNA sequencing was done with the Illumina HiSeq 2000 and 2500. RNA-seq reads were analyzed with JAGuaR (51) to include alignments to a database of exon junction sequences and subsequent repositioning onto the genomic reference. RNA-seq data were processed using in-house pipeline coverage analysis and normalized expression level. Gene expression in the tumor was compared with a compendium of normal tissues and with 1 or more normal libraries from the same tissue type to identify up- and down-regulated genes. Expression levels were also evaluated by comparing against tumor and normal samples from The Cancer Genome Atlas project. Both genomic and RNA-seq tumor data were also assembled using Trans-ABySS (v1.4.3) (52) to identify structural variants and fusion genes. Differential expression analysis was performed using DESeq2 (v1.18.1) (53). Differentially expressed genes were called based on a minimum log2 fold change of 2.0 and a Benjamini–Hochberg adjusted P-value cutoff of 0.01. Datasets S1, S2, and S4 contain the mean of counts, log2 fold change, SE of the log2 fold change, Wald statistic, P value, and adjusted P value for all differentially expressed genes. Gene ontology enrichment analysis was performed using DAVID 6.8 (54).
We used 3 approaches to determine the subtype of each sample: gene enrichment analysis, Spearman correlation, and heuristic clustering. Approach 1: For each of our samples, we collected the RPKM values of 800 genes that determine GBM subtypes as described previously (11), and used the single-sample gene set enrichment analysis (ssGSEA) algorithm from the Broad Institute to determine the subtype of our samples (55). Approach 2: We collected all of the RNA-seq data as well as the subtype of GBM samples from TCGA. For each sample, we conducted a Spearman correlation with all GBM samples using the same 800 genes, and assigned the subtype of TCGA GBM sample with the highest correlation to our sample. Approach 3: We performed a heuristic clustering of all samples using the 800 signature genes. Samples within the same cluster were assigned the same subtype. We determined the subtype of each sample by a majority-win voting scheme based on the 3 prediction approaches described above.
Methylation Profiling and Analysis.
Approximately 750 ng of DNA was used for bisulfite conversion using the EZ DNA Methylation Kit (Zymo Research). Following bisulfite conversion, samples were randomized and 160 ng of bisulfite-converted DNA was applied to the Infinium HumanMethylation450 BeadChip array, as per the manufacturer’s protocols (Illumina) (56). Illumina GenomeStudio v2011.1 software was used for initial quality control checks followed by color correction and background adjustment using control probes contained on the 450K array. Subsequent processing and analysis were performed in R (v3.1.1) (57). More details on probe processing are included in SI Appendix. Methylation subtypes were determined using the methylation profiling classifier previously published (14). Methylation status of the MGMT promoter was determined from Illumina 450K data by implementation of the MGMT-STP27 model, as previously described (58, 59).
Data Availability Statement.
All genome and transcriptome sequencing datasets in this study have been deposited in the European Genome-phenome Archive; the accession number is EGAS00001002709. The methylation data have been deposited in the Gene Expression Omnibus; the accession number is GSE128654.
Supplementary Material
Acknowledgments
We thank the Calgary Brain Tumour and Tissue Bank for providing patient samples, and the sequencing, bioinformatics, and project teams at the Michael Smith Genome Sciences Centre for data processing and management. This study was funded by the Terry Fox Research Institute (TFRI GBM Project 2009–2020), and was supported in part by funding provided by the Canadian Institutes for Health Research (CIHR Award FDN-143288) (to M.A.M.). Methylation profiling was funded by the Stem Cell Network of Canada.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: All genome and transcriptome sequencing datasets reported in this study have been deposited in the European Genome-phenome Archive, https://www.ebi.ac.uk/ega/home (accession no. EGAS00001002709). The methylation data reported in this study have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE128654).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1813495116/-/DCSupplemental.
