Abstract
Background:
Epigenetic age, as defined by DNA methylation, may be influenced by air pollution exposure.
Objective:
To evaluate the relationship between NO2, particulate matter (PM), PM components and accelerated epigenetic age.
Methods:
In a sample of non-Hispanic white women living in the contiguous U.S. (n = 2747), we estimated residential exposure to PM2.5, PM10 and NO2 using a model incorporating land-use regression and kriging. Predictive k-means was used to assign participants to clusters representing different PM2.5 component profiles. We measured DNA methylation (DNAm) in blood using the Illumina's Infinium HumanMethylation450 BeadChip and calculated DNAm age using the Hannum, Horvath and Levine epigenetic clocks. Age acceleration was defined based on residuals after regressing DNAm age on chronological age. We estimated associations between interquartile range (IQR) increases in pollutants and age acceleration using linear regression. For PM2.5, we stratified by cluster membership. We examined epigenome-wide associations using robust linear regression models corrected with false discovery rate q-values.
Results:
NO2 was inversely associated with age acceleration using the Hannum clock (β = −0.24, 95% Cl: − 0.47, −0.02). No associations were observed for PM10. For PM2.5, the association with age acceleration varied by PM2.5 component cluster. For example, with the Levine clock, an IQR increase in PM2.5 was associated with an over 6-year age acceleration in a cluster that has relatively high fractions of crustal elements relative to overall PM2.5 (β = 6.57, 95% CI: 2.68, 10.47), and an almost 2-year acceleration in a cluster characterized by relatively low sulfur fractions (β = 1.88, 95% CI: 0.51, 3.25). In a cluster distinguished by lower relative nitrate concentrations, PM2.5 was inversely associated with age acceleration (β = −1.33, 95% CI: −2.43, −0.23). Across the epigenome, NO2 was associated with methylation at 2 CpG sites.
Conclusion:
Air pollution was associated with epigenetic age, a marker of mortality and disease risk, among certain PM2.5 component profiles.
Keywords: Breast cancer, Air pollution, Particulate matter, Clustering, Mixtures
1. Introduction
Air pollution is a global public health issue that is estimated to contribute to 7 million deaths worldwide per year (World Health Organization, 2016). Air pollution is associated with a higher risk of aging-related health outcomes including cardiovascular disease and cancer (Loomis et al., 2013). Biologic age metrics have been demonstrated to be biomarkers of risk for cardiovascular disease, cancer and mortality (Blackburn et al., 2015; Hertel et al., 2016; Holly et al., 2013; Klemera and Doubal, 2006; Kresovich et al., 2019; Levine, 2013; Peters et al., 2015). Biologic changes stemming from prolonged air pollution exposure may be detectable in DNA methylation (DNAm) patterns, as suggested by both epigenome-wide association studies (Chi et al., 2016; de FC Lichtenfels et al., 2018; Lee et al., 2019; Panni et al., 2016; Plusquin et al., 2017) as well studies that incorporate epigenetic aging metrics (Nwanaji-Enwerem et al., 2016; Nwanaji-Enwerem et al., 2017; Ward-Caviness et al., 2016).
Estimates of epigenetic age, or “epigenetic clocks”, are derived from DNAm at a small set of CpG sites that predict chronological age (Hannum et al., 2013; Horvath, 2013). Levine et al., extended this approach by designing an epigenetic clock that predicted “PhenoAge”, a biologic age metric determined from both chronological age and mortality-related blood parameters (Levine et al., 2018). “Age acceleration” is defined as having an estimated epigenetic age that exceeds an individual’s chronological age. Epigenetic age acceleration has been associated with lifestyle and environmental factors including cigarette smoking (Gao et al., 2016) and socioeconomic status (Dhingra et al., 2018; Hughes et al., 2018). Some evidence has suggested that air pollution may be associated with age acceleration (Nwanaji-Enwerem et al., 2016; Nwanaji-Enwerem et al., 2017; Ward-Caviness et al., 2016). However, these studies did not evaluate the Levine epigenetic clock, which, because it is influenced by clinical aging-related phenotypes, may be a better marker of disease-risk than prior epigenetic clocks.
Particulate matter < 10 μm (PM10) and < 2.5 μm in diameter (PM2.5) are measures of total particle mass per unit volume defined by size and do not distinguish the actual composition of those particles. Due to differences in sources of air pollution exposure, meteorology, and other factors, PM2.5 varies in composition by geographic region (Bell et al., 2007). Associations between PM2.5 and health outcomes including cardiovascular disease (Brook et al., 2010) and mortality (Franklin et al., 2008) have been shown to vary by PM2.5 component profiles. In this study, we evaluated the hypothesis that air pollution is associated with epigenetic age acceleration, allowing for possible heterogeneity in the effect of PM2.5 on epigenetic age by clustering individuals according to compositional profiles determined using predictive k-means clustering (Keller et al., 2017). Age acceleration was primarily defined using the Levine clock (Levine et al., 2018) although we considered associations with two other established epigenetic clocks, the Hannum and Horvath clocks (Hannum et al., 2013; Horvath, 2013). Using an epigenome-wide association study (EWAS), we also examined whether air pollution was associated with individual CpG sites.
2. Methods
2.1. Study population
The Sister Study is a nationwide prospective cohort study of women recruited between 2003 and 2009 who were living in the United States, including Puerto Rico (Sandler et al., 2017). Women ages 35–74 were eligible for this study if they had a sister with breast cancer but no history of breast cancer themselves. Participants completed a two-part computer-assisted telephone interview and had a home visit with a trained study examiner who collected a fasting blood sample. Study participants are contacted annually for health updates and complete detailed follow-up questionnaires every 2–3 years. Questionnaires included information on residential history, demographics and lifestyle factors among other topics. Written informed consent was obtained from all study participants. The Sister Study was approved by the National Institute of Environmental Health Institutional Review Board.
