Abstract
Post-translational modifications (PTMs) are important regulators of protein function, and integrate metabolism with physiological and pathological processes. Phosphorylation and acetylation are particularly well studied PTMs. A relatively recently discovered novel PTM is succinylation in which metabolically derived succinyl CoA modifies protein lysine groups. Succinylation causes a protein charge flip from positive to negative and a relatively large increase in mass compared to other PTMs. Hundreds of protein succinylation sites are present in proteins of multiple tissues and species, and the significance is being actively investigated. The few completed studies demonstrate that succinylation alters rates of enzymes and pathways, especially mitochondrial metabolic pathways. Thus, succinylation provides an elegant and efficient mechanism to coordinate metabolism and signaling by utilizing metabolic intermediates as sensors to regulate metabolism. Even though the brain is one of the most metabolically active organs, an understanding of the role succinylation in the nervous system is largely unknown. Data from other tissues and other PTMs suggest that succinylation provides a coupling between metabolism and protein function in the nervous system and in neurological diseases. This review provides a new insight into metabolism in neurological diseases and suggests that the drug development for these diseases requires a better understanding of succinylation and desuccinylation in the brain and other tissues.
Keywords: Succinlyation, mitochondria, metabolism, sirtuins
1. Importance of post-translational modifications (PTMs) (Overview).
PTMs are important regulators of protein function that modulate many physiological and pathological processes (Table 1). PTMs integrate cellular processes and diversify the proteome. They are significant for activity, stability, protein folding and cellular localization. Since the modifiers are derived from metabolism, they often link metabolism to altered function of proteins and pathways. Common PTMs include acetylation, methylation, biotinylation, ubiquitination, butyrylation, propionylation, crotonylation, glutarylation, malonylation, long-chain fatty acylation, ubiquitination and 2-hydroxyisobutyrylation, phosphorylation and succinylation. Comprehensive identification of the many of the targets of these PTMs and their functional impact remain at an early stage, particularly in the nervous system. The relation of a number of covalent modifications affecting enzymes related to glutamate and glutamine metabolism, including covalent attachment of an acetyl-, palmitoyl-, succinyl-, or small ubiquitin-like modifiers (SUMO) moiety, and the role(s) of sirtuins in these modifications of other proteins has been described in detail [1].
Table 1.
Common PTMs involving addition of functional groups.
| Amino acid | PTMs | |
|---|---|---|
| lysine | side chains | Acetylation, succinylation, methylation, biotinylation, butyrylation, propi onylati on, crotonylation, glutarylation, malonylation, long-chain fatty acylation, ubiquitination, 2-hydroxyisobutyrylation |
| serine | phosphorylation | |
| threonine | ||
| tyrosine | ||
| histidine | Phosphoramidate bonds | |
| lysine | ||
| arginine | ||
| aspartic acid | mixed anhydride linkages | |
| glutamic acid | ||
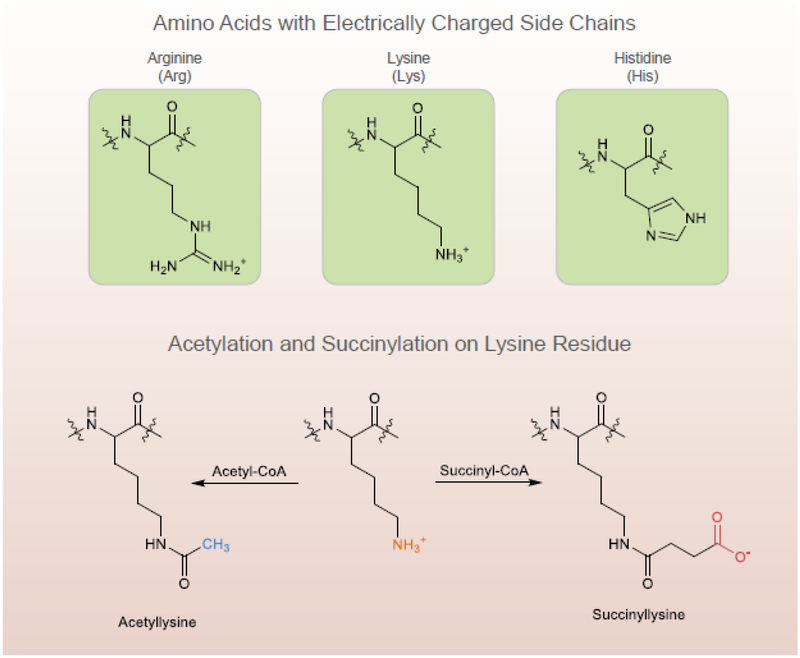
Lysine residues are targets of numerous PTMs (Table 1). Lysine is one of only three amino acid residues with positively charged side chains at physiological pH (Figure 1). The side chains can be involved in multiple noncovalent interactions including van der Waals interactions, hydrogen bonds and electrostatic interactions with negatively charged residues [2]. For example, salt bridge formation between lysine residues and acidic residues are important in forming leucine zipper structures. Lysine residues play key roles in acid-base catalyzed enzymatic reactions in which proton transfer is required. A large literature shows that a charge neutralization of the basic side chain of lysine by acetylation, ubiquitination or methylation changes protein function significantly.
Figure 1.
Succinylation changes protein size and charge, and increases mass by 100 Da and changes charge from a single positive to a single negative
Succinylation is a unique, relatively recently discovered [2, 3], meagerly studied PTM (Figure 1). Compared to methylation (14 Da) or acetylation (40 Da), succinylation (100 Da) causes larger mass change, and alters a positively charged side chain to a negatively charged one causing a two-unit charge shift in the modified residues (Figure 1). The importance of succinylation in cell function and disease in the CNS is an emerging area, and surprisingly, only two papers on succinylation in the CNS have been published [4, 5].
2. Mechanisms for succinylation.
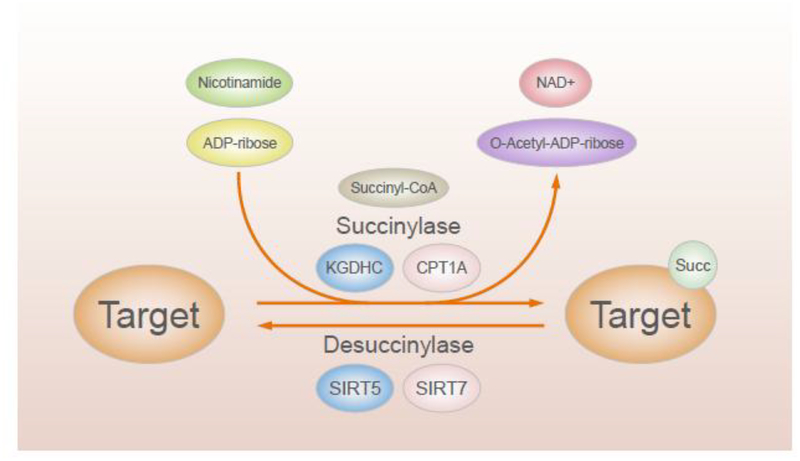
Whether putative succinyl transferases catalyze succinylation or if it occurs passively by reaction of succinyl CoA with surface-exposed lysin is unknown ((Figure 2) [2, 3, 6]. While classes of drugs are known for manipulation of other PTMs such as acetylation [(histone acetyl transferases) and deacetylases (HDAC inhibitors)], efforts to do this for succinylation or desuccinylation are in their infancy because fundamental mechanisms are unknown [7]. Since the mechanisms for succinylation are still under investigation, more clear evidence based on novel molecular mechanisms for succinylation and de-succinylation are necessary to address the unmet needs of novel therapies and related drug discovery and development.
Figure 2.
Little is known about regulation of succinylation and desuccinylation
2.1. Non-enzymatic succinylation by succinyl CoA.
Recent experiments suggest that several of the larger acyl modifications such as succinylation, malonylation, crotonylation, glutarylation, and β-hydroxybutyrylation can occur predominantly by non-enzymatic mechanisms [8, 9]. This suggests that the concentrations of the reactants govern the rates of attachment for the various acyl CoA species to lysine residues. Like acetyl CoA, succinyl CoA is an inherently reactive short chain CoA thioester that maintains steady-state concentrations in the mitochondrial matrix in the low mM range (0.1–0.6 mM). Mixing succinyl CoA with albumin or isocitrate dehydrogenase (ICDH) increases succinylation in a pH and dose-dependent manner [5, 10–12] and mitochondrial pH can regulate non-enzymatic succinylation. The data suggest that protein acylation in mitochondria may be a chemical event facilitated by the alkaline pH and high concentrations of the reactive acyl-CoAs present in the mitochondrial matrix [9, 12].
