Abstract
Purpose:
A number of institutions have clinically implemented CYP2D6 genotyping to guide drug prescribing. We compared implementation strategies of early adopters of CYP2D6 testing, barriers faced by both early adopters and institutions in the process of implementing CYP2D6 testing, and approaches taken to overcome these barriers.
Methods:
We surveyed eight early adopters of CYP2D6 genotyping and eight institutions in the process of adoption. Data were collected on testing approaches, return of results procedures, applications of genotype results, challenges faced, and lessons learned.
Results:
Among early adopters, CYP2D6 testing was most commonly ordered to assist with opioid and antidepressant prescribing. Key differences among programs included test ordering and genotyping approaches, result reporting, and clinical decision support. However, all sites tested for copy number variation and 9 common variants, and reported results in the medical record. Most sites provided automatic consultation and had designated personnel to assist with genotype-informed therapy recommendations. Primary challenges were related to stakeholder support, CYP2D6 gene complexity, phenotype assignment, and sustainability.
Conclusion:
There are specific challenges unique to CYP2D6 testing given the complexity of the gene and its relevance to multiple medications. Consensus lessons learned may guide those interested in pursuing similar clinical pharmacogenetic programs.
Keywords: pharmacogenetics, implementation, CYP2D6, opioids, antidepressants
INTRODUCTION
The cytochrome P450 2D6 (CYP2D6) enzyme metabolizes approximately 25% of prescription drugs and nearly 40% of drugs frequently cited in adverse drug reaction studies1. The CYP2D6 enzyme is the primary metabolic enzyme for select opioids, antidepressants, anti-emetics, and tamoxifen. The CYP2D6 gene is highly polymorphic, which can have important consequences for drug safety and effectiveness.
The CYP2D6 gene has over 100 allelic variants and subvariants, which introduces a high degree of complexity for gene assays and interpretation2–4. Approximately 5 to 10 percent of individuals inherit two nonfunctional CYP2D6 alleles (e.g. *3, *4, *5) and are poor metabolizers (PMs) with no enzyme activity3. Two to 11 percent are intermediate metabolizers (IMs) with a combination of nonfunctional and decreased function (e.g. *41, *17) alleles and significant reductions in enzyme activity. An additional 1 to 2 percent are ultra-rapid metabolizers (UMs) with multiple CYP2D6 gene copies (e.g. *1/*1×2) and increased enzyme activity.
The consequences of CYP2D6 variation on drug response vary depending on whether the enzyme biotransforms the parent drug into a more active or less active metabolite. The prodrugs codeine, tramadol, and tamoxifen are biotransformed via CYP2D6 into metabolites with greater pharmacologic activity than their parent compounds. Compared to normal metabolizers (NMs), PMs and IMs have lower concentrations of the more potent metabolites and may fail to receive therapeutic benefit from these medications5–10. On the other hand, UMs produce higher concentrations of the more potent metabolites of codeine and tramadol compared to NMs 11,12, which increases risk for serious adverse events including respiratory depression and death secondary to toxic concentrations of more potent metabolites13,14.
The implications of CYP2D6 polymorphisms are different when CYP2D6 biotransforms drugs to less active metabolites, as with some selective serotonin reuptake inhibitors (SSRIs), tricyclic antidepressants, ondansetron, and tropisetron. Poor metabolizers treated with paroxetine, fluvoxamine, and tricyclic antidepressants are at increased risk for adverse drug effects secondary to higher plasma concentrations of the active parent drugs, whereas UMs are at risk for subtherapeutic concentrations of the parent drug and therapeutic failure15,16. While there is insufficient evidence on the consequences of reduced metabolism for ondansetron and tropisetron, reduced anti-emetic effects have been observed in UMs17–19.
Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines are available for each of the drugs discussed above3,15–17,20. Many institutions within the NIH-funded Implementing GeNomics In pracTicE (IGNITE) Network have clinically implemented, or are in the process of implementing, CYP2D6 genotyping to assist with drug prescribing21. We previously described experiences and challenges with clinical implementation of CYP2C19 genotype-guided antiplatelet therapy22. Given CYP2D6 gene complexity and relevance for multiple medications, it may present unique implementation challenges. The purpose of this paper is to compare goals and strategies for operationalizing CYP2D6 genotype-guided therapy among early test adopters and summarize challenges faced by both early adopters and those in the process of adoption and approaches to overcome key implementation barriers.
METHODS
Institutions within the IGNITE Network Pharmacogenetics Working Group, a collaboration of pharmacogenetic researchers and implementers who share data and real‐world experiences to advance genomic medicine, were invited to participate. All participating institutions had implemented CYP2D6 testing into practice or were in the process of implementation.
An existing data collection tool was adapted to capture experiences of sites implementing CYP2D6 genotyping22. Data elements included the testing landscape, stakeholder involvement, testing application, genotyping approaches, return of results procedures, clinical decision support (CDS), education strategies, challenges faced and lessons learned. A second tool was designed to collect data on the testing landscape, planned test applications, and initial challenges from sites in the process of establishing CYP2D6 testing. Data elements were selected and refined through discussions at in-person group meetings and teleconferences from September 2017 to December 2017 for the initial tool and January 2018 to May 2018 for the second tool. Challenges faced across sites and recommendations for overcoming challenges were aggregated to a consensus list through teleconference discussions. This research was approved as exempt by the University of Florida Institutional Review Board.
