Significance
The nonsynonymous substitutions (dN)-to-synonymous substitutions (dS) ratio in protein-coding genes is commonly used to study the mechanisms of gene evolution. To understand why RNA viruses show large variations in dN/dS, we studied the dN/dS ratios in 21 human RNA viruses, 8 human DNA viruses, and 17 mammals. Eighteen RNA viruses, but only 2 DNA viruses and no mammals, showed a genome-average dN/dS < 0.10. Thus, many human RNA viruses exhibited extraordinarily stringent selective constraints on protein evolution. Our among-gene and among-species comparisons revealed that both positive selection and population size play significant roles in the dN/dS variation among genes and species. This study clarified several controversial issues and increased our understanding of the mechanisms of RNA virus evolution.
Keywords: picornaviruses, flaviviruses, influenza A viruses, selective constraints, positive selection
Abstract
How negative selection, positive selection, and population size contribute to the large variation in nucleotide substitution rates among RNA viruses remains unclear. Here, we studied the ratios of nonsynonymous-to-synonymous substitution rates (dN/dS) in protein-coding genes of human RNA and DNA viruses and mammals. Among the 21 RNA viruses studied, 18 showed a genome-average dN/dS from 0.01 to 0.10, indicating that over 90% of nonsynonymous mutations are eliminated by negative selection. Only HIV-1 showed a dN/dS (0.31) higher than that (0.22) in mammalian genes. By comparing the dN/dS values among genes in the same genome and among species or strains, we found that both positive selection and population size play significant roles in the dN/dS variation among genes and species. Indeed, even in flaviviruses and picornaviruses, which showed the lowest ratios among the 21 species studied, positive selection appears to have contributed significantly to dN/dS. We found the view that positive selection occurs much more frequently in influenza A subtype H3N2 than subtype H1N1 holds only for the hemagglutinin and neuraminidase genes, but not for other genes. Moreover, we found no support for the view that vector-borne RNA viruses have lower dN/dS ratios than non–vector-borne viruses. In addition, we found a correlation between dN and dS, implying a correlation between dN and the mutation rate. Interestingly, only 2 of the 8 DNA viruses studied showed a dN/dS < 0.10, while 4 showed a dN/dS > 0.22. These observations increase our understanding of the mechanisms of RNA virus evolution.
Rates of nucleotide substitution can be up to 1 million-fold higher in RNA viruses than in their cellular hosts (1–3). This rapid evolution is mainly due to high mutation rates (4, 5), while natural selection occurs mostly as purifying selection (5, 6). Selection is usually measured by the dN/dS ratio, where dS (dN) is the number of synonymous (nonsynonymous) substitutions per synonymous (nonsynonymous) site between 2 sequences. Although dN/dS has been studied in many RNA viruses (7), some important issues remain unresolved. One question is the relative contributions of natural selection and effective population size (Ne) to differences in dN/dS among viral species. Positive (Darwinian) selection increases, while negative (purifying) selection decreases, dN/dS. Unfortunately, it is difficult to determine whether an instance of elevated dN/dS is due to positive selection or relaxed negative selection. Positive selection has been found in viruses such as influenza A viruses (8, 9) and HIV-1 (10–12). However, the contribution of positive selection to the genomic mean dN/dS has not been evaluated. Because natural selection is more effective in large populations and negative selection predominates (7), an increase in Ne would be expected to reduce the mean dN/dS. Unfortunately, Ne is usually unknown.
To address the above questions, we have developed an approach. Specifically, we propose 5 rules to infer the roles of positive selection, negative selection, and Ne in the dN/dS variation among genes in the same genome and among species (or strains) (Results and Materials and Methods).
Another issue is whether evolutionary rates are correlated with mutation rates, as previous studies yielded conflicting results (5, 13). One major difficulty is that mutation rate is measured per cell infection cycle, whereas evolutionary rate is measured per year (5). Moreover, previous studies did not separate nonsynonymous and synonymous rates, so the observed correlation could be mainly due to the correlation between synonymous rate and mutation rate. We address these issues by computing the correlation between dN and dS, using dS as a proxy for mutation rate (14).
We focus on human RNA viruses, which are better studied than nonhuman viruses. For comparison, we also include human DNA viruses and mammalian genes.
Results
dN/dS Ratios in Mammals.
We first obtained the dN/dS ratios of mammalian genes, which are relatively well studied, so that the ratios can serve as a reference for human RNA viruses. Nikolaev et al. (15) estimated the dN and dS values for 17 mammalian lineages using 218 protein-coding genes. The dN/dS ratios vary from 0.155 to 0.351, with an average of 0.219 (Table 1 and Fig. 1), which is similar to the ratio (0.211) obtained from the pairwise dN and dS values between human and mouse genes in table 7.1 of ref. 3. The data in Table 1 suggest an important role of population size in the dN/dS variation among species (Discussion).
Table 1.
dN, dS, and dN/dS values in mammalian genes
| Lineage name | Scientific name | dN* | dS* | dN/dS | Population size or density | Body mass,* g |
| Shrew | Sorex araneus | 0.053 | 0.338 | 0.155 | 200 to 1,750 per km2† | 10 |
| Mouse | Mus musculus | 0.012 | 0.077 | 0.159 | NA | 18 |
| Dog | Canis lupus familiaris | 0.023 | 0.142 | 0.162 | NA | 40,000 |
| Rabbit | Oryctolagus cuniculus | 0.037 | 0.229 | 0.162 | NA | 1,820 |
| Rat | Rattus norvegicus | 0.015 | 0.092 | 0.165 | >200 million† | 340 |
| Galago | Otolemur garnetti | 0.027 | 0.160 | 0.168 | NA | 760 |
| Cow | Bos taurus | 0.034 | 0.181 | 0.187 | NA | 890,000 |
| Tenrec | Echinops telfairi | 0.054 | 0.281 | 0.193 | NA | 126 |
| Gray short-tailed opossum | Monodelphis domestica | 0.070 | 0.346 | 0.201 | NA | 71 |
| Bat | Rhinolophus ferrumequinum | 0.029 | 0.142 | 0.204 | ∼10,000 to 100,000‡ | 21 |
| Marmoset | Callithrix jacchus | 0.015 | 0.064 | 0.226 | >10,000§ | 300 |
| Armadillo | Dasypus novemcinctus | 0.042 | 0.177 | 0.236 | 13 per km2† | 4,200 |
| Elephant | Loxodonta africana | 0.027 | 0.101 | 0.268 | 625,000† | 3,980,000 |
| Human | Homo sapiens | 0.002 | 0.006 | 0.285 | 70,000 | |
| Baboon | Papio anubis | 0.003 | 0.009 | 0.289 | 1 to 63 per km2† | 21,400 |
| Macaque | Macaca mulatta | 0.005 | 0.017 | 0.309 | 5 to 15 or 57 per km2 in high or low forests† | 6,000¶ |
| Chimpanzee | Pan troglodytes | 0.003 | 0.008 | 0.351 | 192,500† | 45,000 |
| Average (SD) | 0.026 (0.019) | 0.140 (0.107) | 0.219 (0.059) |
NA, not available.
The dN and dS values and the body mass (g) data were obtained from Nikolaev et al. (15).
From ref. 49 (pp. 207, 1,520, 66, 1,003, 588, 583, and 625).
From https://www.iucnredlist.org/species/19517/21973253#population (accessed 6 December 2018).
From https://www.iucnredlist.org/species/41518/17936001 (accessed 6 December 2018).
From Wikipedia.
Fig. 1.
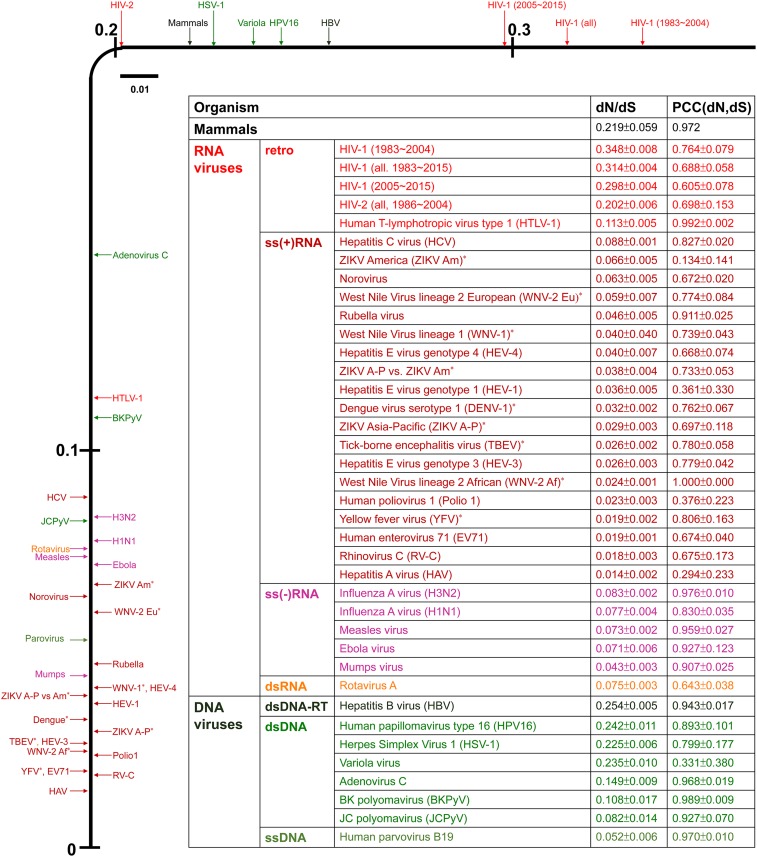
The dN/dS ratios in viruses and mammals. For each virus, the dN/dS ratio was calculated for the entire coding region of the genome. The values for each virus are the mean ratio of whole-genome dN and dS values reported in Dataset S3. Each dN/dS ratio is shown on the y axis of the figure. The flaviviruses are indicated by an asterisk. RT, retrotranscribing.
