Abstract
In yeast and higher eukaryotes, phospholipids and triacylglycerol are derived from phosphatidate at the nuclear/endoplasmic reticulum membrane. In de novo biosynthetic pathways, phosphatidate is channeled into membrane phospholipids via its conversion to CDP-diacylglycerol. Its dephosphorylation to diacylglycerol is required for the synthesis of triacylglycerol as well as for the synthesis of phosphatidylcholine and phosphatidylethanolamine via the Kennedy pathway. In addition to the role of phosphatidate as a precursor, it is a regulatory molecule in the transcriptional control of phospholipid synthesis genes via the Henry regulatory circuit. Pah1 phosphatidate phosphatase and Dgk1 diacylglycerol kinase are key players that function counteractively in the control of the phosphatidate level at the nuclear/endoplasmic reticulum membrane. Loss of Pah1 phosphatidate phosphatase activity not only affects triacylglycerol synthesis but also disturbs the balance of the phosphatidate level, resulting in the alteration of lipid synthesis and related cellular defects. The pah1Δ phenotypes requiring Dgk1 diacylglycerol kinase exemplify the importance of the phosphatidate level in the misregulation of cellular processes. The catalytic function of Pah1 requires its translocation from the cytoplasm to the nuclear/endoplasmic reticulum membrane, which is regulated through its phosphorylation in the cytoplasm by multiple protein kinases as well as through its dephosphorylation by the membrane-associated Nem1-Spo7 protein phosphatase complex. This article is part of a Special Issue entitled Endoplasmic Reticulum Platforms for Membrane Lipid Dynamics.
Keywords: phospholipid, triacylglycerol, phosphatidate, diacylglycerol, phosphatidate phosphatase, diacylglycerol kinase
Graphical abstract
Yeast cell lacking the Pah1 phosphatidate phosphatase enzyme
1. Introduction
Biological membranes are composed of many lipid species in which proteins are embedded. The eukaryotic cell membranes contain three major classes of lipids: phospholipids, sphingolipids, and sterols [1]. Phospholipids, amphipathic molecules consisting of hydrophobic tails (i.e., fatty acids) and hydrophilic heads, are major constituents of the membrane [2]. They also function as reservoirs of lipid mediators and serve to tether proteins at the membrane surface. Sphingolipids, which are also amphipathic in nature, are composed of a lipophilic amino alcohol (sphingosine) and hydrocarbon chains [1]. Additionally, they are involved in membrane structure, function, and signalling [1]. A subgroup of sphingolipids are glycosphingolipids and gangliosides, which play a key role in cell recognition and signalling [3]. Sterols are mainly structured by sterol rings: they contribute to membrane structure/fluidity and are precursors of fat-soluble vitamins and steroid hormones [1]. Lipids are not homogeneously distributed in cellular membranes and their composition within the membranes is highly regulated and mediated by proteins, ATP, or spontaneous diffusion [4,5]. The lipid composition has a strong influence on physical properties of the membrane such as fluidity, curvature, and lipid packing. It also plays a crucial role in the formation of the lipid domains [6], the membrane trafficking, fusion and fission events [7], formation of protein-protein and protein-lipid complexes [8]. Aberrant lipid composition or metabolism affects the function of cell organelles (e.g., endoplasmic reticulum stress [9]) or leads towards severe diseases (e.g., cancer, obesity, non-alcoholic fatty liver disease or fatty-acid induced lipotoxicity [10,11]).
Lipids in eukaryotic cells are primarily synthesized at the nuclear/endoplasmic reticulum (ER) membrane [1]. The ER is a dynamic network of flattened membrane, connected sacs and tubules, surrounding the nucleus and spreading across the cytoplasm [12]. The diverse structure of the ER allows the organelle to perform specialized functions such as the synthesis of lipids [13] and proteins [14–16], and the regulation of calcium levels {3613}. The diverse functions of the ER are performed in different subdomains. The ER is continuous from the outer membrane of the nuclear envelope, which separates genetic material from the cell milieu. Proteins associated with the ER are synthesised, folded and post-translationally modified in the rough ER sheets embedded on the cytoplasmic surface [18,19]. Tomographic studies have revealed an additional portion of highly curved and smooth ribosomes on the membrane part of ER tubules [19]. The tubular system is highly dynamic, and its structure grows and continuously rearranges. Regions associated with ribosomes at the low density and smooth ER are considered as transitional ER. Due to its network structure, the ER associates/communicates with other organelle membranes that include those of the Golgi, lysosome/vacuole, and mitochondria [20–26]. The cortical ER, which is a part of the peripheral ER, interacts with the plasma membrane [27–29].
Much insight into the synthesis, regulation, and function of lipids has been garnered through studies using the yeast Saccharomyces cerevisiae [26,30–34]. Lipid synthesis in the model eukaryote is mostly typical of that found in more complex higher eukaryotes [30,31]. Yeast are easy to culture in large quantities for biochemical studies and their tractable genetics facilitates a molecular characterization of lipid synthesis and its regulation [30,31]. The wealth of genomic, proteomic, and metabolic information is available in the Saccharomyces Genome Database (https://www.yeastgenome.org/), providing knowledge of genetic and biochemical interactions of lipid synthesis with other metabolic pathways that impact on cell physiology.
All membrane phospholipids, as well as the storage lipid triacylglycerol (TAG), are derived from phosphatidate (PA) [1,33]. When yeast cells grow under nutrient-rich conditions, the PA that is normally synthesized from glycolysis-derived glycerol-3-P, is channeled into membrane phospholipids required for cell proliferation [35,36]. During nutrient limitation, the growth of yeast cells is reduced, and PA is channeled more into TAG [35,36]. PA is not only an intermediate in lipid synthesis, but also a regulator in the expression of phospholipid synthesis genes [37]. It is also implicated as an activator in cell growth and proliferation, vesicular trafficking, secretion, and endocytosis [38–44]. Of the enzymes that produce PA (e.g., 1-acylglycerol-3-P (lysoPA) acyltransferase, diacylglycerol (DAG) kinase, phospholipase D) and use it as a substrate (e.g., CDP-diacylglycerol (CDP-DAG) synthase, PA phosphatase), DAG kinase and PA phosphatase have emerged as key regulators in PA homeostasis [32,33,45]. The two enzymes also control whether PA is utilized for the synthesis of membrane phospholipids or TAG [32,33,45]. Here, we review the basic steps of lipid synthesis from PA that occurs at the nuclear/ER membrane and summarize the PA-mediated transcriptional regulation for the expression of phospholipid synthesis genes. The genetic and biochemical evidence for the roles of Pah1 PA phosphatase and DAG kinase in regulating PA and lipid homeostasis, as well as the modes and regulation of these enzymes are also discussed.
2. PA-derived synthesis of lipids at the nuclear/ER membrane
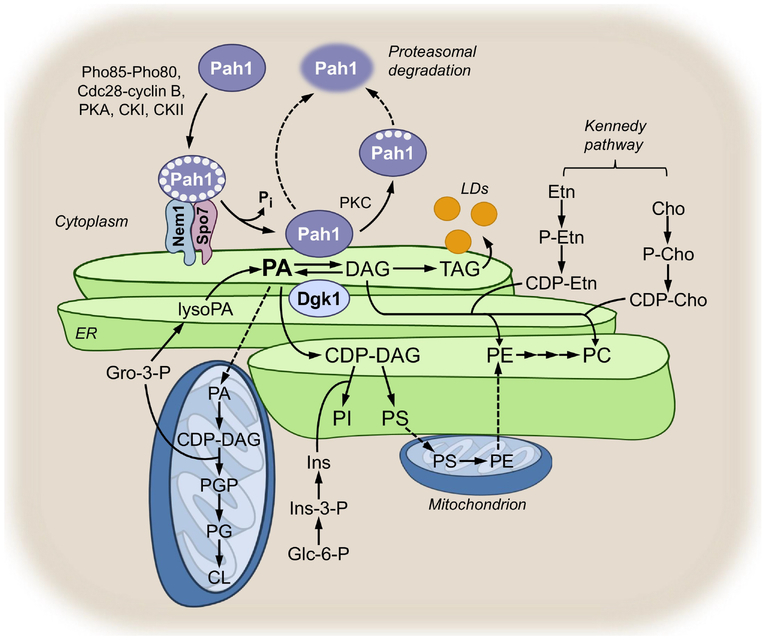
The major steps of lipid synthesis occurring at the nuclear/ER membrane are summarized in Fig. 1. Details on the synthesis of lipid precursors such as fatty acids, inositol, and nucleotides, the remodelling of the acyl groups on lipids, and the degradation of lipids may be found elsewhere [2,33,46,47]. In the de novo pathway, glycerol-3-P is acylated at the 1-position to produce lysoPA by the Gpt2 and Sct1 glycerol-3-P acyltransferase enzymes [48,49]. These enzymes also acylate dihydroxyacetone-P to produce 1-acyl dihydroxyacetone phosphate, which is converted to lysoPA by Ayr1 reductase (not shown in Fig. 1) [50]. The lysoPA is then acylated at the 2-position to produce PA by the Ale1, Loa1, and Slc1 lysoPA/lysophospholipid acyltransferase enzymes [51–54]. The PA produced by the acyltransferase reactions is then partitioned into CDP-DAG and DAG as catalyzed by Cds1 CDP-DAG synthase [55] and Pah1 PA phosphatase [56], respectively. Although both enzymes function at the nuclear/ER membrane, Cds1 is an integral membrane protein whereas Pah1 is a peripheral membrane protein (see below). In wild type cells growing in the absence of choline and ethanolamine supplementation, phosphatidylcholine (PC) and phosphatidylethanolamine (PE) are produced from CDP-DAG in the de novo biosynthetic pathway (referred to as the CDP-DAG pathway). CDP-DAG is first converted by Cho1 PS synthase to phosphatidylserine (PS) [57–61], which is then decarboxylated to produce PE by Psd1 PS decarboxylase [62,63]. The most abundant membrane phospholipid PC is produced from PE via the three sequential methylations using AdoMet as a methyl donor [64–66] by Cho2 PE methyltransferase [64,65] (first methylation step to produce phosphatidylmonomethylethanolamine) and Opi3 phospholipid methyltransferase [65,66] (second and third methylation steps to produce phosphatidyldimethylethanolamine and then PC). Whereas the reactions catalyzed by Cho1, Cho2, and Opi3 take place at the nuclear/ER membrane, PS decarboxylation by Psd1 occurs at the mitochondrial membrane [33]. Thus, the PS synthesized in the nuclear/ER membrane is transferred to the mitochondrial membrane for its decarboxylation to PE, which is then transferred to the nuclear/ER membrane for its conversion to PC (Fig. 1) [33]. The ER and mitochondria are tethered to each other by the ER-mitochondria encounter structure (ERMES) [67–69]. The synthesis of PS occurring at the ER-mitochondrial contact site facilitates its transfer to the mitochondria [70], which is mediated by the Ups2-Mdm35 complex [71]. Psd2 is a second PS decarboxylase enzyme [72] associated with endosomes/vacuoles, but its contribution to PC synthesis via the CDP-DAG pathway is unclear.
Fig. 1.
Synthesis of phospholipids and TAG and model for the regulation of Pah1 PA phosphatase. The figure depicts the pathways for the synthesis of lipids and their precursors that occur in the nuclear/ER membrane (green), in the mitochondria (blue), and in the cytoplasm (tan). A greater detail of lipid synthetic pathways may be found elsewhere [32,33,176,234]. The phosphorylated form of Pah1 is indicated by the white circles within the ellipse surrounding Pah1 and lipid droplets are indicated by the yellow circles emanating from the nuclear/ER. Gro, glycerol; Ins, inositol; Glc, glucose; Etn, ethanolamine; P-Etn, phosphoethanolamine; Cho, choline; P-Cho, phosphocholine; LDs, lipid droplets.
