Abstract
Contamination of recreational beaches due to fecal waste from gulls complicates beach monitoring and may pose a risk to public health. Gulls that feed at human waste sites may ingest human fecal microorganisms associated with that waste. If these gulls also visit beaches, they may serve as vectors, transporting fecal microorganisms to the beach where they may subsequently contaminate sand and water. In this study, samples collected from landfills, treated wastewater storage lagoons, and public beaches demonstrated a spatial and temporal overlap of markers for gull and human-associated microorganisms. In addition, markers for gull, fecal indicator bacteria, and the human-associated marker, HF183, were detected in gull feces and cloacae samples. Further, HF183 was detected in cloacae samples from gulls that were documented by radio-telemetry traveling between human waste sites and public beaches. This study highlights the potential for gulls that visit human waste sites to disperse human-associated microorganisms in the beach landscape.
Keywords: Microbial source tracking, Fecal pollution, Gull transport, Beaches, Fecal indicator bacteria
Graphical Abstract

1. Introduction
Contamination of recreational beaches with fecal waste poses a risk to human health due to the potential occurrence of human pathogens. Despite increased recognition of this risk, outbreaks of illness associated with exposure to contaminated recreational water continue (Hlavsa et al., 2015). Fecal waste coming from human sources is thought to present the greatest risk to human health, while waste from wildlife, including shore birds, is thought to be of lower risk (Schoen and Ashbolt, 2010, Soller et al., 2010). Therefore, the identification of sources of fecal contamination to recreational beaches is often included as a component of beach monitoring and mitigation efforts (Byappanahalli et al., 2015, Edge and Hill, 2007, Goodwin et al., 2016, Goodwin et al., 2017, Noble et al., 2006).
Several studies have revealed gulls as a significant source of contamination at beaches (Araújo et al., 2014, Converse et al., 2012, Edge and Hill, 2007, Haack et al., 2003, Lu et al., 2011a, Staley and Edge, 2016). Indeed, gull waste contains high levels of the traditional fecal indicator bacteria (FIB) used in beach monitoring (Alderisio and DeLuca, 1999, Fogarty et al., 2003, Meerburg et al., 2011), and several studies have correlated gull numbers with FIB densities in water (Converse et al., 2012, Kirschner et al., 2004, Lu et al., 2011a) and sand (Edge and Hill, 2007, Edge et al., 2010, Whitman and Nevers, 2003). More importantly, gulls are relevant to public health because they may shed bacterial pathogens (Ebert et al., 2016, Kinzelman et al., 2008, Lévesque et al., 2000, Lu et al., 2011b, Quessy and Messier, 1992, Whelan et al., 1988), antibiotic resistant bacteria (Bonnedahl et al., 2009, Dolejská et al., 2009), and viruses such as avian influenza virus (Alexander, 2000). Some of these microorganisms (e.g., campylobacters and avian influenza virus) may be endemic in gulls (Kapperud and Rosef, 1983, Webster et al., 1992), but gulls may acquire others from their environment.
Gulls are opportunistic feeders and are often attracted to easily accessible food sources linked to human activity (Ferns and Mudge, 2000). For example, in the Great Lakes region, Ring-billed gulls were found to travel up to 25 km to landfills for foraging, and anthropogenic components made up a substantial portion of their diet (Belant et al., 1998). Investigations have pointed to the potential for gulls to acquire human-associated microorganisms while foraging at sites of human refuse or waste. The prevalence of specific serotypes of Salmonella, including rare types, was associated with feeding at sewage treatment works (Butterfield et al., 1983, Fenlon, 1981, Fricker, 1984). Campylobacter occurrence, including C. jejuni, was directly related to refuse consumption by juvenile gulls (Ramos et al., 2010). Other studies have highlighted the potential for gulls to transport these pathogens to sites where they may then be transferred to other species. Gulls have been associated with outbreaks of salmonellosis in cattle and sheep (Butterfield et al., 1983, Coulson et al., 1983, Johnston et al., 1979) and in humans (Aavitsland and Hofshagen, 1999). Recent studies implicated gulls as vectors in the movement of Salmonella enterica and antibiotic-resistant E. coli between environmental and clinical reservoirs (Hernandez et al., 2013, Retamal et al., 2015, Toro et al., 2016, Varela et al., 2015).
The fact that wild birds, including migratory birds, have been implicated in the transmission of human pathogens (Tsiodras et al., 2008) suggests that gulls are potential reservoirs of bacterial groups used as the targets of fecal source tracking assays. However, evidence for the latter is very scarce, perhaps because most studies have been conducted in areas where larger reservoirs of human fecal waste (e.g., landfills, wastewater treatment plants) are not easily accessible to gulls. In this study, we sought to investigate whether gulls could acquire human-associated microorganisms from human waste sites and whether gulls could serve as transport vectors to recreational beaches. We examined gulls that visited, as determined by radio-telemetry, two different recreational areas that were located nearby a wastewater lagoon and two landfills, and tested them for the presence and abundance of FIB (i.e., enterococci and E. coli), gull- and human-associated fecal markers, and potential pathogens using qPCR assays.
2. Materials and methods
2.1. Sample sites
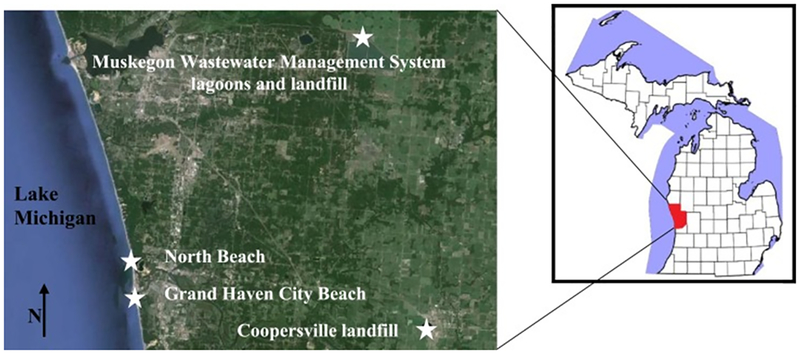
All sites sampled in this study were on the east coast of Lake Michigan in Ottawa and Muskegon counties, Michigan, USA (Fig. 1) and were sampled between May and August 2013. Nearshore lake water samples were collected at two public recreation beaches in Ottawa County, MI: North Beach (NB, n = 10) and Grand Haven City Beach (GHCB, n = 10). Samples were also collected at three municipal waste sites. The Muskegon County Wastewater Management System uses a land treatment process of aeration, settling, and storage on 4452 ha for treatment of domestic and industrial waste. Within this system are two, 344 ha, 5.1 billion gallon, first-stage treated wastewater storage lagoons (MWM) and the 45 ha Muskegon County Solid Waste Management System landfill (ML), which accepts septic tank, grease trap, and agricultural processing waste. The Ottawa County Farms Landfill, Coopersville, MI (CL) is a 112 ha compost facility accepting mixed domestic residential waste. All facilities are uncovered and accessible to gulls. Run-off water (CL, n = 2; ML, n = 9) was collected at both landfills. Run-off water gathered in a shallow pool at the base of the solid waste mound at ML and in a field adjacent to the solid waste mound at CL. When standing water was not present, run-off water-wetted soil slurry was collected (n = 10). Water samples were also collected from the wastewater storage lagoons at MWM (n = 24).
Fig. 1.
Map of study sites in western Michigan, USA. Red shading marks the outline of Muskegon and Ottawa counties of western Michigan (right map). White stars mark the site locations (left map). The Muskegon Wastewater Management System is located in Muskegon County, MI. The Muskegon Solid Waste landfill is on the property of the Muskegon Wastewater Management System. North Beach, Grand Haven City Beach, and Coopersville landfill are located in Ottawa County, MI. This map was generated using Google Earth software.
