Abstract
Influenza viruses antagonize key immune defense mechanisms via the virulence factor NS1 protein. A major NS1 strategy is to block nuclear export of host mRNAs including those encoding immune factors1–3; however, the direct cellular target of NS1 and the mechanism of host mRNA export inhibition are not known. Here, we identify the target of NS1 as the mRNA export receptor NXF1•NXT1, which is the principal receptor to mediate docking and translocation of mRNAs through the nuclear pore complex by interacting with nucleoporins4,5. We determined the crystal structure of NS1 in complex with NXF1•NXT1 at 3.8 Å resolution. The structure reveals that NS1 prevents binding of NXF1•NXT1 to nucleoporins, thereby inhibiting mRNA export through the nuclear pore complex into the cytoplasm for translation. We demonstrate that a mutant influenza virus deficient in binding NXF1•NXT1 does not block host mRNA export and is attenuated. This attenuation is marked by the release of mRNAs encoding immune factors from the nucleus. Together, our study uncovers the molecular basis of a major nuclear function of influenza NS1 protein that causes potent blockage of host gene expression and contributes to inhibition of host immunity.
This year (2018) marks the 100th anniversary of the deadly “Spanish flu” pandemic that caused ~30 million deaths worldwide. However, influenza virus remains a major public health threat killing ~250,000–500,000 people yearly6–8. Influenza A virus is a negative stranded RNA virus with an eight-segmented genome. Transcription and genome replication of influenza A virus take place in the host cell nucleus. Accordingly, influenza A virus has evolved strategies to exploit host nuclear functions. A prominent example is that influenza A infection inhibits export of host mRNAs from the nucleus to the cytoplasm1,2, but the underpinning mechanism is largely unexplored. Importantly, virus virulence depends on inhibition of mRNA export, as this prevents expression of mRNAs encoding antiviral factors1,2.
mRNA nuclear export through the nuclear pore complex (NPC) is the culmination of the nuclear phase of eukaryotic gene expression4,5. To become export competent, mRNA needs to acquire the principal mRNA export receptor, the NXF1•NXT1 heterodimer, whose role is to facilitate mRNA translocation through the NPC. Binding and release of NXF1•NXT1 governs the direction of the mRNA transport, and these events are spatiotemporally regulated through two DEAD-box ATPases, UAP56 in the nucleus and DDX19 at the cytoplasmic face of the NPC. Moreover, NXF1•NXT1 interacts with phenylalanine-glycine (FG) repeats of nucleoporins. Binding of NXF1•NXT1 to FG-nucleoporins is required for NPC docking and translocation of mRNAs through the highly concentrated FG milieu occupying the central NPC channel4,5,9. The virulence factor non-structural protein 1 (NS1) of influenza A virus inhibits host mRNA nuclear export1–3. This effect contributes to NS1-mediated inhibition of host immunity1,2. NS1 suppresses host antiviral response by inhibiting signal transduction and gene expression at multiple levels10. It has been shown to target the host mRNA 3’ processing machinery including CPSF30 and PABII11,12. With respect to inhibition of mRNA nuclear export, we have previously shown NS1 interaction with the mRNA export machinery, including NXF1•NXT1, Rae1, and E1B-AP5. The blockage of mRNA export by NS1 was rescued by ectopic expression of NXF1•NXT11. However, the direct target of NS1 within the mRNA export machinery and the molecular mechanism involved in the mRNA nuclear export blockage have not been resolved.
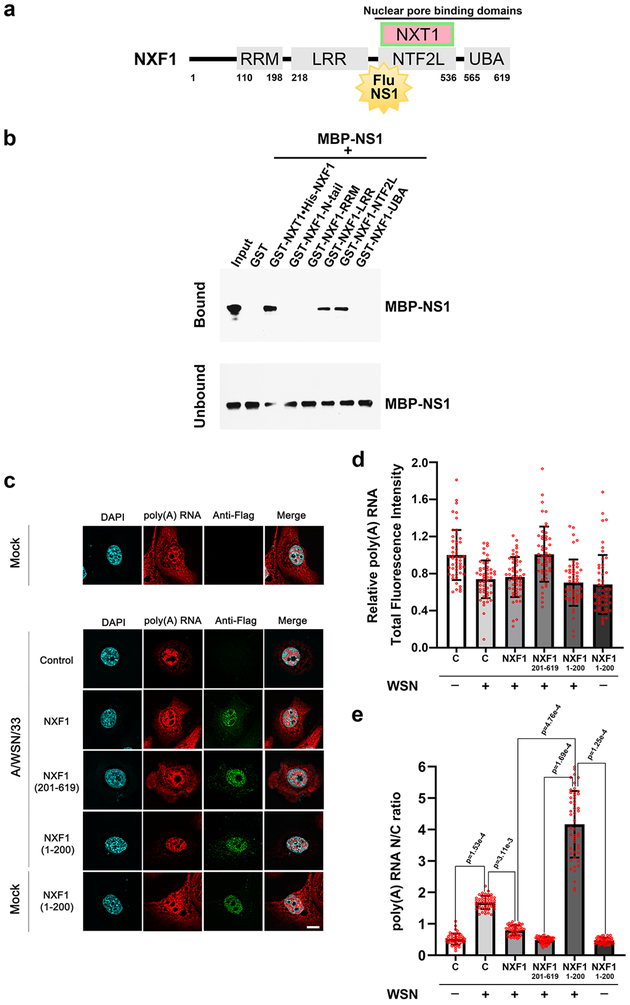
Using recombinant purified proteins in an in vitro binding assay, we show that NS1 from influenza A/Texas/36/91 virus, a human seasonal H1N1 strain, directly interacts with two domains of the mRNA export receptor NXF1: the nucleoporin-binding domain (NTF2L) and the leucine-rich repeat domain (LRR) (Fig. 1a, 1b and Supplementary Fig. 1). Ectopic expression of NXF1 domains containing the NS1 binding regions (NXF1 residues 201–619) or lacking the NS1 binding site (NXF1 residues 1–200) was performed in human bronchial epithelial cells (HBEC) infected with influenza virus to determine their impact on poly(A) RNA distribution. Immunofluorescence microscopy detected NXF1 proteins and RNA-FISH monitored poly(A) RNA and viral M mRNA to select infected cells (Fig. 1c to 1e, and Supplementary Fig. 2). As expected, influenza virus infection retained poly(A) RNA in the nucleus, shown by the increase in nuclear to cytoplasmic ratios of poly(A) RNA without major changes in total poly(A) RNA levels (Fig. 1c to 1e). We then showed that expression of full-length NXF1 or the NS1 binding region of NXF1 (residues 201–619) rescued the mRNA export block induced by influenza virus infection (Fig. 1c to 1e). However, NXF1 lacking the NS1 binding site (residues 1–200) had no effect on this viral-mediated block and did not alter poly(A) RNA distribution when expressed in the absence of infection (Fig. 1c to 1e). Together, these results suggest that interaction of NS1 with overexpressed NXF1 containing the NS1 binding site released endogenous NXF1 to exert its function on nuclear export of poly(A) RNA.
