Abstract
Clostridium difficile toxin A (TcdA) is a major exotoxin contributing to disruption of the colonic epithelium during C. difficile infection. TcdA contains a carbohydrate-binding CROPs (combined repetitive oligopeptides) domain that mediates its attachment to cell surfaces, but recent data suggest the existence of CROPs-independent receptors. Here we carried out genome-wide CRISPR-Cas9-mediated screens using a truncated TcdA lacking the CROPs and identified sulfated glycosaminoglycans (sGAGs) and low-density lipoprotein receptor (LDLR) as host factors contributing to binding and entry of TcdA. TcdA recognizes the sulfation group in sGAGs. Blocking sulfation and GAG synthesis reduces TcdA binding and entry into cells. Binding of TcdA to the colonic epithelium can be reduced by surfen, a small molecule that masks sGAGs, by GM-1111, a sulfated heparan sulfate analog, and by sulfated cyclodextrin, a sulfated small molecule. Cells lacking LDLR also show reduced sensitivity to TcdA, although binding between LDLR and TcdA are not detected, suggesting that LDLR may facilitate endocytosis of TcdA. Finally, GM-1111 reduces TcdA-induced fluid accumulation and tissue damage in the colon in a mouse model of injecting TcdA into the cecum. These data demonstrate in vivo and pathological relevance of TcdA-sGAGs interactions, and reveal a potential therapeutic approach of protecting colonic tissues by blocking TcdA-sGAGs interactions.
Introduction
C. difficile is a spore-forming opportunistic pathogen and one of the three “urgent threats” classified by the Centers for Disease Control and Prevention (CDC) of the United States. Disruption of gut flora by antibiotics allows C. difficile to colonize the colon, leading to diarrhea and life-threatening pseudomembranous colitis1. The occurrence of C. difficile infection (CDI) is exacerbated by the emergence of hyper-virulent and antibiotic-resistant strains2–4. It is now the most common cause of antibiotic-associated diarrhea and gastroenteritis-associated death in developed countries, accounting for a half million cases and ~29,000 deaths annually in the United States5.
Two homologous exotoxins, TcdA and TcdB, which target and disrupt the colonic epithelium, are the major virulent factors of C. difficile6–10. In addition, some hypervirulent strains also express the third toxin known as C. difficile transferase (CDT), which suppresses host eosinophilic responses11. TcdA (~308 kDa) and TcdB (~270 kDa) consist of four functional domains10,12: the N-terminal glucosyltransferase domain (GTD), a cysteine protease domain (CPD) that mediates auto-cleavage and releases the GTD into the host cytosol, a central part containing both the transmembrane delivery domain and receptor-binding domain, and finally a C-terminal CROPs (combined repetitive oligopeptides) domain. The GTD glucosylates small GTPases of the Rho family, including Rho, Rac, and CDC42, and inhibits their function, resulting in cytopathic cell-rounding and ultimately cell death.
The CROPs domains of TcdA and TcdB bear similarity with carbohydrate-binding proteins and may mediate toxin attachment to cell surfaces through various carbohydrate moieties. Particularly, CROPs from TcdA was shown to bind the trisaccharide Galα1,3Galβ1,4GlcNAc13. It has since been shown to also broadly recognize human I, Lewis X, and Lewis Y antigens, as well as glycosphingolipids, which all contain the Galβ1,4GlcNAc motif14,15.
Recent studies showed that truncating the CROPs only modestly reduces the potency of TcdA and TcdB on cultured cells, suggesting the existence of CROPs-independent receptors16,17. Three candidate receptors have been reported for TcdB: chondroitin sulfate proteoglycan 4 (CSPG4), poliovirus receptor-like 3 (PVRL3), and the Wnt receptor frizzled proteins (FZDs)18–21. Two proteins have been previously suggested as potential receptors for full-length TcdA: sucrase-isomaltase and glycoprotein 96 (Gp96)22,23. However, sucrase-isomaltase is not expressed in the colon epithelium and Gp96 resides mainly in the endoplasmic reticulum (ER).
Here we utilized a truncated TcdA lacking the majority of the CROPs domain and carried out genome-wide CRISPR-Cas9-mediated knockout (KO) screens, which identified sulfated glycosaminoglycans (sGAGs) and low-density lipoprotein receptor (LDLR) as CROPs-independent host factors mediating binding and entry of TcdA.
Results
CRISPR screens identify host factors for TcdA
To identify the CROPs-independent receptors involved in TcdA actions, we utilized a truncated TcdA (TcdA1–1874) lacking the majority of the CROPs domain (Supplementary Fig. 1a), which has been previously shown to retain high levels of toxicity on multiple cell lines17. We first validated the toxicity of TcdA1–1874 on various human cell lines using the standard cytopathic cell-rounding assay, which measures the percentages of rounded cells after incubation with a series of concentrations of toxins for 24 h (Supplementary Fig. 1b, c). The toxin concentration that induces 50% of cells to become round is defined as CR50, which is utilized to compare the sensitivity of different cell lines to TcdA1–1874. HeLa cells, which is one of the most sensitive human cell lines to TcdA1–1874, was selected to carry out genome-wide CRISPR-Cas9 mediated KO screens.
HeLa cells stably expressing Cas9 were transduced with a lentiviral sgRNA library (GeCKO v2) targeting 19,052 human genes24. The cells were subjected to three rounds of selection with TcdA1–1874 (40, 80, and 160 pM, Fig. 1a). The genes targeted by sgRNAs in surviving cells were identified via next-generation sequencing (NGS). We ranked the target genes based on the number of unique sgRNAs (y-axis) and the total NGS reads (x-axis) (Fig. 1b). All top ranked genes were enriched over the three rounds, suggesting that mutations in these genes offered survival advantages in the presence of TcdA1–1874 (Fig. 1c).
Figure 1. Genome-wide CRISPR-Cas9-mediated screen identifies host factors for TcdA.
a. Schematic illustration of the screen process utilizing TcdA1–1874 on HeLa cells. Round zero (R0) represents cells at the beginning of the screen. Round 1, 2, 3 (R1, R2, and R3) represent surviving cells after exposure to TcdA1–1874 sequentially at indicated toxin concentrations. NGS: next-generation sequencing.
b. Genes identified after R3 were ranked and plotted. The y axis is the number of unique sgRNA for each gene. The x axis represents the number of sgRNA reads for each gene. The top-ranking genes are color-coded and grouped based on their functions.
c. The NGS reads from R0 to R3 for the top-20 ranked (ordered by NGS reads) genes in R3 were color-coded and plotted. The diameter of the circle represents the number of unique sgRNA detected for the gene. All these top-20 ranked genes progressively enriched from R0 to R3.
The top-ranked gene encodes LDLR, a well-known receptor for low-density lipoproteins. Many other top ranked genes encode key players in heparan sulfate (HS) biosynthesis and sulfation pathways25, including the glycosyltransferases Exostosin-2 (EXT2) and Exostosin like-3 (EXTL3), sulfotransferases Heparan Sulfate 6-O-Sulfotransferase 1 (HS6ST1) and N-Deacetylase And N-Sulfotransferase 1 (NDST1), and Solute Carrier Family 35 Member B2 (SLC35B2), which transports the activated form of sulfate into Golgi. Several other enzymes involved in glycosaminoglycan (GAG) synthesis were also identified (Supplementary Fig. 2a). HS is usually attached to core proteins as heparan sulfate proteoglycans (HSPGs). Both HSPGs and LDLR are widely expressed on the surface of various cells, and thereby promising receptor candidates for TcdA.