References
- 1.Ramirez Y. P., Weatherbee J. L., Wheelhouse R. T., Ross A. H., Glioblastoma multiforme therapy and mechanisms of resistance. Pharmaceuticals (Basel) 6, 1475–1506 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kelly J. J., et al. , Proliferation of human glioblastoma stem cells occurs independently of exogenous mitogens. Stem Cells 27, 1722–1733 (2009). [DOI] [PubMed] [Google Scholar]
- 3.Lee H. W., Lee K., Kim D. G., Yang H., Nam D. H., Facilitating tailored therapeutic strategies for glioblastoma through an orthotopic patient-derived xenograft platform. Histol. Histopathol. 31, 269–283 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Schonberg D. L., Lubelski D., Miller T. E., Rich J. N., Brain tumor stem cells: Molecular characteristics and their impact on therapy. Mol. Aspects Med. 39, 82–101 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xie Y., et al. , The Human Glioblastoma Cell Culture resource: Validated cell models representing all molecular subtypes. EBioMedicine 2, 1351–1363 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singh S. K., et al. , Identification of human brain tumour initiating cells. Nature 432, 396–401 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Reynolds B. A., Weiss S., Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science 255, 1707–1710 (1992). [DOI] [PubMed] [Google Scholar]
- 8.Davis B., et al. ; Terry Fox Research Institute Glioblastoma Consortium , Comparative genomic and genetic analysis of glioblastoma-derived brain tumor-initiating cells and their parent tumors. Neuro Oncol. 18, 350–360 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brennan C. W., et al. ; TCGA Research Network , The somatic genomic landscape of glioblastoma. Cell 155, 462–477 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cancer Genome Atlas Research Network , Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455, 1061–1068 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Verhaak R. G., et al. ; Cancer Genome Atlas Research Network , Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 17, 98–110 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yip S., et al. , MSH6 mutations arise in glioblastomas during temozolomide therapy and mediate temozolomide resistance. Clin. Cancer Res. 15, 4622–4629 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mazor T., et al. , Clonal expansion and epigenetic reprogramming following deletion or amplification of mutant IDH1. Proc. Natl. Acad. Sci. U.S.A. 114, 10743–10748 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Capper D., et al. , DNA methylation-based classification of central nervous system tumours. Nature 555, 469–474 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sturm D., et al. , Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell 22, 425–437 (2012). [DOI] [PubMed] [Google Scholar]
- 16.Newman A. M., et al. , Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 12, 453–457 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aran D., Hu Z., Butte A. J., xCell: Digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 18, 220 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lun X., et al. , Disulfiram when combined with copper enhances the therapeutic effects of temozolomide for the treatment of glioblastoma. Clin. Cancer Res. 22, 3860–3875 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Kim M., et al. , Mutation in TET2 or TP53 predicts poor survival in patients with myelodysplastic syndrome receiving hypomethylating treatment or stem cell transplantation. Bone Marrow Transplant. 50, 1132–1134 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Brachova P., et al. , TP53 oncomorphic mutations predict resistance to platinum- and taxane-based standard chemotherapy in patients diagnosed with advanced serous ovarian carcinoma. Int. J. Oncol. 46, 607–618 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Q., et al. , Tumor evolution of glioma-intrinsic gene expression subtypes associates with immunological changes in the microenvironment. Cancer Cell 32, 42–56.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu A., et al. , Glioma cancer stem cells induce immunosuppressive macrophages/microglia. Neuro Oncol. 12, 1113–1125 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trylcova J., et al. , Effect of cancer-associated fibroblasts on the migration of glioma cells in vitro. Tumour Biol. 36, 5873–5879 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Chen J., et al. , Suppression of T cells by myeloid-derived suppressor cells in cancer. Hum. Immunol. 78, 113–119 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Haabeth O. A., et al. , Inflammation driven by tumour-specific Th1 cells protects against B-cell cancer. Nat. Commun. 2, 240 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bowles A. P. Jr, Perkins E., Long-term remission of malignant brain tumors after intracranial infection: A report of four cases. Neurosurgery 44, 636–642, discussion 642–643 (1999). [DOI] [PubMed] [Google Scholar]
- 27.Dunn G. P., Dunn I. F., Curry W. T., Focus on TILs: Prognostic significance of tumor infiltrating lymphocytes in human glioma. Cancer Immun. 7, 12 (2007). [PMC free article] [PubMed] [Google Scholar]
- 28.Komohara Y., Ohnishi K., Kuratsu J., Takeya M., Possible involvement of the M2 anti-inflammatory macrophage phenotype in growth of human gliomas. J. Pathol. 216, 15–24 (2008). [DOI] [PubMed] [Google Scholar]
