Abstract
Background/Aim: Neurological symptoms (neuro-Behçet’s disease; NBD) occur in a fraction of Behçet’s disease (BD) patients and often present with parenchymal brain lesions and clinical exacerbations. Our aim was to identify genes associated with attack and remission periods of NBD. Materials and Methods: Microarray analysis was performed using peripheral blood mononuclear cell (PBMC) samples obtained during attack and remission periods of five NBD patients. Expression levels of the most significantly up-regulated genes were measured with real-time PCR using PBMC samples of 15 NBD patients and 20 healthy controls. Results: During NBD attacks, the most remarkably up-regulated genes were defensin alpha 1B (DEFA1B) and NLR family, pyrin domain containing 3 (NLRP3). Real time PCR studies showed significantly increased DEFA1B and NLRP3 expression levels during attacks. Conclusion: Immunological factors showing the most significant increase in expression during NBD attacks were primarily associated with innate immunity functions. DEFA1B and NLRP3 can be used as biomarkers for estimation of disease activity in NBD.
Keywords: Behçet’s disease, neuro-Behçet’s disease, microarray, innate immunity, inflammation
Behçet’s disease (BD) is a multisystem inflammatory disorder characterized by recurrent oral aphthae, genital ulcerations and ocular vasculitis or uveitis (1). The clinical features of BD are believed to be triggered by environmental factors, such as mucosal microorganisms in individuals with a strong genetic susceptibility to yield exaggerated inflammatory responses (1,2). Thus, enhanced activity of innate immunity components, such as neutrophils, natural killer (NK) cells and endothelial cells, is a prominent feature of BD (3-5).
Central nervous system involvement (neuro-Behçet’s disease, NBD) is encountered in 5-15% of patients and manifests mostly in parenchymal and less frequently in vascular forms. NBD causes parenchymal lesions primarily in brainstem, basal ganglia and diencephalon regions of the brain and is typified with attack and remission periods (6). Parenchymal NBD symptoms are mostly caused by infiltrating macrophages, neutrophils and lymphocytes (7,8). Very few studies have addressed the gene expression profiles of BD patients (9,10). There are currently no studies comparing immunological gene expression patterns of attack and remission periods of NBD patients and physio-pathological factors leading to clinical exacerbation in NBD are largely unknown. Identification of mechanisms specific to the attack and remission periods might shed light on disease pathogenesis and aid in innovation of specific treatment methods.
To contribute to the identification of molecular pathways engaged in attack and remission periods of NBD, gene expression profiles were examined in the peripheral blood mononuclear cells (PBMC) obtained during attack and remission periods of the same NBD patients, using microarray analysis. Differentially up-regulated genes were validated with-real time PCR analysis.
Materials and Methods
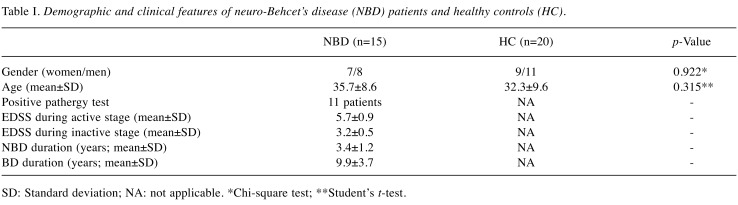
Patients. Parenchymal NBD patients (n=15) fulfilling the diagnostic criteria for BD (11) and age-gender matched healthy controls (n=20) were enrolled. Demographical and clinical features of all participants are shown in Table I. Blood samples were obtained from all patients first during the presence of active clinical findings (active stage) and then 3 months after the clinical episode (inactive stage). During the active stage, all patients displayed the typical parenchymal brain lesions on cranial magnetic resonance imaging (MRI) studies. None of the patients showed MRI lesions during the second blood sampling. To avoid the confounding effects of systemic inflammation, NBD patients with active non-neurological symptoms during blood sampling were not included. None of the patients were under immunosuppressive or immunomodulating drug treatment during blood sampling. NBD patients with other existing disorders were excluded. The study was approved (approval number: 2018/112) by Ethics Committee of Istanbul University and an informed consent was received from each participant.
