Abstract
The transcription factor CREB (cyclic AMP response element (CRE)-binding protein) is implicated in the pathophysiology and treatment of depression. Structural and functional studies in both animals and humans suggest that abnormalities of the hippocampus may play a role in depression. CREB regulates thousands of genes, yet to date, only a handful that mediate depression or antidepressant response have been identified as relevant CREB targets. In order to comprehensively identify genes regulated by CREB in the hippocampus, we employed translating ribosome affinity purification (TRAP) to detect actively translating mRNAs in wild type and CREB-deficient mice. Using CrebloxP/loxP; RosaLSL-GFP-L10a mice, we conducted whole genome sequencing to identify transcripts only in cells that lack CREB, as introduction of Crerecombinase simultaneously deleted CREB and expressed GFP-tagged L10a ribosomes that enabled TRAP. We identified over 200 downregulated genes predominantly associated with inflammation and the immune system, including toll-like receptor 1 (TLR1). To determine if baseline disruption in gene expression in the hippocampus of CREB-deficient mice can modulate behavior, we used unpredictable chronic mild stress (UCMS) to produce a set of behavioral alterations with strong validity for depression.
We found that CREB-deficient mice demonstrated resilience to the physiological effects of UCMS and also showed changes in affective behaviors specifically in the presence of stress. TLR1 expression was increased following UCMS in control but not in CREB-deficient mice. The results suggest that CREB-mediated regulation of immune system and inflammatory factors may provide additional targets for the treatment of depression.
1. Introduction
Alterations in CREB activity and expression levels have been associated with depression-like phenotypes and response to antidepressant treatment (Conti et al., 2002; Nibuya et al., 1996; Thome et al., 2000; Young et al., 1998). CREB regulates expression of genes implicated in depression and antidepressant response, including brain derived neurotrophic factor (BDNF), nuclear receptor 4A transcription factor (NR4A), and other neurotrophic factors (Dias et al., 2003; Gass and Riva, 2007; Nair and Vaidya, 2006; Tardito et al., 2006; Volakakis et al., 2010). Many potential CREB target genes have been identified based on the presence of a CRE element in their promoter. Indeed, the CREB “regulon” has been bioinformatically (Zhang et al., 2005) and biochemically (Impey et al., 2004) mapped, with over 4,000 loci identified within 1kb of a putative CRE site. However, to date, no study has comprehensively identified the specificity of CREB deletion in the hippocampus, a brain region best associated with depression, and evaluated transcriptional profiles. Using a novel strategy to both delete CREB and express a ribosome-specific fusion protein, GFP-L10a (CrebloxP/loxP; RosaLSLGFP-L10a), we employed the translating ribosome affinity purification (TRAP) technique to evaluate baseline changes in gene expression due to CREB deletion. We found predominantly downregulated genes that grouped into functional clusters and networks associated with inflammation and the immune system, including TLR1.
We and others have shown that manipulating CREB levels results in behavioral changes relevant to the development of depression and effectiveness of antidepressant treatments. CREB can have a prodepressive or antidepressant effect, depending upon the species, method of altering CREB levels, region of the brain where CREB is being manipulated, and tests used to evaluate behavioral responses. CREBαΔ mutant mice exhibit antidepressant-like behavior, and both CREBαΔ mice or mice with a specific CREB deletion in the hippocampus results in accelerated response to antidepressant drugs, and increased neurogenesis (Conti et al., 2002; Gundersen et al., 2013; Gur et al., 2007; Mombereau et al., 2010). CREB activity in the hippocampus is upregulated in the presence of stress (Böer et al., 2007), and depressive behaviors induced by inflammatory stress are dependent upon hippocampal CREB (Ni et al., 2019). Viral overexpression of CREB in the hippocampus can produce antidepressant behaviors (Chen et al., 2001), while overexpression of CREB in other brain regions, such as the nucleus accumbens (Pliakas et al. 2001) and amygdala (Wallace et al., 2004) have prodepressive effects. Thus, disrupting CREB homeostasis can result in behavioral changes relevant to depression, yet the role of CREB in a model of depression has not been characterized.
Stress is a causal factor for major depressive disorder (MDD) (Kessler, 1997), although the mechanisms linking the two phenomena have yet to be elucidated. Several paradigms have been developed to model stress-induced depression in rodents, including UCMS (Schmidt and Duman, 2010; Yohn and Blendy, 2017). To evaluate the impact of CREB-dependent gene expression changes on stress-mediated behaviors, we exposed mice with and without hippocampal CREB deletion, to UCMS. The UCMS paradigm has excellent validity as it induces the development of physiological and behavioral effects that are reminiscent of the symptoms used in the DSM-5 to diagnose major depressive disorder (American Psychiatric Association, 2013), including effects on mood, motivation, appetite, and anhedonia. We found that CREB-deficient mice developed resilience to the physiological effects of stress and some affective responses. These differences were associated with the blockade of stress-induced TLR1.
2. Materials and methods
2.1. Animals
Mice containing the Creb1 gene with exon 10/11 flanked by loxP sites (Shin et al., 2014) were crossed with Rosa26LSL-GFP-L10a (Liu et al., 2014) and maintained on a C57BL/6J background. For all behavioral studies, CrebloxP/loxP; Rosa26LSL-GFP-L10a were injected with AAV-Cre virus in the hippocampus to simultaneously delete CREB, and express GFP-L10a in the same cell. Control mice from the same line were injected with AAV-GFP.
All studies were conducted in male mice which were group housed with food and water available ad libitum (except as noted) and were maintained on a 12 h light/dark cycle, lights on at 6:00am, (except as noted during the UCMS paradigm) according to the University of Pennsylvania Animal Care and Use Committee. Mice weighed 20–30 g and were 8–14 weeks old at the time of injection. The four groups tested are referred to as: stress-GFP (N =14); stress-Cre (N =12); no stress-GFP (N =7); and no stress-Cre (N =6); see (Supplementary Table 1) for further details. For molecular studies, AAVCre was injected into CrebloxP/loxP; RosaLSL-GFP-L10a mice for CREB deletion. For convenience, these mice are referred to as CrebloxP/loxP. AAV-Cre was injected into RosaLSL-GFP-L10a mice to serve as control and are referred to as Creb+/+.
2.2. Adeno-associated virus production
The University of Pennsylvania Vector Core generated AAV constructs expressing Cre recombinase [AAV-Cre; AAV2/9.CMV.PI.Cre, titer 2.84X1013 genome copies (gc)/ml], or GFP [AAV-GFP; AAV2/9.CMV.eGFP, titer 1.5X1013 (gc)/ml], as previously described (Brynildsen et al., 2018; Gundersen et al., 2013). Each expression cassette contained AAV2 terminal repeats flanking the cytomegalovirus (CMV) promoter-PI-Cre recombinase or CMV promoter GFP. Vector purification was performed using a CsCl sedimentation method, and quantification of vector genome copies was performed by an RT-PCR method. AAVs were diluted in sterile PBS for microinjections.
2.3. Stereotaxic surgery and intrahippocampal microinjection
Surgery was performed on adult mice (8–14 weeks). Mice were initially anesthetized with 3% isoflurane, secured in a stereotaxic frame (Kopf), and maintained at 1–2% isoflurane for the duration of the procedure. Holes were drilled bilaterally in the skull at the injection sites (four total). Stereotaxic coordinates used for intrahippocampal injections were as follows: anteroposterior −2.1; lateral ±1.4; dorsoventral −2.0 (dorsal hippocampi) and anteroposterior −2.9; lateral ±3.0; dorsoventral −3.8 (ventral hippocampi). A 33-gauge needle attached to a 5μl Hamilton syringe, mounted to the stereotaxic frame and under control of KDS310 Nano Pump (KD Scientific) was used to inject 0.5μl of 3.92X1010 gc/μl AAV at each site. Injections occurred at a rate of 0.1 μl/min, after which the needle was left in place for an additional 3 min. After injections were completed, the skin was sutured, and the animals were treated with 5 mg/kg Meloxicam (s.c) and allowed to recovered for 1 hour on a heating pad before returning to the home cage. Mice remained in the home cage for an additional 6 weeks before the start of behavioral testing. Confirmation of successful injection into the hippocampus and expression of GFP-L10a was conducted in a subset of mice. Mice were transcardially perfused with 30 ml PBS followed by 40 ml 4% paraformaldehyde. Brains were dissected and post fixed overnight before placing in a 30% sucrose solution for three days. Brains were sliced on a cryostat at a 30 μm thickness and GFP expression was imaged using a Leica wide-field epifluorescence microscope.