Blood DNAm was originally assessed as part of a case-cohort study of women with breast cancer (Xu et al., 2019). The DNAm case-cohort was limited to women who were non-Hispanic white and had an available blood sample collected at baseline, when they were breast cancer-free. The case-cohort consisted of a total of 2878 women (including 1542 who developed breast cancer during follow-up and a subcohort composed of a random sample of 1336 women, 74 of who had also become incident cases). The data used here was from data release version 5.0.1.
2.2. Air pollution exposure assessment
Annual average outdoor air pollution concentrations (PM2.5, PM10, NO2) were estimated for study participants' residential addresses during the 12 months prior to enrollment. Air pollution concentrations were estimated by a cross-validated universal kriging models using US Environmental Protection Agency (EPA) Air Quality System (AQS) monitoring data from 2006 (PM2.5 and NO2) and 2000 (PM10) (Sampson et al., 2013; Young et al., 2016). The years of monitoring data were selected as they were either prior to or during the enrollment period and these are consistent with previous research in this study population (Reding et al., 2015). These models incorporate spatial smoothing and information from geographic covariates, some of which were determined using satellite observations. We excluded 6 women who lived outside the contiguous US and 4 whose addresses were not successfully geocoded. For NO2, air pollution estimates were missing for 2 additional women whose addresses had incomplete satellite coverage.
The PM2.5 component clusters were derived using covariate-adapted predictive k-means clustering, as previously described (Keller et al., 2017). 2010 PM2.5 component data were obtained from 130 US EPA AQS monitoring locations that measured the mass concentrations for 22 PM2.5 component species including elemental carbon (EC), organic carbon (OC), NO3−, SO42−, Al, As, Br, Cd, Ca, Co, Cr, Cu, Fe, K, Mn, Na, S, Si, Se, Ni, V and Zn. This year of data was selected due to improved precision and decreased bias with new instrumentation and analysis methods (Spada and Hyslop, 2018). Most components were measured once every six days. An annual average was computed for each component at each monitoring location. Mass concentrations were then converted to mass fractions by dividing the annual average of each species by the annual average PM2.5 at that location. Component mass fractions were log transformed. Clustering was used here to categorize monitor locations into a pre-defined number of (k) clusters based upon similarities in particulate matter component observations. Geographic covariates (such as measures of land-cover, road networks, vegetative index and population density, etc.) were included in the model to improve prediction of the clusters. This approach then predicts cluster membership at individual participant residential location using geographic covariates. The final selected model had 8 clusters, although Cluster 8 was not included in the effect measure modification analysis due to small sample size. These clusters identify groups of women expected to be similar with regards to residential ambient PM2.5 composition.
2.3. DNA methylation assessment
DNA processing procedures have been described previously (O'Brien et al., 2018; Xu et al., 2019). The DNA was analyzed on Illumina HumanMethylation450 BeadChips following the manufacturer's protocol and using high throughput robotics to minimize batch effects. Methylation data preprocessing and quality control using the ENmix R software package (Xu et al., 2016) has been described. After quality control and preprocessing measures, the final number of included CpGs was 423,500. β-Values for methylation were calculated using the fluorescence intensities for unmethylated (U) and methylated (M) alleles and the formula β = M/(M + U + 100). β-Values were then transformed on the logit scale to be M-values. M-values were used in all statistical tests. 102 samples failed methylation quality control leaving 2776 women with available genome-wide methylation data. This resulted in a sample size of 2764 women with available air pollution and DNA methylation data.
2.4. Statistical analysis
We conducted a cross-sectional analysis to evaluate the association between air pollution and epigenetic age acceleration. We considered interquartile range increases in air pollutants based on specific years (2006 PM2.5 IQR = 3.5 μg/m3, 2000 PM10 IQR = 5.7 μg/m3, 2006 NO2 IQR = 6.3 ppb) as our exposure measure of interest. All models were adjusted for a priori selected covariates including future case status (invasive breast cancer or ductal carcinoma in situ, no breast cancer), education (high school or less, some college or technical school, college degree or more), smoking status (current, former, never), hormone therapy use (ever, never). For hormone therapy use, 7 participants with missing data were excluded from the analysis.
Although women were free of breast cancer at the time of blood draw, we previously observed that age acceleration was related to breast cancer risk (Kresovich et al., 2019). Therefore, in addition to adjustment for future case status, we conducted a sensitivity analysis using inverse probability weighted sampling fractions to account for the sampling design.
We calculated DNAm age using the weights provided by the clock developers, as previously described (Kresovich et al., 2019; White et al., 2019). We regressed DNAm age measures on the participant's chronological age at blood draw and used the resulting residuals as the metric of biologic age acceleration, which are, by design, independent of chronological age. Age acceleration was calculated with and without adjustment for blood cell composition (BCC). The Houseman method was used to estimate BCC, which was used in sensitivity analyses to control for effects on methylation due to differences in the relative sizes of blood cell type populations (Houseman et al., 2012). We used linear regression to estimate the association between an IQR increase in air pollutants and epigenetic age acceleration, adjusting for the covariates listed above. We excluded women who had age acceleration estimates that were ≥ 4 standard deviations above or below the mean for any of the epigenetic clocks (N = 10), resulting in an analytic sample size of 2747.
To evaluate effect measure modification by PM2.5 components, we estimated associations between PM2.5 and epigenetic age acceleration metrics stratified by membership in predicted spatial clusters defined by differences in PM2.5 components. We tested for effect modification using a likelihood ratio test to compare models with and without interaction terms between PM2.5 concentrations and PM2.5 component clusters.
As a sensitivity analysis to address potential residual confounding by socioeconomic status, we additionally adjusted our analyses for household income per person living in the home, and census-tract-level education (defined as the percentage of adults above the age of 25 with a bachelor's degree) and census-tract-level income (defined as median family income). We also stratified the association between PM2.5 and epigenetic aging within cluster by study enrollment year (2003–2005, 2006–2009).