Table I. Demographic and clinical features of neuro-Behcet’s disease (NBD) patients and healthy controls (HC).
SD: Standard deviation; NA: not applicable. *Chi-square test; **Student’s t-test.
Separation of PBMC and RNA extraction. Ten milliliters of peripheral blood from participants were withdrawn and placed into EDTA vacutainer tubes. PBMC were isolated by density gradient separation. Briefly, anticoagulated blood was mixed with an equal volume of Dulbecco’s phosphate buffered saline (DPBS, ThermoFisher Scientific, MA, USA) and slowly layered over Ficoll-Hypaque solution (GE Healthcare, Chicago, IL, USA) up to twice the amount of blood. The mixture was centrifuged at 400 × g for 30 min and PBMC layer at the interphase was collected. Cells were washed twice with DPBS at 400 × g for 10 min and counted in a Neubauer chamber. 5×106 cells were lysed in 600 μl RLT Buffer (Qiagen, Hilden, Germany) and total RNA from PBMCs was purified by RNeasy Mini Kit according to the manufacturer’s recommendations (Qiagen). RNA concentration was measured by NanoDrop 1000 spectrophometer (ThermoFisher Scientific). RNA quality was evaluated by Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA) and samples with an RNA integrity number (RIN) greater than 7 were included in the study.
Microarray analysis. Blood samples of five out of 15 NBD patients obtained during active and inactive stages of the disease (a total of 10 samples) were used for microarray analysis. cRNA was amplified and biotin labeled by TotalPrep™ RNA Amplification Kit (Illumina, San Diego, CA, USA) and quantified by NanoDrop 1000 spectrophometer (ThermoFisher Scientific). cRNA samples were hybridized to HumanHT-12v4 Expression BeadChip (Illumina) consisting 47323 probes. Whole genome expression profiles were determined using an Illumina iScan platform (Illumina). Raw data were processed by GenomeStudio software (Illumina). Quality control was assessed by signal intensities and the background noise was subtracted from each probe. Data were log transformed and normalized by quantile normalization. The probes were filtered below the level of 20% of signal intensity and above 0.1 standard deviation. Unpaired t-test was applied for statistical analysis and asymptotic p<0.05 was accepted as significant. Probes were considered differentially expressed when the fold change was greater than 2.
Real time qPCR. Blood samples of all healthy controls and NBD patients collected during active and inactive stages of the disease were used in real time qPCR experiments for validation of the microarray results. Following total RNA extraction, the RNA quality was measured by the A260/A280 ratio and a NanoDrop Spectrophotometer. RNA was then converted to cDNA by using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA). Quantitative real-time PCR reactions were performed in a Stratagene Mx3005P analyser (Agilent Technologies) by using SYBR green master mix (Roche, Basil, Switzerland) and primers designed for the NLR family, pyrin domain containing 3 (NLRP3) gene (forward: 5’-CGTGAGT CCCATTAAGATGGAGT-3’ and reverse: 5’-CCCGACAGTGGAT ATAGAACAGA-3’) and defensin alpha 1B (DEFA1B) gene (forward: 5’-TCCTTGCTGCCATTCTCCTG-3’ and reverse: 5’-TGAGCCTGGATGCTTTGGAG-3’). The thermal profile consisted of an initial 2-min step at 50˚C and 2-min melting step at 95˚C followed by 40 cycles at 95˚C for 15 sec, 59˚C for 20 sec and 72˚C for 30 sec. Melting curve analyses were completed by 1 cycle at 95˚C for 1-min, 55˚C for 30 sec and 95˚C for 30 sec. The relative mRNA expression levels were normalized to GAPDH expression using the simplified comparative threshold cycle delta, cycle threshold (CT) method [2−(ΔCT gene of interest−ΔCT GAPDH)]. Significance was estimated by analysis of variance (one-way ANOVA).
