Significance
Recurrent respiratory papillomatosis is a disease caused by the human papillomavirus that leads to growth of warts in the throat. We report 2 brothers with a form of this disease that involves a mutation in the NLRP1 gene. This study provides a genetic explanation for our patients’ disease and suggests that other people may suffer from the same genetic disease. It also expands our understanding of diseases caused by mutations in NLRP1.
Keywords: inflammasome, NLRP1, recurrent respiratory papillomatosis, genetics, human papillomavirus
Abstract
Juvenile-onset recurrent respiratory papillomatosis (JRRP) is a rare and debilitating childhood disease that presents with recurrent growth of papillomas in the upper airway. Two common human papillomaviruses (HPVs), HPV-6 and -11, are implicated in most cases, but it is still not understood why only a small proportion of children develop JRRP following exposure to these common viruses. We report 2 siblings with a syndromic form of JRRP associated with mild dermatologic abnormalities. Whole-exome sequencing of the patients revealed a private homozygous mutation in NLRP1, encoding Nucleotide-Binding Domain Leucine-Rich Repeat Family Pyrin Domain-Containing 1. We find the NLRP1 mutant allele to be gain of function (GOF) for inflammasome activation, as demonstrated by the induction of inflammasome complex oligomerization and IL-1β secretion in an overexpression system. Moreover, patient-derived keratinocytes secrete elevated levels of IL-1β at baseline. Finally, both patients displayed elevated levels of inflammasome-induced cytokines in the serum. Six NLRP1 GOF mutations have previously been described to underlie 3 allelic Mendelian diseases with differing phenotypes and modes of inheritance. Our results demonstrate that an autosomal recessive, syndromic form of JRRP can be associated with an NLRP1 GOF mutation.
Juvenile-onset recurrent respiratory papillomatosis (JRRP) is a rare childhood disease characterized by the recurrent growth of papillomas in the respiratory tract. The epidemiology of JRRP varies slightly across studies, with an incidence of ∼0.2 to 4/100,000 children and a prevalence of ∼1 to 4/100,000 children depending on the study (1–4). Papillomas are most commonly found in the larynx but may occur anywhere from the mouth to the bronchi (5). Children typically present within the first years of life (most commonly between age 2 and 4 y) with hoarseness or, in more severe cases, respiratory distress or stridor and airway obstruction, and endoscopic examination reveals papillomatous growths in the upper airway (3, 6). The clinical course varies widely in terms of the location and extent of papillomas, speed of recurrence following resection, and duration of the illness (median 4.4 y) (4, 5).
JRRP is associated with infection of the upper airway by human papillomaviruses (HPVs) of the α genus, with infection thought to occur by vertical transmission at birth and with a childhood onset of the lesions (7, 8). Indeed, an adult-onset form of this disease also exists (referred to as adult-onset recurrent respiratory papillomatosis) with a similar but usually milder phenotype; in these cases, HPV is probably acquired by sexual transmission (9, 10). The standard of care remains surgical removal of the papillomas (usually by laser ablation) to keep the airway patent, with patients requiring a median of 4.4 (range 0.2 to 19.3) surgical procedures per year, attesting to the medical burden of this disease (4, 11). Spontaneous remission occurs in approximately one-half of children by adulthood (4, 12, 13). In rare cases, papillomas may progress to malignant squamous cell carcinoma, which can be fatal (14, 15).
HPV-6 and HPV-11 are the strains of HPV usually detected in papillomas, although other rare α-HPVs have also been reported (16–19). HPV-6 and HPV-11 are very closely related α-HPVs and are also the most common cause of genital warts (20). The reported incidence of detectable HPVs in cohorts of patients with JRRP varies from 53% to 100% (16–19). Exposure to HPV in the birth canal is common, while JRRP is rare, suggesting the importance of host factors in the disease. Indeed, cross-sectional studies of cervical HPV carriage rates demonstrate an HPV-6 + HPV-11 prevalence of 6.4% in women age 14–19 y (21), and a longitudinal study (mean follow-up, 5.8 y) found a cumulative prevalence of 91.1% of women with detectable cervical HPV-6 or -11 DNA at some point during the study period (22). Moreover, patients with JRRP do not exhibit increased susceptibility to other types of infectious agents, including viruses, and are not more susceptible to HPV infections at other body sites. Similarly, JRRP has not been reported to be a feature of any wider syndrome and is not seen in primary immunodeficiencies that cause susceptibility to HPVs at other body sites, such as DOCK8 deficiency and other T cell defects (α-HPVs), WHIM (α-HPVs), and epidermodysplasia verruciformis (β-HPVs) (23–25). However, analysis of patients with JRRP has revealed mild immunologic anomalies, including Th2 polarization, restricted Vβ TCR repertoires in CD4 and CD8 T cells, and natural killer cell dysfunction (26), as well as Langerhans cell hyporesponsiveness to IL-36γ (27), suggesting immunologic susceptibility to JRRP. This rare manifestation of infection by a common virus is reminiscent of other viral diseases striking otherwise healthy children, including conditions caused by other HPVs, which can be due to inborn defects of immunity (24, 28–31). We studied 2 patients with JRRP, testing the hypothesis that monogenic inborn errors of immunity could underlie their JRRP.
Results
Clinical Phenotype.
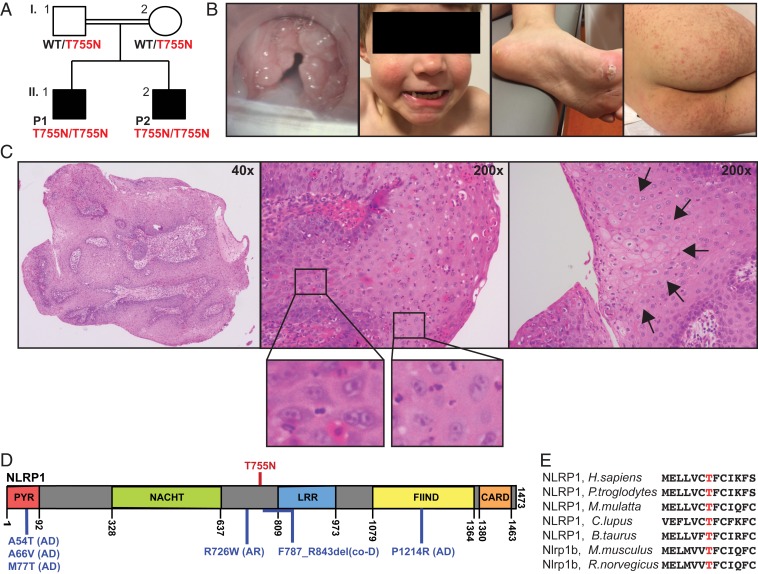
We studied 2 brothers with JRRP, herein identified as patient 1 (P1) and patient 2 (P2), of Belgian ancestry born to consanguineous (first-cousin) parents (Fig. 1A). There were no other siblings. P1 developed hoarseness and recurrent laryngitis at age 5 y. Direct laryngoscopy revealed papillomas in the glottis and supraglottis (Fig. 1B). He required 8 surgical ablations of laryngeal lesions over the next year and continues to require multiple ablations each year, with decreasing frequency. P2 developed hoarseness shortly after birth, and laryngeal papillomatosis was diagnosed at age 20 mo. His disease has been less severe than that of P1, and he has required 2 to 3 ablations per year. Careful retrospective review of their medical history revealed the same mild dermatologic abnormalities in both brothers, including a small number of palmar and plantar warts; keratosis pilaris on the lower back, buttocks, and thighs; and atrophoderma vermiculata on the face, none of which required medical treatment (Fig. 1B); see the case reports in SI Appendix for full details. Dermatologic abnormalities are not typically seen in other patients with JRRP, which is isolated; therefore, these 2 patients had a syndromic form of JRRP. The parents did not have any notable medical history, specifically no history of RRP or dermatologic disease. Histologically, larynx lesions exhibited a papillomatous morphology with focal areas of koilocytosis and scattered binucleated cells, typical of lesions in RRP and pathognomonic of HPV infection (32). (Fig. 1C). Papillomas from P1 (9 specimens) and P2 (2 specimens) tested negative for HPV-6 and HPV-11, consistent with studies of cohorts in which HPV is not detected in every patient (16–19).
