Abstract
Point-scanning two-photon microscopy enables high-resolution imaging within scattering specimens such as the mammalian brain, but sequential acquisition of voxels fundamentally limits its speed. We developed a two-photon imaging technique that scans lines of excitation across a focal plane at multiple angles and computationally recovers high-resolution images, attaining voxel rates of over 1 billion Hz in structured samples. Using a static image as a prior for recording neural activity, we imaged visually-evoked and spontaneous glutamate release across hundreds of dendritic spines in mice at depths over 250 μm and frame-rates over 1 kHz. Dendritic glutamate transients in anaesthetized mice are synchronized within spatially-contiguous domains spanning tens of microns at frequencies ranging from 1-100 Hz. We demonstrate millisecond-resolved recordings of acetylcholine and voltage indicators, 3D single-particle tracking, and imaging in densely-labeled cortex. Our method surpasses limits on the speed of raster-scanned imaging imposed by fluorescence lifetime.
Introduction
The study of brain activity relies on tools with high spatial resolution over large volumes at high rates1. Fluorescent sensors enable recordings from large numbers of individual cells or synapses over scales ranging from micrometers to millimeters2-5. However, the intact brain is opaque, so high-resolution imaging deeper than ~50 μm requires techniques that are insensitive to light absorption and scattering. Two-photon imaging uses nonlinear absorption to confine fluorescence excitation to the high-intensity focus of a laser even in the presence of scattering. Since fluorescence is generated only at the focus, scattered emission light can be collected without forming an optical image. Instead, images are computationally assembled by scanning the focus in space. However, this serial approach creates a tradeoff between achievable frame-rates and pixels per frame. Common fluorophores have fluorescence lifetimes of approximately 3 ns, requiring approximately 10 ns between consecutive measurements to avoid crosstalk6. The maximum achievable frame-rate for a 1-megapixel raster-scanned fluorescence image is therefore approximately 100 Hz, but neuronal communication via action potentials and neurotransmitter release occurs on millisecond timescales3,7,8. Kilohertz megapixel in vivo imaging would enable monitoring of these signals at speeds commensurate with signaling in the brain.
The pixel rate limit can be circumvented by efficient sampling1,4,9. When recording activity, raster images are usually reduced to a lower-dimensional space after acquisition10, e.g. by analyzing only regions of interest. Random-Access Microscopy11,12 samples this lower-dimensional space more directly by only scanning targeted pixel sets, with an access time cost to move between targets. When targets are sparse, time saved by not sampling intervening areas significantly exceeds access times. Another efficient sampling approach is to mix multiple pixels into each measurement. Multifocal Multiphoton Microscopy scans an array of foci through the sample, generating an image that is the sum of those produced by each focus13. Axially-elongated foci such as Bessel beams14-17 generate projections of a volume at the rate of two-dimensional images, and can be tilted to obtain depth information stereoscopically15,17. We refer to these and related approaches4,18 as “Projection Microscopy” because they deliberately project multiple resolution elements into each measurement. Projection Microscopy samples space densely, providing benefits over random-access microscopy in specimens that move unpredictably (e.g. awake animals or moving particles) or when targets are difficult to select rapidly (e.g. dendritic spines). However, recovering sources from projected measurements requires computational unmixing9,13.
Computational unmixing is used across imaging modalities to remove blurring19,20, combine images having distinct measurement functions21-23, or separate overlapping signals10, and is essential to emerging computational imaging techniques such as light field microscopy24-26. Unmixing is usually posed as an optimization problem, using regularization to impose prior knowledge on the structure of recovered sources, such as independence27 or sparsity10. If sources and measurements are appropriately structured, many sources can be recovered from relatively few mixed measurements28 in a framework called Compressive Sensing9,29-31, potentially improving measurement efficiency in optical physiology9. Highly coherent measurements (i.e. that always mix sources together the same way) make recovery ambiguous, while incoherent measurements can guarantee accurate recovery28. Tomographic measurements, which record linear projections of a sample along multiple angles, have low coherence because lines of different angles overlap at only one point. In medical imaging, compressive sensing using several tomographic angles has increased imaging speed and reduced radiation dose31.
Results
Scanned Line Angular Projection Microscopy
We developed a microscope that combines benefits of random-access imaging and projection microscopy by performing tomographic measurements from targeted sample regions. This microscope scans line foci across a 2D sample plane at four different angles (Fig. 1). Line foci achieve the same lateral resolution as points (Fig. S1), efficiently sample a compact area when scanned, produce two-photon excitation more efficiently than non-contiguous foci of the same volume, and perform low-coherence measurements. The resulting measurements are angular projections analogous to Computed Tomography31.
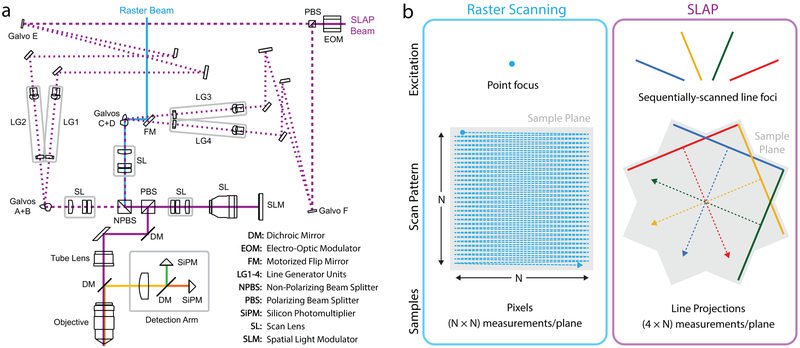
Figure 1. Scanned Line Angular Projection Microscopy.
a) Optical schematic. The SLAP microscope directs a laser beam through four distinct excitation paths in sequence during each frame. Rapid switching between paths is achieved by an electro-optic modulator and polarizing beam splitter. Galvo mirrors (E,F) switch each beam again, producing four paths. Each path contains optics that redistribute the beam intensity into a uniform line of a different angle. Paths are recombined and scanned using two 2D galvo pairs (galvos A+B and C+D), and combined again at a nonpolarizing beamsplitter. The fully recombined beam is relayed and forms an image on a reflective SLM, which selects pixels in the sample to be illuminated, discarding excess light. SLM-modulated light returns through the relay and is imaged into the sample. A separate beam is used for raster scans. b) Raster scanning an (N-by-N)-pixel image requires N2 measurements. SLAP scans line foci to produce 4 tomographic views of the sample plane, requiring 4*N measurements.
This approach, which we call Scanned Line Angular Projection (SLAP) imaging, samples the entire field of view (FOV) with four line scans. Frame time is proportional to the FOV diameter in pixels d, compared to d2 for a raster scan, resulting in greatly increased frame rates. For example, SLAP images a 250×250 μm FOV with 200 nm spacing at a frame-rate of 1016 Hz.
To reduce sample heating and the number of pixels mixed into each measurement, we included a spatial light modulator (SLM) in an amplitude-modulation geometry. This configuration selects a user-defined pattern within the FOV for imaging and discards remaining excitation light, making SLAP a random-access technique (Video 1). This avoids illumination of unlabeled regions, reducing excitation power in sparse samples (Fig. S2), and allows users to artificially introduce sparsity into densely-labeled samples. Unlike other random-access methods, SLAP’s frame-rate is independent of the number of pixels imaged, up to the entire FOV.
Particle Localization and Tracking
Localization and tracking of isolated fluorescent particles are used to monitor organelle dynamics, detect molecular interactions, measure forces, and perform super-resolution imaging32. We demonstrated direct SLAP imaging of sparse samples by localizing and tracking particles. Each particle produces a bump of signal on each scan axis corresponding to its position, and each frame consists of the superposition of signals for all particles in view (Fig. 2a,b). Using the microscope’s empirical measurement function, maximum likelihood reconstruction using Richardson-Lucy deconvolution19,20 produces accurate high-resolution images and error-free localization of sparse particles (Fig. 2c,d). At higher particle densities, maximum likelihood reconstruction produces spurious peaks in recovered images, which can be reduced by regularizing for sparsity (Fig. 2e).
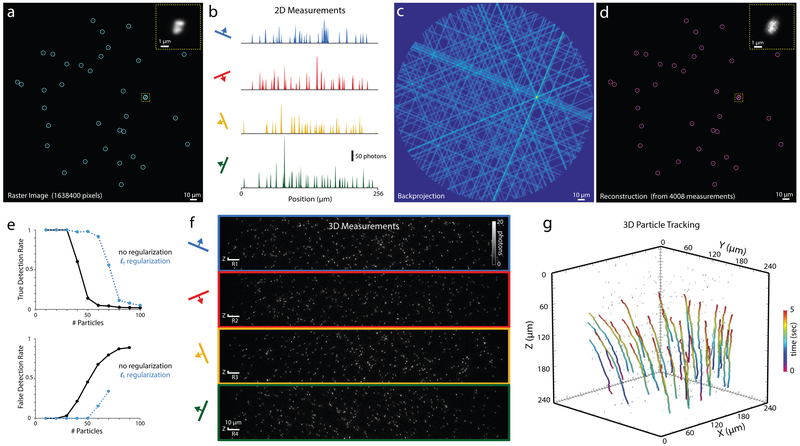
Figure 2. Tomographic particle tracking.
a) 2D raster image of fluorescent beads on glass. Circles denote detected particles (local maxima with intensity >20% of the global maximum). Inset shows zoom of a cluster of beads. b) SLAP measurement of the same sample, consisting of projections along the four scan axes. c) Back-projection of the measurements in b. d) Reconstructed SLAP image with detections circled, demonstrating 100% detection with no false positives. Experiment was repeated 3 times with similar results. e) Simulated detection performance for fields of view containing various numbers of point sources, with and without l1 regularization. (top) detection rate at 5% false positives; (bottom) false positives at 95% detection rate. f) 3D SLAP measurement of a 250×250×250 μm volume (90 planes) obtained in 89 ms. g) Paths of flowing particles in reconstructed 3D SLAP video, tracked using ARIVIS software (Video 3). Tracking experiment was performed once.