2.4. Tissue collection and western blot
Mice were cervically dislocated, rapidly decapitated, and brains were removed immediately. The hippocampus was dissected and each hemisphere was flash frozen for storage. Unilateral hippocampal samples were homogenized in 200 μl RIPA buffer (Abcam, cat. no. ab156034, Cambridge, MA) with protease and phosphatase inhibitors (ThermoFisher Scientific, cat. no. 78440, Waltham, MA). Protein concentration was quantified in a BCA assay with BCA reagents (Pierce, cat. no. 23228 and 1859078, Rockford, IL). For blotting, 50 μg of protein was loaded per sample into a 4–15% Mini-PROTEAN TGX precast gel (Biorad, cat. no. 456–1083) for electrophoretic separation and run at 100V for 1 hour. Transfer to 0.45 μm nitrocellulose membrane (GVS Life sciences, cat. no. 1213888, Sanford, ME) was conducted overnight at 4°C at 20V. Membranes were blocked for 1 hour with Odyssey blocking buffer (Licor, cat. no. 927–40000, Lincoln, NE) then incubated at 4°C overnight with rabbit anti-CREB (Cell Signaling, cat. no. 9197S) at 1:500 and goat anti-GAPDH (Abcam cat. no. ab9483–200) at 1:3000 each. Membranes were washed in PBS with 0.1% Tween-20 (Sigma-Aldrich, cat. no. P9416, St. Louis, MO) 5×5min, then incubated for 1h at room temperature with secondary antibodies donkey anti-rabbit 680RD (Licor, cat. no. 926–68073) and donkey anti-goat 800 CW (Licor, cat. no. 926–32214) at 1:15000. Membranes were washed 4×5min in PBS with 0.1% tween, then 1×5min in PBS. Images of fluorescent bands were captured with an Odyssey scanner (Licor) and Odyssey Clx imaging system (Licor). CREB expression was normalized to GAPDH for each sample.
2.5. Behavioral studies
All behavioral evaluations occurred between the hours of 8:00AM and 3:00PM. Treatment conditions were assigned randomly, and animals were tested in counterbalanced order to account for the effect of time of day on behavior. Animals were allowed to acclimate to testing rooms for 1 hour prior to behavioral testing. All experimental procedures were approved by the University of Pennsylvania’s Animal Care and Use Committee and were conducted in compliance with the National Institute of Health’s Guide for the Care and Use of Laboratory Animals (National Research Council, 2010).
2.5.1. UCMS
Mice underwent unpredictable chronic mild stress for 44 days starting 6 weeks post AAV-Cre or AAV-GFP injection. The UCMS paradigm was adapted from previous studies (Schmidt and Duman, 2010; Yohn and Blendy, 2017). Briefly, animals were exposed to three stressors each day, in the morning, afternoon, and overnight, for 44 consecutive days in dedicated procedure rooms. Stressors lasted from 1–3 hours in the morning and afternoon, and 12–14 hours for overnight (Supplementary Table 2). The UCMS protocol contained both physical and social stressors, including, restraint, isolation, disruption of light dark cycle, wet bedding, tilted cage, water restriction, shaking cage, cold, new cage partners, static noise, and strobe light. Throughout the UCMS protocol, stressors remained unpredictable by pairing stressors, such as wet bedding plus static noise. Mice were returned to the animal colony between stressors. Further details on the UCMS protocol can be found in the supplementary methods section.
2.5.2. Body weight
The body weight of each mouse was evaluated the day prior to the UCMS procedure and weekly thereafter. The change from initial body weight was reported.
2.5.3. Nestlet Shredding Test
This test was conducted as described (Angoa-Pérez et al., 2013) on Day 10 of the UCMS procedure. Mice were isolated in cages containing bedding and nestlets of known weight. After 30 min, the nestlet was removed and the amount of shredding that occurred was evaluated by recording change in nestlet weight.
2.5.4. Sucrose Consumption Test
To confirm the UCMS procedure led to anhedonia in each group, mice underwent a sucrose consumption test (SCT) adapted from (Schmidt and Duman, 2010; Yohn and Blendy, 2017) on Day 26 of UCMS. Prior to testing, mice were habituated to a 1% sucrose solution for 48 hours and then gradually water restricted for 4, 14, and 19 hours to prevent neophobia during testing. On the test day, mice are individually housed in a cage containing home bedding for 1 hour to acclimate and then are allowed access to a 1% sucrose solution for 1 hour. Testing was repeated on the following day, except that mice were given access to water instead of sucrose solution in order to verify that hippocampal CREB deletion did not influence the amount of water consumed.
2.5.5. Elevated Zero Maze
Mice were tested on the elevated zero maze (EZM) on Day 29 or 30 of UCMS in order to test for an anxiety-like phenotype as described in previous research (Cleck et al., 2008). The maze consisted of four quadrants: two closed areas with walls that extended 30.5cm above the maze’s surface and two open areas with lips that extended 1.3 cm above the maze’s surface. Each mouse was placed facing the closed arm quadrant and allowed unlimited exploration of all 4 quadrants for 5 minutes under dim lighting (100 lux). Mice were video recorded for the duration of the test. The amount of time spent in the open area was determined by a trained, blinded observer. Three mice did not enter the open area were not included for statistical analysis.
2.5.6. Locomotor Test
Mice underwent locomotor testing on Day 31 or 32 of UCMS as previously described (Hankenson et al., 2011). Individual mouse assessments were made in standard mouse cages with a single layer of bedding, inserted into an adjustable height frame (30 X 24 X 8 cm) to permit infrared motion detection by 2 levels of sensors. The sensors were arranged in an 8-beam array strip with approximately 1.5 cm spacing. Mice were recorded over a 60-minute period where beam break data were collected directly by using specialty Med Associates Software (St Albans VT). The total number of beam breaks is reported as total activity. A malfunction of a locomotor box occurred and no data could be collected for 1 no stress-Cre mouse.
2.5.7. Novelty Suppressed Feeding
The novelty suppressed feeding (NSF) protocol was used to measure the anxiety-like behaviors in mice on Day 36 of UCMS as adapted from (Deacon, 2011). After 24 hours of food deprivation, the latency to consume a highly palatable food (50% sweetened condensed milk solution) was measured. Mice were placed in a 1 L beaker with a small container of milk solution (3 ml). Mice were video recorded for the duration of the test and latency to consumption, which lasted for at least 3 seconds, was recorded.
2.5.8. Core body temperature
Core body temperature was evaluated on Day 37 at 10:00 am immediately following the morning stressor. A digital thermometer with a rectal probe was lubricated with petroleum jelly and inserted 2.0 cm into the rectum and held in place for 10 seconds. Temperature was determined once per mouse.
2.5.9. Forced Swim Test
On Day 43, mice were exposed to a swim stressor. Mice were placed in plastic cylinders (23 cm tall × 14 cm diameter) containing 15 cm of water (22–24°C) for 6 minutes. Time spent passively floating, (immobility) versus active escape behaviors such as swimming and climbing was scored by a blinded observer.
2.6. Statistical Evaluation
Statistical analysis was conducted using the GraphPad Prism software. Two-way ANOVA was used to evaluate the interaction of stress and genotype on SCT, temperature, NSF, and qPCR of TLR1 expression. A two-way repeated measures ANOVA that included time (day) as the repeated, within-subjects factor was used to evaluate changes in weight. All post-hoc analysis were conducted using the Bonferoni of Tukey’s test for multiple comparisons. In tests where only two groups were included, western blotting, TRAP-qPCR, and nestlet shredding, an unpaired two-tailed Student’s t-test was conducted. In some cases, mice were excluded from the analysis if they were determined to be statistical outliers using the two-sided Grubbs’ test (alpha = 0.05). In the Nestlet Shredding Test, 1 mouse was excluded from each group. In the NSF test, 1 no stress-GFP, 1 no stress-Cre, 1 stress-GFP were excluded.