For the EWAS, to examine associations between air pollution and DNAm at individual CpG sites we conducted robust linear regression. We adjusted for confounders (age at baseline, education, smoking status, hormone therapy use, future breast cancer case status), as well as blood cell composition. Technical variation and batch effects were controlled for with six surrogate variables based on non-negative control probes, plate, and DNA extraction method. We conducted a sensitivity analysis limiting the EWAS to include only the CpG sites that were used to estimate the three epigenetic clocks (n = 870). To correct for multiple testing, we estimated the false discovery rate (FDR) (Storey and Tibshirani, 2003). We considered findings to be notable with a q < 0.05. All analyses were conducted using SAS 9.3 (Cary, NC) and R (R Core Team, 2013).
3. Results
Study participant characteristics are shown in Table 1. Briefly, women were on average 57 years old, over half had a college degree or higher and only 7% were current smokers. The average exposure was 8.8 μg/m3 for PM2.5, 21.9 μg/m3 for PM10 and 9.9 ppb for NO2.
Table 1.
Baseline study population characteristics stratified by PM2.5 component clusters, Sister Study 2003–2009.
| All study participants (N = 2747) |
Cluster identifier |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 (N = 751) |
2 (N = 850) |
3 (N = 523) |
4 (N = 238) |
5 (N =259) |
6 (N = 52) |
7 (N = 74) |
||||||||||
| N | % | N | % | N | % | N | % | N | % | N | % | N | % | N | % | |
| Age at baseline | ||||||||||||||||
| ≤45 | 262 | 10 | 85 | 11 | 80 | 9 | 45 | 9 | 21 | 9 | 20 | 8 | 4 | 8 | 7 | 9 |
| 46–49 | 350 | 13 | 94 | 13 | 115 | 14 | 63 | 12 | 27 | 11 | 37 | 14 | 5 | 10 | 9 | 12 |
| 50–54 | 526 | 19 | 139 | 19 | 172 | 20 | 108 | 21 | 44 | 18 | 38 | 15 | 10 | 19 | 15 | 20 |
| 55–59 | 569 | 21 | 179 | 24 | 160 | 19 | 105 | 20 | 51 | 21 | 49 | 19 | 9 | 17 | 16 | 22 |
| 60–64 | 470 | 17 | 120 | 16 | 149 | 18 | 91 | 17 | 41 | 17 | 49 | 19 | 10 | 19 | 10 | 14 |
| ≥65 | 570 | 21 | 134 | 18 | 174 | 20 | 111 | 21 | 54 | 23 | 66 | 25 | 14 | 27 | 17 | 23 |
| Education | ||||||||||||||||
| High school degree, equivalent or less | 423 | 15 | 400 | 53 | 431 | 51 | 264 | 50 | 133 | 56 | 136 | 53 | 32 | 62 | 40 | 54 |
| Some college/technical school | 888 | 32 | 128 | 17 | 127 | 15 | 90 | 17 | 29 | 12 | 31 | 12 | 5 | 10 | 13 | 18 |
| 4-year college degree or higher | 1436 | 52 | 223 | 30 | 292 | 34 | 169 | 32 | 76 | 32 | 92 | 36 | 15 | 29 | 21 | 28 |
| Smoking status | ||||||||||||||||
| Never smoker | 1430 | 52 | 53 | 7 | 59 | 7 | 42 | 8 | 18 | 8 | 19 | 7 | 4 | 8 | 4 | 5 |
| Past smoker | 1118 | 41 | 418 | 56 | 410 | 48 | 288 | 55 | 124 | 52 | 125 | 48 | 26 | 50 | 39 | 53 |
| Current smoker | 199 | 7 | 280 | 37 | 381 | 45 | 193 | 37 | 96 | 40 | 115 | 44 | 22 | 42 | 31 | 42 |
| Ever HRT | ||||||||||||||||
| No | 1388 | 51 | 395 | 53 | 495 | 58 | 218 | 42 | 110 | 46 | 112 | 43 | 26 | 50 | 32 | 43 |
| Yes | 1359 | 49 | 356 | 47 | 355 | 42 | 305 | 58 | 128 | 54 | 147 | 57 | 26 | 50 | 42 | 57 |
| Air pollutants (mean, SD) | ||||||||||||||||
| PM2.5 (μg/m3) | 10.4 | 2.4 | 11.5 | 1.8 | 9.2 | 2.1 | 11.6 | 1.8 | 11.1 | 2.4 | 8.00 | 1.8 | 12.3 | 1.3 | 7.6 | 1.2 |
| PM10 (μg/m3) | 21.9 | 5.8 | 22.1 | 3.3 | 18.8 | 4.5 | 21.4 | 3.1 | 27.1 | 6.8 | 26.4 | 10.5 | 24.5 | 3.9 | 24.7 | 4.3 |
| NO2 (ppb) | 9.9 | 4.8 | 10.5 | 4.0 | 8.6 | 5.3 | 7.6 | 2.8 | 13.6 | 4.9 | 11.7 | 4.5 | 12.5 | 3.0 | 15.2 | 5.1 |
An IQR increase in NO2 was inversely associated with age acceleration defined by the Hannum clock (β = −0.24, 95% CI: −0.47, − 0.02), although this association was not evident for either the Levine or Horvath clocks. We observed little to no epigenetic age acceleration for PM2.5 or PM10 across the three clocks (Table 2). Although after adjustment for BCC, for an IQR increase in PM10, we observed evidence of epigenetic age acceleration using the Levine clock (β = 0.22, 95% CI: 0.01, 0.43) (Supplemental Table 1). In a sensitivity analysis, the association for PM10 with the Levine clock was unchanged with adjustment for age acceleration defined using the Hannum and Horvath clocks. Point estimates and directions of associations were similar when applying sampling weights (Supplemental Table 2).
Table 2.
Residential air pollution measures and epigenetic age acceleration in the Sister Study, 2003–2009.
| Epigenetic clocka | N | PM2.5 | PM10 | No2 |
|---|---|---|---|---|
| β (95% CI)b | β (95% CI)b | β (95% CI)b | ||
| Hannum | 2747 | −0.14 (−0.40, 0.11) | −0.06 (−0.23, 0.11) | −0.24 (−0.47, −0.02) |
| Horvath | 2747 | −0.13 (−0.41, 0.14) | 0.07 (−0.11, 0.26) | 0.06 (−0.18, 0.30) |
| Levine | 2747 | 0.06 (−0.28, 0.40) | 0.19 (−0.03, 0.42) | −0.05 (−0.35, 0.25) |
Epigenetic age acceleration estimated from the Hannum, Horvath and Levine epigenetic clocks.