Tissues with high acyl CoA also have a high level of post-translational modification. The parallel tissue distribution of acyl CoA and extent of that form of PTMs in some tissues support the suggestion that concentrations of acyl CoA in tissues control succinylation [10]. Profiling acyl CoA concentrations in major mouse organs including liver, heart, kidney, brain and muscle reveal that different tissues have unique acyl CoA profiles. For example, succinyl-CoA is the most abundant acyl CoA in the heart [13]. In the liver, the absolute concentration of succinyl CoA is similar to that in the heart, but acetyl CoA and free CoA are more abundant than succinyl CoA [13]. The brain is lower in acetyl CoA and succinyl CoA than heart or liver [13–15]. Succinylation in wild type mouse or SIRT5 (a proposed de-succinylase as described below) knockout (KO) is also higher in the heart. However, acetylation is much higher in the brain even though acetyl CoA levels are lower [13]. Furthermore, SIRT5 KO in vivo changes succinylation considerably, but does not change the levels of succinyl CoA [13]. An understanding of the tissue-specific control of acyl CoA concentrations will lead to a better understanding of the regulation of succinylation and the development of new therapeutic approaches.
2.2. Enzymatic succinylation (The sources of succinyl CoA for succinylation).
Distinguishing between enzymatic and non-enzymatic succinylation in cells or animals is difficult because the enzymes producing the succinyl groups and controlling the cellular levels may also serve as succinyl-transferases (Figure 3). Altering the availability of succinyl CoA (by genetically ablating specific enzymes of the tricarboxylic acid (TCA) cycle affects the global succinylation pattern in yeast, suggesting that succinyl CoA used for succinylation comes from mitochondria [10]. Succinyl CoA can come from amino acid metabolism or from the TCA cycle. The TCA cycle multi-protein complex α-ketoglutarate dehydrogenase complex (KGDHC) consists of three components: E1k [α- Ketoglutarate Dehydrogenase (KGDH) (EC 1.2.4.2)], E2k [Dihydrolipoyl Succinyltransferase (EC 2.3.1.61)] and E3 [Dihydrolipoyl Dehydrogenase (EC 1.8.1.4)]. The E1k subunit of KGDHC catalyzes a rate-controlling step in the TCA cycle and lies far from equilibrium. The significant energy change makes it a crucial point of regulation not only for the TCA cycle, but also for the entire cellular respiration pathway [16]. E1k and thus KGDHC may also control succinylation, which expands the role of KGDHC to integrate the metabolism of the whole cell. Purified KGDHC can succinylate and modify the activity of multiple proteins including albumin and other enzymes of the TCA cycle including the pyruvate dehydrogenase complex (PDHC) and ICDH. The greater effectiveness of KGDHC than succinyl CoA suggests that the catalysis owing to the E2k succinyltransferase is important [5].
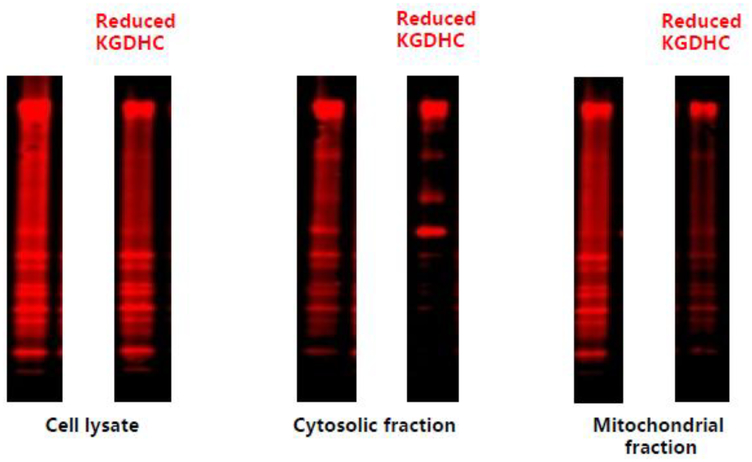
Figure 3.
Inhibition of KGDHC with CESP reduces succinylation in cultured neurons. Neurons were incubated with or without a KGHDC antibody, immuno-precipitated and then stained with antisuccinyl-lysine antibody
KGDHC regulates succinylation either by regulating levels of succinyl CoA or by direct succinylation. In yeast, the mutated loss of E1k leads to a six-fold reduction in global succinylation. On the other hand, the induction of E1k enhances succinylation by three- to five-fold [10]. Transferring yeast to galactose media induces E1k and enhances global lysine succinylation by 1.7-fold, and elevates succinylation of mitochondrial proteins more robustly with a 2.7- to 4.7-fold increase [10]. KGDHC is also critical for succinylation in neurons. Succinylation is high in mature cultured mouse neurons and specific inhibition of E1k of KGDHC diminishes succinylation in cell lysates (26.9%), the cytosol (47%) and mitochondria (51.2%) [5] (Figure 3).
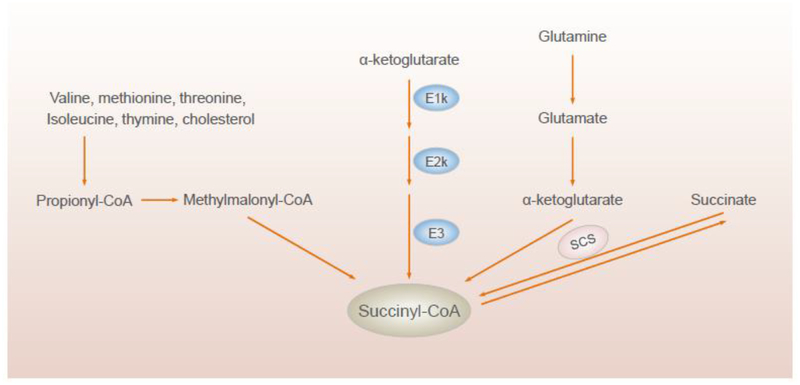
KGDHC mediates succinylation under multiple conditions. Inhibition of respiration by the complex I inhibitor rotenone decreases the concentration of succinyl CoA in the mitochondria by inhibiting aconitase and reducing ketoglutarate for KGDHC [17]. Amino acids are an important source of succinyl CoA for heme synthesis and this is mediated by KGDHC [18]. The TCA cycle cannot provide enough succinyl CoA, so glutamine without equilibration with the TCA cycle provides the alpha-ketoglutarate precursor [19]. Thus, the conversion of glutamine to glutamate to α-ketoglutarate to succinyl CoA provides the succinyl CoA for heme synthesis (Figure 4). This route of succinyl CoA formation has only been demonstrated in murine erythroleukemia cells tissues producing heme, but may be critical in other tissues as well [19].
Figure 4.
Sources of succinyl CoA
Inhibition of KGDHC also reduces succinylation in the cytosol (Figure 3). This suggests that succinylation of cytosolic proteins depends upon succinyl CoA in the mitochondria, but the mechanism is not clear [5, 10]. One possibility is that a product of KGDHC is a required substrate for an alternative succinyl CoA transferase in the cytosol. A second possibility is that succinyl-CoA moves from the mitochondria to the cytosol. A third possibility is that the cytosol has functional KGDHC that is also sensitive to the inhibitor that was used to impair KGDHC [5].
KGDHC is also important for succinylation in the nucleus. Inhibition of the conversion of succinyl CoA to succinate increases succinyl CoA concentrations by as much as four fold and induces protein succinylation and the increased succinylation leads to a nearly linear increase in transcription [20]. Histone modifications including succinylation are central to the regulation of chromatin-based processes. KGDHC binds to lysine acetyltransferase 2A (KAT2A) in the promoter regions of genes [21]. Site-directed mutagenesis indicates a selective binding of succinyl-CoA over acetyl CoA. KAT2A then succinylates histone lysines with a maximum frequency around the transcription start sites of genes. Preventing the KGDHC complex from entering the nucleus, or reducing the expression of KAT2A diminishes gene expression and inhibits tumor growth [21]. Manipulations of E2k alters histone succinylation without altering overall KGDHC activities [21]. Manipulations of E2k can affect the response of cells to oxidative stress independently of KGDHC activity [22]. These findings reveal an important mechanism of histone modification and demonstrate that local generation of succinyl CoA by the nuclear α-KGDHC complex coupled with the succinyltransferase activity of KAT2A is instrumental in histone succinylation [21].