We performed 3D SLAP recordings of thousands of 500 nm fluorescent beads flowing in a 250 μm (diameter) × 250 μm (depth) volume (87 planes; 1016 Hz frame rate, 5080 measurements per plane, 10 Hz volume rate, 200 nm measurement spacing), corresponding to 1.4 billion voxels per second (Fig. 2f, Videos 2,3). Particles were readily tracked in the resulting volume videos with existing commercial software (Fig. 2g).
Computational Recovery of Neural Activity
SLAP achieves high frame rates by performing only a few thousand measurements per frame. Recovering images therefore requires a spatial prior, i.e. a constrained sample representation with fewer unknowns than measurements. For neuronal activity imaging, we adopted a sample representation in which spatial components (dendritic segments) vary in brightness over time10. The components are obtained from a raster-scanned reference volume, which we segment using a trained pixel classifier and a skeletonization-based algorithm that groups voxels into ≤1000 contiguous segments per plane (Fig. 3a, S3). Source recovery consists of determining the intensities X of each segment at each frame according to the following model:
Y = [y1,y2,…,yT] are the measurements, each yi is a frame, P is the microscope’s measurement (projection) matrix, S encodes the segmented reference image, each xi is the vector of segment intensities on frame i, b is a rank-1 baseline, and θ implements the indicator dynamics.
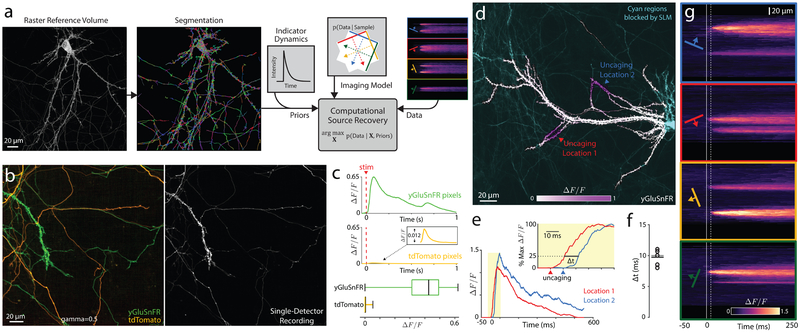
Figure 3. In vitro validation of SLAP activity imaging.
a) Schematic of activity reconstruction. A segmented structural image and model of indicator dynamics are used as priors to constrain image recovery from tomographic measurements. b) Raster image of cultured neurons expressing either yGluSnFR or tdTomato. Only yGluSnFR responds to stimulation. (right) To test activity reconstruction, SLAP imaging was performed with one detector channel. c) (top) Mean temporal responses assigned to yGluSnFR- and tdTomato- expressing pixels. (bottom) Boxplot (vertical lines denote quartiles) of mean ΔF/F0 0-50 ms after stimulus. N = 1707 segments; 3 FOVs. (d-g) Glutamate was uncaged at 2 locations on a yGluSnFR-expressing neuron, 10 ms apart. d) Saturation denotes maximum ΔF/F0 following uncaging. e) Responses at the two uncaging locations. Inset: Responses normalized to peak, demonstrating delay (Δt) between measured signals. Arrowheads denote uncaging times at each location. f) Measured delays for 5 individual trials, black line denotes mean, gray line denotes ground truth. g) SLAP measurements (smoothed in time), showing time delay between the two locations, and diffusion following uncaging. Solid and dashed white lines denote onset of the two uncaging pulses. Experiments are single trials without averaging. Excitation power: (b-c) 36 mW. (d-g) 38 mW. (d-g) Experiment was performed on 8 FOVs with similar results.
This model imposes a strong prior on the space of recovered signals. It enforces that the only changes in the sample are fluctuations in brightness of the segments, with positive spikes and exponential decay dynamics. Motion registration is performed on Y and S before solving. The term that must be estimated is the spikes, W = [w1,w2,…,wT]. We estimate W by maximizing the likelihood of Y using a multiplicative update algorithm related to Richardson-Lucy deconvolution33. Importantly, regularization is unnecessary because S has low rank by design. The solution is nearly always well-determined by the measurements, but adversarial arrangements of segments are possible.
We simulated SLAP imaging and source recovery while varying parameters such as sample brightness and number of sources, assessed sensitivity to systematic modeling errors, and explored performance with different numbers of tomographic angles (Fig. S4). Simulated recovery was highly accurate at signal-to-noise ratios that match our experiments, and robust to errors in model kinetics, segment boundaries, or unexpected sources, but sensitive to motion registration errors. Correlations in source activity were recovered with low bias (Pearson r=0.66; bias −0.0003 +/− 0.0002, 95% ci; Fig. S4g).
Validation in Cultured Neurons
We performed SLAP imaging in rat hippocampal cultures, which facilitate precise optical and electrical stimulation in space and time. First, we co-cultured cells expressing the cytosolic fluorophore tdTomato with cells expressing the glutamate sensor SF-Venus-iGluSnFR34 (yGluSnFR; Fig. 3b). We imaged these cultures using one detector channel in which both fluorophores are bright, while electrically stimulating neurons to trigger yGluSnFR transients. TdTomato does not respond to stimulation, and any transients assigned to tdTomato-expressing cells must be spurious. This experiment tests the solver’s ability to accurately assign fluorescence in space, which we quantified using a separate two-channel raster image. Mean fluorescence transients assigned to yGluSnFR-expressing pixels were more than 52 times greater than transients for tdTomato pixels (tdTomato: mean 0.0078, maximum 0.0125, yGluSnFR: mean 0.4192, maximum 0.6525; Fig. 3c), and no tdTomato pixels showed transients larger than 3% of the mean yGluSnFR amplitude, indicating that both frequency and amplitude of misassigned signals is low even in densely intertwined cultures.
Second, we imaged yGluSnFR-expressing neurons while sequentially uncaging glutamate for 10 ms at each of two locations, to assess timing precision of recovered signals (Fig. 3d-g, Video 4). Even though uncaging produced slow temporally-overlapping transients, SLAP recordings reliably reported the 10 ms delay between these events (mean delay 9.7 +/− 1.1 ms std., N=5 recordings; Fig. 3f). Raw recordings clearly showed this delay and the lateral diffusion of glutamate signals34 (Fig. 3g).
Third, we imaged electrically-evoked action potentials in neurons labeled with the voltage-sensitive dye RhoVR.pip.sulf35 (Fig. S5). We compared 1016 Hz SLAP to widefield imaging of the same FOVs. Suprathreshold field stimulation (90 V/cm) elicited spikes reliably detected by both methods, while weaker stimulation (50 V/cm) stochastically elicited spike patterns that differed across simultaneously-recorded neurons, demonstrating SLAP’s ability to record spikes from individual neurons at millisecond frame rates.
In Vivo Glutamate Imaging
We next recorded activity in dendrites and spines of pyramidal neurons in mouse visual cortex. Pyramidal neuron dendrites are densely decorated with spines receiving excitatory glutamatergic input5,36,37. Spines are often separated from each other and parent dendrites by less than 1 μm36, making spine imaging particularly sensitive to resolution and sample movement. SLAP’s high resolution (432 nm lateral, 1.62 μm axial, Fig. S1) and high frame-rate allow precise (mean error <100 nm) post-hoc motion registration ideal for spine imaging (Fig. S6).
Calcium transients within spines report NMDA receptor activation at synapses37. However, NMDA receptors are activated by concurrent synaptic transmission and postsynaptic depolarization, not necessarily by synaptic transmission alone38. Spine calcium transients are also triggered by other events, such as backpropagating action potentials. Nonlinearities in calcium currents and fluorescent sensors39, and their slow timecourse, further complicate measurement of synaptic inputs using calcium sensors. iGluSnFR enables detection of glutamate transients at individual spines in cortex34, potentially providing a complementary method to record synaptic activity and compare presynaptic to postsynaptic signals.
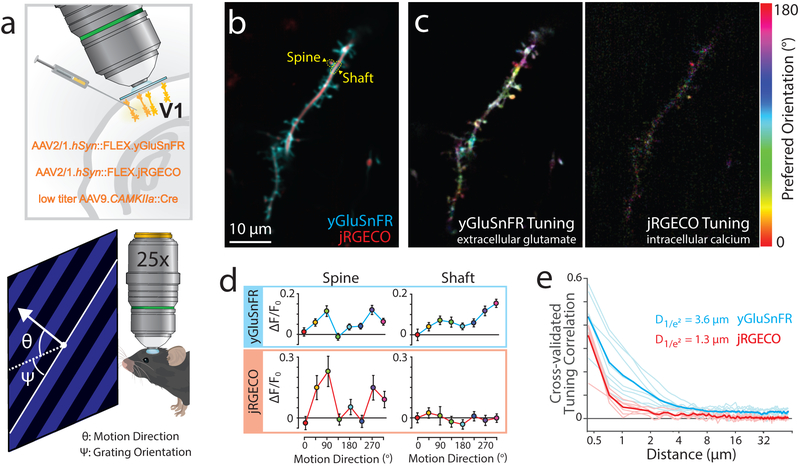
To investigate the relationship between spine glutamate and calcium transients, we imaged dendrites of pyramidal neurons co-expressing yGluSnFR.A184S and the calcium indicator jRGECO1a40, while presenting lightly-anaesthetized (0.75-1% v/v isoflurane) mice with visual motion stimuli in eight directions (Fig. 4a). Vertebrate visual systems show orientation tuning, with similar response amplitudes for a particular motion direction and its opposite41. Raster imaging indicated overlapping but different response patterns for the two indicators (Fig. 4b,c). With trials containing dendrite-wide calcium transients removed, calcium responses were localized to a subset of spines, which showed large orientation selectivity indices (OSIs). At pixels with strongly-tuned calcium responses, glutamate and calcium tuning curves were highly correlated, indicating that yGluSnFR can detect synaptic glutamate that drives calcium transients (Fig. S7). However, strongly-tuned glutamate responses also occurred at spines with no calcium responses, and on dendritic shafts where no spine was visible (Fig. 4c,d, S7). The spatial scale of glutamate responses was larger than that of calcium (1/e2 decay: 3.6 μm yGluSnFR.A184S; 1.3 μm jRGECO1a, Fig. 4e), indicating that yGluSnFR.A184S is sensitive to glutamate that escapes synaptic clefts, similar to its green parent sensor iGluSnFR.A184S42. The affinity of yGluSnFR.A184S (7.5 μM34) is similar to that of the NMDA receptor (1.7 μM43). We therefore reasoned it could be used to investigate patterns of glutamate release relevant to NMDA receptor activation, but distinct from postsynaptic calcium transients.