β estimates are the result of linear regression for the association between an interquartile range (IQR) increase in air pollutants (2006 PM2.5 = 3.5 μg/m3, 2000 PM10 = 5.7 μg/m3, 2006 NO2 = 6.3 ppb) and epigenetic age acceleration adjusting for education, smoking status, postmenopausal hormone use and breast cancer case status.
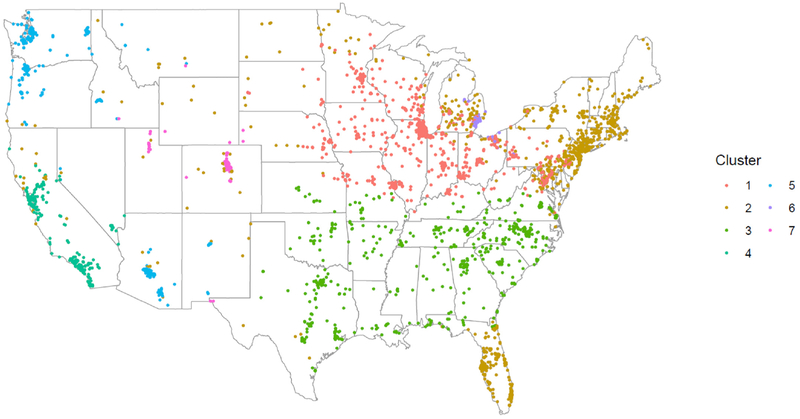
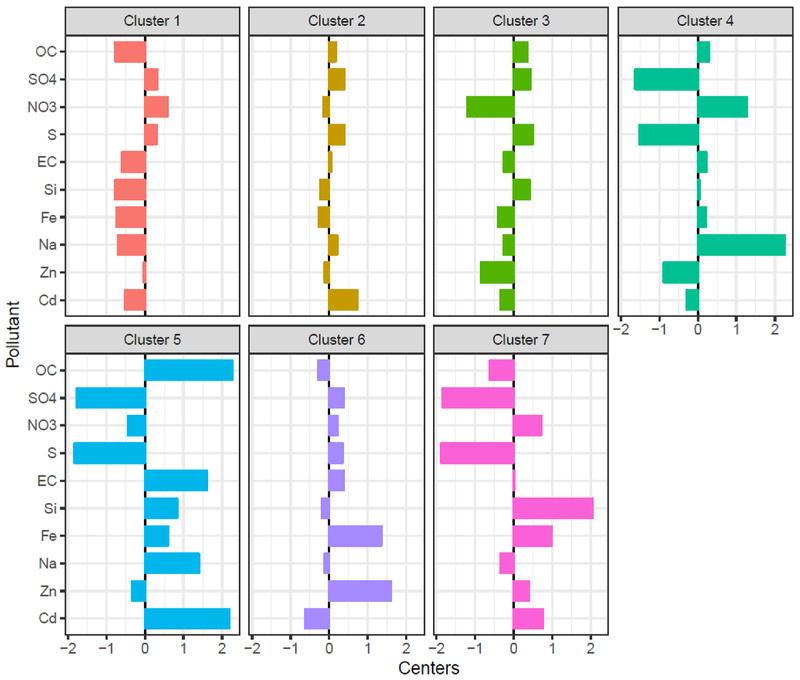
We next considered the relationship between PM2.5 and age acceleration by PM2.5 component cluster. Study population characteristics by PM2.5 component cluster identifiers are also provided in Table 1. Most women were assigned to cluster 1 or 2. There was some variability in demographic and lifestyle factors across clusters. For example, women in Clusters 5 and 6 were slightly older than women in the other clusters. Women in Cluster 7 were less likely to be current smokers and Cluster 3 women were more likely to use postmenopausal hormones compared to the other clusters. The geographic location of women by cluster assignment is shown in Fig. 1. Profiles of the relative composition of selected PM2.5 components are shown in Fig. 2.
Fig. 1.
Sister Study participant enrollment locations by PM2.5 component clusters. Adapted from Keller et al.(2017).
Fig. 2.
Relative composition (standardized log transformed species mass fractions) of selected PM2.5 components by predictive k-means cluster centers identified in the 2010 annual average PM25 component data.
Adapted from Keller et al.(2017).
We observed substantial effect measure modification by PM2.5 component clusters for the association between PM2.5 and age acceleration defined using both the Levine clock (Table 3; p for heterogeneity = < 0.001) and the Hannum clock (p for heterogeneity = 0.03). Using the Levine clock, age acceleration was observed to be associated with PM2.5 for women who were members of Cluster 5 (β = 1.88, 95%CI: 0.51, 3.25) and Cluster 7 (β = 6.57, 95% CI: 2.68, 10.47). Women in cluster 5 were located in both in the Pacific Northwest and the Southwest and the cluster profile was characterized by relatively high mass fraction proportions across most components, except for sulfur. Cluster 7 was a Central-Western cluster, characterized by high proportions of crustal elements including silicon which is representative of the surface soil in that region. An elevated, but imprecise, estimate was also observed for Cluster 6 (β = 3.96, 95% CI: −1.24, 9.16), which is characterized by high fractions of iron and other metals, indicative of industrial emissions. Using the Hannum clock, accelerated epigenetic age was observed for women who were members of Cluster 4 (β = 0.74, 95% CI: −0.14, 1.63). Cluster 4 encompasses monitors located in California and is characterized by having low relative sulfur levels and large fractions of sodium and nitrate (agricultural emissions).
Table 3.