Five Rules for Inferring the Mechanisms of RNA Virus Evolution.
We proposed 5 rules for inferring the roles of positive selection, negative selection, and Ne in RNA virus evolution when dN/dS values are available for 2 or more species (or strains) from the same viral family. These rules are based on 2 rationales. First, positive selection increases, whereas negative selection decreases, the dN/dS ratio. Second, in RNA virus evolution, negative selection is much more prevalent than positive selection (7), so our interpretation of dN/dS is largely based on the slightly deleterious mutation hypothesis of Ohta (16). Under this hypothesis, an increase in the Ne tends to decrease the dN/dS ratio. Note that the genes in a genome share the same Ne.
The 5 rules are described below:
Rule 1: If a species shows low dN/dS ratios for all or most genes in the genome compared with those in other species, that species likely had a larger Ne than the other species. Alternatively, one may assume that these genes were subject to stronger negative selection in that species than in the other species, but this assumption is unlikely to hold for most genes in the genome.
Rule 2: If a gene shows a high dN/dS ratio in a species compared with both other genes in the same genome and the same gene in other species, it has likely undergone positive selection in that species. Alternatively, one may assume that the elevated dN/dS was due to relaxation of negative selection, but relaxed negative selection is less effective than positive selection in increasing the dN/dS value.
Rule 3: If the dN/dS ratio for a gene is low both among genes in the same species (genome) and among species, the gene was likely subject to stronger negative selection than other genes. The low dN/dS ratio could not be due to a larger Ne; otherwise, the other genes in the same species should also tend to show a low dN/dS.
Rule 4: If a gene shows a high dN/dS in all species, it is likely subject to weaker negative selection than other genes. There can be exceptions to this rule; for example, the HA (hemagglutinin) gene can potentially be subject to positive selection and show a high dN/dS in different influenza A strains. Therefore, some caution is needed when applying this rule.
Rule 5: If a strain (or species) shows high dN/dS ratios for all or most of the genes in the genome compared with those in other strains, then that strain likely has had a relatively small Ne and/or the effects of negative selection have not yet fully accumulated [e.g., when closely related viral isolates are compared (17)]. In contrast to rule 1, the dN/dS ratios are elevated rather than decreased, implying a smaller Ne. An elevated dN/dS can occur in a new population (strain) (i.e., the virus has not been found before in that locality) if it has undergone a population bottleneck, so that it has a small Ne, or if the new locality represents a new niche for the virus.
In the above, rule 2 is for inferring positive selection. We did not use any of the standard methods for detecting positive selection, such as that of the PAML program package (18), because most of those tests require dN/dS > 1, which is difficult to meet in RNA viruses because of the prevalence of negative selection (deleterious mutations) in RNA viruses.
dN/dS Ratios in RNA and DNA Viruses.
We studied 21 human RNA viruses, including 13 positive-sense, single-stranded [ss(+)] RNA viruses; 4 negative sense, single-stranded [ss(−)] RNA viruses; 3 ss RNA retrotranscribing (retro) viruses; and 1 double-stranded (ds) RNA virus (Fig. 1 and Dataset S1). For comparison, we also included 8 DNA viruses: 1 ds retro DNA virus, 6 ds DNA viruses, and 1 ss DNA virus (Fig. 1 and Dataset S1).
A striking observation is that 18 of the 21 RNA viruses studied show a dN/dS ratio between 0.01 and 0.10, implying that more than 90% (in some cases, close to 99%) of nonsynonymous mutations are eliminated by negative selection in these species (Fig. 1). The picornaviruses show the lowest dN/dS ratios, with 0.014 for hepatitis A virus, 0.018 for rhinovirus, 0.019 for human enterovirus 71, and 0.022 for human poliovirus 1. The flaviviruses, which include the Zika virus (ZIKV), the West Nile virus (WNV), the dengue virus (DENV), the yellow fever virus (YFV), and the tick-borne encephalitis virus (TBEV), also show very low dN/dS ratios, ranging from 0.019 to 0.066. HIV-1 is an outstanding exception, with a dN/dS ratio (∼0.314) that is much higher than that for mammals (0.219). HIV-2 shows a dN/dS ratio (0.202) close to that for mammals. Human T-lymphotropic virus type 1 (HTLV-1) shows a moderate value of 0.113. Among the 21 RNA viruses studied, only HIV-1 and HIV-2 showed a dN/dS ratio higher than the observed smallest mammalian dN/dS ratio (0.155).
The 8 DNA viruses studied tend to show a higher dN/dS ratio than the RNA viruses (Fig. 1), as found by Hughes and Hughes (19). Indeed, 4 species (hepatitis B virus, human papillomavirus type 16, herpes simplex virus type 1, and variola virus) show a dN/dS higher than that for mammals. However, 2 (JC polyomavirus and human parvovirus B19) show a dN/dS ratio < 0.1.
Flaviviruses.
In this and the next subsections, we examine the dN/dS ratios in RNA viruses in detail, trying to understand the roles of positive selection, negative selection, and Ne in the dN/dS variation among genes within and among species. For this purpose, each dS value is computed from the entire coding region of the genome under study to reduce the effect of stochastic variation in dS on the variation in dN/dS among genes (Dataset S3).
Table 2 shows the dN/dS ratios for 8 flavivirus strains (or species). In WNV-1, the NS4A gene shows the highest dN/dS among the genes in the genome and among the 8 flaviviruses, so it likely has undergone positive selection (rule 2) (20–23). We divide WNV-2 into WNV-2 Africa and WNV-2 Europe. WNV-2 Africa shows the lowest average dN/dS among the 8 strains compared; indeed, all genes except NS5 show a lower dN/dS in WNV-2 Africa than in WNV-1. Thus, WNV-2 Africa likely has a larger Ne than the other strains (rule 1). The dN/dS ratios for all genes in WNV-2 Europe are higher than those in WNV-1 except NS4A and also those in WNV-2 Africa except NS5, suggesting that WNV-2 European has a smaller Ne than WNV-1 and WNV-2 Africa and/or the effect of negative selection has not been fully accumulated (rule 5). Note that WNV-2 Europe is likely a young strain, as it was transmitted to Europe probably in early 21st century (24).
Table 2.