Results
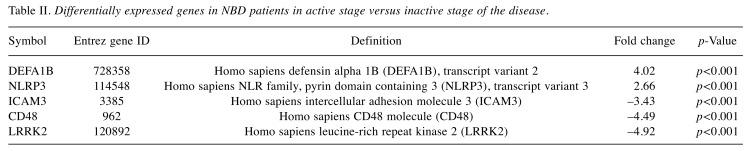
Whole-genome expression profiles of NBD patients. Differential gene expression analysis was performed between two groups (active vs. inactive stage) of NBD patients. Analysis resulted in 433 differentially expressed genes that remained statistically significant after false discovery rate (FDR) correction (FDR<0.1). Among these, 346 genes were up-regulated and 87 genes were down-regulated in the inactive group according to the active group. Differentially expressed genes were sorted according to their fold change values. Up- and down-regulated genes in active and inactive NBD patients with a fold change >2 are listed in Table II.
Table II. Differentially expressed genes in NBD patients in active stage versus inactive stage of the disease.
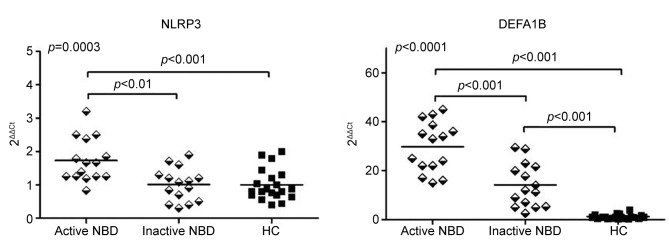
Expression levels of NLRP3 and DEFA1B. For validation of microarray results, expression levels of the most significantly up-regulated genes (DEFA1B and NLRP3) during the attack period were assessed by real time qPCR. Expression level measurements were performed using PBMC samples of healthy controls and NBD patients. Statistical assessment was performed by ANOVA and Tukey’s post-hoc analysis. NBD patients showed significantly increased NLRP3 expression levels during the active stage of the disease (p=0.0003 by ANOVA). In two-group comparisons, active stage samples of NBD patients showed significantly enhanced NLRP3 expression levels as compared to the inactive stage samples of NBD patients (p<0.01 by Tukey) and healthy controls (p<0.001 by Tukey) (Figure 1). There was no significant difference among NLRP3 expression levels of inactive NBD samples and healthy control samples. Likewise, PBMC samples obtained during clinical exacerbation of NBD showed significantly enhanced DEFA1B expression levels (p<0.0001 by ANOVA) compared to those obtained during remission (p<0.001 by Tukey) and those obtained from healthy controls (p<0.001 by Tukey). NBD patients also showed increased DEFA1B expression levels during remission as compared to healthy controls (p<0.001 by Tukey) (Figure 1). Correlation analysis done with Pearson and Spearman tests, as required, failed to show any significant correlations between NLRP3 and DEFA1B levels (measured during active and inactive stages) vs. parameters of age, expanded disability status scale (EDSS) scores, NBD duration and BD duration (not shown).
Figure 1. Peripheral blood mononuclear cell NLRP3 and DEFA1B mRNA expression levels of healthy controls (HC) and parenchymal neuro-Behçet’s disease (NBD) patients during active and inactive stages of the disease. Horizontal lines indicate mean values. p-Value depicted on the upper left corner of the panel is obtained by ANOVA and p-values above the horizontal lines are obtained by the Tukey’s post-hoc test.
Discussion
Gene expression levels of attack and remission PBMC samples of NBD patients were examined to retrieve clues about the pathogenesis of NBD and to identify biomarkers and potential target molecules for treatment of the disease. In the active stage samples, DEFA1B and NLRP3 were the most up-regulated genes. These results were confirmed with real-time PCR, which also showed that DEFA1B expression is enhanced in NBD patients even during the remission period.