Fig. 1.
A private homozygous missense mutation in siblings with JRRP and dermatologic abnormalities. (A) Pedigree showing NLRP1 genotype of individuals. (B) Clinical images of P1 showing (from left to right) larynx papillomas, atrophoderma vermiculata on cheeks, plantar warts, and keratosis pilaris on buttocks and thighs. (C) Micrographs of a larynx papilloma from P1 showing (from left to right) gross morphology of papillomas, areas of binucleated cells (Insets: enlarged), and focal areas of koilocytosis (arrows). (D) Schematic representation of NLRP1 protein showing functional domains, location of patients’ T755N mutations (red), and location of previously described NLRP1 mutations (blue) and their mode of inheritance (AD, AR, or codominant [CoD]). (E) Protein sequencing alignment of human NLRP1 to known orthologs, showing conservation of T755.
Genetic Analysis.
We performed whole-exome sequencing (WES) on P1, P2, and both parents (I.1 and I.2; Fig. 1A). WES showed a high homozygosity rate in P1 (3.56%) and P2 (5.13%) (33), consistent with the known parental consanguinity. Principal component analysis confirmed the patients’ European ancestry (33). In light of this consanguinity, we hypothesized that a rare variant, homozygous in both patients and heterozygous in both parents, might be responsible for the patients’ phenotype. We selected variants predicted to result in a missense, nonsense, indel, or splice site mutation with a minor allele frequency of <0.01 in public databases (ExAC, 1000 Genomes, and NHLBI-ESP6500). Finally, we excluded variants in genes with a Gene Damage Index (GDI) >13.38 (34), variants with a combined annotation-dependent depletion (CADD) score less than the mutation significance cutoff (MSC) (35), and variants in our blacklist with an in-house frequency of >0.01 (36) (SI Appendix, Fig. S1A). This yielded 5 homozygous variants across 5 genes (SI Appendix, Fig. S1B). Two of these were present in homozygosity in healthy individuals in ExAC, suggesting they are unrelated to the patient’s phenotype; another was in an unknown gene (ZNF417); and another was in a gene implicated in cardiac conduction defects (KCNH2) (37). The best candidate was a homozygous missense mutation in Nucleotide-Binding Domain, Leucine-Rich Repeat Family Pyrin Domain-Containing 1 (NLRP1), c.2819C > A (for transcript variant 1; NBCI NM_033004), p.T755N (herein T755N). NLRP1 isoform 1, composed of 1,473 amino acids, acts as a sensor for the innate immune complex known as the inflammasome (38) and is expressed across a variety of tissues and cell types (https://www.proteinatlas.org/ENSG00000091592-NLRP1/). T755N is slightly N-terminal to the leucine-rich repeat (LRR) domain (Fig. 1D). Homozygosity of the NLRP1 T755N allele in P1 and P2 and its familial segregation with the disease was confirmed by Sanger sequencing (SI Appendix, Fig. S1C). Using a similar variant-filtering strategy for the less likely X-linked pattern of inheritance did not yield any candidate variants (SI Appendix, Fig. S1D). Similarly, there were no de novo mutations shared by the 2 patients. Thus, these findings suggested that homozygosity for NLRP1 T755N might be the genetic etiology of JRRP in P1 and P2.
Population Genetics of NLRP1.
The NLRP1 variant T755N is not found in any public database (gnomAD, Bravo/TOPMED) or in our in-house cohort of >5,000 unrelated individuals with a variety of infectious diseases. T755N is predicted to be damaging by CADD, with a high score of 23.1, above the 99% confidence interval MSC value of 3.313 (35). The T755 residue of NLRP1 is highly conserved across species (Fig. 1E). NLRP1 has a GDI of 9.374, indicating a medium amount of mutational burden in the general population (34), and is under low to moderate negative selection, with a McDonald–Kreitman Neutrality Index of 0.400 and a residual variation intolerance score in the 95th percentile of the least intolerant genes (39); however, previous studies have shown that autosomal recessive disease-causing genes are not under purifying selection (40). In gnomAD, there are 40 missense mutations found in homozygosity in 1 or more individuals, 23 of which have a CADD score greater than the MSC. No predicted loss-of-function (LOF) variants are found in homozygosity in gnomAD, and the pRec (probability of being intolerant of homozygous, but not heterozygous, LOF variants) is 0.95 (41). Collectively, these findings suggest that T755N is likely to be damaging to NLRP1 protein function.
NLRP1 T755N Is Gain of Function and Reduces the Threshold for Inflammasome Activation In Vitro.
Germline gain-of-function (GOF) mutations in NLRP1 have been recently discovered to cause 3 Mendelian diseases of the skin. Multiple self-healing palmoplantar carcinoma (MSPC), described in 3 families, follows an autosomal dominant (AD) pattern of inheritance, with all 3 mutations (A54T, A66V, and M77T) in the pyrin (PYR) domain (42, 43). Familial keratosis lichenoides chronica (FKLC), described in 1 family, follows a codominant pattern of inheritance with a mutation (F787_R843del) immediately N terminal to the LRR domain (Fig. 1D) (43). Autoinflammation with arthritis and dyskeratosis (AIADK), described in 2 families, follows autosomal recessive (AR) (R726W) or AD (P1214R) inheritance, with mutations in the N-terminal LRR and function-to-find (FIIND) domains, respectively (44). In vitro, MSPC and FKLC disease-causing alleles display a similar GOF magnitude despite their different modes of inheritance (AD vs. codominant) (43). Because P1 and P2 had skin abnormalities similar to those seen in FKLC, we hypothesized that NRLP1 T755N would also be GOF.