PM2.5, particulate matter component clusters and epigenetic age accelerationa in the Sister Study, 2003–2009.
| PM2.5 component clusters | N | Hannum | Horvath | Levine |
|---|---|---|---|---|
| β (95% CI)b | β (95% CI)b | β (95% CI)b | ||
| Overall | 2747 | −0.14 (−0.40, 0.11) | −0.13 (−0.41, 0.14) | 0.06 (−0.28, 0.40) |
| 1 | 751 | −0.40 (−1.05, 0.25) | −0.16 (−0.83, 0.52) | −0.59 (−1.42, 0.25) |
| 2 | 850 | −0.45 (−0.96, 0.07) | −0.31 (−0.88, 0.27) | −0.42 (−1.15, 0.30) |
| 3 | 523 | −1.10 (−1.90, −0.31) | −1.20 (−2.08, −0.32) | −1.33 (−2.43, −0.23) |
| 4 | 238 | 0.74 (−0.14, 1.63) | 0.22 (−0.77, 1.20) | −0.04 (−1.20, 1.13) |
| 5 | 259 | −0.06 (−1.11, 1.00) | −0.22 (−1.42, 0.99) | 1.88 (0.51, 3.25) |
| 6 | 52 | 2.24 (−1.68, 6.17) | 2.57 (−1.84, 6.97) | 3.96 (−1.24, 9.16) |
| 7 | 74 | 1.92 (−1.12, 4.97) | 2.71 (−1.22, 6.64) | 6.57 (2.68, 10.47) |
| p valuec | 0.03 | 0.1 | < 0.0001 |
Epigenetic age acceleration estimated from the Hannum, Horvath and Levine epigenetic clocks.
β estimates are the result of linear regression for the association between an interquartile range (IQR) increase in air pollutants (2006 PM2.5 = 3.5 μg/m3) and epigenetic age acceleration adjusting for education, smoking status, postmenopausal hormone use and breast cancer case status.
p-Value from a likelihood ratio test comparing models with and without interaction terms between PM2.5 and the clusters.
In contrast, an inverse association with epigenetic age acceleration using the Levine clock was observed in relation to an IQR increase in PM2.5 among women who were in Cluster 3 (β = −1.33, 95% CI: −2.43, −0.23). This inverse association among women in Cluster 3 was evident across all three clocks. This cluster was located in the Southeastern US and characterized by low fractions of nitrate. Effect estimates by cluster were similar although slightly less pronounced with adjustment for blood cell composition (data not shown). There was no evidence of heterogeneity by component cluster membership for the associations between PM2.5 and epigenetic age acceleration using the Horvath clock. Our results did not notably change with further adjustment for socioeconomic status indicators, including household income and census-tract-level education and income (Supplemental Table 3).
When stratifying by enrollment year (Supplemental Table 4), we observed some consistent patterns of associations despite small sample sizes in certain clusters. For example, the inverse association between PM2.5 and epigenetic aging among women in Cluster 3 for all three clocks was consistent across enrollment years. For the Levine clock, the p-value for interaction term remained statistically significant and the positive associations among women in Clusters 5 and 7 were evident for both 2003–2005 and 2006–2009. However, for the Hannum clock, the p-value for the interaction term was not significant for women who enrolled from 2003 to 2005.
In the EWAS analysis, no CpGs were identified to be associated with either PM10 or PM2.5. We observed only 2 CpGs to be associated with an IQR increase in NO2 (Supplemental Table 5). The CpG with the lowest p-value for NO2 was in a gene an intergenic region on chromosome 10 (cg06544185). An IQR increase in NO2 was associated with slightly higher methylation at this site (β = 0.003, p = 1.95 × 10−7 and q = 0.04). The other CpG identified was on chromosome 7 in the ACHE gene in a CpG island region upstream of the transcription start site (cg02607340). At this CpG, an IQR increase in NO2 was associated with lower methylation (β = −0.0074, p = 2.23 × 10−7 and q = 0.04). These two CpGs were not members of any of the three epigenetic clocks. In our sensitivity analysis, limiting the EWAS to only CpGs that were included in the calculation of the epigenetic clocks, we observed only a single CpG to be inversely associated with an IQR increase in PM10 (cg22920873; β = −0.002, p = 1.22 × 10−5 and q = 0.009). This CpG is on chromosome 7 in the C7orf55 gene in a CpG island region downstream from the transcription start site. The cg22920873 is involved in the Horvath clock with a coefficient of 0.1143748. In this sensitivity analysis, we observed no associations with exposure to either PM2.5 or NO2. An exploratory enrichment analysis among the clock CpGs that were associated (p < 0.05) with PM10 suggested > 40 enriched (FDR < 0.05) Gene Ontology biological process pathways, several of which included > 10 genes with significant CpGs (anatomical structure development, regulation of developmental process and regulation of cell communication).
4. Discussion
There was no overall association between PM2.5 and age acceleration, but we did observe significant heterogeneity in the association between PM2.5 and age acceleration across clusters defined by PM2.5 component profiles: for example, among women in two distinct PM2.5 component clusters, an IQR increase in PM2.5 was associated with a 2 and 6-year higher age acceleration using the Levine clock. Unexpectedly, PM2.5 was also inversely associated with age acceleration as estimated by all three clocks among women in another cluster. We observed very little evidence of age acceleration in relation to exposure to PM10 or NO2, although we did observe an inverse association for NO2 exposure in relation to age acceleration defined by the Hannum clock. Although our epigenome-wide analysis found very few significant CpGs, our epigenetic age results suggest that integrated measures of DNAm, such as epigenetic age clocks, may identify methylation differences in relation to exposures such as air pollution that would not be detected at an individual CpG level.
Our findings provide a more detailed and complex characterization of the possible association between air pollution exposure and epigenetic age acceleration. The Cooperative Health Research in the Region of Augsburg (KORA) study (n = 1777) observed that an IQR increase in PM2.5 (0.97 μg/m3) was associated with ~0.3 year higher age acceleration defined using the Horvath clock (Ward-Caviness et al., 2016). When they restricted analyses to women, exposure to both NOx and black carbon, but not PM10, were associated with advanced epigenetic age (Ward-Caviness et al., 2016). In the Normative Aging Study (NAS; n = 589), an elderly cohort of men, a 1 μg/m3 increase in PM2.5 was associated with accelerated epigenetic age characterized by the Horvath clock (Nwanaji-Enwerem et al., 2016). In contrast, we did not observe associations in our study population for overall PM2.5 exposure and age acceleration calculated using either the Horvath clock or the Hannum clock. This lack of consistency may be due to lower exposure levels in our population. For example, our mean exposure levels (PM2.5 mean = 10.4 μg/m3) are lower than in the KORA population (PM2.5 mean = 14 μg/m3).