From May to July 2013, gulls were abundant at all sites and were nesting along a cement dyke that separates the two storage lagoons at MWM. Gulls began nesting in this region in 1974 (Ponshair, 1974) and gull nests have been counted annually on the dyke at the MWM system since 1997 (Ponshair, 2006). In June of 2013, 5516 Ring-billed gull and 11 Herring gull nests were counted (Ponshair, personal communication). Recently deposited, still moist gull feces were collected along the dyke at MWM (n = 14) and at NB (n = 5). Gull feces were collected using sterile polyester tip swabs and stored in 2 ml centrifuge tubes on ice. Additionally, gulls were captured on both beaches as part of a radio-telemetry study of gull habitat use (Jordan, 2014) and cloacal samples were collected from individual gulls (n = 27). To sample the cloaca (internal cavity in the digestive system), the vent was spread and a sterile polyester tip swab saturated in 1 × phosphate buffered saline (PBS) solution (pH 7.5) was gently inserted up into the cloaca. The cloacal swab was then withdrawn and immediately re-submerged and stored in 1 ml PBS in 2 ml cryovials on dry ice. Feces were also collected from some of the captured gulls creating 11 paired samples of cloacal/fecal material acquired from 11 individual gulls (two captured on NB and nine captured on GHCB). Water samples were collected into sterile Whirl-Pak bags attached to a 2 m collection pole. Landfill soil slurry samples were collected into sterile 50 ml centrifuge tubes attached to a 2 m collection pole, or in some cases, soil was collected using sterile spatulas and stored in 50 ml centrifuge tubes. Field blanks during each sample collection trip included 100 ml distilled water and PBS saturated swabs.
All samples were immediately placed on ice after collection and during transportation to the laboratory. Within 6 h of collection, all water samples (30–100 ml for landfill run-off or wastewater; 600–700 ml for beach water) were processed by membrane filtration (Alm et al., 2003) and membrane filters, soil, gull feces, and gull cloacae samples were stored at − 80 °C until used in DNA extractions.
2.2. DNA extraction
DNA from filtered water or from landfill soil (0.25 g wet weight) was extracted using the PowerSoil™ DNA Isolation Kit (MO BIO Laboratories, Inc., Carlsbad, CA) according to the manufacturer’s instructions. For cloacae samples, each were centrifuged at 16,000 × g for 9 min to concentrate the suspension, then 200–400 μl of the cloacae material were used for total DNA extraction using the PowerSoil™ DNA Isolation Kit. DNA from gull feces (0.25 g wet weight) was extracted using the UltraClean™ Fecal DNA Kit (MO BIO Laboratories) according to the manufacturer’s instructions. Field blanks were also extracted.
2.3. Quantitative PCR (qPCR) assays
TaqMan-based quantitative PCR assays (Table 1) were used to test water samples (landfill run-off water, first-stage treated lagoon wastewater, and beach water), landfill run-off soil slurry, gull feces, and gull cloacae samples. TaqMan assays were performed in 25 μl containing 1 × TaqMan universal PCR master mix with AmpErase uracil-N-glycosylase (Applied Biosystems, Foster City, CA), 0.2 μg/μl bovine serum albumin, 0.2 μM (final concentration) primers, 6-carboxyfluorescein (FAM)-labeled hydrolysis probe, and 2 μl of DNA template. The amplification protocol conditions included an initial incubation at 50 °C for 2 min, followed by 10 min of incubation at 95 °C, and then 40 cycles of 95 °C for 15 s and at optimum annealing temperature for each assay for 1 min. All qPCR assays were performed using a 7900 HT fast real-time sequence detector (Applied Biosystems, Foster City, CA). All assays were performed in duplicate in MicroAmp Optical 96-well reaction plates with MicroAmp Optical Caps (Applied Biosystems, Foster City, CA, USA). PCR data were analyzed using ABI’s Sequence Detector software (version 2.2.2). Four independent standard curves for each qPCR assay were generated by plotting threshold cycle (CT) values against the number of target copies corresponding to serially diluted plasmid standards purchased from IDT integrated DNA technologies (Coralville, Iowa, USA). The target copy numbers (T) were estimated by the following equation: T = [D / (PL × 660)] × 6.022 × 1023, where D (g/μl) is plasmid DNA concentration, and PL (bp) is plasmid length in base pairs. Each standard curve was generated from at least five 10-fold plasmid dilutions in triplicates. Percent amplification efficiencies were calculated by the instrument manufacturer’s instructions (Applied Biosystems). Two no-template controls per PCR plate were used to check for cross-contamination. Relative copy numbers (signal intensity values) were recorded for all TaqMan assays. DNA copy numbers were standardized per respective unit for sample types (per 100 ml for water samples; per 1 g (wet weight) for landfill run-off soil slurry and gull feces; per 1 ml for cloacae suspensions).
Table 1.
Summary of oligonucleotide primers and probes for all qPCR assays (TaqMan and SYBR green).
| Assay | Primer/probe sequence (5′ to 3′) | T (°C)a | Size (bp) | Reference |
|---|---|---|---|---|
| General Bacteroidetes(GenBac3) TaqMan | GenBactF3: GGGGTTCTGAGAGGAAGGT GenBactR4: CCGTCATCCTTCACGCTACT GenBactP2: 6FAM-CAATATTCCTCACTGCTGCCTCCCGTA-TAMRA |
60 | 129 | Siefring et al. (2008) |
| Human-associated Bacteroidetes (HF183) TaqMan | HF183–1F: ATCATGAGTTCACATGTCCG BthetR1: CGTAGGAGTTTGGACCGTGT BthetP1: 6FAM-CTGAGAGGAAGGTCCCCCACATTGGA-TAMRA |
60 | 167 | Haugland et al. (2010) |
| Gull2 SYBR green | F: TGCATCGACCTAAAGTTTTGAG R: GTCAAAGAGCGAGCAGTTACTA |
64 | 412 | Lu et al. (2008) |
| Gull4 TaqMan | qGull7F: CTTGCATCGACCTAAAGTTTGAG qGull8R: GGTTCTCTGTATTATGCGGTATTAGCA qGull7Pb: FAM-ACACGTGGGTAACCTGCCCATCAGA-TAMRA |
60 | 116 | Ryu et al. (2012) |
| Escherichia coli(EC23S857) TaqMan | F: GGTAGAGCACTGTTTtGGCAc R: TGTCTCCCGTGATAACtTTCTCc P: 6FAM-TCATCCCGACTTACCAACCCG-TAMRA |
60 | 88 | Chern et al. (2011) |
| General Enterococcus(Entero1) TaqMan | ECST748F: AGAAATTCCAAACGAACTTG ENC854R: CAGTGCTCTACCTCCATCATT GPL813TQ: 6FAM-TGGTTCTCTCCGAAATAGCTTTAGGGCTA-TAMRA |
60 | 92 | Ludwig and Schleifer (2000) |
| Campylobacter spp. (Camp2) Taqman | campF2: CACGTGCTACAATGGCATAT campR2: GGCTTCATGCTCTCGAGTT campP2: FAM-CAGAGAACAATCCGAACTGGGACA-BHQ1–3 |
58 | 108 | Lund et al. (2004) |
| Enterococcus faecalis(Faecalis1) Taqman | FaecalF: CGCTTCTTTCCTCCCGAGT FaecalR: GCCATGCGGCATAAACTG FaecalP: 6FAM-GAGGAGTGGCGGACG-TAMRA |
60 | 143 | Santo Domingo et al. (2003) |
| Enterococcus faecium(Faecium1) Taqman | CiumF: TTCTTTTTCCACCGGAGCTT CiumR: AACCATGCGGTTTYGATTG CiumP: 6FAM-AGTAACACGTGGGTAACCTGCCCATCAGA-TAMRA |
60 | 141 | Ryu et al. (2013) |
F, forward; R, reverse; P, probe.
Optimum PCR annealing temperatures were determined using temperature gradients.
FAM, 6-carboxyfluorescein, fluorescence reporter dye; TAMRA, 6-carboxytetramethylrhodamine, fluorescence quencher dye.
