Significance
The extent to which the fossil record provides an accurate picture of past life is an important issue that is often difficult to assess. We genetically sexed 277 mammalian subfossils using high-throughput sequencing of ancient DNA, and found a strong male bias (∼75%) in Pleistocene bison (n = 186) and brown bears (n = 91), matching signals previously reported for mammoth. Similarly, a male bias was also found in species of nearly all mammal orders in 4 large museum collections. For mammals, we suggest both male behavior and appearance can lead to increased chances of representation in fossil and museum collections, and this previously unrecognized sex bias could have substantial implications for views of past population and ecological processes.
Keywords: sex ratio, sex bias, bison, brown bears, ancient DNA
Abstract
A recent study of mammoth subfossil remains has demonstrated the potential of using relatively low-coverage high-throughput DNA sequencing to genetically sex specimens, revealing a strong male-biased sex ratio [P. Pečnerová et al., Curr. Biol. 27, 3505–3510.e3 (2017)]. Similar patterns were predicted for steppe bison, based on their analogous female herd-based structure. We genetically sexed subfossil remains of 186 Holarctic bison (Bison spp.), and also 91 brown bears (Ursus arctos), which are not female herd-based, and found that ∼75% of both groups were male, very close to the ratio observed in mammoths (72%). This large deviation from a 1:1 ratio was unexpected, but we found no evidence for sex differences with respect to DNA preservation, sample age, material type, or overall spatial distribution. We further examined ratios of male and female specimens from 4 large museum mammal collections and found a strong male bias, observable in almost all mammalian orders. We suggest that, in mammals at least, 1) wider male geographic ranges can lead to considerably increased chances of detection in fossil studies, and 2) sexual dimorphic behavior or appearance can facilitate a considerable sex bias in fossil and modern collections, on a previously unacknowledged scale. This finding has major implications for a wide range of studies of fossil and museum material.
Most mammal species have a sex ratio of 1:1 at birth (1), but this may shift demographically according to differential patterns of mortality between the sexes across various life stages. A variety of factors have been identified that may affect sex ratios in mammal populations from birth to adulthood, including competition for mates and local resources, or the physiological condition of mothers (1–3). The sex ratios in natural populations are helpful in evaluating the impact of these and other factors, and to illuminate aspects of life history and comparative demographics within and across species. However, it is important that field-based studies of sex ratios capture real, rather than biased, information for both sexes. Pečnerová et al. (4) recently demonstrated that males are overrepresented in the fossil record of mammoths, and suggested that this also may be the case for the fossil record of other female herd-based mammal species, such as bison. To explore the extent of this problem, we examined the relative representation of males and females in the fossil record of 2 Late Pleistocene and Holocene megafauna, bison (Bison spp.) and brown bears (Ursus arctos), as well as in museum collections of a range of extant mammals.
Morphological sex determination of fossil and subfossil remains is generally reliable only where sexual dimorphism is apparent, but has been widely used despite this limitation (5, 6). However, it is also possible to genetically sex subfossil specimens using ancient DNA, either by direct PCR of a sex-linked gene or, more powerfully, via shotgun sequencing data (7, 8). In the latter approach, mammalian sex may be inferred by calculating the ratio of the number of reads that map to the Y versus X chromosomes (7), although, because many genome reference assemblies lack a Y chromosome, it is often better to calculate the ratio of reads mapping to the X versus nonsex chromosomes (8). The 2 X chromosomes in female mammals result in approximately double X chromosomal “read dosage” compared with males. Read dosage for both X and Y has also been evaluated using ancient DNA nuclear single-nucleotide polymorphism capture data (9). The use of read dosage is very convenient for ancient DNA studies, as the method requires relatively little sequencing effort, and is typically generated as part of routine DNA quality screening.
The read dosage approach was recently used to show that male specimens are overrepresented (72%) in Holarctic mammoth remains (4). This was suggested to result from the “lone-male model,” originally proposed to explain the excess of young adult males in the Hot Springs mammoth assemblage (10). This model proposes that, after subadult males are expelled from their familial group, they lose the protection of a large herd and experienced group leaders, and consequently engage in riskier behavior or enter more dangerous territory. As a result, the excess of males in the fossil record is caused by segregation of sexes due to their social behavior leading to differential mortality, including at taphonomically favorable sites which preserve fossils (such as bogs and tarpits). Morphological age profiling has provided support for this model at specific mammoth mass death sites (reviewed by ref. 11), but it has not previously been suggested to be a more widespread pattern across the fossil record. Furthermore, the model is not readily falsifiable without the ability to profile age at death, and other possible causes for a male bias also remain untested.
To investigate this issue further, we examined large collections of several other Late Quaternary Holarctic megafauna, bison (Bison spp.) and brown bears (U. arctos) from across Europe, Beringia, and North America (SI Appendix, Fig. S1), along with the original mammoth dataset (4) and a small dataset of the extinct Balearic bovid Myotragus balearicus. Most of the specimens were collected by the authors either directly from the field (most of the North American samples) or from existing museum collections (the majority of the European and Russian samples), providing some level of control against collection biases. We used these datasets to investigate a number of aspects of sample taphonomy and collection activities that might influence their observed sex ratios.