We observed considerable heterogeneity across PM2.5 component cluster membership. PM2.5 is a heterogeneous mixture that varies geographically due to varying sources of PM and other factors. The clusters provide a more comprehensive exposure assessment by incorporating additional information regarding the components of the complex PM2.5 mixture. Age acceleration was observed for exposure to PM2.5 among women assigned to Clusters 5 and 7 (Levine clock) and Cluster 4 (Hannum clock). Women in Cluster 5, a Pacific Northwest and Western-based cluster, would be expected to have higher organic carbon and lower sulfate exposure, which is likely indicative of wood smoke, a predominant heating source in the region, and wild fires. The component profile of Cluster 7, another Western-based cluster, is driven by the PM2.5 components indicative of surface soil. Women in Cluster 4, which encompasses the California monitors, would be expected to have exposures with lower relative sulfur levels and larger fractions of sodium and nitrate (agricultural emissions, marine aerosols and traffic). For women in Cluster 3, PM2.5 was associated with consistent inverse relationships with age acceleration across all three epigenetic clocks. Women in this Southeast-based cluster would be expected to have lower nitrate relative to sulfate levels. In a subsequent study within the NAS, the authors evaluated five PM2.5 component species (EC, OC, sulfate, nitrate and ammonium) using a LASSO model and concluded that their association with Horvath-defined accelerated epigenetic age was driven by the components sulfate and ammonium (Nwanaji-Enwerem et al., 2017). Our findings likely differ from Nwanji-Enwerem et al., due to the inclusion of many more PM components and the use of clustering to identify component profiles rather than focusing on identifying the individual drivers of the association.
In our analyses, we observed both positive and inverse associations with epigenetic age acceleration. In the KORA study, air pollution was positively associated with epigenetic age acceleration in women, but inversely related to age acceleration in men (Ward-Caviness et al.,2016. Although positive age acceleration is more consistently associated with negative health effects, inverse associations with epigenetic aging may also be indicative of adverse outcomes, as has been observed with psychosocial stress (Boks et al., 2015).
Different markers of air pollution have been previously related to locus-specific and global measures of DNA methylation (Martin and Fry, 2018). Previous studies have also used the Illumina's Infinium HumanMethylation450 BeadChip array to evaluate the relationship between ambient air pollution and methylation across the epigenome in adults (Chi et al., 2016; de FC Lichtenfels et al., 2018; Lee et al., 2019; Panni et al., 2016; Plusquin et al., 2017). Both short-term exposure to PM (Panni et al., 2016) and exposure in the prior year (Chi et al., 2016) has been found to be associated with methylation at individual CpGs. Long-term NO2, but not PM, exposure was reported to be associated with overall global hypomethylation (Plusquin et al., 2017) and individual CpG sites (de FC Lichtenfels et al., 2018; Plusquin et al., 2017), but individual CpG site associations did not persist after meta-analysis or in validation datasets (de FC Lichtenfels et al., 2018; Plusquin et al.,2017. In a population of Korean adults, both PM10 and NO2 were associated with a number of methylated sites, including some previously published CpGs (Lee et al., 2019). The two CpGs identified in our population to be associated with NO2 in our study were not reported by these prior studies.
Our study population was limited to non-Hispanic women, each of whom had a first-degree family history of breast cancer. Thus, the magnitude of the associations with epigenetic age may not be fully generalizable to all women. However, women did not have breast cancer at the time of blood draw and adjusting for future development of breast cancer and using sampling weights had little impact on our results.
We estimated average exposures for the geographic location of the address where the study participants lived at during the 12 months prior to enrollment. The high-resolution exposure model is a strength of the analysis as it incorporated both monitored data and geographic variables including those obtained from satellite data using a land-use regression and kriging model approach. We relied on annual average estimates of air pollution exposure that incorporated monitoring data for a given year. Previous studies have suggested that a one-year average estimate of air pollution exposure is an adequate proxy for long-term exposure (Hart et al., 2015), but it is possible that the year selected may not have best represented the relevant exposure period. However, over half of our participants lived at this residence for at least ten years. Thus, exposure estimates may well represent longer-term exposure to the extent that PM2.5 components do not change dramatically over time.
Women were classified into PM25 component clusters based on data from 2010, a year after baseline. However, PM25 component levels have changed over time in some areas (Blanchard et al., 2013) and thus, it is unclear whether these PM2.5 component clusters adequately represent the PM2.5 mixture that women were exposed to at the time of baseline interview. If that were the case, we would expect the associations to be less likely to hold for women who enrolled earlier in the cohort. When stratifying by enrollment year, we observed that in general associations remained similar, especially for the Levine clock, suggesting that the role of these clusters as modifiers of the association between PM2.5 and age acceleration may be consistent over time. Additionally, many of the clusters are defined by components that confirm well-established regional features that would likely not vary substantially over a few years (e.g., marine aerosols and agricultural emissions for California-based Cluster 4 and western-soil trace elements for Cluster 7). However, we did note some variation in estimates across the enrollment period, especially for the Hannum clock and for the clusters with smaller sample sizes, suggesting some caution is necessary in interpreting these results.
There are some other noteworthy limitations to this analysis. Exposure measurements were estimated for the residential ambient outdoor levels and cannot incorporate the variability in individual exposure due to personal activity patterns, including commuting, time at work in a different environment, and time spent outdoors. The clusters varied notably by geographic region; there may be systematic differences across geographic regions in addition to the characteristic composition of their particulate air pollution, and some of those differences could confound our results by themselves being effect modifiers for particulate air pollution. PM10 components also vary geographically; we did not have information on PM10 component data and thus were unable to consider heterogeneity by PM10 components. We attempted to address residual confounding by socioeconomic status by adjusting for census-tract and individual-level education and income. Although our conclusions remained the same, we cannot rule out the possibility of residual confounding. Both the air pollution exposure model and the k-means clustering approaches incorporate geographic covariates to improve prediction and these may have influenced the clusters that were identified. Finally, these are cross-sectional data and we do not have information on changes in epigenetic aging over time.