2.7. Actively translating mRNA isolation
RNA was isolated from hippocampal cells expressing GFP-L10a by TRAP (Heiman et al., 2014). Briefly, CrebloxP/loxP; RosaLSL-GFP-L10a mice or RosaLSL-GFP-L10a mice were sacrificed and the hippocampus was isolated, homogenized with lysis buffer, and incubated with magnetic beads that were pre-conjugated to anti-GFP antibodies (clones htz-GFP-19F and htz-GFP-19C8, Memorial Sloan-Kettering Monoclonal Antibody Facility, New York, New York, USA) to affinity purify RNA that was bound by GFP-L10a fusion protein.
2.8. RNA-seq
RNA integrity was measured using the Agilent RNA 6000 Bioanalyzer (Agilent Technologes). cDNA libraries were made from isolated RNA with the TruSeq Stranded Total RNA Gold Library Prep Kit for Illumina (New England BioLabs) according to the manufacturer’s instructions. Library quality was measured with an Agilent High Sensitivity DNA Bioanalyzer, cDNA libraries were purified, and qPCR quantified (Kapa Biosystems). Samples of equimolar libraries were pooled and sequenced on the Illumina HiSeq 2500. Sample preparation and sequencing was conducted at the University of Pennsylvania’s Next Generation Sequencing Core (https://ngsc.med.upenn.edu).
2.9. RNA-seq data analysis
Fastq files of RNA-seq were processed using the RUM algorithm (Grant et al., 2011). Differential gene expression analysis was performed using R software and the package edgeR (Robinson 2010). Differentially expressed genes (DEGs) were identified by setting a fold change cutoff of >1.2 and an FDR of <0.01. Gene ontology was performed using the Database for Annotation, Visualization, and Integrated Discover DAVIDv6.8 (Huang da et al., 2009; Huang et al., 2008). All 205 downregulated genes defined as DEGs were uploaded to DAVID and analyzed using the functional annotation clustering tool. The top 5 annotation clusters were highlighted and sorted according to enrichment score. String database (Szklarczyk et al., 2016) was used to conduct network analysis of downregulated genes and identified immune system networks. Genes were only included in network analysis if the interaction score was 0.4 or higher, and active interaction sources were limited to experiments and databases.
2.10. qpcr
qPCR was conducted on mRNA isolated by TRAP in the hippocampus, or total RNA from a gross hippocampal dissection using the Absolutely RNA Miniprep RNA isolation kit (Agilent Technologies), with RNase free DNase inhibitors. 200 ng of RNA was reverse transcribed to cDNA with SuperScript II Reverse Transcriptase (Invitrogen, Thermo Fisher Scientific) and qRT-PCR was performed with TLR1 (Mm00446095_m1), multiplexed with primer limited GAPDH (Mm99999915_g1). Relative expression levels were normalized to GAPDH and control samples using the 2−ΔΔct method.
3. Results
3.1. Hippocampal injection of adeno-associated virus expressing Cre recombinase in CrebloxP/loxP, RosaLSL-GFP-L10a mice induces GFP-L10a expression and deletion of CREB expression
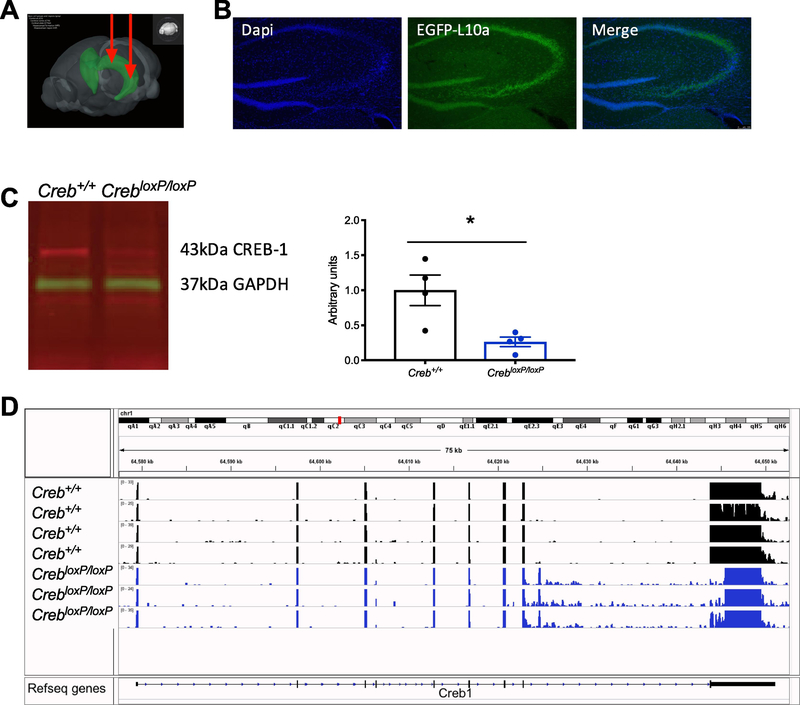
To specifically evaluate the role of CREB in the hippocampus, an AAV virus expressing Cre recombinase was bilaterally injected into the dorsal and ventral hippocampus (Fig. 1a), in a mouse line where deletion of CREB and expression of GFP-L10a was inducible (CrebloxP/loxP; RosaLSL-GFP-L10a). For molecular studies, AAV-Cre was injected into CrebloxP/loxP; RosaLSL-GFP-L10a for CREB deletion, or RosaLSL-GFP-L10a to serve as control mice. GFP-L10a expression was induced in the hippocampus, which served to indicate the placement and spread of virus, and successful recombination (Fig. 1b). As previously described (Gundersen et al., 2013) reduction in CREB protein expression was observed 6 weeks after administration of AAV-Cre as measured by western blotting (Fig. 1c, Student’s t-test P = 0.018, N = 4 Creb+/+, N = 4 CrebloxP/loxP).
Figure 1. Simultaneous deletion of CREB and expression of GFP-L10a.
A, schematic of bilateral hippocampal injections of AAV2/9-Cre expressing virus into the dorsal and ventral hippocampus. Image made from the Allen Institute for Brain Science Atlas (Allen Brain Institue, 2015). B, 6 weeks after injection of AAV-Cre, there is robust expression of GFP-L10a specifically in the hippocampus. C, western blotting of hippocampal lysate 6 weeks after injection of AAV-Cre. CREB1 was normalized to GAPDH, N =4, Student’s t-test P =0.018. D, RNA-sequencing read alignment to the C57BL6 reference genome showed a reduction in transcripts at the floxed region of the AAV-Cre injected mice.
3.2. CREB deletion primarily results in the downregulation of genes
We used TRAP followed by whole genome sequencing to identify cell specific changes in gene expression following CREB deletion. Tissues were harvested 6 weeks after AAV-injection and GFP-tagged polysomes were immunoprecipitated to isolate actively translating mRNAs specifically from CREB-deficient hippocampal cells. Less than 10 pg of RNA was recovered when TRAP was conducted on mice that were not injected with Cre virus, indicating the specificity of TRAP isolation.