Means (SEs) of dN/dS values for genes in flaviviruses
| dN/dS (SE)* | |||||||||||||||||
| Gene (no. of codons) | WNV-1 | WNV-2 Africa | WNV-2 Europe | YFV | TBEV | ZIKV A-P | ZIKV Am | DENV | Average (SE) | ||||||||
| Capsid (118) | 0.047 | (0.016) | 0.000 | (0.000) | 0.057 | (0.031) | 0.050 | (0.009) | 0.083 | 0.012) | 0.119 | (0.044) | 0.104 | (0.030) | 0.051 | (0.010) | 0.064 (0.035) |
| prM (167) | 0.036‡ | (0.012) | 0.008† | (0.006) | 0.085‡ | (0.033) | 0.012† | (0.003) | 0.044‡ | (0.007) | 0.083‡ | (0.022) | 0.035† | (0.014) | 0.030† | (0.005) | 0.041 (0.027) |
| E (498) | 0.030 | (0.006) | 0.013 | (0.005) | 0.064 | (0.014) | 0.010 | (0.002) | 0.018 | (0.003) | 0.034 | (0.012) | 0.033 | (0.010) | 0.035 | (0.005) | 0.029 (0.016) |
| NS1 (352) | 0.037† | (0.010) | 0.015† | (0.008) | 0.073 | (0.019) | 0.014† | (0.002) | 0.022† | (0.005) | 0.034† | (0.007) | 0.137‡ | (0.015) | 0.037† | (0.005) | 0.046 (0.039) |
| NS2A (226) | 0.061 | (0.011) | 0.023† | (0.005) | 0.066 | (0.018) | 0.041 | (0.007) | 0.051 | (0.009) | 0.038† | (0.014) | 0.039† | (0.012) | 0.080‡ | (0.013) | 0.050 (0.017) |
| NS2B (130) | 0.049 | (0.015) | 0.013 | (0.009) | 0.104 | (0.029) | 0.034 | (0.008) | 0.015 | (0.005) | 0.039 | (0.022) | 0.066 | (0.021) | 0.044 | (0.007) | 0.046 (0.028) |
| NS3 (620) | 0.026 | (0.005) | 0.016 | (0.000) | 0.030 | (0.010) | 0.010 | (0.002) | 0.020 | (0.002) | 0.019 | (0.005) | 0.057 | (0.008) | 0.024 | (0.004) | 0.025 (0.013) |
| NS4A (131) | 0.073‡ | (0.018) | 0.000† | (0.000) | 0.037† | (0.025) | 0.022† | (0.005) | 0.020† | (0.005) | 0.012† | (0.008) | 0.038‡ | (0.021) | 0.040‡ | (0.007) | 0.030 (0.021) |
| NS4B (251) | 0.071 | (0.012) | 0.013 | (0.007) | 0.106 | (0.031) | 0.018 | (0.005) | 0.022 | (0.005) | 0.028 | (0.013) | 0.051 | (0.014) | 0.028 | (0.005) | 0.042 (0.030) |
| NS5 (903) | 0.036† | (0.007) | 0.052 | (0.005) | 0.049 | (0.012) | 0.022† | (0.004) | 0.023† | (0.002) | 0.019† | (0.005) | 0.085‡ | (0.008) | 0.026† | (0.004) | 0.039 (0.021) |
| Average (SE) | 0.046 | (0.016) | 0.015 | (0.014) | 0.067 | (0.024) | 0.023 | (0.013) | 0.032 | (0.021) | 0.042 | (0.032) | 0.064 | (0.033) | 0.039 | (0.016) | |
| Effect of positive selection removed | 0.041 | 0.015 | 0.061 | 0.023 | 0.032 | 0.036 | 0.048 | 0.035 | |||||||||
Boldface (underlined) indicates a significantly higher (lower) dN/dS ratio than those ratios in other genes in the same genome.
The gene has a significantly lower dN/dS ratio in the strains (or species) indicated than those in some other strains (species).
The gene has a significantly higher dN/dS ratio in the strains (species) indicated than those in some other strains (species).
Like WNV-2 Africa, YFV and TBEV show low average dN/dS ratios, so these 2 species likely have relatively larger Nes (rule 1).
For ZIKV, we consider ZIKV Asia-Pacific (ZIKV A-P) and ZIKV America (ZIKV Am) separately. In ZIKV A-P, the dN/dS ratio for the prM gene is the second highest among the genes in the genome and is significantly higher than those in the other flaviviruses except WNV-2 Europe, suggesting that this gene in ZIKV A-P has undergone positive selection (rule 2). The dN/dS ratios, except those for Capsid, prM, and E, are higher in ZIKV Am than in ZIKV A-P, suggesting that ZIKV Am has a smaller Ne than ZIKV A-P and/or the effect of negative selection has not been fully accumulated in ZIKV Am because it is a new population (25) (rule 5). In ZIKV Am, the dN/dS ratios for the NS1 and NS5 are high compared with other genes in the genome and higher than those dN/dS ratios in the other flaviviruses, suggesting that these 2 genes have undergone positive selection in America (rule 2).
In DENV, the NS2A gene shows strong evidence of positive selection because its dN/dS (0.080) is the highest among all genes in the genome and among all of the flaviviruses in Table 2 (rule 2).
Picornaviruses and Hepatitis E Virus.
Table 3 shows the dN/dS ratios for 4 picornaviruses. In hepatitis A virus, 6 genes (VP1, VP2, VP3, 3B, 3C, and 3D) show the lowest dN/dS ratios among the 11 genes in the genome and 3 genes (VP1, 3C, and 3D) show the lowest dN/dS ratios among the 4 species studied, suggesting that hepatitis A virus had a larger Ne than the other 3 species (rule 1) and VP1, VP2, VP3, 3B, 3C, and 3D were subject to stronger selective constraint (negative selection) than the other genes in the genome (rule 3). In rhinovirus C, 4 genes (2B, 2C, 3A, and 3B) show the lowest dN/dS ratios among the 4 species, suggesting it likely had a larger Ne than poliovirus 1 and enterovirus 71. On the other hand, rhinovirus C VP1 likely has undergone positive selection because it shows the highest dN/dS ratio among the 4 species and the second highest dN/dS among the genes in the same genome (rule 2). In poliovirus 1, 3C and 3D show relatively higher dN/dS values among the genes in the genome and the highest dN/dS among species, so these 2 genes likely have undergone positive selection in poliovirus 1. The 3C and 3D genes in enterovirus 71 might have undergone positive selection because their values are significantly higher than those in the other species, except poliovirus 1.
Table 3.
Means (SEs) of dN/dS values for genes in picornaviruses
| dN/dS (SE)* | |||||||||
| Gene (no. of codons) | Hepatitis A virus | Rhinovirus C | Poliovirus 1 | Enterovirus 71 | Average (SE) | ||||
| VP1 (293) | 0.009† | (0.003) | 0.030‡ | (0.008) | 0.012 | (0.002) | 0.019‡ | (0.001) | 0.018 (0.008) |
| VP2 (252) | 0.003 | (0.002) | 0.017‡ | (0.008) | 0.002† | (0.001) | 0.010‡ | (0.003) | 0.008 (0.006) |
| VP3 (240) | 0.002 | (0.001) | 0.013 | (0.006) | 0.006 | (0.002) | 0.005 | (0.001) | 0.007 (0.004) |
| VP4 (57) | 0.024 | (0.010) | 0.010 | (0.004) | 0.014 | (0.006) | 0.011 | (0.003) | 0.014 (0.006) |
| 2A (158) | 0.031 | (0.005) | 0.037 | (0.007) | 0.035 | (0.012) | 0.023 | (0.002) | 0.032 (0.005) |
| 2B (101) | 0.022 | (0.008) | 0.008 | (0.004) | 0.017 | (0.006) | 0.014 | (0.002) | 0.015 (0.005) |
| 2C (330) | 0.020 | (0.006) | 0.009 | (0.003) | 0.020 | (0.006) | 0.011 | (0.001) | 0.015 (0.005) |
| 3A (81) | 0.050 | (0.008) | 0.015 | (0.007) | 0.030 | (0.008) | 0.036 | (0.004) | 0.033 (0.013) |
| 3B (22) | 0.009 | (0.008) | 0.008 | (0.009) | 0.043 | (0.017) | 0.051 | (0.008) | 0.028 (0.019) |
| 3C (192) | 0.007† | (0.003) | 0.021‡ | (0.004) | 0.031‡ | (0.005) | 0.027‡ | (0.002) | 0.022 (0.009) |
| 3D (468) | 0.013† | (0.002) | 0.016 | (0.005) | 0.044‡ | (0.008) | 0.028‡ | (0.001) | 0.025 (0.012) |
| Average over genes (SE) | 0.017 | (0.014) | 0.017 | (0.009) | 0.023 | (0.014) | 0.021 | (0.013) | |
Boldface (underlined) indicates a significantly higher (lower) dN/dS ratio than those ratios in other genes in the same genome.
The gene has a significantly lower dN/dS ratio in the species indicated than those in some other species.
The gene has a significantly higher dN/dS ratio in the species indicated than those in the other species.
Table 4 shows the dN/dS ratios for 3 genotypes of hepatitis E virus (HEV-1, HEV-3, and HEV-4). In HEV-4, 2 of the 3 genes have higher dN/dS ratios than those in the other 2 strains (e.g., 0.047 and 0.031 in HEV-4 vs. 0.028 and 0.024 in HEV-3). We propose that HEV-4 had a substantially smaller Ne than HEV-1 and HEV-3 (rule 5); indeed, a study suggested that the population size of HEV-4 started to decline in the 1990s (26).
Table 4.
Means (SEs) of dN/dS values for genes in HEVs
| dN/dS (SE)* | ||||
| Gene (no. of codons) | HEV-1 | HEV-3 | HEV-4 | Average (SE) |
| ORF1 (1,693) | 0.043 (0.006) | 0.028 (0.002) | 0.047 (0.009) | 0.039 (0.008) |
| ORF3 (114) | 0.052 (0.014) | 0.040 (0.006) | 0.045 (0.016) | 0.046 (0.005) |
| C (660) | 0.016 (0.004) | 0.024 (0.008) | 0.031 (0.006) | 0.024 (0.006) |
| Average over genes (SE) | 0.037 (0.015) | 0.031 (0.007) | 0.041 (0.007) | |
Boldface (underlined) indicates a significantly higher (low) dN/dS ratio than those ratios in other genes in the same genome.