An important strength of this study is the inclusion of the k-means clusters to evaluate the role of PM2.5 component profiles as a modifier of the association between PM2.5 and epigenetic age. PM is a complex mixture and it is important to consider its components, as evidenced by the significant heterogeneity in our effect estimates across component clusters. It is not feasible to estimate all of the individual PM2.5 component levels at a participant's residence. By using k-means clustering to predict cluster membership, this approach allows for the consideration of the components in relation to health outcomes. These clusters were developed using an unsupervised method that identifies exposure mixtures without regard to a specific health outcome or study population, so there may be clusters that were not identified that are even stronger modifiers of the association between PM and DNAm. Another strength of this study was the use of multiple epigenetic clocks to define epigenetic age acceleration. Previous studies have suggested that associations may not be consistent across clocks (Carroll et al., 2017; Levine et al., 2018; Quach et al., 2017). We have previously demonstrated that the Levine clock shows particularly stronger associations with occupational shift working (White et al., 2019) consistent with our findings here where associations with PM2.5 component clusters were strongest for the Levine clock. The sensitivity of the Levine clock may be due to the use of mortality-associated clinical biomarkers in addition to chronological age in the predictive models.
In conclusion, we observed notable heterogeneity in the association between age acceleration and PM2.5 air pollution exposure by PM2.5 component cluster membership. Overall, these findings support a relationship between air pollution and epigenetic age, a marker of mortality and chronic disease risk. Our results suggest that the effects of environmental exposures such as air pollution may be better captured using integrated methylation measures, such as epigenetic age acceleration, rather than considering individual CpG sites. Further, these results underscore the importance of considering the variability in air pollution composition and support the use of epigenetic clocks to assess epigenetic modifications related to environmental exposures.
Supplementary Material
Acknowledgements
This research was supported by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences (Z01-ES044005).
Footnotes
Declaration of Competing Interest
The authors report no conflicts of interest.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.envint.2019.105071.
References
- Bell ML, Dominici F, Ebisu K, Zeger SL, Samet JM, 2007. Spatial and temporal variation in PM2.5 chemical composition in the United States for health effects studies. Environ. Health Perspect. 115, 989–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn EH, Epel ES, Lin J, 2015. Human telomere biology: a contributory and interactive factor in aging, disease risks, and protection. Science 350, 1193–1198. [DOI] [PubMed] [Google Scholar]
- Blanchard C, Hidy G, Tanenbaum S, Edgerton E, Hartsell B, 2013. The Southeastern Aerosol Research and Characterization (search) study: temporal trends in gas and pm concentrations and composition, 1999–2010. J. Air Waste Manage. Assoc. 63, 247–259. [DOI] [PubMed] [Google Scholar]
- Boks MP, van Mierlo HC, Rutten BP, Radstake TR, De Witte L, Geuze E, et al. , 2015. Longitudinal changes of telomere length and epigenetic age related to traumatic stress and post-traumatic stress disorder. Psychoneuroendocrinology 51, 506–512. [DOI] [PubMed] [Google Scholar]
- Brook RD, Rajagopalan S, Pope CA 3rd, Brook JR, Bhatnagar A, Diez-Roux AV, et al. , 2010. Particulate matter air pollution and cardiovascular disease: an update to the scientific statement from the American Heart Association. Circulation 121, 2331–2378. [DOI] [PubMed] [Google Scholar]
- Carroll JE, Irwin MR, Levine M, Seeman TE, Absher D, Assimes T, et al. , 2017. Epigenetic aging and immune senescence in women with insomnia symptoms: findings from the Women's Health Initiative Study. Biol. Psychiatry 81, 136–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi GC, Liu Y, MacDonald JW, Barr RG, Donohue KM, Hensley MD, et al. , 2016. Long-term outdoor air pollution and DNA methylation in circulating monocytes: results from the Multi-Ethnic Study of Atherosclerosis (mesa). Environ. Health 15, 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de FC Lichtenfels AJ, van der Plaat DA, de Jong K, van Diemen CC, Postma DS, Nedeljkovic I, et al. , 2018. Long-term air pollution exposure, genome-wide DNA methylation and lung function in the lifelines cohort study. Environ. Health Perspect. 126, 027004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhingra R, Nwanaji-Enwerem JC, Samet M, Ward-Caviness CK, 2018. DNA methylation age—environmental influences, health impacts, and its role in environmental epidemiology. Curr. Environ. Health Rep 5, 317–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin M, Koutrakis P, Schwartz J, 2008. The role of particle composition on the association between PM2.5 and mortality. Epidemiology (Cambridge, Mass) 19, 680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Zhang Y, Breitling LP, Brenner H, 2016. Relationship of tobacco smoking and smoking-related DNA methylation with epigenetic age acceleration. Oncotarget 7, 46878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannum G, Guinney J, Zhao L, Zhang L, Hughes G, Sadda S, et al. , 2013. Genomewide methylation profiles reveal quantitative views of human aging rates. Mol. Cell 49, 359–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart JE, Puett RC, Rexrode KM, Albert CM, Laden F, 2015. Effect modification of long-term air pollution exposures and the risk of incident cardiovascular disease in US women. J. Am. Heart Assoc. 4, e002301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertel J, Friedrich N, Wittfeld K, Pietzner M, Budde K, Van der Auwera S, et al. , 2016. Measuring biological age via metabonomics: the metabolic age score. J. Proteome Res. 15, 400–410. [DOI] [PubMed] [Google Scholar]