Fig. 1 |. Influenza A virus NS1 protein binds the nucleoporin-binding domain of NXF1 to block host mRNA nuclear export.
a, Schematic representation of NXF1 domains: N-terminal RNA recognition motif (RRM), leucine-rich repeat (LRR) domain, nuclear transport factor 2-like domain (NTF2L) and ubiquitin-associated (UBA) domain. Also shown is the NXT1 protein that forms a heterodimer with NXF1. b, In vitro GST-pull down assays show that purified NS1 protein directly binds to the LRR and NTF2L domains of NXF1. n=3. c-e, HBECs were transfected with vector control, or Flag-NXF1, or Flag-NXF1(1–200), or Flag-NXF1(201–619) and mock infected or infected with A/WSN/1933. Samples were subjected to immunofluorescence with anti-Flag antibody to label expressed proteins and RNA-FISH to detect poly(A) RNA (c). Viral M mRNA was also labeled by RNA-FISH to identify infected cells (Supplementary Fig. 2). Fluorescence intensity of poly(A) RNA was quantified in whole cells (d) or in the nucleus and cytoplasm to determine nuclear to cytoplasmic ratios (e). Scale bar =10 μM. Values (d-e) are mean ± sd of 50 cells that were counted in three independent experiments. The p-values were calculated using the unpaired, two-tailed Student’s t-test.
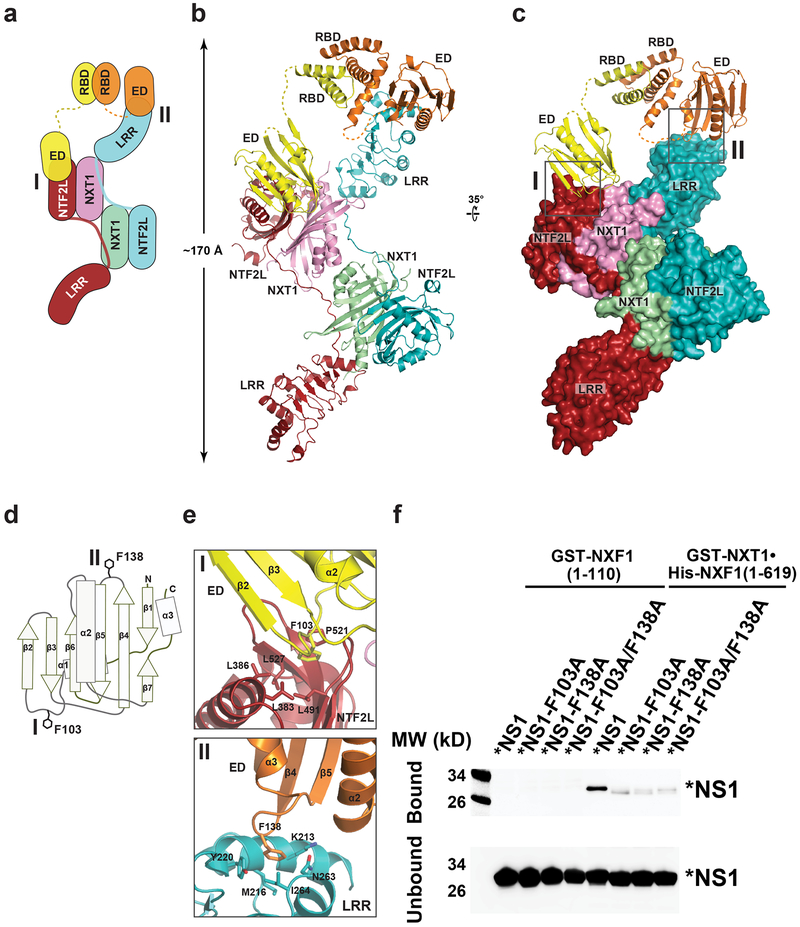
To then investigate the molecular determinants of NS1-NXF1•NXT1 recognition, we solved a crystal structure of their complex. Untagged full length NS1 is prone to aggregation in solution. A mutant NS1 (R38A and K41A), which suppresses NS1 aggregation but lacks RNA-binding activity13,14, was employed for crystallization, as the interaction between NS1 and NXF1•NXT1 does not require RNA (Fig. 1b). We initially obtained poorly diffracting crystals using NS1-R38A/K41A and a truncated NXF1117–619•NXT1 heterodimer that encompasses all annotated domains (NXF1 residues 117–619, NXT1 residues 1–140). In light of the observation that the flexible linker connecting the RNA-binding domain (RBD) and the effector domain (ED) of NS1 causes structural polymorphism13,14, this linker in NS1-R38A/K41A was shortened (residues 80–85 replaced by a proline residue) to obtain diffraction-quality crystals. We refer to the resulting NS1 mutant in the text as *NS1 (Supplementary Fig. 1). Importantly, the *NS1 protein remained competent to bind NXF1•NXT1 (Fig. 2f, lane 6). The *NS1•NXF1117–619•NXT1 complex crystallized in space group P65. The structure was determined by molecular replacement and refined at 3.8 Å resolution with Rwork and Rfree values of 23.6% and 28.3%, respectively (Supplementary Fig. 3 and Supplementary Table 1). The structure reveals a 2:2:2 complex of *NS1•NXF1117–619•NXT1 (Fig. 2a to 2c). Two *NS1 molecules dimerize through their RBD domains. The mutated R38 and K41 residues in *NS1 are located in the RBD domain, which does not contact NXF1117–619•NXT1 directly. The *NS1 dimer, via the ED domains, caps on one end of an elongated platform generated by a domain swapped dimer of NXF1117–619•NXT1. In this domain swapped dimer, two NXT1 molecules are juxtaposed; NXF1-LRR and NXF1-NTF2L of each NXF1 are connected by a linker that traverses along the surface of both NXT1. The RRM and UBA domains of NXF1 were not well ordered, and thus not built in the present structure. Of note, the elongated NXF1117–619•NXT1 platform is symmetrical. While one end of the platform is capped by a NS1 dimer, the other end is not occupied. It is therefore plausible that another NS1 dimer could bind to the unoccupied end in solution.
Fig. 2 |. Structure of a 2:2:2 complex of *NS1•NXF1117–619•NXT1.
a, Schematic diagram of the *NS1•NXF1117–619•NXT1 complex. *NS1 binds to NXF1117–619•NXT1 through two interfaces, I and II. b, c, Overall structure of *NS1•NXF1117–619•NXT1 in two orientations, colored as in (a). Insets illustrate the *NS1-NXF1117–619•NXT1 binding interfaces, and are expanded in (e). d, Schematic diagram of the NS1-ED domain. e, Expanded views of Interface I and II, viewed as in (c). f, In vitro GST-Pull down assays with NXF1•NXT1 and * NS1 or *NS1 with mutations on the NXF1 binding site shown in (e). Purified GST-NXT1•His-NXF1 was incubated with purified *NS1, *NS1-F103A, *NS1-F138A, or *NS1-F103A/F138A. NS1 was detected by western blot and showed diminished interaction upon mutations on the NXF1 binding site. The N-terminal domain of GST-NXF1(1–110) was used as control. n=3.