The cho1 [58], psd1 psd2 [72], and cho2 opi3 [66,73] mutants defective in the CDP-DAG pathway are auxotrophic for choline or ethanolamine. The choline and ethanolamine supplemented to the cells are channeled into PC and PE, respectively, by way of the CDP-choline and CDP-ethanolamine branches of the Kennedy pathway (Fig. 1). In yeast, the Kennedy pathway is a salvage or auxiliary pathway that permits the synthesis of PC and PE when the primary CDP-DAG pathway is blocked [30]. In the Kennedy pathway, choline or ethanolamine in growth medium is transported into the cell by the Hnm1 choline/ethanolamine transporter [74]. The water-soluble lipid precursors are then phosphorylated in the cytoplasm to form phosphocholine and phosphoethanolamine by Cki1 choline kinase [75] and Eki1 ethanolamine kinase [76], respectively. The lipid intermediates are then activated with CTP to form CDP-choline and CDP-ethanolamine by Pct1 phosphocholine cytidylyltransferase[77] and Ect1 phosphoethanolamine cytidylyltransferase [78], respectively. The locations of the enzyme reactions are ambiguous; they are reported in the cytoplasm, nuclear periphery, or within the nucleus [79–81]. The last steps in the Kennedy pathway, namely the conversions of CDP-choline and CDP-ethanolamine to PC and PE, respectively, are catalyzed by the Cpt1 choline phosphotransferase [82,83] and Ept1 ethanolamine phosphotransferase, respectively [84,85]. The locations of Cpt1 and Ept1 are also unclear; they are reported to localize in the nuclear/ER membrane or the mitochondrial membrane [79,80]. The DAG required in the reactions is derived from the Pah1 PA phosphatase reaction [56] that takes place at the nuclear/ER membrane [36,86]. Whereas the CDP-DAG pathway is the predominant one used by wild type cells in the absence of choline and ethanolamine supplementation, they utilize the Kennedy pathway when supplemented with choline or ethanolamine [30,32,33]. The choline required for the CDP-choline branch of the pathway may also be provided from the Spo14 phospholipase D-mediated turnover of the PC that is synthesized via the CDP-DAG pathway [76,87,88]. Yeast mutants (e.g., cki1 eki1 and cpt1 ept1) defective in both branches of the Kennedy pathway can synthesize PC only via the CDP-DAG pathway [76,89–92]. These mutants, unlike those defective in the CDP-DAG pathway, do not exhibit any auxotrophic requirements [76,92].
It should be noted that mutants defective in PC or PE synthesis via the CDP-DAG pathway are not strict choline/ethanolamine auxotrophs. Their growth is also supported by supplementation of lysoPE, lysoPC, or PC with short acyl chains. Inside the cell, lysoPC and lysoPE are acylated by the Ale1 lysophospholipid acyltransferase [51,52,93,94], whereas the short chain PC is remodeled for its membrane incorporation with long fatty acyl chains (16 and 18 carbons) by phospholipase B and lysophospholipid acyltransferase enzymes [95–98].
The CDP-DAG used for the synthesis of PS is also converted to PI by the nuclear/ER membrane-associated Pis1 PI synthase [99,100] (Fig. 1). The PI derived from CDP-DAG also serves as a precursor of the D-3, D-4, and D-5 phosphoinositides [101–105], glycosyl PI anchors [106,107], and sphingolipids {1478, 2318}, which are synthesized in other cellular compartments {2805, 2975} (not shown in Fig. 1). The headgroup of these inositol-containing lipids is synthesized in the cytoplasm from glucose-6-P via the reactions catalyzed by Ino1 inositol-3-P synthase [111] and Inm1 inositol-3-P phosphatase [112].
In addition to being the source of PS and PI, the CDP-DAG molecule donates its phosphatidyl moiety to glycerol-3-P and phosphatidylglycerol (PG) to form phosphatidylglycerophosphate (PGP) and cardiolipin (CL), respectively. These two reactions occurring in the mitochondria are catalyzed by the membrane-associated Pgs1 PGP synthase [113,114] and Crd1 CL synthase [115–117] (Fig. 1). The PG used for the CL synthesis is derived from PGP by Gep4 PGP phosphatase [118]. After its synthesis, CL is remodeled with respect to its fatty acyl composition by the Cld1 CL-specific phospholipase A [119] and the Taz1 monolysoCL-specific acyltransferase [120,121] (not shown in Fig. 1). The CDP-DAG used for the synthesis of the mitochondrial phospholipids differs in its origin from that used for the synthesis PS and PI. The mitochondrial CDP-DAG is produced in the organelle by Tam41 CDP-DAG synthase [122] using PA that is translocated from the nuclear/ER membrane by the Ups1 lipid transfer protein [123].
As yeast cells progress from the exponential to the stationary phase of growth due to the depletion of nutrients in the medium, the utilization PA shifts from phospholipid synthesis to TAG synthesis [35,124,125]. The DAG produced by Pah1 PA phosphatase is converted to TAG by the ER-associated Dga1 acyl-CoA dependent DAG acyltransferase [126] and/or the Lro1 phospholipid-dependent DAG acyltransferase [127]. The Are1 and Are2 sterol acyltransferase enzymes also acylate DAG to produce TAG, but their contributions are minor when compared with those of Dga1 and Lro1 [128]. The TAG synthesized at the nuclear/ER membrane is then packaged into lipid droplets [129,130], an organelle that emerges from the nuclear/ER membrane [131,132] (Fig. 1). Indeed, nuclear/ER membrane-associated enzymes as well as Pah1 PA phosphatase, which are responsible for TAG synthesis, may be found at the ER-lipid droplet contact sites [131]. While lipid droplets are known to be produced from the outer nuclear/ER membrane, a recent study has shown that they are also produced at a low level from the inner nuclear membrane [133].
3. PA-mediated regulation of lipid synthesis at the nuclear/ER via the Henry regulatory circuit
The synthesis of lipids is regulated by controlling the expression of lipid synthesis genes and by biochemical modulation of the gene products. The gene expression is strongly influenced by the availability of fatty acids, phospholipid precursors (e.g., inositol, serine, choline, ethanolamine), and nucleotides (e.g., CTP) [30–33,45]. Inositol auxotrophy of the ino2 and ino4 mutants [134] and inositol excretion of the opi1 mutant [135] are linked to the misregulation of INO1 expression through a UASINO element (also known as inositol response element) {786, 561, 1193}. Moreover, mutations in any step of the CDP-DAG pathway (e.g., cds1, cho1, psd1, cho2, or opi3) result in the elevation of the PA level and the excretion of inositol because of the derepression of INO1 expression [30–33,45]. These findings led Henry and Patton-Vogt [138] to propose that the phospholipid PA is a major component in transcriptional regulation of the UASINO-containing genes. Many of the genes involved in phospholipid synthesis contain the UASINO element in the promoter, which include those encoding enzymes responsible for fatty acid synthesis (e.g., ACC1, FAS1, FAS2), phospholipid synthesis (e.g., CDS1, CHO1, PSD1, CHO2, OPI3, EKI1, EPT1, CKI1, CPT1), and inositol synthesis (e.g., INO1) [37,139]. The UASINO element is also present in genes encoding the choline/ethanolamine (HNM1) and inositol (ITR1) permeases that are responsible for the uptake of phospholipid precursors into the cell [74,140].
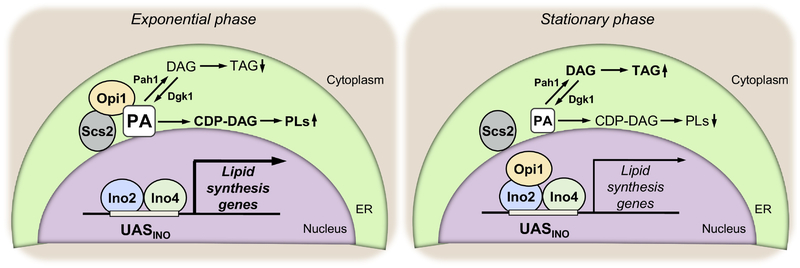
The transcriptional regulation of the UASINO-containing genes (Fig. 2), called “Henry regulatory circuit”, includes the Ino2-Ino4 transcriptional activator complex, the Opi1 repressor, and the Scs2 protein [141,142,142–146]. In the nucleus, transcription of the UASINO-containing genes is activated by the Ino2-Ino4 complex through its binding to the cis-acting element {786, 561, 1193}. This transcriptional activation is negatively controlled by the Opi1 repressor through its interaction with Ino2 [37,139]. The nuclear localization of Opi1 is crucial for its repressor function and is controlled by PA and the Scs2 protein. The Opi1 repressor interacts with Scs2 at the nuclear/ER membrane, and its protein interaction is stabilized by its interaction with PA. When the level of PA is high, Opi1 is tethered to the nuclear/ER membrane through its protein and lipid interactions [146,147]. When the PA level is lowered, Opi1 loses its interaction with the phospholipid and it dissociates from Scs2 and enters the nucleus for its repressor function [33,37,146]. The interaction of Opi1 with PA is mediated by its amphipathic helix [148], and the protein-lipid interaction is favored when the fatty acyl chain of PA is 16 carbons long [149]. The interaction of Opi1 with Scs2 is mediated by its FFAT (two phenylalanines (FF) in an Acidic Tract) motif [145]. As expected, the interaction of Opi1 with Scs2 is essential for the PA-mediated regulation of gene expression in the Henry regulatory circuit [150]. The Opi1-Scs2 interaction also governs the transfer of PA from the nuclear/ER membrane to the mitochondrial membrane where it reacts with CTP to form CDP-DAG for the synthesis of CL [150] (see above). Opi1 repressor function is also controlled by its phosphorylation. For example, the phosphorylations by casein kinase (CK) II [151] and protein kinase A (PKA) [152] stimulate the repressor function, whereas phosphorylation by protein kinase C (PKC) [153] inhibits repressor function. The mechanistic basis for phosphorylation-mediated regulations of Opi1 is unknown.
Fig. 2.
Henry regulatory circuit for the PA-mediated regulation of UASINO-containing lipid synthesis genes during growth. The model depicts the regulation that occurs in the exponential and stationary phases of growth. Details are described in the text and elsewhere [32,33,37].
As indicated above, the PA-mediated regulation of lipid synthesis genes is triggered by growth phase, nutrient availability, and the gene mutations [30,31,33,37,139,154]. For example, the expression of the CHO1 gene, which encodes PS synthase, is derepressed in the exponential phase of cells grown in the absence of phospholipid precursors such as inositol, choline, ethanolamine, and serine [135,155–158] as well as in the presence of the essential nutrient zinc [159]. Conversely, the CHO1 expression is repressed in the exponential phase of cells grown with inositol supplementation, and the gene repression is enhanced by the supplementation of choline, ethanolamine, or serine [135,155–158]. The gene expression is also repressed by the depletion of zinc in the growth medium [159] or when cells progress from the exponential to the stationary phase of growth [160,161].
Unlike the partitioning of CDP-DAG to PS, its partitioning to PI is not regulated by controlling the expression of the enzyme responsible for the reaction step, but by controlling the production of its water-soluble substrate inositol. The INO1 gene encoding inositol-3-P synthase, a key enzyme for the de novo synthesis of inositol, is coordinately regulated with CHO1 and other UASINO-containing genes in the CDP-DAG pathway [162]. The PIS1 gene, which encodes PI synthase, is subject to a different transcriptional control, and its expression is induced under zinc-limited conditions by the Zap1 transcriptional activator [163].
Interestingly, the expression of the PAH1 gene encoding PA phosphatase is regulated by some of the same growth conditions that control gene expression via the Henry regulatory circuit, but with an opposite effect [124,164]. The regulation of PAH1 expression involves the transcription factors Ino2/Ino4/Opi1, Gis1/Rph1 (for inositol and growth phase regulation), and the transcription factor Zap1 (for zinc-mediated regulation) [124,164]. The induction of PAH1 expression in the stationary phase or in response to zinc depletion correlates with the elevation of PA phosphatase activity [124,164]. On the one hand, the elevation of PA phosphatase activity in zinc-replete stationary phase cells is responsible for the increase of TAG synthesis at the expense of phospholipid synthesis [124]. On the other hand, the reduction of PA phosphatase activity in zinc-deplete exponential phase cells is responsible for increased synthesis of PC via the CDP-choline branch of the Kennedy pathway [164].
4. Pah1 PA phosphatase and Dgk1 DAG kinase are key enzymes that control the PA level and lipid synthesis at the nuclear/ER membrane
Pah1 PA phosphatase [56] and Dgk1 DAG kinase [165] have emerged as key enzymes that control PA content and lipid synthesis regulation at the nuclear/ER membrane. As discussed above, Pah1 PA phosphatase dephosphorylates PA to produce DAG [56], whereas DAG kinase phosphorylates DAG to produce PA [165]. Dgk1, a nuclear/ER integral membrane protein [80], is an unconventional DAG kinase in that it utilizes CTP instead of ATP as a phosphate donor [165]. Pah1 functions as a peripheral membrane enzyme, but it is largely localized in the cytoplasm [80] as a highly phosphorylated form (see below). Fluorescence imaging of Pah1 localization on the nuclear/ER membrane is difficult because of its low abundance on the membrane, transient interaction with the substrate PA, inherent protein instability, and susceptibility to proteasomal degradation [166,167]. However, the membrane localization of the GFP-tagged Pah1 was shown by overexpression of Dgk1 [86] or the Nem1-Spo7 phosphatase complex [36]. The higher level of Dgk1 DAG kinase increases the production of PA [165] that interacts with Pah1 [168], whereas the higher level of Nem1-Spo7 facilitates the dephosphorylation of Pah1 for its localization to the nuclear/ER membrane [36,169,170]. The catalytically inactive form of Pah1 [171], which is stable to proteasomal degradation [166] and binds non-productively to PA, was shown to localize at the nuclear/ER membrane, the nuclear/ER membrane-lipid droplet contact site, or the nuclear-vacuole junction [125]. Notwithstanding the fluorescence imaging of its nuclear/ER membrane localization, Pah1 is known to associate with the membrane for its cellular function through the control of the membrane-associated Nem1-Spo7 phosphatase [172]; its dephosphorylation by the phosphatase complex is essential for its membrane localization and catalytic function [36,169,170].