Figure 4. Comparison of calcium and glutamate responses in cortical dendrites.
a) Experimental design. Viruses encoding Cre-dependent yGluSnFR or yGluSnFR and jRGECO1a (b-e) mixed with low titer Cre were injected into visual cortex to sparsely label pyramidal neurons. We recorded responses to 8 directions of drifting grating motion stimuli. b) Example raster image of dendrite in visual cortex co-expressing jRGECO1a (red) and SF-Venus.iGluSnFR.A184S (cyan). c) Pixelwise orientation tuning maps for each sensor. Hue denotes preferred orientation. Saturation denotes OSI. Intensity denotes response amplitude. d) Tuning curves for ROIs outlined in (b), showing similar tuning at the spine, but distinct responses in the shaft. N=20 trials per direction. Markers denote mean +/− S.E.M. 7 FOVs (3 mice) were recorded with similar results. e) Spatial scale of tuning similarity for simultaneously-recorded jRGECO and yGluSnFR signals. N=7 FOVs, 3 mice, 20-25 repetitions of each stimulus/session. Dark lines denote means, light lines denote sessions.
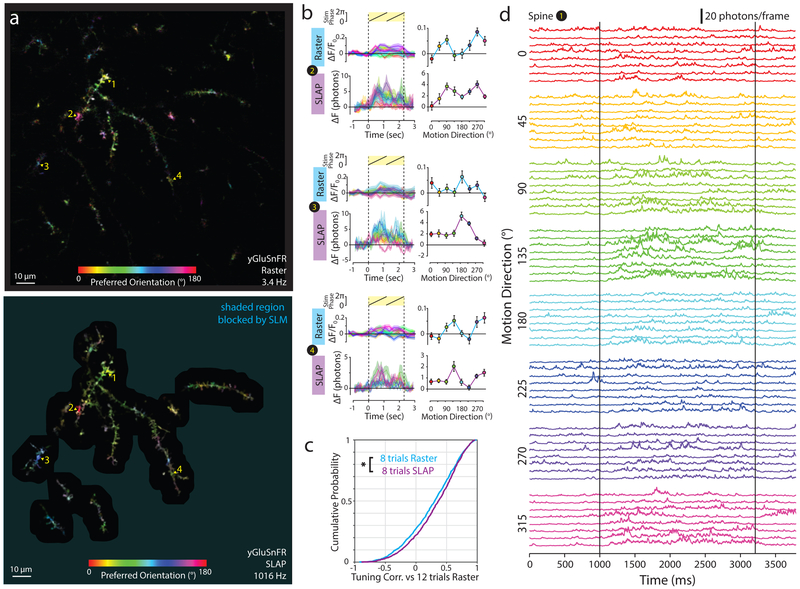
We interleaved SLAP (1016 Hz) and raster (3.41 Hz) recordings of FOVs containing hundreds of spines of isolated yGluSnFR.A184S-labeled layer 2/3 pyramidal neurons in visual cortex while presenting drifting grating stimuli (Videos 5,6). The two methods showed similarly-tuned yGluSnFR responses (Fig. 5a,b). SLAP was more accurate than raster scanning at predicting the tuning of dendritic segments as measured with larger numbers of raster trials (Fig. 5c; SLAP r=0.35+/−0.02, Raster r=0.28+/−0.03; mean+/−SEM, p=0.015). Individual iGluSnFR.A184S transients rise in less than 5 ms, and decay with time constant less than 100 ms34,42. At orientation-tuned spines, SLAP recordings exhibited numerous high-speed transients (Fig. 5d), with power spectral density greater than chance at all frequencies below 137 Hz (Fig. S8). To control for possible artefacts in imaging or source recovery, we generated a ‘dead sensor’ variant of yGluSnFR (yGluSnFR-Null) that lacks a periplasmic glutamate-binding protein domain. yGluSnFR-Null showed equivalent brightness and localization to yGluSnFR, but did not show stimulus responses or significant high-frequency power (N=4 sessions; Fig. S8, Video 7).
Figure 5. High-speed imaging of visually-evoked glutamate activity in cortical dendrites.
a) Pixelwise tuning maps for raster imaging (left; 20 repetitions per stimulus) and SLAP (right; 8 repetitions per stimulus) in yGluSnFR-expressing dendrites. Shaded regions were blocked by the SLM during SLAP imaging. b) Tuning curves and mean temporal responses for 4 spines recorded with raster imaging (20 trials per stimulus) and SLAP (8 trials per stimulus). Markers denote mean +/− S.E.M. c) Cumulative probability of correlation of tuning curve measured from 8 trials per stimulus of raster or SLAP with tuning curve from 12 raster trials per stimulus. N = 1215 segments. *p<0.05, two-sided t-test. d) SLAP recording (1016 Hz) for a single spine over all trials grouped by stimulus type. Excitation power: 96 mW, 110 μm below dura
Spatiotemporal patterns of brain activity encode sensory information, memories, decisions, and behaviors44-46. Neuronal activity exhibits transient synchronization at frequencies ranging from 0.01 to 500 Hz, giving rise to macroscale phenomena associated with distinct cognitive states47. In cortex, synchronous activity reflects attention44,48, task performance48,49, and gating of communication between brain regions45,50. Synchronized input is prominent during specific phases of sleep and anaesthesia51, and is present in awake animals45,52. Such patterns have been explored at the regional scale53, but activity patterns sampled by dendrites of individual neurons have not been recorded with high spatiotemporal resolution.
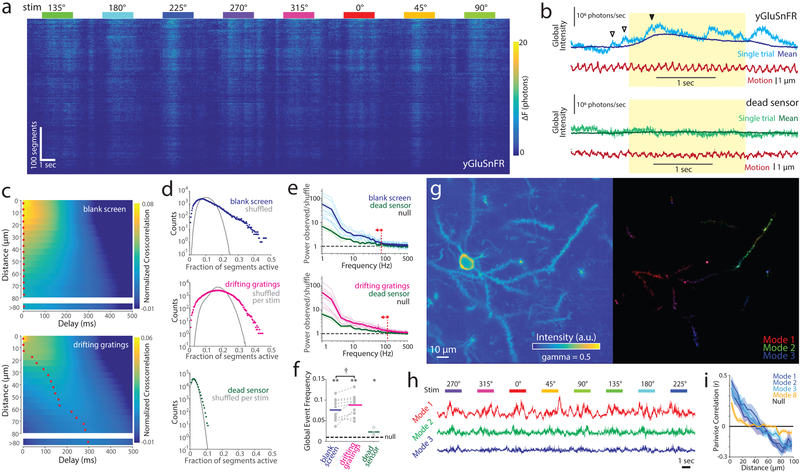
Simultaneous multi-site recordings are essential for studying spatio-temporal activity patterns53. SLAP recordings revealed spatio-temporally patterned dendritic glutamate transients lasting tens to hundreds of milliseconds, occurring both in the presence and absence of motion stimuli (Fig. 6a). These transients were visible in raw yGluSnFR recordings before source recovery, but absent in yGluSnFR-Null (Fig. 6b). In the absence of motion stimuli, cross-correlations in yGluSnFR activity peaked at delays less than 10 ms regardless of distance. During drifting grating stimuli, cross-correlations peaked at delays proportional to the distance between segments (linear fit 0.28+/−0.02 μm/ms 95% c.i.; p<1e-11, N=10 sessions), indicating that activity patterns travel across cortical space as stimuli travel across the retina (Fig. 6c). This cortical magnification (12.7+/−0.7 μm/degree of visual field) was consistent with previous measurements over larger spatial scales54. Cross-correlations in yGluSnFR-Null recordings peaked weakly at a delay of 0 ms regardless of distance or stimuli.
Figure 6. Spatiotemporal structure of cortical glutamate activity.
a) Recording of 460 dendritic segments of a single layer 2/3 neuron over 8 consecutive stimuli. b) Photon counts integrated across the FOV in a single trial, illustrating synchronous activity in yGluSnFR.A184S-labeled dendrites (top), absent in yGluSnFR-Null controls (bottom). Darker traces denote mean response (8 trials) for that stimulus direction. Red trace shows estimated sample motion. Yellow shading denotes stimulus period. Arrows indicate stimulus-evoked (filled) and spontaneous (hollow) global transients. c) Mean cross-correlations between segment pairs (normalized by segment brightness; Methods), binned by distance, in the absence (top) and presence (bottom) of motion stimuli. N=10 sessions. Red stars denote cross-correlation maximum for each distance bin. d) Distribution of count of simultaneously active segments in the absence (top) or presence (center) of motion stimuli, and (bottom) corresponding distribution for dead sensor control. Nulls (gray) are distributions after shuffling across trials of the same stimulus type. e) PSD of number of active segments, divided by corresponding PSD for the shuffled null, during blank screen and motion stimuli, for yGluSnFR and yGluSnFR-Null. Dark lines denote means, light lines denote individual sessions. Vertical red line denotes the minimum frequency bin at which yGluSnFR does not differ from yGluSnFR-Null (two-sided t-test, p>0.05; N=10 sessions (yGluSnFR); 4 sessions (yGluSnFR-Null)). f) Frequency of highly-synchronous activity (p<0.01 under null). Horizontal lines denote mean. *:p<0.05, **:p<0.01, one sample t-test vs. null; †: p<0.05 paired two-sided t-test. N=10 sessions (yGluSnFR), 4 sessions (yGluSnFR-Null). g) Reference image (left), and spatial weights of the largest 3 NMF factors (right) for one session. h) Temporal components for the NMF factors in (g), over 8 consecutive stimulus presentations. i) Pearson correlations between spatial component weights, binned by distance. Line denotes mean, shading denotes S.E.M. N=10 sessions. Excitation power and depth: (a,b(top)): 96 mW, 110 um (bottom) 82 mW, 85 um
To quantify synchrony, we selected dendritic segments with stimulus-evoked responses, and counted the proportion that were significantly active in each frame. The distribution of these counts can be compared to the null distribution obtained by shuffling traces for each segment among trials of the same stimulus type (Fig. 6d). Synchronization results in a higher probability of many segments all being active or all being inactive at the same time. yGluSnFR activity was synchronized across segments both in the presence and absence of motion stimuli in all sessions recorded (N=10 sessions; Kolmogorov-Smirnov test, p<0.001). We compared the frequency spectrum of population fluctuations to the corresponding spectrum for the per-stimulus shuffled null. Activity was significantly more synchronized than yGluSnFR-Null controls at all frequencies below 117 Hz (during motion stimuli), or 73 Hz (blank screen) (two-sided t-tests, p<0.05; Fig. 6e). Highly synchronous events (p<0.01 under the null) were slightly more frequent in the presence versus absence of motion stimuli (8.8% vs. 7.6% of frames; N=10 sessions; two-sided paired t-test, p=0.03; Fig. 6f).