RESULTS
Early adopters of CYP2D6 testing
Institutional landscape and CYP2D6 test implementation planning
As of July 2018, 8 institutions in the IGNITE Network had implemented CYP2D6 genotyping and contributed information on implementation strategies (Table 1). Two institutions (Cincinnati Children’s Hospital Medical Center [CCHMC] and Indiana University School of Medicine [IU]) launched CYP2D6 testing in conjunction with testing for multiple other genes. All others had previously implemented testing for other genes – most commonly for CYP2C19 to guide antiplatelet therapy after coronary intervention and TPMT to guide thiopurine dosing. Four of the programs were solely clinical implementations, while 4 included research components. At 7 institutions, a formal precision medicine team, usually including a pharmacist, health informatics expert(s), and physician stakeholders, led the design and initiation of CYP2D6 testing and provided program oversight (Figure 1A, Table S1). An individual physician champion specializing in psychiatry led the initiative at the University of Alabama at Birmingham (UAB).
Table 1.
Landscape of early adopters of CYP2D6 testing and implementation design
| Institution name | Institution type | Previous gene-drug pairs implemented into practice | CYP2D6 Testing | |
|---|---|---|---|---|
| Year of launch | Model | |||
| Cincinnati Children’s Hospital Medical Center | Nonprofit academic | CYP2C19-select SSRIs, TCAs; CYP2C9-warfarin; and TPMT-thiopurinesa | 2004 | Clinical |
| Indiana University | Academic | CYP2C19-clopidogrel, proton pump inhibitors, SSRIs, voriconazole; TPMT-thiopurines; IFNL3-peg-interferon; SLCO1B1-simvastin; CYP2C9/VKORC1/CYP4F2-warfarin; G6PD-rasburicase; CYP3A4/CYP3A5-tacrolimus; CYP2C9-phenytoin; CYP2D6-codeine, SSRIs, atomoxetine, tramadol; CYP2B6-efavirez; ITPA-thiopurines; DPYD-fluoropyrimidinesa | 2014 | Clinical and Research |
| Sanford Health | Integrated Health System; Not-for-profit | CYP2C19-clopidogrel; TPMT-thiopurines; SLCO1B1-simvastatin; CYP2C9/VKORC1-warfarin | 2014 | Clinical |
| University of Florida Health | Academic | CYP2C19-clopidogrel; TPMT-thiopurines; IFNL3-peg-interferon | 2015 | Clinical and Research |
| Mission Health | Community Health System | HLA-B*15:02-carbamazepine; TPMT-thiopurines | 2015 | Clinical and Research |
| Mount Sinai Hospital | Academic | CYP2C19-clopidogrel, escitalopram, citalopram, sertraline; SLCO1B1-simvastatin; CYP2C9/VKORC1-warfarin | 2016 | Research |
| University of Alabama, Birmingham | Academic | CYP2C19-clopidogrel | 2016 | Clinical |
| Vanderbilt University Medical Center | Academic | CYP2C19-clopidogrel; TPMT-thiopurines; SLCO1B1-simvastatin; CYP2C9/VKORC1-warfarin; CYP3A5-tacrolimus | 2017 | Clinical |
CYP2D6 testing implemented at the same time as testing for other gene-drug pairs
CYP, cytochrome P450; DPYD, dihydropyrimidine dehydrogenase; FCAMR, Fc Fragment Of IgA And IgM Receptor; G6PD, glucose-6-phosphate dehydrogenase; HLA-B, human leucocyte antigen subtype B; IFNL3, interferon lambda 3; ITPA, inosine triphosphate pyrophosphatase; RARG, retinoic acid receptor gamma; SLCO1B1, solute carrier organic anion transporter family member 1B1; SSRIs, selective serotonin receptor inhibitors; SV2C, synaptic vesicle glycoprotein 2A; TCAs, tricyclic antidepressants; TPMT, thiopurine methyltransferase; VKORC1, vitamin K oxidoreductase complex 1
Figure 1A.
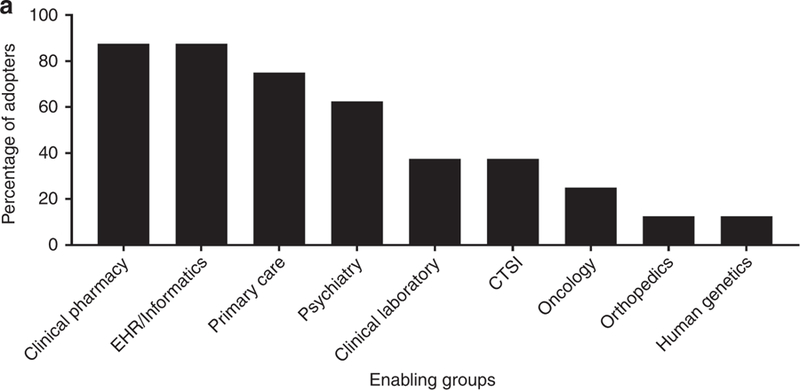
Groups enabling CYP2D6 genotype implementation at 8 early adopters of testing.