Late Pleistocene bison thrived on the vast mammoth steppe, leaving a substantial fossil record across Eurasia and North America. Modern bison are polygynous and gregarious, forming large herds comprising mostly female adults and young of both sexes. Adult males are solitary or form small bachelor groups, joining with the female groups for only 1 to 2 mo of the year. Similar structures have been implied for Pleistocene steppe bison (12), and this has led to predictions that, like mammoth, steppe bison remains would also exhibit a pronounced male bias (4). We examined this by genetically sexing 188 subfossil bison specimens from across Europe, Beringia, and North America, mostly recovered from alluvial sediments.
Both modern and Late Pleistocene brown bears have a Holarctic distribution, and individuals are typically either solitary or form small family groups, only congregating in large numbers under atypical circumstances of highly abundant food. Dispersal of extant brown bears is density-dependent (13), with more than one-third of females and 80 to 90% of males dispersing before adulthood (13, 14). As a result, while the lone-male model doesn’t apply to brown bears as there is no female-herd structure, the more generic model that greater landscape ranging in males might produce a male sex bias in fossil records can be examined. Given that brown bears are facultative carnivores, both their ecology and social structure are clearly different than mammoths and bison and provide a strong comparison to examine biased sex ratios. We genetically sexed 91 brown bear subfossils from Europe, Russia, and North America, recovered from caves and alluvial sediments.
Results
Shotgun sequencing data were used to confidently assign sex to 186 of the 188 subfossil bison and all of the 91 brown bear specimens, using the ratio of reads mapping to the X chromosome versus nonsex chromosomes (Materials and Methods and Table 1). A pronounced male sex bias close in size to that of mammoths (72%) was observed across all bison (75%) and the vast majority of the brown bear specimens (75%) (Table 1). Interestingly, in the more limited sets of cave-preserved bones, a contradictory signal of female bias was observed for bison (4 males, 8 females), and for brown bears from the Alps region (8 males, 16 females). However, the dominance of female brown bears has previously been noted for Austrian caves (15), and is thought to relate to behavioral differences in the Alps region, where female bears hibernate in caves, whereas males do not. Outside of the Alps, both male and female brown bears hibernate, and a strong male sex bias was observed in cave sites (50 males, 26 females), while open sites showed a more equal ratio (8 males, 7 females).
Table 1.
Male and female sample counts
| Bison | Brown bears | ||||||
| Variable | All | Postcrania | Noncave | All | Alps | Non-Alps | Mammoths* |
| Males | 139 | 72 | 135 | 58 | 8 | 50 | 67 |
| Females | 47 | 31 | 39 | 33 | 16 | 17 | 26 |
| Total | 186 | 103 | 174 | 91 | 24 | 67 | 93 |
| % male | 74.73 | 69.90 | 77.59 | 63.74 | 33.33 | 74.63 | 72.04 |
| Unassigned | 2 | 0 | 2 | 0 | 0 | 0 | 5 |
Mammoth data are from ref. 4.
To test whether additional information about the samples might explain the excess male ratio, we used an intercept-only logistic regression, as a null model, for comparison with logistic regression models containing explanatory variables. Intuitively, this null model can be interpreted as “there is a fixed ratio of males to females,” while the alternative models that we construct should be interpreted as “the sex ratio changes as the explanatory variable changes.” Alternative models were compared with the null using a likelihood ratio test (LRT). Logistic regression models with univariate predictors of sex were constructed for a variety of explanatory variables.
Bison.
For the bison, only the type of site (cave vs. noncave) was found to be significantly better than the intercept-only model, due to the female bias in the 12 cave specimens noted above (Table 2). We searched for site-specific factors that might contribute to differential mortality of males and females, but we rejected univariate models with the following explanatory variables: latitude, longitude, and altitude. Univariate models may not reveal differences that arise only when jointly considering latitude and longitude, so we implemented a Gaussian kernel 2-sample test (16), for more-complex spatial differences between the sexes. This multivariate test has good sensitivity to detect such differences (SI Appendix), but was unable to reveal any sex-specific patterns for bison remains (, ).
Table 2.