We investigated the spatial structure of synchronous activity by using principal component analysis (PCA) and nonnegative matrix factorization (NMF) to identify spatial modes (i.e. weighted groups of segments) that account for a large fraction of variance in our recordings. We used PCA to factorize recordings for each stimulus direction separately, after subtracting variance attributable to trial time, segment, or stimulus identities. The strongest resulting spatial modes were similar across all stimuli, and were not correlated to brain movement (Fig. S9), indicating that a few motifs account for most fluctuations in glutamate release across stimulus contexts. The largest PCA mode accounted for more variance in glutamate transients than did visual stimuli (adjusted R2 10.3% vs 7.3%; p=0.049, two-sided paired t-test, N=10 sessions). Using NMF, we found that the strongest modes in each recording consisted of spatially-contiguous patches spanning 20-50 μm of dendrite (Fig. 6g,h). We quantified this spatial organization by calculating the correlation in mode weights between pairs of segments as a function of their distance. Across recordings, the strongest modes were spatially structured on a scale of approximately 50 μm, while weaker modes showed less spatial structure (Fig. 6i). We investigated these correlations by raster scanning strips of cortex densely expressing yGluSnFR under the same experimental conditions. The spatial profile of correlations in these recordings matched that of single-neuron SLAP recordings (Fig. S10), suggesting that spatial modes represent regions of active neuropil. These regions may arise from local branching of single axons, or multiple synchronously-active axons. Some spatial modes were strongly orientation-tuned while others were not; across sessions, the largest mode was significantly tuned (Fig. S11; p=2.5e-4, Fisher’s combined test, N=10 sessions). Together, these results indicate that cortical glutamate activity during anaesthesia is organized into synchronously-active spatial domains spanning tens of microns, and that individual pyramidal neurons sample several such domains with their dendritic arbors.
Discussion
Two-photon laser scanning microscopy enables imaging of submicron structures within scattering tissue, but its maximum pixel rate is limited by the sequential recording of pixels. Sophisticated raster scanning techniques55 now perform frame scanning at the limit imposed by fluorescence lifetime. To exceed this limit, random access imaging and projection microscopy can exploit prior knowledge of structured samples. SLAP combines these two approaches to enable diffraction-limited kilohertz frame rate recordings of FOVs spanning hundreds of micrometers. Compared to acousto-optic deflector (AOD) random-access imaging, SLAP allows post-hoc motion compensation and higher frame-rates for large numbers of targets, but AOD-based imaging enables higher frame-rates for small numbers of targets. Compared to multifocal and Bessel beam microscopy, SLAP benefits from lower coherence, higher frame-rates, and random-access excitation, but requires axial scanning to record volumes, whereas the other projection methods can simultaneously record from multiple axial coordinates. Compared to compressive sensing methods using sparsity priors, SLAP activity imaging avoids explicit regularization parameters, which can strongly impact reconstructed images and be difficult to set. Instead, SLAP uses indicator dynamics and a segmented sample volume as clearly-interpretable domain-specific regularizers. As with any projection microscopy method, adversarial arrangements of sources can create ambiguities. In particular, densely packed sources or dim sources neighbored by very bright sources may be recovered less precisely. SLM masking allows dense or bright regions to be blocked or dimmed, ameliorating these issues.
The maximum number of sources for accurate recovery is approximately 1000 per plane for the FOVs imaged here. This condition is readily achieved in sparsely-labeled samples or with SLM masking in densely-labeled samples. SLAP can record high-resolution signals in densely-labeled samples without SLM masking if activity is sparse. For example, we used SLAP to detect and localize activity 300 μm deep within cortex densely labeled with the acetylcholine sensor SF-Venus.iAChSnFR.V9, triggered by electrical stimulation of the cholinergic nucleus basalis (Fig. S12). Punctate (<2 μm extent) and rapid (<30 ms) stimulation-induced transients could be identified and backprojected to their origin in pixel space. Such brief and localized signals would be extremely difficult to detect with other methods without prior knowledge of their location, highlighting the combination of spatial resolution, sampling volume, and speed achieved by SLAP.
Potential applications of SLAP include in vivo neurotransmitter imaging, voltage imaging, volumetric calcium imaging, recording movements in brain, muscle, or vasculature, and tracking organelles or other particles within thick tissue.
Online Methods
Optics
The microscope (Fig. 1) directs a single laser source (Yb:YAG, 1030 nm, 190 fs, tunable repetition rate 1-10 MHz; BlueCut, Menlo Systems, Germany) through four distinct excitation paths during each frame. An electro-optic modulator (EOM) rotates the beam polarization, enabling rapid switching between two paths at a polarizing beam splitter. A galvanometer mirror on each path again switches the beam between two paths. This configuration allows the beam to be sequentially directed onto the four paths with switching time limited by the EOM (<2 μs per switch). Only one path is illuminated at a time.
Each path contains a line generator assembly, consisting of an aperture followed by a series of custom cylindrical lenses, which redistributes the Gaussian input beam into a uniform line of tunable aspect ratio. The two pairs of paths are recombined at 2-dimensional galvo pairs. The resulting paths each pass through a scan lens and are combined using a nonpolarizing beamsplitter. This results in a loss of 50% of the beam but allows the transmitted beam to have a single polarization required for amplitude modulation by the SLM. The beam is transmitted by a polarizing beam splitter and is relayed by scan lenses to form an intermediate image on the surface of a reflective liquid crystal spatial light modulator. Reflected light returns through the relay and the SLM-modulated pattern is directed to the sample at a polarizing beam splitter. The spatially modulated image is relayed by a tube lens and objective to the sample. Fluorescence emission is collected through the objective and reflected by a dichroic mirror to a non-imaging, non-descanned detection arm, as in conventional two-photon microscopes. We detect light with customized silicon photomultiplier modules (Hamamatsu). These detectors have high gain, high linearity, and exceptionally low multiplicative noise (Supplemental Methods, Fig. S13), making them ideal for parallelized imaging, where large numbers of detected photons arrive simultaneously. Wide-field epifluorescence illumination and camera detection (not shown) are coupled into the objective via a shortpass dichroic. For raster imaging, a different beam is introduced into the optical path by a flip mirror. The beam is scanned by one of the 2D galvo pairs as in conventional two-photon microscopes.
The number of paths was chosen to best suit tradeoffs between optomechanical complexity and acquisition speed (favoring fewer paths), versus benefits to source recovery (favoring more paths). For example, three measurement axes are necessary and sufficient to localize particles at low density, but additional axes are needed to resolve ambiguities at higher densities. For a fixed photon budget, increasing the number of tomographic angles improved source recovery in activity imaging simulations in a sample-density dependent manner (Fig. S4h).
The line generator units (LG1-4) are used to impart a 1D angular range onto the incoming 2D Gaussian beam, such that when that beam is focused by a subsequent laser scanning microscope it makes a line focus with substantially uniform intensity distribution along the line, and diffraction-limited width in the focused dimension. The LG uses 3 custom-made cylindrical lenses (ARW Optical, Wilmington, NC). The first lens (convex-concave) imparts a large, negative spherical aberration (in 1D) to the beam, which transforms the initial spatial intensity distribution into a uniform angular intensity distribution. The two subsequent cylindrical lenses (concave-plano, plano-convex) refocus the light in 1D onto the 2D galvanometer mirrors. Thereafter, the microscope transforms the uniform 1D angular intensity distribution at the pupil into a uniform 1D spatial intensity distribution at the line focus at the sample. The relative lateral and axial translations of the first two cylindrical lenses allow an adjustment of the transform between the initial spatial intensity profile and subsequent angular intensity profile. Because spherical aberration alone cannot linearize an initial peaked power distribution over the entire initial distribution, a 1D mask is used at the beginning of the LG to remove the tails from the intensity distribution of the incoming beam. Because a sharp-edged mask at this location would create a 1D symmetric diffraction pattern, a mask with toothed edge profile was utilized. Alternately, the first two cylindrical lenses could be replaced by a suitable acylinder lens. Such a system would have fewer degrees of freedom to actively adjust the final line intensity distribution, but would allow an intensity transform function that can utilize the full tails of the initial spatial intensity distribution.
Detectors
The parallel excitation used in SLAP can result in large numbers of emitted photons (from 0 up to ~400 in our experiments) arriving simultaneously in response to a single laser pulse. This necessitates a detector with large dynamic range, and higher photon rates favor detectors with low multiplicative noise. SLAP uses Silicon Photomultiplier (SiPM) detectors (MPPCs; Hamamatsu, C13366-3050GA-SPL and C14455-3050SPL) for both raster scanning and fast line scan acquisitions. These detectors have extremely low multiplicative noise, sufficient gain to easily detect single photons, and are highly linear in their response within the range of photon rates we encounter, making the integrated photocurrent a precise measure of the number of incident photons (Fig. S13). The measured quantum efficiency of our detection path (for light originating from the objective pupil) is 32% at 525 nm and 20% at 625 nm, comparable to other microscopes at Janelia with the same detection geometry using photomultiplier tubes. Detector voltage is digitized at 250 MHz, synchronized to laser emission via a phase-locked loop. Photon counts are estimating by integrating photocurrent within a time window following each laser pulse and normalizing to the integrated current for a single photon.