Lower case denotes deliberately mismatched base.
A gull-associated SYBR green-based qPCR assay (Gull2; Lu et al., 2008), widely used in MST studies, was also used to test water sources, landfill run-off soil slurry, gull feces, and gull cloacae samples (Table 1). Reaction mixtures (25 μl total volume) for the SYBR green assay contained 1 × Power SYBR green master mix (Applied Biosystems, Foster City, CA), 0.2 μg/μl bovine serum albumin, and 0.2 μM (final concentration) of each primer, and 2 μl of DNA template. Ten-fold dilutions of each DNA extract (2 μl, final volume) were used as templates to test for PCR inhibition. The amplification protocol conditions included an initial incubation at 50 °C for 2 min, followed by 95 °C for 10 min and 40 cycles each at 95 °C for 15 s, and 64 °C annealing temperature for 1 min, followed by a melting curve analysis (i.e., from 60 to 90 °C in increments of 0.1 °C). Equipment and data analysis were performed as described above. Presence/absence readings were assigned to data generated from the Gull2 SYBR green assay based on signal intensity values.
The range of quantification for all assays, including TaqMan and SYBR green, was 101 to 105 DNA copies per reaction. Untreated domestic influent to the MWM served as a positive control for detection of HF183. For both TaqMan and SYBR green assays, all of the field blanks and no-template controls were negative, indicating no evidence of cross-contamination. A McNemar’s test (Zar, 2009) on the paired gull samples was used to compare the proportion of positive HF183 samples from cloacae vs. feces. A Fisher exact test (Zar, 2009) was used to compare the proportion of positive HF183 samples among beaches, human waste sites, and gulls.
3. Results and discussion
3.1. The human-marker, HF183, was detected in gull waste samples
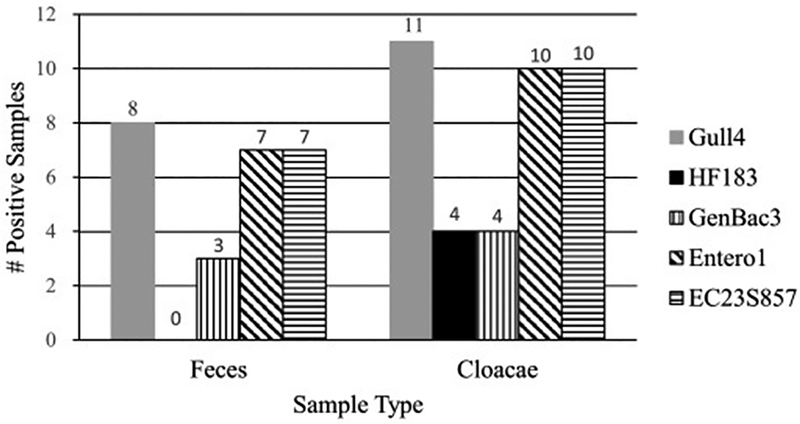
An objective of this study was to investigate the potential for gulls that visit human waste sites to acquire human fecal bacteria. To assess this objective the human-associated marker HF183 was used as a proxy for human fecal bacteria. Gulls at public beaches and at human waste sites were sampled to test for the presence of markers for gull, FIB, and HF183. Feces can be contaminated after excretion from the bird (IDL, 2016), and other studies have shown higher concentrations of microorganisms in cloacal material as compared to feces (Sarker et al., 2012, Tracey, 2010), therefore, we considered cloacal material to be more representative of the presence of markers in gull digestive contents and collected cloacae samples when possible. All of the markers including HF183 were detected in gull feces and cloacae samples (Table 2). From 11 individual gulls captured for radio-telemetry, both cloacal material and feces were collected (n = 11 pairs). Cloacae from paired collections returned samples that were positive for FIB (10/11) and HF183 (4/11) markers (Fig. 2). Interestingly, while feces from paired collections also had positive samples for FIB markers (7/11), none were positive for HF183. In paired collections, fewer fecal samples were positive for markers, including the gull markers, than were cloacae samples (P = 0.016, McNemar’s test). These results suggest that including cloacae samples may provide information that could be missed by evaluating feces alone (Van Hoorebeke et al., 2009, Krauss et al., 2013).
Table 2.
Prevalence of FIB, human- and gull-associated and pathogen markers in water, soil, gull feces, and gull cloacae samples.
| Sample | n = | EC23S857 | Entero1 | GenBac3 | HF183 | Gull4 | Gull2 | Camp2 | Faecalis1 | Faecium1 |
|---|---|---|---|---|---|---|---|---|---|---|
| Human waste | ||||||||||
| CL-water | 2 | 100% | 100% | 100% | 50% | 50% | 50% | 100% | 100% | 50% |
| ML-water | 9 | 100% | 78% | 100% | 56% | 67% | 44% | 100% | 67% | 56% |
| ML-soil | 10 | 90% | 70% | 90% | 30% | 20% | 0% | 90% | 10% | 10% |
| MWM-lagoon | 24 | 100% | 50% | 100% | 38% | 75% | 58% | 96% | 17% | 0% |
| Total | 45 | 98% | 62% | 98% | 40% | 60% | 42% | 96% | 29% | 16% |
| Beach | ||||||||||
| NB-water | 10 | 100% | 40% | 100% | 60% | 100% | 80% | 70% | 0% | 0% |
| GHCB-water | 10 | 100% | 20% | 100% | 50% | 100% | 90% | 80% | 0% | 0% |
| Total | 20 | 100% | 30% | 100% | 55% | 100% | 85% | 75% | 0% | 0% |
| Gull feces | ||||||||||
| MWM-dyke | 14 | 100% | 100% | 57% | 36% | 93% | 57% | 71% | 57% | 64% |
| NB | 7 | 57% | 43% | 14% | 43% | 86% | 86% | 43% | 29% | 0% |
| GHCB | 9 | 67% | 67% | 22% | 0% | 78% | 67% | 67% | 44% | 11% |
| Total | 30 | 80% | 77% | 37% | 27% | 87% | 67% | 63% | 53% | 67% |
| Gull cloacae | ||||||||||
| NB | 4 | 50% | 50% | 25% | 50% | 100% | 75% | 50% | 75% | 0% |
| GHCB | 23 | 100% | 96% | 30% | 13% | 100% | 74% | 57% | 65% | 0% |
| Total | 27 | 93% | 89% | 30% | 19% | 100% | 74% | 56% | 67% | 0% |
EC23S857, E. coli; Entero1, enterococci; GenBac3, general Bacteroidetes; Camp2, campylobacter; Faecalis1, Enterococcus faecalis; Faecium1, Enterococcus faecium; CL, Coopersville Landfill; ML, Muskegon County Landfill; MWM, Muskegon County Waste Management; NB, North Beach; GHCB, Grand Haven City Beach.
Fig. 2.
Paired feces and cloacae samples (n = 11 pairs) that had positive assays with gull-associated (Gull4), human-associated (HF183), general Bacteroidetes (GenBac3), enterococci (Entero1), and E. coli (EC23S857) markers.
The proportion of positive HF183 samples among sites differed (P = 0.020), with higher prevalence at beaches and human waste sites than within gulls. Further, the proportion of positive HF183 samples at beaches and human waste sites was equal. Concentrations of HF183 in beach water, landfills, and treated wastewater were low in comparison to concentrations in untreated influent (Table 3).
Table 3.