Enzymatic TCA cycle steps beyond KGDHC can also regulate succinylation. Succinyl CoA ligase (or succinyl-CoA synthetase, SCS) can regulate succinylation (Figure 4) [10, 11, 23]. Succinyl CoA ligase, which is composed of Lsc1 and Lsc2, converts succinyl CoA to succinate. It is critical in the TCA cycle and is involved in the ketone-body breakdown in animals (Figure 4). Lsc1 is required for succinyl CoA ligase activity and like E1k is induced by growth on galactose-containing media [24]. Loss of lsc1 blocks the conversion of succinyl CoA to succinate, causes accumulation of succinyl CoA, and increases succinylation levels throughout the cell. This suggests that that succinyl CoA is the succinyl donor to lysine and that succinyl CoA may traverse the mitochondrial membrane or be generated outside of mitochondria [10]. Moreover, inactivating succinate dehydrogenase (SDH) by depleting the SDHA or SDHB subunits of the SDH complex using shRNAs increases succinyl CoA levels and hypersuccinylation by 500% and 290%, respectively. Furthermore, inhibiting SDH with 3-nitropropionic acid increases succinyl CoA and hypersuccinylation in HEK293T cells. These results confirm that SDH inactivation induces hypersuccinylation by causing the accumulation of succinyl CoA [23]. E. Coli converts added succinate to succinyl CoA by succinyl CoA synthetase, which increases succinylation [2].
Another transferase with lysine succinyltransferase activity in mammalian cells is carnitine palmitoyltransferase 1A (CPT1A) (Figure 2)[25][26]. CPT1A is a mitochondrial enzyme responsible for the formation of acyl carnitines by catalyzing the transfer of the acyl group of a long-chain fatty acyl CoA from coenzyme A to L-carnitine. CPT1A associates with the mitochondrial outer membrane through transmembrane region in the peptide chain [27]. Both the N- and C- terminal domains are exposed to the cytosolic side of the membrane. CPT1A succinylates proteins using succinyl CoA as a substrate [26]. CPT1A increases lysine succinylation in cells without altering succinyl CoA levels. Quantitative succinylation proteomics analysis identified 171 lysine sites on 101 proteins (out of 550 lysine sites on 247 proteins total) that were succinylated in a CPT1A expression dependent manner in cells. Mutation studies show that the carnitine palmitoyltransferase 1A activity and the lysine succinyltransferase activity of CPT1A are separate. Subcellular distribution studies show that almost 50% of the proteins that are succinylated by CPT1A are cytosolic proteins [26].
2.3. Enzymatic desuccinylation by desuccinylases (Figure 2).
Succinylated proteins clearly reflect a balance of succinylation and de-succinylation (Figure 2) [28]. The only recognized desuccinylase to act in all cells compartments is NAD+ dependent Sirtuin 5 (SIRT 5), although other sirtuins exhibit limited desuccinylase activity (e.g. SIRT 7 in the nucleus). Succinylation is sensitive to both increases and decreases of SIRT5 [3, 6]. SIRT5 possesses unique enzymatic activity on hydrolyzing negatively charged lysine modifications such as lysine succinylation. These properties these also make SIRT5 effective for demalonylation, and deglutarylation [3]. Thus, discussions that attribute actions to succinylation based only on SIRT5 KO or knockdown (KD) are oversimplified.
Sirtuin proteins are detectable in human brain, cerebral spinal fluid and plasma, as well as in guinea pig and mouse tissues [29]. SIRTs 3,4 and 5 are primarily localized to the mitochondria [29, 30]. SIRT5 is present in all cell compartments, and is particularly enriched in mitochondria [6]. Different forms of SIRT5 occur in the mitochondria, cytosol and nucleus and their stability is regulated differently [31]. The subcellular localization and C-termini of two SIRT5 isoforms (i.e., SIRT5iso1 and SIRT5iso2) encoded by the human SIRT5 gene differ from each other. Although both isoforms contain cleavable mitochondrial targeting signals at their N-termini, the cleaved SIRT5iso2 is localized mainly in mitochondria, whereas the cleaved SIRT5iso1 is localized in both mitochondria and cytoplasm [32]. SIRT7 is a histone desuccinylase that functionally links to chromatin compaction and genome stability [33]. SIRT7-catalysed desuccinylation is primarily nuclear and critically implemented in DNA-damage response and cell survival [33].
KO of SIRT5 increases succinylation of specific proteins. For example, deletion of SIRT5 increases succinylation of mitochondrial hydroxyl CoA dehydrogenase by more than 200 fold [34]. SIRT5 also regulates cytosolic PTMs. SIRT5 does not act on all succinlyated lysines. For example, many of the proteins succinylated by CPT1A are not desucinylated by SIRT5. Among 32 mitochondria CPT1A dependent lysine succinylated proteins, only eight are potential SIRT5 substrates [26]. SIRT5 controls cytosolic and mitochondrial protein malonylation with glycolysis as a major target [35].
The cellular localization is also critical. In the central nervous system, SIRT5 is primarily expressed in neurons and endothelial cells with mitochondrial and extra-mitochondrial localization. Immunostaining for SIRT5 in adult mice reveals a predominantly neuronal-like expression pattern in hippocampus and cortex. SIRT5 staining is present within micro-vessel structures with endothelial like morphology and significant co-localization of SIRT5 and the endothelial marker Tie-2. The lack of SIRT5 co-localization with oligodendrocytes (Olig2) or microglia (Iba1) indicates these cells lack SIRT5 [31].
2.4. Motifs (Table 2).
Table 2.
Motif analysis
| Motif | Species | Reference |
|---|---|---|
| GK | WT and SIRT5 KO mouse MEFs | [34] |
| AK | ||
| FK | ||
| KA | ||
| K.L | WT and CPT1A KD HEK293T cell | [26] |
| L.K | ||
| L…K | ||
| L….K | ||
| I.K | ||
| I….K | ||
| I……K | ||
| V.K | ||
| V..K | ||
| V…..K | ||
| V……..K | ||
| K……..V | ||
| K………G | ||
| K…K | Toxoplasma gondii | [38] |
| I…K | ||
| LK | ||
| KG | ||
| QK | ||
| R……K | Marine Bacterium Vibrio Parahemolyticus | [39] |
| K……K | ||
| K…….K | ||
| K……K | ||
| K……R | ||
| EK | Mycobacterium tuberculosis | [59] |
| K…..K |
A unique sequence motif may be required for the substrate of either succinylase or desuccinylase enzymes. The observation that fold changes for the lysine succinylation sites in the same protein range from 1.0 to 11.4 suggests particular motifs are important [36].
Identified succinylation motifs in tomato [37], protozoan [38], bacteria [39] and mammalian systems are postulated (Table 2), but no experiments verify the function of these motifs or the relationship with succinylases or desuccinylases. The sequences do not reveal a strong bias for a particular amino acid. However, aspartic acid, glutamic acid, and lysine occurred most frequently at the +1 position, while leucine and alanine occur frequently at the −1 position [10]. Succinylated lysine sites targeted by SIRT5 tend to be near glycine, alanine, serine, or threonine residues [34, 40]. Succinylation with CPT1A is near leucine, valine, and isoleucine and these sites are not sensitive to SIRT5 [26]. Among 32 mitochondrial CPT1A-dependent lysine-succinylated proteins only eight proteins are identified potential SIRT5 substrates [26]. Thus, strong evidence suggests that different desuccinylases have a varied sequence of bias towards specific lysine-succinylation sites.
Modification of catalysis or cofactor binding sites would provide a likely explanation of the ability of succinylation to alter enzyme activities. Sixteen lysine succinylation sites overlap with known cofactor binding or catalytic sites, and 74 lysine succinylation sites are located within five residues in flanking distance to enzymatic activity sites [34]. Although succinylation of ICDH alters activity, it does not occur at lysines directly involved in substrate binding or catalysis. However, mutations at the succinylation sites diminish activity and change the alpha helices [2]. Hydroxyl-coenzyme A lysine residue 81, which is adjacent to a CoA binding site at lysine residue 80, is succinylated, and its modification increases by more than 200-fold in SIRT5 KO fibroblasts [34].
3. Variations in succinylation between proteins, tissues and species is informative about the role of succinylation.
Succinylation has been assessed multiple organisms and tissues (Table 3). The reported results vary considerably between reports in part because different approaches were utilized, but clear patterns emerge. All reports suggest extensive succinylation and major agreements are summarized in the following sections.