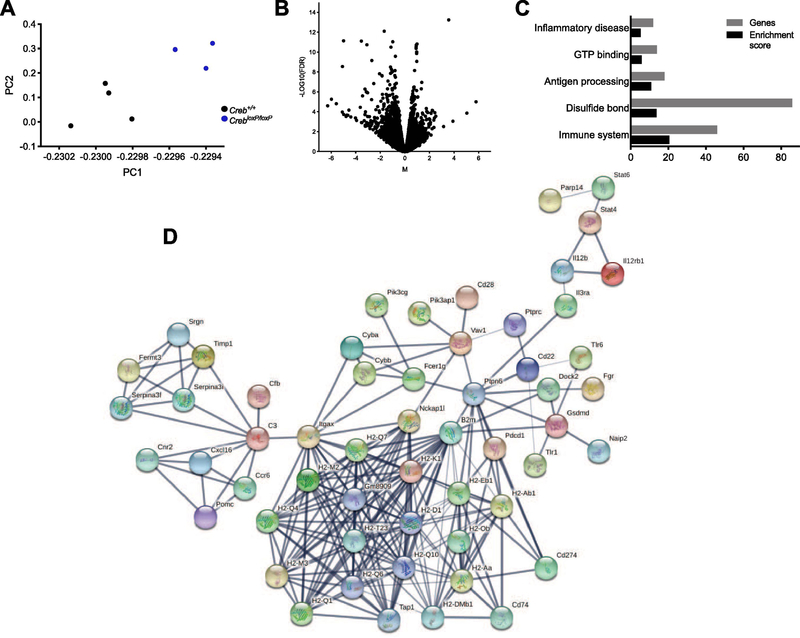
cDNA libraries were generated from seven samples, N = 4 Creb+/+ and N = 3 CrebloxP/loxP, and sequencing yielded an average of 16.8 million uniquely mapped reads. As expected, mice containing CrebloxP/loxP had reduced reads aligning to the loxP flanked region of the Creb gene as visualized by integrative genomics viewer (IGV) software version 2.4.16 (Fig. 1d). Additionally, a nonlinear principle component analysis (PCA) found linear combinations of the Creb+/+ and CrebloxP/loxP samples (Fig 2a). CREB is best characterized as a transcriptional activator, therefore, it is not surprising that deletion of CREB lead to >200 downregulated genes as compared to only 35 upregulated genes (Fig. 2b, FDR < 0.01, Log2FC > 1.2).
Figure 2. TRAP-sequencing was conducted on the hippocampus of CREB deletion and control mice.
A, nonlinear principle component analysis (PCA) of the complete RNA-seq data set found linear combinations of the 4 Creb+/+ and 3 CrebloxP/loxP samples. B, volcano plot from RNA-seq graphically shows the Log10FDR (False Discovery Rate) and the Fold Change (FC), CREB+/+FC-log2 CrebloxP/loxP / Creb+/+ FC. Genes with the largest fold change were downregulated. C, the top 205 downregulated genes were evaluated using DAVIDv6.8 for functional annotation clustering and listed according to enrichment score and the number of genes associated with the cluster. D, network analysis was conducted using STRING database. A network of 56 genes including cytokines, interleukins, and histocompatibility factors was identified.
3.3. CREB deletion decreases expression of inflammation and immune system related genes
Functional annotation was conducted on the 205 downregulated genes using DAVIDv6.8 (Huang da et al., 2009; Huang et al., 2008). Hierarchical clustering of these genes is listed by highest enrichment score. Of the top five clusters, three are associated with inflammation and the immune system (Fig. 2c). These genes were further evaluated for network connectivity using STRING database (Szklarczyk et al., 2016). The downregulated DEGs grouped into a single 56-gene network, largely containing immune system and inflammatory factors including cytokines, interleukins, and histocompatibility factors (Fig. 2d). A complete list of DEGs is listed in (Supplementary Table 3).
3.4. TLR1 actively translating mRNA is downregulated in CREB-deficient mice
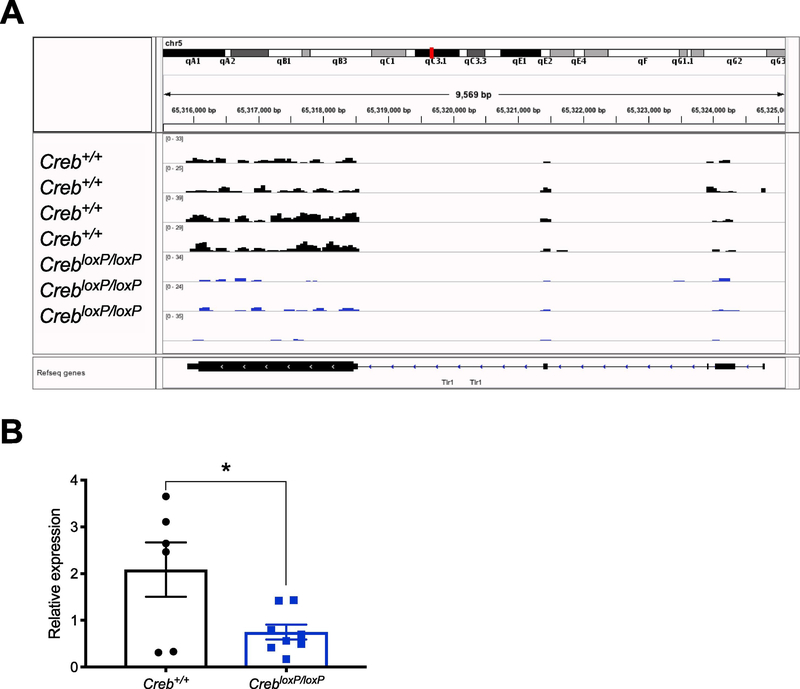
One of the top DEGs that was identified in the immune system-related hierarchical clustering analysis and 56-gene network, was TLR1 (FDR = 3.5×10−5, log2 Fold Change = −2.2). A visual representation of the TRAP-seq read alignment to the reference sequence indicated that there were reduced reads throughout the entire TLR1 gene body in the CrebloxP/loxP group compared to the Creb+/+(Fig. 3a). TLR1 has been well studied in the inflammation field, and while it has been linked to depression (Hung et al., 2016; Pandey et al., 2014) very little is known about the role of TLR1 in stress, depression, and antidepressant response. TRAP-qPCR was conducted in a second independent cohort of mice and confirmed our initial finding that actively translating TLR1 mRNA was reduced in the hippocampus after CREB deletion (Fig. 3b, Student’s t-test P = 0.0273, N = 6 Creb+/+ and N = 8 CrebloxP/loxP).
Figure 3. Hippocampal CREB deficiency decreases TLR1.
A, TRAP-sequencing read alignment to the Reference sequence (Refseq) Tlr1 genome. There were fewer reads aligned in the CrebloxP/loxP samples throughout the gene body. B, TRAP-qPCR confirmed reduced expression of TLR1 after CREB deletion in an independent cohort of mice Student’s t-test P = 0.0273, N = 6 Creb+/+ and N = 8 CrebloxP/loxP. Relative expression was reported using the 2−ΔΔct method where TLR1 CT values were normalized to GAPDH, which served as an internal control in each qPCR reaction.
3.5. UCMS produces anhedonia regardless of CREB deletion
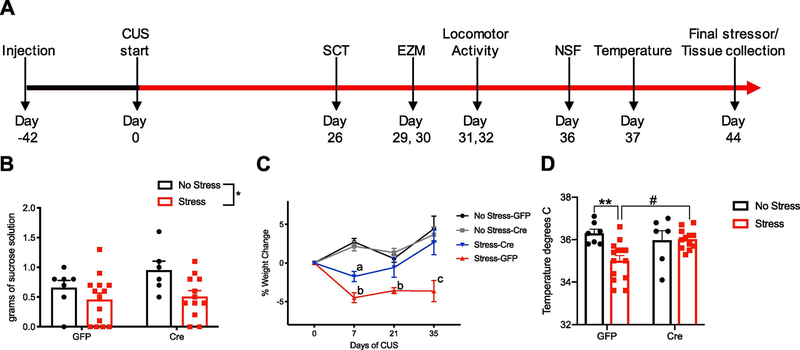
Deletion of CREB has been associated with decreased depression-like behavior in mice (Conti et al., 2002; Gur et al., 2007), and accelerated response to antidepressant treatment (Gundersen et al., 2013). Here we evaluated if deletion of hippocampal CREB results in resilience to the effects of stress. The timeline of the injections, experimentation, and behavioral analysis was conducted as depicted in (Fig. 4a). Mice were injected with either AAV-Cre or AAV-GFP 6 weeks prior to the beginning of UCMS. Stressors were conducted three times daily. Multiple non-invasive behavioral assessments were conducted on the same animals to minimize animal numbers. After 26 days of UCMS, mice were evaluated for anhedonia by measuring sucrose consumption as previously described (Schmidt and Duman, 2010; Yohn and Blendy, 2017). The deletion of CREB in the hippocampus did not alter sucrose consumption as both GFP- and Cre- injected UCMS mice consumed less sucrose than their no stress control counterparts. A two-way ANOVA revealed a significant effect of stress (Fig. 4b, F1,35=6.445, P =0.016).