4.1. Phenotypes of the pah1Δ mutant
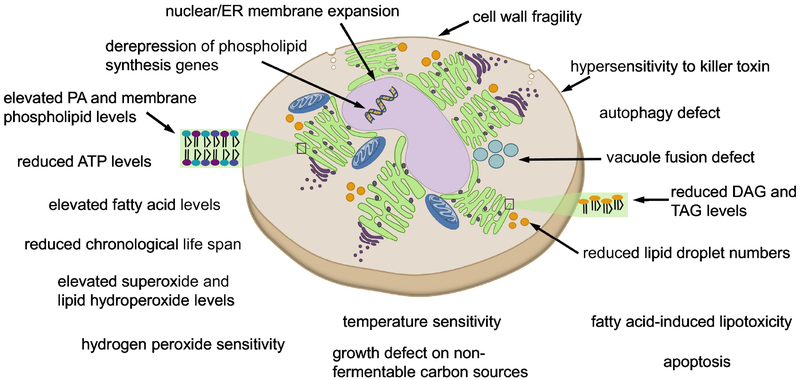
The loss of Pah1 PA phosphatase activity in the cell affects many aspects of lipid metabolism and cell physiology (Fig. 3). Due to the lack of DAG production, the pah1Δ mutant exhibits a great reduction in the level and synthesis of TAG [56,124]. In addition, the mutant cells exhibit a significantly lower number of lipid droplets, which seems to be related to the effect of DAG limitation on the organelle formation [173]. The lack of DAG production and thereby its acylation defect causes the accumulation of fatty acids [56], which renders the mutant cells susceptible to fatty acid-induced lipotoxicity [56,174]. Indeed, TAG synthesis is important to mitigate the toxic effects of fatty acids [175–177].
Fig. 3.
Phenotypes and cellular defects caused by the pah1Δ mutation.
The lack of DAG production in the pah1Δ mutant causes the accumulation of its precursor PA at the nuclear/ER membrane [178], drastically affecting phospholipid synthesis and related cellular processes. The mutant cells exhibit a high increase in the level and synthesis of phospholipids [56,124,174], the PA-mediated derepression of the UASINO-containing phospholipid synthesis genes [169,179], and a reduction in the ATP level [180]. The notable phenotype of the pah1Δ mutant in relation to the increased level of phospholipids is its aberrant nuclear morphology with the expansion of the nuclear/ER membrane [169]. Data indicate that the membrane expansion phenotype is caused by increased phospholipid synthesis in conjunction with the elevated PA content [165,170].
In addition to its role as a precursor in phospholipid synthesis, PA exhibits a strong stimulatory effect on PS synthase [181], which catalyzes a committed step in PC synthesis via the CDP-DAG pathway (Fig. 1). The accumulation of fatty acids in pah1Δ mutant cells, which is caused by a defect in TAG synthesis, may also be attributed to an increase in their synthesis through the derepression of fatty acid synthesis genes (e.g., FAS1, FAS2, and ACC1) that are regulated via the Henry regulatory circuit [182–185] (Fig. 2). The reduced level of ATP in the pah1Δ mutant is not due to a defect in its production by mitochondria (i.e., oxidative phosphorylation), but instead attributed to its exhaustive use for the synthesis of fatty acids and phospholipids [180]. Interestingly, the increased synthesis of phospholipids in the pah1Δ mutant alleviates growth inhibition caused by the infection of brome mosaic virus, which utilizes host phospholipid resources for its replication [186]. Moreover, the pah1Δ mutation confers increased stability to the yeast and heterologous plasmids [187].
The pah1Δ mutant, which contains the elevated levels of superoxides and lipid hydroperoxides, is hypersensitive to hydrogen peroxide, [180]; it is defective in growth on non-fermentable carbon sources (e.g., glycerol) and has a shorter chronological life span [180], exhibiting apoptotic cell death in the stationary phase [174]. The elevated levels of superoxides and lipid hydroperoxides in the mutant cells may be attributed to the increased level of phospholipids as well as to the reduced activity of Sod2 superoxide dismutase and catalase [180]. The apoptotic cell death may be attributed to the increased fatty acid content [188]. It is unclear why the mutant cells are defective in growth on non-fermentable carbon sources.
Other phenotypes of the pah1Δ mutant include its weakened cell wall [189,190], vacuole fragmentation [191], and defect in autophagy induction after TORC1 inactivation [192]. The loss of cell wall strength might be connected to a defect in the cell wall integrity pathway that is controlled by Pkc1 PKC {3478, 3477}. The enzyme that is stimulated by the lipids DAG and PS [195] would be expected to be less active in the pah1Δ mutant. Vacuole fusion is controlled not only by protein factors but also by lipid factors that include PA, DAG, phosphoinositides, and sterols [196]. The increased level of PA in the pah1Δ mutant has an inhibitory effect on SNARE priming through its interaction with Sec18 [197]. Conversely, the defect of PA production in the dgk1Δ mutant, which lacks Dgk1 DAG kinase catalyzing DAG phosphorylation [165], has a positive effect on SNARE priming and vacuole fusion [198]. The elevated level of PA in pah1Δ cells also causes the derepression of the UASINO-containing V-ATPase genes that affect vacuole acidification [199], and the acidification of the vacuole has a negative effect on membrane fusion [200]. The underpinning of why the pah1Δ mutation has a negative impact on autophagy is unclear, but it might be related to a defect in processes involving the membrane fission/fusion events of vesicles.
Finally, the pah1Δ mutant exhibits both heat sensitivity [56,165,169,187] and cold sensitivity [201]. Since the lack of growth at the elevated temperature (e.g., 37°C) is characteristic of cell wall mutants, the growth defect of the pah1Δ mutant is likely to be caused by its cell wall defect. Although the basis for growth defect at the reduced temperature (e.g., 15°C) is not known, the altered levels and composition of lipids in the pah1Δ mutant might have a detrimental effect on its cold-growth capability.
4.2. Role of Dgk1 DAG kinase in pah1Δ phenotypes
The phenotypes of the pah1Δ mutant, which include the elevated PA content and phospholipid synthesis along with the aberrant nuclear morphology [124,165,174], are suppressed (or at least partially suppressed) by the deletion of the DGK1 gene. The dgk1Δ effects indicate that DAG kinase activity plays a major role in the elevation of the PA level to elicit the alteration of lipid synthesis and cellular processes [165]. Other pah1Δ phenotypes that are suppressed by the dgk1Δ mutation and thus thought to be related to the increased PA level rather than the decreased DAG level include its reduced lipid droplet numbers, hypersensitivity to hydrogen peroxide, growth defect on non-fermentable carbon sources, and shortened chronological life span [165,173,180]. As expected, the dgk1Δ mutation does not suppress the pah1Δ phenotypes that are caused by the lack of DAG production, which include the reduced levels of DAG and TAG [165] and vacuole fragmentation [198].
An important question that deserves attention is the source of DAG utilized by Dgk1 at the nuclear/ER membrane. In pah1Δ mutant cells, DAG can be produced from PA by other PA phosphatase enzymes such as App1 [202], Dpp1 [203], and Lpp1 [204]. However, these enzymes, which are localized to other subcellular compartments, have not been shown to be involved in lipid synthesis or its regulation [205]. During growth resumption from stasis, Dgk1 utilizes the DAG derived from TAG [206] by the Tgl3 {2063} and Tgl4 {2834} TAG lipase enzymes. However, this source of DAG is not likely in the pah1Δ mutant, which is already defective in the synthesis of TAG [56]. Other potential sources of DAG production include the reactions catalyzed by Plc1 PI-4,5-P2-specific phospholipase C [209], Pgc1 PG-specific phospholipase C [210], and Aur1 inositol phosphorylceramide synthase {3654}. Plc1 [212] and Pgc1 [80] are localized to the cytoplasm/nucleus and cytoplasm/mitochondria, respectively, and do not appear to be in a right place to provide DAG for Dgk1 activity at the nuclear/ER. The more likely source of DAG is considered to be the reaction catalyzed by Aur1 inositol phosphorylceramide synthase {3654}, an early Golgi-associated enzyme [80] that catalyzes the conversion of PI and phytoceramide to phosphorylceramide and DAG.
Cells carrying the dgk1Δ mutation alone do not exhibit any distinct phenotypes under optimal growth conditions [165]. However, the mutational effect becomes evident on quiescent cells in the stationary phase that resume growth in the presence of cerulenin, an inhibitor of de novo fatty acid synthesis [206]. Under this condition, the Dgk1-mediated production of PA from DAG, which is generated from TAG hydrolysis [213], is essential for the synthesis of membrane phospholipids (e.g., PC) via the CDP-DAG pathway [206]. Accordingly, the dgk1Δ mutant, which cannot produce PA for phospholipid synthesis, fails to resume growth from the stationary phase in the absence of fatty acid synthesis. Yet, the growth defect of the dgk1Δ mutant is partially ameliorated by supplementation of choline that drives PC synthesis using DAG via the Kennedy pathway [206].
5. Regulation of Pah1 PA phosphatase and Dgk1 DAG kinase by phosphorylation/dephosphorylation
5.1. Phosphorylation and dephosphorylation of Pah1
Pah1, which functions at a location different from that of its synthesis, is regulated for its translocation by phosphorylation and dephosphorylation (Fig. 1). To date, 38 sites of its phosphorylation have been identified [214], and about half of them have been ascribed to specific protein kinases [214]. We do know that the phosphorylations by Pho85-Pho80 [215], Cdc28-cyclin B [169,216], and PKA [217] prevent Pah1 function by causing its retention in the cytoplasm apart from the substrate PA present at the nuclear/ER membrane (Fig. 1). The phosphorylations of Pah1 by Pho85-Pho80 [215] and PKA [217] also reduce its PA phosphatase activity. Additionally, Pah1 is phosphorylated by CKII [218] and PKC [219], but these phosphorylations do not have direct effects on the localization of Pah1 or its catalytic activity. Instead, the phosphorylation of Pah1 by CKII inhibits its phosphorylation by protein kinase A [218], and the phosphorylation by PKC, which is favored without phosphorylation by Pho85-Pho80, promotes the degradation of Pah1 by the 20S proteasome [167,219] (Fig. 1). Pah1 is also phosphorylated by CKI, whose sites of phosphorylation overlap with some of the sites phosphorylated by Pho85-Pho80, PKA, and PKC.
The dephosphorylation of Pah1, which is catalyzed by the nuclear/ER membrane-associated Nem1 (catalytic subunit)-Spo7 (regulatory subunit) phosphatase complex [169,172], facilitates its association with the membrane and the substrate PA [86], and at the same time has the effect of stimulating its catalytic activity [170,215,216,220]. The Nem1-Spo7 protein phosphatase removes all phosphates on Pah1 [169], and its dephosphorylation efficiency on the kinase-specific phosphorylation sites is in the order Pho85-Pho80 > PKA = CKII > Cdc28-cyclin B > PKC [218,220]. The highest affinity of Nem1-spo7 for the sites of Pah1 phosphorylated by Pho85-Pho80 is consistent with the strongest effects of the non-phosphorylatable alanine mutations on bypassing the requirement of the phosphatase complex for its function on the membrane (e.g., increase in nuclear/ER membrane association and TAG synthesis) [170,215,216]. Nem1-Spo7 phosphatase activity has the pH optimum of 5 [220], which is like the intracellular pH of yeast cells in the stationary phase when Pah1 activity and TAG level are maximal and the conversion of PA to the lipid storage lipid is favored over membrane phospholipid synthesis [124,125,220,221].
Cells carrying the nem1Δ or spo7Δ mutation lack a functional phosphatase complex and exhibit the abnormal expansion of the nuclear/ER membrane, a phenotype shown by the lack of Pah1 PA phosphatase. Likewise, the mutants lacking the phosphatase complex exhibit a defect in balancing the coordinated synthesis of TAG and membrane phospholipids [124], and exhibit defects in phospholipid synthesis gene regulation [169], lipid droplet formation [222], vacuole morphology [192], and autophagy [192,222]. Given the requirement of dephosphorylation for the function of Pah1, it is not surprising that the nem1Δ and/or spo7Δ mutant exhibits the same phenotypes as the pah1Δ mutant.