Table 3.
Succinylation varies by tissue and investigation (need to check the SIRT5−/− numbers they seem backwards)
3.1. Variation by protein.
The number of succinylation sites per protein varies from 1 to 28 depending on the protein. In the SIRT5 KO heart, ECHA, a protein involved in fatty acid oxidation, has the most succinylation sites (at 28 Lys residues). Among the 66 lysine residues of ECHA, 28 are succinylated and the majority of succinylated residues (26 out of 28) are only found in SIRT5 KO heart [13]. In SIRT5 deficient mouse liver carbamyl phosphate synthase (CPS1) is succinylated at 47 sites and hydroxyacyl-coenzyme A dehydrogenase at 32 sites [34]. In SIRT5-deficient mouse embryonic fibroblasts (MEFs), 28 lysine succinlyation sites increase more than 10-fold. Succinylation of hydroxyl-coenzyme A amino acid residue lysine 81 increase by more than 200-fold and mitochondrial acetyl CoA acetyltransferase increases 120 fold in SIRT 5 KO fibroblasts [34]. Whether increased number of succinylation sites indicates greater functional consequences is unknown.
3.2. Variation between tissues and species (Table 3).
Succinylation varies between tissues and species. Unless the results are from the same laboratory at the same time, mass spectrometry technical variation makes comparisons difficult. Comparison of succinylation in various tissues will provide insight into the regulation of succinylation and enable studies to discover succinylases and succinyltransferases. Mouse heart is particularly high in succinylation, has the greatest increase with KO of SIRT5 and the greatest abundance of succinyl CoA, but this relationship is not true in other tissues [13].
3.3. Variation by cell compartment.
Except in yeast, all experiments agree that succinylation is highly concentrated in the mitochondria. In yeast, only 8% of succinylation sites occurs on mitochondrial proteins [10]. In HeLa cells and mouse liver, the proportion of succinylation occurring on mitochondrial proteins is significantly larger than in yeast, with the greatest proportion found in mouse liver [10]. The occurrence of frequent lysine succinylation outside of mitochondria suggests that succinyl CoA, succinate, or another succinyl-metabolite drives succinylation in the cytoplasm and nucleus [10]. In mouse liver, as much as 70% of succinylation sites are on mitochondrial proteins, and mitochondrial proteins are more frequently succinylated at multiple sites than non-mitochondrial proteins. A total of 32% of the proteins designated as mitochondrial are succinylated and every enzyme of the TCA cycle is succinylated [10]. Similarly, in heart among all of the identified succinylated proteins, more than 75% were mitochondrial proteins. More than 90% of succinylation sites show increased abundance in SIRT5 KO heart with an average increase of 8.37 and a median of 1.64. Over 25% of the sites show over threefold greater abundance in SIRT5 KO heart [13].
Succinylation also occurs in the nucleus [20]. Chromatin immuno-precipitation sequencing suggests that more than one-third of all nucleosomes contain lysine succinylation marks and demonstrate a potential role of chromatin succinylation in modulating gene expression [20]. This striking prevalence of histone succinylation suggests that succinylation of chromatin components may be important for regulating nuclear functions. Forty-five percent of hypersuccinylated peaks and 30% of hyposuccinylated peaks map to promoters [20]. As described above KGDHC localizes to the nucleus in human cell lines and binds to KAT2A in the promoter regions of genes [21]. Nuclear protein complexes that are enriched among lysine succinylated proteins have not been identified [34]. The nucleus is positive for SIRT5, which further implicates succinylation/desuccinylases [31, 34]. Lysine succinylation sites on histones primarily localize to the C-terminal globular domains. This is in contrast to lysine acetylation, which primarily occupies the N-terminal tails of histones, potentially suggesting a distinct epigenetic role of histone lysine succinylation in comparison to lysine acetylation [34]. Thirteen, 7, 10 and 7 histone lysine succinylation sites occur in HeLa cells, MEFs, Drosophila S2 and S. cerevisiae cells, respectively [41].
4. Succinylation alters select metabolic pathways and enzymes.
4.1. Changes in succinylation with altered metabolism (Table 4).
Table 4.
Succinylation alters select metabolic pathways and enzymes.
| Pathway | Affected enzymes and products | Model | Reference |
|---|---|---|---|
| TCA cycle | PDHC, SDH | SIRT5 KO mouse liver and SIRT5 KO MEFs | [34] |
| ATP level | SIRT5 KO mouse heart | [13] | |
| IDH | E. coli | [2] | |
| IDH | Pure protein | [62] | |
| Redox state | SOD1 | 293T cells | [42] |
| Respiratory Chain | F1F0 ATP synthase, respiratory chain complexes (complexes I, III, and IV) | SIRT5 KO mouse liver and SIRT5 KO MEFs | [34] |
| membrane proteins (Complex I and II), ATP synthase | SIRT5 KO mouse | [43] | |
| Complex II (SDH) | HT1080 cells stably overexpressing SIRT5 | [23] | |
| Val-Leu-Ile degradation pathway | - | SIRT5 KO mouse liver and SIRT5 KO MEFs | [34] |
| Fatty acid metabolism | - | SIRT5 KO mouse liver and SIRT5 KO MEFs | [34] |
| ECHA | SIRT5 KO mouse heart | [13] | |
| Ketogenesis | HMGCS2, HMG CoA | SIRT5 KO mouse liver mitochondria | [44] |
| Mitochondrial acetyl CoA acetyltransferase | SIRT5 KO MEFs | [34] | |
| UREA cycle | CPS1 | SIRT5 KO mouse | [6] |
Succinylation responds to changes in metabolism, which suggests that succinylation may help to integrate the responses to the metabolic challenges (Table 4). The data is consistent with the suggestion that changes in metabolism alter succinylation which then provides feedback on metabolism but also provides crosstalk with other proteins that are important to pathology. Post-translational modifications in general, and specifically succinylation provide new levels of integration of classical metabolic pathways. High glucose conditions lead to more lysinesuccinylated proteins, and enhance the abundance of succinyl-lysine peptides more significantly than acetyl-lysines which suggests that glucose has a more profound impact on succinylation than on acetylation [36]. Succinylation levels in whole-cell extracts and mitochondria vary in the liver and kidney under fed and fasting conditions vary. The changes are more marked in the liver than the kidney [34]. In neuronal cells, mitochondrial succinylation is dynamic and changes in response to metabolic perturbations [4]. In cultured mouse neurons, reduced glycolysis (2-deoxyglucose), and/or glutathione depletion (iodoacetic acid), depressed TCA cycle activity (carboxyethyl ester of succinyl phosphonate), and impairment of electron transport (antimycin), ATP synthase (oligomycin), uncouplers of oxidative phosphorylation (carbonyl cyanide m-chlorophenyl hydrazine or tyrphostin) decreased succinylation. In contrast, reducing the oxygen from 20% to 2.4% increased succinylation [4]. In cells with reduced succinic dehydrogenase, succinate and succinyl CoA increase and succinylation of histones increase [20]. Analysis of mitochondrial proteins in this differentially succinylated subset revealed dramatic effects for proteins involved in glycolysis/TCA cycle, fatty acid catabolism, ketone body metabolism, heat shock response, solute transport, ATP synthesis, amino acid synthesis, and the electron transport chain [20]. The results demonstrate that succinylation varies with metabolic states and may help coordinate the response to the metabolic challenge [4, 5].
In E.Coli, KEGG pathway evaluation, GO annotation, and Pfam domain analyses suggest that lysine succinylation substrates associate with the ribosome and protein expression/translation-associated events. Lysine succinylated proteins are also highly enriched in purine/pyrimidine metabolism, glycolysis/gluconeogenesis, pyruvate metabolism [36].
A second approach to test the role of succinylation is to evaluate changes in enzymes or pathways in cells or animals in which SIRT5 is diminished or abolished. However, SIRT5 is also a deglutarylase and demalonase, so this tests SIRT5’s actions rather than the role of succinylation. Furthermore, just the presence of succinylation does not necessarily indicate altered activity. For example, SIRT5 KO induced succinylation of citrate synthase or ATP synthase does not alter the activities of these targets [13]. Nevertheless, the activity of enzymes and pathways can be modulated by succinylation. Lysine succinylated proteins are significantly enriched in the cellular metabolic process with specific enrichment in oxoacid metabolism, oxidation reduction processes, and coenzyme metabolism [3, 6, 34]. In SIRT5 KO mice a number of highly connected subnetworks among Lys-succinylated proteins are apparent including the F1F0 ATP synthase, respiratory chain super-complexes (complexes I, III, and IV), glutathione S transferases, ribosomes and the chaperonin-containing TCP1 complex [34].