Figure 4. The physiological effects of UCMS are decreased with hippocampal CREB deletion.
A, experimental timeline for viral injections, UCMS protocol, behavioral assays, and physiological evaluations. B, sucrose consumption during a 1-hour SCT. Reported as amount of sucrose drunk in grams. Two-way ANOVA, F1,35=6.445, P=0.016. C, percent weight change from baseline during the UCMS paradigm was measured on days 7, 21, and 35. Two-way repeated measures ANOVA, F9,105 = 4.99 P<0.0001 (a= P < 0.05 for stress-Cre compared to all other groups, b = P < 0.05 for stress-GFP compared to all other groups, and c = P < 0.0001 for stress-GFP compared to all other groups). D, the core body temperature was decreased in the stress-GFP group compared to the no stress-GFP control group (P = 0.005) and the stress-Cre groups (P = 0.01), but there was no difference between the stress- Cre and no stress-Cre groups. Two-way ANOVA, F1, 35 = 6.546, P = 0.0150.
3.6. CREB-deficient mice are resilient to the physiological effects of UCMS
Chronic stress typically results in weight loss, and after one week we see a significant body weight reduction in stress groups compared to no stress groups regardless of CREB deletion. However, CREB-deficient mice recovered lost weight after 3 weeks and gained weight at levels comparable to the no stress groups. A two-way repeated measures ANOVA revealed an interaction of time and genotype (Fig. 4c, F9,105 = 4.99 P <0.0001). Tukey’s multiple comparisons test revealed that after seven days of UCMS, stress-GFP mice lost more weight compared to all other groups, P < 0.05, and stress-Cre mice lose less weight than stress-GFP mice, P < 0.05. At 21 days P < 0.05, and 35 days, P < 0.0001, the stress-GFP group remained lower in weight, while stress-Cre mice recovered weight lost and even increased from their baseline weight. Chronic stress can also decrease core body temperature. A two-way ANOVA determined a significant interaction of stress and genotype, driven by the stress-GFP group. (Fig. 4d, F1, 35 = 6.546, P = 0.0150). UCMS decreased the core body temperature of the stress-GFP group compared to the no stress-GFP control (P = 0.005), and the stress-Cre groups (P = 0.01), but there was no change in body temperature in the stress-Cre group compared to the no stress-Cre. Therefore, weight loss due to stress could be due to the metabolic or thermal regulatory state of the stress-Cre mice. Together, this indicates that CREB-deficient mice are resilient to physiological effects of stress.
3.7. CREB deletion decreases shredding behavior during UCMS, but does not affect FST or food consumption
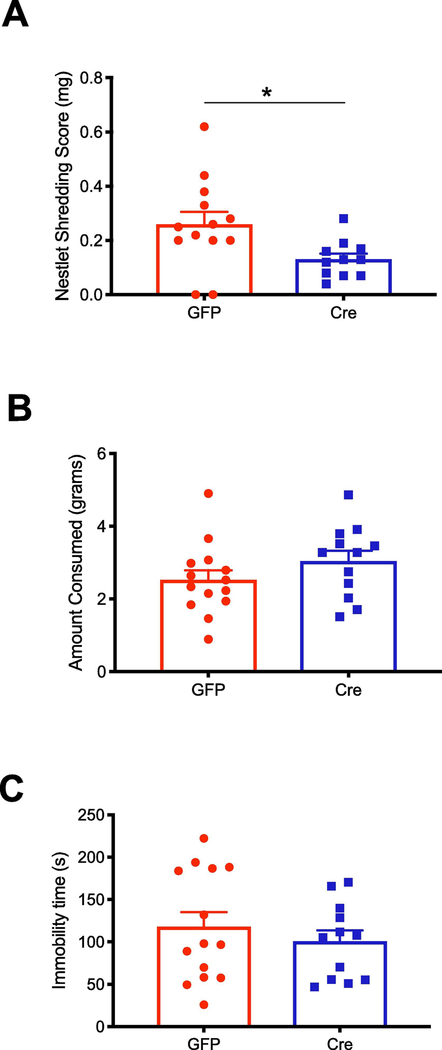
We measured behavioral changes due to CREB deletion during the first isolation and swimming stressors conducted as part of the UCMS protocol. Increased nestlet shredding in mice has been associated with compulsive behaviors (Angoa-Pérez et al., 2013). Here, we observed a decrease in the amount of nestlet shredded in the stress-Cre mice compared to stress-GFP mice during a 30-minute isolation stress (Fig. 5a, Student’s t-test, P = 0.017, N = 13 GFP, N = 11 Cre). There was no change in food consumption during an overnight isolation (Fig. 5b) or immobility time during the swimming stressor (Fig. 5c).
Figure 5. CREB deficiency affects stress specific behavior.
All behaviors were measured only in mice receiving UCMS A, the amount of nestlet shredded for Cre- and GFP- injected mice was quantified by weight (N = 13 GFP, N = 11 Cre), Student’s t-test P =0.041. B, the amount of food consumed during a routine isolation stressor was not changed due to CREB deficiency. C, time spent immobile in a swimming stressor did not change due to CREB deficiency.
3.8. Hippocampal CREB deletion alters the psychological effects of UCMS
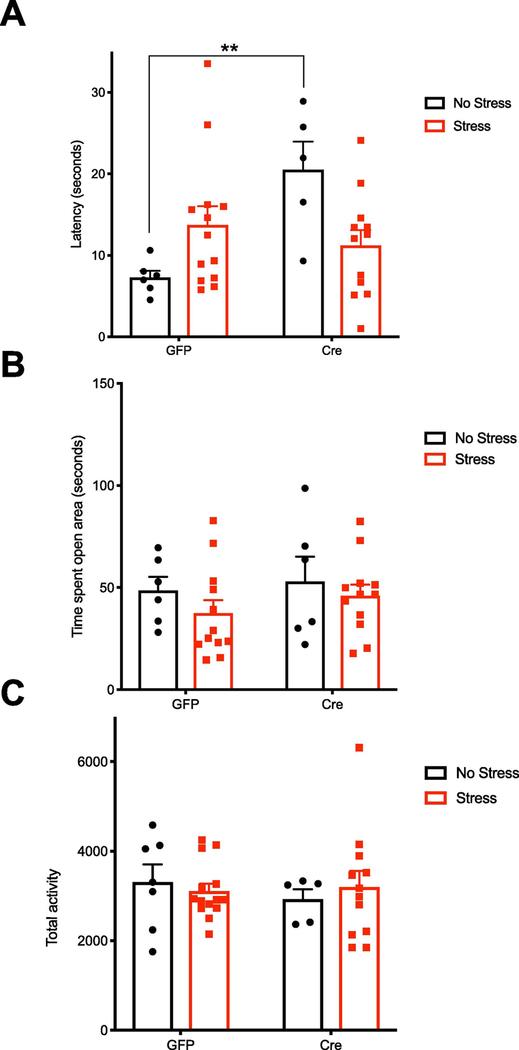
Previously, we reported that deletion of CREB in the hippocampus increased baseline latency in the novelty induced hypophagia (NIH) test (Gundersen et al., 2013). We sought to determine if this phenotype affected behavior after UCMS. We conducted the NSF test, a variation of the NIH, to evaluate hyponeophagia in a novel, anxiogenic environment. In a two-way ANOVA, there was a significant interaction (Fig. 6a, F1, 32 = 9.803, P =0.0037, stress-GFP N =13; stress-Cre N =12; no stress-GFP N =6; and no stress-Cre N =5), driven by the increased latency in no stress-Cre mice (P = 0.007). This recapitulated the previous NIH test findings that CREB deletion increases baseline anxiety-like behavior (Gundersen et al., 2013; Gur et al., 2007). Though not significant, stress increased latency for control mice to consume food, while in contrast, stress decreased latency in the CREB-deficient group. There were no changes in the time spent in the open area in a 5 minute elevated zero maze test (EZM) (Fig. 6b). To rule out overall activity as a factor, we evaluated locomotor activity for 60 minutes and there were no changes between groups (Fig. 6c).