5.2. Phosphorylation of the Nem1-Spo7 protein phosphatase
Like its substrate Pah1, the Nem1-Spo7 phosphatase is a phosphoprotein [223,224] that is subject to phosphorylation by PKA [225] and PKC [195]. Wild type cells expressing the Nem1-Spo7 complex deficient its phosphorylation by PKA show reduced phospholipid synthesis but increased TAG synthesis [225]. This finding suggests that the PKA phosphorylation of Nem1-Spo7 is inhibitory on its function to dephosphorylate and activate Pah1 PA phosphatase. The effect of the PKC-mediated phosphorylation of the phosphatase complex is yet unknown. The catalytic subunit Nem1 is also phosphorylated in wild type cells when supplemented with rapamycin [226], an inhibitor of the TORC1 protein kinase complex [227]. The rapamycin-induced Nem1 phosphorylation correlates with the dephosphorylation of Pah1 and an increase in TAG content [226], indicating that the phosphorylation of Nem1 occurring under nutrient limitation is stimulatory on its catalytic function to activate Pah1 PA phosphatase. The protein kinase involved in this regulation has yet to be identified [226].
5.3. Phosphorylation of Dgk1
Dgk1 treated with alkaline phosphatase exhibits a great reduction in DAG kinase activity, indicating that its phosphorylation is stimulatory on catalytic activity. The N-terminus of Dgk1 contains at least 5 phosphorylation sites [223,224], two of which are target sites for CKII [228]. The CKII-mediated phosphorylation of the dephosphorylated Dgk1 restores most of its enzyme activity that is lost by treatment with alkaline phosphatase [228]. The site-specific mutational analysis of the phosphorylation sites indicates that the CKII-mediated phosphorylation of Dgk1 is required for its function in eliciting the pah1Δ mutant phenotypes such as PA accumulation, nuclear/ER membrane expansion, and reduced lipid droplet formation [228].
6. Summary and perspectives
In this review, we have discussed the PA-mediated regulation of lipid synthesis that occurs at the nuclear/ER membrane (Fig. 1). The importance of Pah1 PA phosphatase in the production of DAG and simultaneous control of the PA level is manifested by a multitude of defects in cells lacking the enzyme activity (Fig. 3). Dgk1 DAG kinase and Pah1 PA phosphatase, which counteract each other in the production of PA and DAG, play a major role in the control of the lipid levels. The PA-mediated regulation of phospholipid synthesis is largely governed by Pah1 (Fig. 1) and Opi1 (Fig. 2), which translocate to the nuclear/ER membrane with opposite effects on their function. Whereas Pah1 is active on the membrane, Opi1 becomes inactive. In addition, Pah1 PA phosphatase activity, which reduces the level of PA by converting it to DAG, has a positive regulatory effect in dissociating Opi1 from the membrane for its repressor function. The membrane localization of Pah1 is controlled by its phosphorylation and dephosphorylation; its phosphorylated form is cytoplasmic, whereas its unphosphorylated form (by dephosphorylation) is associated with the membrane. The dephosphorylation of Pah1 by the Nem1-Spo7 protein phosphatase complex is required for its localization and function at the nuclear/ER membrane. However, it is unknown how the enzyme and substrate interact with each other, and what effects the phosphorylations of the Nem1 and Spo7 subunits have on the formation of the phosphatase complex and its activity on Pah1. The phosphorylation and dephosphorylation of Pah1 also govern its susceptibility to degradation by the 20S proteasome. However, it is unknown whether Pah1 is also subject to the ubiquitin-mediated proteasomal degradation. Like Pah1, Opi1 is subject to multiple phosphorylations, but the effect of phosphorylation on its localization is unclear.
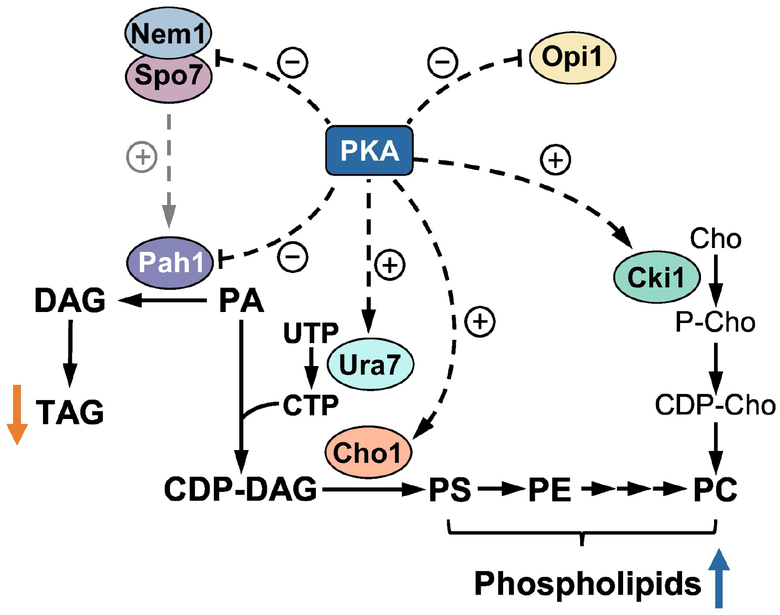
Key enzymes in phospholipid synthesis (Ura7 CTP synthetase, Cho1 PS synthase, and Cki1 choline kinase) are also subject to multiple phosphorylations [32]. One of the protein kinases common to the phosphorylation of the enzymes is PKA [152,229–231] (Fig. 4). In yeast, PKA is a principal protein kinase that transmits signals through the RAS/cAMP pathway; the stimulation of PKA is coupled with enhanced metabolic activity and active cell growth [232,233]. The PKA-mediated phosphorylation of the Nem1-Spo7 complex, Pah1, and Opi1 has an inhibitory effect on their function, whereas the phosphorylation of Ura7, Cho1, and Cki1 stimulate their function (Fig. 4). The net effect of the phosphorylation-mediated regulation is to channel PA for the synthesis of phospholipids rather than TAG.
Fig. 4.
Model for the regulation of lipid synthesis by PKA. The phosphorylations of the Nem1-Spo7 protein phosphatase complex, Pah1, and Opi1 by PKA have negative effects on their functions. The positive effect Nem1-Spo7 has on Pah1 function is attenuated (indicated by the dashed grey arrow) by its phosphorylation. The phosphorylations of Ura7, Cho1, and Cki1 have positive effects on their functions. The net effect of these phosphorylations is an increase in phospholipids (blue arrow) and a decrease in TAG (orange arrow).
Acknowledgments
This work was supported in part by United States Public Health Service Grants GM028140 and GM050679 from the National Institutes of Health.
Abbreviations:
- ER
endoplasmic reticulum
- PC
phosphatidylcholine
- PI
phosphatidylinositol
- PS
phosphatidylserine
- PE
phosphatidylethanolamine
- TAG
triacylglycerol
- PA
phosphatidate
- lysoPA
1-acylglycerol-3-P
- DAG
diacylglycerol
- CDP-DAG
CDP-diacylglycerol
- PG
phosphatidylglycerol
- CL
cardiolipin
- PGP
phosphatidylglycerophosphate
- PKA
protein kinase A
- PKC
protein kinase C
- CK
casein kinase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
This article is part of a Special Issue entitled Endoplasmic Reticulum Platforms for Membrane Lipid Dynamics.
References
- [1].Vance DE. Glycerolipid biosynthesis in eukaryotes In Biochemistry of Lipids, Lipoproteins and Membranes, (Vance DE and Vance J, eds) 5th Ed, Elsevier Science Publishers B.V., Amsterdam: (2004) pp.153–181. [Google Scholar]
- [2].Renne MF, de Kroon AIPM, The role of phospholipid molecular species in determining the physical properties of yeast membranes, FEBS Lett 592 (2018) 1330–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Hakomori S, Igarashi Y, Functional role of glycosphingolipids in cell recognition and signaling, J. Biochem 118 (1995) 1091–1103. [DOI] [PubMed] [Google Scholar]
- [4].van MG, de Kroon AI, Lipid map of the mammalian cell, J. Cell Sci 124 (2011) 5–8. [DOI] [PubMed] [Google Scholar]
- [5].Pomorski T, Hrafnsdottir S, Devaux PF, van MG, Lipid distribution and transport across cellular membranes, Semin. Cell Dev. Biol 12 (2001) 139–148. [DOI] [PubMed] [Google Scholar]
- [6].Rajendran L, Simons K, Lipid rafts and membrane dynamics, J. Cell Sci 118 (2005) 1099–1102. [DOI] [PubMed] [Google Scholar]
- [7].van Meer G, Voelker DR, Feigenson GW, Membrane lipids: where they are and how they behave, Nat. Rev. Mol. Cell Biol 9 (2008) 112–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Suzuki KG, Kasai RS, Hirosawa KM, Nemoto YL, Ishibashi M, Miwa Y, Fujiwara TK, Kusumi A, Transient GPI-anchored protein homodimers are units for raft organization and function, Nat. Chem. Biol 8 (2012) 774–783. [DOI] [PubMed] [Google Scholar]
- [9].Volmer R, Ron D, Lipid-dependent regulation of the unfolded protein response, Curr. Opin. Cell Biol 33 (2015) 67–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Leamy AK, Egnatchik RA, Young JD, Molecular mechanisms and the role of saturated fatty acids in the progression of non-alcoholic fatty liver disease, Prog. Lipid Res 52 (2013) 165–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Fu S, Yang L, Li P, Hofmann O, Dicker L, Hide W, Lin X, Watkins SM, Ivanov AR, Hotamisligil GS, Aberrant lipid metabolism disrupts calcium homeostasis causing liver endoplasmic reticulum stress in obesity, Nature 473 (2011) 528–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Matsuura S, Masuda R, Sakai O, Tashiro Y, Immunoelectron microscopy of the outer membrane of rat hepatocyte nuclear envelopes in relation to the rough endoplasmic reticulum, Cell Struct. Funct 8 (1983) 1–9. [DOI] [PubMed] [Google Scholar]
- [13].Fagone P, Jackowski S, Membrane phospholipid synthesis and endoplasmic reticulum function, J. Lipid Res 50 Suppl (2009) S311–S316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Reid DW, Nicchitta CV, Diversity and selectivity in mRNA translation on the endoplasmic reticulum, Nat. Rev. Mol. Cell Biol 16 (2015) 221–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Rapoport TA, Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes, Nature 450 (2007) 663–669. [DOI] [PubMed] [Google Scholar]
- [16].Braakman I, Hebert DN, Protein folding in the endoplasmic reticulum, Cold Spring Harb. Perspect. Biol 5 (2013) a013201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Rizzuto R, Pinton P, Carrington W, Fay FS, Fogarty KE, Lifshitz LM, Tuft RA, Pozzan T, Close contacts with the endoplasmic reticulum as determinants of mitochondrial Ca2+ responses, Science 280 (1998) 1763–1766. [DOI] [PubMed] [Google Scholar]
- [18].Shibata Y, Shemesh T, Prinz WA, Palazzo AF, Kozlov MM, Rapoport TA, Mechanisms determining the morphology of the peripheral ER, Cell 143 (2010) 774–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].West M, Zurek N, Hoenger A, Voeltz GK, A 3D analysis of yeast ER structure reveals how ER domains are organized by membrane curvature, J. Cell Biol 193 (2011) 333–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Mannella CA, Buttle K, Rath BK, Marko M, Electron microscopic tomography of rat-liver mitochondria and their interaction with the endoplasmic reticulum, Biofactors 8 (1998) 225–228. [DOI] [PubMed] [Google Scholar]
- [21].Meier PJ, Spycher MA, Meyer UA, Isolation and characterization of rough endoplasmic reticulum associated with mitochondria from normal rat liver, Biochim. Biophys. Acta 646 (1981) 283–297. [DOI] [PubMed] [Google Scholar]
- [22].Shore GC,Tata JR, Two fractions of rough endoplasmic reticulum from rat liver. I. Recovery of rapidly sedimenting endoplasmic reticulum in association with mitochondria, J. Cell Biol 72 (1977) 714–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Csordas G, Renken C, Varnai P, Walter L, Weaver D, Buttle KF, Balla T, Mannella CA, Hajnoczky G, Structural and functional features and significance of the physical linkage between ER and mitochondria, J. Cell Biol 174 (2006) 915–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Breuza L, Halbeisen R, Jeno P, Otte S, Barlowe C, Hong W, Hauri HP, Proteomics of endoplasmic reticulum-Golgi intermediate compartment (ERGIC) membranes from brefeldin A-treated HepG2 cells identifies ERGIC-32, a new cycling protein that interacts with human Erv46, J. Biol. Chem 279 (2004) 47242–47253. [DOI] [PubMed] [Google Scholar]
- [25].Hauri HP, Kappeler F, Andersson H, Appenzeller C, ERGIC-53 and traffic in the secretory pathway, J. Cell Sci 113 (Pt 4) (2000) 587–596. [DOI] [PubMed] [Google Scholar]
- [26].Yang Y, Lee M, Fairn GD, Phospholipid subcellular localization and dynamics, J. Biol. Chem 293 (2018) 6230–6240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Block BA, Imagawa T, Campbell KP, Franzini-Armstrong C, Structural evidence for direct interaction between the molecular components of the transverse tubule/sarcoplasmic reticulum junction in skeletal muscle, J. Cell Biol 107 (1988) 2587–2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Takeshima H, Komazaki S, Nishi M, Iino M, Kangawa K, Junctophilins: a novel family of junctional membrane complex proteins, Mol. Cell 6 (2000) 11–22. [DOI] [PubMed] [Google Scholar]