Quantification of mean positive signals for FIB, human-associated marker, and gull-associated markers in water, soil, gull feces, and gull cloacae samples.
| Mean positive signal concentration (DNA/copies/unit)a (± standard error) | |||||
|---|---|---|---|---|---|
| Site-sample | EC23S857 | Entero1 | GenBac3 | HF183 | Gull4 |
| Waste sites | |||||
| CL-water | 2.1 × 104(± 1.0 × 104) | 2.4 × 104(± 2.0 × 104) | 6.2 × 103(± 2.0 × 103) | 3.8 × 101 (NA) | 5.3 × 103(NA) |
| ML-water | 4.6 × 103(± 9.0 × 102) | 5.2 × 105(± 2.0 × 105) | 2.0 × 105(± 6.0 × 104) | 1.4 × 102(± 1.0 × 101) | 2.0 × 103(± 1.0 × 103) |
| ML-soil | 1.3 × 104(± 6.0 × 103) | 1.9 × 106(± 2.0 × 106) | 7.7 × 104(± 6.0 × 104) | 1.3 × 102(± 2.0 × 101) | 7.3 × 102(± 2.0 × 102) |
| MWM-lagoon | 2.7 × 104(± 1.0 × 104) | 2.5 × 104(± 8.0 × 103) | 2.7 × 104(± 5.0 × 103) | 1.7 × 102(± 6.0 × 101) | 1.3 × 104(± 5.0 × 103) |
| MWM-influent | – | – | – | 3.0 × 106(single sample) | < LOQ |
| Beach water | |||||
| NB | 3.9 × 102(± 1.0 × 102) | 1.1 × 103(± 8.0 × 102) | 2.4 × 103(± 1.0 × 103) | 5.1 × 101(± 2.0 × 101) | 3.6 × 103(± 3.0 × 103) |
| GHCB | 3.1 × 102(± 1.0 × 102) | 8.2 × 102(± 2.0 × 102) | 2.8 × 103(± 2.0 × 103) | 4.0 × 101(± 3.0 × 101) | 7.9 × 102(± 2.0 × 102) |
| Gull feces | |||||
| MWM-lagoon | 6.5 × 106(± 3.0 × 106) | 3.5 × 107(± 2.0 × 107) | 7.8 × 104(± 5.0 × 104) | 1.3 × 102(± 2.0 × 101) | 8.7 × 104(± 3.0 × 104) |
| NB | 5.3 × 103(± 2.0 × 103) | 4.2 × 105(± 4.0 × 105) | 1.7 × 103(NA) | 1.2 × 102(± 3.0 × 101) | 2.9 × 106(± 1.0 × 106) |
| GHCB | 5.5 × 105(± 4.0 × 105) | 3.2 × 105(± 2.0 × 105) | 5.1 × 103(± 4.0 × 103) | < LOQ | 2.4 × 107(± 1.0 × 107) |
| Gull cloacae | |||||
| NB | 3.9 × 103(± 2.0 × 103) | 5.7 × 104(± 1.0 × 104) | 1.3 × 103(NA) | 4.3 × 101(± 5.0 × 101) | 2.8 × 106(± 2.0 × 106) |
| GHCB | 8.5 × 103(± 6.0 × 103) | 1.0 × 105(± 1.0 × 105) | 2.1 × 103(± 2.0 × 103) | 5.5 × 101(± 3.0 × 101) | 5.3 × 105(± 2.0 × 105) |
EC23S857, E. coli; Entero1, enterococci; GenBac3, general Bacteroidetes. NA, only one sample of replicate collections yielded a positive signal; < LOQ, below limit of quantification. LOQs and range of amplification efficiency for each qPCR are noted in Table S1. Total number of samples (n =) are listed in Table 2.
Sample units; water – 100 ml; soil and feces – 1 g (wet weight); cloacae – 1 ml.
The finding of HF183 in gull cloacal material is especially interesting. The cloaca (internal cavity) is the terminus of the gull digestive system and a repository for digested material prior to expulsion as feces. Cloacal material is less likely to be contaminated by contact with the environment of the gull. Detection of HF183 in the cloaca suggests that it was ingested. To our knowledge, this is the first reported detection of HF183 in gull cloacae material. It is unlikely that HF183 can colonize gulls, but rather is present only transiently following ingestion at a contaminated site. It is possible that the detection of HF183 in gull cloacae is related to marker specificity, as cross-reactivity has been reported with nonhuman targets such as dog, chicken, and duck (Staley et al., 2012, Boehm et al., 2013). However, the HF183 TaqMan assay used in this study is considered highly specific for human source. For example, Ahmed et al. (2016) reported a 94.6% specificity for HF183 when tested against 2966 individual nonhuman samples.
Detection of HF183 in gull cloacae and feces suggests that under some circumstances gulls may acquire human fecal bacteria. Surveys in Europe and North America have pointed to the ability of gulls to acquire potentially pathogenic bacteria including Campylobacter and Salmonella (Kinzelman et al., 2008, Lévesque et al., 2000, Quessy and Messier, 1992, Tizard, 2004). Gulls have also been shown to carry strains of E. coli resistant to antibiotics important in human medicine (Bonnedahl et al., 2009). Further, correlations between FIB and gull markers have implicated gulls as potential sources of FIB and pathogens to beaches (Goodwin et al., 2016). While gulls may not be common vectors of human fecal bacteria, in scenarios where there are large concentrations of gulls, or where interactions between gulls and people are likely, the public health relevance increases, particularly if the prevalence of human fecal bacteria at the sites gulls visit is high (Quessy and Messier, 1992).
3.2. Radio-telemetry corroborates potential for gulls to disperse human-associated microorganisms
To further assess the possibility of gulls acquiring and transporting human-associated bacteria, gulls (n = 27 gulls) were captured on public beaches and fit with radio-telemetry transmitters to track their movements. Following release, Jordan (2014) located gulls by radio-telemetry inland at the waste sites (24 km from capture site to CL and 38 km to MWM), as far north as Muskegon State Park (22 km from capture), and as far south as Benton Harbor, MI (105 km from capture). Five gulls were documented traveling between human waste sites (landfills or wastewater lagoons) and beaches. All samples collected from these five gulls tested positive for enterococci, E. coli, and Gull4 markers (Table 4). Cloacae samples from two of the five (40%) radio-tagged gulls that traveled between waste sites and beaches also tested positive for HF183, whereas only two of 22 (11%) radio-tagged gulls that did not visit waste sites during the study tested positive for HF183 (Table 4). The two cloacae samples from the tagged gulls that were positive for HF183 were negative for Campylobacter species, while for the other tagged gulls (negative for HF183), Campylobacter signals were detected.
Table 4.
Quantification of positive signals for FIB, human- and gull-associated and pathogen markers detected in single feces and cloacae samples from five individual gulls tracked with radio telemetry traveling between waste sites and beaches.
| Mean positive signal concentration (DNA copies/unit) | ||||||||
|---|---|---|---|---|---|---|---|---|
| EC23S857 | Entero1 | GenBac3 | HF183 | Gull4 | Camp2 | Faecalis1 | Faecium1 | |
| Gull feces (1 g) | ||||||||
| aGHCB to CL & GHCB to MWMb | 1.8 × 103 | 6.5 × 104 | 6.5 × 102 | < LOQ | 8.9 × 107 | 3.8 × 106 | < LOQ | < LOQ |
| Gull cloacae (1 ml) | ||||||||
| GHCB to CL & GHCB to MWMb | 2.0 × 103 | 1.1 × 104 | 1.8 × 102 | 6.3 × 101 | 7.0 × 103 | < LOQ | < LOQ | < LOQ |
| GHCB to CL | 8.3 × 102 | 4.5 × 103 | < LOQ | < LOQ | 2.3 × 105 | 1.2 × 106 | < LOQ | < LOQ |
| GHCB to MWM & GHCB to ML | 9.7 × 102 | 4.2 × 104 | < LOQ | < LOQ | 3.4 × 106 | 3.7 × 104 | < LOQ | < LOQ |
| GHCB to CL | 1.3 × 103 | 2.9 × 104 | < LOQ | 5.8 × 101 | 9.0 × 105 | < LOQ | 6.7 × 104 | < LOQ |
| GHCB to CL | 1.3 × 103 | 7.4 × 105 | < LOQ | < LOQ | 4.0 × 104 | 8.8 × 104 | 3.2 × 106 | < LOQ |
< LOQ, below limit of quantification. LOQs and range of amplification efficiency for each qPCR are noted in Table S1.