Influenza A, Mumps, and Measles Viruses.
Table 5 shows the dN/dS ratios for influenza A virus subtypes H1N1 and H3N2. It is well known that the HA and NA (neuraminidase) genes often undergo positive selection, and Table 5 shows that the dN/dS ratios for their encoding genes are indeed high, especially in H3N2. The M2 (matrix protein 2) and NS1 (nonstructural protein 1) genes also have higher dN/dS ratios. The dN/dS ratio for the M2 gene is significantly higher in H1N1 than in H3N2, suggesting that this gene in H1N1 has undergone positive selection. The NS1 and NEP (nuclear export protein) genes also show substantially higher dN/dS ratios in H1N1 than in H3N2. Thus, the average dN/dS for all genes is virtually the same for H1N1 (0.092) and H3N2 (0.088) and is substantially higher for H1N1 (0.076) than for H3N2 (0.062) if the HA and NA genes are excluded from comparison (Table 5). Therefore, positive selection in H1N1 might have been as frequent as in H3N2. The Ne has been suggested to be both larger [Volz et al. (27)] and smaller [Rambaut et al. (28)] in H1N1 than in H3N2. The data in Table 5, however, give no evidence for a substantial difference in Ne between H1N1 and H3N2 because the dN/dS ratios for the PB2, PA, and M1 genes are similar for H1N1 and H3N2 (i.e., 0.041 vs. 0.033, 0.039 vs. 0.047, 0.041 vs. 0.046). The low dN/dS ratios for these 3 genes suggest that they are subject to strong negative selection. Therefore, a significantly smaller Ne should lead to weaker negative selection and a higher dN/dS ratio (rule 4), but no such difference is observed between H1N1 and H3N2.
Table 5.
Means (SEs) of dN/dS values for genes in influenza A viruses
| dN/dS (SE)* | ||||
| Gene (no. of codons) | H1N1 | H3N2 | ||
| PB2 (759) | 0.041 | (0.004) | 0.033 | (0.002) |
| PB1 (758) | 0.041‡ | (0.003) | 0.028† | (0.002) |
| PA (716) | 0.039 | (0.003) | 0.047 | (0.005) |
| HA (563) | 0.147† | (0.017) | 0.202‡ | (0.009) |
| NP (498) | 0.055 | (0.006) | 0.072 | (0.006) |
| NA (468) | 0.169 | (0.015) | 0.180 | (0.008) |
| M2 (97) | 0.159‡ | (0.018) | 0.097† | (0.016) |
| M1 (252) | 0.041 | (0.008) | 0.046 | (0.006) |
| NS1 (230) | 0.152 | (0.017) | 0.131 | (0.019) |
| NEP (121) | 0.078 | (0.010) | 0.046 | (0.009) |
| Average (SE) | 0.092 | (0.054) | 0.088 | (0.060) |
| Average (SE) (excluding HA and NA) | 0.076 | (0.048) | 0.062 | (0.033) |
Boldface (underlined) indicates a significantly higher (lower) dN/dS ratio than those ratios in other genes in the same genome.
The gene has a significantly lower dN/dS ratio in the strain indicated than that in the other strain.
The gene has a significantly higher dN/dS ratio in the strain indicated than that in the other strain.
Although the measles and mumps viruses (Paramyxoviridae) are not related to influenza A virus, we include them here so that their estimated Nes (29) may be compared (Discussion). Table 6 shows the dN/dS ratios for the mumps and measles viruses. As the dN/dS ratios tend to be higher in the measles virus than in the mumps virus, the Ne is likely smaller in the measles virus (rule 5). For the N, P/V, and L genes, the dN/dS ratios are considerably higher in the measles virus, suggesting that these genes have undergone positive selection in the measles virus (rule 2). Thus, in this virus, positive selection may have occurred rather frequently, although it is not known for frequent positive selection.
Table 6.
Means (SEs) of dN/dS values for genes in mumps and measles viruses
| dN/dS (SE) | ||||
| Gene (no. of codons) | Mumps | Measles§ | ||
| N (537) | 0.030† | (0.008) | 0.076‡ | (0.008) |
| P/V (449) | 0.097† | (0.008) | 0.169‡ | (0.011) |
| M (355) | 0.022 | (0.005) | 0.025 | (0.006) |
| F (550) | 0.072 | (0.006) | 0.047 | (0.005) |
| H/HN (600) | 0.074 | (0.005) | 0.097 | (0.006) |
| L (2,222) | 0.019† | (0.002) | 0.058‡ | (0.003) |
| Average (SE) | 0.052 | (0.030) | 0.079 | (0.046) |
*Boldface (underlined) indicates a significantly higher (lower) dN/dS ratio than those ratios in other genes in the same genome.
The gene has a significantly lower dN/dS ratio in the species indicated than that in the other species.
The gene has a significantly higher dN/dS ratio in the species indicated than that in the other species.
The SH gene in mumps was excluded because it is absent in measles.
Retroviruses.
Table 7 shows the dN/dS ratios for HIV-1 and HIV-2. For HIV-1, we separated the isolates into 2 groups, one from 1983 to 2004 and the other from 2005 to 2015, because AIDS drugs have become increasingly effective. We note that for all genes, the dN/dS ratios are higher in the first group than in the second group of HIV-1 isolates. This difference could be because more effective drug treatments after 2004 have put a stronger negative selection pressure on the virus. Note that the difference is larger for the ENV (envelope), TAT (transactivator), and REV (regulator of expression of virion proteins) genes; TAT and REV both partially overlap ENV. Our result is in agreement with the proposal that positive selection on the ENV gene was stronger in the 1980s than in the 2000s (30). Compared with HIV-1, HIV-2 shows a lower dN/dS ratio for all genes except the VPR gene. In particular, the ratio for the ENV gene is almost 2-fold higher in HIV-1 (1983 to 2004) than in HIV-2. This is consistent with the observation that in intrapatient viral evolution, the ENV C2V3 regions evolved faster in patients infected with HIV-1 than in those infected with HIV-2 (31, 32). The POL and GAG genes show the lowest and the second lowest dN/dS among the genes in the genome in both HIV-1 and HIV-2, so they are likely subjected to stronger negative selection than the other genes (rule 3).
Table 7.
Means (SEs) of dN/dS values for genes in retroviruses
| dN/dS (SE)* | ||||||||||
| HIVs | HTLV-1 | |||||||||
| Genes (no. of codons) | HIV-1 (1983 to 2015)§ | HIV-1 (1983 to 2004)§ | HIV-1 (2005 to 2015)§ | HIV-2 (1985 to 2004)¶ | Average (SE) | |||||
| GAG (510, 429) | 0.204 | (0.004) | 0.216‡ | (0.009) | 0.199‡ | (0.005) | 0.121† | (0.006) | 0.185 (0.037) | 0.081 (0.009) |
| POL (1,057, 864) | 0.141 | (0.003) | 0.142‡ | (0.005) | 0.140‡ | (0.003) | 0.110† | (0.004) | 0.133 (0.014) | 0.086 (0.007) |
| VIF (204) | 0.329 | (0.007) | 0.355‡ | (0.011) | 0.312‡ | (0.009) | 0.185† | (0.009) | 0.295 (0.066) | NA |
| VPR (92) | 0.266 | (0.009) | 0.279 | (0.018) | 0.260 | (0.010) | 0.308 | (0.028) | 0.278 (0.019) | NA |
| TAT (108) | 0.466 | (0.013) | 0.524‡ | (0.024) | 0.432 | (0.014) | 0.358† | (0.020) | 0.445 (0.060) | NA |
| REV (110) | 0.479 | (0.013) | 0.557‡ | (0.023) | 0.437‡ | (0.013) | 0.312† | (0.030) | 0.446 (0.089) | NA |
| ENV (858, 488) | 0.561 | (0.010) | 0.643‡ | (0.023) | 0.523‡ | (0.009) | 0.315† | (0.013) | 0.511 (0.121) | 0.149 (0.012) |
| NEF (232) | 0.452 | (0.010) | 0.501‡ | (0.021) | 0.426 | (0.010) | 0.385† | (0.019) | 0.441 (0.042) | NA |
| REX (372)# | NA | NA | NA | NA | NA | 0.131 (0.011) | ||||
| TAX (353)# | NA | NA | NA | NA | NA | 0.134 (0.011) | ||||
| PRO (229) | NA | NA | NA | NA | NA | 0.201 (0.019) | ||||
| Average (SE) | 0.362 | (0.140) | 0.402 | (0.168) | 0.341 | (0.126) | 0.262 | (0.100) | 0.130 (0.040) | |
Boldface (underlined) indicates a significantly higher (lower) dN/dS ratio in this (or these) gene(s) than those ratios in other genes in the same genome. NA indicates the gene is absent in the genome.