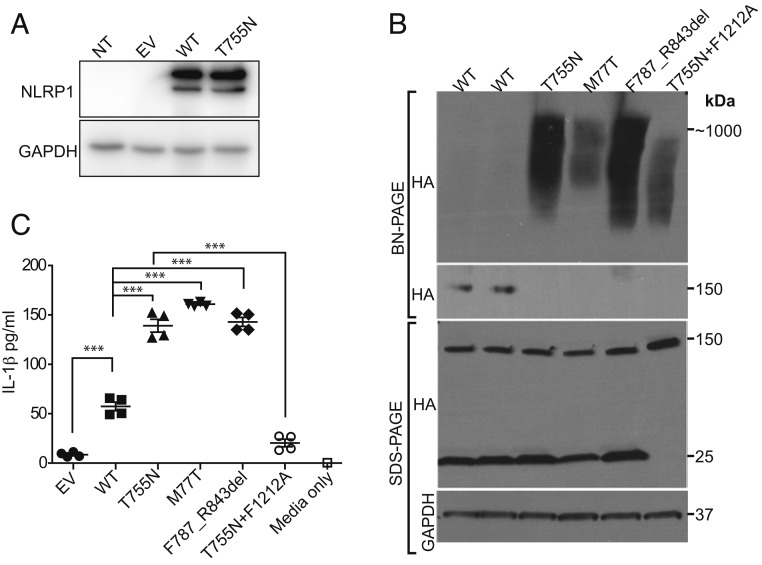
We first confirmed that both NLRP1 wild type (WT) and T755N cDNAs were expressed normally by transfecting them in HEK293T cells (Fig. 2A). Published NLRP1 GOF alleles spontaneously oligomerize and induce secretion of IL-1β in keratinocytes (43). When NLRP1 T755N was overexpressed in HEK293T cells that do not express other inflammasome components, it spontaneously oligomerized, similar to previously described NLRP1 GOF variants M77T and F787_R843del and in contrast to the WT NLRP1 (Fig. 2B). This oligomerization of T755N is partially dependent on the autocleavage site, amino acid F1212 within the FIIND domain, as the noncleavable mutation F1212A reduced the amount of oligomerized NLRP1 T755N (Fig. 2B, lane 6). Taken together, these results suggest that the T755N mutant behaves in a similar fashion biochemically with the other NLRP1 GOF mutants in causing increased inflammasome activation via spontaneous oligomerization. In addition, overexpression of NLRP1 T755N in immortalized keratinocytes led to elevated production of secreted IL-1β, similar to that in previously described GOF alleles (Fig. 2C), which was dependent on cleavage, as seen in other NLRP1 GOF alleles (Fig. 2C). The magnitude of functional gain was similar in alleles that follow AD (M77T), codominant (F787_R843del), and AR inheritance (T755N) (Fig. 2 B and C). In summary, these findings demonstrate that NLRP1 T755N can cause increased inflammasome activation in vitro, suggesting that this allele is GOF and thus probably pathogenic.
Fig. 2.
NLRP1 T755N is GOF for inflammasome activation. (A) Western blot showing similar expression of NLRP1 WT and T755N protein in HEK293T cells. A GAPDH Western blot is shown as a loading control. The image is representative of 3 independent experiments. (B) Western blot for HA-tagged NLRP1 after BN-PAGE or conventional SDS-PAGE and lysates from HEK293T cells overexpressing cDNA of NLRP1 WT (2 replicates), T755N, previously published GOF alleles (M77T and F878_R843del), or noncleavable NLRP1 T755N (T755N+F1212A), demonstrating oligomerization of T755N NLRP1 similar to other GOF mutations. GAPDH Western blot is shown as a loading control. (C) ELISA of IL-1β in supernatants of keratinocytes after transfection with NLRP1 alleles demonstrating that T755N is GOF for IL-1β production and that cleavage at the F1212 position is required for IL-1β production. NT, nontransfected cells; EV, empty vector. Data are an average of 4 replicates. ***P < 0.001, 1-way ANOVA.
Primary Keratinocytes from P1 and P2 Demonstrate Spontaneous Activation of the Inflammasome.
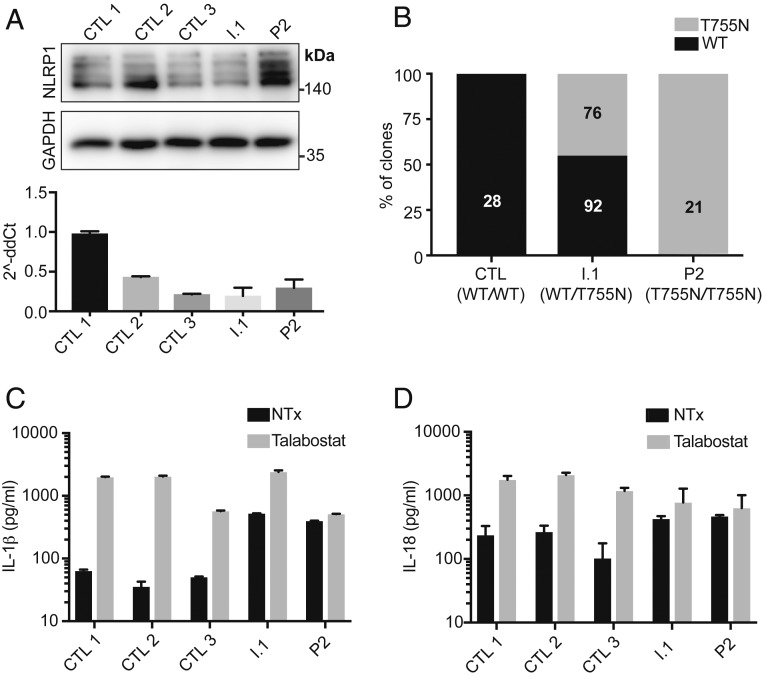
We derived primary keratinocyte cell lines from skin biopsy specimens taken from P2 and I.1 (NLRP1 genotypes T755N/T755N and WT/T755N, respectively). NLRP1 mRNA and protein were expressed to similar levels in P2, I.1, and 3 healthy control primary keratinocyte lines (Fig. 3A), confirming that the NLRP1 T755N allele is expressed at normal levels in healthy heterozygous and patient-derived cells. We next confirmed that T755N and WT NLRP1 mRNA are expressed in keratinocytes in proportion to their genotype. Cloning of a partial NLRP1 cDNA encompassing T755 showed that in heterozygous cells from I.1, ∼50% of the transcripts were WT and ∼50% were T755N (Fig. 3B), suggesting that mRNA of the T755N NLRP1 allele is expressed at levels equal to WT. Keratinocytes from P2 and I.1 released IL-1β into the supernatants, suggesting baseline activation of the inflammasome at a functional level (Fig. 3C). In contrast, basal IL-1β release was not seen in control cells (Fig. 3C). When these cells were stimulated with Val-boroPro (talabostat, a DPP9 inhibitor shown to activate the NLRP1 inflammasome) (45, 46), control and heterozygous keratinocytes released large amounts of IL-1β, while release of IL-1β in keratinocytes from P2 was unchanged (Fig. 3C). Similar results were observed for IL-18 (Fig. 3D). The abrogation of talabostat responsiveness in keratinocytes from P2 suggests that the mechanism of GOF in this allele is due to a decrease in DPP9 inhibition. Taken together, these results demonstrate that keratinocytes homozygous for NLRP1 T755N display inflammasome activation at the basal level.
Fig. 3.
Patient-derived keratinocytes show normal expression of NLRP1 protein, baseline inflammasome activation, and unresponsiveness to further NLRP1 activation. (A) Western blot (Top) and qPCR (Bottom) of NLRP1 expression in keratinocytes from P2, heterozygous father (I.1), and 3 controls. The image is representative of 3 independent experiments. (B) Relative expression of NLRP1 WT, T755N transcripts as assessed by TA cloning and Sanger sequencing of an NLPR1 cDNA from keratinocytes from control (CTL), heterozygous father (I.1), and P2. (Inset) Numbers correspond to the number of unique clones sequenced. (C) IL-1β ELISA of supernatants from keratinocytes that were untreated or treated with 3 μM of talabostat (Val-boroPro) for 16 h. (D) IL-18 ELISA of supernatants from keratinocytes that were untreated or treated with 3 μM of talabostat for 16 h. Bars represent mean ± 1 SD. The results are representative of 3 independent experiments.
P1 and P2 Display Elevated Serum Cytokine Levels Consistent with Spontaneous Inflammasome Activation In Vivo.