DEFA1B belongs to the greater family of defensins, antimicrobial peptides produced predominantly by neutrophils. Defensins exhibit a great functional diversity and play important roles in innate immune defense against infectious pathogens through disintegration of target cell membrane, release of chemokines, induction of reactive oxygen species and attraction of lymphocytes to the inflammation site (12). In addition to immune cells, β-defensins are also produced by epithelial cells including those of oral mucosa (13). Polymorphisms of the DEFA1B gene have been associated with periodontitis emphasizing its role in host defense at mucosal surfaces (14). In likeness to our study, β-defensin 1 was found to be increased in plasma samples of BD and NBD patients and a polymorphism of the DEFA1B gene was associated with increased BD risk (15). Moreover, mRNA and protein levels of α-defensin 1 were increased in active and inactive stages of BD (16). No correlation could be determined among defensin levels and clinical features of BD (15,16). Notably, defensin gene variants have not been determined in any of the previous genome-wide association studies of BD patients, underlining the distinction between causative and attack-inducing factors in NBD.
NLRP3 is a component of the inflammasome complex, which primarily controls IL-1β and IL-18 production (17,18). Mutations of NLRP3 may induce inflammation attacks characterized with enhanced signaling of IL-1β, which is profoundly involved in neutrophil recruitment and BD pathogenesis (19,20). NLRP3 genetic variants have been previously associated with both BD and hereditary periodic fever syndrome, another autoinflammatory condition induced by overactive innate immunity (19,21,22). Furthermore, in concordance with our study, NLRP3 expression levels were found to be increased in PBMCs and skin lesions of BD patients, particularly during the active stage of the disease (23).
Both genes that were up-regulated during NBD attacks were closely associated with innate immunity and particularly neutrophil functions. DEFA1B expression levels of NBD patients were higher than those of healthy controls even during the inactive stage, emphasizing the constant over-activation of innate immunity in this disease. The innate immune system is centrally involved in BD pathogenesis, as evidenced by mucocutaneous symptoms, increased expression of pattern recognition receptors and abundance of active neutrophils in affected organs including the central nervous system (1,2,24). Moreover, both β-defensins and NLRP3 are expressed by glial cells, which are also members of the innate immunity, and contribute to neuroinflammation and neurodegeneration (25,26). Therefore, these two molecules might also be associated with occurrence of neurological symptoms in NBD attacks. In brief, our results suggest that innate immunity is primarily involved in acute attacks of NBD and treatment strategies specifically targeting neutrophils might prove useful in the attack treatment of NBD and putatively replace steroids that are currently used for this purpose.
Interestingly, neither DEFA1B nor NLRP3 expression was associated with clinical parameters including disability scores in both our study and previous ones (15,16,23). Putatively, while these two factors may be important in lesion formation and attack initiation, additional factors might be more intimately associated with disability progression. Therefore, investigation of additional genes in larger NBD cohorts are recommended for better understanding of the emergence of neurological symptoms in BD.
Expression levels of intercellular adhesion molecule 3 (ICAM3), CD48 and leucine-rich repeat kinase 2 (LRRK2) were significantly reduced in active stage samples of NBD patients. Intriguingly, all of these molecules are involved in inflammation and immune responses and their expression is expected to be increased during NBD attacks. However, a distinctive feature of these factors is that they are centrally involved in functions of both innate and adaptive immunity cells. ICAM3, CD48 and LRRK2 are abundantly expressed in lymphocytes, and contribute to T cell activation and intracellular signaling events triggered by receptor activation (27-29). A potential explanation for reduced expression of these factors is compensatory inhibition of adaptive immune system to restrict the hazardous consequences of inflammation during an attack. This assertion needs to be further scrutinized and expression levels of these three molecules should also be determined in future experiments.
Studies investigating factors underlying clinical activation and progression of BD and NBD provide clues for treatment and management of all autoinflammatory disorders. In this context, our findings suggest that DEFA1B and NLRP3 gene expression levels can be used as valuable biologic markers for estimation of disease activity in NBD and as potential targets for future therapeutic trials for inflammatory disorders.
Conflicts of Interest
The Authors declare no conflict of interest regarding this study.