Figure 6. Hippocampal CREB deficiency changes affective behavior.
A, CREB-deficient mice had a baseline increase in the latency to eat palatable food in the NSF test (F1, 32 = 9.803, P =0.0037, stress-GFP N =13; stress-Cre N =12; no stress-GFP N =6; and no stress-Cre N =5). The non-significant trend toward an increase in latency due to stress in the GFP group was not observed in the Cre group. C, CREB deficiency or stress did not change time spent in the open area in the elevated zero maze test, D, or in the overall locomotor activity of mice during 60 minutes of monitoring.
3.9. Hippocampal CREB deletion blunts stress-induced upregulation of TLR1
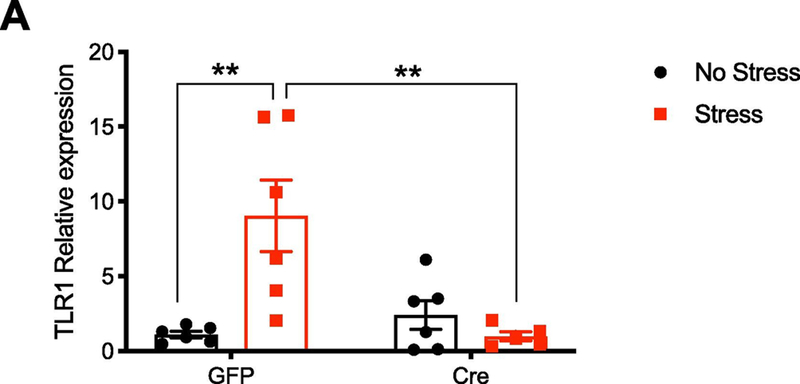
After 44 days of UCMS, mice were sacrificed and the hippocampus was dissected for the no stress groups, and following restraint stress for the stressed groups. In a two-way ANOVA, there was a significant interaction (Fig. 7, F1, 19 = 11.59, P =0.003, N =6 per group) driven by stress (P =0.003) and genotype (P = 0.004). As expected, stress significantly increased TLR1 levels. However, this effect was not observed in CREB-deficient mice.
Figure 7. TLR1 increase due to stress is diminished by CREB deletion.
qPCR was conducted to evaluate changes in TLR1 levels due to stress. Relative expression was reported using the 2−ΔΔct method, relative to GAPDH levels. Stress increased TLR1 levels in GFP-injected mice, and this effect was blunted in the absence of CREB (F1, 19 = 11.59, P =0.003, N =6 per group).
4. Discussion
To determine the specificity of hippocampal CREB deletion in gene expression, we used a mouse in which we induced expression of GFP-L10a and deleted CREB specifically in the same cells. Loss of hippocampal CREB decreased baseline levels of over 200 genes. Many of these downregulated genes were clustered into inflammatory and immune system-related networks, and the downregulation of TLR1 was further recapitulated in an independent cohort of mice. We next evaluated if this baseline decrease in inflammation-related gene expression in the hippocampus promotes resilience to the effects of stress-induced physiological and depressive-like behaviors. We observed that in the UCMS paradigm, CREB-deficient mice lost less weight, were resilient to hypothermia, and were resistant to increased latency in the NSF test. These physiological and behavioral changes paired with the finding that CREB deletion diminished the increased expression of TLR1 due to stress. Together, these data suggest that CREB-dependent regulation of immune system factors should be further evaluated for antidepressant treatment.
4.1. Hippocampal CREB deficiency leads to decreased baseline expression of inflammation and immune response genes
Genes subjected to direct regulation by CREB have been defined bioinformatically, by locating putative binding sites throughout the genome (Zhang et al., 2005), and biochemically, by using chromatin immunoprecipitation to identify the location of bound CREB at CRE regulatory elements (Impey et al., 2004). In order to comprehensively identify genes directly and indirectly regulated by CREB in the hippocampus, we employed the TRAP-seq method to specifically identify actively translating mRNA. This technique ensured that our RNA-seq analysis was limited to cells that effectively incorporated viral expression of Cre recombinase and successfully deleted CREB. CREB is known to initiate transcription, therefore, we anticipated finding primarily downregulated genes, and indeed we found over 200 downregulated DEGs. Functional clusters and networks associated with inflammatory disease and the immune system were identified.
The activity of the immune system has been strongly linked to the pathophysiology of depression and antidepressant medications. MDD patients who are otherwise healthy have increased activation of inflammatory pathways, see for review (Raison et al., 2006). This also occurs in animal models of stress. For example, inescapable shock stressor increases proinflammatory cytokines in the hippocampus (O’Connor et al., 2003). Additionally, there is evidence that peripheral immune activation and neuroinflammation may contribute to the development of depression (Dantzer et al., 2008). This has been explored in animal models where lipopolysaccharide (LPS) is used to induce a state of sickness which results in behaviors reminiscent of depression (Frenois et al., 2007). Many cytokines and inflammatory response factors identified in our network analysis should be considered for future studies. In this work, we focused on a top DEG, TLR1 which was identified in our gene ontology cluster and network analysis, yet has not been well characterized in depression. In an independent cohort of mice, we confirmed TLR1 to be significantly downregulated due to CREB deletion.
TLRs are part of the family of pattern recognition receptors (PPR) of the innate immune system, and are commonly found on the surface of monocytes, macrophages, neutrophils, etc. TLR1 recognizes lipids and lipopeptides and can heterodimerize with TLR2 which together recognizes triacyl lipopetides (Takeuchi et al., 2002), and a wide variety of pathogen-associated molecular patterns (PAMPs) including peptidoglycan, lipopeptides, lipoproteins of gram positive bacteria, mycoplasma lipopeptides, and fungal zymosan (Yu et al., 2010). In a recent review, by Timberlake and Dwivedi, it was proposed that psychological stress can evoke cellular responses similar to an infectious assault such as LPS, through the unfolded protein response pathway, which upregulates TLRs and results in increased inflammation (Timberlake and Dwivedi, 2018). Interestingly, TLRs are increased in the PFC of depressed patients and suicide victims (Pandey et al., 2014) and many of the commonly prescribed antidepressants decrease TLR1 expression (Hung et al., 2016). Activation of the NFkb signaling pathway through TLRs during an immune challenge leads to the release of proinflammatory factors including cytokines and interleukins, but it also provides a link between the neuroendocrine response to stressful psychosocial events and changes at the cellular response (Bierhaus et al., 2003). Here, we show increased TLR1 transcripts after UCMS, however, this induction is blocked when CREB is deleted. CREB is likely a critical component to increasing TLR1 expression following stress, and inhibiting this induction could be associated with resilience or adaptation to stress. The AAV2/9- Cre virus used to express Cre recombinase in the hippocampus can transduce different cell types. Thus, it is not possible to determine if the effects of hippocampal CREB deletion on the hippocampal transcriptome represent effects of immune cells, glial cells, neurons, or a consortium of cells.
4.2. Deletion of CREB in the hippocampus confers resilience to the effects of UCMS
The UCMS paradigm was chosen to model depression because it has strong validity, and the development of depressive related behaviors and physiological changes can be evaluated over time. The UCMS paradigm decreased sucrose consumption, which is typically interpreted as a measurement of anhedonic behavior in mice (Muscat and Willner, 1992). We also observed initial weight loss and an inhibition of weight gain over time, another common measure of the effects of UCMS (Matthews et al., 1995; Pothion et al., 2004). Remarkably, mice with hippocampal CREB deletion lost less weight in the first week of UCMS, and then proceeded to gain weight similarly to the no stress mice by the completion of the study.