Optimizations for Excitation Efficiency
Economy of illumination power is critical for biological imaging56, and conventional two-photon imaging is limited by brain heating under common configurations57. We were concerned that sample heating from light absorption could limit the practicality of two-photon projection microscopy methods such as SLAP, because higher degrees of parallelization require a linear proportional increase in power to maintain two-photon excitation efficiency. Higher degrees of parallelization also make source recovery more challenging by increasing background excitation and mixing between sources. In general, parallel two-photon imaging methods benefit by using the lowest degree of parallelization compatible with an experiment’s required frame-rate2,57.
SLAP uses a customized Yb:YAG amplifier delivering high pulse energies (>9W @ 5MHz) at 1030 nm. The laser’s pulse compressor was tuned to achieve the minimum pulsewidth at the sample, ~190 fs at 5 MHz. To maximize excitation efficiency, each measurement consists of a single laser pulse. We performed simulations to optimize the laser repetition rate given estimated nonlinear and measured thermal57 damage thresholds of the sample. The optimal rate (data available on request) is approximately 5 MHz for single-plane imaging and 1 MHz for volume imaging in simulations we performed. The optimum pulse rate differs between single-plane and 3D imaging because the spatial extent of heating is broad (several mm), while nonlinear photodamage is local. When scanning in 3D, this local component is diluted over a larger volume while heating remains roughly constant, thus favoring higher pulse energies.
For a given focal area within the sample plane, coherent line foci produce more efficient two-photon excitation than arrays of isolated points. This effect is partly because the edges of isolated points produce weak excitation. Line foci have a lower perimeter-to-area ratio than isolated points, making excitation more efficient.
The microscope contains a Spatial Light Modulator (ODPDM512-1030, Meadowlark Optics) that is used as an amplitude modulator to reject unnecessary excitation light. Cortical dendrites fill only a small fraction of their enclosing volume (<3% of voxels in our segmentations of single labeled neurons), allowing the majority of excitation light to be discarded. We retain a buffer of several micrometers surrounding all points of interest to guard against brain movement. The fraction of the SLM that is active depends on the amplitude of motion and the sample structure but is approximately 10% in most of our dendritic imaging experiments, substantially decreasing average excitation power. For sufficiently sparse samples, the SLM and other optimizations outlined above allow SLAP to use less laser power than full-field raster scanning (Fig. S2). Powers used in experiments involving SLM masking were well below thermal damage thresholds for continuous imaging57 (Supplementary Table 1). One experiment did not use SLM masking (Fig. S12) and was conducted with a low duty cycle to avoid damage57. We observed no signs of photodamage or phototoxicity in any of our experiments, and neuronal tuning and morphology were unaffected when reimaged 6 days after initial SLAP imaging (Fig. S14).
The combination of SLM masking and scanning used to vary the excitation pattern in SLAP produces much higher pattern rates (5 MHz) than can be achieved with SLMs alone (<1 kHz), and more control over those patterns than scanning alone. This approach could be used similarly with other projection microscopy approaches, and is easiest to implement if all excitation foci can be made to lie in the plane of the SLM.
Scan Patterns
In the simplest scanning scheme, each frame consists of a single scan of each line orientation across the FOV. The maximum frame rate is determined by the cycle rate of the galvanometers, ~1300 Hz for a 250 μm FOV, limited by heat dissipation in the galvo servo controllers. We routinely image at 1016 Hz, to synchronize frames with the refresh rate of our SLM, which would otherwise produce a mild artifact. We also use tiled scanning patterns for efficiently scanning larger fields of view at only slightly reduced frame-rates (e.g. a 500 μm FOV at 800 Hz for tiling factor 2). The frame-rates achieved here could in principle be improved without substantial changes in the design, and are limited by a combination of scanner cycle rates and laser power. For an ideal scanner, the maximum frame-rate at any resolution would be achieved at a laser repetition rate of ~100 MHz (a 20x improvement over this work), above which the fluorescence lifetime would substantially mix consecutive measurements.
Raster imaging is performed using linear galvos (Fig. 1, galvos C+D), shared with the line scanning path, by inputting a Gaussian beam that bypasses the line generator units. Galvo command waveforms were optimized for all scans using Scanimage’s waveform optimization feature to maximize frame rate while maintaining a linear scan path. Linear galvos enable the alternating fast-axis scans used to produce unwarped reference images. An additional resonant scanner could, in principle, be added to the raster scan system.
Axial scanning is performed using a piezo objective stage (PIFOC P-725K085, Physik Instrumente). 3D SLAP imaging in Fig. 2f-g was performed at a volume rate of 10 Hz using a continuous (rather than stepped) unidirectional scan. When performing source recovery for 3D imaging data, we correct the microscope’s projection function for the continuous linear movement of the objective. SLAP is also compatible with a remote focusing system placed after the SLM in the optical path.
To maximize the cycle rate of the galvanometers and piezo objective mount during SLAP imaging, we optimized the control signals used to command the manufacturer-supplied servo controllers for these devices. We optimized command waveforms iteratively, by measuring the error in the response waveform on each iteration, and using second-derivative regularized deconvolution to update the command signal in a direction that reduces that error, while maintaining smoothness. These calibrations take less than 30 seconds and are performed once per set of imaging parameters. With this approach, we were able to drive our actuators over 50% faster within our position error tolerance (<0.1% RMS error) than with methods that account only for actuator lag.
Particle Tracking Experiments
Fluorescent beads (red fluorescent FluoSpheres, 0.5 μm; Molecular Probes, diluted 1:10000 in water) were sealed in a 2 mm diameter × 2 mm deep well capped by a coverslip. A 250×250×250 μm volume centered 150 μm below the coverslip was imaged by sequentially scanning 2D planes while axially translating the objective in a unidirectional sawtooth scan, at 10 Hz volume rate, 1016 Hz frame rate. We reconstructed volumes at a pixel spacing equal to the measurement spacing (200 nm), which matches the Nyquist criterion (187nm) for the system’s point spread function (440nm FWHM). This lateral pixel spacing corresponds to a voxel rate of 1.49 billion voxels/second. Reconstructions were performed using 30 iterations of 3D Richardson-Lucy deconvolution, with a projection matrix that accounts for the movement of the piezo objective stage. Tracking was performed with a linear assignment problem (LAP) tracking method in the commercial software ARIVIS v2.12.4.
Hippocampal Culture Experiments
Rat hippocampal primary cultures were imaged 19 to 20 days after plating. For uncaging experiments, 3% of cells were nucleofected with a plasmid encoding CAG-SF.Venus-GluSnFR.A184V at time of plating. For experiments with tdTomato-labeled cells, a separate 3% of cells were nucleofected with a plasmid encoding CAG-tdTomato. In both cases, SLAP imaging was performed with a single channel using a 540/80nm filter. For uncaging experiments, 10 μM NBQX and 150 μM Rubi-Glutamate (Tocris) were added to the imaging buffer (145 mM NaCl, 2.5 mM KCl, 10 mM glucose, 10 mM HEPES, pH 7.4, 2 mM CaCl2, 1 mM MgCl2). Two-photon excitation power for culture experiments ranged from 39 to 42 mW at the sample. One-photon glutamate uncaging was performed with 420 nm fiber-coupled LEDs (Thorlabs M420F2). The tips of the fibers were imaged onto the sample plane through the same objective used for activity imaging. The two LEDs were activated at a current of 600 mA for 10 ms each in sequence.
For voltage imaging experiments (Fig. S5), hippocampal cultures were plated on glass-bottom dishes (Mattek) and bath labelled 7-9 days later with RhoVR.pip.sulf (SHA02-059; Fig. S15, Supplemental Information), a rhodamine-based PeT voltage indicator derivative of RhoVR35. RhoVR.pip.sulf was added to media at a final concentration of 0.5 μM (from 1000x stock in DMSO) for 20 minutes at 37°C, then replaced with imaging buffer for recording. Platinum wire stimulation electrodes (1 cm spacing) were placed into the bath. Action potentials were triggered by an SD-9 stimulator (Grass Instruments) with 1 ms, 90V pulses. To generate asynchronous stochastic activity (Fig. S5b) the voltage was reduced to 50V, and the following buffer was used (in mM): 145 NaCl, 2.5 KCl, 10 glucose, 10 HEPES, pH 7.4, 2 CaCl2, 1 MgCl2. We performed SLAP and one-photon widefield imaging over the same fields of view. One-photon widefield imaging (Excitation filter: AT540/25X, Dichroic: 565DCXR, Emission filter: ET570LP, Chroma Technology; LED light source: MCWHL5, Thorlabs) was performed with an ORCA Flash 4.0 camera (Hamamatsu). For voltage imaging analysis, regions of interest (ROIs) were drawn manually. For one-photon recordings, traces were obtained by averaging intensity within the region of interest, and F0 was defined as the average of each trace in the 100 ms prior to stimulus onset. For two-photon recordings, the two-dimensional regions of interest were projected into the measurement space (using the microscope measurement matrix P) to produce expected measurements. F0 was defined by the lower convex hull of the measurements low-pass filtered in time, and subtracted from the measurements. The F0-subtracted data were unmixed using the ROI expected measurements using frame-independent non-negative least squares (Matlab lsqnonneg). Excitation powers (SLAP: 31-33 mW, Widefield: 0.5-0.7 mW, 250 μM diameter illuminated area) were chosen to maximize signal-to-noise ratios while minimizing photobrightening of the voltage indicator, which occurs at higher excitation powers. Wavesurfer software was used to synchronize imaging system components and stimulation.