Flight path, gulls were tracked flying from site A to B using radio telemetry technology where GHCB to CL is Grand Haven City Beach to Coopersville Landfill; GHCB to MWM is Grand Haven City Beach to Muskegon Wastewater Management lagoon; GHCB to ML is Grand Haven City Beach to Muskegon Landfill.
Feces and cloacae samples were collected from the same gull.
3.3. The relationship between markers for gull and human contamination suggests a potential for gulls to act as a transport vector of human pathogens
This study hypothesizes that gulls that frequent sites with human waste can acquire human-associated bacteria and thus represent a potential vector for subsequent transport. To test this, samples were collected from sites of human waste and from public beaches and evaluated for an overlap in distribution of human-fecal bacteria and gull markers. Forty-five samples were taken of landfill run-off and wastewater storage lagoon water. E. coli and Bacteroidetes markers were frequently detected in these samples (i.e., 98%). The human marker, HF183, was found in 40% of the samples (Table 2). The campylobacter marker was also frequently detected in these samples (i.e., 96%), whereas two species of enterococci, E. faecalis and E. faecium were found in 29% and 16% of samples, respectively (Table 2). All recreational beach water samples (n = 20) tested positive for E. coli and Bacteroidetes markers while 55% of the samples tested positive for HF183 (Table 2). No recreational beach water samples (n = 20) tested positive for the two enterococci species, while 75% of the samples tested positive for campylobacter marker and tested positive for each E. coli and general bacteroidetes (Table 2). These sites were also tested for the presence of gull markers. Gull2 and Gull4 were present in 42% and 60% of the samples collected at human waste sites, respectively. Most beach water samples were positive for gull markers (85% for Gull2 and 100% for Gull4). These abundances and concentrations of gull marker (Table 3) are consistent with other studies that examine waters thought to be contaminated with gull fecal waste (Cloutier and McLellan, 2017, Russell et al., 2013, Ryu et al., 2012). Our study is in agreement with the high prevalence of E. faecalis and E. faecium noted by Ryu et al. (2013) for 24 different animals including gulls, suggesting that these species are cosmopolitan (i.e., present in various hosts). Campylobacter species were also detected in the majority of the gull samples as well samples from the human waste sites. When coupled with the radio-telemetry findings, the qPCR data suggest that gulls traveling between human waste sites and public beaches may be dispersing human-associated microbes. Surprisingly, the prevalence of the enterococci marker and both Enterococcus species markers was relatively low (i.e., 30% and 0%, respectively) in the beach water samples. In contrast, 55% and 100% of the beach samples tested positive for HF183 and Gull4 markers respectively, suggesting that human and gull feces appear to be two dominant fecal contamination sources in the study area.
The presence, abundance, and distribution of gull and human markers suggest a possible role for gulls as transport vectors of human fecal bacteria. In the study area, which included both human waste sites and recreational beaches, markers associated with human fecal contamination and with gulls overlapped geographically and temporally. Other studies have also reported the presence of gull and human markers in the same environmental water samples (Byappanahalli et al., 2015, Green et al., 2012, Lauer, 2015, Russell et al., 2013). When looking at the presence of Gull4 and HF183 in waste site samples, both markers were found together in 11 samples, Gull4 alone was found in 16 samples, HF183 alone was found in 7 samples, and 11 samples did not have either marker. For public beach samples, HF183 was never found in a water sample that did not also have Gull4. Gulls are opportunistic scavengers, and it is speculated that gulls that feed at sites of human waste may acquire human-associated microbes and subsequently transport them to beach sites (Converse et al., 2012). These results are consistent with other studies that show relatedness between bacteria present in gull feces and at waste sites (Nelson et al., 2008) and with studies that indicate gulls foraging at waste sites carry potential human pathogens (Fricker, 1984, Quessy and Messier, 1992, Ramos et al., 2010). Our data do not suggest gulls are the only source of FIB or human marker in the sites sampled. However, combined with the results showing HF183 in gull cloacae, they further support that gulls that frequent human waste sites may acquire human-associated bacteria.
3.4. The Gull4 marker was found to be a more sensitive marker when compared to Gull2
The presence of gulls was evaluated in this study with two different assays (Gull2 and Gull4). The marker Gull4 was found in 100% of the cloacae samples and 87% of the feces, but Gull2 was found in only 74% of cloacae and 67% of feces (Table 2). When looking at the relationship between both markers, Gull2 was not found without Gull4. Additionally, all of the cloacae samples from paired collections tested positive for the Gull4 marker whereas only eight (73%) of feces were positive for Gull4.
Ryu et al. (2012) also observed a high incidence of the Gull2 and Gull4 markers when tested against the feces of 255 individual gulls. Both assays also generated false positive signals when tested against non-target feces, but these were mostly associated to other avian species, suggesting that C. marimammalium prefers the avian gut environment. The genome of C. marimammalium has revealed a reduced metabolic network and several functions that are linked to a symbiotic lifestyle (Weigand et al., 2013). It should also be noted that Catellicoccus spp. have also been seen in migratory birds from stopover beaches simultaneously colonized by gulls (Grond et al., 2014), suggesting that fecal bacteria can be transferred between different co-inhabiting avian species.
3.5. Implications for beach management
Gulls that visit landfills, wastewater management facilities, and beaches for foraging and loafing are a possible reservoir and transport vector of FIB and human-associated bacteria. This study integrated tools of wildlife ecology and molecular microbiology to evaluate this potential. The detection of low levels of the human-associated marker, HF183, in the feces and cloacae of gulls in this study implies that gulls are able to acquire bacteria associated with human fecal waste. Further, detecting HF183 in samples collected from radio-tagged gulls with documented travel between human waste sites and public beaches suggest the potential for gulls to disperse human-associated microorganisms in the beach landscape.
Gull acquisition and transport of human bacteria to beaches may be an infrequent occurrence, however, the potential should be considered when gull abundance is high and when human waste sites are nearby and are accessible to gulls. To reduce this potential, mechanisms should be employed at waste sites to exclude gulls and should be considered at public beaches to reduce gull visitation.
Supplementary Material
Highlights.
Gull cloacae and feces samples contained the human specific marker, HF 183.
Markers for gull and human contamination showed spatial and temporal overlap.
Radio-telemetry supports potential for gulls to disperse human-associated microbes.
Gulls may act as transport vectors of human pathogens.
Gull4 was a more sensitive source-tracking marker when compared to Gull2.
Acknowledgements
We would like to thank the Muskegon County Wastewater Management System for site access, Lucas Calloway (CMU) for field assistance, and Michael Elk (EPA) for laboratory assistance. We would also like to thank Jim Ponshair, Feller DeWitt, and Richard Pedler of the Muskegon County Nature Club for gull nest data. Funding: Ottawa County, MI funded gull telemetry and a graduate student award from ORGS, CMU supported work by Q.D-W. Our research was approved by the Institutional Animal Care and Use Committee at Central Michigan University (IACUC no. 12–14A). This is contribution #88 of the Central Michigan University Institute for Great Lakes Research.
The U. S. Environmental Protection Agency, through its Office of Research and Development, partially funded and collaborated in the research described herein. Any opinions expressed in this paper are those of the authors and do not necessarily reflect the views of the agency; therefore, no official endorsement should be inferred. Any mention of trade names or commercial products does not constitute endorsement or recommendation for use.