EHR, electronic health records; CTSI, Clinical Translational Science Institute
Clinical applications of CYP2D6 testing and genotyped populations
Most institutions reported multiple indications for which CYP2D6 genotyping was ordered (Figure 1B, Table S1), with antidepressant and opioid prescribing being the most common. Other indications included prescribing of antiemetic and antipsychotic agents. Three sites reported limited instances of CYP2D6 testing to assist with tamoxifen prescribing.
Figure 1B.
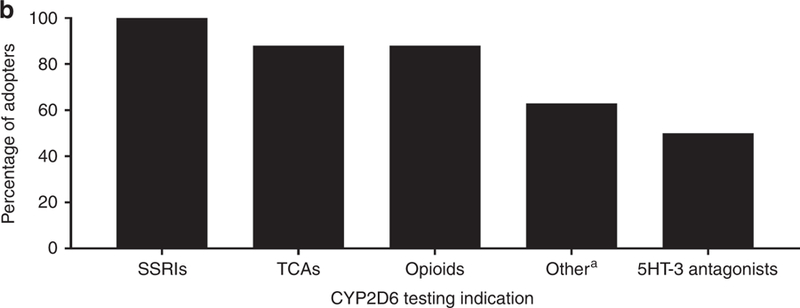
Target drugs for CYP2D6 genotyping among 8 early adopters.
SSRI, selective serotonin reuptake inhibitor; TCA, tricyclic antidepressant
*aripiprazole, haloperidol, olanzapine, perphenazine, risperidone, thioridazine, venlafaxine, atomoxetine
Most institutions used both reactive (e.g., test ordered at time of drug prescribing) and pre-emptive (e.g., test ordered to assist with future prescribing) genotyping models (Table S1). At IU, the CYP2D6 test is part of a multi-gene panel ordered for patients prescribed any drug with CPIC Level A evidence23. In this regard, if the prescription triggering the genotype order was for a CYP2D6 substrate drug (e.g. opiate, SSRI), CYP2D6 genotyping would be reactive. Otherwise, CYP2D6 testing would be done pre-emptively as part of the panel to inform future prescribing of CYP2D6 substrates. Similar models were in place at Vanderbilt University Medical Center (VUMC) and Sanford Health. Other pre-emptive models included testing of patients with depressive symptoms to assist with initial prescribing of antidepressants or antipsychotics, testing patients pre-operatively to assist with opioid prescribing, and testing for primary care patients as part of a pilot pharmacogenetic implementation project. One site had a reactive only testing model where genotyping was primarily reserved for patients with depression not responding to current therapy.
Genetic testing
The process for CYP2D6 test ordering varied among programs (Table S2). Prescribers electively placed genotype orders at 7 of 8 sites. Three of these sites also ordered clinical genotyping for participants in pharmacogenetic studies. At one site, CYP2D6 genotyping was preselected on the order set for patients admitted to a psychiatric unit as part of routine care.
Six sites performed testing onsite in a College of American Pathologists (CAP)/Clinical Laboratory Improvement Act (CLIA)-certified laboratory (Table S2). Deciding factors in choosing in-house testing (versus outsourcing) included having existing expertise and infrastructure for clinical genotyping, desire to have the ability to alter variants tested as evidence evolves, and in the case of very early adopters, the limited availability of commercial testing when the program began. While genotyping platforms varied across these sites, all tested and reported allele-defining variants for *2, *3, *4, *6, *9, *10, *17, and *41, at minimum, in addition to gene deletion (*5) and copy number variation (Figure 2). Four institutions genotyped CYP2D6 as part of a multi-gene panel. Factors considered in genotyping platform selection were workflow; turn-around time; ability to batch samples, detect full gene deletion and duplication, and customize variants tested; capability for multiplexed targeted genotyping; and FDA clearance. Four sites had validated assays for blood and either buccal cell or saliva samples. Genotype test turnaround time ranged from 2 to 14 business days across sites.
Figure 2.
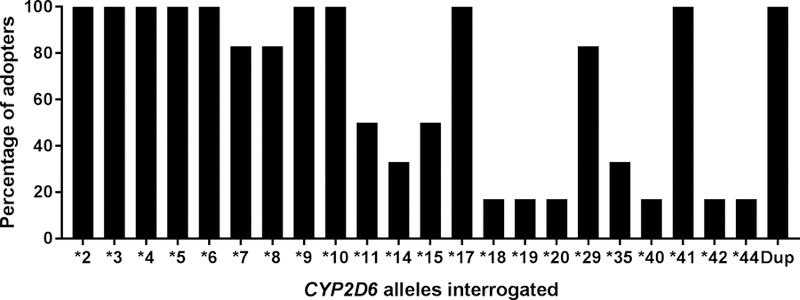
CYP2D6 variations tested across sites
Dup, duplications
Two sites relied solely on commercial or reference laboratories for genotyping. Two sites with in-house genotyping capabilities also reported use of commercial laboratories in select cases, such as when patients preferred noninvasive sample collection and in-house genotyping was validated for blood samples only or when patient payment assistance, available through some commercial laboratories, was desired. Factors influencing the choice of outside laboratory included which genes were tested besides CYP2D6, cost of testing, characteristics of patient assistance programs, and provider knowledge/awareness of a specific laboratory.