Surgical Procedures
All animal procedures were in accordance with protocols approved by the HHMI Janelia Research Campus Institutional Animal Care and Use Committee (IACUC 17–155). We performed experiments with 19 C57Bl/6NCrl mice (females, 8-10 weeks at the time of the surgery). Each mouse was anaesthetized using isoflurane in oxygen (3-4% for induction, 1.5-2% for maintenance), placed on a 37°C heated pad, administered Buprenorphine HCl (0.1 mg/kg) and ketoprofen (5 mg/kg), and its head gently affixed by a toothbar. A flap of skin and underlying tissue covering the parietal bones and the interparietal bone was removed. The sutures of the frontal and parietal bones were covered with a thin layer of cyanoacrylate glue. A titanium headbar was glued over the left visual cortex. We carefully drilled a ~4.5 mm craniotomy (centered ~3.5 mm lateral, ~0.5 mm rostral of lambda) using a high-speed microdrill (Osada, EXL-M40) and gently removed the central piece of bone. The dura mater was left intact. Glass capillaries (Drummond Scientific, 3-000-203-G/X) were pulled and bevelled (30° angle, 20 μm outer diameter). Using a precision injector (Drummond Scientific, Nanoject III) we performed injections (30 nl each, 1 nl/s, 300 μm deep) into 6-8 positions within the left visual cortex. For sparse labeling experiments, the viral suspension was composed of AAV9.CaMKII.Cre.SV40 (Penn Vector Core, final used titer 1×108 genome counts/ml) mixed with a virus encoding the reporter at 5×1011 gc/ml (one of: AAV2/1.hSyn.FLEX.GCaMP6f.WPRE.SV40 (Penn Vector Core); AAV2/1.hSyn.FLEX.iGluSnFR-SF-Venus.A184S (‘yGluSnFR.A184S’; Janelia Vector Core), AAV2/1.hSyn.FLEX.iGluSnFR-SF-Venus.NULL (‘yGluSnFR-Null’; Janelia Vector Core); AAV2/1.hSyn.FLEX.iGluSnFR-SF-Venus.A184V (‘yGluSnFR.A184V’; Janelia Vector Core); AAV2/1.hSyn.FLEX.NES-jRGECO1a (‘jRGECO’; GENIE Project vector 1670-115); AAV2/1.hSyn.FLEX.SF-Venus.iAChSnFR (‘yAChSnFR’; Janelia Vector Core). For coexpression of jRGECO and yGluSnFR, we included both viruses at 5×1011 gc/ml each. The craniotomy was closed with a 4 mm round #1.5 cover glass that was fixed to the skull with cyanoacrylate glue. Animals were imaged 5-15 weeks after surgery.
Visual Stimulation Experiments
A vertically-oriented screen (ASUS PA248Q LCD monitor, 1920×1200 pixels), was placed 17 cm from the right eye of the mouse, centered at 65 degrees of azimuth and −10 degrees of elevation. A high-extinction 500 nm shortpass filter (Wratten 47B-type) was affixed to the screen. Stimuli were drifting square-wave gratings (0.153 cycles per cm, 1 cycle per second, 22 degrees of visual field per second, 2 second duration) in 8 equally-spaced directions, spaced by periods of mean luminance, as measured using the microscope detection path. 0° denotes a horizontal grating moving upwards, 90° denotes a vertical grating moving rearwards. Stimuli were created and presented using Psychtoolbox-358. Mice were weakly anaesthetized with isoflurane (0.75-1% vol/vol) during recording. Anaesthetized mice were heated to maintain a body temperature of 37°C.
Reference Images
Accurate registration and source recovery require minimally warped reference raster images. The SLAP user interface communicates with ScanImage (Vidrio Technologies), which is used to perform all point-scanning imaging. Raster images can be warped by many factors, including nonlinearity in the scan pattern, sample motion, and the ‘rolling shutter’ artifact of the raster scan. These errors must be corrected in the reference stacks to perform accurate source recovery. We compensate for warping by collecting two sets of reference images interleaved, one with each of the two galvos acting as the fast axis. We assume that motion is negligible during individual lines along the fast axis (~300 μs), and that the galvo actuators track the command accurately along the slow axis. These assumptions allow recovery of unwarped 2D images by a series of stripwise registrations. The axial position of the image planes is estimated by alignment to a consensus volume.
Segmentation
SLAP performs relatively few measurements per frame, which must be combined with some form of structural prior to obtain pixel-space images. For activity imaging, we obtain that structural prior by segmenting a detailed raster reference image. The goal of segmentation is to divide the labeled sample volume into compartments such that the number of compartments in any imaging plane is less than the number of measurements, and the activity within each compartment is homogeneous. The segmentation forms the spatial basis for recovered signals; each segment corresponds to one source. We use a manually trained pixel classifier (Ilastik59 in autocontext mode) to label each voxel in the volume as belonging to one of four categories: dendritic shaft, spine head, other labeled region, or unlabeled background. We do not deconvolve the reference stack or attempt to label features finer than the optical resolution. Instead, we label features at the optical resolution and design the projection matrix P such that it accounts for the transformation from the point spread function of the raster scan to that of each line scan (Fig. S1). We use a skeletonization-based algorithm to agglomerate labeled voxels into short segments of roughly 1.4 μm, which approximately correspond to individual spines or short segments of dendritic shafts. Segments that are predicted to produce very few photon counts (after considering SLM masking) are merged with their neighbors. Most fields of view contained 400 to 600 segments in the focal plane. In simulations with similar sample brightness and density to our in vivo recordings, source recovery performs well when there are less than 1000 segments in the focal plane (Fig. S4b). Some analyses can be performed without a segmentation (e.g. Fig. S12).
Measurement Matrix
The microscope’s measurement (projection) matrix P is measured in an automated process using a thin (<<1 μm) fluorescent film (see measure_PSF in the software package). Images of the excitation focus in the film, collected by a camera, allow a correspondence to be made between the positions of galvanometer scanners and the location of the resulting line focus. The raster scanning focus is also mapped, allowing us to create a model of the line foci transformed into the space of the sample image obtained by the raster scan. The camera is unable to resolve the shape of the excitation focus of the line along the shortest axis (the R-axis). The point spread function on the R and Z axes is measured by scanning sparse fluorescent beads, but this measurement is not needed to generate P because we model SLAP measurements as a fixed convolution of the raster point spread function, which is already encoded in the raster reference image. SLAP achieves the same lateral resolution as point scanning two-photon, but the focus is extended slightly axially because light is focused on only one axis to produce each line (full-width-half-maximum 1.62 μm vs. 1.46 μm for point focus, Fig. S1). This increased depth of field can in some cases be of benefit as it reduces effects of axial motion.
SLAP Data Reduction
Data acquisition and signal generation were performed with a National Instruments PXI data acquisition system incorporating an 8-channel oscilloscope/FPGA module (NI PXIe-5170R / Kintex-7 325T FPGA), and four other input/output DAQs (2x PXIe-6341, 1x PXIe-6738, 1x PXIe-6363). Detector voltages and galvo position sensor voltages were digitized at 250 MHz, synchronized to the laser output clock via phase-locked-loop. Galvo positions were downsampled to 5 MHz on the FPGA by averaging. Detector voltages are recorded at the full rate and the FPGA also generates a concurrent 5MHz data stream for live display. Auxiliary analog inputs (such as photodiode voltages to monitor visual stimulus retrace times, and piezo objective stage position sensors for 3D experiments) were digitized at up to 10 MHz, synchronized to the laser output clock. The data rate recorded to disk is ~1.1 Gigabytes/second, streamed to a RAID 0 array of SCSI hard drives.
SLAP data reduction converts recorded raw voltages into frames, by estimating the number of photons detected at each laser pulse, and interpolating these intensities onto a set of reference galvo positions, based upon simultaneously-recorded galvo position sensor data. This registration compensates for changes in the galvo path during high-speed recordings that would otherwise contribute to measurement error. For details, see SLAP_reduce in the software package. SLAP data reduction requires approximately 5 seconds per second of data on a single CPU.
Motion Registration
Recorded SLAP data are spatially registered to compensate for sample motion. As with raster imaging, translations of a single resolution element can be sufficient to impact recovery of activity in fine structures, necessitating precise registration (Fig. S4e). Motion registration (Fig. S6) is performed by identifying the 3D translation of the sample that maximizes the sum of correlations (or optionally, minimizes Dynamic Time Warping distances60) between the recorded signal and the expected measurements on each of the four scan axes. If the SLM is not used, this objective is well approximated using cross-correlations that can be rapidly computed. If the SLM is used, cross-correlations may not be effective, so we perform a coarse-to-fine grid search using the full measurement matrix (i.e. we evaluate the alignment loss on a coarse grid, followed by a finer grid in the vicinity of the initial optimum). Under these conditions, motion registration requires approximately 50 seconds per second of data on a single CPU. For anaesthetized recordings with little motion, we performed alignments in blocks, by first performed cross-correlation-based alignment of the measurements on each projection axis to correct for rapid in-plane motion of the sample, then averaging over 4-second blocks and performing the above alignment process on each block. All alignments were evaluated for goodness-of-fit, and activity imaging trials containing frames that exceeded a threshold alignment error were discarded (2 trials over all datasets recorded).
When imaging at 1016 Hz, frame-to-frame brain motion is significantly smaller than the optical resolution, even during large-amplitude motions in awake mice (Fig. S6f-I; Mean and maximum displacements of 60nm and 200 nm lateral, 36 nm and 375 nm axial, respectively. Maximum displacements reflect the alignment resolution and are upper bounds). This allows the SLAP solver to treat all measurements within a single frame as simultaneous.
Analyses of in vitro recordings
In Fig. 3c, source recovery was performed on Channel 1 of the SLAP recording and projected into two dimensions using the axial mean projection of the segmentation matrix S. ΔF/F0 was fit for each voxel in this projection. Fig. 3c shows the mean ΔF/F0 for the period 0-50ms after the stimulus, for the 1/3 most green (Ch1>Ch2) and the 1/3 most red (Ch2>Ch1) pixels of the reference image within the segmentation. In Figure 3e-f, we selected the dendritic segment closest to each uncaging location, which were registered relative to the scanner coordinates by imaging in a thin fluorescent sample. Δt was calculated as the difference between times at which each trace crossed 25% of its maximum within the 150 ms period following the first uncaging event. Figure 3f shows Δt values from single trials across 5 experiments.