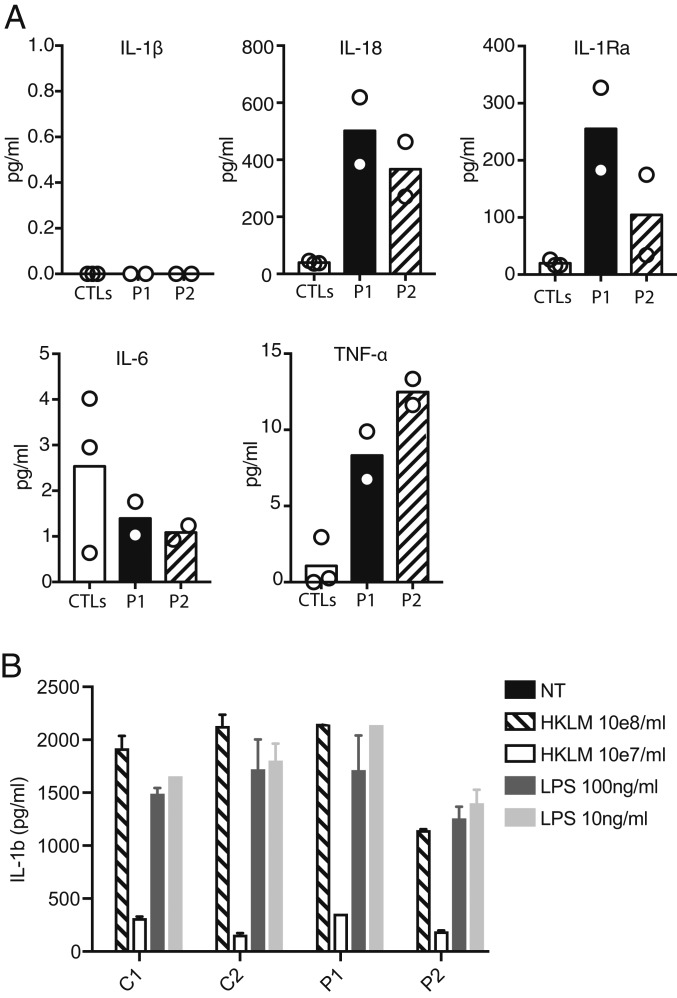
We tested whether P1 and P2 had any clinical markers of spontaneous inflammasome activation. Patient serum was first tested for elevation of IL-1β and IL-18, the 2 cytokines that may be produced on activation of the inflammasome (47, 48). Both patients showed elevation of IL-18, but not of IL-1β, in repeated analyses (Fig. 4A), similar to previously reports of patients with homozygous NLRP1 GOF mutations near the LRR region of the protein (43, 44). Such a divergence of IL-18 and IL-1β elevation in the blood is also seen in other inflammasome activation disorders, such as NLRC4-mediated autoinflammation (49), and may underlie the phenotypic differences between inflammasome disorders and disease stage (50). Both P1 and P2 also showed elevated TNF-α, which is induced by IL-1β and IL-18 in many cell types and may mediate further up-regulation of inflammasome components (51), although IL-6 was not elevated (Fig. 4A), as was also seen in the previously described FKLC patient with a homozygous NLRP1 F787_R853del GOF mutation (43). Serum IL-1RA was also elevated in P1 and P2 (Fig. 4A), consistent with chronic inflammasome activation. Serum cytokine levels were not elevated in the heterozygous parents (SI Appendix, Fig. S2), consistent with the absence of any clinical manifestations. Stimulation of patient and healthy control peripheral blood mononuclear cells (PBMCs) with lipopolysaccharide (LPS) or heat-killed Listeria monocytogenes (HKLM), 2 Toll-like receptor agonists that trigger IL-1β production in an NLRP1-independent manner (52), led to similar levels of IL-1β production in patients and controls (Fig. 4B), suggesting a normal response to TLR ligands. These data demonstrate that the sera of P1 and P2 showed signs of inflammasome activation in vivo.
Fig. 4.
Inflammasome activation in P1 and P2. (A) Luminex measurements of serum levels of IL-1β, IL-18, IL-1Ra, IL-6, and TNF-α from P1, P2, and 3 healthy controls. Results are representative of 2 independent experiments. (B) ELISA for IL-1β after stimulation of PBMCs from P1, P2, and 2 controls with TLR ligands that induce IL-1β in an NLRP1-independent manner demonstrating normal regulation of production. Bars represent mean ± 1 SD. The results are representative of 2 independent experiments.
Discussion
Here we have described a homozygous NLRP1 mutation in siblings with a syndromic form of JRRP. Six other NLRP1 GOF mutations were previously shown to underlie 3 allelic conditions—MSPC, FKLC, and AIADK—which have overlapping but distinct phenotypes that all include dermatologic abnormalities. MSPC presents with severe dermatologic abnormalities, including palmoplantar keratoacanthomas, and corneal lesions (42, 43); FKLC manifests as palmoplantar warts and lichenoid papules on the lower trunk and extremities (43); and AIADK involves variable dermatologic abnormalities accompanied by systemic autoinflammation with arthritis (44). Phenotypically, P1 and P2 do not have evidence of the systemic inflammatory symptoms of AIADK or the severe dermatologic manifestations of MSPC. They have dermatologic abnormalities at the mild end of the spectrum, most similar to those in FKLC, and their most notable clinical feature is JRRP. There is a degree of variable expressivity between P1 and P2, with differing JRRP severity despite their close genetic relatedness and shared environment. The variability of disease in the 6 previously described kindreds, with at least 3 phenotypes, is unexplained and may result from differences between the GOF alleles themselves or modifier genes. Conversely, the absence of overt JRRP in other NLRP1 GOF syndromes, with the possible exception of 1 patient with AIADK with uncharacterized larynx lesions (44), suggests that there is incomplete penetrance.
Genetically, the NLPR1 GOF-mediated diseases vary in mode of inheritance. MSPC is AD, due to mutations in the PYR domain, while FKLC follows codominant inheritance due to a mutation N terminal to the LRR domain (hereinafter N-LRR) and AIADK follows an AD or AR inheritance depending on the mutation. As we have shown here, the T755N mutation in the N-LRR region underlies an AR pattern of inheritance. Interestingly, JRRP was apparently described in 1 AIADK patient harboring a biallelic mutation in the N-terminal LRR domain (R726W) (44). In mice, NLRP1B is activated after cleavage by bacterial proteases, leading to degradation of the N-terminal fragment of NLRP1B and activation of inflammasomes by the disinhibited C-terminal fragment (53, 54), suggesting that NLRP1 GOF mutations may perturb this process. However, the mechanism whereby these human NLRP1 mutations lead to GOF is not fully understood, and neither is how certain alleles underlie dominant, codominant, or recessive phenotypes. Although the mutations cluster to different regions of the protein, the cellular and biochemical experiments reported here and elsewhere (43) show no differences between GOF alleles in the magnitude of functional gain, suggesting that other, perhaps domain-specific, aspects of NLRP1 function contribute to the diversity of phenotypes. Indeed, all mutations described to date fit a domain-specific hypothesis, with variants underlying AD inheritance in the PYR and FIIND domains and those with codominant or AR inheritance restricted to the N-LRR region (Fig. 1C and Table 1). It is also interesting that NLRP1 GOF can underlie an AR phenotype, as human GOF mutations underlying a recessive phenotype are extremely rare. To our knowledge, this has only been described in CASR mutations in hypocalcemic hypoparathyroidism, where GOF mutations may underlie both AD and AR patterns of inheritance (55). Interestingly, the AR and AD inherited mutations lead to the same disease, although like NLRP1, the AD and AR mutations localize to different regions of the protein (55). Recent findings suggest that familial Mediterranean fever (FMF) caused by mutations in the MEFV gene and long thought to be exclusively AR, can also be AD, as evidenced by knock-in mice engineered to express the human B30.2 domain of MEFV harboring mutations found in patients with AR FMF (56). The mechanism whereby heterozygous carriers of the T755N allele remain clinically unaffected and exhibit normal serum cytokines (SI Appendix, Fig. S2) while displaying GOF in cellular assays (Fig. 3 C and D) is unclear. This may be due to the sensitivity of the in vitro assays, the impact of other negative regulatory signals present in vivo, or perhaps a result of the remaining WT allele maintaining responsiveness to DPP9-mediated inhibition of NLRP1 activation.