The gene has a significantly lower dN/dS ratio in HIV-2 than those in HIV-1 (1983 to 2004) and HIV-1 (2005 to 2015).
The gene has a significantly higher dN/dS ratio in the strain(s) indicated than that (or those) in the other strain(s). HIV-1 (1983 to 2015) was not included in the tests.
The extra gene in HIV-1, viral protein U (VPU), is not included.
The extra gene in HIV-2, viral protein X (VPX), is not included. After 2004, only 2 isolates for HIV-2 were available.
The TAX coding region is contained in the coding region of REX.
Table 7 also shows the dN/dS ratios for HTLV-1, also a retrovirus. This virus shares 3 genes (GAG, POL, and ENV) with HIV-1 and HIV-2, and all of them show a much lower dN/dS in HTLV-1, suggesting a larger Ne for HTLV-1 (rule 1). The much lower dN/dS values in HTLV-1 suggest that it undergoes much less frequent adaptive evolution than HIV-1 and HIV-2, as proposed previously (33). However, in HTLV-1, the dN/dS ratios for PRO and ENV (0.201 and 0.149, respectively) are considerably higher than those for the other genes in HTLV-1. This observation suggests that PRO and ENV have undergone positive selection or have been subjected to weaker selective constraint than the other genes.
Correlation between dN and dS.
As the dN and dS values were computed from each isolate pair within a species/strain and no isolate was used more than once (Materials and Methods), pairwise comparisons between isolates could be used to compute the Pearson correlation coefficient (PCC) between dN and dS for each species/strain. Among the 30 PCC values for the RNA viruses studied, PCC ≥ 0.70 for 20 cases, 0.64 < PCC < 0.70 for 6 cases, and PCC < 0.036 for 4 cases (Fig. 1). The evolutionary implications of these data will be discussed in Discussion.
Discussion
In this study, the dN/dS ratios for the viruses were computed using the Li–Wu–Luo method (34), while those for the mammals in Table 1 were cited from a study by Nikolaev et al. (15), which used the method of Goldman and Yang (35). In table 2 of ref. 35, it is indicated that the method of Nei and Gojobori (36) gave higher dN/dS ratios for mammalian α- and β-globin genes than the method of Goldman and Yang (35). This is because the method of Nei and Gojobori (36) assumes equal likelihoods for dN and dS, so that it tends to overestimate dN and underestimate dS. The Li–Wu–Luo method (34) would not have this problem because it gives higher weights for dS than dN. Note that as mentioned in the first subsection of Results, the mean dN/dS (0.211) between human and mouse genes computed by the Li–Wu–Luo method (34) was very close to the mean dN/dS (0.219) for mammalian lineages shown in Table 1, which was computed by the method of Goldman and Yang (35). Thus, the mean ratio of 0.219 seems to be a reasonable mean value for mammalian genes.
The dN/dS ratios in mammals showed a large variation, ranging from 0.155 to 0.351 (Table 1). Small mammals such as the shrew, mouse, rat, and rabbit, which are 4 of the most common mammals, tend to have large population sizes and also have the lowest dN/dS ratios. The galago, which is a small lower primate and likely has a large population size, has a lower dN/dS than the other primates (human, chimpanzee, baboon, macaque, and marmoset). Although the African elephant is much larger than the chimpanzee, the estimated census population size (625,000) is much larger than that of the chimpanzee (192,500), probably because the elephant has a larger territory. Again, this may explain why it has a lower dN/dS (0.269) than the chimpanzee (0.351). Thus, it seems that the difference in population size is an important factor for the variation in dN/dS among mammals, and these comparisons suggest that the dN/dS ratios in Table 1 may be used to infer the relative long-term values of Ne in these mammals. Note that although mammals show a large variation in dN/dS, their dN/dS ratios are far less variable than those of viruses (2-fold vs. 20-fold) and that only HIV-1 and HIV-2 showed a ratio higher than the lowest ratio (0.155) in mammals. We speculate that one reason for the much larger dN/dS ratios in mammals is that they have a smaller Ne than RNA viruses.
Among the 21 human RNA viruses studied, 18 showed a dN/dS ratio <0.10. This observation supports the view that natural selection plays mostly a negative role in RNA virus evolution (4, 5). However, it does not imply that positive selection plays an insignificant role. Indeed, we found that positive selection plays a significant role even in the evolution of picornaviruses and flaviviruses, which showed the lowest dN/dS ratios among the RNA viruses studied.
Estimating the contribution of positive selection to genome-wide dN/dS is a complex problem and does not seem to have been attempted before. However, it may be roughly evaluated as follows, using the flaviviruses as an example. In Table 2, the dN/dS for NS4A in WNV-1 is 0.073, while the mean for the 7 other strains is (0.012 + 0.038 + 0.000 + 0.037 + 0.040 + 0.022 + 0.020)/7 = 0.024. Thus, we might predict that the dN/dS ratio for NS4A in WNV-1 would be 0.024 instead of 0.073 in the absence of positive selection. Under this assumption, the average dN/dS for WNV-1 becomes 0.041 instead of 0.046, resulting in a >10% reduction. WNV-1 NS2A also shows a relatively high dN/dS, but it is lower than those in WNV-2 and DENV NS2A; thus, whether WNV-1 NS2A has undergone positive selection is uncertain. In a similar manner, we obtain the new ratios for the other strains in Table 2. Note that we have made no change in the average dN/dS ratios in WNV-1 Africa, YFV, and TBEV because no gene in these 3 species shows clear evidence of positive selection. However, on average, positive selection has contributed ∼10% to the dN/dS ratios in flaviviruses (Table 2). Thus, when several species from a virus family or several strains from a species are available, one may be able to make a crude estimate of the contribution of positive contribution to the dN/dS ratio. This approach likely tends to give an underestimate if only clear cases of positive selection are used to estimate the contribution. A more rigorous method is needed to estimate the contribution of positive selection to dN/dS.
The 5 rules we proposed have facilitated data interpretation. In particular, using these rules, we have inferred the significant roles of both positive selection and Ne in RNA virus evolution. Moreover, we found that although the HA and NA genes are more often subject to positive selection in influenza A subtype H3N2 than subtype H1N1, the opposite is true for the M2 and PB1 genes (Table 5) and that there seems to be no substantial difference in Ne between H3N2 and H1N1.
It is interesting to note that RNA viruses from the same family tend to have similar dN/dS ratios (Tables 2–4). This might be because they experience similar transmission dynamics, live in similar intrahost environments, and may have similar genome structures and Nes. However, the ratio tends to be higher for a new population or strain (as discussed above). That might be because the virus has recently experienced a population bottleneck, it may have a selective advantage in a new niche and/or the effect of negative selection has not been fully accumulated (7, 17).
It has been suggested that vector-borne RNA viruses have lower dN/dS ratios than non–vector-borne RNA viruses (7). However, the majority of the strains used to draw this conclusion were flaviviruses (figure 3.8 of ref. 7), and, as mentioned above, these viruses belong the same family, so they would tend to have similar dN/dS ratios. Moreover, many non–vector-borne RNA viruses showed lower or similar ratios as vector-borne RNA viruses (figure 3.8 of ref. 7). Among the RNA viruses examined in this study, the 4 picornaviruses, which are non–vector-borne, showed the lowest dN/dS ratios and the 3 HEV strains showed similar ratios as the flaviviruses studied (Fig. 1). Vector-borne RNA viruses indeed tend to have low dN/dS ratios, and the proposed hypothesis that there are inherent difficulties for a virus to cyclically infect hosts that are phylogenetically divergent (e.g., from mosquitoes to humans) is attractive. However, there are other determinants of dN/dS. For example, a very large Ne would likely lead to a low dN/dS.
Bedford et al. (29) estimated Ne = 526 for influenza A H3N2 and Ne = 4,135 for the measles virus, a 7.86-fold difference. If Ne in H3N2 is indeed only 526, both negative and positive selection would be ineffective for those mutations with a fitness effect of <(1/526) = 0.0019, much higher than the selection threshold (1/4,135 = 0.0002) for the measles virus. However, despite this implied relaxed negative selection and frequent positive selection in H3N2, it has an average dN/dS ratio for all genes similar to that for the measles virus (0.088 vs. 0.079). Thus, if H3N2 has an 8-fold smaller Ne than the measles virus, this observation implies much more stringent functional constraints on influenza A virus genes except HA and NA. Note, however, that the estimate of Ne = 526 for influenza A H3N2 was based on HA gene sequences. The other genes are unlinked to HA (37), so their Ne would be larger. However, because a substantial number of mutations have small fitness effects in RNA viruses (38), the question remains how to explain the low average dN/dS over genes (0.062, when HA and NA are excluded; Table 5) if Ne is not considerably larger than 526. On the other hand, although it is not certain if the Ne values of H3N2 and measles viruses really differ by 8-fold, the study by Bedford et al. (29) did suggest a considerably smaller Ne in H3N2 than in the measles virus. Therefore, the similar dN/dS ratios for the PB1 and PB2 genes in H3N2 (0.28 and 0.33, respectively) and for the M gene in the measles virus (0.22) suggest much more stringent selective constraints on the PB1 and PB2 genes than on the M gene.