Among the top 50 ranked genes, three (UGP2, PI4KB, and ATP6V0D1) were also found in the top list of genes in our previous genome-wide CRISPR-Cas9 screen using TcdB1–1830 (Supplementary Fig. 2b). UGP2 encodes UDP-glucose pyrophosphorylase, which synthesizes UDP-glucose, a co-factor required for TcdA and TcdB to glucosylate small GTPases26. ATP6V0D1 is a component of vacuolar-type H+-ATPase for acidification of endosomes, which is an essential condition to trigger translocation of TcdA and TcdB27,28. PI4KB is a key player in phospholipid metabolism/signaling and its role in toxin action remains to be established.
Other notable top hits include COG5, COG7, TMEM165, and RIC8A. COG5 and COG7 are members of the conserved oligomeric Golgi (COG) complex29. In fact, all eight COG members were identified in the final round of screening (Supplementary Fig. 2c). TMEM165 is a multi-pass transmembrane protein localized to the Golgi. Although the exact function of the COG complex and TMEM165 remains to be fully established, mutations in COG complex and TMEM165 both result in congenital disorders of glycosylation29,30, and affect multiple glycosylation pathways including biosynthesis of HS31–33. RIC8A is a guanine nucleotide exchange factor and its role in TcdA action remains to be validated.
We also performed a genome-wide CRISPR-Cas9-mediated KO screen using full-length TcdA in parallel on HeLa cells (Supplementary Fig. 2d). However, this screen only yielded UGP2 as the top hit. Two other hits, SGMS1 and ZNF283, are barely over our threshold. SGMS1 regulates lipid raft formation and may affect the endocytosis process. ZNF283 is a cytosolic protein, and its role in TcdA action remains to be validated. Lack of potential receptor candidates in the top hits suggests that full-length TcdA may utilize multiple receptors and entry pathways.
sGAGs contribute to cellular entry of TcdA1–1832
TcdA1–1874 still contains a short fragment of the CROPs domain, we further generated a truncated TcdA (TcdA1–1832) that deletes the entire CROPs in order to exclude any potential contribution from the residual CROPs domain (Supplementary Fig. 1a). TcdA1–1874 and TcdA1–1832 showed similar potency on HeLa cells in the cytopathic cell-rounding assays (Supplementary Fig. 1b).
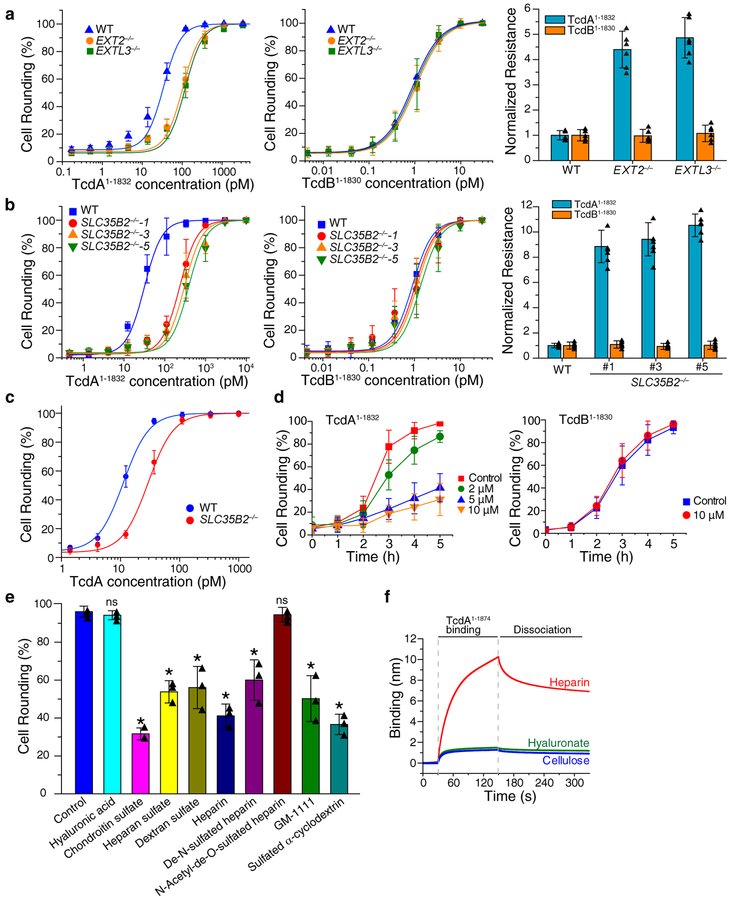
Using TcdA1–1832, we first validated the role of EXT2 and EXTL3, as they are specifically required for the elongation of the HS chain, but not other types of GAGs. We generated EXT2 and EXTL3 KO HeLa cell lines using the CRISPR-Cas9 system. Both cell lines showed a reduction of cell surface HS levels compared to wild type (WT) cells, measured by flow cytometry analysis using an HS antibody (Supplementary Fig. 3a). Both EXT2 and EXTL3 KO cells showed a modest 4 to 5-fold reduction in sensitivity to TcdA1–1832 compared with wild type (WT) cells, while their sensitivities toward TcdB1–1830 remained the same as WT cells (Fig. 2a).
Figure 2. sGAGs contribute to cellular entry of TcdA1–1832.
a. The sensitivities of EXT2−/− and EXTL3−/− HeLa cells to TcdA1–1832 (left panel) and TcdB1–1830 (middle panel) were quantified using the cytopathic cell-rounding assay. The percentage of rounded cells were quantified, plotted, and fitted. The toxin concentrations that resulted in 50% cell-rounding is defined as CR50 and is utilized for comparison by normalizing to the level of WT HeLa cells as normalized resistance (right panel, y axis, each data point was also shown as triangle in the bar graph). Centre values represent mean. Error bars represent ± s.d. (standard deviation).
b. The sensitivities of three SLC35B2−/− HeLa cell lines to TcdA1–1832 and TcdB1–1830 were quantified using the cytopathic cell-rounding assay and normalized to the level of WT HeLa cells. Each data point was also shown as triangle in the bar graph (right panel).
c. The sensitivities of HeLa WT and SLC35B2−/− cells (clone #5) to full-length TcdA were evaluated using the cytopathic cell-rounding assays. The percentage of rounded cells were quantified, plotted, and fitted.
d. Pre-incubation of surfen in the medium reduced the potency of TcdA1–1874 but not TcdB1–1830 in a concentration-dependent manner on HeLa cells, as measured by the cytopathic cell-rounding assay over time.
e. Competition assay on HeLa cells by pre-incubating TcdA1–1874 (2 nM) with the indicated GAGs, polysaccharides and synthetic sulfated molecules (all at 1 mg/mL). The degrees of protection were evaluated by the cytopathic cell-rounding assay 4 h later. (* p < 0.005, p > 0.05 is considered as non-significant (ns), two-side Student’s t test, n=3). Each data point was also shown as triangle in the bar graph.
f. TcdA1–1874 (1 μM) strongly bound to biotin-heparin but not to biotin-hyaluronate or biotin-cellulose, measured by BLI assays. Experiments were repeated three times.
For a-d, n=6, error bar represents mean ± s.d.. The experiments were repeated three times independently with similar results.
Several top-ranked genes identified in our screen, including SLC35B2, NDST, HS6ST, HS2ST, and HS3ST, are involved in sulfation of GAGs25 (Supplementary Fig. 2a). To examine the role of sulfation, we generated three single clones of SLC35B2 KO HeLa cells using the CRISPR-Cas9 approach. Reduction of HS in these cells was confirmed by flow cytometry analysis (Supplementary Fig. 3b). These cell lines all showed ~10-fold reduction in sensitivity toward TcdA1–1832 compared with WT cells, while their sensitivities toward TcdB1–1830 were not changed (Fig. 2b). The reduced sensitivity of SLC35B2 KO cells to TcdA1–1832 was further confirmed by immunoblot analysis detecting RAC1 glucosylation (Supplementary Fig. 4a). Finally, SLC35B2 KO cells also showed ~3-fold reduction in sensitivity to full-length TcdA (Fig. 2c).