Phenotype assignment
All but one site reported CYP2D6 phenotype assignment based on activity scores as described in the current CPIC guidelines3; IU classified patients with one no-function and one normal function allele (e.g. *1/*4) as NM for codeine, per CPIC guidelines, and as IM for tramadol, per Dutch Pharmacogenetic Working Group guidelines24. Three sites reported that either the laboratory report or patient-specific clinical consult note mentioned of use of CYP2D6 inhibitors that can cause phenoconversion (i.e. change the phenotype from that predicted based on genotype alone).
Communication of results, approach to therapy modification, and education strategies
All institutions reported CYP2D6 test results in the EHR, usually within the “laboratory results” section (Table S3). Six sites reported both CYP2D6 genotype and predicted phenotype (metabolizer status) in the laboratory report, and two sites (CCHMC and IU) additionally provided genotype-specific drug therapy recommendations. All but one institution had a designated service or pharmacist available to provide genotype-informed drug therapy recommendations, and 4 institutions provided an automatic consultation via personal communication or an electronic consult note. Five of 8 sites provided downstream alerting or electronic CDS triggered with future drug orders within the EHR, most commonly with codeine, tramadol, and antidepressants. A sixth site was in the process of building electronic CDS.
Five programs provided patients with their test results via a patient web portal. Most programs provided education to patients about CYP2D6 testing through disseminated brochures, pamphlets, or other literature, or in-person education. Authors reported that providers were most engaged when education was delivered through focused discussions with providers who prescribe the target drugs, particularly when directly related to a patient case, and in-services to specialty groups.
Institutions in the process of implementing CYP2D6 testing into practice
Eight institutions were in the process of CYP2D6 implementation (Table S4). All but one had previously implemented testing of other genes, with CYP2C19 being most common. Precision medicine “teams” or “steering committees” were the drivers of these efforts at most sites. Four institutions were in the process of engaging stakeholders and leadership support. Development of informatics-related support and resources and selection or validation of the genotyping platform were also commonly identified as a current step in the implementation process. Nearly all locations were weighing considerations related to panel (multi-gene) versus single gene testing and whether to conduct CYP2D6 testing internally versus through a reference laboratory. All but two institutions were planning to implement CYP2D6 testing as part of clinical practice (as opposed to a research protocol). Varying therapeutic areas were identified for implementation, although pain management and psychiatry were specifically noted by 6 and 5 institutions, respectively. Institutions anticipated deploying CYP2D6 testing in settings ranging from targeted populations and specific clinical care locations to a larger, institution-wide general patient population.
Implementation challenges and lessons learned
Table 2 summarizes challenges reported and lessons learned across sites. A major challenge was the inability to determine which allele was duplicated or multiplicated for heterozygous genotypes with copy number variation (CNV), which often led to the inability to unambiguously assign phenotype. Further challenges were variable phenotype assignment based on the CYP2D6 substrate, and the potential for phenoconversion with concomitant CYP2D6 inhibitor use. An additional challenge reported by sites in the process of adopting testing was related to potential risk management issues in the event that appropriate action was not taken in response to genotype results. Sites reported that providers were especially concerned given the multiple drug substrates for CYP2D6 whose response may be impacted by genotype. This concern was not shared by sites that had already adopted testing, though they reported it was discussed prior to implementation.
Table 2.