The NS1 dimer in the present structure is asymmetrically arranged: two NS1-ED domains are oriented differently with respect to the RBD dimer (Fig. 2a to 2c). It makes direct contacts with NXF1117–619•NXT1 via both ED domains. One NS1-ED domain binds to the NXF1-NTF2L domain (interface I), whereas the other NS1-ED domain binds to the NXF1-LRR domain of the neighboring NXF1 (interface II). Importantly, the observed binding sites are fully consistent with our above-mentioned studies for mapping the NS1-NXF1•NXT1 interacting domains (Fig. 1b). The *NS1•NXF1117–619•NXT1 structure shows that NS1-ED employs two key phenylalanine residues, F103 and F138, to engage NXF1117–619•NXT1 (Fig. 2d and 2e). NS1-ED adopts an α-helix βcrescent fold, with the β-strands forming a crescent-like shape around the central α2-helix (Fig. 2d). Both NS1-ED and NXF1117–619•NXT1 do not undergo significant conformational changes upon interaction13,15. At interface I, F103 on the α1-β2 loop of NS1-ED inserts into a hydrophobic pocket on the NXF1-NTF2L surface generated by conserved residues L383, L386, L491, P521, and L527 (Fig. 2e and Supplementary Fig. 4). Whereas at interface II, F138 on the β4-β5 loop of NS1-ED binds to the edge of NXF1-LRR lined by conserved residues K213, M216, Y220, N263, and I264 (Fig. 2e and Supplementary Fig. 4). When F103 and F138 of *NS1 were mutated to alanine, individually or in combination (Supplementary Fig. 5), and subjected to in vitro binding assays with NXF1 lacking the NS1 binding site (NXF1 residues 1–110) or full length NXF1 (1–619), the results show dramatic reduction of the interaction between *NS1 and NXF1•NXT1 (Fig. 2f). These findings corroborate the structural results on the critical role of these NS1 residues in the interaction with NXF1•NXT1. Importantly, F103 and F138 are highly conserved in influenza A viruses NS1 proteins and the majority of strains have at least one of these residues (Supplementary Table 2).
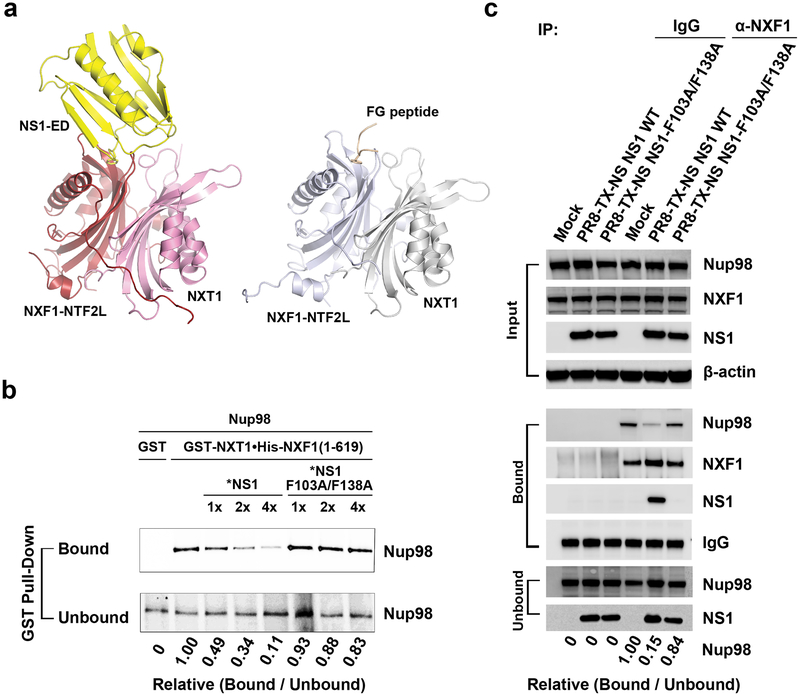
Comparison of the *NS1•NXF1117–619•NXT1 structure and the previously reported NXF1-NTF2L•NXT1 in association with a nucleoporin FG peptide reveals that NS1-ED and FG peptide bind to the same site on NXF1-NTF2L (Fig. 3a)16. This observation predicts that NS1 inhibits binding of FG peptide to NXF1•NXT1. We tested this hypothesis by determining whether NS1 would compete with an FG-nucleoporin, Nup98, for NXF1 binding. In vitro transcribed and translated Nup98 was incubated with purified NXF1•NXT1 and increasing concentrations of purified *NS1 or *NS1-F103A/F138A (Fig. 3b). Indeed, *NS1 protein displaced Nup98 binding to NXF1 while *NS1-F103A/F138A, which did not interact properly with NXF1 (Fig. 2e), was unable to alter the interaction (Fig. 3b). We then generated a recombinant influenza virus carrying the same F103A/F138A mutations on NS1 and infected cells with this mutant virus to compare with the wild-type virus. Cell extracts were subjected to immunoprecipitation with IgG or anti-NXF1 antibodies followed by western blot to detect NS1, NXF1 and Nup98 (Fig. 3c). Again, NS1 mutant on F103A/F138A could not compete as wild-type NS1 for NXF1 interaction in infected cells. Similarly, the interaction of the FG-nucleoporin Nup153 with NXF1 was diminished in the presence of NS1, and their interaction required the C-terminal FG-repeat domain of Nup153 (Supplementary Fig. 6). These findings demonstrate a key role for NS1 F103 and F138 residues on the interaction with NXF1 in vitro and in infected cells, which prevents NXF1 docking to a subset of FG-Nups involved in mRNA nuclear export17–21.
Fig. 3 |. NS1 blocks binding of the FG nucleoporin Nup98 to NXF1•NXT1 in vitro and in influenza infected cells.
a, Structural comparison of the binding of NS1 and nucleoporin FG peptide (PDB ID 1JN5) to the NTF2L domain. b, Competition between NS1 and Nup98 for NXF1•NXT1 binding in vitro. In vitro binding assays were performed with purified recombinant GST-NXT1•His-NXF1 and *NS1 or *NS1-F103A/F138A, and in vitro transcribed and translated Nup98 labeled with 35S-Met. Relative bound proteins were determined by densitometry. n=3. c, Competition between NS1 and Nup98 for NXF1•NXT1 binding in influenza infected cells. Total cell lysates from A549 cells non-infected or infected with PR8-TX-NS NS1 WT or PR8-TX-NS NS1-F103A/F138A for 6h were immunoprecipitated with anti-NXF1 antibody or mouse IgG as control. Western blot was performed to detect the depicted proteins. Relative bound proteins were determined by densitometry. n=3.