References
- Aavitsland and Hofshagen, 1999.Aavitsland P, Hofshagen T Salmonella outbreak in Herøy municipality News from Environmental and Social Medicine, 3, National Institute of Public Health, Norway: (1999), p. 12 [Google Scholar]
- Ahmed et al., 2016.Ahmed W, Hughes B, Harwood VJ Current status of marker genes of Bacteroides and related taxa for identifying sewage pollution in environmental waters Water, 8 (6) (2016), p. 231, 10.3390/w8060231 [DOI] [Google Scholar]
- Alderisio and DeLuca, 1999.Alderisio KA, DeLuca N Seasonal enumeration of fecal coliform bacteria from the feces of ring-billed gulls (Larus delawarensis) and Canada Geese (Branta canadensis) Appl. Environ. Microbiol, 65 (12) (1999), pp. 5628–5630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander, 2000.Alexander DJ A review of avian influenza in different bird species Vet. Microbiol, 74 (2000), pp. 3–13 (PII: S0378–1135(00)00160–7) [DOI] [PubMed] [Google Scholar]
- Alm et al., 2003.Alm EW, Burke J, Spain A Fecal indicator bacteria are abundant in wet sand at freshwater beaches Water Res, 37 (2003), pp. 3978–3982, 10.1016/S0043-1354(03)00301-4 [DOI] [PubMed] [Google Scholar]
- Araújo et al., 2014.Araújo S, Henriques IS, Leandro SM, Alves A, Pereira A, Correia A Gulls identified as major source of fecal pollution in coastal waters: a microbial source tracking study Sci. Total Environ, 470–471 (2014), pp. 84–91, 10.1016/j_scitotenv.2013.09.075 [DOI] [PubMed] [Google Scholar]
- Belant et al., 1998.Belant JL, Ickes SK, Seamans TW Importance of landfills to urban-nesting herring and ring-billed gulls Landsc. Urban Plan, 43 (1998), pp. 11–19 (PII S0169–2046(98)00100–5) [Google Scholar]
- Boehm et al., 2013.Boehm AB, van de Werfhorst LC, Griffith JF, Holden PA, Jay JA, Shanks OC, Wang D, Weisberg SB Performance of forty-one microbial source-tracking methods: a twenty-seven lab evaluation study Water Res, 47 (2013), pp. 6812–6828, 10.1016/j/waters.2012.12.046 [DOI] [PubMed] [Google Scholar]
- Bonnedahl et al., 2009.Bonnedahl J, Drobni M, Gauther-Clerc M, Hernandez J, Granholm S, Kayser Y, Melhus A, Kahlmeter G, Waldenström JA, Olsen B Dissemination of Escherichia coli with CTX-M type ESBL between humans and yellow-legged gulls in the south of France Plos One, 4 (6) (2009), pp. 1–6, 10.1371/journal.pone.0005958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butterfield et al., 1983.Butterfield J, Coulson JC, Kearsey SV, Monaghan P, McCoy JH, Spain GE The herring gull Larus argentatus as a carrier of salmonella J. Hyg. Camb, 91 (1983), pp. 429–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byappanahalli et al., 2015.Byappanahalli MN, Nevers MB, Whitman RL, Ge Z, Shively D, Spoljaric A, Przybyla-Kelly Wildlife K, urban inputs, and landscape configuration are responsible for degraded swimming water quality at an embayed beach J. Great Lakes Res, 41 (2015), pp. 156–163, 10.1016/j.jglr.2014.11.027 [DOI] [Google Scholar]
- Chern et al., 2011.Chern EC, Siefring S, Paar J, Doolittle M, Haugland RA Comparison of quantitative PCR assays for Escherichia coli targeting ribosomal RNA and single copy genes Lett. Appl. Microbiol, 52 (2011), pp. 298–306, 10.1111/j.1472-765X.2010.03001.x [DOI] [PubMed] [Google Scholar]
- Cloutier and McLellan, 2017.Cloutier DD, McLellan SL Distribution and differential survival of traditional and alternative indicators of fecal pollution at freshwater beaches Appl. Environ. Microbiol, 83 (4) (2017), pp. e02811–16, 10.1128/AEM.02881-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Converse et al., 2012.Converse RR, Kinzelman JL, Sams EA, Hudgens E, Dufour AP, Ryu H, Santo-Domingo JW, Kelty CA, Shanks OC, Siefring SD, Haugland RA, Wade T Dramatic improvements in beach water quality following gull removal Environ. Sci. Technol, 46 (2012), pp. 10206–10213, 10.1021/es302306b [DOI] [PubMed] [Google Scholar]
- Coulson et al., 1983.Coulson JC, Butterfield J, Thomas C The herring gull Larus argentatus as a likely transmitting agent of Salmonella montevideo to sheep and cattle J. Hyg. Camb, 91 (1983), pp. 437–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolejská et al., 2009.Dolejská M, Bierošová B, Kohoutová L, Literák I, Ćížek A Antibiotic-resistant Salmonella and Escherichia coli isolates with integrons and extended-spectrum beta-lactamases in surface water and sympatric black-headed gulls J. Appl. Microbiol, 106 (6) (2009), pp. 1941–1950, 10.1111/j.1365-2672.2009.04155.x [DOI] [PubMed] [Google Scholar]
- Ebert et al., 2016.Ebert LA, Schlemper JC, Pelisser MR, Pereira BA, da Silva MAC, Branco JO Pathogenic bacteria associated with kelp gull Larus dominicanus (Charadriiformes, Laridae) on the coast of Santa Catarina State – Brazil Int. J. Curr. Microbiol. App. Sci, 5 (5) (2016), pp. 458–473, 10.20546/ijcmas.2016.505.048 [DOI] [Google Scholar]
- Edge and Hill, 2007.Edge TA, Hill S Multiple lines of evidence identify the sources of fecal pollution at a freshwater beach in Hamilton Harbour, Lake Ontario Water Res, 41 (2007), pp. 3585–3594, 10.1016/j.watres.2007.05.012 [DOI] [PubMed] [Google Scholar]
- Edge et al., 2010.Edge TA, Hill S, Seto P, Marsalek J Library-dependent and library-independent microbial source tracking to identify spatial variation in faecal contamination sources along a Lake Ontario beach (Ontario, Canada) Water Sci. Technol, 62 (3) (2010), pp. 719–727, 10.2166/wst.2010.335 [DOI] [PubMed] [Google Scholar]
- Fenlon, 1981.Fenlon DR Seagulls (Larus spp.) as vectors of salmonellae: an investigation into the range of serotypes and numbers of salmonellae in gull feces J. Hyg. Camb, 86 (2) (1981), pp. 195–202, 10.1017/S0022172400068911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferns and Mudge, 2000.Ferns PN, Mudge GP Abundance, diet and Salmonella contamination of gulls feeding at sewage outfalls Water Res, 34 (10) (2000), pp. 2653–2660 (PII S0043–1354(99)00427–3) [Google Scholar]
- Fogarty et al., 2003.Fogarty LR, Haack SK, Wolcott MJ, Whitman RL Abundance and characteristics of the recreational water quality indicator bacteria Escherichia coli and enterococci in gull faeces J. Appl. Microbiol, 94 (5) (2003), pp. 865–878, 10.1046/j.1365-2672.2003.01910.x [DOI] [PubMed] [Google Scholar]
- Fricker, 1984.Fricker CR A note on salmonella excretion in the black headed gull (Larus ribibundus) feeding at sewage treatment works J. Appl. Bacteriol, 56 (1984), pp. 499–502 [DOI] [PubMed] [Google Scholar]
- Goodwin et al., 2016.Goodwin KD, Gruber S, Vondrak M, Crumpacker A Watershed assessment with beach microbial source tracking and outcomes of resulting gull management Environ. Sci. Technol, 50 (2016), pp. 9900–9906, 10.1021/acs.est.6b02564 [DOI] [PubMed] [Google Scholar]
- Goodwin et al., 2017.Goodwin KD, Schriewer A, Jirik A, Curtis K, Crumpacker A Consideration of natural sources in a bacteria TMDL - Lines of evidence, including beach microbial source tracking Environ. Sci. Technol, 51 (2017), pp. 7775–7784, 10.1021/acs.est.6b05886 [DOI] [PubMed] [Google Scholar]