Characterizing the specificity of TcdA-sGAGs interactions
We next carried out competition assays to further validate the role of sGAGs. We first utilized surfen (bis-2-methyl-4-amino-quinolyl-6-carbamide), which is a small molecule that binds to and neutralizes negative charges on all sGAGs34. Pre-incubation of cells with surfen protected HeLa cells from TcdA1–1832 in a concentration dependent manner, while it offered no protection from TcdB1–1830 (Fig. 2d, Supplementary Fig. 5a). Similar results were observed with Huh7 cells as well (Supplementary Fig. 5b).
To understand the selectivity of TcdA-GAG interactions, we next carried out competition assays using a panel of GAGs including HS, heparin, De-N-sulfated heparin, N-acetyl-de-O-sulfated heparin, chondroitin sulfate (CS), and dextran sulfate (DS). Heparin is a highly sulfated variant of HS and it is widely utilized as an anticoagulant. In addition, we also tested synthetic sulfated molecules GM-1111 and sulfated cyclodextrin. GM-1111 contains the same carbohydrate moieties and sulfation groups as HS, but with distinct glycosidic bonds. It has been developed as a HS mimic with reduced anti-coagulation activities35. Sulfated cyclodextrin is a small molecule distinct from GAGs. Non-sulfated GAG hyaluronic acid (HA) and polysaccharide cellulose were also examined. The schematic drawings of these molecules are illustrated in Supplementary Fig. 6.
Pre-incubation of TcdA1–1832 with HS, heparin, CS, DS, GM-1111, and sulfated cyclodextrin all reduced the level of cell-rounding, while HA showed no effect (Fig. 2e). These results suggest that TcdA may not recognize HS specifically, but rather interacts mainly with the sulfation group. Furthermore, the finding that De-N-sulfated heparin protected cells from TcdA1–1832, whereas N-Acetyl-de-O-sulfated heparin did not offer any protection (Fig. 2e), suggests that TcdA preferentially recognizes O-sulfation.
To further characterize direct TcdA-sGAG interactions, we utilized bio-layer interferometry (BLI) assay by immobilizing biotinylated heparin onto the probe. Binding of TcdA to the immobilized heparin would result in a shift in the light interference pattern that can be monitored in real-time. Biotinylated hyaluronate and cellulose were analyzed in parallel as controls. Both full-length TcdA and TcdA1–1874 showed robust binding to biotin-heparin, but not to biotin-hyaluronate and biotin-cellulose (Fig. 2f, Supplementary Fig. 7a). TcdA-heparin interactions appear to be influenced by ionic strength of the buffer: higher salt concentrations reduce heparin-TcdA interactions (Supplementary Fig. 7b). At 150 mM salt concentrations, the apparent dissociation constants (KD) for TcdA-heparin and TcdA1–1874-heparin are at similar levels (85.5 nM for TcdA1–1874 versus 23.2 nM for full-length TcdA, Supplementary Fig. 7c–e).
LDLR contributes to cellular entry of TcdA1–1832
To validate the role of LDLR, we generated LDLR KO HeLa cells using the CRISPR-Cas9 system. Three single KO clones were established and the loss of LDLR expression were confirmed by immunoblot analysis (Fig. 3a). All three KO lines showed reduced sensitivity by ~ 7-fold to TcdA1–1832, while their sensitivity to TcdB1–1830 remained the same as WT cells (Fig. 3b). The reduced sensitivity of LDLR KO cells to TcdA1–1832 was also confirmed by immunoblot analysis detecting RAC1 glucosylation (Supplementary Fig. 4b). LDLR−/− cells also showed ~3-fold reduction in sensitivity to full-length TcdA, thus validating the role of LDLR for full-length TcdA (Fig. 3c). The sensitivity to TcdA was restored when LDLR KO cells were transfected with mouse Ldlr (Fig. 3d), which is not targeted by the sgRNA. Furthermore, Huh7 LDLR−/− cells, which were previously generated and validated36, also showed reduced sensitivity to TcdA1–1832 compared to WT Huh7 cells (Supplementary Fig. 8).
Figure 3. LDLR contributes to cellular entry of TcdA1–1832.
a. The absence of LDLR expression in three LDLR−/− HeLa cell lines was confirmed by immunoblot analysis. Actin served as a loading control. The experiments were repeated three times independently with similar results.
b. The sensitivities of three LDLR−/− HeLa cell lines to TcdA1–1832 and TcdB1–1830 were quantified using the cytopathic cell-rounding assay and normalized to the levels of WT HeLa cells (n=6). Each data point was also shown as triangle in the bar graph.
c. The sensitivities of HeLa WT and LDLR−/− cells to full-length TcdA were evaluated using the cytopathic cell-rounding assays (n=6). The percentage of rounded cells were quantified, plotted, and fitted.
d. Ectopic expression of a mouse Ldlr in LDLR−/− (#4) cells restored the sensitivity of these cells to TcdA1–1832 and resulted in cell rounding under our assay conditions (2 nM, 4 h). Green fluorescent protein (GFP) was co-transfected to mark transfected cells. Representative fluorescence images of transfected cells were shown on the left side, while the percentage of rounded cells were quantified and shown on the right side (n=6). Each data point was also shown as triangle in the bar graph.
e. Pre-incubation of the ecto-domain of LDLR (residues 22–788, 400 nM) with TcdA1–1832 (2 nM, 4 h) protected HeLa cells from the toxin and prevented cell-rounding. The experiments were repeated three times independently with similar results.
f. Pre-incubation of RAP (4 μM) in culture medium further protected LDLR−/− (#4) cells from TcdA1–1832 (10 nM) as measured by the cell rounding assay over time (n=6).
For d and e, scale bar represents 50 μm.
Error bar represents mean ± s.d.. Experiments were repeated two times independently with similar results.
We further carried out a competition assay using the soluble extracellular domain of LDLR (residues 22–788, LDLR22–788). Co-incubation of LDLR22–788 with TcdA1–1832 (200:1) reduced the percentage of rounded cells (Fig. 3e). LDLR belongs to a large family of proteins including VLDLR, LRP1, LRP1b, LRP2 (Megalin), LRP5, LRP6, and LRP8 (ApoER2), which share similar domains with LDLR and often act as redundant receptors for many LDLR ligands. RAP (receptor associated protein) binds tightly to most LDLR family members and its binding inhibits binding of LDL and many other ligands37–39. Adding RAP to the medium further reduced the sensitivity of LDLR KO cells to TcdA1–1832 (Fig. 3f), suggesting that other LDLR family members also contribute to TcdA1–1832 entry into cells.
To examine binding of TcdA1–1874 to LDLR in vitro, we utilized purified Fc-tagged extracellular domain of LDLR (LDLR22–788-Fc) produced in HEK293 cells. This LDLR22–788-Fc mediated strong binding of RAP, but we did not detect direct binding of TcdA1–1874 to LDLR22–788-Fc in either BLI assays or an alternative dot blot assay (Supplementary Fig. 9). These results suggest that either TcdA1–1874 binding to LDLR is weak or their interactions may require additional cellular factors.
sGAGs are major cellular attachment factors for TcdA1–1874
To further understand the role of LDLR and sGAGs, we generated LDLR−/−/SLC35B2−/− double KO cell lines by knocking out LDLR from HeLa SLC35B2−/− cells using the CRISPR-Cas9 approach. Two single cell clones were established and lack of LDLR expression was confirmed by immunoblot (Fig. 4a). However, these two double-KO cell lines did not further increase the resistance to TcdA1–1832 compared with LDLR and SLC35B2 single KO cells (Fig. 4b, Supplementary Fig. 4c). Moreover, overexpression of exogenous mouse Ldlr by transient transfection did not increase the sensitivity of SLC35B2−/− cells to TcdA1–1832 (Fig. 4c). These data suggest that LDLR and sGAGs are not redundant receptors, and they could act cooperatively. We then examined binding of TcdA1–1874 to WT versus LDLR−/− and SLC35B2−/− HeLa cells, utilizing TcdA1–1874 directly labelled with a fluorescent dye. As shown in Fig. 4d, LDLR−/− cells showed similar overall TcdA1–1874 binding as WT cells. In contrast, binding of TcdA1–1874 to SLC35B2−/− cells was diminished. These results suggest that sGAGs are the major attachment factor mediating TcdA1–1874 binding on cell surfaces under our assay conditions.