Others have shown that chronic inescapable stress can decrease core body temperature (Oka, 2018), and repeated restraint stress or immobilization causes hypothermia (Amar and Sanyal, 1981; Van Eijl et al., 2006). Consistent with these reports, UCMS decreased core body temperature in the stress-GFP group, however CREB-deficient mice were resilient to this effect. Temperature regulation is typically thought to be controlled by the hypothalamus, therefore we were surprised that CREB deletion in the hippocampus affected this physiological stress response. Interestingly, it has been shown that inhibiting the breakdown of serotonin abolishes stress-induced hypothermia (Amar and Sanyal, 1981) indicating a link between the serotonin system and temperature regulation. Given the strong connection between CREB levels and the antidepressant activity of selective serotonin reuptake inhibitors (Conti et al., 2002; Nibuya et al., 1996; Thome et al., 2000; Young et al., 1998), it is possible that CREB deletion in the hippocampus confers resilience to stress-induced hypothermia due to changes in the breakdown of serotonin. Further experiments should evaluate the relevance of this mechanism.
Hippocampal CREB deletion increased baseline latency in the NSF test, which is consistent with previous results in a comparable assay (Gundersen et al., 2013). Generally, increased latency is associated with increased anxiety, but we did not see increased anxiety-like behavior in the EZM, which is also a measure of anxiety. Stress increased latency to consume in the NSF by 50% in the wildtype mice compared to their no stress counterpart, while in contrast, stress decreased latency to consume by 50% in CREB deletion mice compared to their no stress counterpart. These data further suggest that CREB deletion results in resilience to the effects of stress. Of note, mice were exposed to sucrose two weeks prior to the NSF test. This previous exposure to sweet food may have cause an overall reduction in the baseline latency to consume in the NSF test, however we were still able to identify a difference between groups. We also observed decreased nestlet shredding in the first 30 minutes of isolation in the CREB deletion mice, which has been interpreted as decreased compulsive-like behaviors (Angoa-Pérez et al., 2013). However, hippocampal lesions have been reported to slow the initiation of behavior in novel surroundings, independent of locomotor activity, presumably due to changes in emotionality (Deacon et al., 2002). Thus, reduced shredding of an unfamiliar object and increased baseline latency in NSF due to CREB deletion could mean that these behaviors are specific to the activity of the hippocampus and reflect a lack of engagement in a novel environment; a behavior that might be appropriated to depression. Taken together, these data suggest that hippocampal CREB deletion confers resilience and promotes adaptation to the physiological and behavioral effects of stress that are relevant to symptoms of MDD. Overexpression of CREB prior to the stressful learned helplessness paradigm resulted in prodepressive behaviors (Wallace et al., 2004), while we show that deletion of CREB prior to stress promotes resilience to the development of depressive effects. These correlating results indicate that homeostasis of CREB is critical to resilience or adaptation to stress. This could be further explored by conducting stressors, such as social defeat stress, that separate resilient versus susceptible groups. Decreased CREB and TLR1 levels in resilient compared to susceptible mice would provide more evidence of the importance of CREB and TLR1 in resilience to the physiological and psychological effects of stress.
5. Conclusions
This was the first study conducted where CREB deletion in the hippocampus was associated with resilience to the development of phenotypes analogous to symptoms of major depressive disorder. Selective CREB deletion within the hippocampus of adult mice decreased actively translating mRNA from genes related to the immune system and inflammation. In the UCMS paradigm, mice with a hippocampal CREB deletion conferred resilience to several of the physiological and behavioral effects of UCMS. CREB deletion ablated weight loss, decreased core body temperature and inhibited increased latency due to stress in the NSF paradigm. Stress also induced expression of TLR1 in the hippocampus, an effect that was blunted by CREB deletion. This study links the importance of CREB-mediated regulation of immune system factors in the hippocampus with the physiological and behavioral effects of stress. Future work targeting inflammatory factors regulated by CREB could provide a path forward in antidepressant treatment.
Supplementary Material
Highlights.
CREB was specifically deleted in the hippocampus and cells were simultaneously tagged with a GFP-L10a fusion protein.
TRAP determined that genes controlling inflammation were CREB dependent.
Hippocampal CREB deletion conferred resilience to physiological and behavioral effects of stress-induced depressive phenotypes.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- Amar A, Sanyal A, 1981. Immobilization stress in rats: Effect on rectal temperature and possible role of brain monoamines in hypothermia. Psychopharmacology 73, 157–160. [DOI] [PubMed] [Google Scholar]
- Angoa-Pérez M, Kane MJ, Briggs DI, Francescutti DM, Kuhn DM, 2013. Marble burying and nestlet shredding as tests of repetitive, compulsive-like behaviors in mice. Journal of visualized experiments: JoVE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Psychiatric Association, 2013. Diagnostic and statistical manual of mental disorders (DSM-5®). American Psychiatric Pub. [Google Scholar]
- Bierhaus A, Wolf J, Andrassy M, Rohleder N, Humpert PM, Petrov D, Ferstl R, von Eynatten M, Wendt T, Rudofsky G, 2003. A mechanism converting psychosocial stress into mononuclear cell activation. Proceedings of the National Academy of Sciences 100, 1920–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böer U, Alejel T, Beimesche S, Cierny I, Krause D, Knepel W, Flügge G, 2007. CRE/CREB-driven up-regulation of gene expression by chronic social stress in CRE-luciferase transgenic mice: reversal by antidepressant treatment. Plos one 2, e431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brynildsen JK, Lee BG, Perron IJ, Jin S, Kim SF, Blendy JA, 2018. Activation of AMPK by metformin improves withdrawal signs precipitated by nicotine withdrawal. Proceedings of the National Academy of Sciences 115, 4282–4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen AC-H, Shirayama Y, Shin K-H, Neve RL, Duman RS, 2001. Expression of the cAMP response element binding protein (CREB) in hippocampus produces antidepressant effect. Biological psychiatry 49, 753–762. [DOI] [PubMed] [Google Scholar]
- Cleck JN, Ecke LE, Blendy JA, 2008. Endocrine and gene expression changes following forced swim stress exposure during cocaine abstinence in mice. Psychopharmacology 201, 15–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti AC, Cryan JF, Dalvi A, Lucki I, Blendy JA, 2002. cAMP response element-binding protein is essential for the upregulation of brain-derived neurotrophic factor transcription, but not the behavioral or endocrine responses to antidepressant drugs. Journal of Neuroscience 22, 3262–3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Research Council, 2010. Guide for the care and use of laboratory animals. National Academies Press. [Google Scholar]
- Dantzer R, O’Connor JC, Freund GG, Johnson RW, Kelley KW, 2008. From inflammation to sickness and depression: when the immune system subjugates the brain. Nature reviews neuroscience 9, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deacon RM, 2011. Hyponeophagia: a measure of anxiety in the mouse. Journal of visualized experiments: JoVE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deacon RM, Croucher A, Rawlins JNP, 2002. Hippocampal cytotoxic lesion effects on species-typical behaviours in mice. Behavioural brain research 132, 203–213. [DOI] [PubMed] [Google Scholar]
- Dias BG, Banerjee SB, Duman RS, Vaidya VA, 2003. Differential regulation of brain derived neurotrophic factor transcripts by antidepressant treatments in the adult rat brain. Neuropharmacology 45, 553–563. [DOI] [PubMed] [Google Scholar]
- Frenois F, Moreau M, O’Connor J, Lawson M, Micon C, Lestage J, Kelley KW, Dantzer R, Castanon N, 2007. Lipopolysaccharide induces delayed FosB/DeltaFosB immunostaining within the mouse extended amygdala, hippocampus and hypothalamus, that parallel the expression of depressive-like behavior. Psychoneuroendocrinology 32, 516–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gass P, Riva MA, 2007. CREB, neurogenesis and depression. Bioessays 29, 957–961. [DOI] [PubMed] [Google Scholar]