Table 1.
Summary of pathogenic NLRP1 GOF mutations described to date
| NLRP1 mutation | Protein domain | Inheritance | Disease | Phenotype | JRRP | References |
| A54T | PYR | AD | MSPC | Palmoplantar keratoacanthomas | No | 42, 43 |
| Conjunctival keratoacanthomas | ||||||
| A66V | PYR | AD | MSPC | Palmoplantar keratoacanthomas | No | 43 |
| Conjunctival keratoacanthomas | ||||||
| M77T | PYR | AD | MSPC | Palmoplantar keratoacanthomas | No | 43 |
| Conjunctival keratoacanthomas | ||||||
| P1214R | FIIND | AD | AIADK | Hepatosplenomegaly with extramedullary hematopoiesis | No | 44 |
| Candidiasis | ||||||
| Vitamin A deficiency | ||||||
| Follicular hyperkeratosis, palmoplantar hyperkeratosis, corneal dyskeratosis | ||||||
| Hemolytic anemia, periodic fever, inflammatory arthritis | ||||||
| Mucocutaneous candidiasis | ||||||
| R726W | N-LRR | AR | AIADK | Follicular hyperkeratosis, palmoplantar wart-like hyperkeratotic lesions | Yes? | 44 |
| Inflammatory arthritis | ||||||
| Uveitis (1 of 2 patients) | ||||||
| Vitamin A deficiency | ||||||
| Respiratory papillomatosis (1 of 2 patients) | ||||||
| F787_R843del | N-LRR | Codominant | FKLC | Lichenoid papules on arms, legs, lower trunk | No | 43 |
| Palmoplantar wart-like hyperkeratotic papules | ||||||
| T755N | N-LRR | AR | JRRP | Recurrent respiratory papillomatosis | Yes | This study |
| Keratosis pilaris on legs and lower trunk | ||||||
| Palmoplantar wart-like hyperkeratotic papules | ||||||
| Atrophoderma vermiculata on cheeks |
Beyond NLRP1, the role of HPV in our patients’ JRRP remains unclear, as we did not detect the typical causative strains HPV-6 and -11. JRRP without detectable HPV-6 or -11 has been described previously (16–19), but whether our present findings are due to failed detection or truly HPV-6/11–negative disease is unclear. One hypothesis is that infection of our patients’ respiratory epithelial keratinocytes by HPV triggers exaggerated activation of the NLRP1 inflammasome due to their GOF allele, leading to IL-1 family cytokine-mediated keratinocyte proliferation and papilloma formation (43, 57) and also increased inflammasome-mediated suppression of the virus, leading to undetectable viral levels or even viral clearance. Indeed, there is evidence of host–pathogen interaction between the IL-1β axis and HPV (58, 59). In this model, NLRP1 GOF underlies JRRP mainly because of its expression in keratinocytes. Consistently, none of the many inborn errors of leukocytes underlie JRRP (23, 60). Nevertheless, we do not exclude the possibility that other cell types contribute to HPV-driven JRRP in patients with GOF mutations in NLRP1. The alternative hypothesis is that certain NLRP1 GOF alleles, especially in homozygosity, lead to hyperproliferative lesions in the upper airway by themselves, manifesting as HPV-negative JRRP. The genetic diagnosis of an NLRP1 GOF syndrome should be considered in patients presenting with JRRP (regardless of HPV status), especially in those with a syndromic form with concurrent skin abnormalities. Although there are no current therapies for these NLRP1-mediated diseases, anti–IL-1 or anti–IL-18 (e.g., IL-18BP) therapies have demonstrated promise for other autoinflammatory inflammasome disorders (61–63) and may be a reasonable therapeutic approach for these patients.
In addition to RRP, isolated susceptibility to HPV has also been described in epidermodysplasia verruciformis (EV) and “tree man” syndrome (TMS; a syndrome distinct from but frequently incorrectly referred to as EV). Interestingly, these syndromes vary in their anatomic site of disease, morphology of warts, and dominant causative strains of HPV. While RRP displays isolated susceptibility to α-HPVs (most commonly HPV-6 and -11) in the upper airway, EV is characterized by susceptibility to β-HPVs in the skin manifesting as flat warts (24, 25, 64), and TMS is characterized by susceptibility to α-HPVs (usually HPV-2) and manifests as bulky hyperkeratotic “cutaneous horns” primary on the hands and feet (65–67). Despite the shared HPV susceptibility, a combination of these phenotypes has never been described in a single individual, suggesting that the mechanism of disease are distinct, likely due to the diversity of HPV strains, their tissue tropisms, and the strain- and tissue-specific mechanisms of host immunity to these viruses. This is consistent with the differences between the known underlying genotypes. Indeed, EV is due to biallelic null mutations in TMC6, TMC8, and CIB1, the products of which form a complex that governs keratinocyte-intrinsic immunity to β-HPVs (24, 25, 64). It is intriguing that GOF mutations in NLRP1 and LOF mutations in EVER1-EVER2-CIB1 disrupt immunity to α-HPVs and β-HPVs in respiratory and cutaneous keratinocytes, respectively. It will be important to decipher more genetic defects underlying these and other isolated susceptibilities to HPVs, including α-HPV–driven cervical cancer (68).
Methods
Patient Recruitment and Human Subject Protections.
All studies were performed in accordance with institutional and municipal guidelines. Approval for this study was obtained from the French Comité de Protection des Personnes, L'Agence Nationale de Sécurité du Médicament et des Produits de Santé, INSERM (protocol C10-13), and The Rockefeller University Institutional Review Board (protocol JCA-0700). Patient consent was obtained for use of clinical information and specimens.
WES and Sanger Sequencing.
Genomic DNA was extracted from blood. WES was performed at the New York Genome Center. Alignment to the GRCh37 reference genome was done with BWA-MEM 0.7.12 (69), and variant calling was done using Genome Analysis Toolkit Unified Genotyper version 3.4–46 (70). Annotated variants were filtered by allele frequency in public databases (1000 Genomes [http://www.1000genomes.org], NHLBI-EVS [https://evs.gs.washington.edu/EVS/], ExAC [http://exac.broadinstitute.org], and gnomAD [https://gnomad.broadinstitute.org/ and with criteria developed in the laboratory) (34–36). In silico prediction of deleterious of variants was performed with CADD (http://genetics.bwh.harvard.edu/pph2/). Estimated homozygosity from WES data were performed as described previously (33). The NLRP1 variant c.2819C > A was confirmed by Sanger sequencing. PCR amplification was performed with gene-specific primers flanking the variant of interest (forward: acaaaacgttcctgacacaagtg; reverse: ctcaggtcactcgggcttatg). Sequencing was performed with BigDye Terminator V1.1 chemistry (Thermo Fisher Scientific) and analyzed on an ABI 3730 DNA sequencer (Thermo Fisher Scientific). Sequence alignment was performed using SnapGene software.