Analyses of in vivo recordings
Raster activity recordings were aligned with custom MATLAB code that compensates for bidirectional scanning artifacts, warping of galvo scan paths, and rolling shutter movement artifacts using a series of stripwise registrations, included in the published software package (alignScanImageData and related functions). For simultaneous recordings of jRGECO and yGluSnFR (Fig. 4), the two recording channels (540/80 and 650/90 bandpass filters) were linearly unmixed with the mixing coefficients 0.069 (yGluSnFR->jRGECO bleedthrough) and 0.058 (jRGECO->yGluSnFR bleedthrough), estimated using robust regression against pixel intensities in the aligned average images.
The method of calculating F0 was different for different imaging methods. In raster recordings, F0 for a region of interest was calculated for each stimulus presentation as the mean intensity in that region over 3 frames prior to the stimulus. For SLAP recordings, the terms X in the activity model can be directly interpreted as the ΔF/F0 values for each segment. The baseline fluorescence is accounted for by the time-varying baseline term b, which was fit in the space of the measurements based on the cumulative minimum of the regression of the observed against the expected measurements (see SLAP_solve). For plots of ΔF/F0 in the measurement space (Fig. 3g), a time-varying F0 was fit based on the cumulative minimum of the raw intensity for each row of Y, and ΔF/F0was computed, temporally smoothed using non-negative deconvolution61 and downsampled for display.
In Fig. 4e, correlations in pixelwise tuning curves were calculated by cross-validation; the dataset for each session was dividing into two halves and tuning curves were calculated for each pixel. For each pixel, the tuning curve from one half of the data was correlated against tuning curves obtained from the other half of the data for all other pixels. This was done for 5 different random splittings of the data, and results averaged, for each session. This approach avoids introducing correlations due to trial-to-trial variations.
Orientation selectivity indexes (OSIs) (Fig. 4c,5a, S11) were calculated from the 8-point tuning curves (i.e. the mean response during the stimulus period for the 8 stimuli, minus the mean response during 1 second prior to the stimulus) for SLAP and raster recordings in the same way. The 8 stimulus angles were mapped onto four equally-spaced angles (i.e. 0, 90, 180, 270, 0, 90, 180, 270 degrees), multiplied by response amplitude, and vector summed. The preferred orientation (hues in Fig. 4c,5a) is the angle of the sum vector. The OSI is the length of the sum vector, divided by the sum of the lengths of the input vectors. It has a value in [0 1] which is 1 when there is a response to only one of four orientations, and 0 when responses are uniform. The response amplitude is the 2-norm of the 8-point tuning curve.
Videos 5, 6, and 7 are rendered with saturations as ΔF/F0 conditioned on the Z-score, computed as ΔF/sqrt(F0+3/8), with F in photons. This was calculated by dividing each activity by a maximum saturation level (for SLAP recordings ΔF/F0:1.8, Z:3.5, for raster ΔF/F0:1.4, Z:4), and taking the minimum of the two. This ensures that every rendered transient has high confidence, suppressing ΔF/F0 fluctuations of low statistical significance in pixels with low baseline rates. The samples in Videos 6 and 7 had similar labeling brightness and were imaged at the same laser powers prior to the SLM, produced similar mean photon rates per dendritic segment (14.5 photons/segment, Video 6; 18.2 photons/segment, Video 7), and were reconstructed using the same solver parameters. The raster recording (Video 5) was spatially smoothed by summing within a σ=1-pixel Gaussian window prior to computing Z-scores.
In Figure 5c, Raster recordings were aligned to the SLAP reference image, and dendritic segments were projected into the plane of the raster recording to generate regions of interest for correlation analyses. Only segments with components within the plane of raster imaging were included in analysis. Tuning curves were obtained for 8 randomized trails, and compared to the remaining 12 trials of the raster recording, for 100 randomizations. SLAP tuning curves (8 trials) were compared to the corresponding 12 trials for each randomization, and the distribution of correlations for each method is plotted.
For SLAP imaging population analyses (Fig. 6), we analyzed all segments in the sample volume with mean F0 greater than 1.5 photons per frame that were within the imaging plane in at least all but 3 trials (“selected segments”). Motion censoring is performed on the basis of goodness-of-fit of the SLAP 3D alignment method (SLAP_PSXdata and related functions). In the experiments shown here, only two trials across all datasets contained unacceptable sample motion and were censored. Additionally, for calculating stimulus tuning, we censored the first three stimulus presentations of SLAP recordings in each session.
In Fig. 6c cross-correlations in activity traces were calculated between all pairs among selected segments in each trial, normalized to the geometric mean of the variances for each pair, and averaged across trials and sessions for each distance bin weighted by the product of the two segment brightnesses. In Fig. 6d-f, we labeled each segment in each frame as active or inactive by high-pass filtering at 4 Hz and thresholding at 3.5 standard deviations of the bottom 80% of the data above the median. We analyzed the distribution of the number of active segments across frames. The null distribution was calculated by randomly shuffling each trial for each segment across the 8 trials of the same stimulus type for the same segment, and calculating the same distribution. This method controls for synchronization due to structure in the stimulus and shared stimulus tuning. In Fig. 6f, we calculated observed rates of events exceeding the p=0.01 level of the null distribution, and used two-sided Z-tests to compare to the known null rate (0.01). We used two-sided paired t-tests to compare the two stimulus conditions to each other.
In Fig. S9, spatial modes were calculated by PCA over concatenated trials of each stimulus after subtracting the mean response to that stimulus for each recorded segment. We report the correlations in the spatial weights of the modes across segments. Note that the precise ordering of modes can vary slightly across stimuli despite a one-to-one correspondence, such that e.g. the second largest mode of a given stimulus might correlate highly to the largest mode of all others. For visualization in Fig. 6g spatial modes were calculated by NMF, to overcome the orthogonality constraint imposed by PCA. Because NMF performs a strictly positive factorization, subtraction of mean responses was not performed, and modes were instead calculated only from fluctuations during the blank screen period. The spatial appearance of modes obtained by NMF was qualitatively similar to the positive components of modes obtained by PCA from the complete datasets after stimulus subtraction. The NMF modes were then used to obtain temporal components for all frames in the stimulus-subtracted data by regression (Fig. 6h). In Fig. 6i, we report the correlation in the spatial weights of pairs of segments, where the distance between the segments falls into the specified distance bin, for each mode. This correlation is positive for a given distance if segments spaced by that distance tend to have the same weight. It is zero if weights are independent and identically distributed (i.e. unstructured in space).
In Fig. S12, events were localized manually on the four measurement axes and the measurements corresponding to the selected locations were backprojected into the pixel space, to identify the spatial origin of the transients.
Visualizations
All videos and traces shown are of single-trial recordings without averaging unless explicitly stated. Where noted, raster images have been square-root transformed (“gamma=0.5”) to better display dim features.
Statistics
Statistical tests were performed using Matlab (R2018b). Code used for bootstrap sampling, simulations, and generation of null distributions is available at www.github.com/KasparP/SLAP. Additional code is available from the corresponding author.
Data Availability Statement
Optical and mechanical designs and instructions for use are available through the Janelia Open Science portal at www.janelia.org/open-science. Example datasets, demonstrations, and code used for analysis and simulations are available at www.github.com/KasparP/SLAP. All other resources and data are available from the corresponding author upon reasonable request.
Code Availability Statement
Software was written in Matlab (R2015b - R2018b), C, and LabView FPGA. Custom code used for microscope control, data processing, analysis and simulations are available at www.github.com/KasparP/SLAP. All other custom code is available from the corresponding author upon reasonable request.
Supplementary Material
Acknowledgements
Work by SA, PED, and EWM was funded by the following sources: NIH / NIGMS R35GM119855
NIH / NINDS R01NS098088, NSF NeuroNex 1707350, Klingenstein Simons Foundation. Work of the remaining authors was funded by the Howard Hughes Medical Institute.
The authors thank Heather Davies, Salvatore Dilisio, Vasily Goncharov, Jenny Hagemeier, Amy Hu, Na Ji, Aaron Kerlin, Justin Little, Christopher McRaven, Brett Mensh, Boaz Mohar, Manuel Mohr, Marius Pachitariu, Steven Sawtelle, Brenda Shields, Carsen Stringer, Srinivas Turaga, Deepika Walpita, and Ondrej Zelenka for contributions to this work.
The mouse image in Fig. 4a was obtained from the Database Center for Life Sciences (DBCLS TogoTV, © 2016; CC-BY-4.0 license).
Footnotes
Competing Financial Interests Statement
AK, DF,and KP are listed as inventors on a patent application No. 62/502,643 claiming Scanned Line Angular Projection Microscopy, filed by HHMI.