Authors’ Contributions
Ayca Olcay, Elif Sanlı and Ece Akbayır conducted real-time PCR and microarray experiments. Elif Ugurel and Ece Erdag were responsible for data collection and statistics analysis. Murat Kurtuncu, Cem İsmail Kucukali and Tuncay Gunduz were responsible for the clinical care and follow-up visits provided to the patients. Vuslat Yilmaz was responsible for planning experiments and interpretation of data. Erdem Tuzun and Burcak Vural supervised all experiments and drafted the manuscript.
Acknowledgements
This study was funded by Scientific Research Projects Coordination Unit of Istanbul University. Project number: TDK-2016-22073.
References
- 1.Leccese P, Alpsoy E. Behçet’s Disease: An overview of etiopathogenesis. Front Immunol. 2019;10:1067–1067. doi: 10.3389/fimmu.2019.01067. PMID: 31134098. DOI: 10.3389/fimmu.2019.01067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zierhut M, Mizuki N, Ohno S, Inoko H, Gül A, Onoé K, Isogai E. Immunology and functional genomics of Behçet’s disease. Cell Mol Life Sci. 2003;60:1903–1922. doi: 10.1007/s00018-003-2333-3. PMID: 14523551. DOI: 10.1007/s00018-003-2333-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cosan F, Aktas Cetin E, Akdeniz N, Emrence Z, Cefle A, Deniz G. Natural killer cell subsets and their functional activity in Behçet’s disease. Immunol Invest. 2017;46:419–432. doi: 10.1080/08820139.2017.1288240. PMID: 28388249. DOI: 10.1080/08820139.2017.1288240. [DOI] [PubMed] [Google Scholar]
- 4.Neves FS, Spiller F. Possible mechanisms of neutrophil activation in Behçet’s disease. Int Immunopharmacol. 2013;17:1206–1210. doi: 10.1016/j.intimp.2013.07.017. PMID: 23973446. DOI: 10.1016/j.intimp. 2013.07 .017. [DOI] [PubMed] [Google Scholar]
- 5.Butta NV, Fernández-Bello I, López-Longo FJ, Jiménez-Yuste V. Endothelial dysfunction and altered coagulation as mediators of thromboembolism in Behçet disease. Semin Thromb Hemost. 2015;41:621–628. doi: 10.1055/s-0035-1556727. PMID: 26276934. DOI: 10.1055/s-0035-1556727. [DOI] [PubMed] [Google Scholar]
- 6.Kürtüncü M, Tüzün E, Akman-Demir G. Behçet’s disease and nervous system involvement. Curr Treat Options Neurol. 2016;18:19–19. doi: 10.1007/s11940-016-0405-6. PMID: 27007988. DOI: 10.1007/s11940-016-0405-6. [DOI] [PubMed] [Google Scholar]
- 7.Totsuka S, Hattori T, Yazaki M, Nagao K, Mizushima S. Clinicopathologic studies on neuro-Behçet’s disease. Folia Psychiatr Neurol Jpn. 1985;39:155–166. doi: 10.1111/j.1440-1819.1985.tb02899.x. PMID: 4065759. [DOI] [PubMed] [Google Scholar]
- 8.Arai Y, Kohno S, Takahashi Y, Miyajima Y, Tsutusi Y. Autopsy case of neuro-Behçet’s disease with multifocal neutrophilic perivascular inflammation. Neuropathology. 2006;26:579–585. doi: 10.1111/j.1440-1789.2006.00734.x. PMID: 17203596. [DOI] [PubMed] [Google Scholar]