Figure 4. sGAGs are major attachment factors for TcdA.
a. The absence of LDLR expression in two SLC35B2−/−/LDLR−/− HeLa cell lines was confirmed by immunoblot analysis. The experiments were repeated three times independently with similar results.
b. The sensitivities of two SLC35B2−/−/LDLR−/− HeLa cell lines and their parental cell line SLC35B2−/− (#5) to TcdA1–1832 were quantified using the cytopathic cell-rounding assay, and normalized to the level of WT HeLa cells.
c. Ectopic expression of a mouse Ldlr did not restore TcdA1–1832 (2 nM, 4 h) entry into SLC35B2−/− cells under our assay conditions. GFP marked transfected cells. Representative images were shown on the left side. Cell rounding was quantified and shown on the right side.
d. Immunofluorescence analysis showed that Alexa555-labelled TcdA1–1874 (5 nM) robustly bound to HeLa WT and LDLR−/− (#4) cells, but not SLC35B2−/− (#5) cells. Cell nuclei were labeled with Hoechst dye. DIC: differential interference contrast image. The experiments were repeated three times.
e. Co-injection of surfen (50 μM) with Alexa555-labelled TcdA1–1874 (5 nM, red) into ligated colon prevented TcdA1–1874 binding to the colonic epithelium. Cell nuclei were labeled with DAPI dye (blue). Ep: epithelial cells; SM: smooth muscles.
f. Co-injection of heparin, GM-1111, or sulfated cyclodextrin, but not HA (all at 1 mg/mL) with Alexa555-labelled TcdA1–1874 (5 nM) into the ligated colon reduced TcdA1–1874 binding to the colonic epithelium. Cell nuclei were labeled with DAPI dye (blue). Ep: epithelial cells; SM: smooth muscles.
For b and c, n = 6, n.s. (p > 0.05). Mann-Whitney Test (two-sided). For e and f, n = 3, binding of TcdA was quantified using ImageJ and statistical analysis is two-sided Student’s t test. Each data point was also shown as triangle in the bar graph. Centre values represent mean and error bars represent ± s.d..
sGAGs are attachment factors for TcdA1–1874 in the colonic epithelium
The colonic epithelium is the pathologically relevant target of TcdA. sGAGs are abundant both in the intestinal mucosa and on the basolateral side of the epithelium40–42. To examine the contribution of sGAGs to TcdA binding to the colonic epithelium, we utilized a colon loop ligation assay previously established20. Briefly, fluorescence-labelled TcdA1–1874 was injected into a ligated colon segment and incubated for 30 min. Colon tissues were then dissected and fixed. TcdA1–1874 showed strong binding to the apical side of the colonic epithelium and binding appears to extend into the lumen (Fig. 4e). Co-injecting surfen reduced binding of TcdA1–1874 (Fig. 4e). Similarly, heparin, GM-1111, and sulfated cyclodextrin all reduce binding of TcdA1–1874, whereas HA showed no effect (Fig. 4f). These results suggest that sGAGs are major attachment factors in the colon epithelium for TcdA1–1874.
Blocking sGAG-TcdA interactions reduces TcdA toxicity in the colon
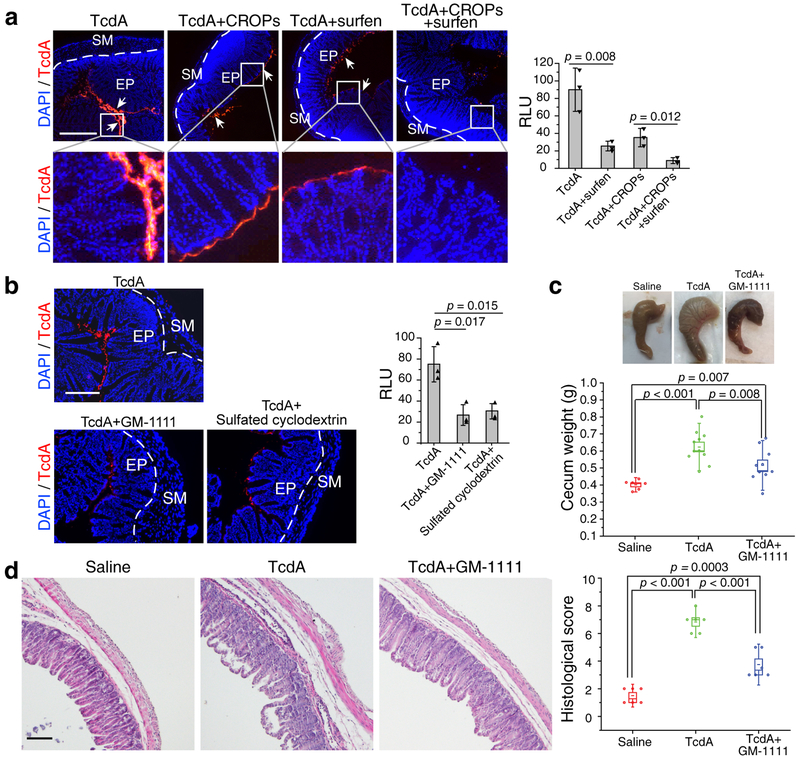
We next examined the contribution of sGAGs-mediated binding in the context of full-length TcdA in vivo. Injecting fluorescence-labelled full-length TcdA into the ligated colon loop for 30 min resulted in robust binding to the apical side of the colonic epithelium (Fig. 5a). Co-injecting recombinantly produced CROPs fragment reduced binding of TcdA, consistent with the finding that CROPs region mediates TcdA binding to cells43. Co-injecting surfen with TcdA reduced binding of TcdA, confirming that sGAGs contribute to binding of full-length TcdA to the colonic epithelium (Fig. 5a). Similarly, co-injection with GM-1111 or sulfated cyclodextrin reduced TcdA binding to the colonic epithelium as well (Fig. 5b). Interestingly, combining CROPs and surfen together largely abolished binding of TcdA to the colonic epithelium (Fig. 5a). Thus, both CROPs-mediated and sGAGs-mediated binding contributes to TcdA binding to the colonic epithelium.
Figure 5. Blocking sGAG-TcdA interactions reduces TcdA toxicity in the colon.
a. Co-injecting either surfen (50 μM) or TcdA CROPs (150 nM) with Alexa555-labelled full-length TcdA (5 nM) partially reduces TcdA binding to the colonic epithelium. Co-injecting both surfen and TcdA CROPs with TcdA largely abolished toxin binding. Representative images were shown on the left, and quantification of binding is shown in the right panel (n=3).
b. Co-injection of GM-1111 or sulfated cyclodextrin with TcdA both reduced TcdA binding to the colonic epithelium (n=3).
c. TcdA (4 μg), TcdA premixed with GM-1111(0.5 mg/ml), or saline was injected into the cecum of mice. After 6 h, the cecum tissue was excised. The representative cecum tissues were shown, and the weight of each cecum was measured and plotted. Boxes represent mean ± SEM; the bars represent s.d.; two-sided Student’s t test. N = 6 (saline), 11 (TcdA), or 10 (TcdA+GM-1111).
d. Cecum tissues from panel c were sectioned and subjected to H&E staining. The representative images are shown and the histological scores were assessed on the basis of disruption of the epithelia, hemorrhagic congestion, mucosal edema, and inflammatory cell infiltration. Boxes represent mean ± s.e.m; the bars represent s.d.; Student’s t test.