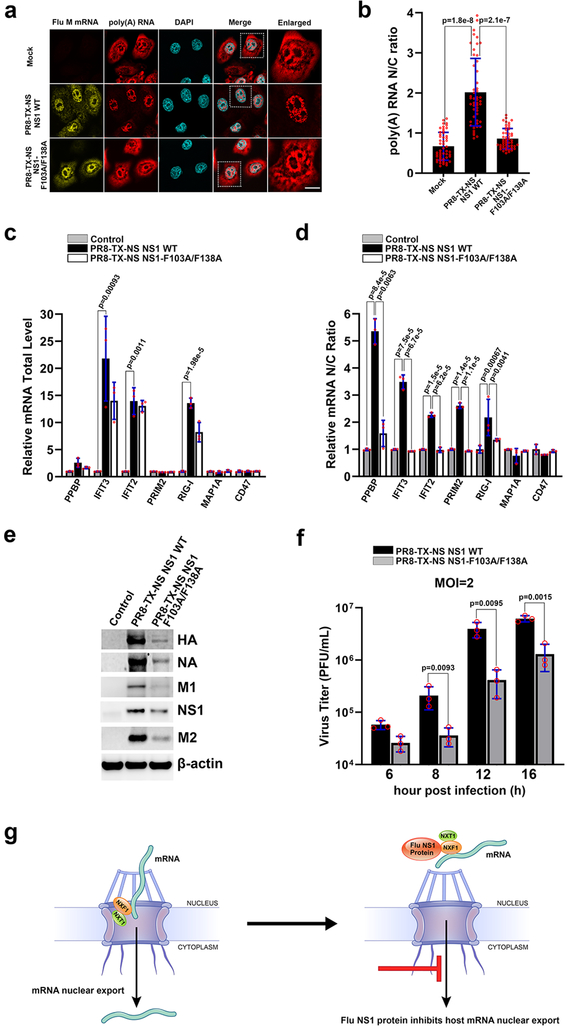
To determine the effect of the virus carrying NS1-F103A/F138A on the intracellular distribution of poly(A) RNA, RNA-FISH was performed in infected HBECs to monitor poly(A) RNA and viral M mRNA simultaneously. Both wild-type and mutant viruses did not significantly alter total poly(A) RNA levels (Fig. 4a and Supplementary Fig. 7a). The virus carrying wild-type NS1 induced nuclear retention of poly(A) RNA, as compared to mock infected cells, while the NS1-F103A/F138A mutant deficient in NXF1 binding was unable to efficiently block poly(A) RNA in the nucleus, and significant amounts of poly(A) RNA were exported (Fig. 4a and 4b). We have also monitored the intracellular distribution of individual host mRNAs in A549 cells, which are less robust on inhibiting nuclear export of bulk poly(A) RNA upon infection as compared to HBEC. In A549 cells infected with the influenza virus A/WSN/33 strain, which is like the Texas strain an H1N1 virus, 1,223 mRNAs are blocked in the nucleus at 6h post-infection as determined by transcriptome analysis of nuclear and cytoplasmic fractions as well as of total cell extracts in comparison to mock-infected cells (Supplementary Table 3). Selected mRNA hits were then validated by qPCR in total cell extracts and nuclear and cytoplasmic fractions of A549 cells infected with recombinant virus expressing wild-type NS1 or NS1-F103A/F138A. As shown in Fig. 4c, the total levels of a subset of mRNAs were up-regulated by viral infection in both wild-type and mutant viruses. However, the mRNAs blocked in the nucleus during infection by the virus expressing wild-type NS1 were exported upon infection with virus encoding NS1-F103A/F138A. Of note, NS1 is known to be a multifunctional protein. Residue F103 also contributes to NS1 interaction with the polyadenylation factor CPSF3022,23. As cellular mRNAs have different dynamics during processing, one can see the differential effects of NS1 in inhibiting biogenesis/processing and/or nuclear export of individual mRNAs at a specific time during infection. For example, at 6h post-infection, the total mRNA levels of selective transcripts such as IFIT2 are not altered by wild-type or mutant viruses (Fig. 4c). However, IFIT2 mRNA is retained in the nucleus by the wild-type virus and is released in the presence of mutant virus (Fig. 4d). These results suggest that the total levels of IFIT2 mRNA have not yet been affected/detected by a defect in poly(A) processing via CPSF30, which would likely target it to degradation, but NS1 has already inhibited IFIT2 mRNA nuclear export at this time point during infection. Among the mRNAs released by the mutant virus are several that encode antiviral factors or are regulated by interferon, including RIG-I24, IFIT2, and IFIT325 (Fig. 4d). As a result of unleashing an antiviral response, viral protein levels (Fig. 4e and Supplementary Fig. 7b) and virus replication (Fig. 4f) are reduced in cells infected with the virus expressing the NS1 mutant deficient on NXF1•NXT1 binding.
Fig. 4 |. Influenza virus mutant on the NXF1 binding site of NS1 allows nuclear export of host mRNAs and is attenuated.
a, HBECs were mock infected or ~ 100% infected with PR8-TX-NS NS1 WT or PR8-TX-NS NS1-F103A/F138A for 30h. Samples were subjected to RNA-FISH to detect viral M mRNA and poly(A) RNA. White square marks the enlarged panel on the right. Scale bar =10 μM. b, Quantification of fluorescence intensity in the nucleus and cytoplasm of samples in (a) followed by determination of nuclear to cytoplasmic ratios. Values are mean ± sd of 50 cells counted for each condition in three independent experiments. c, d, A549 cells were mock infected or infected with PR8-TX-NS NS1 WT or PR8-TX-NS NS1-F103A/F138A at MOI 2 for 6h. RNA was purified from total cell lysates (c) or from nuclear or cytoplasmic fractions (d). mRNAs selected based on transcriptome analysis shown in Supplementary Table 3 were quantified by qPCR. n=3. e, Cells were infected as in (c) except that infection lasted for 8h. Total cell lysates were subjected to western blot to detect the depicted proteins. n=3. f, Cells were infected as in (c) except that infection was performed for the indicated time points. Viral titers were measured in culture supernatants by plaque assay. n=3. The p-values (b, c, d, and f) were calculated using the unpaired, two-tailed Student’s t-test. g, Model for NS1-mediated inhibition of cellular mRNA nuclear export.
In sum, we uncovered the structural and biochemical basis of a key mechanism by which influenza A virus NS1 protein counteracts host antiviral defense. We establish that NS1 targets the principal mRNA export receptor NXF1•NXT1 to block nuclear export of host mRNAs. The crystal structure of NS1•NXF1•NXT1 reveals that NS1 occupies the nucleoporin binding site on NXF1•NXT1 thereby abrogating the ability of NXF1•NXT1 to escort host mRNAs through the NPC (Fig. 4g). Because of the critical importance of mRNA nuclear export in eukaryotic gene expression, this pathway is exploited by a broad spectrum of viruses26. We show that transcripts of interferon or immune regulated genes are among the top transcripts retained in the host nucleus upon influenza A infection. Importantly, a mutant virus that is unable to inhibit NXF1•NXT1 is significantly attenuated, reinforcing the importance of NS1 mediated host mRNA export blockage in virus replication. Our work therefore provides new mechanistic insights into influenza pathogenesis that may help in therapeutic strategies.