- Green et al., 2012.Green HC, Dick LK, Gilpin B, Samadpour M, Field KG Genetic markers for rapid PCR-based identification of gull, Canada goose, duck, and chicken fecal contamination in water Appl. Environ. Microbiol, 503–510 (2012), 10.1128/AEM.05734-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grond et al., 2014.Grond K, Ryu H, Baker A, Santo Domingo J, Buehler D Gut microbiota of two migratory shorebird species during spring-migration staging in Delaware Bay, USA J. Ornithol, 155 (2014), pp. 969–977 [Google Scholar]
- Haack et al., 2003.Haack SK, Fogarty LR, Wright C Escherichia coli and enterococci at beaches in the Grand Traverse Bay, Lake Michigan; Sources, characteristics, and environmental pathways Environ. Sci. Technol, 37 (2003), pp. 3275–3282, 10.1021/es021062n [DOI] [PubMed] [Google Scholar]
- Haugland et al., 2010.Haugland RA, Varma M, Sivaganesan M, Kelty C, Peed L, Shanks OC Evaluation of genetic markers from the 16S rRNA gene V2 region for use in quantitative detection of selected Bacteroidales species and human fecal waste by qPCR Syst. Appl. Microbiol, 33 (2010), pp. 348–357, 10.1016/j.syapm.2010.06.001 [DOI] [PubMed] [Google Scholar]
- Hernandez et al., 2013.Hernandez J, Johansson A, Stedt J, Bengtsson S, Porczak A, Granholm S, González-Acuña D, Olsen B, Bonnedahl J, Drobni M Characterization and comparison of extended-spectrum β-lactamase (ESBL) resistance genotypes and population structure of Escherichia coli isolated from Franklin’s gulls (Leucophaeus pipixan) and humans in Chile Plos One, 8 (9) (2013), pp. 1–9, 10.1371/journal.pone.0076150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hlavsa et al., 2015.Hlavsa MC, Roberts VA, Kahler AM, Hilborn ED, Mecher TR, Beach MJ, Wade TJ, Yoder JS Outbreaks of illness associated with recreational water – United States, 2011–2012 Morb. Mortal. Wkly Rep, 64 (24) (2015), pp. 668–672 [PMC free article] [PubMed] [Google Scholar]
- IDL. Infectious Diseases Laboratory, 2016.IDL. Infectious Diseases Laboratory Instructions for Avian Sample Collection and Submission www.vet.uga.edu/idl/pdf/Instructions_for_Sample_Collection_and_Submission-1.pdf (2016)
- Johnston et al., 1979.Johnston WS, MacLachlan GK, Hopkins GE The possible involvement of seagulls (Larus sp) in the transmission of salmonella in dairy cattle Vet. Rec, 105 (23) (1979), pp. 526–527 [PubMed] [Google Scholar]
- Jordan, 2014.Jordan DW Exclusion of Gulls From Recreational Beaches Using Border Collies and Monitoring Gull Habitat Use Relative to Human Infrastructure Master’s Thesis Central Michigan University, Mt. Pleasant, MI: (2014) Retrieved 01/31/2017 from https://scholarly.cmich.edu/cgi-bin/imageserver.pl?oid=CMUGR2014-30&getpdf=true [Google Scholar]
- Kapperud and Rosef, 1983.Kapperud G, Rosef O Avian wildlife reservoir of Campylobacter fetus subsp. jejuni, Yersinia spp., and Salmonella spp. in Norway Appl. Environ. Microbiol, 45 (2) (1983), pp. 375–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinzelman et al., 2008.Kinzelman J, McLellan SL, Amick A, Preedt J, Scopel CO, Olapade O, Gradus S, Singh A, Sedmak G Identification of human enteric pathogens in gull feces at Southwestern Lake Michigan bathing beaches Can. J. Microbiol, 54 (2008), pp. 1006–1015, 10.1139/W08-096 [DOI] [PubMed] [Google Scholar]
- Kirschner et al., 2004.Kirschner AKT, Zechmeister TC, Kavka GG, Beiwl C, Herzig A, Mach RL, Farnleitner AH Integral strategy for evaluation of fecal indicator performance in bird-influenced saline inland waters Appl. Environ. Microbiol, 70 (12) (2004), pp. 7396–7403, 10.1128/AEM.70.12.7396-7403.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauss et al., 2013.Krauss S, Pryor SP, Raven G, Danner A, Kayali G, Webby RJ, Webster RG Respiratory tract versus cloacal sampling of migratory ducks for influenza A viruses: are both ends relevant? Influenza Other Respir. Viruses, 7 (2013), pp. 93–96, 10.1111/j.1750-2659.2012.00359.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauer, 2015.Lauer KC Quantitative Analysis of Fecal Contamination in Stormwater Conveyance Systems and the Effects of Storm Drain Discharge on Beach Water Quality in Wrightsville Beach, NC Master’s Thesis University of North Carolina, Chapel Hill, NC: (2015) Retrieved 01/31/2017 from https://cdr.lib.unc.edu/record/uuid:22fdd6d8-be3e-40a6-b837-854b25d38943 [Google Scholar]
- Lévesque et al., 2000.Lévesque B, Brousseau P, Dewailly E, Joly J Study of the bacterial content of ring-billed gull droppings in relation to recreational water quality Water Res, 34 (4) (2000), pp. 1089–1096 [Google Scholar]
- Lu et al., 2008.Lu J, Santo Domingo JW, Lamendella R, Edge T, Hill S Phylogenetic diversity and molecular detection of bacteria in gull feces Appl. Environ. Microbiol, 74 (13) (2008), pp. 3969–3976, 10.1128/AEM.00019-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu et al., 2011a.Lu J, Ryu H, Hill S, Schoen M, Ashbolt N, Edge TA, Santo Domingo J Distribution and potential significance of a gull fecal marker in urban coastal and riverine areas of southern Ontario, Canada Water Res, 45 (2011), pp. 3960–3968, 10.1016/j.watres.2011.05.003 [DOI] [PubMed] [Google Scholar]
- Lu et al., 2011b.Lu J, Ryu H, Santo Domingo J, Griffith JF, Ashbolt N Molecular detection of Campylobacter spp. in California gull (Larus californicus) excreta Appl. Environ. Microbiol, 77 (14) (2011), pp. 5034–5039, 10.1128/AEM.00018-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig and Schleifer, 2000.Ludwig W, Schleifer K-H How quantitative is quantitative PCR with respect to cell counts? Syst. Appl. Microbiol, 23 (4) (2000), pp. 556–562, 10.1016/S0723-2020(00)80030-2 [DOI] [PubMed] [Google Scholar]
- Lund et al., 2004.Lund M, Nordentoft S, Pedersen K, Madsen M Detection of Campylobacter spp. in chicken fecal samples by real-time PCR J. Clin. Microbiol, 42 (11) (2004), pp. 5125–5132, 10.1128/JCM.42.11.5125-5132.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meerburg et al., 2011.Meerburg BG, Koene GJ, Kleijn D Escherichia coli concentrations in feces of geese, coots, and gulls residing on recreational water in The Netherlands Vector Borne Zoonotic Dis., 11 (5) (2011), pp. 601–603, 10.1089/vbz.2010.0218 [DOI] [PubMed] [Google Scholar]
- Nelson et al., 2008.Nelson M, Jones SH, Edwards C, Ellis JC Characterization of Escherichia coli populations from gulls, landfill trash, and wastewater using ribotyping Dis. Aquat. Org, 81 (2008), pp. 53–63, 10.3354/dao01937 [DOI] [PubMed] [Google Scholar]
- Noble et al., 2006.Noble RT, Griffith JF, Blackwood AD, Fuhrman JA, Gregory JB, Hernandez X, Liang X, Bera AA, Schiff K Multitiered approach using quantitative PCR to track sources of fecal pollution affecting Santa Monica Bay, California Appl. Environ. Microbiol, 72 (2) (2006), pp. 1604–1612, 10.1128/AEM.72.2.1604-1612.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponshair, 1974.Ponshair JF Ring-billed gulls nesting in Muskegon County The Jack-Pine Warbler, 52 (4) (1974), pp. 194–195 [Google Scholar]