One intriguing question is why only 1 (HIV-1) of the 21 RNA viruses studied, but 4 of the 8 DNA viruses studied, showed a ratio higher than that (0.22) for mammals. It is possible that most RNA viruses have a larger Ne than DNA viruses and mammals, so that negative selection is more effective. As an RNA virus replicates rapidly, it can quickly recover from a bottleneck, so that its effect on Ne would be much less severe than that in mammals. HIV-1 shows an exceptionally high dN/dS, probably because positive selection is prevalent. Indeed, evidence for positive selection in HIV-1 has been found for the ENV, NEF, and GAG genes (12, 39–41).
ZIKV Am is a new strain and shows a ratio (0.066) considerably higher than that (0.029) for the ZIKV A-P strain, which is older. It is unlikely that this is due entirely to small dS values for the ZIKV Am isolates, because a higher average dN/dS was also seen when the dN and dS values were computed between ZIKV A-P vs. ZIKV Am (Fig. 1). Note also that almost all genes in WNV-2 Europe, a new population, showed a higher dN/dS ratio than the corresponding ratio in WNV-2 Africa, an old population. When a new virus emerges or when a virus enters a new territory, it may enjoy some selective advantages, which increases the dN/dS ratio. Also, a new strain may have recently gone through a severe bottleneck in population size, so that slightly deleterious mutations may become fixed in the population, which might later be subject to reverse and/or compensatory mutation. Additionally, the effect of purifying selection may not have fully accumulated in an emerging strain (population), so that the dN/dS ratio would tend to be higher than that of a well-established strain (17).
As RNA viruses have been found to evolve rapidly despite being subject to strong negative selection, the question arose as to whether the rapid evolution is almost completely due to high mutation rates and whether there exists a positive correlation between the rate of evolution and the rate of mutation. A weak or no correlation would mean that the rate of evolution has been strongly distorted by positive selection. Some previous studies found a correlation (5), while others did not (13, 17). However, as mentioned in the Introduction, while mutation rate is measured in terms of per cell generation, evolutionary rate is measured in terms of per year, making it difficult to compute their correlation. Moreover, previous studies did not separate synonymous and nonsynonymous rates, so it was not clear if an observed correlation was largely due to the correlation between synonymous rate and mutation rate. We therefore studied the correlation between dN and dS, because dS can be used as a proxy of mutation rate. Since dN is more strongly affected by positive selection than dS, a weak correlation between dN and dS would imply a strong effect of positive selection. We did find a positive correlation between dN and dS in the majority of the species studied, but it varied considerably among species (Fig. 1). There are 3 possible reasons for the large variation: statistical fluctuations, estimation errors, and variation in the intensity of positive selection among species. The first 2 factors can be important when dN and dS are small. To see this, let us consider the case of ZIKV Am, which has a very small PCC, only 0.13. The dN and dS values were very small (dS ranging from 0.010 to 0.025, with first, second, and third quartiles of 0.013, 0.015, and 0.017, respectively), so they were subject to strong statistical fluctuations, and even a small estimation error in dS or dN can have a strong effect on PCC. In comparison, the PCC values for ZIKV A-P and ZIKV A-P vs. ZIKV Am were 0.70 and 0.73, respectively, much higher than that (0.13) for ZIKV Am, suggesting that a positive PCC indeed exists for long-term evolution of ZIKV. For HEV genotype 1, dS ranged from 0.101 to 0.412, which is a suitable range for computing dS, so it is not clear why the PCC was only 0.36. It is also not clear why the PCC was low for human poliovirus 1 and hepatitis A virus (PCC = 0.38 and 0.29, respectively), because the ranges of dS used for these 2 cases were [0.102, 0.489] and [0.102, 0.330], respectively. Thus, although a positive correlation generally exists between dN and dS, a substantial fraction of cases show low or no correlation and the reason is unknown, although one may speculate it is, in part, due to positive selection. In conclusion, the relationship between dN and dS (or mutation rate) in RNA viruses is more complex than that in mammals (Fig. 1). Further research is required to have a good understanding of this relationship and the factors that affect this relationship.
The dN/dS values of ss(−)RNA, ss(+)RNA, and dsRNA viruses are intermingled (Fig. 1). The dN/dS values of ss(−)RNA viruses are similar to those of the rotavirus (a dsRNA virus). The retrovirus HTLV-1 has an intermediate dN/dS, whereas the retrovirus HIV-1 has the highest dN/dS. Thus, there seems to be no strong relationship between the type of replication mechanism and dN/dS, although this conclusion is difficult to assess for retroviruses, for which our sample size was small.
Materials and Methods
Data Collection and Preprocessing.
We first collected the data for the 21 RNA viruses that infect humans and have at least 10 distinct genome sequences curated by the National Center for Biotechnology Information (NCBI) Viral Genomes browser (https://www.ncbi.nlm.nih.gov/genomes/GenomesGroup.cgi?taxid=10239, accessed 13 September 2018) (Dataset S1). For DENV, we selected serotype 1 because it has more genomes available than the other serotypes. For the same reason, rotavirus A was selected to represent rotaviruses, HIV-1 group M subtype B was selected to represent HIV-1, and HIV-2 group A was selected to represent HIV-2. For influenza viruses, we selected influenza A H1N1 and H3N2 because their data were most abundant and there were disagreements about which of them had a larger population size (28, 42). For comparison, we also included 8 DNA viruses. The genome annotation and genome sizes of the viruses under study were obtained from RefSeq (43).
For each virus, we first collected the available genome sequences for isolates with a clearly labeled collection year and location (country). For HIV-1 and HIV-2, we first downloaded the codon-based multiple sequence alignments (MSAs) of the protein-coding genes of HIV-1 and HIV-2 from the HIV Sequence Database (https://www.hiv.lanl.gov/content/index) (44). An HIV-1 or HIV-2 genome was selected if the sequences of all its genes could be found in the downloaded alignments. For the viruses that were specifically curated by the NCBI Virus Variation Resource, we excluded Middle East respiratory syndrome-related coronavirus because more than half of the isolate pairs showed dS < 0.01; when dS < 0.01, the dN/dS ratio can be overestimated because an underestimation of dS can substantially inflate dN/dS. For each of the remaining viruses (ZIKV, DENV, WNV, rotavirus A, Ebola virus, and influenza A virus H1N1 and H3N2) (https://www.ncbi.nlm.nih.gov/genome/viruses/variation/), we first collected a set of genomes in which all of the genomes had distinct protein sequences in at least 1 protein-coding gene. For the case where more than 1 strain had the same protein sequences for all protein-coding genes, we chose that with the earliest isolation date according to the NCBI Virus Variation Resource. For ZIKV, we excluded the strains isolated in Africa because almost all African strains were not isolated from humans. For the viruses that were not specifically curated by the HIV sequence database and/or the NCBI Virus Variation Resource, we collected all available genomes from GenBank.
After the data collection, we first tried to eliminate closely related sequences to reduce statistical correlations. For a virus with >1,000 available genomes, we randomly selected only 1 genome per year in 1 country. For a virus with ≤1,000 genomes, we selected the genomes that had the complete set of protein-coding genes. A genome was considered to have a complete protein-coding gene if we could identify at least 90% of its coding region in the reference genome of the virus. We discarded a genome if not all of the genes were found. The genomes chosen for our analysis are indicated in red in Dataset S2.
MSA.
For HIV-1 and HIV-2, we used the codon-based MSAs we obtained in our preprocessing steps. For each of the other viruses, we first constructed the codon-based MSA for each of its protein-coding genes from the selected genomes using MUSCLE (45). Then, we constructed a codon-based MSA of the entire coding region of each virus by concatenating the codon-based MSAs of its protein-coding genes. In the case of 2 overlapping genes, we kept the overlapping region if it was <10% of both genes; otherwise, the overlapped region on the shorter gene was cleaved.
Calculation of dN/dS Ratios.