References
- 1.Yang W & Yuste R In vivo imaging of neural activity. Nat. Methods 14, 349–359 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sofroniew NJ, Flickinger D, King J & Svoboda K A large field of view two-photon mesoscope with subcellular resolution for in vivo imaging. eLife 5, e14472 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peterka DS, Takahashi H & Yuste R Imaging voltage in neurons. Neuron 69, 9–21 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prevedel R et al. Fast volumetric calcium imaging across multiple cortical layers using sculpted light. Nat. Methods 13, 1021–1028 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen X, Leischner U, Rochefort NL, Nelken I & Konnerth A Functional mapping of single spines in cortical neurons in vivo. Nature 475, 501–505 (2011). [DOI] [PubMed] [Google Scholar]
- 6.Strickler SJ & Berg RA Relationship between Absorption Intensity and Fluorescence Lifetime of Molecules. J. Chem. Phys. 37, 814–822 (1962). [Google Scholar]
- 7.Sobczyk A, Scheuss V & Svoboda K NMDA Receptor Subunit-Dependent [Ca2+] Signaling in Individual Hippocampal Dendritic Spines. J. Neurosci. 25, 6037–6046 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hao J & Oertner TG Depolarization gates spine calcium transients and spike-timing-dependent potentiation. Curr. Opin. Neurobiol. 22, 509–515 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Pnevmatikakis EA & Paninski L Sparse nonnegative deconvolution for compressive calcium imaging: algorithms and phase transitions in Advances in Neural Information Processing Systems 26 (eds. Burges CJC, Bottou L, Welling M, Ghahramani Z & Weinberger KQ) 1250–1258 (Curran Associates, Inc., 2013). [Google Scholar]
- 10.Pnevmatikakis EA et al. Simultaneous Denoising, Deconvolution, and Demixing of Calcium Imaging Data. Neuron 89, 285–299 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Szalay G et al. Fast 3D Imaging of Spine, Dendritic, and Neuronal Assemblies in Behaving Animals. Neuron 92, 723–738 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bullen A, Patel SS & Saggau P High-speed, random-access fluorescence microscopy: I. High-resolution optical recording with voltage-sensitive dyes and ion indicators. Biophys. J. 73, 477–491 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang W et al. Simultaneous Multi-plane Imaging of Neural Circuits. Neuron 89, 269–284 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu R et al. Video-rate volumetric functional imaging of the brain at synaptic resolution. Nat. Neurosci. 20, 620–628 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song A et al. Volumetric Two-photon Imaging of Neurons Using Stereoscopy (vTwINS). Nat. Methods 14, 420–426 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Botcherby EJ, Juškaitis R & Wilson T Scanning two photon fluorescence microscopy with extended depth of field. Opt. Commun. 268, 253–260 (2006). [Google Scholar]
- 17.Thériault G, Cottet M, Castonguay A, McCarthy N & De Koninck Y Extended two-photon microscopy in live samples with Bessel beams: steadier focus, faster volume scans, and simpler stereoscopic imaging. Front. Cell. Neurosci. 8, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Field JJ et al. Superresolved multiphoton microscopy with spatial frequency-modulated imaging. Proc. Natl. Acad. Sci. 113, 6605–6610 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Richardson WH Bayesian-Based Iterative Method of Image Restoration*. JOSA 62, 55–59 (1972). [Google Scholar]
- 20.Lucy LB An iterative technique for the rectification of observed distributions. Astron. J. 79, 745 (1974). [Google Scholar]
- 21.Neil M. a. A., Juškaitis R & Wilson T Method of obtaining optical sectioning by using structured light in a conventional microscope. Opt. Lett. 22, 1905–1907 (1997). [DOI] [PubMed] [Google Scholar]
- 22.Gustafsson MG Extended resolution fluorescence microscopy. Curr. Opin. Struct. Biol. 9, 627–628 (1999). [DOI] [PubMed] [Google Scholar]
- 23.Preibisch S et al. Efficient Bayesian-based multiview deconvolution. Nat. Methods 11, 645–648 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Broxton M et al. Wave optics theory and 3-D deconvolution for the light field microscope. Opt. Express 21, 25418–25439 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Antipa N et al. DiffuserCam: lensless single-exposure 3D imaging. Optica 5, 1–9 (2018). [Google Scholar]
- 26.Prevedel R et al. Simultaneous whole-animal 3D imaging of neuronal activity using light-field microscopy. Nat. Methods 11, 727–730 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brown GD, Yamada S & Sejnowski TJ Independent component analysis at the neural cocktail party. Trends Neurosci. 24, 54–63 (2001). [DOI] [PubMed] [Google Scholar]
- 28.Candes EJ The restricted isometry property and its implications for compressed sensing. Comptes Rendus Math. 346, 589–592 (2008). [Google Scholar]
- 29.Lustig M, Donoho D & Pauly JM Sparse MRI: The application of compressed sensing for rapid MR imaging. Magn. Reson. Med. 58, 1182–1195 (2007). [DOI] [PubMed] [Google Scholar]
- 30.Pégard NC et al. Compressive light-field microscopy for 3D neural activity recording. Optica 3, 517–524 (2016). [Google Scholar]
- 31.Chen G-H, Tang J & Leng S Prior image constrained compressed sensing (PICCS): A method to accurately reconstruct dynamic CT images from highly undersampled projection data sets. Med. Phys. 35, 660–663 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.von Diezmann A, Shechtman Y & Moerner WE Three-Dimensional Localization of Single Molecules for Super-Resolution Imaging and Single-Particle Tracking. Chem. Rev. 117, 7244–7275 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kazemipour A, Babada B, Wu M, Podgorski K & Druckmann S Multiplicative Updates for Optimization Problems with Dynamics. in Asilomar Conference on Signals, Systems, and Computers (2017). [Google Scholar]
- 34.Marvin JS et al. Stability, affinity, and chromatic variants of the glutamate sensor iGluSnFR. Nat. Methods 15, 936 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deal PE, Kulkarni RU, Al-Abdullatif SH & Miller EW Isomerically Pure Tetramethylrhodamine Voltage Reporters. J. Am. Chem. Soc. 138, 9085–9088 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gray EG Electron Microscopy of Synaptic Contacts on Dendrite Spines of the Cerebral Cortex. Nature 183, 1592–1593 (1959). [DOI] [PubMed] [Google Scholar]
- 37.Mainen ZF, Malinow R & Svoboda K Synaptic calcium transients in single spines indicate that NMDA receptors are not saturated. Nature 399, 151–155 (1999). [DOI] [PubMed] [Google Scholar]
- 38.Mayer ML, Westbrook GL & Guthrie PB Voltage-dependent block by Mg2+ of NMDA responses in spinal cord neurones. Nature 309, 261–263 (1984). [DOI] [PubMed] [Google Scholar]
- 39.Chen T-W et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dana H et al. Sensitive red protein calcium indicators for imaging neural activity. eLife 5, e12727 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tan AYY, Brown BD, Scholl B, Mohanty D & Priebe NJ Orientation Selectivity of Synaptic Input to Neurons in Mouse and Cat Primary Visual Cortex. J. Neurosci. 31, 12339–12350 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jensen TP et al. Multiplex imaging of quantal glutamate release and presynaptic Ca2+ at multiple synapses in situ. bioRxiv 336891 (2018). doi: 10.1101/336891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nahum-Levy R, Tam E, Shavit S & Benveniste M Glutamate But Not Glycine Agonist Affinity for NMDA Receptors Is Influenced by Small Cations. J. Neurosci. 22, 2550–2560 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harris KD & Thiele A Cortical state and attention. Nat. Rev. Neurosci. 12, 509–523 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Luczak A, Bartho P & Harris KD Gating of Sensory Input by Spontaneous Cortical Activity. J. Neurosci. 33, 1684–1695 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stringer C et al. Spontaneous behaviors drive multidimensional, brain-wide population activity. bioRxiv 306019 (2018). doi: 10.1101/306019 [DOI] [Google Scholar]
- 47.Buzsáki G, Logothetis N & Singer W Scaling Brain Size, Keeping Timing: Evolutionary Preservation of Brain Rhythms. Neuron 80, 751–764 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cohen MR & Maunsell JHR Attention improves performance primarily by reducing interneuronal correlations. Nat. Neurosci. 12, 1594–1600 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamamoto J, Suh J, Takeuchi D & Tonegawa S Successful Execution of Working Memory Linked to Synchronized High-Frequency Gamma Oscillations. Cell 157, 845–857 (2014). [DOI] [PubMed] [Google Scholar]
- 50.Zucca S et al. An inhibitory gate for state transition in cortex. eLife 2017. May 16;6 pii: e26177. doi: 10.7554/eLife.26177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Constantinople CM & Bruno RM Effects and Mechanisms of Wakefulness on Local Cortical Networks. Neuron 69, 1061–1068 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Okun M et al. Population Rate Dynamics and Multineuron Firing Patterns in Sensory Cortex. J. Neurosci. 32, 17108–17119 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xie Y et al. Resolution of High-Frequency Mesoscale Intracortical Maps Using the Genetically Encoded Glutamate Sensor iGluSnFR. J. Neurosci. 36, 1261–1272 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schuett S, Bonhoeffer T & Hübener M Mapping Retinotopic Structure in Mouse Visual Cortex with Optical Imaging. J. Neurosci. 22, 6549–6559 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu J et al. Kilohertz in vivo imaging of neural activity. bioRxiv 543058 (2019). doi: 10.1101/543058 [DOI] [Google Scholar]
Methods-Only References
- 56.Laissue PP, Alghamdi RA, Tomancak P, Reynaud EG & Shroff H Assessing phototoxicity in live fluorescence imaging. Nat. Methods 14, 657–661 (2017). [DOI] [PubMed] [Google Scholar]
- 57.Podgorski K & Ranganathan G Brain heating induced by near-infrared lasers during multiphoton microscopy. J. Neurophysiol. 116, 1012–1023 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brainard DH The Psychophysics Toolbox. Spat. Vis. 10, 433–436 (1997). [PubMed] [Google Scholar]
- 59.Sommer C, Straehle C, Köthe U & Hamprecht FA Ilastik: Interactive learning and segmentation toolkit. in 2011 IEEE International Symposium on Biomedical Imaging: From Nano to Macro 230–233 (2011). doi: 10.1109/ISBI.2011.5872394 [DOI] [Google Scholar]
- 60.Sakoe H & Chiba S Dynamic programming algorithm optimization for spoken word recognition. IEEE Trans. Acoust. Speech Signal Process. 26, 43–49 (1978). [Google Scholar]
- 61.Podgorski K & Haas K Fast non-negative temporal deconvolution for laser scanning microscopy. J. Biophotonics 6, 153–162 (2013). [DOI] [PubMed] [Google Scholar]
- 62.Friedrich J, Zhou P & Paninski L Fast online deconvolution of calcium imaging data. PLOS Comput. Biol. 13, e1005423 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.White R Image restoration using the damped Richardson-Lucy iteration. in Astronomical Data Analysis Software and Systems III 61, 292 (1994). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Optical and mechanical designs and instructions for use are available through the Janelia Open Science portal at www.janelia.org/open-science. Example datasets, demonstrations, and code used for analysis and simulations are available at www.github.com/KasparP/SLAP. All other resources and data are available from the corresponding author upon reasonable request.