Methods
Reagents
50xAdvantage Polymerase mix (Clontech (EMD), 639202); dNTP’s (Clontech (EMD), 639125); BamHI-HF (New England Biolabs (NEB), R3136T); NotI (New England Biolabs (NEB), R0189S); SmaI (New England Biolabs (NEB), R0141); XhoI (New England Biolabs (NEB), R0146S); T4 DNA Ligase (New England Biolabs (NEB), M0202); QIAquick Gel Extraction Kit (Qiagen, 28704); NEB® 5-alpha Competent E. coli (New England Biolabs (NEB), C29871); Rosetta (DE3) Competent Cells (EMD Millipore, 70954); SOC Outgrowth Medium (New England Biolabs (NEB), B9020); QIAprep Spin Miniprep Kit (250) (Qiagen, 27106); Lipofectamine® RNAiMAX Transfection Reagent (Thermo Fisher Scientific, 13778150); RNeasy Plus Mini Kit (Qiagen, 74134); Random Hexamers (50 μM) (Thermo Fisher Scientific, N8080127); Protector RNase Inhibitor (Roche, 03335402001); SuperScript II Reverse Transcriptase (Thermo Fisher Scientific, 18064014); LIGHTCYCLER 480 SYBR GREEN I MASTER (Roche, 04707516001); LightCycler® 480 Multiwell Plate 96, White (Roche, 04729692001); NE-PER™ Nuclear and Cytoplasmic Extraction Reagents (Thermo Fisher Scientific, 78833); Complete EDTA-free protease inhibitor tablets (Sigma-Aldrich, 11873580001); TransIT-X2® Dynamic Delivery System (Mirus Bio, MIR6000); SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific, 34096); L-Glutathione reduced (Sigma-Aldrich, G4251–25G); Ampicillin (Sigma-Aldrich, A-9518); Kanamycin Monosulfate (Gold Biotechnology, K-120–10); IPTG (Gold Biotechnology, I2481C50); Imidazole (Sigma-Aldrich, 56750); PMSF (RPI, P20270); Aprotinin (Santa Cruz Biotechnology, sc-3595); Leupeptin (Santa Cruz Biotechnology, sc295358); Pepstatin A (Thermo Fisher Scientific, BP267110); Glutathione Sepharose 4B (GE Healthcare, 17–0756-01); Amylose Resin (New England Biolabs (NEB), E8021S); Ni-NTA Agarose (Qiagen, 30210); Mono Q 5/50 GL (GE Healthcare, 17-5166-01); HiTrap SP HP (GE Healthcare, 17-1151-01); Superdex 200 HR 10/30 (GE Healthcare, 17-1088-01); EasyTag EXPRESS35S Protein Labeling Mix, [35S]-, 7mCi (PerkinElmer, NEG772007MC); T7 RiboMAX™ Express Large Scale RNA Production System (Promega, P1320); Protein G Sepharose® 4 Fast Flow (GE Healthcare, 17-0618-01); Hoechst 33258 (Molecular Probes/Life Technologies); M mRNA probes (Biosearch Technologies)27; ProLong Gold antifade reagent (Life Technologies, P36930).
Antibodies
Influenza A virus NS1 antibody, a gift from J.A. Richt (National Animal Disease Center, Iowa)28 was used at a 1:10000 dilution. β-actin monoclonal antibody (Sigma-Aldrich, clone AC-74, A5528) was used at a 1:5000 dilution. His tag monoclonal antibody (TAKARA & Clontech, 631210) was used at a 1:5000 dilution. ANTI-FLAG® M2 monoclonal antibody (Sigma-Aldrich, F1804) was used at a 1:1000 dilution. Horseradish peroxidase (Hrp)-conjugated secondary antibodies included donkey anti-rabbit and sheep anti-mouse (GE Healthcare, NA934V and NA931V, respectively). Polyclonal rabbit anti-Nup98 antibody29 was used at a 1:1000 dilution. Mouse anti-nuclear pore complex proteins antibody MAB414 (Abcam, ab24609) was used at a 1:1000 dilution. Mouse anti-NXF1 antibody (Sigma-Aldrich, T1076–200UL) was used at a 1:2000 dilution. Mouse anti-M2 antibody (Thermo Fisher Scientific, MA1–082) was used at a 1:1000 dilution. Rabbit anti-HA antibody (Genetex, GTX127357) was used at a 1:1000 dilution. Rabbit anti-NA antibody (Genetex, GTX125974) was used at a 1:1000 dilution. Goat anti-influenza A virions antibody (Meridian Life Science, B65141G) was used at a 1:1000 dilution. Mouse IgG (Thermo Fisher Scientific, 31903) was diluted to 0.4 mg/ml.
Cell culture
Human lung adenocarcinoma epithelial cells (A549) were cultured in DMEM media supplemented with 10% heat inactivated fetal bovine serum (Sigma-Aldrich) and penicillin-streptomycin (Gibco). Human bronchial epithelial cells (HBEC) were cultured in Keratinocyte-SFM supplied with human recombinant epidermal growth factor, bovine pituitary extract (Invitrogen), and penicillin-streptomycin. All cells were maintained at 37°C with 5% CO2.
Viruses
Influenza A virus A/WSN/1933 (WSN) was propagated in MDCK cells and titered as previously described30. Influenza viruses expressing either NS1 wild type or mutant (F103A/F138A) proteins from A/Texas/36/1991 were rescued on an A/Puerto Rico/8/1934 (PR8) backbone and subsequently propagated in MDCK cells after plaque purification. Viral titers were determined by plaque assay in the presence of tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK)-trypsin. WT or mutant NS1 sequences were synthesized through GeneArt Strings DNA fragments (Invitrogen) and cloned into SAPI sites into the pDZ vector, respectively. Recombinant Influenza viruses, PR8-TX-NS NS1 WT and PR8-TX-NS NS1-F103A/F138A, were generated using plasmid-based reverse genetics as previously described31. All virus work was performed in strict accordance with CDC guidelines for biosafety level 2.
Plaque assay
A549 cells were infected with PR8-TX-NS NS1 WT or PR8-TX-NS NS1-F103A/F138A at MOI 2 for 6 h, 8 h, 12 h, and 16 h. The culture supernatants were collected and titer was determined by plaque assay, as previously described30.
Plasmids
Full length NS1 (UniProtKB accession number Q9WPI6, residues 1–230) from A/Texas/36/91 was cloned into Not I and BamH I sites of the pMALTEV vector (pMAL with TEV cleavage site) with MBP fusion at the amino-terminus. A mutant NS1 (UniProtKB accession number I7CAR2, residues 1–230, R38A/K41A, and residues 80–85 replaced by a proline residue), termed *NS1, was cloned into BamHI and NotI sites of a pGEX-4T-1 vector modified to contain a TEV cleavable N-terminal GST tag. Plasmids GST-*NS1-F103A, GST-*NS1-F138A, and GST-*NS1-F103A/F138A were generated from the GST-*NS1 template above, using QuikChange II Site-Directed Mutagenesis Kit. Human NXF1 (UniProtKB accession number Q9UBU9, residues 117–619) was cloned into BamHI and NotI sites of the same modified pGEX-4T-1 vector described above. Full length human NXT1 (UniProtKB accession number Q9UKK6, residues 1–140) was cloned into NdeI and EcoRI sites of the pBB75 vector for co-expression with NXF1 (residues 117–619). Full length human NXF1 (residues 1–619) was cloned into BamH I and Xhol I sites of the pET32a vector to generate His-NXF1. Full length human NXT1 was cloned into BamH I and Xhol I sites of pGEX-6P-1 vector to generate GST-NXT1. GST-NXF1-N-Tail (residues 1–109), GST-NXF1-RRM (residues 110–200), GST-NXF1-LRR (residues 201–371), GST-NXF1-NTF2L (residues 372–564), GST-NXF1-UBA (residues 565–619) were cloned into pGEX-4T-2. Full length human NXF1 (residues 1–619) and its mutants NXF1 (residues 1–200), NXF1 (residues 201–619) were cloned into the Xho I and Smal I sites of the pCI-neo-3xFlag vector with the 3xFlag tag at the amino-terminus, generating plasmids Flag-NXF1, Flag-NXF1(1–200), and Flag-NXF1(201–619). Plasmid encoding Nup98 was generated as described32. Full length human Nup153 (residues 1–1475) and its mutant Nup153 (residues 1–879) were cloned into the Xho I and Smal I sites of the pCI-neo-3xFlag vector with the 3xFlag tag at the amino-terminus to generate plasmids encoding Flag-Nup153 and Flag-Nup153(1–879).