TCA cycle.
In SIRT5 KOs, over 80% of all proteins in the TCA cycle are succinylated [34]. Succinylation reduces mitochondrial function by actions on PDHC and SDH. SIRT5 knockdown substantially increases SDH activity and elevates cellular respiration in the presence of the SDH substrate succinate. SIRT5 knockdown in cells also increased respiration in the presence of the PDHC substrates [34]. Lack of SIRT5 lowers cardiac ATP Levels [13]. One of the first proteins shown to be succinylated is isocitrate dehydrogenase, [2], and succinylation of isocitrate dehydrogenase inhibits it activity [5]. Mutation of critical lysines also suggest succinylation to be inhibitory [2].
Redox state.
Enzymes in the cytosol and mitochondria may respond differently to changes in SIRT5. SOD1 is a cytosolic enzyme that is partially regulated by succinylation. Succinylation decreases SOD1 activity, while SIRT5 binds to, desuccinylates and activates SOD1. SOD1-mediated ROS reduction increases when SIRT5 is co-expressed [42].
Respiratory Chain.
Analysis of succinylated proteins reveals a number of highly connected subnetworks among Lys-succinylated proteins, including the F1F0 ATP synthase, respiratory chain complexes (complexes I, III, and IV) [34]. Several SIRT5-targeted lysine residues lie at the protein-lipid interface of SDH subunit B. Succinylation at these sites may disrupt Complex II subunit-subunit interactions and electron transfer. Two studies has explored the effects of SIRT5 on Complex II SDH activity in cell culture models. Park et al. [34] observe higher SDH activity in cultured mouse embryonic fibroblasts following SIRT5 KD and suggest that succinylation activates SDH. Li et al. observed a lower SDH activity in succinylation mimetic SDHB mutant (K53E) cells [23], which is diametrically opposed to Park’s result [34]. The hypersuccinylation caused by IDH1 overexpression that decreased the activities of SDH is reversed by SIRT5 overexpression, suggesting that succinylation suppresses, whereas SIRT5-related desuccinylation promotes SDH activity [23]. Research in SIRT5 KO liver further documents compromised Complex II and ATP synthase function in vivo [43], which results agree with those of Li et al.[23] These two pieces of evidence in vitro and vivo support the view of SIRT5 as a promoter of mitochondrial energy metabolism rather than a suppressor.
Val-Leu-Ile degradation pathway.
In SIRT5 KOs, 37 of 51 proteins in the Val-Leu-Ile degradation pathway are succinylated [34].
Fatty acid metabolism.
SIRT5 KO reduces fatty acid oxidation and accumulation of long-chain fatty acyl CoAs. In SIRT5 KOs, 60% of all proteins in fatty acid metabolism are succinylated [34]. ECHA is the α-subunit of the mitochondrial trifunctional enzyme, which is important for fatty acid β-oxidation [13]. ECHA is regulated by SIRT5 and affects heart function [13]. Succinylation inhibits ECHA, and SIRT5 mediated desuccinylation activates ECHA. As a result, SIRT5-deficient mice exhibit defective fatty acid metabolism and decreased ATP production [13].
Ketogenesis.
Loss of SIRT5 leads to accumulation of medium- and long-chain acylcarnitines and decreases β-hydroxybutyrate production in vivo. SIRT5 regulates succinylation of the rate-limiting ketogenic enzyme 3-hydroxy-3-methylglutaryl-CoA synthase 2 (HMGCS2) both in vivo and in vitro [44]. SIRT5 up-regulates hepatic ketogenesis through activation of 3-hydroxy-3-methylglutaryl-CoA synthase (HMG-CoA) [44]. HMG-CoA in extracts of rat liver mitochondria is inactivated by succinyl CoA and activated by incubation in a medium designed to cause desuccinylation [44, 45]. Similarly, activity could be manipulated by changing succinyl CoA content of mitochondria. In vivo, mitochondrial HMG CoA synthase in fed rats is normally substantially succinylated (about 40%) and inactivated, and that glucagon increases the activity of HMG CoA synthase by lowering the concentration of succinyl CoA and thus decreasing the extent of succinylation of the enzyme. Finally, mutation of hypersuccinylated residues lysine 83 and lysine 310 on HMGCS2 to glutamic acid strongly inhibits enzymatic activity [44, 45]. Mitochondrial acetyl CoA acetyltransferase is a critical enzyme in ketone body metabolism that conjugates acetyl CoA into acetoacetyl CoA. Succinylation at lysine 265, located near a CoA-binding site (lysine 260) increases in abundance by more than 120-fold in SIRT5 KO cells [34], Thus, succinylation clearly alters metabolism.
Urea cycle.
SIRT5 regulates the urea cycle by regulation of the succinylation of carbamoyl phosphate synthetase (CPS1) [6]. CPS1 has 47 succinylation sites [46].
Other pathways and enzymes that have been shown to be regulated by succinylation are propionate metabolism [34], glyceraldehyde phosphate dehydrogenase [2], glutathione S transferase [34], ribosomal proteins, TCP1 [34] and purine biosynthesis [34].
5. Overlap of succinylation with other PTMs.
The differential regulation of proteins in common pathways by phosphorylation, acetylation and succinylation suggest crosstalk of these modifiers in regulating mitochondrial metabolic networks. Consensus about the cross talk is just evolving[10, 47–51]. The differences between groups may be technical as well as biological. Succinylation/acetylation overlap appears to be low (~8–10%) in MEFS as well as HeLa cells (12.6%) [10, 34]. By contrast, lysine-succinylation/acetylation overlap is higher in mouse liver (24%) and reaches a maximum (38.5%) in purified liver mitochondria [10, 44]. Systematic profiling of the mammalian succinylome identifies 2,565 succinylation sites on 779 proteins and most do not overlap with acetylation sites [41]. Minimal overlap of succinylation and acetylation sites suggests differential regulation of succinylation and acetylation [34]. However, others report that lysine succinylation extensively overlaps with acetylation in prokaryotes and eukaryotes [10]. Malonylation also targets a different set of proteins than acetylation and succinylation [35].
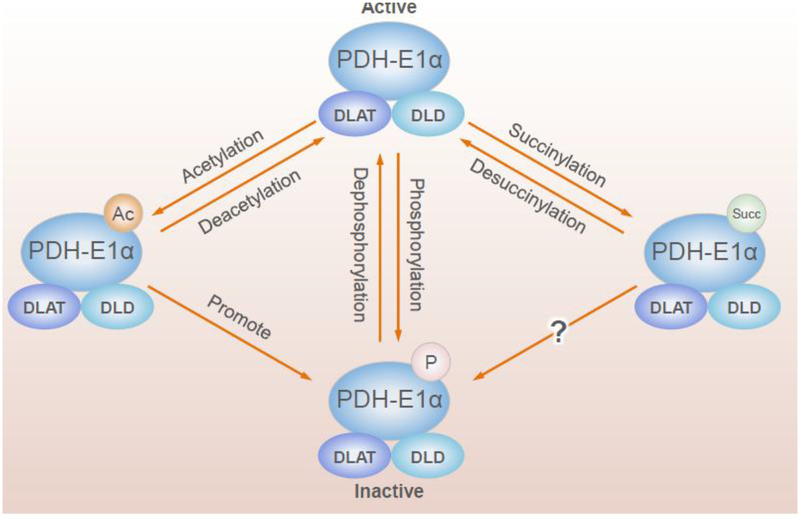
The complexity of the interactions of PTMs is apparent with the PDHC (Figure 5). Phosphorylation, succinylation, acetylation and likely other PTMs regulate PDHC. PDHC consists of three primary enzymes (E1p, E2p and E3) and many modulatory phosphatases, kinases, succinylases, desuccinylases, acetylases and deacetylases. E1α, a key regulatory subunit of E1p, is regulated by many post-translational mechanisms. Phosphorylation of E1α by pyruvate dehydrogenase kinases (PDK) suppresses activity and dephosphorylation by pyruvate dehydrogenase phosphatases (PDP) activates PDHC [52]. A recent global acetyl-proteomic analysis reveals novel interactions of PTMs including acetylation and phosphorylation [53]. Mitochondrial ACAT1 and SIRT3 are the upstream acetyltransferase and deacetylase, respectively. Tyrosine 381 phosphorylation of E1p provokes SIRT3 dissociation and ACAT1 recruitment. ACAT1 acetylates E1p at lysine 202 and E1α at lysine 321, which results in the dissociation of PDP1 from E1α and the recruitment of active PDK1 to E1α. Thus, the changes in the acetylation status of E1α alter the activity of PDHC and promote glycolysis. Global analysis of lysine succinylation identifies several sites of succinylation on the E1α, E1β, E2 and E3 subunits, which, at least for PDHE1α, do not overlap with the acetylation sites that are regulated by SIRT3 [34]. In addition, SIRT5 suppresses PDHC activity by desuccinylation of PDHC subunits [34]. Multiple subunits of E1p are hypersuccinylated in SIRT5 knockout MEFs, as well as liver from SIRT5 knockout mice. Immunoblotting of commercially available E1p confirms succinylation on multiple subunits [34]. Treatment of purified E1p with SIRT5 decreases holoenzyme activity in vitro. SIRT5 depletion in two independent human embryonic kidney (HEK) 293T cell lines results in elevated E1p activity [34]. Additional data provide evidence that acetylation and succinylation constitute novel mechanisms by which E1p activity can be regulated and provide complementary insights into our current understanding of the regulation of E1a through the phosphorylation/dephosphorylation cycle [53, 54].