- [29].Quon E, Sere YY, Chauhan N, Johansen J, Sullivan DP, Dittman JS, Rice WJ, Chan RB, Di PG, Beh CT, Menon AK, Endoplasmic reticulum-plasma membrane contact sites integrate sterol and phospholipid regulation, PLoS. Biol 16 (2018) e2003864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Carman GM, Henry SA, Phospholipid biosynthesis in yeast, Annu. Rev. Biochem 58 (1989) 635–669. [DOI] [PubMed] [Google Scholar]
- [31].Carman GM, Henry SA, Phospholipid biosynthesis in the yeast Saccharomyces cerevisiae and interrelationship with other metabolic processes, Prog. Lipid Res 38 (1999) 361–399. [DOI] [PubMed] [Google Scholar]
- [32].Carman GM, Han G-S, Regulation of phospholipid synthesis in the yeast Saccharomyces cerevisiae, Ann. Rev. Biochem 80 (2011) 859–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Henry SA, Kohlwein S, Carman GM, Metabolism and regulation of glycerolipids in the yeast Saccharomyces cerevisiae, Genetics 190 (2012) 317–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Fernandez-Murray JP, McMaster CR, Lipid synthesis and membrane contact sites: a crossroads for cellular physiology, J. Lipid Res 57 (2016) 1789–1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Taylor FR, Parks LW, Triacylglycerol metabolism in Saccharomyces cerevisiae relation to phospholipid synthesis, Biochim. Biophys. Acta 575 (1979) 204–214. [DOI] [PubMed] [Google Scholar]
- [36].Karanasios E, Barbosa AD, Sembongi H, Mari M, Han G-S, Reggiori F, Carman GM, Siniossoglou S, Regulation of lipid droplet and membrane biogenesis by the acidic tail of the phosphatidate phosphatase Pah1p, Mol. Biol. Cell 24 (2013) 2124–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Carman GM, Henry SA, Phosphatidic acid plays a central role in the transcriptional regulation of glycerophospholipid synthesis in Saccharomyces cerevisiae, J. Biol. Chem 282 (2007) 37293–37297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Waggoner DW, Xu J, Singh I, Jasinska R, Zhang QX, Brindley DN, Structural organization of mammalian lipid phosphate phosphatases: implications for signal transduction, Biochim. Biophys. Acta 1439 (1999) 299–316. [DOI] [PubMed] [Google Scholar]
- [39].Sciorra VA, Morris AJ, Roles for lipid phosphate phosphatases in regulation of cellular signaling, Biochim. Biophys. Acta 1582 (2002) 45–51. [DOI] [PubMed] [Google Scholar]
- [40].Testerink C, Munnik T, Phosphatidic acid: a multifunctional stress signaling lipid in plants, Trends Plant Sci 10 (2005) 368–375. [DOI] [PubMed] [Google Scholar]
- [41].Wang X, Devaiah SP, Zhang W, Welti R, Signaling functions of phosphatidic acid, Prog. Lipid Res 45 (2006) 250–278. [DOI] [PubMed] [Google Scholar]
- [42].Brindley DN, Lipid phosphate phosphatases and related proteins: signaling functions in development, cell division, and cancer, J. Cell Biochem 92 (2004) 900–912. [DOI] [PubMed] [Google Scholar]
- [43].Howe AG, McMaster CR, Regulation of phosphatidylcholine homeostasis by Sec14, Can. J. Physiol. Pharmacol 84 (2006) 29–38. [DOI] [PubMed] [Google Scholar]
- [44].Foster DA, Regulation of mTOR by phosphatidic acid?, Cancer Res 67 (2007) 1–4. [DOI] [PubMed] [Google Scholar]
- [45].Carman GM, Han G-S, Regulation of phospholipid synthesis in yeast, J. Lipid Res 50 (2009) S69–S73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Case KC, Salsaa M, Yu W, Greenberg ML, Regulation of inositol biosynthesis: balancing health and pathophysiology, Handb. Exp. Pharmacol (2018) [DOI] [PubMed] [Google Scholar]
- [47].Chang Y-F, Carman GM, CTP synthetase and its role in phospholipid synthesis in the yeast Saccharomyces cerevisiae, Prog. Lipid Res 47 (2008) 333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Zheng Z, Zou J, The initial step of the glycerolipid pathway: identification of glycerol 3-phosphate/dihydroxyacetone phosphate dual substrate acyltransferases in Saccharomyces cerevisiae, J. Biol. Chem 276 (2001) 41710–41716. [DOI] [PubMed] [Google Scholar]
- [49].Matsushita M, Nikawa J, Isolation and characterization of a SCT1 gene which can suppress a choline-transport mutant of Saccharomyces cerevisiae, J. Biochem 117 (1995) 447–451. [DOI] [PubMed] [Google Scholar]
- [50].Athenstaedt K, Daum G, 1-Acyldihydroxyacetone-phosphate reductase (Ayr1p) of the yeast Saccharomyces cerevisiae encoded by the open reading frame YIL124w is a major component of lipid particles, J. Biol. Chem 275 (2000) 235–240. [DOI] [PubMed] [Google Scholar]
- [51].Riekhof WR, Wu J, Jones JL, Voelker DR, Identification and characterization of the major lysophosphatidylethanolamine acyltransferase in Saccharomyces cerevisiae, J. Biol. Chem 282 (2007) 28344–28352. [DOI] [PubMed] [Google Scholar]
- [52].Jain S, Stanford N, Bhagwat N, Seiler B, Costanzo M, Boone C, Oelkers P, Identification of a Novel Lysophospholipid Acyltransferase in Saccharomyces cerevisiae, J. Biol. Chem 282 (2007) 30562–30569. [DOI] [PubMed] [Google Scholar]
- [53].Ayciriex S, Le GM, Camougrand N, Velours G, Schoene M, Wattelet-Boyer V, Dupuy JW, Shevchenko A, Schmitter JM, Lessire R, Bessoule JJ, Testet E, YPR139c/LOA1 encodes a novel lysophosphatidic acid acyltransferase associated with lipid droplets and involved in TAG homeostasis, Mol. Biol. Cell 23 (2011) 233–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Benghezal M, Roubaty C, Veepuri V, Knudsen J, Conzelmann A, SLC1 and SLC4 Encode Partially Redundant Acyl-Coenzyme A 1-Acylglycerol-3-phosphate O-Acyltransferases of Budding Yeast, J. Biol. Chem 282 (2007) 30845–30855. [DOI] [PubMed] [Google Scholar]
- [55].Shen H, Heacock PN, Clancey CJ, Dowhan W, The CDS1 gene encoding CDP-diacylglycerol synthase in Saccharomyces cerevisiae is essential for cell growth, J. Biol. Chem 271 (1996) 789–795. [DOI] [PubMed] [Google Scholar]
- [56].Han G-S, Wu W-I, Carman GM, The Saccharomyces cerevisiae lipin homolog is a Mg2+-dependent phosphatidate phosphatase enzyme, J. Biol. Chem 281 (2006) 9210–9218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Atkinson K, Fogel S, Henry SA, Yeast mutant defective in phosphatidylserine synthesis, J. Biol. Chem 255 (1980) 6653–6661. [PubMed] [Google Scholar]
- [58].Atkinson KD, Jensen B, Kolat AI, Storm EM, Henry SA, Fogel S, Yeast mutants auxotropic for choline or ethanolamine, J. Bacteriol 141 (1980) 558–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Letts VA, Klig LS, Bae-Lee M, Carman GM, Henry SA, Isolation of the yeast structural gene for the membrane-associated enzyme phosphatidylserine synthase, Proc. Natl. Acad. Sci. USA 80 (1983) 7279–7283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Kiyono K, Miura K, Kushima Y, Hikiji T, Fukushima M, Shibuya I, Ohta A, Primary structure and product characterization of the Saccharomyces cerevisiae CHO1 gene that encodes phosphatidylserine synthase, J. Biochem 102 (1987) 1089–1100. [DOI] [PubMed] [Google Scholar]
- [61].Nikawa J, Tsukagoshi Y, Kodaki T, Yamashita S, Nucleotide sequence and characterization of the yeast PSS gene encoding phosphatidylserine synthase, Eur. J. Biochem 167 (1987) 7–12. [DOI] [PubMed] [Google Scholar]
- [62].Clancey CJ, Chang S-C, Dowhan W, Cloning of a gene (PSD1) encoding phosphatidylserine decarboxylase from Saccharomyces cerevisiae by complementation of an Escherichia coli mutant, J. Biol. Chem 268 (1993) 24580–24590. [PubMed] [Google Scholar]
- [63].Trotter PJ, Pedretti J, Voelker DR, Phosphatidylserine decarboxylase from Saccharomyces cerevisiae. Isolation of mutants, cloning of the gene, and creation of a null allele, J. Biol. Chem 268 (1993) 21416–21424. [PubMed] [Google Scholar]
- [64].Gaynor PM, Gill T, Toutenhoofd S, Summers EF, McGraw P, Homann MJ, Henry SA, Carman GM, Regulation of phosphatidylethanolamine methyltransferase and phospholipid methyltransferase by phospholipid precursors in Saccharomyces cerevisiae, Biochim. Biophys. Acta 1090 (1991) 326–332. [DOI] [PubMed] [Google Scholar]
- [65].Kodaki T, Yamashita S, Yeast phosphatidylethanolamine methylation pathway: Cloning and characterization of two distinct methyltransferase genes, J. Biol. Chem 262 (1987) 15428–15435. [PubMed] [Google Scholar]
- [66].McGraw P, Henry SA, Mutations in the Saccharomyces cerevisiae OPI3 gene: Effects on phospholipid methylation, growth, and cross pathway regulation of phospholipid synthesis, Genetics 122 (1989) 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Kornmann B, Currie E, Collins SR, Schuldiner M, Nunnari J, Weissman JS, Walter P, An ER-mitochondria tethering complex revealed by a synthetic biology screen, Science 325 (2009) 477–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Jeong H, Park J, Jun Y, Lee C, Crystal structures of Mmm1 and Mdm12-Mmm1 reveal mechanistic insight into phospholipid trafficking at ER-mitochondria contact sites, Proc. Natl. Acad. Sci. U. S. A 114 (2017) E9502–E9511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Kawano S, Tamura Y, Kojima R, Bala S, Asai E, Michel AH, Kornmann B, Riezman I, Riezman H, Sakae Y, Okamoto Y, Endo T, Structure-function insights into direct lipid transfer between membranes by Mmm1-Mdm12 of ERMES, J. Cell Biol 217 (2018) 959–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Kannan M, Lahiri S, Liu LK, Choudhary V, Prinz WA, Phosphatidylserine synthesis at membrane contact sites promotes its transport out of the ER, J. Lipid Res 58 (2017) 553–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Miyata N, Watanabe Y, Tamura Y, Endo T, Kuge O, Phosphatidylserine transport by Ups2-Mdm35 in respiration-active mitochondria, J. Cell Biol 214 (2016) 77–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Trotter PJ, Voelker DR, Identification of a non-mitochondrial phosphatidylserine decarboxylase activity (PSD2) in the yeast Saccharomyces cerevisiae, J. Biol. Chem 270 (1995) 6062–6070. [DOI] [PubMed] [Google Scholar]
- [73].Summers EF, Letts VA, McGraw P, Henry SA, Saccharomyces cerevisiae cho2 mutants are deficient in phospholipid methylation and cross-pathway regulation of inositol synthesis, Genetics 120 (1988) 909–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Nikawa J, Tsukagoshi Y, Yamashita S, Cloning of a gene encoding choline transport in Saccharomyces cerevisiae, J. Bacteriol 166 (1986) 328–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Hosaka K, Kodaki T, Yamashita S, Cloning and characterization of the yeast CKI gene encoding choline kinase and its expression in Escherichia coli, J. Biol. Chem 264 (1989) 2053–2059. [PubMed] [Google Scholar]
- [76].Kim K, Kim K-H, Storey MK, Voelker DR, Carman GM, Isolation and characterization of the Saccharomyces cerevisiae EKI1 gene encoding ethanolamine kinase, J. Biol. Chem 274 (1999) 14857–14866. [DOI] [PubMed] [Google Scholar]
- [77].Tsukagoshi Y, Nikawa J, Yamashita S, Molecular cloning and characterization of the gene encoding cholinephosphate cytidylyltransferase in Saccharomyces cerevisiae, Eur. J. Biochem 169 (1987) 477–486. [DOI] [PubMed] [Google Scholar]
- [78].Min-Seok R, Kawamata Y, Nakamura H, Ohta A, Takagi M, Isolation and characterization of ECT1 gene encoding CTP:phosphoethanolamine cytidylyltransferase of Saccharomyces cerevisiae, J. Biochem 120 (1996) 1040–1047. [DOI] [PubMed] [Google Scholar]
- [79].Natter K, Leitner P, Faschinger A, Wolinski H, McCraith S, Fields S, Kohlwein SD, The spatial organization of lipid synthesis in the yeast Saccharomyces cerevisiae derived from large scale green fluorescent protein tagging and high resolution microscopy, Mol. Cell Proteomics 4 (2005) 662–672. [DOI] [PubMed] [Google Scholar]