Protein expression and purification
GST-*NS1 and GST-NXF1117–619•NXT1 were expressed in E. coli Rosetta cells. Protein expression was induced at an OD600 of 0.8 with 0.5 mM IPTG at 20°C and cells were incubated for 16 h. Cells were harvested and lysed with a cell disruptor (Avestin) in a wash buffer (50 mM Tris, pH 8.0, 500 mM NaCl, 0.5 mM TCEP) supplemented with a protease inhibitor mix containing 1 mM PMSF, 5 mg/L aprotinin, 1 mg/L pepstatin, and 1 mg/L leupeptin. Proteins were purified using a Glutathione Sepharose column, followed by overnight digestion with TEV protease at 4°C. *NS1 was applied to a mono Q column and eluted using a 100–300 mM NaCl gradient in a buffer containing 10 mM Tris, pH 8.0. Pooled NS1 fractions were loaded onto a Glutathione Sepharose column to remove residual GST that was eluted along with *NS1 on mono Q, recovering *NS1 in the flow through. NXF1117–619•NXT1 was applied to a HiTrap SP column and eluted using a 200–400 mM NaCl gradient in a buffer containing 10 mM HEPES, pH 7.0. *NS1 and NXF1117–619•NXT1 were further purified using a Superdex 200 column equilibrated with 10 mM HEPES, pH 7.0, 150 mM NaCl, and 0.5 mM TCEP. Purified proteins were concentrated and stored at −80°C. Purification of GST-*NS1-F103A, GST-*NS1-F138A, and GST-*NS1-F103A/F138A followed the same protocol as GST-*NS1. GST-NXF1-N-Tail, GST-NXF1-RRM, GST-NXF1-LRR, GST-NXF1-NTF2L, GST-NXF1-UBA, His-NXF1, GST-NXT1, MBP, and MBP-NS1 were expressed in BL21(DE3). Protein expression was induced by 0.5 mM IPTG at 16°C overnight. Bacteria expressing His-NXF1 and GST-NXT1 were harvested together, and were first purified by Ni2+ ion affinity chromatography to generate the GST-NXT1•His-NXF1 complex. GST tagged NXF1 domains were first purified by glutathione affinity chromatography. MBP and MBP-NS1 were first purified by Amylose resin. All the proteins were further purified by gel filtration in a buffer containing 20 mM Tris, 150 mM NaCl, 1 mM EDTA, 1 mM DTT, 1x complete protease inhibitor cocktail at pH 7.5.
Crystallization and structure determination
Crystals of *NS1•NXF1117–619•NXT1 were obtained at 20°C by vapor diffusion in sitting drops using 1 μL protein mixture (NXF1117–619•NXT1 at 4 mg/mL, *NS1 at 8 mg/mL) and 1 μL reservoir solution consisting of 50 mM Sodium Citrate (pH 5.6), 6.5% PEG 3350, 0.1 M NaCl, and 0.1 M KCl. The crystals were transferred in three steps of increasing glycerol concentration to a cryoprotectant solution containing 50 mM Sodium Citrate (pH 5.6), 6.5% PEG 3350, 0.1 M NaCl, 0.1 M KCl, and 22% glycerol. X-ray diffraction data were collected at the 21-ID-F Beamline at the Advanced Photon Source, Argonne National Laboratory. The crystals exhibited a long unit cell axis of ~ 950 Å, which caused severe overlapping of diffraction spots. To resolve this issue, we adjusted the kappa axis to position the crystals in an orientation so that the long axis was approximately parallel to the rotation axis. X-ray intensities were processed using the HKL2000 package33. The best diffracting crystal yielded a 3.8 Å resolution dataset in space group P65.
The asymmetric unit contains two copies of a 2:2:2 complex of *NS1•NXF1117–619•NXT1. Initial electron density map was obtained by molecular replacement using the coordinates of NXF1•NXT1 (PDB ID 4WYK for the NXF1-NTF2L domain and NXT1; 3RW6 for the NXF1-LRR domain) and NS1 (PDB ID 4OPA for NS1-RBD and NS1-ED domains) as search models in Phaser34. The structure was built with Coot35 and refined with phenix.refine36. NCS restraints, secondary structure restraints, Ramachandran restraints, and TLS (with each of the NS1-RBD, NS1-ED, NXF1-LRR, NXF1-NTF2L, and NXT1 domains assigned to one group) were applied during the refinement. The *NS1•NXF1117–619•NXT1 complex consists of NXF1 (residues 205–423 and 430–549), NXT1 (residues 3–140), and NS1 (residues 2–72 and 88–202 for one molecule; residues 8–45, 88–136, and 142–202 for the other molecule). In the *NS1•NXF1117–619•NXT1 structure, NS1 molecules exhibit higher B factors than NXF1 and NXT1 molecules. In particular, the NS1-RBD dimer appears to be largely positioned by crystal packing. It is notable that a helix (residues 46–72) belonging to the yellow RBD (Fig. 2b) is not observed in the structure. Potential steric clashes between this helix and a symmetry-related molecule due to crystal packing likely caused deviation of this helix from the expected position. Details of the data collection and refinement statistics are in Supplementary Table 1. Molecular graphics were rendered using PyMOL (Schrödinger, http://www.pymol.org/).
GST pull-down assays
GST or GST-NXT1•His-NXF1, or GST tagged NXF1 domains were incubated with MBP or MBP-NS1 and GST-NXF1(1–110) or GST-NXT1•His-NXF1 were incubated with *NS1 or its mutants (*NS1-F103A, *NS1-F138A, *NS1-F103A/F138A) in the binding buffer (20 mM Tris, 150 mM NaCl, 1 mM DTT, 1 mM EDTA, pH 7.5) at room temperature for 30 min. 1 μM of each protein was loaded into the binding assay. Beads were pelleted by centrifugation at 5,000 rpm for 5 min and washed 5x with 1 ml of binding buffer. Proteins remaining on the resin were extracted by sample buffer, resolved in SDS-PAGE, and then detected by western blot using anti-NS1 antibody.