- 29.Moulignier A., et al. , Cerebral glial tumors and human immunodeficiency virus-1 infection. More than a coincidental association. Cancer 74, 686–692 (1994). [DOI] [PubMed] [Google Scholar]
- 30.Pereira M. B., et al. , Transcriptional characterization of immunological infiltrates and their relation with glioblastoma patients overall survival. OncoImmunology 7, e1431083 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang I., et al. , CD8+ T-cell infiltrate in newly diagnosed glioblastoma is associated with long-term survival. J. Clin. Neurosci. 17, 1381–1385 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Felsberg J., et al. ; German Glioma Network , Promoter methylation and expression of MGMT and the DNA mismatch repair genes MLH1, MSH2, MSH6 and PMS2 in paired primary and recurrent glioblastomas. Int. J. Cancer 129, 659–670 (2011). [DOI] [PubMed] [Google Scholar]
- 33.van Thuijl H. F., et al. , Evolution of DNA repair defects during malignant progression of low-grade gliomas after temozolomide treatment. Acta Neuropathol. 129, 597–607 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Friedmann-Morvinski D., Glioblastoma heterogeneity and cancer cell plasticity. Crit. Rev. Oncog. 19, 327–336 (2014). [DOI] [PubMed] [Google Scholar]
- 35.Patel A. P., et al. , Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 344, 1396–1401 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang J., et al. , Clonal evolution of glioblastoma under therapy. Nat. Genet. 48, 768–776 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee J. K., et al. , Spatiotemporal genomic architecture informs precision oncology in glioblastoma. Nat. Genet. 49, 594–599 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maleki Vareki S., Garrigós C., Duran I., Biomarkers of response to PD-1/PD-L1 inhibition. Crit. Rev. Oncol. Hematol. 116, 116–124 (2017). [DOI] [PubMed] [Google Scholar]
- 39.Huang J., et al. , Immune checkpoint in glioblastoma: Promising and challenging. Front. Pharmacol. 8, 242 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Campbell B. B., et al. , Comprehensive analysis of hypermutation in human cancer. Cell 171, 1042–1056.e10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang L., et al. , Gamma-secretase represents a therapeutic target for the treatment of invasive glioma mediated by the p75 neurotrophin receptor. PLoS Biol. 6, e289 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li H., Durbin R., Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Picard , Version 1.71. http://broadinstitute.github.io/picard. Accessed 1 August 2012.
- 44.Li H., et al. ; 1000 Genome Project Data Processing Subgroup , The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jones S. J., et al. , Evolution of an adenocarcinoma in response to selection by targeted kinase inhibitors. Genome Biol. 11, R82 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ha G., et al. , Integrative analysis of genome-wide loss of heterozygosity and monoallelic expression at nucleotide resolution reveals disrupted pathways in triple-negative breast cancer. Genome Res. 22, 1995–2007 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ding J., et al. , Feature-based classifiers for somatic mutation detection in tumour-normal paired sequencing data. Bioinformatics 28, 167–175 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saunders C. T., et al. , Strelka: Accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics 28, 1811–1817 (2012). [DOI] [PubMed] [Google Scholar]
- 49.Cingolani P., et al. , A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6, 80–92 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Flicek P., et al. , Ensembl 2013. Nucleic Acids Res. 41, D48–D55 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Butterfield Y., et al. , JAGuaR: Junction alignments to genome for RNA-seq reads. PLos One 9, e102398 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Birol I., et al. , De novo transcriptome assembly with ABySS. Bioinformatics 25, 2872–2877 (2009). [DOI] [PubMed] [Google Scholar]
- 53.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang W., Sherman B. T., Lempicki R. A., Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Subramanian A., et al. , Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bibikova M., et al. , High density DNA methylation array with single CpG site resolution. Genomics 98, 288–295 (2011). [DOI] [PubMed] [Google Scholar]
- 57.R, Version 3.1.1. https://www.r-project.org. Accessed 28 March 2018.
- 58.Bady P., Delorenzi M., Hegi M. E., Sensitivity analysis of the MGMT-STP27 model and impact of genetic and epigenetic context to predict the MGMT methylation status in gliomas and other tumors. J. Mol. Diagn. 18, 350–361 (2016). [DOI] [PubMed] [Google Scholar]
- 59.Bady P., et al. , MGMT methylation analysis of glioblastoma on the Infinium methylation BeadChip identifies two distinct CpG regions associated with gene silencing and outcome, yielding a prediction model for comparisons across datasets, tumor grades, and CIMP-status. Acta Neuropathol. 124, 547–560 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All genome and transcriptome sequencing datasets in this study have been deposited in the European Genome-phenome Archive; the accession number is EGAS00001002709. The methylation data have been deposited in the Gene Expression Omnibus; the accession number is GSE128654.