For panels a and b: binding of TcdA was quantified using ImageJ and statistical analysis is Student’s t test (two-sided, multiple comparisons). Centre values represent mean and error bars represent ± s.d.. Each data point was also shown as triangle in the bar graph.
To further examine the relevance of sGAG-TcdA interactions for TcdA-induced pathogenesis in vivo, we utilized a mouse cecum injection model previously established to assess pathogenesis of TcdA and TcdB44. Briefly, TcdA or TcdA pre-mixed with inhibitors was injected into the cecum. Mice were allowed to recover for 6 h before euthanization. The cecum plus the ascending colon was harvested and weighted to measure the degree of fluid accumulation. The cecum tissue was also fixed and subjected to hematoxylin and eosin (H&E) staining and histological score analysis based on four criteria (disruption of the epithelium, hemorrhagic congestion, mucosal edema, and inflammatory cell infiltration) on a scale of 0–3 (normal, mild, moderate, or severe). Injection of TcdA induced fluid accumulation in the colon tissues, severe mucosal edema, as well as mild to moderate disruption of the epithelium, hemorrhagic congestion, and inflammatory cell infiltration (Fig. 5c, d).
Finding a suitable inhibitor however was challenging as HS and many sGAG mimics induced hemorrhages in the intestine and colon after incubation for 6 h. This is likely due to their anticoagulation activity. Surfen alone at the concentration required to reduce TcdA binding also induced damage to colonic tissues after incubation for 6 h. After surveying many different sGAG mimics, we found that GM-1111, which is specifically developed to reduce the anticoagulation activity, can be utilized at the dose that reduces TcdA binding without inducing visible tissue damage by itself. Co-injecting GM-1111 with TcdA significantly reduced fluid accumulation in the colon (cecum weight, Fig. 5c) and overall tissue damage as evidenced by histological scoring (Fig. 5d).
Discussion
The presence of numerous negatively charged sulfate groups in sGAGs provides an ideal multi-valent landing pad for proteins and macromolecules through electrostatic interactions. They are known to interact with a large array of endogenous ligands, such as fibroblast growth factors (FGFs), vascular endothelial growth factor (VEGF), transforming growth factor β (TGFβ), chemokines, and cytokines45. Not surprisingly, these proteoglycans are also exploited by a long list of viral, bacterial, and parasitic pathogens as attachment factors46. As TcdA is capable of binding to isolated sGAGs, it should be able to bind to both proteoglycans containing sGAGs as well as free sGAGs on the cell surface and in the extracellular matrix. The exact binding sites for sGAGs in TcdA remain to be determined and it is possible that multiple positively charged surface regions of TcdA are involved.
LDLR belongs to a family of structurally related receptors and many of them act as redundant receptors for various ligands and viruses47. Interestingly, a LDLR family member LRP1 was previously established as the receptor for TpeL toxin39, which belongs to the same toxin family as TcdA but naturally lacks the CROPs domain. It is likely that LDLR family members other than LDLR can also contribute to TcdA1–1832 entry, as RAP further reduces the sensitivity of LDLR KO cells.
LDLR family of receptors are known to rapidly and constitutively recycle between cell membranes and endosomes. This provides an ideal portal to mediate endocytosis into cells. Indeed, LDLR has been exploited by many viruses as their receptors, such as vesicular stomatitis virus (VSV), hepatitis C virus, and the minor group common cold virus36,38,48. Although it remains unknown whether TcdA is capable of recognizing LDLR family members directly on cell surfaces, the major contribution of LDLR members is likely through facilitating endocytosis of TcdA bound to sGAGs. Similar synergistic actions between proteoglycan and LDLR family members are common for endogenous ligands. For instance, it was shown that HSPG contributes to the capture of PCSK9 onto cell surfaces and subsequently presents PCSK9 to LDLR49. Furthermore, many viruses that utilize HSPG as the initial attachment factor recruit additional protein receptors to mediate their endocytosis50. For instance, respiratory syncytial virus (RSV) utilizes HSPG as an attachment factor and ICAM1 and VLDLR as additional protein receptors50. Such a “two-step” model allows the pathogens and toxins to both maximize their chance of landing onto the cell surface, as well as taking advantage of rapid endocytosis and recycling of LDLR family members.
A combination of surfen and the CROPs domain protein largely abolished binding of full-length TcdA to the colonic epithelium, demonstrating that TcdA attaches to the colonic epithelium via at least two independent binding interfaces: interactions with sGAGs in a CROPs-independent manner and interactions with carbohydrate moieties by the CROPs. These results are consistent with the previous finding that TcdA1–1874 and TcdA1875–2710 do not compete with each other, while both can reduce binding of full-length TcdA to cells17. These results further support a previously proposed “two-receptor” model for TcdA10,16,39. Finally, GM-1111 alone reduced the toxicity of TcdA in the cecum injection model in vivo, demonstrating the therapeutic potential of protecting colonic tissues from TcdA by targeting TcdA-sGAGs interactions.
Methods
Materials.
HeLa (H1, #CRL-1958), HT-29 (#HTB-38), CHO-C6, and 293T (#CRL-3216) cells were originally obtained from ATCC. They tested negative for mycoplasma contamination, but have not been authenticated. Huh7 and Huh7 LDLR−/− cells were kindly provided by Y. Matsuura36. The following mouse monoclonal antibodies were purchased from the indicated vendors: RAC1 (23A8, Abcam), non-glucosylated RAC1 (Clone 102, BD Biosciences), β-actin (AC-15, Sigma), and HS (F58–10E4, mouse IgM, Amsbio). Rabbit monoclonal IgG against LDLR (EP1553Y) was purchased from Abcam. Chicken polyclonal IgY (#753A) against TcdA was purchased from List Biological Labs. Statistical analysis was performed using OriginPro 8 (V8.0724, OriginLab Corp.) software.
Protein purification.
Recombinant TcdA (from C. difficile strain VPI 10463), TcdA1–1874, TcdA1–1832, and CROPs (TcdA1875–2710) were cloned into modified pWH1520 vector, and TcdB1–1830 into pHIS1522 vector and expressed in Bacillus megaterium and purified as His6-tagged proteins. The expression plasmid pQTEV-LRPAP1 (#31327) encoding RAP was obtained from Addgene and RAP was purified as a His6-tagged protein. Genes encoding the ecto-domain of human LDLR (residues 22–788) and IgG1-Fc were fused and cloned into pHLsec vector (kindly provided by A. Jonathan). For the expression of Fc-tagged LDLR22–788, HEK293T cells were transfected with Lipofectamine™ 3000 (Invitrogen). Transfected cells were grown for 5 h, followed by replacing the culture medium with serum-free medium for 4 days. LDLR22–788-Fc in the culture medium was collected and purified.
Genome-wide CRISPR-Cas9 screening with TcdA1–1874.
The HeLa CRISPR genome-wide knockout library was generated as previously described20. In short, the GeCKO v2 library is composed of two sub-libraries. Each sub-library contains three unique sgRNA per gene and was independently prepared and screened. HeLa-Cas9 Cells were transduced with sgRNA lentiviral library at a MOI (multiplicity of infection) of 0.2. For each CRISPR sub-library, 7.9×107 cells were plated onto three 15-cm cell culture dishes to ensure sufficient sgRNA coverage, with each sgRNA being represented around 1200 times. These cells were exposed to TcdA1–1874 for 48 h. Cells were then washed three times to remove loosely attached cells. The remaining cells were cultured with toxin-free medium to ~70% confluence and subjected to the next round of screening with higher concentrations of toxins. Three rounds of screenings were performed with TcdA1–1874 (40, 80, and 160 pM). Remaining cells from each round were harvested and their genomic DNA extracted using the Blood and Cell Culture DNA mini kit (Qiagen). DNA fragments containing the sgRNA sequences were amplified by PCR using primers lentiGP-1_F (AATGGACTATCATATGCTTACCGTAACTTGAAAGTATTTCG) and lentiGP-3_R (ATGAATACTGCCATTTGTCTCAAGATCTAGTTACGC). Next-generation sequencing (Illumina MiSeq) was performed by a commercial vendor (Genewiz).