Figure 5.
Regulation of PDHC
6. Modification of transcription by acetylation and succinylation.
Early reports suggest that succinylated nucleosomes have potent transcription-activating properties [55]. Initial reports of succinylation PTMs also included lysine succinylation of histones [41]. These early studies reveal specific succinylation sites and characterize loss of yeast viability resulting from mutation of histone residues that are normally highly succinylated. These results suggested that histone succinylation is important for cell viability. Studies in immortalized mouse fibroblasts, chromatin immuno-precipitation sequencing (ChIP-seq) suggests that more than one-third of all nucleosomes contain lysine succinylation marks and demonstrate a potential role of chromatin succinylation in modulating gene expression [20]. This striking prevalence of histone succinylation suggests that succinylation of chromatin components may be important for regulating nuclear functions. 45% of hypersuccinylated peaks and 30% of hyposuccinylated peaks map to promoters. These results suggest that either perturbation of chromatin succinylation causes altered gene expression or that the succinylation status of specific genomic loci dynamically changes to reflect perturbed gene expression patterns [20]. Defective TCA cycle metabolism perturbs the succinyllysine distribution in chromatin. This is consistent with previous observations linking nucleosome succinylation with enhanced in vitro transcription [20]. Abnormal TCA cycle metabolism results in defective DNA repair and increased sensitivity to genotoxic agents, consistent with previously reported chromatin hypersuccinylation effects observed with SIRT7 (nuclear desuccinylase) depletion [20]. Chromatin succinylation may thus represent a mechanism by which metabolism modulates both genome-wide transcription and DNA repair activities. Chromatin succinylation is a link between the TCA cycle status and epigenetic transcriptional control [20].
7. Succinylation and Disease.
Succinylation appears important in in cancer, but the roles of succinylation in other diseases is unknown. Hypersuccinylation (i.e., SIRT5 knockout) leads to hypertrophic cardiomyopathy, as evident from the increased heart weight relative to body weight, as well as reduced shortening and ejection fractions [13]. The extensive overlap between acetylation and succinylation suggests that succinylation, like acetylation, may be implicated in multiple disorders [46]. Tau acetylation inhibits tau function via impaired tau-microtubule interactions and promotes pathological tau aggregation [56, 57]. Immunohistochemical and biochemical studies of brains from tau transgenic mice and patients with AD and related tauopathies show that acetylated tau pathology is specifically associated with insoluble, thioflavin-positive tau aggregates. The close overlap suggests that succinylation is also likely to be involved in such diseases. Hypoxia increase succinylation [4] and succinylation may also alter the response to injury.
Succinylation alters outcome from ischemia in heart and brain. Hearts in SIRT5 KO mice are more susceptible to ischemia-reperfusion injury compared with wild type [58]. TCA cycle defects associated with SDH loss results in increased concentrations of succinyl CoA and bulk protein hypersuccinylation affecting multiple subcellular compartments [20]. Protein kinase C epsilon (PKCε)-induced protection from focal ischemia is lost in SIRT5 KO mice. Purine metabolism is a common metabolic pathway regulated by SIRT5, PKCε and ischemic preconditioning. Thus, SIRT5 is linked to the regulation of pathways central to brain metabolism with links to ischemic tolerance. Together, these data suggest that SIRT5 regulates mitochondrial bioenergetics to promote ischemic tolerance [31]. No other neurological diseases have yet been associated with alterations in succinylation.
8. Conclusions.
Although research on succinylation is in its infancy, the data are clear that succinylation can have widespread consequences in health and disease. The ability of succinylation to modify many pathways suggest that it is critical to cell integration. The regulation of succinylation by metabolic enzymes suggest succinylation provides a mechanism to extend the influence of metabolic enzymes far beyond just maintaining ATP or redox state. The findings imply that lysine succinylation modulates enzyme activity in a previously unappreciated layer of biological regulation. The nearly complete lack of studies in the nervous system or in neurological disease is a gap in our knowledge.
Acknowledgements.
NIH AG014930 and the Burke Neurological Institute supported this research.
We dedicate this manuscript to Dr. Vera Adam-Vizi. Our work has complemented her outtanding research in many ways for many years. Her research on KGDHC and regulation of mitochondrial bioenergetics has been particularly insightful for our own research. Succinlyation is another way that KGDHC alters all aspects of cellular function. We are thankful that she has led the way for so long.
References.
- 1.McKenna MC, Ferreira GC (2016) Enzyme Complexes Important for the Glutamate–Glutamine Cycle In: Schousboe A, Sonnewald U (eds) The Glutamate/GABA-Glutamine Cycle: Amino Acid Neurotransmitter Homeostasis. Springer International Publishing, Cham, pp 59–98 [DOI] [PubMed] [Google Scholar]
- 2.Zhang Z, Tan M, Xie Z, Dai L, Chen Y, Zhao Y (2010) Identification of lysine succinylation as a new post-translational modification. Nature Chemical Biology 7:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peng C, Lu Z, Xie Z, Cheng Z, Chen Y, Tan M, Luo H, Zhang Y, He W, Yang K, Zwaans BMM, Tishkoff D, Ho L, Lombard D, He T-C, Dai J, Verdin E, Ye Y, Zhao Y (2011) The first identification of lysine malonylation substrates and its regulatory enzyme. Molecular & Cellular Proteomics [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen H, Xu H, Potash S, Starkov A, Belousov VV, Bilan DS, Denton TT, Gibson GE (2017) Mild metabolic perturbations alter succinylation of mitochondrial proteins. Journal of Neuroscience Research 95:2244–2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gibson GE, Xu H, Chen H-L, Chen W, Denton TT, Zhang S (2015) Alpha-ketoglutarate dehydrogenase complex-dependent succinylation of proteins in neurons and neuronal cell lines. Journal of Neurochemistry 134:86–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Du J, Zhou Y, Su X, Yu JJ, Khan S, Jiang H, Kim J, Woo J, Kim JH, Choi BH, He B, Chen W, Zhang S, Cerione RA, Auwerx J, Hao Q, Lin H (2011) Sirt5 Is a NAD-Dependent Protein Lysine Demalonylase and Desuccinylase. Science 334:806–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maurer B, Rumpf T, Scharfe M, Stolfa DA, Schmitt ML, He W, Verdin E, Sippl W, Jung M (2012) Inhibitors of the NAD+-Dependent Protein Desuccinylase and Demalonylase Sirt5. ACS Medicinal Chemistry Letters 3:1050–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simithy J, Sidoli S, Yuan Z-F, Coradin M, Bhanu NV, Marchione DM, Klein BJ, Bazilevsky GA, McCullough CE, Magin RS, Kutateladze TG, Snyder NW, Marmorstein R, Garcia BA (2017) Characterization of histone acylations links chromatin modifications with metabolism. Nature Communications 8:1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wagner GR, Bhatt DP, O’Connell TM, Thompson JW, Dubois LG, Backos DS, Yang H, Mitchell GA, Ilkayeva OR, Stevens RD, Grimsrud PA, Hirschey MD (2017) A Class of Reactive Acyl-CoA Species Reveals the Non-enzymatic Origins of Protein Acylation. Cell Metabolism 25:823–837.e828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weinert Brian T, Schölz C, Wagner Sebastian A, Iesmantavicius V, Su D, Daniel Jeremy A, Choudhary C (2013) Lysine Succinylation Is a Frequently Occurring Modification in Prokaryotes and Eukaryotes and Extensively Overlaps with Acetylation. Cell Reports 4:842–851 [DOI] [PubMed] [Google Scholar]