- [80].Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O’Shea EK, Global analysis of protein localization in budding yeast, Nature 425 (2003) 686–691. [DOI] [PubMed] [Google Scholar]
- [81].Haider A, Wei YC, Lim K, Barbosa AD, Liu CH, Weber U, Mlodzik M, Oras K, Collier S, Hussain MM, Dong L, Patel S, varez-Guaita A, Saudek V, Jenkins BJ, Koulman A, Dymond MK, Hardie RC, Siniossoglou S, Savage DB, PCYT1A regulates phosphatidylcholine homeostasis from the inner nuclear membrane in response to membrane stored curvature elastic stress, Dev. Cell 45 (2018) 481–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Hjelmstad RH, Bell RM, Mutants of Saccharomyces cerevisiae defective in sn-1,2-diacylglycerol cholinephosphotransferase: Isolation, characterization, and cloning of the CPT1 gene, J. Biol. Chem 262 (1987) 3909–3917. [PubMed] [Google Scholar]
- [83].Hjelmstad RH, Bell RM, The sn-1,2-diacylglycerol cholinephosphotransferase of Saccharomyces cerevisiae. Nucleotide sequence, transcriptional mapping, and gene product analysis of the CPT1 gene, J. Biol. Chem 265 (1990) 1755–1764. [PubMed] [Google Scholar]
- [84].Hjelmstad RH, Bell RM, The sn-1,2-diacylglycerol ethanolaminephosphotransferase of Saccharomyces cerevisiae. Isolation of mutants and cloning of the EPT1 gene, J. Biol. Chem 263 (1988) 19748–19757. [PubMed] [Google Scholar]
- [85].Hjelmstad RH, Bell RM, sn-1,2-diacylglycerol choline- and ethanolaminephosphotransferases in Saccharomyces cerevisiae. Nucleotide sequence of the EPT1 gene and comparison of the CPT1 and EPT1 gene products, J. Biol. Chem 266 (1991) 5094–5103. [PubMed] [Google Scholar]
- [86].Karanasios E, Han G-S, Xu Z, Carman GM, Siniossoglou S, A phosphorylation-regulated amphipathic helix controls the membrane translocation and function of the yeast phosphatidate phosphatase, Proc. Natl. Acad. Sci. U. S. A 107 (2010) 17539–17544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Patton-Vogt JL, Griac P, Sreenivas A, Bruno V, Dowd S, Swede MJ, Henry SA, Role of the yeast phosphatidylinositol/phosphatidylcholine transfer protein (Sec14p) in phosphatidylcholine turnover and INO1 regulation, J. Biol. Chem 272 (1997) 20873–20883. [DOI] [PubMed] [Google Scholar]
- [88].Xie Z, Fang M, Rivas MP, Faulkner AJ, Sternweis PC, Engebrecht J, Bankaitis VA, Phospholipase D activity is required for suppression of yeast phosphatidylinositol transfer protein defects, Proc. Natl. Acad. Sci. USA 95 (1998) 12346–12351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Rose K, Rudge SA, Frohman MA, Morris AJ, Engebrecht J, Phospholipase D signaling is essential for meiosis, Proc Natl Acad Sci U S A 92 (1995) 12151–12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Waksman M, Eli Y, Liscovitch M, Gerst JE, Identification and characterization of a gene encoding phospholipase D activity in yeast, J Biol Chem 271 (1996) 2361–2364. [DOI] [PubMed] [Google Scholar]
- [91].McMaster CR, Bell RM, Phosphatidylcholine biosynthesis in Saccharomyces cerevisiae. Regulatory insights from studies employing null and chimeric sn-1,2-diacylglycerol choline- and ethanolaminephosphotransferases, J. Biol. Chem 269 (1994) 28010–28016. [PubMed] [Google Scholar]
- [92].Morash SC, McMaster CR, Hjelmstad RH, Bell RM, Studies employing Saccharomyces cerevisiae cpt1 and ept1 null mutants implicate the CPT1 gene in coordinate regulation of phospholipid biosynthesis, J. Biol. Chem 269 (1994) 28769–28776. [PubMed] [Google Scholar]
- [93].Tamaki H, Shimada A, Ito Y, Ohya M, Takase J, Miyashita M, Miyagawa H, Nozaki H, Nakayama R, Kumagai H, LPT1 encodes a membrane-bound O-acyltransferase involved in the acylation of lysophospholipids in the yeast Saccharomyces cerevisiae, J. Biol. Chem 282 (2007) 34288–34298. [DOI] [PubMed] [Google Scholar]
- [94].Riekhof WR, Wu J, Gijon MA, Zarini S, Murphy RC, Voelker DR, Lysophosphatidylcholine Metabolism in Saccharomyces cerevisiae: The role of P-type ATPases in transport and a broad specificity acyltransferase in acylation, J. Biol. Chem 282 (2007) 36853–36861. [DOI] [PubMed] [Google Scholar]
- [95].Tanaka K, Fukuda R, Ono Y, Eguchi H, Nagasawa S, Nakatani Y, Watanabe H, Nakanishi H, Taguchi R, Ohta A, Incorporation and remodeling of extracellular phosphatidylcholine with short acyl residues in Saccharomyces cerevisiae, Biochim. Biophys. Acta 1781 (2008) 391–399. [DOI] [PubMed] [Google Scholar]
- [96].Stålberg K, Neal AC, Ronne H, Stahl U, Identification of a novel GPCAT activity and a new pathway for phosphatidylcholine biosynthesis in S. cerevisiae, J. Lipid Res 49 (2008) 1794–1806. [DOI] [PubMed] [Google Scholar]
- [97].Patton-Vogt J, Transport and metabolism of glycerophosphodiesters produced through phospholipid deacylation, Biochim. Biophys. Acta 1771 (2007) 337–342. [DOI] [PubMed] [Google Scholar]
- [98].Fernandez-Murray JP, McMaster CR, Phosphatidylcholine synthesis and its catabolism by yeast neuropathy target esterase 1, Biochim. Biophys. Acta 1771 (2007) 331–336. [DOI] [PubMed] [Google Scholar]
- [99].Nikawa J, Yamashita S, Molecular cloning of the gene encoding CDP-diacylglycerol-inositol 3-phosphatidyl transferase in Saccharomyces cerevisiae, Eur. J. Biochem 143 (1984) 251–256. [DOI] [PubMed] [Google Scholar]
- [100].Nikawa J, Kodaki T, Yamashita S, Primary structure and disruption of the phosphatidylinositol synthase gene of Saccharomyces cerevisiae, J. Biol. Chem 262 (1987) 4876–4881. [PubMed] [Google Scholar]
- [101].Paltauf F, Kohlwein SD, and Henry SA. Regulation and Compartmentalization of Lipid Synthesis in Yeast In The Molecular and Cellular Biology of the Yeast Saccharomyces: Gene Expression (Jones EW, Pringle JR, and Broach JR, eds), Cold Spring Harbor Laboratory, Cold Spring Harbor, NY: (1992) pp.415–500. [Google Scholar]
- [102].Fruman DA, Meyers RE, Cantley LC, Phosphoinositide kinases, Annu. Rev. Biochem 67 (1998) 481–507. [DOI] [PubMed] [Google Scholar]
- [103].Balla T, Phosphatidylinositol 4-kinases, Biochim. Biophys. Acta 1436 (1998) 69–85. [DOI] [PubMed] [Google Scholar]
- [104].Gehrmann T, Heilmayer LG Jr., Phosphatidylinositol 4-kinases, Eur. J. Biochem 253 (1998) 357–370. [DOI] [PubMed] [Google Scholar]
- [105].Odorizzi G, Babst M, Emr SD, Phosphoinositide signaling and the regulation of membrane trafficking in yeast, Trends Biochem. Sci 25 (2000) 229–235. [DOI] [PubMed] [Google Scholar]
- [106].Leidich SD, Drapp DA, Orlean P, A conditionally lethal yeast mutant blocked at the first step in glycosyl phosphatidylinositol anchor synthesis, J Biol. Chem 269 (1994) 10193–10196. [PubMed] [Google Scholar]
- [107].Leidich SD, Orlean P, Gpi1, a Saccharomyces cerevisiae protein that participates in the first step in glycosylphosphatidylinositol anchor synthesis, J Biol. Chem 271 (1996) 27829–27837. [DOI] [PubMed] [Google Scholar]
- [108].Dickson RC, Sphingolipid functions in Saccharomyces cerevisiae: Comparison to mammals, Annu. Rev. Biochem 67 (1998) 27–48. [DOI] [PubMed] [Google Scholar]
- [109].Dickson RC, Lester RL, Sphingolipid functions in Saccharomyces cerevisiae, Biochim. Biophys. Acta 1583 (2002) 13–25. [DOI] [PubMed] [Google Scholar]
- [110].Strahl T, Thorner J, Synthesis and function of membrane phosphoinositides in budding yeast, Saccharomyces cerevisiae, Biochim. Biophys. Acta 1771 (2007) 353–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Donahue TF, Henry SA, Myo-inositol-1-phosphate synthase: characteristics of the enzyme and identification of its structural gene in yeast, J. Biol. Chem 256 (1981) 7077–7085. [PubMed] [Google Scholar]
- [112].Murray M, Greenberg ML, Expression of yeast INM1 encoding inositol monophosphatase is regulated by inositol, carbon source and growth stage and is decreased by lithium and valproate, Mol. Microbiol 36 (2000) 651–661. [DOI] [PubMed] [Google Scholar]
- [113].Chang YY, Kennedy EP, Biosynthesis of phosphatidyl glycerolphosphate in Escherichia coli, J. Lipid Res 8 (1967) 447–455. [PubMed] [Google Scholar]
- [114].Chang SC, Heacock PN, Clancey CJ, Dowhan W, The PEL1 gene (renamed PGS1) encodes the phosphatidylglycerophosphate synthase of Saccharomyces cerevisiae, J. Biol. Chem 273 (1998) 9829–9836. [DOI] [PubMed] [Google Scholar]
- [115].Chang SC, Heacock PN, Mileykovskaya E, Voelker DR, Dowhan W, Isolation and characterization of the gene (CLS1) encoding cardiolipin synthase in Saccharomyces cerevisiae, J. Biol. Chem 273 (1998) 14933–14941. [DOI] [PubMed] [Google Scholar]
- [116].Jiang F, Rizavi HS, Greenberg ML, Cardiolipin is not essential for the growth of Saccharomyces cerevisiae on fermentable or non-fermentable carbon sources, Mol. Microbiol 26 (1997) 481–491. [DOI] [PubMed] [Google Scholar]
- [117].Tuller G, Hrastnik C, Achleitner G, Schiefthaler U, Klein F, Daum G, YDL142c encodes cardiolipin synthase (Clslp) and is non-essential for aerobic growth of Saccharomyces cerevisiae, FEBS LETTERS 421 (1998) 15–18. [DOI] [PubMed] [Google Scholar]
- [118].Osman C, Haag M, Wieland FT, Brugger B, Langer T, A mitochondrial phosphatase required for cardiolipin biosynthesis: the PGP phosphatase Gep4, EMBO J 29 (2010) 1976–1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Beranek A, Rechberger G, Knauer H, Wolinski H, Kohlwein SD, Leber R, Identification of a cardiolipin-specific phospholipase encoded by the gene CLD1 (YGR110W) in yeast, J. Biol. Chem (2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Gu Z, Valianpour F, Chen S, Vaz FM, Hakkaart GA, Wanders RJ, Greenberg ML, Aberrant cardiolipin metabolism in the yeast taz1 mutant: a model for Barth syndrome, Mol. Microbiol 51 (2004) 149–158. [DOI] [PubMed] [Google Scholar]
- [121].Testet E, Laroche-Traineau J, Noubhani A, Coulon D, Bunoust O, Camougrand N, Manon S, Lessire R, Bessoule JJ, Ypr140wp, ‘the yeast tafazzin’, displays a mitochondrial lysophosphatidylcholine (lyso-PC) acyltransferase activity related to triacylglycerol and mitochondrial lipid synthesis, Biochem. J 387 (2005) 617–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Tamura Y, Harada Y, Nishikawa S, Yamano K, Kamiya M, Shiota T, Kuroda T, Kuge O, Sesaki H, Imai K, Tomii K, Endo T, Tam41 is a CDP-diacylglycerol synthase required for cardiolipin biosynthesis in mitochondria, Cell Metab 17 (2013) 709–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Connerth M, Tatsuta T, Haag M, Klecker T, Westermann B, Langer T, Intramitochondrial transport of phosphatidic acid in yeast by a lipid transfer protein, Science 338 (2012) 815–818. [DOI] [PubMed] [Google Scholar]
- [124].Pascual F, Soto-Cardalda A, Carman GM, PAH1-encoded phosphatidate phosphatase plays a role in the growth phase- and inositol-mediated regulation of lipid synthesis in Saccharomyces cerevisiae, J. Biol. Chem 288 (2013) 35781–35792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Barbosa AD, Sembongi H, Su W-M, Abreu S, Reggiori F, Carman GM, Siniossoglou S, Lipid partitioning at the nuclear envelope controls membrane biogenesis, Mol. Biol. Cell 26 (2015) 3641–3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Oelkers P, Cromley D, Padamsee M, Billheimer JT, Sturley SL, The DGA1 gene determines a second triglyceride synthetic pathway in yeast, J Biol. Chem 277 (2002) 8877–8881. [DOI] [PubMed] [Google Scholar]
- [127].Oelkers P, Tinkelenberg A, Erdeniz N, Cromley D, Billheimer JT, Sturley SL, A lecithin cholesterol acyltransferase-like gene mediates diacylglycerol esterification in yeast, J Biol. Chem 275 (2000) 15609–15612. [DOI] [PubMed] [Google Scholar]