The dN and dS values between each isolate pair were computed for each gene by the Li–Wu–Luo method (34), using MEGA6.0 (46). These values were then used to compute the dN/dS ratios (Dataset S3). However, the dN and dS values in Fig. 1 were computed for the entire (concatenated) coding region of each genome because the dS value fluctuates among genes and because if the dS value for a gene is small, the estimate may have a large SE relative to the mean. Also, we avoided using any isolate more than once to reduce the correlation between isolate pairs.
For the ZIKV, the WNV, and the HEV, we classified the isolates in a species into subgroups by constructing a neighbor-joining (NJ) tree of the isolates in the species using the dS values for the entire genome. For the ZIKV, our NJ tree (SI Appendix, Fig. S1) exhibited a clear separation of the American isolates from the non-American isolates similar to that of Metsky et al. (25). For the WNV, our NJ tree exhibited a clear separation between lineage 1 and lineage 2 (SI Appendix, Fig. S2), similar to the tree of Lanciotti et al. (47). For the HEV, the genotypes of the isolates we selected were determined by comparing our NJ tree (SI Appendix, Fig. S3) with the phylogenies of the HEVs reported by Smith et al. (48).
Virus isolates are often collected from the same patients or from the same local area. Such closely related isolates usually have very small dS values, which are not suitable for computing the dN/dS ratio because the ratio can be overestimated. We therefore tried to select isolate pairs with suitable dS values. We first studied the dS distribution of all isolate pairs in a species. We then focused on the species whose median of the dS values was ≥0.1 (rhinovirus C, human poliovirus 1, human enterovirus 71, hepatitis A virus, HEV, rubella virus, norovirus, hepatitis C virus, YFV, DENV, TBEV, influenza virus A H1N1 and H3N2, measles virus, rotavirus A, HIV-1, HIV-2, and hepatitis B virus). For each of these species, we first selected a set of isolates with the criterion that all selected genome pairs have a dS ≥ 0.05. This step is performed to reduce the chance that 2 selected isolates are very closely related to each other. Then, we started the set construction by first randomly picking up 1 genome from the species under study. Additional genomes were added 1 at a time into the set only if its dS to all of the genomes already in the set was ≥0.05. After we finished constructing the set, we selected genome pairs for estimating dS and dN values. For this purpose, we required the dS value for each pair to be in the range [0.1, 0.5] because the estimation of dN/dS could be inflated if dS < 0.1 and might not be accurate if dS > 0.5. In this way, we collected a set of isolate pairs to be used for computing the dS, dN, and dN/dS values as follows. First, we randomly chose 1 pair from the set of collected pairs and removed all pairs in the set that contained either of the 2 isolates, so that no isolate was selected more than once. We continued this process until no pair remained in the set. Second, we computed the dS and dN and recorded the number of pairs that satisfied the criterion of 0.1 ≤ dS ≤ 0.5. This procedure was repeated 5,000 times to obtain an empirical distribution of the number of nonoverlapping pairs we could select. Let M be the median of the numbers of nonoverlapping pairs in the 5,000 rounds. Third, we repeated 1,000 rounds of selecting M random pairs from the collected pairs; in each round, we estimated the dN, dS, and dN/dS for each protein-coding gene and the entire genome, and also the PCC between dN and dS [PCC(dN, dS)] for the entire genome. Finally, we computed the averages and the SEs of dN, dS, dN/dS, and PCC(dN, dS) from the 1,000 rounds.
For the viruses whose median of the dS values was <0.1 (WNV, ZIKV, mumps virus, HTLV-1, and all of the dsDNA and ssDNA viruses), we followed the above procedure, but we defined the threshold for set construction as 0.005 and the dS range for collecting a set of genome pairs as [0.01, 0.5].
There were 3 cases whose M value was <4. Therefore, we relaxed the selection conditions, so that we could choose more pairs. For the WNV-2 African strains, we skipped the step for selecting the subset of strains and instead used all strains available because there are only 4 strains available. For rhinovirus C, we skipped the step for selecting the subset of strains and instead used all strains available because its dS values were generally high (median dS ≈ 2.33), and we used the range [0.1, 0.5]. For the variola virus, we defined the threshold for set construction as 0.001 and the dS range for collecting genome pairs as [0.005, 0.5] because its median dS was only ∼0.002. As the genome size of the variola virus is ∼185 kilobases, lowering the threshold to 0.005 would not severely compromise the dN/dS calculation, for the following reason. For the variola virus genome, the length of the coding region was 164,451 nucleotide sites and the number of synonymous sites is ∼32,000 according to the Li–Wu–Luo method (34). Therefore, for dS = 0.005, the SD of dS is ∼0.0004, which is much smaller than the mean.
Statistical Tests.
To compare the dN/dS ratios of a gene with the other genes in the same genome or its orthologs in the other species (strains), we first collected the 1,000 sets of dN/dS ratios of random pairs of the genes, which were generated in the preceding subsection when we calculated the averages and the SEs of dN, dS, and dN/dS.
We first compare the dN/dS ratios of the genes in the same genome. Let G be the set of n genes g1, . . ., gn in a genome that are sorted in the increasing order of the dN/dS ratio. When there are only 2 genes in G, we use the Wilcoxon rank-sum test to assess whether the distribution of the dN/dS ratios is significantly different between the 2 genes using the 1,000 sets of random pairs. The null hypothesis is that the dN/dS ratios for the 2 genes are equal, while the alternative hypothesis is that the 2 genes have different dN/dS ratios. We say that the 2 genes differ significantly in dN/dS if ≥950 tests with a P value <0.05 are observed among the 1,000 tests.
When there are more than 2 genes in G, we use the Kruskal–Wallis H test, a nonparametric and rank-based variant of ANOVA. If the null hypothesis that all genes in G have the same dN/dS ratio is rejected (i.e., ≥950 tests with a P value <0.05 among the 1,000 tests), we identify the smallest j such that the null hypothesis of equal dN/dS ratios for all genes in G1,j = (g1, . . ., gj) is rejected. Then, Gj,n = (gj, . . ., gn) represents the set of genes with relatively high dN/dS ratios. Similarly, we obtain the gene set G1,i = (g1, . . ., gi) with relatively low dN/dS ratios. If G1,i and Gj,n overlap, we remove the genes in G1,i (Gj,n) with a dN/dS ratio higher (lower) than the average dN/dS for all genes. In this way, we obtain 2 nonoverlapping gene sets, one with relatively low dN/dS ratios and the other with relatively high dN/dS ratios.
In a similar manner, we compare the dN/dS ratios of a gene among different strains or species.
The results of our analysis are given in Dataset S4.
Explanations for the 5 Rules.
We now provide some arguments for the 5 rules proposed in Results. Rule 1 says, “If a species shows low dN/dS ratios for all or most of the genes in the genome compared with those in other species, then that species likely had a larger Ne than the other species.” This rule is based on the reasoning that in RNA viruses, negative selection is much more prevalent than positive selection, implying that a larger Ne will increase the effectiveness of negative selection, and thus reduce the dN/dS ratio. Note that we do not require a low dN/dS for all genes because a gene could have undergone positive selection and show a relatively high dN/dS. Rule 2 says, “If a gene shows a high dN/dS ratio in a species compared with both the ratios for the other genes in the same genome and the ratios for the same gene in other species, it likely had undergone positive selection in that species.” This rule is based on the following reasoning. If a gene shows a higher dN/dS than some other genes in the genome, it can be because the gene is subject to weaker negative selection or it had undergone positive selection. However, weaker negative selection is not a good explanation if a higher dN/dS is not observed in other species. Rule 3 says, “If the dN/dS ratio for a gene tends to be low both among genes and among species, the gene is likely subject to stronger negative selection than other genes.” The logic for this rule is that it obviously cannot be due to positive selection or to a larger Ne, which should reduce the dN/dS for all genes, except for genes that had undergone positive selection. Rule 4 says, “If a gene shows a high dN/dS in all species, it is likely subject to weaker negative selection than other genes in the genome.” An alternative explanation for the observed high dN/dS in all species is that the gene was subject to positive selection in all species, but this possibility is low if several species (strains) have been studied. Rule 5 says, “If a strain (or species) shows high dN/dS ratios for all or most of the genes in the genome compared with those in other strains (species), then that strain likely had a smaller Ne than the other strains and/or the effect of negative selection in that strain has not been fully accumulated yet if closely related viral isolates are compared.” A smaller Ne is a better explanation for this observation than positive selection because positive selection is unlikely to occur for all or most genes in a genome at the same time. Note that if a gene shows high dN/dS ratios both within the genome and among the species compared, it is not simple to infer if the high dN/dS ratios are due to positive selection, weak negative selection, or both. The 2A gene in picornaviruses (Table 3) is such an example. The dN/dS ratios (0.031, 0.037, 0.035, and 0.023) of this gene in the 4 species studied are not significantly different. In such a case, data from more species can be helpful because if the new data again show no significant difference in dN/dS among species, the higher dN/dS ratios are likely due to weaker negative selection. On the other hand, if the new data reveal significantly lower dN/dS ratios in some species, which would imply strong negative selection (selective constraint), then the higher dN/dS ratios in other species would likely be due to positive selection.