- 11.Wagner GR, Payne RM (2013) Widespread and Enzyme-independent Nϵ-Acetylation and Nϵ-Succinylation of Proteins in the Chemical Conditions of the Mitochondrial Matrix. Journal of Biological Chemistry 288:29036–29045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wagner GR, Payne RM (2013) Widespread and enzyme-independent Nϵ-acetylation and Nϵ-succinylation in the chemical conditions of the mitochondrial matrix. Journal of Biological Chemistry [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sadhukhan S, Liu X, Ryu D, Nelson OD, Stupinski JA, Li Z, Chen W, Zhang S, Weiss RS, Locasale JW, Auwerx J, Lin H (2016) Metabolomics-assisted proteomics identifies succinylation and SIRT5 as important regulators of cardiac function. Proceedings of the National Academy of Sciences 113:4320–4325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shurubor YI, D’Aurelio M, Clark-Matott J, Isakova EP, Deryabina YI, Beal MF, Cooper AJL, Krasnikov BF (2017) Determination of Coenzyme A and Acetyl-Coenzyme A in Biological Samples Using HPLC with UV Detection. Molecules 22:1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Q, Zhang S, Berthiaume JM, Simons B, Zhang G-F (2014) Novel approach in LC-MS/MS using MRM to generate a full profile of acyl-CoAs: discovery of acyl-dephospho-CoAs. Journal of Lipid Research 55:592–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frank RAW, Price AJ, Northrop FD, Perham RN, Luisi BF (2007) Crystal Structure of the E1 Component of the Escherichia coli 2-Oxoglutarate Dehydrogenase Multienzyme Complex. Journal of Molecular Biology 368:639–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Basu SS, Blair IA (2011) Rotenone-Mediated Changes in Intracellular Coenzyme A Thioester Levels: Implications for Mitochondrial Dysfunction. Chemical Research in Toxicology 24:1630–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cavender FL (1971) The metabolic source of the succinyl-CoA moiety of δ-aminolevulinic acid. Biochemical Medicine 5:515–520 [Google Scholar]
- 19.Burch JS, Marcero JR, Maschek JA, Cox JE, Jackson LK, Medlock AE, Phillips JD, Dailey HA (2018) Glutamine via α-ketoglutarate dehydrogenase provides succinyl-CoA for heme synthesis during erythropoiesis. Blood 132:987–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smestad J, Erber L, Chen Y, Maher LJ (2018) Chromatin Succinylation Correlates with Active Gene Expression and Is Perturbed by Defective TCA Cycle Metabolism. iScience 2:63–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y, Guo YR, Liu K, Yin Z, Liu R, Xia Y, Tan L, Yang P, Lee J-H, Li X-j, Hawke D, Zheng Y, Qian X, Lyu J, He J, Xing D, Tao YJ, Lu Z (2017) KAT2A coupled with the α-KGDH complex acts as a histone H3 succinyltransferase. Nature 552:273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi Q, Chen H-L, Xu H, Gibson GE (2005) Reduction in the E2k Subunit of the α-Ketoglutarate Dehydrogenase Complex Has Effects Independent of Complex Activity. Journal of Biological Chemistry 280:10888–10896 [DOI] [PubMed] [Google Scholar]
- 23.Li F, He X, Ye D, Lin Y, Yu H, Yao C, Huang L, Zhang J, Wang F, Xu S, Wu X, Liu L, Yang C, Shi J, He X, Liu J, Qu Y, Guo F, Zhao J, Xu W, Zhao S (2015) NADP+-IDH Mutations Promote Hypersuccinylation that Impairs Mitochondria Respiration and Induces Apoptosis Resistance. Molecular Cell 60:661–675 [DOI] [PubMed] [Google Scholar]
- 24.Przybyla-Zawislak B, Dennis RA, Zakharkin SO, McCammon MT (1998) Genes of succinyl-CoA ligase from Saccharomyces cerevisiae. European Journal of Biochemistry 258:736–743 [DOI] [PubMed] [Google Scholar]
- 25.Wang C, Zhang C, Li X, Shen J, Xu Y, Shi H, Mu X, Pan J, Zhao T, Li M, Geng B, Xu C, Wen H, You Q (2019) CPT1A-mediated succinylation of S100A10 increases human gastric cancer invasion. Journal of Cellular and Molecular Medicine 23:293–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kurmi K, Hitosugi S, Wiese EK, Boakye-Agyeman F, Gonsalves WI, Lou Z, Karnitz LM, Goetz MP, Hitosugi T (2018) Carnitine Palmitoyltransferase 1A Has a Lysine Succinyltransferase Activity. Cell Reports 22:1365–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee K, Kerner J, Hoppel CL (2011) Mitochondrial Carnitine Palmitoyltransferase 1a (CPT1a) Is Part of an Outer Membrane Fatty Acid Transfer Complex. Journal of Biological Chemistry 286:25655–25662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Papanicolaou KN, O’Rourke B, Foster DB (2014) Metabolism leaves its mark on the powerhouse: recent progress in post-translational modifications of lysine in mitochondria. Frontiers in Physiology 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jayasena T, Poljak A, Braidy N, Zhong L, Rowlands B, Muenchhoff J, Grant R, Smythe G, Teo C, Raftery M, Sachdev P (2016) Application of Targeted Mass Spectrometry for the Quantification of Sirtuins in the Central Nervous System. Scientific Reports 6:35391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tan M, Peng C, Anderson Kristin A, Chhoy P, Xie Z, Dai L, Park J, Chen Y, Huang H, Zhang Y, Ro J, Wagner Gregory R, Green Michelle F, Madsen Andreas S, Schmiesing J, Peterson Brett S, Xu G, Ilkayeva Olga R, Muehlbauer Michael J, Braulke T, Mühlhausen C, Backos Donald S, Olsen Christian A, McGuire Peter J, Pletcher Scott D, Lombard David B, Hirschey Matthew D, Zhao Y (2014) Lysine Glutarylation Is a Protein Posttranslational Modification Regulated by SIRT5. Cell Metabolism 19:605–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koronowski KB, Khoury N, Morris-Blanco KC, Stradecki-Cohan HM, Garrett TJ, Perez-Pinzon MA (2018) Metabolomics Based Identification of SIRT5 and Protein Kinase C Epsilon Regulated Pathways in Brain. Frontiers in Neuroscience 12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsushita N, Yonashiro R, Ogata Y, Sugiura A, Nagashima S, Fukuda T, Inatome R, Yanagi S (2011) Distinct regulation of mitochondrial localization and stability of two human Sirt5 isoforms. Genes to Cells 16:190–202 [DOI] [PubMed] [Google Scholar]
- 33.Li L, Shi L, Yang S, Yan R, Zhang D, Yang J, He L, Li W, Yi X, Sun L, Liang J, Cheng Z, Shi L, Shang Y, Yu W (2016) SIRT7 is a histone desuccinylase that functionally links to chromatin compaction and genome stability. Nature Communications 7:12235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park J, Chen Y, Tishkoff Daniel X, Peng C, Tan M, Dai L, Xie Z, Zhang Y, Zwaans Bernadette MM, Skinner Mary E, Lombard David B, Zhao Y (2013) SIRT5-Mediated Lysine Desuccinylation Impacts Diverse Metabolic Pathways. Molecular Cell 50:919–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nishida Y, Rardin Matthew J, Carrico C, He W, Sahu Alexandria K, Gut P, Najjar R, Fitch M, Hellerstein M, Gibson Bradford W, Verdin E (2015) SIRT5 Regulates both Cytosolic and Mitochondrial Protein Malonylation with Glycolysis as a Major Target. Molecular Cell 59:321–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Colak G, Xie Z, Zhu AY, Dai L, Lu Z, Zhang Y, Wan X, Chen Y, Cha YH, Lin H, Zhao Y, Tan M (2013) Identification of lysine succinylation substrates and the succinylation regulatory enzyme CobB in E. coli. Molecular & Cellular Proteomics [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jin W, Wu F (2016) Proteome-Wide Identification of Lysine Succinylation in the Proteins of Tomato (Solanum lycopersicum). PLOS ONE 11:e0147586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li X, Hu X, Wan Y, Xie G, Li X, Chen D, Cheng Z, Yi X, Liang S, Tan F (2014) Systematic Identification of the Lysine Succinylation in the Protozoan Parasite Toxoplasma gondii. Journal of Proteome Research 13:6087–6095 [DOI] [PubMed] [Google Scholar]