- 9.Özdemir FT, Demiralp EE, Aydın SZ, Atagündüz P, Ergun T, Direskeneli H. Immune and inflammatory gene expressions are different in Behçet’s disease compared to those in Familial Mediterranean Fever. Eur J Rheumatol. 2016;3:146–152. doi: 10.5152/eurjrheum.2016.15099. PMID: 28149656. DOI: 10.5152/eurjrheum.2016.15099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okuzaki D, Yoshizaki K, Tanaka T, Hirano T, Fukushima K, Washio T, Nojima H. Microarray and whole-exome sequencing analysis of familial Behçet’s disease patients. Sci Rep. 2016;6:19456–19456. doi: 10.1038/srep19456. PMID: 26785681. DOI: 10.1038/srep19456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.International Study Group for Behçet’s Disease Criteria for diagnosis of Behçet’s disease. Lancet. 1990;335:1078–1080. PMID: 1970380. [PubMed] [Google Scholar]
- 12.Zhao L, Lu W. Defensins in innate immunity. Curr Opin Hematol. 2014;21:37–42. doi: 10.1097/MOH.0000000000000005. PMID: 24275690. DOI: 10.1097/ MOH.0000000000000005. [DOI] [PubMed] [Google Scholar]
- 13.Bogefors J, Kvarnhammar AM, Höckerfelt U, Cardell LO. Reduced tonsillar expression of human β-defensin 1, 2 and 3 in allergic rhinitis. FEMS Immunol Med Microbiol. 2012;65:431–438. doi: 10.1111/j.1574-695X.2012.00959.x. PMID: 22444247. DOI: 10.1111/j.1574-695X.2012. 00959.x. [DOI] [PubMed] [Google Scholar]
- 14.Schaefer AS, Richter GM, Nothnagel M, Laine ML, Rühling A, Schäfer C, Cordes N, Noack B, Folwaczny M, Glas J, Dörfer C, Dommisch H, Groessner-Schreiber B, Jepsen S, Loos BG, Schreiber S. A 3’ UTR transition within DEFB1 is associated with chronic and aggressive periodontitis. Genes Immun. 2010;11:45–54. doi: 10.1038/gene.2009.75. PMID: 19829306. DOI: 10.1038/gene.2009.75. [DOI] [PubMed] [Google Scholar]
- 15.Kallel A, Ben Salem T, Hammami MB, Said F, Jemaa R, Houman MH, Feki M. Association of systemic beta-defensin-1 and -20G/A DEFB1 gene polymorphism with Behçet’s disease. Eur J Intern Med. 2019;S0953-6205(19):30052-4–30052-4. doi: 10.1016/j.ejim.2019.02.008. PMID: 30819604. DOI: 10.1016/j.ejim.2019.02.008. [DOI] [PubMed] [Google Scholar]
- 16.Ahn JK, Hwang JW, Oh JM, Bae EK, Lee J, Lee YS, Koh EM, Cha HS. Increased α-defensin-1 expression in Korean patients with Behcet’s disease. Joint Bone Spine. 2011;78:593–597. doi: 10.1016/j.jbspin.2011.01.012. PMID: 21441057. DOI: 10.1016/j.jbspin.2011.01.012. [DOI] [PubMed] [Google Scholar]
- 17.McGettrick AF, O’Neill LA. NLRP3 and IL-1β in macrophages as critical regulators of metabolic diseases. Diabetes Obes Metab. 2013;15:19–25. doi: 10.1111/dom.12169. PMID: 24003917. DOI: 10.1111/dom.12169. [DOI] [PubMed] [Google Scholar]
- 18.Charmoy M, Hurrell BP, Romano A, Lee SH, Ribeiro-Gomes F, Riteau N, Mayer-Barber K, Tacchini-Cottier F, Sacks DL. The Nlrp3 inflammasome, IL-1β, and neutrophil recruitment are required for susceptibility to a nonhealing strain of Leishmania major in C57BL/6 mice. Eur J Immunol. 2016;46:897–911. doi: 10.1002/eji.201546015. PMID: 26689285. DOI: 10.1002/eji.201546015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rigante D, Frediani B, Cantarini L. A comprehensive overview of the hereditary periodic fever syndromes. Clin Rev Allergy Immunol. 2018;54:446–453. doi: 10.1007/s12016-016-8537-8. PMID: 27068928. DOI: 10.1007/s12016-016-8537-8. [DOI] [PubMed] [Google Scholar]
- 20.Barış S, Akyürek Ö, Dursun A, Akyol M. The impact of the IL-1β, IL-1Ra, IL-2, IL-6 and IL-10 gene polymorphisms on the development of Behcet’s disease and their association with the phenotype. Med Clin (Barc) 2016;146:379–383. doi: 10.1016/j.medcli.2015.09.017. PMID: 26654556. DOI: 10.1016/j.medcli.2015.09.017. [DOI] [PubMed] [Google Scholar]