In vitro binding assays with Nup98 and Nup153
Nup98 protein (UniProtKB accession number P52948) was generated by in vitro transcription and translation using a coupled reticulocyte lysate transcription/translation system, in the presence of [35S]methionine, according to manufacturer’s instructions. Binding reactions between Nup98 and NXF1•NXT1 were carried out using 25μl of in vitro transcribed and translated proteins and 2 μM GST-NXT1•His-NXF1 recombinant protein. Increasing amounts (2 μM, 4 μM, 8 μM) of *NS1 or *NS1-F103A/F138A were added to the binding assay. Bound and unbound fractions were subjected to SDS-PAGE followed by autoradiography.
Flag-Nup153 (UniProtKB accession number P49790) or Flag-Nup153(1–879) proteins were expressed in the rabbit reticulocyte lysate as in the case Nup98 except that they were detected by western blot using anti-Flag antibody. Binding reactions between Nup153 or Nup153(1–879) and NXF1•NXT1 were carried out using 25μl of in vitro transcribed and translated proteins and 2 μM GST-NXT1•His-NXF1 recombinant proteins. Increasing amounts (2 μM, 8 μM) of *NS1 were added to the binding assay. Bound and unbound fractions were subjected to SDS-PAGE followed by western blot using anti-Flag antibody.
Immunoprecipitation and immunoblot
A549 cells grown in 10-cm plates were non-infected or infected with PR8-TX-NS NS1 WT virus or PR8-TX-NS NS1-F103A/F138A virus for 6h. Cells were lysed for 30 min at 4°C in the following buffer: 50 mM Tris, pH 7.5, 150 mM NaCl, 1% IGEPAL CA-630, 0.1 mM Na3VO4, 1 mM NaF, 1 mM DTT, 1 mM EDTA, 1 mM PMSF, 1x complete protease inhibitor cocktail, and 10% glycerol. Cell lysates were centrifuged at 13,000xg for 10 min to remove cellular debris. The supernatant was applied to protein G beads bound to either mouse IgG or mouse anti-NXF1 antibody and incubated for 4h at 4°C. After 5 wash with lysis buffer, the proteins were eluted with 2x sample buffer and subjected to 8% SDS-PAGE.
Cell fractionation
A549 cells were infected with PR8-TX-NS NS1 WT virus or PR8-TX-NS NS1-F103A/F138A virus for 6h. Cells were harvested by trypsinization and collected in 15-ml conical tubes on ice, washed three times with cold PBS, and transferred to microfuge tubes. Cell fractionation was performed using the NE-PER™ Nuclear and Cytoplasmic Extraction Reagents according to the manufacturer’s instructions.
Transcriptome analysis
A549 cells were infected with A/WSN/1933 at an MOI of 5 for 6h. RNA of total cell, or nuclear fraction, or cytoplasmic fraction was prepared using RNeasy Plus mini Kit. Purified RNA was quantified by Nanodrop (Thermo Fisher). The RNA samples were subjected to transcriptome analysis using Illumina Human BeadChip 22k according to manufacturer’s instructions.
qRT-PCR
1 μg of total RNA was used to generate cDNA by reverse transcription using SuperScript II reverse transcriptase. cDNA was diluted at a ratio of 1:5. cDNA samples were mixed with specific primers and Roche 480 SYBR Green I Master real-time PCR reagents, and were subjected to quantitative real-time PCR (qPCR) using the Roche LightCycler 480 system. The program used for Real-time PCR is the same as we previously reported2. Relative RNA level was calculated by ddCt method compared to β-actin control. Primer Sequences used in this study are listed as following (5’−3’):
β-actin (NCBI accession number NM_001101.5, Forward: CCGCGAGAAGATGACCCAGAT, Reverse: CGTTGGCACAGCCTGGATAGCAACG); PPBP (NCBI accession number NM_002704.3, Forward: TCCTCCACCAAAGGACAAAC, Reverse: CCATCCTTCAGTGTGGCTATC); IFIT3 (NCBI accession number NM_001549.6, Forward: TCAGAAGTCTAGTCACTTGGGG, Reverse: ACACCTTCGCCCTTTCATTTC);
IFIT2 (NCBI accession number NM_001547.5, Forward: GACACGGTTAAAGTGTGGAGG, Reverse: TCCAGACGGTAGCTTGCTATT); PRIM2 (NCBI accession number NM_000947.5, Forward: CGGCTTGCTTATTGCCAGTCT, Reverse: CAATCTCCTGTTCTCGAAGAGTC);
RIG-I (NCBI accession number NM_014314.4, Forward: ACGGGTGTATGCGCTTCAC, Reverse: TTGCGGTCATCGAACAGGG); MAP1A (NCBI accession number NM_002373.6, Forward: GCTGAGTTCTCCGAGTATGTCT, Reverse: TGAGTAGCACCGAGTCAATGC); CD47 (NCBI accession number NM_001777.3, Forward: AGAAGGTGAAACGATCATCGAG, Reverse: CTCATCCATACCACCGGATCT)
Western blot
A549 cells were mock infected or infected with PR8-TX-NS NS1 WT, or PR8-TX-NS NS1-F103A/F138A for 8h at MOI=2, lysed in 2x sample buffer, and subjected to 10% SDS-PAGE. Western blot was performed with the following primary antibodies: rabbit anti-HA antibody, rabbit anti-NA antibody, goat anti-M1 antibody, rabbit anti-NS1 antibody, and mouse anti-M2 antibody. Proteins were visualized in the LI-COA system and quantified using imageJ64. The UniProtKB accession numbers for HA, NA, M1, and M2 are B4UPA6, G1UK16, P03485, and Q84028, respectively.
RNA-FISH and immunofluorescence microscopy
In Figures 1c and 1d, HBEC were transfected with pcI-neo-3xFlag vector, or Flag-NXF1, or Flag-NXF1(1–200), or Flag-NXF1(201–619) for 48h, and then ~20% of cells were infected with WSN for 30h. The samples were then subjected to RNA-FISH and immunofluorescence microscopy, using anti-Flag antibody, as we previously described27. In Figures 4a and 4b, ~100% of HBEC cells were infected with PR8-TX-NS NS1 WT or PR8-TX-NS NS1-F103A/F138A for 30h. Cells were then subjected to RNA-FISH as reported27.
Circular Dichroism (CD)
CD spectra were collected on 12 μM of recombinant NS1 proteins (*NS1, *NS1-F103A, *NS1-F138A, and *NS1-F103A/F138A), in CD buffer (20 mM Sodium Phosphate pH 8.0, 50 mM NaF, 0.5 mM TCEP). The spectra were collected in the far-UV range (250–190nm) on a Jasco-815 CD spectrometer in 1 mm path-length cuvette at a scanning rate of 50 nm/min in 0.1 nm data intervals. The final CD spectrum for each sample were obtained by averaging six repeat scans. The resulting spectra is plotted as molar ellipticity against wavelength.
Statistical analysis
Statistical analyses were performed using the two-sample, two-tailed, t-test assuming equal variance. For statistical analysis of the M mRNA imaging study, a minimum of 50 cells were used for analysis in each condition. For all imaging studies, a one-sample Kolmogorov–Smirnov test was conducted. A normal distribution can be assumed for all populations (P > 0.05).