Primary implementation challenges and lessons learned
| Challenges | Lessons learned |
|---|---|
| Obtaining provider/stakeholder buy-in | |
| • Limited provider exposure to pharmacogenetics and awareness about genotyping availability. • Limited data on clinical utility of CYP2D6 genotype-guided drug therapy. |
• Having broad, multidisciplinary program buy-in, including a physician champion, facilitates implementation. • Important to educate providers about testing availability and applications to prescribing decisions and to engage physicians when building CDS language. |
| Sample collection and CYP2D6 genotyping | |
| • Multiple single nucleotide variants, gene deletion, duplication, and multiplication define alleles • Difficult to quantitate duplication/multiplication or determine which allele is duplicated/multiplicated in heterozygotes |
• Noninvasive sample collection method facilitates testing for children and when no phlebotomy services are available on site. • Saliva and buccal cell samples can give inconsistent copy number results • CYP2D6 genotype expertise important for establishing genotyping procedures and interpreting results. • Platform must be able to detect gene deletion and copy number variation, with a process should be in place for indeterminate genotypes due to copy number variation |
| Genotype reporting and phenotype assignment | |
| • Large number of possible CYP2D6 diplotypes complicates automation of EHR processes. • Phenotype assignment may vary depending on drug substrate.a • Moderate to strong CYP2D6 inhibitors can cause phenoconversion • EHR changes may interfere with functioning of electronic CDS. |
• Genotypes should be reported in a consistent location in the EHR. • Entering genotype results as discrete data enables genotype specific electronic CDS. • Designated personnel and/or electronic CDS are important to assist with integrating genotype results into prescribing decisions. • Need CDS system that assigns phenotype based on drug substrate and accounts for phenoconversion • Quality control procedures are important for ensuring electronic CDS maintained with EHR updates |
| Sustainability | |
| • Building a reimbursement model • Updates to genotype reports and automated CDS needed as evidence evolves to support genotype-guided prescribing for additional medications. |
• Clinical outcomes and cost-effectiveness data may support reimbursement and additional stakeholder buy-in. • Personnel need to stay abreast of scientific developments in the field and update reports and decision support as needed. |
| Personnel issuesb | |
| • Having sufficient personnel to direct and support implementation | • Create partnerships with pharmacists or other clinicians on clinical teams through a decentralized model |
| Risk management issuesb | |
| • Concern if there is an actionable variant and no one acts on it | • While risk management issues may be part of the discussion prior to implementation, they were no longer voiced as a concern post-implementation |
Combination of one normal function allele plus one no function allele may be assigned the intermediate metabolizer phenotype for tamoxifen, but the normal metabolizer phenotype for codeine, tricyclic antidepressants, and select selective serotonin reuptake inhibitors).14,15,19
Challenges specific to institutions in the process of implementing
CDS, clinical decision support; EHR, electronic health record
Choosing genotyping methodology that detects copy number variation was felt to be critical in order to identify UMs (due to gene duplication) and some IMs (due to gene deletion). Recognizing that most genotype platforms at present cannot detect which allele is duplicated/multiplicated, sites agreed that a process must be in place for assigning phenotype for heterozygous genotypes with CNV to allow for appropriate reporting and downstream CDS25,26. Most sites used a ranged phenotype (e.g. NM to UM) when this occurred. Also ranked as important was having multidisciplinary stakeholder involvement and in particular, a physician champion, to facilitate implementation and additional stakeholder buy-in. It is in the authors’ consensus opinion, that while not required for implementation, in an ideal future state where feasible to do so, the following would also be in place to support the provision of CYP2D6 genotype-guided therapies: 1) noninvasive methods for DNA sample collection, especially for pediatric patients; 2) assay methods capable of assessing which allele was duplicated or multiplicated; 3) rapid or pre-emptive testing to minimize or eliminate delays in prescribing decisions; 4) CYP2D6 result (genotype and phenotype) reporting in an easily accessible location in the EHR, preferably one for lifetime results, and as discrete data to enable automated CDS; 5) automated CDS that accounts for phenoconversion secondary to use of CYP2D6 inhibitors; and 6) a mechanism for patients and providers to access genotype results throughout the patient’s life.
DISCUSSION
In surveying early adopters of CYP2D6 genotyping to guide prescribing decisions, we found multiple approaches to implementation, even within the same institution, with sites reporting use of both reactive and preemptive genotyping models, single and multi-gene testing, and clinical- and research-based deployment, much like in our previous report of implementation of CYP2C19-guided antiplatelet therapy22. Other similarities between implementing CYP2D6-guided therapies and CYP2C19-guided antiplatelet therapy22 included the role of a physician champion working alongside clinicians who ordered and applied test results clinically and use of a multi-disciplinary team approach, including pharmacists and health informatics experts, to lead implementation. While CYP2D6 testing was most commonly ordered to assist with antidepressant and opioid prescribing, most institutions applied genotype results to multiple therapies. In addition, most sites had electronic CDS tools in place or under development and designated personnel to assist with interpretation and translation of genotype results to inform prescribing decisions. Provider education and building evidence to support the utility and cost effectiveness of testing were recognized as keys to driving greater stakeholder engagement.
There were also notable differences between CYP2D6 testing and CYP2C19-guided antiplatelet therapy22. Whereas CYP2C19-guided antiplatelet therapy was most often the first pharmacogenetic implementation, CYP2D6 genotyping was typically a subsequent deployment or only implemented as part of a larger, more comprehensive pharmacogenetics program. This may be reflective of the relative complexity of the CYP2D6 genotype and resultant challenges related to genotype procedures and phenotype assignment. In contrast to CYP2C19 where all institutions deployed testing on site, half of the early adopters of CYP2D6 testing leveraged external reference labs as primary or supporting CYP2D6 testing facilities. For CYP2C19-guided antiplatelet therapy, institutional efforts were focused on a single use case and engaged the interventional cardiology practitioners. This allowed for highly tailored workflows and targeted educational efforts. In our experiences, CYP2D6 implementation efforts, in contrast, required engaging practitioners across multiple practice areas (e.g. psychiatry, primary care, surgery, and oncology) given the multiple drug substrates for the CYP2D6 enzyme (e.g. antidepressants, opioids, anti-emetics, tamoxifen). This may require a broader strategy and more extensive educational effort to rally the practice groups in acceptance and adoption of CYP2D6 genotype-guided medication therapy. In addition, unlike CYP2C19 testing for antiplatelet guidance where patients are undergoing an invasive procedure and collection of a blood sample for genotyping was not an issue, having a non-invasive sample collection method for CYP2D6 testing was deemed important as it is commonly done in an outpatient setting where phlebotomy may not be available or for children who may be especially averse to blood collection.