- 39.Pan J, Chen R, Li C, Li W, Ye Z (2015) Global Analysis of Protein Lysine Succinylation Profiles and Their Overlap with Lysine Acetylation in the Marine Bacterium Vibrio parahemolyticus. Journal of Proteome Research 14:4309–4318 [DOI] [PubMed] [Google Scholar]
- 40.Rardin MJ, Newman JC, Held JM, Cusack MP, Sorensen DJ, Li B, Schilling B, Mooney SD, Kahn CR, Verdin E, Gibson BW (2013) Label-free quantitative proteomics of the lysine acetylome in mitochondria identifies substrates of SIRT3 in metabolic pathways. Proceedings of the National Academy of Sciences 110:6601–6606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xie Z, Dai J, Dai L, Tan M, Cheng Z, Wu Y, Boeke JD, Zhao Y (2012) Lysine succinylation and lysine malonylation in histones. Molecular & Cellular Proteomics [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin Z-F, Xu H-B, Wang J-Y, Lin Q, Ruan Z, Liu F-B, Jin W, Huang H-H, Chen X (2013) SIRT5 desuccinylates and activates SOD1 to eliminate ROS. Biochemical and Biophysical Research Communications 441:191–195 [DOI] [PubMed] [Google Scholar]
- 43.Zhang Y, Bharathi SS, Rardin MJ, Lu J, Maringer KV, Sims-Lucas S, Prochownik EV, Gibson BW, Goetzman ES (2017) Lysine desuccinylase SIRT5 binds to cardiolipin and regulates the electron transport chain. Journal of Biological Chemistry 292:10239–10249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rardin Matthew J, He W, Nishida Y, Newman John C, Carrico C, Danielson Steven R, Guo A, Gut P, Sahu Alexandria K, Li B, Uppala R, Fitch M, Riiff T, Zhu L, Zhou J, Mulhern D, Stevens Robert D, Ilkayeva Olga R, Newgard Christopher B, Jacobson Matthew P, Hellerstein M, Goetzman Eric S, Gibson Bradford W, Verdin E (2013) SIRT5 Regulates the Mitochondrial Lysine Succinylome and Metabolic Networks. Cell Metabolism 18:920–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Quant PA, Tubbs PK, Brand MD (1990) Glucagon activates mitochondrial 3-hydroxy-3-methylglutaryl-CoA synthase in vivo by decreasing the extent of succinylation of the enzyme. European Journal of Biochemistry 187:169–174 [DOI] [PubMed] [Google Scholar]
- 46.Min S-W, Chen X, Tracy TE, Li Y, Zhou Y, Wang C, Shirakawa K, Minami SS, Defensor E, Mok SA, Sohn PD, Schilling B, Cong X, Ellerby L, Gibson BW, Johnson J, Krogan N, Shamloo M, Gestwicki J, Masliah E, Verdin E, Gan L (2015) Critical Role of Acetylation in Tau-Mediated Neurodegeneration and Cognitive Deficits. Nature medicine 21:1154–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fukushima A, Alrob OA, Zhang L, Wagg CS, Altamimi T, Rawat S, Rebeyka IM, Kantor PF, Lopaschuk GD (2016) Acetylation and succinylation contribute to maturational alterations in energy metabolism in the newborn heart. American Journal of Physiology-Heart and Circulatory Physiology 311:H347–H363 [DOI] [PubMed] [Google Scholar]
- 48.He D, Wang Q, Li M, Damaris RN, Yi X, Cheng Z, Yang P (2016) Global Proteome Analyses of Lysine Acetylation and Succinylation Reveal the Widespread Involvement of both Modification in Metabolism in the Embryo of Germinating Rice Seed. Journal of Proteome Research 15:879–890 [DOI] [PubMed] [Google Scholar]
- 49.Kouzarides T (2000) Acetylation: a regulatory modification to rival phosphorylation? The EMBO Journal 19:1176–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krämer OH, Knauer SK, Greiner G, Jandt E, Reichardt S, Gührs K-H, Stauber RH, Böhmer FD, Heinzel T (2009) A phosphorylation-acetylation switch regulates STAT1 signaling. Genes & Development 23:223–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tseng Anne H-H, Wu L-H, Shieh S-S, Wang Danny L (2014) SIRT3 interactions with FOXO3 acetylation, phosphorylation and ubiquitinylation mediate endothelial cell responses to hypoxia. Biochemical Journal 464:157–168 [DOI] [PubMed] [Google Scholar]
- 52.Patel MS, Nemeria NS, Furey W, Jordan F (2014) The Pyruvate Dehydrogenase Complexes: Structure-based Function and Regulation. Journal of Biological Chemistry 289:16615–16623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fan J, Shan C, Kang H-B, Elf S, Xie J, Tucker M, Gu T-L, Aguiar M, Lonning S, Chen H, Mohammadi M, Britton L-Mae P, Garcia Benjamin A, Alečković M, Kang Y, Kaluz S, Devi N, Van Meir Erwin G, Hitosugi T, Seo Jae H, Lonial S, Gaddh M, Arellano M, Khoury Hanna J, Khuri Fadlo R, Boggon Titus J, Kang S, Chen J (2014) Tyr Phosphorylation of PDP1 Toggles Recruitment between ACAT1 and SIRT3 to Regulate the Pyruvate Dehydrogenase Complex. Molecular Cell 53:534–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Saunier E, Benelli C, Bortoli S (2016) The pyruvate dehydrogenase complex in cancer: An old metabolic gatekeeper regulated by new pathways and pharmacological agents. International Journal of Cancer 138:809–817 [DOI] [PubMed] [Google Scholar]
- 55.Pin˜eiro M, Hernández F, Palacián E (1992) Succinylation of histone amino groups facilitates transcription of nucleosomal cores. Biochimica et Biophysica Acta (BBA) - Gene Structure and Expression 1129:183–187 [DOI] [PubMed] [Google Scholar]
- 56.Cohen TJ, Guo JL, Hurtado DE, Kwong LK, Mills IP, Trojanowski JQ, Lee VMY (2011) The acetylation of tau inhibits its function and promotes pathological tau aggregation. Nature Communications 2:252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Min S-W, Cho S-H, Zhou Y, Schroeder S, Haroutunian V, Seeley WW, Huang EJ, Shen Y, Masliah E, Mukherjee C, Meyers D, Cole PA, Ott M, Gan L (2010) Acetylation of Tau Inhibits Its Degradation and Contributes to Tauopathy. Neuron 67:953–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Boylston JA, Sun J, Chen Y, Gucek M, Sack MN, Murphy E (2015) Characterization of the cardiac succinylome and its role in ischemia–reperfusion injury. Journal of Molecular and Cellular Cardiology 88:73–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xie L, Liu W, Li Q, Chen S, Xu M, Huang Q, Zeng J, Zhou M, Xie J (2015) First Succinyl-Proteome Profiling of Extensively Drug-Resistant Mycobacterium tuberculosis Revealed Involvement of Succinylation in Cellular Physiology. Journal of Proteome Research 14:107–119 [DOI] [PubMed] [Google Scholar]
- 60.Xu H, Chen X, Xu X, Shi R, Suo S, Cheng K, Zheng Z, Wang M, Wang L, Zhao Y, Tian B, Hua Y (2016) Lysine Acetylation and Succinylation in HeLa Cells and their Essential Roles in Response to UV-induced Stress. Scientific Reports 6:30212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meyer JG, Softic S, Basisty N, Rardin MJ, Verdin E, Gibson BW, Ilkayeva O, Newgard C, Kahn CR, Schilling B (2018) Multi-Omic Profiling Reveals the Opposing Forces of Excess Dietary Sugar and Fat on Liver Mitochondria Protein Acetylation and Succinylation. bioRxiv [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gibson GE, Xu H, Chen HL, Chen W, Denton TT, Zhang S (2015) Alpha‐ketoglutarate dehydrogenase complex‐dependent succinylation of proteins in neurons and neuronal cell lines. Journal of neurochemistry 134:86–96 [DOI] [PMC free article] [PubMed] [Google Scholar]