Logistic regression models with sex as the dependent variable
| Bison | Brown bears | ||||||
| Explanatory variable | All | Postcrania | Noncave | All | Alps | Non-Alps | Mammoths* |
| Intercept-only | 1.31E-10 | 8.80E-05 | 8.51E-12 | 0.00973 | 0.110 | 0.000122 | 4.21E-05 |
| Cave/noncave | 0.00176 | 0.00646 | 0.367 | 0.0399 | |||
| Material1 | 0.618 | 0.634 | 0.716 | 0.264 | 0.758 | 0.0695 | 0.132 |
| Material2 | 0.227 | 0.245 | 0.594 | 0.671 | 0.590 | ||
| 14C age | 0.768 | 0.534 | 0.614 | 0.0122 | 0.133 | 0.174 | 0.992 |
| Latitude | 0.954 | 0.657 | 0.682 | 0.619 | 0.494 | 0.0244 | |
| Longitude | 0.490 | 0.527 | 0.965 | 0.0171 | 0.708 | 0.417 | |
| Altitude | 0.676 | 0.802 | 0.847 | 0.0157 | 0.158 | 0.911 | |
| Alps/non-Alps | 0.000363 | ||||||
| Endogenous | 0.707 | 0.790 | 0.941 | 0.137 | 0.521 | 0.439 | |
| GC ratio | 0.312 | 0.625 | 0.468 | 0.723 | 0.386 | 0.168 | |
| DNA fragment length | 0.237 | 0.343 | 0.705 | 0.352 | 0.717 | 0.514 | |
| 5′ deamination (CT) | 0.558 | 0.681 | 0.644 | 0.162 | 0.446 | 0.148 | |
The row corresponding to an intercept-only model shows P values for the intercept term, which tests the null hypothesis that there is a 1:1 male to female ratio. All other cells contain P values from LRTs, comparing a logistic regression model of the form “sex ∼ X,” where X is a single explanatory variable, to the intercept-only model above it. Ps <0:05 are shown in boldface italics. Material1 consists of factors such as tooth, leg, astragalus, foot, petrous, other skull, vertebrae, flat bone, and horn. Material2 collapses factors from Material1 into crania and noncrania. Full model fitting results can be found in SI Appendix, Tables S1 and S2.
Mammoth data are from ref. 4.
To examine whether larger bison might generate a “trophy” collection bias, we searched for an increase in the proportion of male bone samples where sexual dimorphism is more apparent (e.g., skulls). Due to the small sample size of many types of bone used for DNA extraction, we also collapsed the categories into either “crania” or “postcrania,” with teeth placed into the crania category, as they are regularly taken from full or partial skulls. Neither the model containing all bone categories nor that containing collapsed categories was significantly better than the null.
Brown Bears.
While several variables (14C age, longitude, and altitude) explained the brown bear male sex bias better than an intercept-only model (Table 2), these are all related to the strong female bias in the Alps cave samples (). Outside of the Alps region, the only variables significantly better than an intercept-only model were latitude and cave/noncave (Table 2). Importantly, the male bias was less extreme at higher latitudes, where female home ranges are larger due to food scarcity, particularly after emerging from dens (17). This suggests the ratio of male to female landscape ranging may be an important factor. Brown bear bones found in caves outside the Alps showed a male bias, suggesting the female hibernation behavior in the Alps may indeed be producing the female sex bias, while, elsewhere, males dominated caves as preferred denning sites.
The kernel 2-sample test was also applied to brown bears, and identified the sex-specific spatial distribution caused by female-dominated sites in the Alps (, ). However, when applied to only brown bear remains outside the Alps, no spatial differences between the sexes could be identified (, ).
Mammoth.
We also reanalyzed the mammoth samples from the previous study (4) for comparison, using our methods for consistency. Of 98 samples, 93 were unambiguously assigned to a sex (Table 1). We evaluated the 2 variables that were provided, material type and 14C date, as possible explanations for the sex ratio. Neither was significantly better than an intercept-only model (Table 2).
Myotragus.
We sexed 9 bones of the fossil dwarf bovid M. balearicus from several different Mallorcan deposits (Balearic Islands, Spain) that were part of another study (18). Larger bones were deliberately chosen from available collections in an effort to identify specimens with good DNA preservation. All 9 bones were found to be male, suggesting that the deliberate choice of large bones in medium-small size species can result in a substantial male bias for taxa that have obvious sexual size dimorphism.
Modern Mammal Collections.
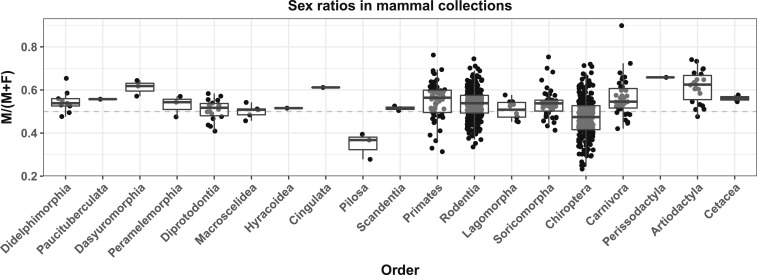
To further explore the potential for biases in museum collections, we counted male and female samples in the online databases of large mammalogy collections from the American Museum of Natural History (AMNH), New York; the Natural History Museum (NHM), London; the Smithsonian Institution National Museum of Natural History (USNM), Washington; and the Royal Ontario Museum (ROM), Ontario. These specimens of modern and historical mammal samples were obtained during the past few hundred years, largely from hunted or trapped individuals. Many were sexed at the time of collection, or subsequently, based on preserved genitalia, or clearly distinguishing secondary sexual characters (such as antlers, for most deer species). The ratio of males was calculated for each species represented by more than 100 individuals (Fig. 1). The male ratio, averaged across species, was greater than 1:1 in most mammalian orders, with notable exceptions for Chiroptera (bats) and Pilosa (sloths and anteaters). However, there was extreme variability across taxa, which may result from the method of collection (hunting vs. trapping), or the source of the samples (zoo vs. wild).