Generating HeLa KO cell lines.
To generate EXT2−/−, EXTL3−/− and LDLR−/− cells, the following sgRNA sequences were cloned into LentiGuide-Puro vectors (Addgene) to target the indicated genes: 5’-CGATTACCCACAGGTGCTAC-3’ (EXT2), 5’-GAGGTGAGCATCGTCATCAA-3’ (EXTL3), 5’-CCAGCTGGACCCCCACACGA-3’ (LDLR). HeLa-Cas9 cells were transduced with lentiviruses that expressing sgRNA. Mixed populations of infected cells were selected with puromycin (2.5 μg/ml). For LDLR knockouts, three single colonies were isolated (LDLR−/−-#4, LDLR−/−-#6 and LDLR−/−-#7). To generate SLC35B2−/− cells, a guide RNA targeting the exon 1 of SLC35B2 (5’-GCTTTATGGTACCTGGCTAC-3’) was cloned into lentiCRISPR v2-Blast (Addgene plasmid #83480). Lentivirus was generated by transfecting 293T cells with lentiCRISPR v2-Blast-SLC35B2sgRNA, pCD/NL-BH*DDD and pCAGGS-VSV-G. Hela-Cas9 cells were transduced with the lentivirus and selected with 10 μg/ml blasticidin. Three single colonies were isolated and validated (SLC35B2−/−-#1, SLC35B2−/−-#3 and SLC35B2−/−-#5). SLC35B2−/−/LDLR−/− double knockout cells were generated based on SLC35B2−/−-#5 by transducing lentiviruses that express LDLR sgRNA (5’-CCAGCTGGACCCCCACACGA-3’). Stable SLC35B2−/−/LDLR−/− cells were selected with puromycin (2.5 μg/ml) and hygromycin B (200 μg/ml). The deficiency of LDLR in LDLR−/− and SLC35B2−/−/LDLR−/− cells were validated by immunoblot.
FACS analysis.
Briefly, cells were collected with 1 mM EDTA in PBS and subsequently re-suspended in PBS with 1% BSA. Cells were incubated with either the 10E4 monoclonal antibody against HS (1:400), or mouse IgM (1:200; ab18401, Abcam, Cambridge, MA) for 1 h on ice. Cells were washed twice with PBS and incubated with goat anti-mouse IgG/IgM Alexa488 (1:1000; A10680, Molecular Probes, Eugene, OR) for 1 h on ice, washed twice, and followed by single-cell sorting using a FACS MoFlo Astrios EQ (Cell sorter-Beckman coulter, Indianapolis, IN). Data was analyzed using FlowJo software (FlowJo Inc, Ashland, OR).
Cytopathic cell rounding assay.
The cytopathic effect of TcdA and TcdB was analyzed using the standard cell-rounding assay. Briefly, cells were exposed to TcdA, TcdA1–1874, TcdA1–1832, or TcdB1–1830 for 24 h, and the phase-contrast images of cells were recorded (Olympus IX51, 10–20X objectives). A zone of 300×300 μm was selected randomly, which usually contains 50–150 cells. The numbers of normal shaped and round-shaped cells were counted manually. The percentage of round-shaped cells was analyzed using the Origin software.
Competition assays with GAGs or ecto-domain of LDLR.
TcdA1–1832 (2 nM) was pre-mixed with or without 1 mg/ml HS (Sigma, H7640), chondroitin sulfate (CS), dextran sulfate (DS, Sigma, D4911), hyaluronic acid (HA, Sigma, 53747), heparin (Fisher Bioreagents, BP252450), De-N-sulfated heparin (Carbosynth, YD58544), N-acetyl-de-O-sulfated heparin (Carbosynth, YD58545), sulfated cyclodextrin (Sigma-Aldrich, 494542–5G), GM-1111 (Glycomira Inc., Salt Lake City, UT), or 400 nM LDLR22–788 in fresh DMEM medium and incubated at 37 °C for 20 min. The mixture was then added to the cells. Cells were further incubated at 37 °C and the percentage of rounded cells over time were recorded and analyzed.
Competition assays with RAP or surfen.
The cells were pre-incubated with RAP or surfen in the medium at indicated concentrations at 37 °C for 20 min. 2 nM TcdA1–1832 was then added to the medium. Cells were further incubated at 37 °C and the percentage of rounded cells over time were recorded and analyzed.
Dot blot assay.
The indicated amounts of RAP, TcdA1–1832, and TcdB1–1830 were spotted onto a nitrocellulose membrane and allowed to dry in the air completely. The membrane was then blocked with 5% skimmed milk for 1 h at room temperature followed by overnight incubation with LDLR22–788-Fc at 4 °C. The bound LDLR22–788-Fc was detected with a monoclonal antibody against human Fc fragment. The experiments were repeated in triplicates.
Surface binding of TcdA1–1874 on HeLa cells.
TcdA and TcdA1–1874 were labeled using an Alexa555 antibody labeling kit (A20187, ThermoFisher Scientific) following manufacturer’s instruction. WT, SLC35B2−/−, or LDLR−/− HeLa H1-Cas9 cells were incubated with 5 nM Alexa555-labeled TcdA1–1874 in PBS for 30 min on ice. Cells were washed three times with ice-cold PBS, fixed with 4% paraformaldehyde. Cell nuclei were labeled with Hoechst dye. Confocal images were captured with the Ultraview Vox Spinning Disk Confocal System.
Biolayer interferometry (BLI) assay.
The binding affinities between TcdA1–1874 and heparin were measured by BLI assay using the Blitz system (ForteBio). Briefly, biotinylated heparin (20 μg/ml, B9806, Sigma-Aldrich), biotin-cellulose (Creative PEGWorks, CE501), or biotin-hyaluronate-biotin (Sigma, B1557) was immobilized onto capture biosensors (Dip and Read Streptavidin, ForteBio) and balanced with indicated buffers. The biosensors were then exposed to TcdA1–1874, followed by washing. Binding affinities (KD) were calculated using the Blitz system software (ForteBio). The experiments were repeated in triplicates.
Colon loop ligation assay.
All animal studies were conducted in accordance with ethical regulations under protocols approved by the Institute Animal Care and Use Committee (IACUC) at Boston Children’s Hospital (#3028). Statistical consideration was not used to determine the sample size of mice. Animals were distributed to each experimental group randomly. Experiments and data analysis were carried out without blinding. Colons from adult CD-1 mice (6–8 weeks, both male and female, from Envigo, NJ) were dissected out and incubated in PBS on ice. A ~2 cm loop in ascending colon was sealed with silk ligatures. 100 μl of Alexa555-labeled TcdA1–1874 or TcdA (5 nM, each) in PBS was injected through an intravenous catheter into the sealed colon segment with and without the TcdA1875–2710 (150 nM) and/or surfen (5 μM), GM-1111 (1 mg/mL), sulfated cyclodextrin (1 mg/mL). The colon segments were incubated on ice for 30 min, then cut open, washed with PBS, fixed with paraformaldehyde, and subjected to cryosectioning into sections of 10 μm thick. Confocal images were captured with the Ultraview Vox Spinning Disk Confocal System. Toxin binding was quantified using ImageJ software. The binding signal intensity was averaged based on the length of the epithelium. Three images were analyzed; the p-value was calculated by Student’s t-test.
Cecum injection assay.