There were several common challenges with CYP2D6-guided therapy. Some were technical in nature and related to CYP2D6 gene complexity, with variation including single nucleotide polymorphisms, deletions and multiple copies of the entire gene, and fusions with downstream pseudogenes4. As the field moves more towards whole genome sequencing, these unique characteristics of CYP2D6 make extracting the accurate information from short read whole genome sequencing difficult and requiring special informatics approaches to accurately call the alleles27,28. Thus, sites agreed that local CYP2D6 expertise was an important asset to implementation, and in the absence of such, sites may prefer to utilize a commercial laboratory for genotyping.
Despite gene complexity and independent assay development, there is remarkable similarity across sites in the variants tested. No site was performing comprehensive analysis of all known CYP2D6 variants, and each site tested for copy number variation and assays enabling the assignment of 9 common alleles. While having a list of “must-test alleles” would be helpful to guide assay development, in the absence of that, the list of variants tested across sites may be useful for other institutions considering CYP2D6 testing.
A second layer of technical challenges surround reporting test results in EHRs and use of results in CDS. Many systems do not have mature EHR systems in place for storing, reporting, and using genetic results. Historically, many genetic testing results have been reported in EHRs in text-based reports or scanned documents. Building the technical capability for genetic results to be stored as discrete data enables downstream applications like genotype specific electronic CDS but requires an initial investment and ongoing maintenance. In addition, accounting for use of CYP2D6 inhibitors is important for refining CYP2D6 phenotype given evidence of altered enzyme activity, disposition of CYP2D6 substrates, and drug response with CYP2D6 inhibitors29–32. Strong inhibitors (e.g. paroxetine) can convert NMs, based on genotype results, to PMs, whereas moderate inhibitors (e.g. duloxetine) can convert NMs, based on genotype, to IMs3,32. More sophisticated decision support may be necessary to account for this phenoconversion with concomitant use of CYP2D6 inhibitors.
Challenges that were specific to the early implementation phase included the unknown ramifications of failure to act on genotype results, especially since CYP2D6 genotype has implications for multiple therapies, and having sufficient personnel in place to facilitate implementation. Interestingly, early test adopters stated that potential risk management issues were discussed prior to implementation but are no longer perceived as a barrier, indicating that potential risk ramifications around pharmacogenetic testing are a common perceived obstacle, but may not be a practical reality at this time. Indeed, there are limited examples of a legal action involving the use or lack of use of pharmacogenetic information in clinical practice33. Regarding personnel, early adopters were able to overcome this obstacle by engaging across departments, teams, and services to build multidisciplinary teams. Of note, the effort of those teams is ongoing, as CYP2D6 testing and pharmacogenetic CDS requires maintenance in the face of laboratory and EHR changes, and continual assessment of new developments in all aspects from variant detection to prescribing advice.
Strengths of this research are the inclusion of multiple institutions and different applications of genotype results, increasing the generalizability of challenges faced and lessons learned. A potential limitation is that all institutions were from a common research network and approaches to implementation may be similar for this reason. In addition, we do not provide data on specific recommendations provided or implementation success metrics as our intent was to survey the landscape of CYP2D6 testing across multiple sites. Future studies may focus on individual applications of CYP2D6 testing. While we provide information on the platforms used for genetic testing, it was beyond the scope of the paper to provide details on genotyping methodology in each laboratory. Finally, we do not address the cost of implementation or genetic testing. The cost of analytical validation can be significant with a new laboratory test (approximately $30,000 in reagents alone based on personal observations [V.M.P.]), and increase further if the laboratory has to invest in new equipment. The cost of genetic testing for institutions and patients or payors is influenced by multiple factors, including technology used for the assay, turn-around time, number of samples, and state-specific rules and regulations. With evolving technology and policies, genotyping costs are also variable over time.
In summary, despite of the complexity of the CYP2D6 genotype and associated challenges with genotype interpretation and phenotype assignment, multiple institutions have prioritized implementation of CYP2D6 genotyping to inform drug prescribing. To our knowledge, this is the first description of strategies for CYP2D6 testing to guide drug prescribing across multiple sites along with challenges faced and important lessons learned. Implementation resources from our sites, including supporting literature, CDS language, and education materials are available through the IGNITE website (https://ignite-genomics.org/). Our experiences and lessons learned may be valuable for other institutions seeking to implement CYP2D6 testing.