Fig. 1.
Box plot summarizing the proportion of male samples for distinct species in modern mammal collections, grouped by order. Black dots represent the proportion of males for a single species, and are jittered horizontally. Boxes show the 25th, 50th (median), and 75th percentiles. Only species with more than 100 sexed samples were included.
Discussion
A bias toward males appears to be a pervasive feature in both subfossil and live-collected mammal collections, and could be due to a range of plausible factors. Perhaps the simplest explanation in the subfossil datasets is a taphonomic artifact, where male bones in sexually dimorphic species such as bison are larger or denser and more likely to be better preserved or identified as likely to contain DNA. If this was the case, male bias might be expected to correlate with factors associated with postmortem DNA preservation, such as sample age, average DNA fragment length, and cytosine deamination rate. Greater bone density might also be expected to inhibit microbial intrusion, and thus increase the proportion of endogenous DNA (host species vs. microbial DNA). However, no such trends were observed here (Table 2), and it is reasonable to conclude that DNA preservation is equal between the sexes.
Given the evidence of equivalent postmortem preservation, the observed male bias could relate to differences in either deposition rates or collection activities. Regarding the latter, we found no evidence of a decreased male sex bias in smaller skeletal elements where sexual dimorphism is less apparent, suggesting that size-biased sampling is unlikely to be a major driver of the observed sex ratios in bison or brown bears. Consequently, our data would appear to support a biased male deposition rate in both bison and brown bears, consistent with the male landscape-ranging hypothesis proposed for mammoth (4), where male deaths are more broadly distributed. This bias is expected to be particularly strong for female-herding taxa, where female ranges are potentially clustered geographically (SI Appendix, Fig. S2). While the latter is likely to change in geographic distribution over time, random sampling across the landscape is still more likely to locate male remains.
A corollary of this model is that locations dominated by large female groups should be encountered occasionally, yielding female-biased ratios for such sites. Our dataset contains very few sites for which we have multiple samples, but we observed one such female-biased region with Alpine brown bears (for which a behavioral explanation is available). In addition, the overall female bias observed in bison cave specimens appears to be driven by 3 Canadian sites, accounting for half our total bison cave specimens (n = 6 out of 12), from which we obtained no male individuals (Extinction Cave and China Bowl Cave, Manitoba; Bison Cave, Yukon Territory), and may therefore represent sites within the core range of female herds. However, while this observation is consistent with the landscape-ranging hypothesis, the low number of cave sites from which we obtained bison remains (n = 6) prevents us from drawing strong conclusions. Finally, the female-biased sex ratios observed for bats may derive from collections dominated by sampling of single roosts, which, at certain times of the year, may be inhabited only by one sex, particularly maternity colonies (19).
Cave sites appear to provide different sex biases from open alluvial systems, possibly related to behavioral traits such as the differential denning activities for bears in the Alps and elsewhere. For example, the dominance of male brown bears in cave sites outside the Alps may reflect the ability of males to drive off females from preferred denning locations such as caves. Since the lone-male model technically only applies to herd animals, the brown bear data support a more generic model where greater landscape ranging in males results in higher average chances of fossil finds. This is supported by the finding that the male bias decreases at higher latitudes where female bear ranges are larger. It would be possible to further investigate specific predictions of the lone-male model by examining the age at death, which should be younger for males than for females, due to lack of experience and herd protection. Age at death can be measured morphologically from factors such as tooth eruption and wear, and, in mammoths, by dating the enamel layers of tusks. However, large collections of subfossil teeth preserving ancient DNA have not, so far, been analyzed. Certain methylomic loci can be used to indicate age in humans (20), so cytosine methylation in ancient DNA (21) could potentially also be used to age subfossil specimens.
Collection Bias.
Where we deliberately sampled thicker and larger M. balearicus bones to maximize DNA preservation in a warm climate, all were found to be male (), indicating that this bias can potentially affect subfossil collections. It is highly likely that a similar collection bias affects modern mammalian collections arising from predominantly hunted and trapped individuals. For modern mammals, this bias need not only be driven by deliberate selection of large “impressive” male specimens, but also due to other factors such as hunters or trappers avoiding females tending young because of legislation or other motivation. At the same time, museum collections do not only represent the choices of collectors and hunters. Museum curators may act judiciously to select materials for accession with a goal of representing both sexes (as well as representing different localities, times, or ages) for species in their collections, a factor that may, in fact, counteract, to some extent, any tendency for extreme male bias in some collections. Whatever the cause, the pervasiveness of male overrepresentation in mammal collections requires attention. The use of museum specimens as the major platform for comparative anatomy, morphological variability, ontogenetic development, parasitology, stable isotope chemistry, stomach contents, and many other aspects of biology in mammalian species (22) raises the question of the extent that previous studies may be impacted by an unrecognized male bias.