Mice (CD1, 6–8 weeks, male and female, Envigo, NJ) were anesthetized with 3% Isoflurane after overnight fasting. A midline laparotomy was performed. TcdA (4 μg in 100 μl saline), TcdA premixed with GM-1111(4 μg TcdA + 0.5 mg/ml GM-1111), or saline was injected into the cecum through the ileocecal junction. The gut was then put back to the abdomen. The incision was closed with stitches and mice were allowed to recover. After 6 h, mice were euthanized and the cecum plus the ascending colon (~1.5 cm) was excised and weighed. The cecum tissue was fixed with 10% Phosphate Buffer Formalin and embedded in paraffin. Tissue sections were subjected to H&E staining for histological score analysis based on four criteria (disruption of the epithelium, hemorrhagic congestion, mucosal edema, and inflammatory cell infiltration) on a scale of 0–3 (normal, mild, moderate, or severe).
Data Availability
The data that support the findings of this study are available from the corresponding authors upon request.
Supplementary Material
Acknowledgments
We thank Y. Matsuura (Osaka University, Japan) and A. Jonathan (Harvard Medical School, U.S.) for providing cDNA and cell lines, H. Tatge (Hannover Medical School, Germany) for toxin purification, J. Savage (Glycomira Inc.) for providing GM-1111, C. Araneo (Harvard Medical School, U.S.) for assisting flow cytometry analysis. This study was partially supported by National Institute of Health (NIH) grants (R01NS080833, R01AI132387, R01AI139087, and R21NS106159 to M.D.). R.G. acknowledges support by the Federal State of Lower Saxony, Niedersächsisches Vorab (VWZN2889, VWZN3215, VWZN3266). M.D. and D.T.B. acknowledges support by the NIH-funded Harvard Digestive Disease Center (P30DK034854) and Boston Children’s Hospital Intellectual and Developmental Disabilities Research Center (P30HD18655). L.T. acknowledges support by the National Natural Science Foundation of China (Grant No. 31800128). M.D. holds the Investigator in the Pathogenesis of Infectious Disease award from the Burroughs Wellcome Fund.
Footnotes
Competing Interests
The authors declare no conflicts of interest.
References
- 1.Theriot CM & Young VB Interactions Between the Gastrointestinal Microbiome and Clostridium difficile. Annu Rev Microbiol 69, 445–461 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Collins J, Robinson C, Danhof H, Knetsch CW, van Leeuwen HC, Lawley TD, Auchtung JM & Britton RA Dietary trehalose enhances virulence of epidemic Clostridium difficile. Nature 553, 291–294 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McDonald LC, Killgore GE, Thompson A, Owens RC Jr., Kazakova SV, Sambol SP, Johnson S & Gerding DN An epidemic, toxin gene-variant strain of Clostridium difficile. N Engl J Med 353, 2433–2441 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Hunt JJ & Ballard JD Variations in virulence and molecular biology among emerging strains of Clostridium difficile. Microbiol Mol Biol Rev 77, 567–581 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lessa FC, Mu Y, Bamberg WM, Beldavs ZG, Dumyati GK, Dunn JR, Farley MM, Holzbauer SM, Meek JI, Phipps EC, Wilson LE, Winston LG, Cohen JA, Limbago BM, Fridkin SK, Gerding DN & McDonald LC Burden of Clostridium difficile infection in the United States. N Engl J Med 372, 825–834 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lyras D, O’Connor JR, Howarth PM, Sambol SP, Carter GP, Phumoonna T, Poon R, Adams V, Vedantam G, Johnson S, Gerding DN & Rood JI Toxin B is essential for virulence of Clostridium difficile. Nature 458, 1176–1179 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carter GP, Chakravorty A, Pham Nguyen TA, Mileto S, Schreiber F, Li L, Howarth P, Clare S, Cunningham B, Sambol SP, Cheknis A, Figueroa I, Johnson S, Gerding D, Rood JI, Dougan G, Lawley TD & Lyras D Defining the Roles of TcdA and TcdB in Localized Gastrointestinal Disease, Systemic Organ Damage, and the Host Response during Clostridium difficile Infections. MBio 6, e00551 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuehne SA, Cartman ST, Heap JT, Kelly ML, Cockayne A & Minton NP The role of toxin A and toxin B in Clostridium difficile infection. Nature 467, 711–713 (2010). [DOI] [PubMed] [Google Scholar]
- 9.Kuehne SA, Collery MM, Kelly ML, Cartman ST, Cockayne A & Minton NP Importance of toxin A, toxin B, and CDT in virulence of an epidemic Clostridium difficile strain. J Infect Dis 209, 83–86 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aktories K, Schwan C & Jank T Clostridium difficile Toxin Biology. Annu Rev Microbiol 71, 281–307 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Cowardin CA, Buonomo EL, Saleh MM, Wilson MG, Burgess SL, Kuehne SA, Schwan C, Eichhoff AM, Koch-Nolte F, Lyras D, Aktories K, Minton NP & Petri WA Jr. The binary toxin CDT enhances Clostridium difficile virulence by suppressing protective colonic eosinophilia. Nat Microbiol 1, 16108 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chumbler NM, Rutherford SA, Zhang Z, Farrow MA, Lisher JP, Farquhar E, Giedroc DP, Spiller BW, Melnyk RA & Lacy DB Crystal structure of Clostridium difficile toxin A. Nat Microbiol 1, 15002 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krivan HC, Clark GF, Smith DF & Wilkins TD Cell surface binding site for Clostridium difficile enterotoxin: evidence for a glycoconjugate containing the sequence Gal alpha 1–3Gal beta 1–4GlcNAc. Infect Immun 53, 573–581 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tucker KD & Wilkins TD Toxin A of Clostridium difficile binds to the human carbohydrate antigens I, X, and Y. Infect Immun 59, 73–78 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Teneberg S, Lonnroth I, Torres Lopez JF, Galili U, Halvarsson MO, Angstrom J & Karlsson KA Molecular mimicry in the recognition of glycosphingolipids by Gal alpha 3 Gal beta 4 GlcNAc beta-binding Clostridium difficile toxin A, human natural anti alpha-galactosyl IgG and the monoclonal antibody Gal-13: characterization of a binding-active human glycosphingolipid, non-identical with the animal receptor. Glycobiology 6, 599–609 (1996). [DOI] [PubMed] [Google Scholar]
- 16.Genisyuerek S, Papatheodorou P, Guttenberg G, Schubert R, Benz R & Aktories K Structural determinants for membrane insertion, pore formation and translocation of Clostridium difficile toxin B. Mol Microbiol 79, 1643–1654 (2011). [DOI] [PubMed] [Google Scholar]
- 17.Olling A, Goy S, Hoffmann F, Tatge H, Just I & Gerhard R The repetitive oligopeptide sequences modulate cytopathic potency but are not crucial for cellular uptake of Clostridium difficile toxin A. PLoS One 6, e17623 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yuan P, Zhang H, Cai C, Zhu S, Zhou Y, Yang X, He R, Li C, Guo S, Li S, Huang T, Perez-Cordon G, Feng H & Wei W Chondroitin sulfate proteoglycan 4 functions as the cellular receptor for Clostridium difficile toxin B. Cell Res 25, 157–168 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.LaFrance ME, Farrow MA, Chandrasekaran R, Sheng J, Rubin DH & Lacy DB Identification of an epithelial cell receptor responsible for Clostridium difficile TcdB-induced cytotoxicity. Proc Natl Acad Sci U S A 112, 7073–7078 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tao L, Zhang J, Meraner P, Tovaglieri A, Wu X, Gerhard R, Zhang X, Stallcup WB, Miao J, He X, Hurdle JG, Breault DT, Brass AL & Dong M Frizzled proteins are colonic epithelial receptors for C. difficile toxin B. Nature 538, 350–355 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen P, Tao L, Wang T, Zhang J, He A, Lam