Supplementary Material
Acknowledgements
This work was supported by the National Institutes of Health (NIH) IGNITE Network (http://ignite-genomics.org/) through grants U01HG007269, U01HG007253, U01HG007762, U01HG007775, and U01HG007278. Additional funding provided by the University of Florida and its Clinical Translational Science Institute (NCATS UL1TR000064 and UL1TR001427) for JAJ, LHC, and KWW; NHLBI R0HL092173 and K24HL133373, NCATS UL1TR000165, University of Alabama Birmingham’s Health Service Foundations’ General Endowment Fund and Hugh Kaul Personalized Medicine Institute to NAL; NIH Common Fund Program in Health Economics and NHLBI U01HL122904 to JFP; Indiana University Health – Indiana University School of Medicine Strategic Research Initiative to VMP and TCS; NHGRI eMERGE Network U01HG8666 and U01HG006828 (Cincinnati Children’s Hospital Medical Center) and CIDR U01HG004438; University of Minnesota Enhance Comprehensive Pharmacist Services to Improve Patient Health Clinical Research Award to JRB; NCI P30CA076292, Cancer Epidemiology Innovation Funds, DeBartolo Family Personalized Medicine Institute Pilot Research Award in Personalized Medicine, ASHP Research and Education Foundation, and OneOme to JKH; NHGRI U01HG008672 and NCATS UL1TR002243; University of Pennsylvania, Center for Precision Medicine Accelerator Fund to ST; NCATS UL1TR001857, an Anonymous Donor, internal funds from the University of Pittsburgh Medical Center, and the Institute for Precision Medicine to PEE; NCBiotech Presidential Grant to GDB; philanthropic donation by T. Denny Sanford IMAGENETICS (Internal Medicine and Genetics) to LJH, and NHGRI 3U01HG008701 to AOO.
Dr. Cavallari has received research funding from Mallinckrodt Pharmaceuticals. Dr. Limdi serves as a consultant for Admera Health. Dr. Pratt is affiliated with the Indiana University School of Medicine Pharmacogenomics Laboratory, which is a fee-for-service clinical laboratory. Dr. Hicks receives clinical trial support from OneOme and serves as a consultant for Quest Diagnostics. Dr. Greg was employed by OneOme, a for-profit genotyping laboratory, at the time of this work.
Footnotes
CONFLICT OF INTEREST: None of the other authors have any conflicts of interest to declare.
References
- 1.Phillips KA, Veenstra DL, Oren E, Lee JK & Sadee W Potential role of pharmacogenomics in reducing adverse drug reactions: a systematic review. JAMA 2001;286:2270–2279 [DOI] [PubMed] [Google Scholar]
- 2.Pharmacogene Variation Consortium (PharmVar, www.PharmVar.org). Accessed September 10, 2018 . Vol. 2018. [Google Scholar]
- 3.Crews KR, Gaedigk A, Dunnenberger HM, et al. Clinical Pharmacogenetics Implementation Consortium guidelines for cytochrome P450 2D6 genotype and codeine therapy: 2014 update. Clin Pharmacol Ther 2014;95:376–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hicks JK, Swen JJ & Gaedigk A Challenges in CYP2D6 phenotype assignment from genotype data: a critical assessment and call for standardization. Curr Drug Metab 2014;15:218–232 [DOI] [PubMed] [Google Scholar]
- 5.Eckhardt K, Li S, Ammon S, et al. Same incidence of adverse drug events after codeine administration irrespective of the genetically determined differences in morphine formation. Pain 1998;76:27–33 [DOI] [PubMed] [Google Scholar]
- 6.Lotsch J, Rohrbacher M, Schmidt H, et al. Can extremely low or high morphine formation from codeine be predicted prior to therapy initiation? Pain 2009;144:119–124 [DOI] [PubMed] [Google Scholar]
- 7.Poulsen L, Arendt-Nielsen L, Brosen K & Sindrup SH The hypoalgesic effect of tramadol in relation to CYP2D6. Clin Pharmacol Ther 1996;60:636–644 [DOI] [PubMed] [Google Scholar]
- 8.Stamer UM, Musshoff F, Kobilay M, et al. Concentrations of tramadol and O-desmethyltramadol enantiomers in different CYP2D6 genotypes. Clin Pharmacol Ther 2007;82:41–47 [DOI] [PubMed] [Google Scholar]
- 9.Schroth W, Goetz MP, Hamann U, et al. Association between CYP2D6 polymorphisms and outcomes among women with early stage breast cancer treated with tamoxifen. JAMA 2009;302:1429–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schroth W, Winter S, Murdter T, et al. Improved Prediction of Endoxifen Metabolism by CYP2D6 Genotype in Breast Cancer Patients Treated with Tamoxifen. Front Pharmacol 2017;8:582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kirchheiner J, Keulen JT, Bauer S, Roots I & Brockmoller J Effects of the CYP2D6 gene duplication on the pharmacokinetics and pharmacodynamics of tramadol. J Clin Psychopharmacol 2008;28:78–83 [DOI] [PubMed] [Google Scholar]
- 12.Kirchheiner J, Schmidt H, Tzvetkov M, et al. Pharmacokinetics of codeine and its metabolite morphine in ultra-rapid metabolizers due to CYP2D6 duplication. Pharmacogenomics J 2007;7:257–265 [DOI] [PubMed] [Google Scholar]