While we have not examined the extent of male bias in modern bird collections, we suspect that the remarkable sexual dimorphism in color in many bird species may lead to similar male bias, as males typically exhibit more visually striking plumage. However, data available for the extinct moas of New Zealand suggest a different pattern for ratite birds, where sex roles are reversed. Moa exhibit pronounced reverse sexual size dimorphism, with females 2 or more times heavier than males (23). Fossil remains of 4 different moa species show heavily female-dominated sex ratios across 2 different deposits, with suggestions that female territoriality led to their abundance near watering holes or other prime sites (24). Importantly, this provides a further indication that differential sexual morphology and behavioral ecology of large vertebrates, rather than being male specifically, may be important drivers of sex ratios observed in the fossil record.
Conclusion
We observed a substantial excess of male bison and brown bear subfossils across a range of Late Quaternary Holarctic deposits, consistent with a model of greater landscape ranging in males. The female-herd structure of bison, like mammoths, explains the high ratio of male subfossils, as females are expected to be clustered geographically, and therefore more heterogeneous on the landscape. In the case of brown bears, the lack of a herd structure leads to a more equal distribution of subfossil remains in alluvial sites, but a notably lower male ratio at higher latitudes where female ranges are larger.
Regardless of the actual mechanisms, a substantial male sex bias exists in both the subfossil record and modern mammalian collections. The biases are highly taxon-specific, and are likely to differ between collections. This has major implications for studies that assume their samples are representative of the whole population under consideration, such as comparisons of taxa or studies of factors such as bone dietary isotopes where sexes differ in their behavior or distribution. Our results suggest that sex biases are ubiquitous in collections, and should not be ignored. The routine application of genetic sexing will allow the possible confounding effects of cryptic sexual dimorphism to be identified when working with subfossils or museum collections.
Materials and Methods
Laboratory Procedures.
All ancient DNA work was performed in the purpose-built isolated ancient DNA facility at the University of Adelaide’s Australian Centre for Ancient DNA, or the Henry Wellcome Ancient Biomolecules Centre at Oxford University, following previously published guidelines (25, 26). DNA was extracted from bison samples using either a phenol–chloroform (27) or in-house silica-based method (28). Brown bear samples were extracted using a phenol–chloroform-based extraction protocol (29) or an in-house silica-based protocol (30). Double-stranded Illumina sequencing libraries were built from 25 L of DNA extract following the partial uracil–DNA–glycosylase treatment protocol (31), modified to include the use of dual 7-mer internal barcode sequences as per ref. 28. The libraries were pooled and sequenced using paired-end reactions on an Illumina MiSeq, NextSeq, or HiSeq.
Alignment and Filtering.
Demultiplexed reads were mapped using the Paleomix pipeline (32) configured to use BWA-aln (33) with typical ancient DNA parameters (-l 16384 -o 2 -n 0.01). Alignments were subsequently filtered to exclude those with mapping quality lower than 30, and fragments longer than 100 base pairs (bp). We considered only samples with at least 5,000 reads mapped to the nuclear genome, and subsampled down to ∼20,000 reads for sex determination.
Bison.
Bison reads were mapped to a composite cattle reference assembly formed by concatenating the assembly UMD3.1 (34), with the Y chromosomal sequence from Btau4.6.1 (35). As very few reads map to this Y sequence, we were unable to do genetic sexing using counts of reads mapping to the Y chromosome vs. counts of those mapping to the X chromosome as in ref. 7. We instead counted reads mapping to the X chromosome vs. the autosome, in an approach similar to ref. 8.
We counted the reads that mapped to the X chromosome, , and the reads that mapped to the autosome, , using samtools idxstats (36). Assuming reads are drawn from the genome uniformly along its length, the observed ratio can be predicted from the length of the X chromosome, , and the length of the autosome, . Conditional on the sex, the expected ratios are
The likelihood of the male ratio given the observed counts and can thus be described using the Binomial probability mass function,
and similarly for the female ratio. We determined whether one sex fit the data best using an LRT, requiring that the LRT result in a P value for one or the other sex, in order that a sex be assigned. Further, we considered
depending on the result of the LRT, to cluster males near 0.5 and females near 1.0. We did not assign a sex to samples that had , under the assumption that they violated both male and female models. Our Python code implementation for the sex assignment is available from https://github.com/grahamgower/sexassign.
Mammoths.
Mammoth sexing was done using the same method as for bison. Read counts and were taken from supplementary table 1 of ref. 4, which also lists material type and 14C age for each sample. and were derived from the African elephant reference loxAfr4. A total of 398,360 mapped reads were reported for sample L285, which is likely missing a digit. We appended a zero, placing this sample into the male range, which matches the inferred sex from ref. 4.
Bears.