- Ponshair, 2006.Ponshair JF Census of breeding Ring-billed gulls at the Muskegon wastewater system 1974–2006 Natural History Survey at the Muskegon Wastewater System, Muskegon County: Report No. 31 (2006) [Google Scholar]
- Quessy and Messier, 1992.Quessy S, Messier S Prevalence of Salmonella spp., Campylobacter spp. and Listeria spp. in ring-billed gulls (Larus delawarensis) J. Wildl. Dis, 28 (4) (1992), pp. 526–531 [DOI] [PubMed] [Google Scholar]
- Ramos et al., 2010.Ramos R, Cerdà-Cuéllar M, Ramírez F, Jover L, Ruiz X Influence of refuse sites on the prevalence of Campylobacter spp. and Salmonella serovars in seagulls Appl. Environ. Microbiol, 76 (9) (2010), pp. 3052–3056, 10.1128/AEM.02524-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Retamal et al., 2015.Retamal P, Fresno M, Dougnac C, Gutierrez S, Gornall V, Vidal R, Vernal R, Pujol M, Barreto M, González-Acuña D, Abalos P Genetic and phenotypic evidence of the Salmonella enterica serotype Enteritidis human-animal interface in Chile Front. Microbiol, 6 (464) (2015), pp. 1–10, 10.3389/fmicb.2015.00464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell et al., 2013.Russell TL, Sassoubre LM, Wang D, Masuda S, Chen H, Soetjipto C, Hassaballah A, Boehm A A coupled modeling and molecular biology approach to microbial source tracking at Cowell Beach, Santa Cruz, CA, United States Environ. Sci. Technol, 47 (2013), pp. 10231–10239, 10.1021/es402303w [DOI] [PubMed] [Google Scholar]
- Ryu et al., 2012.Ryu H, Griffith JF, Khan IUH, Hill S, Edge TA, Toledo-Hernandez C, Gonzalez-Nieves J, Santo J Domingo Comparison of gull feces-specific assays targeting the 16S rRNA genes of Catellicoccus marimammalium and Streptococcus spp. Appl. Environ. Microbiol, 78 (6) (2012), pp. 1909–1916, 10.1128/AEM.07192-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu et al., 2013.Ryu H, Henson M, Elk M, Toledo-Hernandez C, Griffith J, Blackwood D, Noble R, Gourmelon M, Glassmeyer S, Santo JW Domingo Development of quantitative PCR assays targeting the 16S rRNA genes of Enterococcus spp. and their application to the identification of Enterococcusspecies in environmental samples Appl. Environ. Microbiol, 79 (1) (2013), pp. 196–204, 10.1128/AEM.02802-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santo Domingo et al., 2003.Santo Domingo JW, Siefring SC, Haugland RA Real-time PCR method to detect Enterococcus faecalis in water Biotechnol. Lett, 25 (3) (2003), pp. 261–265, 10.1023/A:1022303118122 [DOI] [PubMed] [Google Scholar]
- Sarker et al., 2012.Sarker MAH, Jahan M, Parvin MN, Malek MA, Hossain MT Identification of bacterial flora isolated from apparently healthy water birds of Dhaka Zoo of Bangladesh Bangl. J. Vet. Med, 10 (1&2) (2012), pp. 21–26 (ISSN: 1729–7893 (Print) 2308–0922 (Online)) [Google Scholar]
- Schoen and Ashbolt, 2010.Schoen ME, Ashbolt NJ Assessing the pathogen risk to swimmers at non-sewage impacted recreational beaches Environ. Sci. Technol, 44 (2010), pp. 2286–2291, 10.1021/es903623q [DOI] [PubMed] [Google Scholar]
- Siefring et al., 2008.Siefring S, Varma M, Atikovic E, Wymer L, Haugland RA Improved real-time PCR assays for the detection of fecal indicator bacteria in surface waters with different instrument and reagent systems J. Water Health, 6 (2) (2008), pp. 225–237, 10.2166/wh.2008.022 [DOI] [PubMed] [Google Scholar]
- Soller et al., 2010.Soller JA, Schoen ME, Bartrand T, Ravenscroft JE, Ashbolt NJ Estimated human health risks from exposure to recreational waters impacted by human and non-human sources of faecal contamination Water Res, 44 (2010), pp. 4674–4691, 10.1016/j.watres.2010.06.049 [DOI] [PubMed] [Google Scholar]
- Staley and Edge, 2016.Staley ZR, Edge TA Comparative microbial source tracking methods for identification of fecal contamination courses at Sunnyside Beach in the Toronto region area of concern J. Water Health, 14 (5) (2016), pp. 839–850, 10.2166/wh.2016.296 [DOI] [PubMed] [Google Scholar]
- Staley et al., 2012.Staley C, Gordon KV, Schoen ME, Harwood VJ Performance of two quantitative PCR methods for microbial source tracking of human sewage and implications for microbial risk assessment in recreational waters Appl. Environ. Microbiol, 78 (20) (2012), pp. 7317–7326, 10.1128/AEM.01430-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tizard, 2004.Tizard I Salmonellosis in wild birds Sem. Avian Exotic Pet Med, 13 (2) (2004), pp. 50–66, 10.1053/j.saep.2004.01.008 [DOI] [Google Scholar]
- Toro et al., 2016.Toro M, Retamal P, Ayers S, Barreto M, Allard M, Brown EW, Gonzalez-Escalona N Whole-genome sequencing analysis of Salmonella enterica serovar Enteritidis isolates in Chile provides insights into possible transmission between gulls, poultry, and humans Appl. Environ. Microbiol, 82 (20) (2016), pp. 6223–6232, 10.1128/AEM.01760-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tracey, 2010.Tracey JP Risk-based surveillance of avian influenza in Australia’s wild birds Wildl. Res, 37 (2010), pp. 134–144, 10.1071/WR09152 [DOI] [Google Scholar]
- Tsiodras et al., 2008.Tsiodras S, Kelesidis T, Kelesidis I, Bauchinger U, Falagas ME Human infections associated with wild birds J. Inf. Secur, 56 (2008), pp. 83–98, 10.1016/j.jinf.2007.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hoorebeke et al., 2009.Van Hoorebeke S, Van Immerseel F, De Vylder J, Ducatelle R, Haesebrouck F, Pasmans F, de Kruif A, Dewulf J Faecal sampling underestimates the actual prevalence of Salmonella in laying hen flocks Zoonoses Public Health, 56 (2009), pp. 471–476, 10.1111/j.1863-2378.2008.01211.x [DOI] [PubMed] [Google Scholar]
- Varela et al., 2015.Varela AR, Manageiro V, Ferreira E, Guimarães MA, da Costa PM, Caniça M, Manaia CM Molecular evidence of the close relatedness of clinical, gull and wastewater isolates of quinolone-resistant Escherichia coli J. Glob. Antimicrob. Res, 3 (2015), pp. 286–289, 10.1016/j.jgar.2015.07.008 [DOI] [PubMed] [Google Scholar]
- Webster et al., 1992.Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y Evolution and ecology of Influenza A viruses Microbiol. Rev, 56 (1) (1992), pp. 152–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigand et al., 2013.Weigand MR, Ryu H, Bozcek L, Konstantinidis KT, Santo Domingo J Draft genome sequence of Catellicoccus marimammalium, a novel species commonly found in gull feces Genome Announc., 1 (1) (2013), pp. e00019–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan et al., 1988.Whelan CD, Monaghan P, Girdwood RWA, Fricker CR The significance of wild birds (Larus sp.) in the epidemiology of campylobacter infections in humans Epidemiol. Infect, 101 (1988), pp. 259–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitman and Nevers, 2003.Whitman RL, Nevers MB Foreshore sand as a source of Escherichia coli in nearshore water of a Lake Michigan beach Appl. Environ. Microbiol, 69 (9) (2003), pp. 5555–5562, 10.1128/AEM.69.9.555-5562.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zar, 2009.Zar JH Biostatistical Analysis (fifth ed), Pearson, New York: (2009) [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.