- 13.Gasche Y, Daali Y, Fathi M, et al. Codeine intoxication associated with ultrarapid CYP2D6 metabolism. N Engl J Med 2004;351:2827–2831 [DOI] [PubMed] [Google Scholar]
- 14.Orliaguet G, Hamza J, Couloigner V, et al. A case of respiratory depression in a child with ultrarapid CYP2D6 metabolism after tramadol. Pediatrics 2015;135:e753–755 [DOI] [PubMed] [Google Scholar]
- 15.Hicks JK, Bishop JR, Sangkuhl K, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2D6 and CYP2C19 Genotypes and Dosing of Selective Serotonin Reuptake Inhibitors. Clin Pharmacol Ther 2015;98:127–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hicks JK, Sangkuhl K, Swen JJ, et al. Clinical pharmacogenetics implementation consortium guideline (CPIC) for CYP2D6 and CYP2C19 genotypes and dosing of tricyclic antidepressants: 2016 update. Clin Pharmacol Ther 2017;102:37–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bell GC, Caudle KE, Whirl-Carrillo M, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for CYP2D6 genotype and use of ondansetron and tropisetron. Clin Pharmacol Ther 2017;102:213–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Candiotti KA, Birnbach DJ, Lubarsky DA, et al. The impact of pharmacogenomics on postoperative nausea and vomiting: do CYP2D6 allele copy number and polymorphisms affect the success or failure of ondansetron prophylaxis? Anesthesiology 2005;102:543–549 [DOI] [PubMed] [Google Scholar]
- 19.Kaiser R, Sezer O, Papies A, et al. Patient-tailored antiemetic treatment with 5-hydroxytryptamine type 3 receptor antagonists according to cytochrome P-450 2D6 genotypes. J Clin Oncol 2002;20:2805–2811 [DOI] [PubMed] [Google Scholar]
- 20.Goetz MP, Sangkuhl K, Guchelaar HJ, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2D6 and Tamoxifen Therapy. Clin Pharmacol Ther 2018;103:770–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cavallari LH, Beitelshees AL, Blake KV, et al. The IGNITE Pharmacogenetics Working Group: An Opportunity for Building Evidence with Pharmacogenetic Implementation in a Real-World Setting. Clin Transl Sci 2017;10:143–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Empey PE, Stevenson JM, Tuteja S, et al. Multisite Investigation of Strategies for the Implementation of CYP2C19 Genotype-Guided Antiplatelet Therapy. Clin Pharmacol Ther 2018;104:664–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caudle KE, Klein TE, Hoffman JM, et al. Incorporation of pharmacogenomics into routine clinical practice: the Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline development process. Curr Drug Metab 2014;15:209–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Swen JJ, Nijenhuis M, de Boer A, et al. Pharmacogenetics: from bench to byte--an update of guidelines. Clin Pharmacol Ther 2011;89:662–673 [DOI] [PubMed] [Google Scholar]
- 25.Ramamoorthy A, Flockhart DA, Hosono N, et al. Differential quantification of CYP2D6 gene copy number by four different quantitative real-time PCR assays. Pharmacogenet Genomics 2010;20:451–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramamoorthy A & Skaar TC Gene copy number variations: it is important to determine which allele is affected. Pharmacogenomics 2011;12:299–301 [DOI] [PubMed] [Google Scholar]
- 27.Lee SB, Wheeler MM, Patterson K, et al. Stargazer: a software tool for calling star alleles from next-generation sequencing data using CYP2D6 as a model. Genet Med 2018; [DOI] [PMC free article] [PubMed]
- 28.Numanagic I, Malikic S, Pratt VM, et al. Cypiripi: exact genotyping of CYP2D6 using high-throughput sequencing data. Bioinformatics 2015;31:i27–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Storelli F, Matthey A, Lenglet S, et al. Impact of CYP2D6 Functional Allelic Variations on Phenoconversion and Drug-Drug Interactions. Clin Pharmacol Ther 2018;104:148–157 [DOI] [PubMed] [Google Scholar]
- 30.Kiss A, Menus A, Toth K, et al. Phenoconversion of CYP2D6 by inhibitors modifies aripiprazole exposure. Eur Arch Psychiatry Clin Neurosci 2019; [DOI] [PubMed]
- 31.Monte AA, Heard KJ, Campbell J, et al. The effect of CYP2D6 drug-drug interactions on hydrocodone effectiveness. Acad Emerg Med 2014;21:879–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Borges S, Desta Z, Jin Y, et al. Composite functional genetic and comedication CYP2D6 activity score in predicting tamoxifen drug exposure among breast cancer patients. J Clin Pharmacol 2010;50:450–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu AH, White MJ, Oh S & Burchard E The Hawaii clopidogrel lawsuit: the possible effect on clinical laboratory testing. Per Med 2015;12:179–181 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