Brown bear reads were mapped to the polar bear reference UrsMar1.0 (37), a scaffold-level reference assembly. For sex determination, we counted reads that mapped to X-linked scaffolds as , and applied the same method as for bison. Only scaffolds longer than 1 Mbp were used in calculations of , , , and .
A list of X-linked scaffolds (SI Appendix, Table S3) was obtained by mapping all UrsMar1.0 scaffolds to the dog reference CanFam3.1 (38), with minimap2 (39). The default mapping parameters were used (minimap2 CanFam3.1.fasta UrsMar1.0.fasta > aln.paf), which provides an approximate alignment lacking base-level precision. We retained only UrsMar1.0 scaffolds having more than 100 kbp cumulative matches to the CanFam3.1 chrX, resulting in 28 putatively X-linked scaffolds comprising 102 Mbp of sequence.
Model Violations.
While care was taken to minimize contamination from exogenous sources, such model violations may yet occur due to sample cross-contamination. Other factors that may contribute to sample-specific model violations include chromosome translocations, aneuploidy, and unanticipated postmortem preservation artifacts that (dis)favor one chromosome over another.
Systematic model violations may also be present, such as due to reference assembly errors, or postmortem preservation artifacts. Inactivated copies of chromosome X are heavily methylated, which may lead to additional postmortem DNA fragmentation compared with the active copy and hence fewer reads mapping from the inactivated chromosome. Conversely, an inactivated chromosome is condensed into heterochromatin, which may facilitate greater postmortem preservation than the active copy.
We note that the UrsMar1.0 assembly was derived by sequencing a male, and thus Y-linked scaffolds may be present, while the CanFam3.1 assembly was derived by sequencing a female and thus lacks a chrY. This leaves open the possibility that the pseudoautosomal region (PAR) on Y-linked UrsMar1.0 scaffolds could have mapped to CanFam1.0 chrX. The dog PAR region is 6.6 Mbp (40), small compared with the size of chrX, but this could yet artificially inflate values for males. Nonetheless, we observed a clear separation of values into 2 cohorts, with few intermediate values, suggesting model violations are rare, or do not notably influence sex determination.
GLM.
Logistic regression models were implemented in R (41) using the bayesglm function with default parameters, from the arm package (42). For categorical variables with 3 or more levels, we constructed multiple models, each with different reference levels, to verify this did not have a notable influence on the outcome.
Testing Spatial Distribution.
We implemented the 2-sample kernel test described by ref. 16 with a Gaussian kernel, and obtained a P value by comparing the test statistic to 1,000 permutations. The Gaussian kernel , where is the great circle distance between and , has a scaling parameter , which was chosen to maximize the test statistic in each permutation. More details regarding the test statistic, and validation of its performance for spatial data, can be found in SI Appendix. Our R code implementation for the kernel test is available from https://github.com/grahamgower/kernel-test.
Mammalian Databases.
For mammalian species listed in the PanTHERIA WR05 database (43), we downloaded sample information from 3 museum databases: AMNH (44), NHM (45), and ROM v11.5 (46). In addition, samples for 38 species were manually downloaded from USNM (47). We excluded juveniles and hybrids, and sex ratios were calculated only for species represented by more than 100 samples.
Data Availability.
Mapped reads and sample-associated metadata are available from https://figshare.com/projects/Widespread_male_sex_bias_in_mammal_fossil_and_museum_collections/60446.
Supplementary Material
Acknowledgments
We thank P. Bover, J. Soubrier, S. Bray, J. Austin, J. Metcalf, B. Shapiro, C. Valdiosera, M. Wilson, D. Makowiecki, I. Barnes, and M. T. Rabanus-Wallace for assistance with sample collection and lab work. In addition, we are grateful for the assistance of the many institutions and curators who provided samples for this study: K. Østbye and E. Østbye (University of Oslo); S-E. Lauritzen (University of Bergen); K. Aaris-Sørense (Zoological Museum, University of Copenhagen); M. Pacher (Institute of Palaeontology, University of Vienna); P. Kosintsev (Institute of Plant and Animal Ecology, Russian Academy of Sciences); D. Fedje (Parks Canada); D. Guthrie, R. Gangloff, and R. Stephenson (University of Alaska Fairbanks); D. Harington (Canadian Museum of Nature); J. Storer, P. Matheus, and G. Zazula (Yukon Paleontology Program); A. Kitchener (National Museums Scotland); B. Hockett (Bureau of Land Management); D. Tedford (AMNH); P. Wrinn and S. Vasil’ev (Institute of Archaeology and Ethnography, Russian Academy of Sciences); L. Martin (University of Kansas); G. Storrs (Museum of Natural History & Science, Cincinnati Museum); A. Sher (Paleontological Institute, Russian Academy of Sciences); S. Zimov and S. Davidoff (Northeast Science Station); N. Vereshchagin (Zoological Institute, St. Petersburg); J. Burns (Provincial Museum Alberta); J. Driver (Simon Fraser University); K. Rogers (Bell Museum Natural History); M. Arakelyan (Yerevan State University); M. Križnar (Slovenian Museum of Natural History); N. Spassov (National Museum of Natural History, Sofia); L. Bartosiewicz (Institute of Archeological Science, Hungary); A. Archacka (Nature Museum in