- Grant GR, Farkas MH, Pizarro AD, Lahens NF, Schug J, Brunk BP, Stoeckert CJ, Hogenesch JB, Pierce EA, 2011. Comparative analysis of RNA-Seq alignment algorithms and the RNA-Seq unified mapper (RUM). Bioinformatics 27, 2518–2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundersen BB, Briand LA, Onksen JL, Lelay J, Kaestner KH, Blendy JA, 2013. Increased hippocampal neurogenesis and accelerated response to antidepressants in mice with specific deletion of CREB in the hippocampus: role of cAMP response-element modulator tau. J Neurosci 33, 13673–13685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gur TL, Conti AC, Holden J, Bechtholt AJ, Hill TE, Lucki I, Malberg JE, Blendy JA, 2007. cAMP response element-binding protein deficiency allows for increased neurogenesis and a rapid onset of antidepressant response. Journal of Neuroscience 27, 7860–7868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hankenson FC, Braden-Weiss GC, Blendy JA, 2011. Behavioral and activity assessment of laboratory mice (Mus musculus) after tail biopsy under isoflurane anesthesia. Journal of the American Association for Laboratory Animal Science 50, 686–694. [PMC free article] [PubMed] [Google Scholar]
- Heiman M, Kulicke R, Fenster RJ, Greengard P, Heintz N, 2014. Cell type-specific mRNA purification by translating ribosome affinity purification (TRAP). Nat Protoc 9, 1282–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA, 2009. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA, 2008. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic acids research 37, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung Y-Y, Huang K-W, Kang H-Y, Huang GY-L, Huang T-L, 2016. Antidepressants normalize elevated Toll-like receptor profile in major depressive disorder. Psychopharmacology 233, 1707–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Impey S, McCorkle SR, Cha-Molstad H, Dwyer JM, Yochum GS, Boss JM, McWeeney S, Dunn JJ, Mandel G, Goodman RH, 2004. Defining the CREB regulon: a genome-wide analysis of transcription factor regulatory regions. Cell 119, 1041–1054. [DOI] [PubMed] [Google Scholar]
- Allen Brain Institute, 2015. Allen Brain Atlas API. Allen Institute for Brain Science. Kessler, R.C., 1997. The effects of stressful life events on depression. Annual review of psychology 48, 191–214. [DOI] [PubMed] [Google Scholar]
- Liu J, Krautzberger AM, Sui SH, Hofmann OM, Chen Y, Baetscher M, Grgic I, Kumar S, Humphreys B, Hide WA, 2014. Cell-specific translational profiling in acute idney injury. The Journal of clinical investigation 124, 1242–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews K, Forbes N, Reid IC, 1995. Sucrose consumption as an hedonic measure following chronic unpredictable mild stress. Physiology & behavior 57, 241–248. [DOI] [PubMed] [Google Scholar]
- Mombereau C, Gur TL, Onksen J, Blendy JA, 2010. Differential effects of acute and repeated citalopram in mouse models of anxiety and depression. International Journal of neuropsychopharmacology 13, 321–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muscat R, Willner P, 1992. Suppression of sucrose drinking by chronic mild unpredictable stress: a methodological analysis. Neuroscience & Biobehavioral Reviews 16, 507–517. [DOI] [PubMed] [Google Scholar]
- Nair A, Vaidya V, 2006. Cyclic AMP response element binding protein and brain-derived neurotrophic factor: molecules that modulate our mood? Journal of biosciences 31, 423–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni S, Huang H, He D, Chen H, Wang C, Zhao X, Chen X, Cui W, Zhou W, Zhang J, 2019. AAV-Mediated Overexpression of CRTC1 in the Hippocampal Dentate Gyrus Ameliorates Lipopolysaccharide-Induced Depression-Like Behaviour in Mice. Journal of neurochemistry. [DOI] [PubMed] [Google Scholar]
- Nibuya M, Nestler EJ, Duman RS, 1996. Chronic antidepressant administration increases the expression of cAMP response element binding protein (CREB) in rat hippocampus. Journal of Neuroscience 16, 2365–2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor KA, Johnson JD, Hansen MK, Frank JLW, Maksimova E, Watkins LR, Maier SF, 2003. Peripheral and central proinflammatory cytokine response to a severe acute stressor. Brain research 991, 123–132. [DOI] [PubMed] [Google Scholar]
- Oka T, 2018. Stress-induced hyperthermia and hypothermia Handbook of clinical neurology. Elsevier, pp. 599–621. [DOI] [PubMed] [Google Scholar]
- Pandey GN, Rizavi HS, Ren X, Bhaumik R, Dwivedi Y, 2014. Toll-like receptors in the depressed and suicide brain. Journal of psychiatric research 53, 62–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pothion S, Bizot J-C, Trovero F, Belzung C, 2004. Strain differences in sucrose preference and in the consequences of unpredictable chronic mild stress. Behavioural brain research 155, 135–146. [DOI] [PubMed] [Google Scholar]
- Raison CL, Capuron L, Miller AH, 2006. Cytokines sing the blues: inflammation and the pathogenesis of depression. Trends in immunology 27, 24–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt HD, Duman RS, 2010. Peripheral BDNF produces antidepressant-like effects in cellular and behavioral models. Neuropsychopharmacology 35, 2378–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin S, Le Lay J, Everett LJ, Gupta R, Rafiq K, Kaestner KH, 2014. CREB mediates the insulinotropic and anti-apoptotic effects of GLP-1 signaling in adult mouse β-cells. Molecular metabolism 3, 803–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, 2016. The STRING database in 2017: quality-controlled protein–protein association networks, made broadly accessible. Nucleic acids research, gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi O, Sato S, Horiuchi T, Hoshino K, Takeda K, Dong Z, Modlin RL, Akira S, 2002. Cutting edge: role of Toll-like receptor 1 in mediating immune response to microbial lipoproteins. The Journal of Immunology 169, 10–14. [DOI] [PubMed] [Google Scholar]
- Tardito D, Perez J, Tiraboschi E, Musazzi L, Racagni G, Popoli M, 2006. Signaling pathways regulating gene expression, neuroplasticity, and neurotrophic mechanisms in the action of antidepressants: a critical overview. Pharmacol Rev 58, 115–134. [DOI] [PubMed] [Google Scholar]
- Thome J, Sakai N, Shin K-H, Steffen C, Zhang Y-J, Impey S, Storm D, Duman R, 2000. cAMP response element-mediated gene transcription is upregulated by chronic antidepressant treatment. Journal of Neuroscience 20, 4030–4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timberlake MI, Dwivedi Y, 2018. Linking unfolded protein response to inflammation and depression: potential pathologic and therapeutic implications. Molecular psychiatry, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Eijl S, Van Oorschot R, Olivier B, Nijkamp F, Bloksma N, 2006. Stress and hypothermia in mice in a nose-only cigarette smoke exposure system. Inhalation toxicology 18, 911–918. [DOI] [PubMed] [Google Scholar]
- Volakakis N, Kadkhodaei B, Joodmardi E, Wallis K, Panman L, Silvaggi J, Spiegelman BM, Perlmann T, 2010. NR4A orphan nuclear receptors as mediators of CREB-dependent neuroprotection. Proceedings of the National Academy of Sciences 107, 12317–12322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace TL, Stellitano KE, Neve RL, Duman RS, 2004. Effects of cyclic adenosine monophosphate response element binding protein overexpression in the basolateral amygdala on behavioral models of depression and anxiety. Biological psychiatry 56, 151–160. [DOI] [PubMed] [Google Scholar]
- Yohn NL, Blendy JA, 2017. Adolescent Chronic Unpredictable Stress Exposure Is a Sensitive Window for Long-Term Changes in Adult Behavior in Mice. Neuropsychopharmacology 42, 1670–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young LT, Dowlatshahi D, MacQueen GM, Wang JF, 1998. Increased temporal cortex CREB concentrations and antidepressant treatment in major depression. The Lancet 352, 1754–1755. [DOI] [PubMed] [Google Scholar]
- Yu L, Wang L, Chen S, 2010. Endogenous toll‐like receptor ligands and their biological significance. Journal of cellular and molecular medicine 14, 2592–2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Odom DT, Koo S-H, Conkright MD, Canettieri G, Best J, Chen H, Jenner R, Herbolsheimer E, Jacobsen E, 2005. Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proceedings of the National Academy of Sciences 102, 4459–4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.