Data availability
The coordinates of the NS1•NXF1•NXT1 structure have been deposited in the Protein Data Bank under accession number 6E5U. The data for the transcriptome analysis has been deposited in GEO under accession number GSE129318. All other data that support the findings of this study are available from the corresponding authors upon request.
Supplementary Material
Acknowledgments
We thank Zdzislaw Wawrzak at Advanced Photon Source (APS) beamline 21-ID-F for assistance with X-ray data collection; Phil Jeffrey for suggestions for data processing; and Yuh Min Chook for reagents and assistance on protein purification. Funding was provided by NIH R01 GM113874 to B.M.A.F.; R01 AI125524 to B.M.A.F. and A.G.-S. This work is also partially funded by CRIP (Center for Research on Influenza Pathogenesis), an NIAID funded Center of Excellence for Influenza Research and Surveillance (CEIRS, contract # HHSN272201400008C) and by NIAID grant U19AI135972.
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.Satterly N et al. Influenza virus targets the mRNA export machinery and the nuclear pore complex. Proc Natl Acad Sci U S A 104, 1853–1858, (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang L et al. Inhibition of Pyrimidine Synthesis Reverses Viral Virulence Factor-Mediated Block of mRNA Nuclear Export J. Cell Biol. 196, 315–326, (2012. ). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qiu Y & Krug RM The influenza virus NS1 protein is a poly(A)-binding protein that inhibits nuclear export of mRNAs containing poly(A). J Virol 68, 2425–2432, (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carmody SR & Wente SR mRNA nuclear export at a glance. J Cell Sci 122, 1933–1937, (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stewart M Nuclear export of mRNA. Trends Biochem Sci 35, 609–617, (2010). [DOI] [PubMed] [Google Scholar]
- 6.Nair H et al. Global burden of respiratory infections due to seasonal influenza in young children: a systematic review and meta-analysis. Lancet 378, 1917–1930, (2011). [DOI] [PubMed] [Google Scholar]
- 7.Reed C et al. Estimating influenza disease burden from population-based surveillance data in the United States. PLoS One 10, e0118369, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.WHO. http://www.who.int/mediacentre/factsheets/fs211/en/index.html. Fact sheet Number 211, (April 2009).
- 9.Kim SJ et al. Integrative structure and functional anatomy of a nuclear pore complex. Nature 555, 475–482, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ayllon J & Garcia-Sastre A The NS1 protein: a multitasking virulence factor. Curr Top Microbiol Immunol 386, 73–107, (2015). [DOI] [PubMed] [Google Scholar]
- 11.Chen Z, Li Y & Krug RM Influenza A virus NS1 protein targets poly(A)-binding protein II of the cellular 3’-end processing machinery. The EMBO journal 18, 2273–2283, (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nemeroff ME, Barabino SM, Li Y, Keller W & Krug RM Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3’ end formation of cellular pre-mRNAs. Mol. Cell 1, 991–1000, (1998). [DOI] [PubMed] [Google Scholar]
- 13.Bornholdt ZA & Prasad BV X-ray structure of NS1 from a highly pathogenic H5N1 influenza virus. Nature 456, 985–988, (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carrillo B et al. The influenza A virus protein NS1 displays structural polymorphism. J Virol 88, 4113–4122, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aibara S, Katahira J, Valkov E & Stewart M The principal mRNA nuclear export factor NXF1:NXT1 forms a symmetric binding platform that facilitates export of retroviral CTE-RNA. Nucleic Acids Res 43, 1883–1893, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fribourg S, Braun IC, Izaurralde E & Conti E Structural basis for the recognition of a nucleoporin FG repeat by the NTF2-like domain of the TAP/p15 mRNA nuclear export factor. Mol Cell 8, 645–656, (2001). [DOI] [PubMed] [Google Scholar]
- 17.Blevins MB, Smith AM, Phillips EM & Powers MA Complex formation among the RNA export proteins Nup98, Rae1/Gle2, and TAP. J Biol Chem 278, 20979–20988, (2003). [DOI] [PubMed] [Google Scholar]
- 18.Faria PA et al. VSV disrupts the Rae1/mrnp41 mRNA nuclear export pathway. Mol Cell 17, 93–102, (2005). [DOI] [PubMed] [Google Scholar]
- 19.Bachi A et al. The C-terminal domain of TAP interacts with the nuclear pore complex and promotes export of specific CTE-bearing RNA substrates. RNA 6, 136–158, (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ullman KS, Shah S, Powers MA & Forbes DJ The nucleoporin nup153 plays a critical role in multiple types of nuclear export. Mol Biol Cell 10, 649–664, (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lelek M et al. Chromatin organization at the nuclear pore favours HIV replication. Nat Commun 6, 6483, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Twu KY, Kuo RL, Marklund J & Krug RM The H5N1 influenza virus NS genes selected after 1998 enhance virus replication in mammalian cells. J Virol 81, 8112–8121, (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Das K et al. Structural basis for suppression of a host antiviral response by influenza A virus. Proc Natl Acad Sci U S A 105, 13093–13098, (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chow KT, Gale M Jr. & Loo YM RIG-I and Other RNA Sensors in Antiviral Immunity. Annu Rev Immunol 36, 667–694, (2018). [DOI] [PubMed] [Google Scholar]
- 25.Fensterl V & Sen GC Interferon-induced Ifit proteins: their role in viral pathogenesis. J Virol 89, 2462–2468, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yarbrough ML, Mata MA, Sakthivel R & Fontoura BM Viral Subversion of Nucleocytoplasmic Trafficking. Traffic 15, 127–140, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mor A et al. Influenza Virus mRNA Trafficking Through Host Nuclear Speckles. Nat Microbiol 1, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Solorzano A et al. Mutations in the NS1 protein of swine influenza virus impair anti-interferon activity and confer attenuation in pigs. J Virol 79, 7535–7543, (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Radu A, Moore MS & Blobel G The peptide repeat domain of nucleoporin Nup98 functions as a docking site in transport across the nuclear pore complex. Cell 81, 215–222, (1995). [DOI] [PubMed] [Google Scholar]
- 30.Tsai PL et al. Cellular RNA Binding Proteins NS1-BP and hnRNP K Regulate Influenza A Virus RNA Splicing. PLoS Pathog 9, e1003460, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fodor E et al. Rescue of influenza A virus from recombinant DNA. J Virol 73, 9679–9682, (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fontoura BM, Blobel G & Matunis MJ A conserved biogenesis pathway for nucleoporins: proteolytic processing of a 186-kilodalton precursor generates Nup98 and the novel nucleoporin, Nup96. J Cell Biol 144, 1097–1112, (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Otwinowski Z & Minor W Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276, 307–326, (1997). [DOI] [PubMed] [Google Scholar]
- 34.McCoy AJ et al. Phaser crystallographic software. J Appl Crystallogr 40, 658–674, (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Emsley P, Lohkamp B, Scott WG & Cowtan K Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66, 486–501, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Afonine PV et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr 68, 352–367, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The coordinates of the NS1•NXF1•NXT1 structure have been deposited in the Protein Data Bank under accession number 6E5U. The data for the transcriptome analysis has been deposited in GEO under accession number GSE129318. All other data that support the findings of this study are available from the corresponding authors upon request.