Drozdowo); V. Gedminas (Tadas Ivanauskas Zoological Museum in Kaunas); M. Szymkiewicz (Nature Museum in Olsztyn); E. Keczyńska-Moroz (Białowieża National Park); J. Jastrzȩbski and J. Deptuła (Northern-Mazovian Museum in Łomża); N. Czeremnyh (State Museum of Natural History in Lviv, old Museum Dzieduszyckich); K. Wysocka (Vinnytsia Regional Local History Museum); M. Czarniauski (Institute of History NAS of Belarus in Minsk); B. Studencka (Museum of the Earth PAS, Poland); U. Göhlich, F. Zachos, and E. Pucher (Vienna Natural History Museum); D. Nagel and D. Doeppes (University of Vienna); and staff at the Lietuvos Nacionalinis Muziejus and Universidad de Burgos. This work was supported by Australian Government Research Training Program Scholarships (to G.G., L.F., A.T.S., and A.L.v.L.), University of Adelaide Research Fellowship (to B.L.), Australian Research Council grant and Fellowship support (to A.C., B.L., H.H., and K.J.M.), and Polish National Science Centre grants (nos. 2013/11/B/NZ8/00914 and N N304 301940 [to R.K.] and 2015/17/N/ST10/01707 [to E.H.]).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1903275116/-/DCSupplemental.
References
- 1.Karlin S., Lessard S., Theoretical Studies on Sex Ratio Evolution (Princeton University Press, Princeton, NJ, 1986). [PubMed] [Google Scholar]
- 2.Trivers R. L., Willard D. E., Natural selection of parental ability to vary the sex ratio of offspring. Science 179, 90–92 (1973). [DOI] [PubMed] [Google Scholar]
- 3.Charnov E. L., Sex ratio selection in an age-structured population. Evolution 29, 366–368 (1975). [DOI] [PubMed] [Google Scholar]
- 4.Pečnerová P., et al. , Genome-based sexing provides clues about behavior and social structure in the woolly mammoth. Curr. Biol. 27, 3505–3510.e3 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Frayer D. W., Wolpoff M. H., Sexual dimorphism. Annu. Rev. Anthropol. 14, 429–473 (1985). [Google Scholar]
- 6.Rehg J. A., Leigh S. R., Estimating sexual dimorphism and size differences in the fossil record: A test of methods. Am. J. Phys. Anthropol. 110, 95–104 (1999). [DOI] [PubMed] [Google Scholar]
- 7.Skoglund P., Storå J., Götherström A., Jakobsson M., Accurate sex identification of ancient human remains using DNA shotgun sequencing. J. Archaeol. Sci. 40, 4477–4482 (2013). [Google Scholar]
- 8.Mittnik A., Wang C. C., Svoboda J., Krause J., A molecular approach to the sexing of the triple burial at the upper paleolithic site of Dolní Věstonice. PLoS One 11, e0163019 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fu Q., et al. , The genetic history of Ice Age Europe. Nature 534, 200–205 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Agenbroad L., Mead J., Age structure analyses of Mammuthus columbi, Hot Springs mammoth site, South Dakota. Curr. Res. Pleistocene 4, 101–102 (1987). [Google Scholar]
- 11.Haynes G., Finding meaning in mammoth age profiles. Quat. Int. 443, 65–78 (2017). [Google Scholar]
- 12.Guthrie R. D., Frozen Fauna of the Mammoth Steppe: The Story of Blue Babe (University of Chicago Press, Chicago, IL, 1989). [Google Scholar]
- 13.Støen O. G., Zedrosser A., Sæbø S., Swenson J. E., Inversely density-dependent natal dispersal in brown bears Ursus arctos. Oecologia 148, 356–364 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Zedrosser A., Støen O. G., Sæbø S., Swenson J. E., Should I stay or should I go? Natal dispersal in the brown bear. Anim. Behav. 74, 369–376 (2007). [Google Scholar]
- 15.Döppes D., Pacher M., 10,000 years of Ursus arctos in the Alps – A success story? Analyses of the Late Glacial and Early Holocene brown bear remains from alpine caves in Austria. Quat. Int. 339-340, 266–274 (2014). [Google Scholar]
- 16.Gretton A., Borgwardt K. M., Rasch M. J., Schölkopf B., Smola A., A kernel two-sample test. J. Mach. Learn. Res. 13, 723–773 (2012). [Google Scholar]
- 17.Bunnell F. L., Tait D. E. N., “Population dynamics of bears – implications” in Dynamics of Large Mammal Populations, Fowler C. W., Smith T. D., Eds. (John Wiley, 1981), pp. 75–98. [Google Scholar]
- 18.Bover P., et al. , Unraveling the phylogenetic relationships of the extinct bovid Myotragus balearicus Bate 1909 from the Balearic Islands. Quat. Sci. Rev. 215, 185–195 (2019). [Google Scholar]
- 19.Kunz T. H., Ed. Ecology of Bats (Plenum Press, New York, NY, 1982). [Google Scholar]
- 20.Horvath S., Raj K., DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat. Rev. Genet. 19, 371–384 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Llamas B., et al. , High-resolution analysis of cytosine methylation in ancient DNA. PLoS One 7, e30226 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McLean B. S., et al. , Natural history collections-based research: Progress, promise, and best practices. J. Mammal. 97, 287–297 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bunce M., et al. , Extreme reversed sexual size dimorphism in the extinct New Zealand moa Dinornis. Nature 425, 172–175 (2003). [DOI] [PubMed] [Google Scholar]
- 24.Allentoft M. E., Bunce M., Scofield R. P., Hale M. L., Holdaway R. N., Highly skewed sex ratios and biased fossil deposition of moa: Ancient DNA provides new insight on New Zealand’s extinct megafauna. Quat. Sci. Rev. 29, 753–762 (2010). [Google Scholar]
- 25.Cooper A., Poinar H. N., Ancient DNA: Do it right or not at all. Science 289, 1139–1139 (2000). [DOI] [PubMed] [Google Scholar]
- 26.Shapiro B., Hofreiter M., Eds. Ancient DNA: Methods and Protocols (Methods in Molecular Biology, Humana Press, 2012).