Cloning of NLRP1 Constructs.
Full-length NLRP1 transcript variant 1 cDNA that matched the NCBI reference sequence NM_033004.4 was cloned by first amplifying NLRP1 from multiple in-house made SV40-immortalized fibroblast cDNA preparations using Pfu polymerase (Agilent), addition of A overhangs with Taq polymerase (Thermo Fisher Scientific) (forward: atggctggcggagcctggggccg; reverse: tcagctgctgagtggcaggagtccctttttgctgccc), agarose gel purification of the expected 4.5-kb band using the Qiagen QIAquick Gel Extraction Kit, and TA cloning into pGEM-T Easy plasmid (Promega). Multiple clones were fully Sanger-sequenced to identify a reference sequence transcript variant 1. This was subcloned by PCR amplification (Pfu; Agilent) in 2 pieces (amino acids 1 to 755 and 755 to end) into EcoRI- and XhoI-digested (New England Biolabs) pCDNA3.1(+) plasmid (Invitrogen) using the Cold Fusion Cloning Kit (System Biosciences) with primers that either matched the WT sequence or introduced a T755N mutation using the primers listed in SI Appendix, Table S1. Plasmids were prepared for transfection with the Qiagen Maxiprep Kit. All open reading frames were completely Sanger-sequenced before use with primers listed in SI Appendix, Table S1 using BigDye Terminator V1.1 chemistry (Thermo Fisher Scientific), analyzed on an ABI 3730 DNA sequencer (Thermo Fisher Scientific), and aligned using SnapGene software. For the experiments shown in Fig. 2 B and C, NLRP1 constructs were as described previously (43) and contained a C-terminal HA tag.
Cell Lines, Cell Culture, and Transfections.
HEK293T cells were purchased from American Type Culture Collection (CRL-3216) and were cultured in DMEM and Glutamax (Invitrogen) plus 10% FCS. The patients’ primary keratinocytes were derived from skin biopsy specimen as described previously (24). Control keratinocytes were single- donor adult human epidermal keratinocytes (Lonza; catalog no. 00192627, donors 34014, 30214, and 34015). Keratinocytes were maintained on mitomycin-C–inactivated 3T3-J2 feeder cells in Complete Green medium (DMEM/Ham’s F-12 in a 2:1 ratio supplemented with 10% FBS, 180 nM adenine, 10 ng/mL EGF, 0.4 μg/mL hydrocortisone, 8.47 ng/mL cholera toxin, 5 μg/mL insulin, 1.36 ng/mL triiodothyronine, and 10 μM ROCK inhibitor Y-27632). Immortalized N/TERT-1 keratinocytes (a gift from J. G. Rheinwald to F.L.Z.) were cultured in KSFM media (Life Technologies) supplemented with 300 μM CaCl2 as described previously (43). Transient transfection of HEK293T cells was performed using Lipofectamine 2000 (Life Technologies) at a 2:1 ratio according to the manufacturer’s instructions. Transfection of immortalized keratinocytes was performed using FuGENE HD (Promega) at a 3:1 ratio according to the manufacturer’s instructions. In the indicated experiments, keratinocytes were stimulated with 3 uM talabostat (MedChemExpress) for 16 h before analysis.
Western Blot Analysis.
Cells were lysed in RIPA buffer with cOmplete protease inhibitor (Roche), and protein was quantified using the BCA protein assay (Pierce). For standard denaturing polyacrylamide gel electrophoresis (PAGE), lysate was subjected to reducing sodium dodecyl sulfate (SDS)-PAGE using Tris-glycine buffers and then transferred to PVDF membranes (Immobilon), followed by detection with primary antibody against NLRP1 (R&D Systems; AF6788) and secondary anti–sheep HRP (R&D Systems; HAF016). Anti-GAPDH (Santa Cruz Biotechnology; FL-335) served as a loading control. Membranes were developed and detected using ECL Western blotting substrate (Pierce) and imaged on an Amersham Imager 600 (GE Healthcare). Blue natural PAGE (BN-PAGE) was performed with the Novex NativePAGE Bis-Tris gel system (Thermo Fisher Scientific) as described previously (43).
Quantitative PCR.
RNA was extracted from keratinocytes at indicated time points using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. cDNA was synthesized with the SuperScript III First-Strand Synthesis System (Thermo Fisher Scientific) with random hexamers according to the manufacturer’s instructions. Quantitative PCR (qPCR) was performed using TaqMan Universal PCR Master mix with the following FAM-MGB conjugated TaqMan Gene Expression Assays (Thermo Fisher Scientific) for NLRP1 (Hs00248187_m1) duplexed with VIC-MGB RNase P TaqMan Assay (4403328) as an endogenous control. qPCR was run on an Applied Biosystems 7500 Fast Real-Time PCR system. Gene expression was quantified by the 2-ddCt method.
Cytokine Measurements.
Human IL-1β, ILR1Ra, IL-6, TNF-α, and IL-18 cytokine levels were determined in serum of P1 and P2 and 2 healthy controls by a magnetic bead-based multiplex assay using Luminex technology (Bio-Rad). GraphPad Prism 6.0 software was used for data analysis. ELISA for IL-1β (BD Biosciences) and IL-18 (R&D Systems) was performed on tissue culture supernatants according to the manufacturer’s instructions.
PBMC Purification and Stimulation with TLR Ligands.
PBMCs were isolated using Leucosep tubes (Greiner Bio-One) containing Ficoll density gradient medium. Cells were stored in RPMI-1640 medium supplemented with GlutaMAX (Gibco; 61870044) enriched with 10% FCS (Sigma-Aldrich; F7524) containing 10% DMSO (Sigma-Aldrich; D2650) at −150 °C until further use. PBMCs were thawed in 37 °C preheated complete medium (RPMI-1640 medium supplemented with GlutaMAX, 10% FCS, and 1% penicillin-streptomycin [10,000 U/mL; Gibco; 15140122], 1 mM sodium pyruvate [Gibco; 11360070], 1% nonessential amino acids [Gibco; 11140035], and 50 μM 2-mercaptoethanol [Gibco; 31350010]). In the setting of functional testing, cells were left to recuperate for 30 min at 37 °C and 5% CO2 after removal of DMSO. PBMCs were stimulated for 24 h with TLR ligands, including HKLM (10e8/mL and 10e7/mL) and LPS (100 ng/mL and 10 ng/mL) (InvivoGen).