Supplementary Material
Acknowledgments
We thank Chase Nelson, Rafael Sanjuán, John Wang, Tzi-Yuan Wang, Yi-Ling Lin, and Ming-Hsuan Lee for valuable suggestions and Haipeng Lee for compiling the population size and density data in Table 1. J.-J.L. was supported by an Academia Sinica postdoctoral fellowship.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1907626116/-/DCSupplemental.
References
- 1.Yokoyama S., Gojobori T., Molecular evolution and phylogeny of the human AIDS viruses LAV, HTLV-III, and ARV. J. Mol. Evol. 24, 330–336 (1987). [DOI] [PubMed] [Google Scholar]
- 2.Li W.-H., Tanimura M., Sharp P. M., Rates and dates of divergence between AIDS virus nucleotide sequences. Mol. Biol. Evol. 5, 313–330 (1988). [DOI] [PubMed] [Google Scholar]
- 3.Li W., Molecular Evolution (Sinauer Associates Incorporated, 1997). [Google Scholar]
- 4.Holmes E. C., The evolutionary genetics of emerging viruses. Annu. Rev. Ecol. Evol. Syst. 40, 353–372 (2009). [Google Scholar]
- 5.Sanjuán R., From molecular genetics to phylodynamics: Evolutionary relevance of mutation rates across viruses. PLoS Pathog. 8, e1002685 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duffy S., Shackelton L. A., Holmes E. C., Rates of evolutionary change in viruses: Patterns and determinants. Nat. Rev. Genet. 9, 267–276 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Holmes E. C., The Evolution and Emergence of RNA Viruses (Oxford University Press, 2009). [Google Scholar]
- 8.Fitch W. M., Leiter J. M., Li X. Q., Palese P., Positive Darwinian evolution in human influenza A viruses. Proc. Natl. Acad. Sci. U.S.A. 88, 4270–4274 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shih A. C., Hsiao T. C., Ho M. S., Li W. H., Simultaneous amino acid substitutions at antigenic sites drive influenza A hemagglutinin evolution. Proc. Natl. Acad. Sci. U.S.A. 104, 6283–6288 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maldarelli F., et al. , HIV populations are large and accumulate high genetic diversity in a nonlinear fashion. J. Virol. 87, 10313–10323 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frost S. D., et al. , Evidence for positive selection driving the evolution of HIV-1 env under potent antiviral therapy. Virology 284, 250–258 (2001). [DOI] [PubMed] [Google Scholar]
- 12.Zanotto P. M., Kallas E. G., de Souza R. F., Holmes E. C., Genealogical evidence for positive selection in the nef gene of HIV-1. Genetics 153, 1077–1089 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hicks A. L., Duffy S., Cell tropism predicts long-term nucleotide substitution rates of mammalian RNA viruses. PLoS Pathog. 10, e1003838 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jenkins G. M., Rambaut A., Pybus O. G., Holmes E. C., Rates of molecular evolution in RNA viruses: A quantitative phylogenetic analysis. J. Mol. Evol. 54, 156–165 (2002). [DOI] [PubMed] [Google Scholar]
- 15.Nikolaev S. I., et al. ; National Institutes of Health Intramural Sequencing Center Comparative Sequencing Program , Life-history traits drive the evolutionary rates of mammalian coding and noncoding genomic elements. Proc. Natl. Acad. Sci. U.S.A. 104, 20443–20448 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ohta T., Slightly deleterious mutant substitutions in evolution. Nature 246, 96–98 (1973). [DOI] [PubMed] [Google Scholar]
- 17.Holmes E. C., Patterns of intra- and interhost nonsynonymous variation reveal strong purifying selection in dengue virus. J. Virol. 77, 11296–11298 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang Z., PAML: A program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13, 555–556 (1997). [DOI] [PubMed] [Google Scholar]
- 19.Hughes A. L., Hughes M. A. K., More effective purifying selection on RNA viruses than in DNA viruses. Gene 404, 117–125 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zou G., et al. , Exclusion of West Nile virus superinfection through RNA replication. J. Virol. 83, 11765–11776 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.May F. J., Davis C. T., Tesh R. B., Barrett A. D., Phylogeography of West Nile virus: From the cradle of evolution in Africa to Eurasia, Australia, and the Americas. J. Virol. 85, 2964–2974 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McMullen A. R., et al. , Evolution of new genotype of West Nile virus in North America. Emerg. Infect. Dis. 17, 785–793 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nelson C. W., et al. , Selective constraint and adaptive potential of West Nile virus within and among naturally infected avian hosts and mosquito vectors. Virus Evol. 4, vey013 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zehender G., et al. , Reconstructing the recent West Nile virus lineage 2 epidemic in Europe and Italy using discrete and continuous phylogeography. PLoS One 12, e0179679 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Metsky H. C., et al. , Zika virus evolution and spread in the Americas. Nature 546, 411–415 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Purdy M. A., Khudyakov Y. E., Evolutionary history and population dynamics of hepatitis E virus. PLoS One 5, e14376 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Volz E. M., Koelle K., Bedford T., Viral phylodynamics. PLoS Comput. Biol. 9, e1002947 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rambaut A., et al. , The genomic and epidemiological dynamics of human influenza A virus. Nature 453, 615–619 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bedford T., Cobey S., Pascual M., Strength and tempo of selection revealed in viral gene genealogies. BMC Evol. Biol. 11, 220 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoshida I., et al. , Change of positive selection pressure on HIV-1 envelope gene inferred by early and recent samples. 6, e18630 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.MacNeil A., et al. , Long-term intrapatient viral evolution during HIV-2 infection. J. Infect. Dis. 195, 726–733 (2007). [DOI] [PubMed] [Google Scholar]
- 32.Barroso H., et al. , Evolutionary and structural features of the C2, V3 and C3 envelope regions underlying the differences in HIV-1 and HIV-2 biology and infection. PLoS One 6, e14548 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lemey P., Van Dooren S., Vandamme A.-M., Evolutionary dynamics of human retroviruses investigated through full-genome scanning. Mol. Biol. Evol. 22, 942–951 (2005). [DOI] [PubMed] [Google Scholar]
- 34.Li W.-H., Wu C.-I., Luo C.-C., A new method for estimating synonymous and nonsynonymous rates of nucleotide substitution considering the relative likelihood of nucleotide and codon changes. Mol. Biol. Evol. 2, 150–174 (1985). [DOI] [PubMed] [Google Scholar]
- 35.Goldman N., Yang Z., A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol. Biol. Evol. 11, 725–736 (1994). [DOI] [PubMed] [Google Scholar]
- 36.Nei M., Gojobori T., Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3, 418–426 (1986). [DOI] [PubMed] [Google Scholar]
- 37.Bouvier N. M., Palese P., The biology of influenza viruses. Vaccine 26 (suppl. 4), D49–D53 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanjuán R., Mutational fitness effects in RNA and single-stranded DNA viruses: Common patterns revealed by site-directed mutagenesis studies. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365, 1975–1982 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Piontkivska H., Hughes A. L., Patterns of sequence evolution at epitopes for host antibodies and cytotoxic T-lymphocytes in human immunodeficiency virus type 1. Virus Res. 116, 98–105 (2006). [DOI] [PubMed] [Google Scholar]
- 40.Brumme Z. L., et al. , HLA-associated immune escape pathways in HIV-1 subtype B Gag, Pol and Nef proteins. PLoS One 4, e6687 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Snoeck J., Fellay J., Bartha I., Douek D. C., Telenti A., Mapping of positive selection sites in the HIV-1 genome in the context of RNA and protein structural constraints. Retrovirology 8, 87 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Volz E. M., Koelle K., Bedford T., Viral phylodynamics. PLoS Comput. Biol. 9, e1002947 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haft D. H., et al. , RefSeq: An update on prokaryotic genome annotation and curation. Nucleic Acids Res. 46, D851–D860 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Foley B. T., et al. , “HIV Sequence Compendium 2018” (Tech. Rep. LA-UR 18-25673, Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, NM, 2018).
- 45.Edgar R. C., MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S., MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lanciotti R. S., et al. , Complete genome sequences and phylogenetic analysis of West Nile virus strains isolated from the United States, Europe, and the Middle East. Virology 298, 96–105 (2002). [DOI] [PubMed] [Google Scholar]
- 48.Smith D. B., et al. , Proposed reference sequences for hepatitis E virus subtypes. J. Gen. Virol. 97, 537–542 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nowak R. M., Walker E. P., Walker’s Mammals of the World (Johns Hopkins University Press, Baltimore, MD, 1999). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.