KH, Liu Z, He X, Perry K, Dong M & Jin R Structural basis for recognition of frizzled proteins by Clostridium difficile toxin B. Science 360, 664–669 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pothoulakis C, Gilbert RJ, Cladaras C, Castagliuolo I, Semenza G, Hitti Y, Montcrief JS, Linevsky J, Kelly CP, Nikulasson S, Desai HP, Wilkins TD & LaMont JT Rabbit sucrase-isomaltase contains a functional intestinal receptor for Clostridium difficile toxin A. J Clin Invest 98, 641–649 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Na X, Kim H, Moyer MP, Pothoulakis C & LaMont JT gp96 is a human colonocyte plasma membrane binding protein for Clostridium difficile toxin A. Infect Immun 76, 2862–2871 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanjana NE, Shalem O & Zhang F Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 11, 783–784 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kreuger J & Kjellen L Heparan sulfate biosynthesis: regulation and variability. J Histochem Cytochem 60, 898–907 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chaves-Olarte E, Florin I, Boquet P, Popoff M, von Eichel-Streiber C & Thelestam M UDP-glucose deficiency in a mutant cell line protects against glucosyltransferase toxins from Clostridium difficile and Clostridium sordellii. J Biol Chem 271, 6925–6932 (1996). [DOI] [PubMed] [Google Scholar]
- 27.Barth H, Pfeifer G, Hofmann F, Maier E, Benz R & Aktories K Low pH-induced formation of ion channels by clostridium difficile toxin B in target cells. J Biol Chem 276, 10670–10676 (2001). [DOI] [PubMed] [Google Scholar]
- 28.Qa’Dan M, Spyres LM & Ballard JD pH-induced conformational changes in Clostridium difficile toxin B. Infect Immun 68, 2470–2474 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith RD & Lupashin VV Role of the conserved oligomeric Golgi (COG) complex in protein glycosylation. Carbohydr Res 343, 2024–2031 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Foulquier F, Amyere M, Jaeken J, Zeevaert R, Schollen E, Race V, Bammens R, Morelle W, Rosnoblet C, Legrand D, Demaegd D, Buist N, Cheillan D, Guffon N, Morsomme P, Annaert W, Freeze HH, Van Schaftingen E, Vikkula M & Matthijs G TMEM165 deficiency causes a congenital disorder of glycosylation. Am J Hum Genet 91, 15–26 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jae LT, Raaben M, Riemersma M, van Beusekom E, Blomen VA, Velds A, Kerkhoven RM, Carette JE, Topaloglu H, Meinecke P, Wessels MW, Lefeber DJ, Whelan SP, van Bokhoven H & Brummelkamp TR Deciphering the glycosylome of dystroglycanopathies using haploid screens for lassa virus entry. Science 340, 479–483 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tanaka A, Tumkosit U, Nakamura S, Motooka D, Kishishita N, Priengprom T, Sa-Ngasang A, Kinoshita T, Takeda N & Maeda Y Genome-Wide Screening Uncovers the Significance of N-Sulfation of Heparan Sulfate as a Host Cell Factor for Chikungunya Virus Infection. J Virol 91(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tian S, Muneeruddin K, Choi MY, Tao L, Bhuiyan RH, Ohmi Y, Furukawa K, Boland S, Shaffer SA, Adam RM & Dong M Genome-wide CRISPR screens for Shiga toxins and ricin reveal Golgi proteins critical for glycosylation. PLoS Biol 16, e2006951 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schuksz M, Fuster MM, Brown JR, Crawford BE, Ditto DP, Lawrence R, Glass CA, Wang L, Tor Y & Esko JD Surfen, a small molecule antagonist of heparan sulfate. Proc Natl Acad Sci U S A 105, 13075–13080 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang J, Xu X, Rao NV, Argyle B, McCoard L, Rusho WJ, Kennedy TP, Prestwich GD & Krueger G Novel sulfated polysaccharides disrupt cathelicidins, inhibit RAGE and reduce cutaneous inflammation in a mouse model of rosacea. PLoS One 6, e16658 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamamoto S, Fukuhara T, Ono C, Uemura K, Kawachi Y, Shiokawa M, Mori H, Wada M, Shima R, Okamoto T, Hiraga N, Suzuki R, Chayama K, Wakita T & Matsuura Y Lipoprotein Receptors Redundantly Participate in Entry of Hepatitis C Virus. PLoS Pathog 12, e1005610 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fisher C, Beglova N & Blacklow SC Structure of an LDLR-RAP complex reveals a general mode for ligand recognition by lipoprotein receptors. Mol Cell 22, 277–283 (2006). [DOI] [PubMed] [Google Scholar]
- 38.Finkelshtein D, Werman A, Novick D, Barak S & Rubinstein M LDL receptor and its family members serve as the cellular receptors for vesicular stomatitis virus. Proc Natl Acad Sci U S A 110, 7306–7311 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schorch B, Song S, van Diemen FR, Bock HH, May P, Herz J, Brummelkamp TR, Papatheodorou P & Aktories K LRP1 is a receptor for Clostridium perfringens TpeL toxin indicating a two-receptor model of clostridial glycosylating toxins. Proc Natl Acad Sci U S A 111, 6431–6436 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Griffin CC, Linhardt RJ, Van Gorp CL, Toida T, Hileman RE, Schubert RL 2nd & Brown SE Isolation and characterization of heparan sulfate from crude porcine intestinal mucosal peptidoglycan heparin. Carbohydrate Research 276, 183–197 (1995). [DOI] [PubMed] [Google Scholar]
- 41.Bode L, Salvestrini C, Park PW, Li JP, Esko JD, Yamaguchi Y, Murch S & Freeze HH Heparan sulfate and syndecan-1 are essential in maintaining murine and human intestinal epithelial barrier function. Journal of Clinical Investigation 118, 229–238 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamamoto S, Nakase H, Matsuura M, Honzawa Y, Matsumura K, Uza N, Yamaguchi Y, Mizoguchi E & Chiba T Heparan sulfate on intestinal epithelial cells plays a critical role in intestinal crypt homeostasis via Wnt/beta-catenin signaling. Am J Physiol Gastrointest Liver Physiol 305, G241–249 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sauerborn M, Leukel P & von Eichel-Streiber C The C-terminal ligand-binding domain of Clostridium difficile toxin A (TcdA) abrogates TcdA-specific binding to cells and prevents mouse lethality. FEMS Microbiol Lett 155, 45–54 (1997). [DOI] [PubMed] [Google Scholar]
- 44.Zhang Y, Yang Z, Gao S, Hamza T, Yfantis HG, Lipsky M & Feng H The role of purified Clostridium difficile glucosylating toxins in disease pathogenesis utilizing a murine cecum injection model. Anaerobe 48, 249–256 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lindahl U, Couchman J, Kimata K & Esko JD Proteoglycans and Sulfated Glycosaminoglycans. 207–221 (2015). [Google Scholar]
- 46.Kamhi E, Joo EJ, Dordick JS & Linhardt RJ Glycosaminoglycans in infectious disease. Biol Rev Camb Philos Soc 88, 928–943 (2013). [DOI] [PubMed] [Google Scholar]
- 47.Jeon H & Blacklow SC Structure and physiologic function of the low-density lipoprotein receptor. Annu Rev Biochem 74, 535–562 (2005). [DOI] [PubMed] [Google Scholar]
- 48.Agnello V, Abel G, Elfahal M, Knight GB & Zhang QX Hepatitis C virus and other flaviviridae viruses enter cells via low density lipoprotein receptor. Proc Natl Acad Sci U S A 96, 12766–12771 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gustafsen C, Olsen D, Vilstrup J, Lund S, Reinhardt A, Wellner N, Larsen T, Andersen CBF, Weyer K, Li JP, Seeberger PH, Thirup S, Madsen P & Glerup S Heparan sulfate proteoglycans present PCSK9 to the LDL receptor. Nat Commun 8, 503 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bomsel M & Alfsen A Entry of viruses through the epithelial barrier: pathogenic trickery. Nat Rev Mol Cell Biol 4, 57–68 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding authors upon request.