- 27.Shapiro B., et al. , Rise and fall of the Beringian steppe bison. Science 306, 1561–1565 (2004). [DOI] [PubMed] [Google Scholar]
- 28.Soubrier J., et al. , Early cave art and ancient DNA record the origin of European bison. Nat. Commun. 7, 13158 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bray S. C. E., et al. , Ancient DNA identifies post-glacial recolonisation, not recent bottlenecks, as the primary driver of contemporary mtDNA phylogeography and diversity in Scandinavian brown bears. Divers. Distrib. 19, 245–256 (2013). [Google Scholar]
- 30.Dabney J., et al. , Complete mitochondrial genome sequence of a Middle Pleistocene cave bear reconstructed from ultrashort DNA fragments. Proc. Natl. Acad. Sci. U.S.A. 110, 15758–15763 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rohland N., Harney E., Mallick S., Nordenfelt S., Reich D., Partial uracil–DNA–glycosylase treatment for screening of ancient DNA. Philos. Trans. R. Soc. Lond. B Biol. Sci. 370, 20130624 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schubert M., et al. , Characterization of ancient and modern genomes by SNP detection and phylogenomic and metagenomic analysis using PALEOMIX. Nat. Protoc. 9, 1056–1082 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Li H., Durbin R., Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zimin A. V., et al. , A whole-genome assembly of the domestic cow, Bos taurus. Genome Biol. 10, R42 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Elsik C. G., Tellam R. L., Worley K. C., The genome sequence of taurine cattle: A window to ruminant biology and evolution. Science 324, 522–528 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H., A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27, 2987–2993 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu S., et al. , Population genomics reveal recent speciation and rapid evolutionary adaptation in polar bears. Cell 157, 785–794 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lindblad-Toh K., et al. , Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature 438, 803–819 (2005). [DOI] [PubMed] [Google Scholar]
- 39.Li H., Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Young A. C., Kirkness E. F., Breen M., Tackling the characterization of canine chromosomal breakpoints with an integrated in-situ/in-silico approach: The canine PAR and PAB. Chromosome Res. 16, 1193–1202 (2008). [DOI] [PubMed] [Google Scholar]
- 41.R Core Team , R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna, Austria, 2017). [Google Scholar]
- 42.Gelman A., Jakulin A., Pittau M. G., Su Y. S., A weakly informative default prior distribution for logistic and other regression models. Ann. Appl. Stat. 2, 1360–1383 (2008). [Google Scholar]
- 43.Jones K. E., et al. , PanTHERIA: A species-level database of life history, ecology, and geography of extant and recently extinct mammals. Ecology 90, 2648–2648 (2009). [Google Scholar]
- 44.American Museum of Natural History , AMNH vertebrate zoology database. http://sci-web-001.amnh.org/db/emuwebamnh/index.php. Accessed 29 May 2018.
- 45.London Natural History Museum , Dataset: Collection specimens. Resource: Specimens. Natural History Museum Data Portal. http://data.nhm.ac.uk. Accessed 29 May 2018.
- 46.Royal Ontario Museum , Mammalogy collection - Royal Ontario Museum. http://gbif.rom.on.ca/ipt/resource.do?r=mamm. Accessed 29 May 2018.
- 47.Smithsonian National Museum of Natural History , Mammals collections search. https://collections.nmnh.si.edu/search/mammals/. Accessed 30 May 2018.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Mapped reads and sample-associated metadata are available from https://figshare.com/projects/Widespread_male_sex_bias_in_mammal_fossil_and_museum_collections/60446.