Supplementary Material
Acknowledgments
We thank all the patients and their family members for participating in this study. We thank Yelena Nemirovskaya, Dominick Papandrea, Mark Woollett, and Cécile Patissier for administrative assistance; Benedetta Bigio for bioinformatics assistance; Tatiana Kochetkov for technical assistance; and all lab members for helpful discussions. We also thank David Geneviève for support in investigating the AIADK patient with a JRRP-like phenotype. B.R. is a fellow of the Branco Weiss Foundation, an A*STAR Investigator, a National Research Foundation Singapore and Amsterdam Academic Alliance fellow, and a Young EMBO Investigator and is funded by a Strategic Positioning Fund on Genetic Orphan Diseases from A*STAR, Singapore. F.H. is funded by the Jeffrey Modell Foundation and a Bijzonder Onderzoeksfonds-Tenure Grant. A.W. is funded by the Cure-AID Grant from the European Union ERA-Net for Research Programmes on Rare Diseases. F.L.Z. is a Nanyang Assistant Professor and is supported by National Research Foundation Fellowship (Class 2019). This work was supported by the National Center for Research Resources and National Center for Advancing Translational Sciences (Grant UL1TR001866), the “Investissement d’avenir” program (Grant ANR-10-IAHU-01), the Integrative Biology of Emerging Infectious Diseases Laboratoire d’Excellence (Grant ANR-10-LABX-62-IBEID), the NIH (Grant 5 R21 AI107508-02), the French Cancer Institute (Grant 2013-1-PL BIO-11-1), the St. Giles Foundation, The Rockefeller University, Institut National de la Santé et de la Recherche Médicale, and Paris Descartes University. S.B.D. was supported by the Shapiro–Silverberg Fund for the Advancement of Translational Research and an American Philosophical Society Daland Fellowship in Clinical Investigation.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1906184116/-/DCSupplemental.
References
- 1.Marsico M., Mehta V., Chastek B., Liaw K.-L., Derkay C., Estimating the incidence and prevalence of juvenile-onset recurrent respiratory papillomatosis in publicly and privately insured claims databases in the United States. Sex. Transm. Dis. 41, 300–305 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Derkay C. S., Task force on recurrent respiratory papillomas: A preliminary report. Arch. Otolaryngol. Head Neck Surg. 121, 1386–1391 (1995). [DOI] [PubMed] [Google Scholar]
- 3.Larson D. A., Derkay C. S., Epidemiology of recurrent respiratory papillomatosis. APMIS 118, 450–454 (2010). [DOI] [PubMed] [Google Scholar]
- 4.Armstrong L. R., Derkay C. S., Reeves W. C., Initial results from the national registry for juvenile-onset recurrent respiratory papillomatosis. RRP Task Force. Arch. Otolaryngol. Head Neck Surg. 125, 743–748 (1999). [DOI] [PubMed] [Google Scholar]
- 5.Venkatesan N. N., Pine H. S., Underbrink M. P., Recurrent respiratory papillomatosis. Otolaryngol. Clin. North Am. 45, 671–694, viii–ix (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Derkay C. S., Wiatrak B., Recurrent respiratory papillomatosis: A review. Laryngoscope 118, 1236–1247 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Shah K., et al. , Rarity of cesarean delivery in cases of juvenile-onset respiratory papillomatosis. Obstet. Gynecol. 68, 795–799 (1986). [PubMed] [Google Scholar]
- 8.Shah K. V., Stern W. F., Shah F. K., Bishai D., Kashima H. K., Risk factors for juvenile-onset recurrent respiratory papillomatosis. Pediatr. Infect. Dis. J. 17, 372–376 (1998). [DOI] [PubMed] [Google Scholar]
- 9.Taliercio S., et al. , Adult-onset recurrent respiratory papillomatosis: A review of disease pathogenesis and implications for patient counseling. JAMA Otolaryngol. Head Neck Surg. 141, 78–83 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Ruiz R., et al. , Risk factors for adult-onset recurrent respiratory papillomatosis. Laryngoscope 124, 2338–2344 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Ivancic R., Iqbal H., deSilva B., Pan Q., Matrka L., Current and future management of recurrent respiratory papillomatosis. Laryngoscope Investig. Otolaryngol. 3, 22–34 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruparelia S., Unger E. R., Nisenbaum R., Derkay C. S., Reeves W. C., Predictors of remission in juvenile-onset recurrent respiratory papillomatosis. Arch. Otolaryngol. Head Neck Surg. 129, 1275–1278 (2003). [DOI] [PubMed] [Google Scholar]
- 13.Reeves W. C., et al. , National registry for juvenile-onset recurrent respiratory papillomatosis. Arch. Otolaryngol. Head Neck Surg. 129, 976–982 (2003). [DOI] [PubMed] [Google Scholar]
- 14.Karatayli-Ozgursoy S., Bishop J. A., Hillel A., Akst L., Best S. R. A., Risk factors for dysplasia in recurrent respiratory papillomatosis in an adult and pediatric population. Ann. Otol. Rhinol. Laryngol. 125, 235–241 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Cook J. R., Hill D. A., Humphrey P. A., Pfeifer J. D., El-Mofty S. K., Squamous cell carcinoma arising in recurrent respiratory papillomatosis with pulmonary involvement: Emerging common pattern of clinical features and human papillomavirus serotype association. Mod. Pathol. 13, 914–918 (2000). [DOI] [PubMed] [Google Scholar]
- 16.Sanchez G. I., et al. , Human papillomavirus genotype detection in recurrent respiratory papillomatosis (RRP) in Colombia. Head Neck 35, 229–234 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Omland T., et al. , Recurrent respiratory papillomatosis: HPV genotypes and risk of high-grade laryngeal neoplasia. PLoS One 9, e99114 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Szeps M., et al. , Human papillomavirus, viral load and proliferation rate in recurrent respiratory papillomatosis in response to alpha interferon treatment. J. Gen. Virol. 86, 1695–1702 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Giuliano A. R., et al. , Epidemiology of human papillomavirus infection in men, cancers other than cervical and benign conditions. Vaccine 26 (suppl. 10), K17–K28 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Villiers E.-M., Fauquet C., Broker T. R., Bernard H.-U., zur Hausen H., Classification of papillomaviruses. Virology 324, 17–27 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Dunne E. F., et al. , Human papillomavirus (HPV) 6, 11, 16, and 18 prevalence among females in the United States—National Health and Nutrition Examination Survey, 2003-2006: Opportunity to measure HPV vaccine impact? J. Infect. Dis. 204, 562–565 (2011). [DOI] [PubMed] [Google Scholar]
- 22.Ermel A. C., et al. , DNA detection and seroprevalence of human papillomavirus in a cohort of adolescent women. Sex. Transm. Infect. 90, 64–69 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leiding J. W., Holland S. M., Warts and all: Human papillomavirus in primary immunodeficiencies. J. Allergy Clin. Immunol. 130, 1030–1048 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Jong S. J., et al. , The human CIB1-EVER1-EVER2 complex governs keratinocyte-intrinsic immunity to β-papillomaviruses. J. Exp. Med. 215, 2289–2310 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Jong S. J., et al. , Epidermodysplasia verruciformis: Inborn errors of immunity to human beta-papillomaviruses. Front. Microbiol. 9, 1222 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bonagura V. R., et al. , Recurrent respiratory papillomatosis: A complex defect in immune responsiveness to human papillomavirus-6 and -11. APMIS 118, 455–470 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DeVoti J., et al. , Decreased Langerhans cell responses to IL-36γ: Altered innate immunity in patients with recurrent respiratory papillomatosis. Mol. Med. 20, 372–380 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ciancanelli M. J., et al. , Life-threatening influenza and impaired interferon amplification in human IRF7 deficiency. Science 348, 448–453 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hernandez N., et al. , Life-threatening influenza pneumonitis in a child with inherited IRF9 deficiency. J. Exp. Med. 215, 2567–2585 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Casrouge A., et al. , Herpes simplex virus encephalitis in human UNC-93B deficiency. Science 314, 308–312 (2006). [DOI] [PubMed] [Google Scholar]