- [128].Yang H, Bard M, Bruner DA, Gleeson A, Deckelbaum RJ, Aljinovic G, Pohl TM, Rothstein R, Sturley SL, Sterol esterification in yeast: a two-gene process, Science 272 (1996) 1353–1356. [DOI] [PubMed] [Google Scholar]
- [129].Sorger D, Daum G, Triacylglycerol biosynthesis in yeast, Appl. Microbiol. Biotechnol 61 (2003) 289–299. [DOI] [PubMed] [Google Scholar]
- [130].Kohlwein SD, Triacylglycerol homeostasis: insights from yeast, J. Biol. Chem 285 (2010) 15663–15667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Barbosa AD, Siniossoglou S, Function of lipid droplet-organelle interactions in lipid homeostasis, Biochim. Biophys. Acta Mol. Cell Res 1864 (2017) 1459–1468. [DOI] [PubMed] [Google Scholar]
- [132].Henne WM, Reese ML, Goodman JM, The assembly of lipid droplets and their roles in challenged cells, EMBO J 37 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Romanauska A, Kohler A, The inner nuclear membrane Is a metabolically active territory that generates nuclear lipid droplets, Cell (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Loewy BS, Henry SA, The INO2 and INO4 loci of Saccharomyces cerevisiae are pleiotropic regulatory genes, Mol. Cell. Biol 4 (1984) 2479–2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Klig LS, Homann MJ, Carman GM, Henry SA, Coordinate regulation of phospholipid biosynthesis in Saccharomyces cerevisiae: pleiotropically constitutive opi1 mutant, J. Bacteriol 162 (1985) 1135–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Kodaki T, Hosaka K, Nikawa J, Yamashita S, Identification of the upstream activation sequences responsible for the expression and regulation of the PEM1 and PEM2 genes encoding the enzymes of the phosphatidylethanolamine methylation pathway in Saccharomyces cerevisiae, J. Biochem 109 (1991) 276–287. [PubMed] [Google Scholar]
- [137].Bachhawat N, Ouyang Q, Henry SA, Functional characterization of an inositol-sensitive upstream activation sequence in yeast. A cis-regulatory element responsible for inositol-choline mediated regulation of phospholipid synthesis, J. Biol. Chem 270 (1995) 25087–25095. [DOI] [PubMed] [Google Scholar]
- [138].Henry SA, Patton-Vogt JL, Genetic regulation of phospholipid metabolism: yeast as a model eukaryote, Prog. Nucleic Acid Res 61 (1998) 133–179. [DOI] [PubMed] [Google Scholar]
- [139].Chen M, Hancock LC, Lopes JM, Transcriptional regulation of yeast phospholipid biosynthetic genes, Biochim. Biophys. Acta 1771 (2007) 310–321. [DOI] [PubMed] [Google Scholar]
- [140].Nikawa J, Tsukagoshi Y, Yamashita S, Isolation and characterization of two distinct myo-inositol transporter genes of Saccharomyces cerevisiae, J. Biol. Chem 266 (1991) 11184–11191. [PubMed] [Google Scholar]
- [141].White MJ, Hirsch JP, Henry SA, The OPI1 gene of Saccharomyces cerevisiae, a negative regulator of phospholipid biosynthesis, encodes a protein containing polyglutamine tracts and a leucine zipper, J. Biol. Chem 266 (1991) 863–872. [PubMed] [Google Scholar]
- [142].Nikoloff DM, McGraw P, Henry SA, The INO2 gene of Saccharomyces cerevisiae encodes a helix-loop- helix protein that is required for activation of phospholipid synthesis, Nucleic Acids Res 20 (1992) 3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Ambroziak J, Henry SA, INO2 and INO4 gene products, positive regulators of phospholipid biosynthesis in Saccharomyces cerevisiae, form a complex that binds to the INO1 promoter, J. Biol. Chem 269 (1994) 15344–15349. [PubMed] [Google Scholar]
- [144].Klig LS, Hoshizaki DK, Henry SA, Isolation of the yeast INO4 gene, a positive regulator of phospholipid synthesis, Curr. Genet 13 (1988) 7–14. [DOI] [PubMed] [Google Scholar]
- [145].Loewen CJR, Roy A, Levine TP, A conserved ER targeting motif in three families of lipid binding proteins and in Opi1p binds VAP, EMBO J 22 (2003) 2025–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Loewen CJR, Gaspar ML, Jesch SA, Delon C, Ktistakis NT, Henry SA, Levine TP, Phospholipid metabolism regulated by a transcription factor sensing phosphatidic acid, Science 304 (2004) 1644–1647. [DOI] [PubMed] [Google Scholar]
- [147].Loewen CJR, Levine TP, A highly conserved binding site in vesicle-associated membrane protein-associated protein (VAP) for the FFAT motif of lipid-binding proteins, J Biol. Chem 280 (2005) 14097–14104. [DOI] [PubMed] [Google Scholar]
- [148].Hofbauer HF, Gecht M, Fischer SC, Seybert A, Frangakis AS, Stelzer EHK, Covino R, Hummer G, Ernst R, The molecular recognition of phosphatidic acid by an amphipathic helix in Opi1, J. Cell Biol 217 (2018) 3109–3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Hofbauer HF, Schopf FH, Schleifer H, Knittelfelder OL, Pieber B, Rechberger GN, Wolinski H, Gaspar ML, Kappe CO, Stadlmann J, Mechtler K, Zenz A, Lohner K, Tehlivets O, Henry SA, Kohlwein SD, Regulation of gene expression through a transcriptional repressor that senses acyl-chain length in membrane phospholipids, Dev. Cell 29 (2014) 729–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Gaspar ML, Chang YF, Jesch SA, Aregullin M, Henry SA, Interaction between repressor Opi1p and ER membrane protein Scs2p facilitates transit of phosphatidic acid from the ER to mitochondria and is essential for INO1 gene expression in the presence of choline, J. Biol. Chem 292 (2017) 18713–18728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Chang Y-F, Carman GM, Casein kinase II phosphorylation of the yeast phospholipid synthesis transcription factor Opi1p, J. Biol. Chem 281 (2006) 4754–4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Sreenivas A, Carman GM, Phosphorylation of the yeast phospholipid synthesis regulatory protein Opi1p by protein kinase A, J. Biol. Chem 278 (2003) 20673–20680. [DOI] [PubMed] [Google Scholar]
- [153].Sreenivas A, Villa-Garcia MJ, Henry SA, Carman GM, Phosphorylation of the yeast phospholipid synthesis regulatory protein Opi1p by protein kinase C, J. Biol. Chem 276 (2001) 29915–29923. [DOI] [PubMed] [Google Scholar]
- [154].Carman GM, Han G-S, Regulation of phospholipid synthesis in Saccharomyces cerevisiae by zinc depletion, Biochim. Biophys. Acta 1771 (2007) 322–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Poole MA, Homann MJ, Bae-Lee M, Carman GM, Regulation of phosphatidylserine synthase from Saccharomyces cerevisiae by phospholipid precursors, J. Bacteriol 168 (1986) 668–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Bailis AM, Poole MA, Carman GM, Henry SA, The membrane-associated enzyme phosphatidylserine synthase of yeast is regulated at the level of mRNA abundance, Mol. Cell. Biol 7 (1987) 167–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Bailis AM, Lopes JM, Kohlwein SD, Henry SA, Cis and trans regulatory elements required for regulation of the CHO1 gene of Saccharomyces cerevisiae, Nucleic Acids Res 20 (1992) 1411–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Homann MJ, Bailis AM, Henry SA, Carman GM, Coordinate regulation of phospholipid biosynthesis by serine in Saccharomyces cerevisiae, J. Bacteriol 169 (1987) 3276–3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Iwanyshyn WM, Han G-S, Carman GM, Regulation of phospholipid synthesis in Saccharomyces cerevisiae by zinc, J. Biol. Chem 279 (2004) 21976–21983. [DOI] [PubMed] [Google Scholar]
- [160].Homann MJ, Poole MA, Gaynor PM, Ho C-T, Carman GM, Effect of growth phase on phospholipid biosynthesis in Saccharomyces cerevisiae, J. Bacteriol 169 (1987) 533–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Lamping E, Luckl J, Paltauf F, Henry SA, Kohlwein SD, Isolation and characterization of a mutant of Saccharomyces cerevisiae with pleiotropic deficiencies in transcriptional activation and repression, Genetics 137 (1995) 55–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Henry SA, Gaspar ML, Jesch SA, The response to inositol: regulation of glycerolipid metabolism and stress response signaling in yeast, Chem. Phys. Lipids 180 (2014) 23–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Han S-H, Han G-S, Iwanyshyn WM, Carman GM, Regulation of the PIS1-encoded phosphatidylinositol synthase in Saccharomyces cerevisiae by zinc, J Biol. Chem 280 (2005) 29017–29024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Soto-Cardalda A, Fakas S, Pascual F, Choi HS, Carman GM, Phosphatidate phosphatase plays role in zinc-mediated regulation of phospholipid synthesis in yeast, J. Biol. Chem 287 (2011) 968–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Han G-S, O’Hara L, Carman GM, Siniossoglou S, An unconventional diacylglycerol kinase that regulates phospholipid synthesis and nuclear membrane growth, J. Biol. Chem 283 (2008) 20433–20442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Pascual F, Hsieh L-S, Soto-Cardalda A, Carman GM, Yeast Pah1p phosphatidate phosphatase is regulated by proteasome-mediated degradation, J. Biol. Chem 289 (2014) 9811–9822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Hsieh L-S, Su W-M, Han G-S, Carman GM, Phosphorylation regulates the ubiquitin-independent degradation of yeast Pah1 phosphatidate phosphatase by the 20S proteasome, J. Biol. Chem 290 (2015) 11467–11478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Xu Z, Su W-M, Carman GM, Fluorescence spectroscopy measures yeast PAH1-encoded phosphatidate phosphatase interaction with liposome membranes, J. Lipid Res 53 (2011) 522–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Santos-Rosa H, Leung J, Grimsey N, Peak-Chew S, Siniossoglou S, The yeast lipin Smp2 couples phospholipid biosynthesis to nuclear membrane growth, EMBO J 24 (2005) 1931–1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].O’Hara L, Han G-S, Peak-Chew S, Grimsey N, Carman GM, Siniossoglou S, Control of phospholipid synthesis by phosphorylation of the yeast lipin Pah1p/Smp2p Mg2+-dependent phosphatidate phosphatase, J. Biol. Chem 281 (2006) 34537–34548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Han G-S, Siniossoglou S, Carman GM, The cellular functions of the yeast lipin homolog Pah1p are dependent on its phosphatidate phosphatase activity, J. Biol. Chem 282 (2007) 37026–37035. [DOI] [PubMed] [Google Scholar]
- [172].Siniossoglou S, Santos-Rosa H, Rappsilber J, Mann M, Hurt E, A novel complex of membrane proteins required for formation of a spherical nucleus, EMBO J 17 (1998) 6449–6464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Adeyo O, Horn PJ, Lee S, Binns DD, Chandrahas A, Chapman KD, Goodman JM, The yeast lipin orthologue Pah1p is important for biogenesis of lipid droplets, J. Cell Biol 192 (2011) 1043–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Fakas S, Qiu Y, Dixon JL, Han G-S, Ruggles KV, Garbarino J, Sturley SL, Carman GM, Phosphatidate phosphatase activity plays a key role in protection against fatty acid-induced toxicity in yeast, J. Biol. Chem 286 (2011) 29074–29085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Garbarino J, Padamsee M, Wilcox L, Oelkers PM, D. D’A, Ruggles K, Ramsey N, Jabado O, Turkish A, Sturley SL, Sterol and diacylglycerol acyltransferase deficiency triggers fatty acid mediated cell death, J. Biol. Chem 284 (2009) 30994–31005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Petschnigg J, Wolinski H, Kolb D, Zellnig G, Kurat CF, Natter K, Kohlwein SD, Good fat - essential cellular requirements for triacylglycerol synthesis to maintain membrane homeostasis in yeast, J. Biol. Chem 284 (2009) 30981–30993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Connerth M, Czabany T, Wagner A, Zellnig G, Leitner E, Steyrer E, Daum G, Oleate inhibits steryl ester synthesis and causes liposensitivity in yeast, J. Biol. Chem 285 (2010) 26832–26841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Hassaninasab A, Han G-S, Carman GM, Tips on the analysis of phosphatidic acid by the fluorometric coupled enzyme assay, Anal. Biochem 526 (2017) 69–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Han G-S, Carman GM, Yeast PAH1-encoded phosphatidate phosphatase controls the expression of CHO1-encoded phosphatidylserine synthase for membrane phospholipid synthesis, J. Biol. Chem 292 (2017) 13230–13242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Park Y, Han GS, Mileykovskaya E, Garrett TA, Carman GM, Altered lipid synthesis by lack of yeast Pah1 phosphatidate phosphatase reduces chronological life span, J. Biol. Chem 290 (2015) 25382–25394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Bae-Lee M, Carman GM, Regulation of yeast phosphatidylserine synthase and phosphatidylinositol synthase activities by phospholipids in Triton X-100/phospholipid mixed micelles, J. Biol. Chem 265 (1990) 7221–7226. [PubMed] [Google Scholar]