- Holly AC, Melzer D, Pilling LC, Henley W, Hernandez DG, Singleton AB, et al. , 2013. Towards a gene expression biomarker set for human biological age. Aging Cell 12, 324–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S, 2013. DNA methylation age of human tissues and cell types. Genome Biol. 14, R115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, et al. , 2012. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinf. 13, 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes A, Smart M, Gorrie-Stone T, Hannon E, Mill J, Bao Y, et al. , 2018. Socioeconomic position and DNA methylation age acceleration across the life course. Am. J. Epidemiol. 187, 2346–2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller JP, Drton M, Larson T, Kaufman JD, Sandler DP, Szpiro AA, 2017. Covariate-adaptive clustering of exposures for air pollution epidemiology cohorts. Ann. Appl. Stat. 11, 93–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemera P, Doubal S, 2006. A new approach to the concept and computation of biological age. Mech. Ageing Dev. 127, djz020. [DOI] [PubMed] [Google Scholar]
- Kresovich JK, Xu Z, O'Brien KM, Weinberg CR, Sandler DP, Taylor JA, 2019. Methylation-based biological age and breast cancer risk. J. Natl. Cancer Inst. Ill (10), 240–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MK, Xu CJ, Carnes MU, Nichols CE, Ward JM, Kwon SO, et al. , 2019. Genome-wide DNA methylation and long-term ambient air pollution exposure in Korean adults. Clin. Epigenetics 11, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine ME, 2013. Modeling the rate of senescence: can estimated biological age predict mortality more accurately than chronological age? J. Gerontol. A Biol. Sci. Med. Sci. 68, 667–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine ME, Lu AT, Quach A, Chen BH, Assimes TL, Bandinelli S, et al. , 2018. An epigenetic biomarker of aging for lifespan and healthspan. Aging (Albany NY) 10, 573–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loomis D, Grosse Y, Lauby-Secretan B, El Ghissassi F, Bouvard V, Benbrahim-Tallaa L, et al. , 2013. The carcinogenicity of outdoor air pollution. Lancet Oncol. 14 (13), 1262–1263. [DOI] [PubMed] [Google Scholar]
- Martin EM, Fry RC, 2018. Environmental influences on the epigenome: exposure-associated DNA methylation in human populations. Annu. Rev. Public Health 39, 309–333. [DOI] [PubMed] [Google Scholar]
- Nwanaji-Enwerem JC, Colicino E, Trevisi L, Kloog I, Just AC, Shen J, et al. , 2016. Long-term ambient particle exposures and blood DNA methylation age: findings from the VA normative aging study. Environ. Epigenetics 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nwanaji-Enwerem JC, Dai L, Colicino E, Oulhote Y, Di Q, Kloog I, et al. , 2017. Associations between long-term exposure to PM2.5 component species and blood DNA methylation age in the elderly: the VA normative aging study. Environ. Int. 102, 57–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien KM, Sandler DP, Xu Z, Kinyamu HK, Taylor JA, Weinberg CR, 2018. Vitamin D, DNA methylation, and breast cancer. Breast Cancer Res. 20, 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panni T, Mehta AJ, Schwartz JD, Baccarelli AA, Just AC, Wolf K, et al. , 2016. Genome-wide analysis of DNA methylation and fine particulate matter air pollution in three study populations: KORA F3, KORA F4, and the normative aging study. Environ. Health Perspect. 124, 983–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters MJ, Joehanes R, Pilling LC, Schurmann C, Conneely KN, Powell J, et al. , 2015. The transcriptional landscape of age in human peripheral blood. Nat. Commun. 6, 8570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plusquin M, Guida F, Polidoro S, Vermeulen R, Raaschou-Nielsen O, Campanella G, et al. , 2017. DNA methylation and exposure to ambient air pollution in two prospective cohorts. Environ. Int. 108, 127–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quach A, Levine ME, Tanaka T, Lu AT, Chen BH, Ferrucci L, et al. , 2017. Epigenetic clock analysis of diet, exercise, education, and lifestyle factors. Aging (Albany NY) 9, 419–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team, 2013. R: A Language and Environment for Statistical Computing.
- Reding KW, Young MT, Szpiro AA, Han CJ, DeRoo LA, Weinberg C, et al. , 2015. Breast cancer risk in relation to ambient air pollution exposure at residences in the sister study cohort. Cancer Epidemiol. Biomark. Prev. 24, 1907–1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson PD, Richards M, Szpiro AA, Bergen S, Sheppard L, Larson TV, et al. , 2013. A regionalized national universal kriging model using partial least squares regression for estimating annual PM2. 5 concentrations in epidemiology. Atmos. Environ. 75, 383–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandler Dale P., et al. , 2017. The sister study cohort: baseline methods and participant characteristics. Environmental health perspectives 125.12, 127003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spada NJ, Hyslop NP, 2018. Comparison of elemental and organic carbon measurements between IMPROVE and CSN before and after method transitions. Atmos. Environ. 178, 173–180. [Google Scholar]
- Storey JD, Tibshirani R, 2003. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. 100, 9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward-Caviness CK, Nwanaji-Enwerem JC, Wolf K, Wahl S, Colicino E, Trevisi L, et al. , 2016. Long-term exposure to air pollution is associated with biological aging. Oncotarget 7, 74510–74525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White AJ, Kresovich JK, Xu Z, Sandler DP, Taylor JA, 2019. Shift work, DNA methylation and epigenetic age. Int. J. Epidemiol. dyz027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization, 2016. Ambient Air Pollution: A Global Assessment of Exposure and Burden of Disease.
- Xu Z, Niu L, Li L, Taylor JA, 2016. Enmix: a novel background correction method for Illumina HumanMethylation450 BeadChip. Nucleic Acids Res. 44, e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Sandler DP, Taylor JA, 2019. Blood DNA methylation and breast cancer: a prospective case-cohort analysis in the Sister Study. J. Natl. Cancer Inst. 112 (1), djz065 (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young MT, Bechle MJ, Sampson PD, Szpiro AA, Marshall JD, Sheppard L, et al. , 2016. Satellite-based NO2 and model validation in a national prediction model based on universal kriging and land-use regression. Environ. Sci. Technol. 50, 3686–3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.