- 31.Byun M., et al. , Inherited human OX40 deficiency underlying classic Kaposi sarcoma of childhood. J. Exp. Med. 210, 1743–1759 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abramson A. L., Steinberg B. M., Winkler B., Laryngeal papillomatosis: Clinical, histopathologic and molecular studies. Laryngoscope 97, 678–685 (1987). [DOI] [PubMed] [Google Scholar]
- 33.Belkadi A., et al. ; Exome/Array Consortium , Whole-exome sequencing to analyze population structure, parental inbreeding, and familial linkage. Proc. Natl. Acad. Sci. U.S.A. 113, 6713–6718 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Itan Y., et al. , The human gene damage index as a gene-level approach to prioritizing exome variants. Proc. Natl. Acad. Sci. U.S.A. 112, 13615–13620 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Itan Y., et al. , The mutation significance cutoff: Gene-level thresholds for variant predictions. Nat. Methods 13, 109–110 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maffucci P., et al. , Blacklisting variants common in private cohorts but not in public databases optimizes human exome analysis. Proc. Natl. Acad. Sci. U.S.A. 116, 950–959 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Curran M. E., et al. , A molecular basis for cardiac arrhythmia: HERG mutations cause long QT syndrome. Cell 80, 795–803 (1995). [DOI] [PubMed] [Google Scholar]
- 38.Chavarría-Smith J., Vance R. E., The NLRP1 inflammasomes. Immunol. Rev. 265, 22–34 (2015). [DOI] [PubMed] [Google Scholar]
- 39.Petrovski S., Wang Q., Heinzen E. L., Allen A. S., Goldstein D. B., Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet. 9, e1003709 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blekhman R., et al. , Natural selection on genes that underlie human disease susceptibility. Curr. Biol. 18, 883–889 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lek M., et al. ; Exome Aggregation Consortium , Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mamaï O., et al. , Multiple self-healing palmoplantar carcinoma: A familial predisposition to skin cancer with primary palmoplantar and conjunctival lesions. J. Invest. Dermatol. 135, 304–308 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhong F. L., et al. , Germline NLRP1 mutations cause skin inflammatory and cancer susceptibility syndromes via inflammasome activation. Cell 167, 187–202.e17 (2016). [DOI] [PubMed] [Google Scholar]
- 44.Grandemange S., et al. , A new autoinflammatory and autoimmune syndrome associated with NLRP1 mutations: NAIAD (NLRP1-associated autoinflammation with arthritis and dyskeratosis). Ann. Rheum. Dis. 76, 1191–1198 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Zhong F. L., et al. , Human DPP9 represses NLRP1 inflammasome and protects against auto-inflammatory diseases via both peptidase activity and FIIND domain binding. J. Biol. Chem. 10.1074/jbc.RA118.004350 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Okondo M. C., et al. , Inhibition of Dpp8/9 activates the Nlrp1b inflammasome. Cell Chem. Biol. 25, 262–267.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lamkanfi M., Dixit V. M., Inflammasomes and their roles in health and disease. Annu. Rev. Cell Dev. Biol. 28, 137–161 (2012). [DOI] [PubMed] [Google Scholar]
- 48.Guo H., Callaway J. B., Ting J. P.-Y., Inflammasomes: Mechanism of action, role in disease, and therapeutics. Nat. Med. 21, 677–687 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Canna S. W., et al. , An activating NLRC4 inflammasome mutation causes autoinflammation with recurrent macrophage activation syndrome. Nat. Genet. 46, 1140–1146 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brydges S. D., et al. , Divergence of IL-1, IL-18, and cell death in NLRP3 inflammasomopathies. J. Clin. Invest. 123, 4695–4705 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McGeough M. D., et al. , TNF regulates transcription of NLRP3 inflammasome components and inflammatory molecules in cryopyrinopathies. J. Clin. Invest. 127, 4488–4497 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gaidt M. M., et al. , Human monocytes engage an alternative inflammasome pathway. Immunity 44, 833–846 (2016). [DOI] [PubMed] [Google Scholar]
- 53.Chui A. J., et al. , N-terminal degradation activates the NLRP1B inflammasome. Science 364, 82–85 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sandstrom A., et al. , Functional degradation: A mechanism of NLRP1 inflammasome activation by diverse pathogen enzymes. Science 364, eaau1330 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cavaco B. M., et al. , Homozygous calcium-sensing receptor polymorphism R544Q presents as hypocalcemic hypoparathyroidism. J. Clin. Endocrinol. Metab. 103, 2879–2888 (2018). [DOI] [PubMed] [Google Scholar]
- 56.Chae J. J., et al. , Gain-of-function Pyrin mutations induce NLRP3 protein-independent interleukin-1β activation and severe autoinflammation in mice. Immunity 34, 755–768 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Maas-Szabowski N., Stark H.-J., Fusenig N. E., Keratinocyte growth regulation in defined organotypic cultures through IL-1–induced keratinocyte growth factor expression in resting fibroblasts. J. Invest. Dermatol. 114, 1075–1084 (2000). [DOI] [PubMed] [Google Scholar]
- 58.Niebler M., et al. , Post-translational control of IL-1β via the human papillomavirus type 16 E6 oncoprotein: A novel mechanism of innate immune escape mediated by the E3-ubiquitin ligase E6-AP and p53. PLoS Pathog. 9, e1003536 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Karim R., et al. , Human papillomavirus deregulates the response of a cellular network comprising of chemotactic and proinflammatory genes. PLoS One 6, e17848 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Picard C., et al. , Primary immunodeficiency diseases: An update on the classification from the International Union of Immunological Societies Expert Committee for Primary Immunodeficiency 2015. J. Clin. Immunol. 35, 696–726 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Canna S. W., et al. , Life-threatening NLRC4-associated hyperinflammation successfully treated with IL-18 inhibition. J. Allergy Clin. Immunol. 139, 1698–1701 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Federici S., Martini A., Gattorno M., The central role of anti–IL-1 blockade in the treatment of monogenic and multi-factorial autoinflammatory diseases. Front. Immunol. 4, 351 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.De Benedetti F., et al. , Canakinumab for the treatment of autoinflammatory recurrent fever syndromes. N. Engl. J. Med. 378, 1908–1919 (2018). [DOI] [PubMed] [Google Scholar]
- 64.Ramoz N., et al. , Mutations in two adjacent novel genes are associated with epidermodysplasia verruciformis. Nat. Genet. 32, 579–581 (2002). [DOI] [PubMed] [Google Scholar]
- 65.Alisjahbana B., et al. , Disfiguring generalized verrucosis in an Indonesian man with idiopathic CD4 lymphopenia. Arch. Dermatol. 146, 69–73 (2010). [DOI] [PubMed] [Google Scholar]
- 66.Wang W., et al. , Detection of HPV-2 and identification of novel mutations by whole-genome sequencing from biopsies of two patients with multiple cutaneous horns. J. Clin. Virol. 39, 34–42 (2007). [DOI] [PubMed] [Google Scholar]
- 67.Wang C., et al. , Multiple huge cutaneous horns overlying verrucae vulgaris induced by human papillomavirus type 2: A case report. Br. J. Dermatol. 156, 760–762 (2007). [DOI] [PubMed] [Google Scholar]
- 68.Muñoz N., et al. ; International Agency for Research on Cancer Multicenter Cervical Cancer Study Group , Epidemiologic classification of human papillomavirus types associated with cervical cancer. N. Engl. J. Med. 348, 518–527 (2003). [DOI] [PubMed] [Google Scholar]
- 69.Li H., Durbin R., Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 26, 589–595 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li H., et al. ; 1000 Genome Project Data Processing Subgroup , The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.