- 21.Li L, Yu H, Jiang Y, Deng B, Bai L, Kijlstra A, Yang P. Genetic variations of NLR family genes in Behcet’s Disease. Sci Rep. 2016;6:20098–20098. doi: 10.1038/srep20098. PMID: 26833430. DOI: 10.1038/srep20098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yüksel Ş, Eren E, Hatemi G, Sahillioğlu AC, Gültekin Y, Demiröz D, Akdiş C, Fresko İ, Özören N. Novel NLRP3/cryopyrin mutations and pro-inflammatory cytokine profiles in Behçet’s syndrome patients. Int Immunol. 2014;26:71–81. doi: 10.1093/intimm/dxt046. PMID: 24135410. DOI: 10.1093/intimm/dxt046. [DOI] [PubMed] [Google Scholar]
- 23.Kim EH, Park MJ, Park S, Lee ES. Increased expression of the NLRP3 inflammasome components in patients with Behçet’s disease. J Inflamm (Lond) 2015;12:41–41. doi: 10.1186/s12950-015-0086-z. PMID: 26136643. DOI: 10.1186/s12950-015-0086-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Türe-Özdemir F, Tulunay A, Elbasi MO, Tatli I, Maurer AM, Mumcu G, Direskeneli H, Eksioglu-Demiralp E. Pro-inflammatory cytokine and caspase-1 responses to pattern recognition receptor activation of neutrophils and dendritic cells in Behcet’s disease. Rheumatology (Oxford) 2013;52:800–805. doi: 10.1093/rheumatology/kes399. PMID: 23325038. DOI: 10.1093/rheumatology/kes399. [DOI] [PubMed] [Google Scholar]
- 25.Williams WM, Castellani RJ, Weinberg A, Perry G, Smith MA. Do β-defensins and other antimicrobial peptides play a role in neuroimmune function and neurodegeneration. Scientific WorldJournal. 2012;2012:905785–905785. doi: 10.1100/2012/905785. PMID: 22606066. DOI: 10.1100/2012/905785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee E, Hwang I, Park S, Hong S, Hwang B, Cho Y, Son J, Yu JW. MPTP-driven NLRP3 inflammasome activation in microglia plays a central role in dopaminergic neurode-generation. Cell Death Differ. 2019;26:213–228. doi: 10.1038/s41418-018-0124-5. PMID: 29786072. DOI: 10.1038/s41418-018-0124-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Fougerolles AR, Springer TA. Intercellular adhesion molecule 3, a third adhesion counter-receptor for lymphocyte function-associated molecule 1 on resting lymphocytes. J Exp Med. 1992;175:185–190. doi: 10.1084/jem.175.1.185. PMID: 1730916. DOI: 10.1084/ jem.175.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McArdel SL, Terhorst C, Sharpe AH. Roles of CD48 in regulating immunity and tolerance. Clin Immunol. 2016;164:10–20. doi: 10.1016/j.clim.2016.01.008. PMID: 26794910. DOI: 10.1016/j.clim.2016.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hakimi M, Selvanantham T, Swinton E, Padmore RF, Tong Y, Kabbach G, Venderova K, Girardin SE, Bulman DE, Scherzer CR, LaVoie MJ, Gris D, Park DS, Angel JB, Shen J, Philpott DJ, Schlossmacher MG. Parkinson’s disease-linked LRRK2 is expressed in circulating and tissue immune cells and upregulated following recognition of microbial structures. J Neural Transm (Vienna) 2011;118:795–808. doi: 10.1007/s00702-011-0653-2. PMID: 21552986. DOI: 10.1007/ s00702-011-0653-2. [DOI] [PMC free article] [PubMed] [Google Scholar]