- [182].Schuller HJ, Schorr R, Hoffmann B, Schweizer E, Regulatory gene INO4 of yeast phospholipid biosynthesis is positively autoregulated and functions as a transactivator of fatty acid synthase genes FAS1 and FAS2 from Saccharomyces cerevisiae, Nucleic Acids Res 20 (1992) 5955–5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Schuller HJ, Hahn A, Troster F, Schutz A, Schweizer E, Coordinate genetic control of yeast fatty acid synthase genes FAS1 and FAS2 by an upstream activation site common to genes involved in membrane lipid biosynthesis, EMBO J 11 (1992) 107–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Hasslacher M, Ivessa AS, Paltauf F, Kohlwein SD, Acetyl-CoA carboxylase from yeast is an essential enzyme and is regulated by factors that control phospholipid metabolism, J. Biol. Chem 268 (1993) 10946–10952. [PubMed] [Google Scholar]
- [185].Chirala SS, Zhong Q, Huang W, Al-Feel W, Analysis of FAS3/ACC regulatory region of Saccharomyces cerevisiae: identification of a functional UASINO and sequences responsible for fatty acid mediated repression, Nucleic Acids Res 22 (1994) 412–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].Zhang Z, He G, Han G-S, Zhang J, Catanzaro N, Diaz A, Wu Z, Carman GM, Xie L, Wang X, Host Pah1p phosphatidate phosphatase limits viral replication by regulating phospholipid synthesis, PLoS. Pathog 14 (2018) e1006988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Irie K, Takase M, Araki H, Oshima Y, A gene, SMP2, involved in plasmid maintenance and respiration in Saccharomyces cerevisiae encodes a highly charged protein, Mol. Gen. Genet 236 (1993) 283–288. [DOI] [PubMed] [Google Scholar]
- [188].Rockenfeller P, Ring J, Muschett V, Beranek A, Buettner S, Carmona-Gutierrez D, Eisenberg T, Khoury C, Rechberger G, Kohlwein SD, Kroemer G, Madeo F, Fatty acids trigger mitochondrion-dependent necrosis, Cell Cycle 9 (2010) 2836–2842. [DOI] [PubMed] [Google Scholar]
- [189].Lussier M, White AM, Sheraton J, di PT, Treadwell J, Southard SB, Horenstein CI, Chen-Weiner J, Ram AF, Kapteyn JC, Roemer TW, Vo DH, Bondoc DC, Hall J, Zhong WW, Sdicu AM, Davies J, Klis FM, Robbins PW, Bussey H, Large scale identification of genes involved in cell surface biosynthesis and architecture in Saccharomyces cerevisiae, Genetics 147 (1997) 435–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [190].Ruiz C, Cid VJ, Lussier M, Molina M, Nombela C, A large-scale sonication assay for cell wall mutant analysis in yeast, Yeast 15 (1999) 1001–1008. [DOI] [PubMed] [Google Scholar]
- [191].Sasser T, Qiu QS, Karunakaran S, Padolina M, Reyes A, Flood B, Smith S, Gonzales C, Fratti RA, The yeast lipin 1 orthologue Pah1p regulates vacuole homeostasis and membrane fusion, J. Biol. Chem 287 (2011) 2221–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [192].Rahman MA, Mostofa MG, Ushimaru T, The Nem1/Spo7-Pah1/lipin axis is required for autophagy induction after TORC1 inactivation, FEBS J 285 (2018) 1840–1860. [DOI] [PubMed] [Google Scholar]
- [193].Levin DE, Cell wall integrity signaling in Saccharomyces cerevisiae, Microbiol. Mol. Biol. Rev 69 (2005) 262–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [194].Levin DE, Regulation of cell wall biogenesis in Saccharomyces cerevisiae: the cell wall integrity signaling pathway, Genetics 189 (2011) 1145–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [195].Dey P, Su WM, Han GS, Carman GM, Phosphorylation of lipid metabolic enzymes by yeast Pkc1 protein kinase C requires phosphatidylserine and diacylglycerol, J. Lipid Res 58 (2017) 742–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [196].Starr ML, Fratti RA, The participation of regulatory lipids in vacuole homotypic fusion, Trends Biochem. Sci (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- [197].Starr ML, Sparks RP, Arango AS, Hurst LR, Zhao Z, Lihan M, Jenkins JL, Tajkhorshid E, Fratti RA, Phosphatidic acid induces conformational changes in Sec18 protomers that prevent SNARE priming, J. Biol. Chem (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- [198].Miner GE, Starr ML, Hurst LR, Fratti RA, Deleting the DAG kinase Dgk1 augments yeast vacuole fusion through increased Ypt7 activity and altered membrane fluidity, Traffic 18 (2017) 315–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [199].Sherr GL, LaMassa N, Li E, Phillips G, Shen CH, Pah1p negatively regulates the expression of V-ATPase genes as well as vacuolar acidification, Biochem. Biophys. Res. Commun 491 (2017) 693–700. [DOI] [PubMed] [Google Scholar]
- [200].Desfougeres Y, Vavassori S, Rompf M, Gerasimaite R, Mayer A, Organelle acidification negatively regulates vacuole membrane fusion in vivo, Sci. Rep 6 (2016) 29045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].Corcoles-Saez I, Hernandez ML, Martinez-Rivas JM, Prieto JA, Randez-Gil F, Characterization of the S. cerevisiae inp51 mutant links phosphatidylinositol 4,5-bisphosphate levels with lipid content, membrane fluidity and cold growth, Biochim. Biophys. Acta 1861 (2016) 213–226. [DOI] [PubMed] [Google Scholar]
- [202].Chae M, Han G-S, Carman GM, The Saccharomyces cerevisiae actin patch protein App1p is a phosphatidate phosphatase enzyme, J. Biol. Chem 287 (2012) 40186–40196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [203].Toke DA, Bennett WL, Dillon DA, Wu W-I, Chen X, Ostrander DB, Oshiro J, Cremesti A, Voelker DR, Fischl AS, Carman GM, Isolation and characterization of the Saccharomyces cerevisiae DPP1 gene encoding for diacylglycerol pyrophosphate phosphatase, J. Biol. Chem 273 (1998) 3278–3284. [DOI] [PubMed] [Google Scholar]
- [204].Toke DA, Bennett WL, Oshiro J, Wu W-I, Voelker DR, Carman GM, Isolation and characterization of the Saccharomyces cerevisiae LPP1 gene encoding a Mg2+-independent phosphatidate phosphatase, J. Biol. Chem 273 (1999) 14331–14338. [DOI] [PubMed] [Google Scholar]
- [205].Carman GM, Discoveries of the phosphatidate phosphatase genes in yeast published in the Journal of Biological Chemistry, J. Biol. Chem 294 (2018) 1681–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [206].Fakas S, Konstantinou C, Carman GM, DGK1-encoded diacylglycerol kinase activity is required for phospholipid synthesis during growth resumption from stationary phase in Saccharomyces cerevisiae, J. Biol. Chem 286 (2011) 1464–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [207].Athenstaedt K, Daum G, YMR313c/TGL3 Encodes a Novel Triacylglycerol Lipase Located in Lipid Particles of Saccharomyces cerevisiae, J. Biol. Chem 278 (2003) 23317–23323. [DOI] [PubMed] [Google Scholar]
- [208].Athenstaedt K, Daum G, Tgl4p and Tgl5p, two triacylglycerol lipases of the yeast Saccharomyces cerevisiae are localized to lipid particles, J. Biol. Chem 280 (2005) 37301–37309. [DOI] [PubMed] [Google Scholar]
- [209].Flick JS, Thorner J, Genetic and biochemical characterization of a phosphatidylinositol-specific phospholipase C in Saccharomyces cerevisiae, Mol. Cell Biol 13 (1993) 5861–5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [210].Simockova M, Holic R, Tahotna D, Patton-Vogt J, Griac P, Yeast Pgc1p (YPL206c) controls the amount of phosphatidylglycerol via a phospholipase C-type degradation mechanism, J. Biol. Chem 283 (2008) 17107–17115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [211].Nagiec MM, Nagiec EE, Baltisberger JA, Wells GB, Lester RL, Dickson RC, Sphingolipid synthesis as a target for antifungal drugs. Complementation of the inositol phosphorylceramide synthase defect in a mutant strain of Saccharomyces cerevisiae by the AUR1 gene, J. Biol. Chem 272 (1997) 9809–9817. [DOI] [PubMed] [Google Scholar]
- [212].Nadler-Holly M, Breker M, Gruber R, Azia A, Gymrek M, Eisenstein M, Willison KR, Schuldiner M, Horovitz A, Interactions of subunit CCT3 in the yeast chaperonin CCT/TRiC with Q/N-rich proteins revealed by high-throughput microscopy analysis, Proc. Natl. Acad. Sci. U. S. A 109 (2012) 18833–18838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [213].Kurat CF, Natter K, Petschnigg J, Wolinski H, Scheuringer K, Scholz H, Zimmermann R, Leber R, Zechner R, Kohlwein SD, Obese yeast: triglyceride lipolysis is functionally conserved from mammals to yeast, J. Biol. Chem 281 (2006) 491–500. [DOI] [PubMed] [Google Scholar]
- [214].Carman GM, Han GS, Fat-regulating phosphatidic acid phosphatase: a review of its roles and regulation in lipid homeostasis, J. Lipid Res 60 (2019) 2–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [215].Choi H-S, Su W-M, Han G-S, Plote D, Xu Z, Carman GM, Pho85p-Pho80p phosphorylation of yeast Pah1p phosphatidate phosphatase regulates its activity, location, abundance, and function in lipid metabolism, J. Biol. Chem 287 (2012) 11290–11301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [216].Choi H-S, Su W-M, Morgan JM, Han G-S, Xu Z, Karanasios E, Siniossoglou S, Carman GM, Phosphorylation of phosphatidate phosphatase regulates its membrane association and physiological functions in Saccharomyces cerevisiae: identification of Ser602, Thr723, and Ser744 as the sites phosphorylated by CDC28 (CDK1)-encoded cyclin-dependent kinase, J. Biol. Chem 286 (2011) 1486–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [217].Su W-M, Han G-S, Casciano J, Carman GM, Protein kinase A-mediated phosphorylation of Pah1p phosphatidate phosphatase functions in conjunction with the Pho85p-Pho80p and Cdc28p-cyclin B kinases to regulate lipid synthesis in yeast, J. Biol. Chem 287 (2012) 33364–33376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [218].Hsieh L-S, Su W-M, Han G-S, Carman GM, Phosphorylation of yeast Pah1 phosphatidate phosphatase by casein kinase II regulates its function in lipid metabolism, J. Biol. Chem 291 (2016) 9974–9990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [219].Su W-M, Han G-S, Carman GM, Cross-talk phosphorylations by protein kinase C and Pho85p-Pho80p protein kinase regulate Pah1p phosphatidate phosphatase abundance in Saccharomyces cerevisiae, J. Biol. Chem 289 (2014) 18818–18830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [220].Su W-M, Han G-S, Carman GM, Yeast Nem1-Spo7 protein phosphatase activity on Pah1 phosphatidate phosphatase is specific for the Pho85-Pho80 protein kinase phosphorylation sites, J. Biol. Chem 289 (2014) 34699–34708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [221].Orij R, Urbanus ML, Vizeacoumar FJ, Giaever G, Boone C, Nislow C, Brul S, Smits GJ, Genome-wide analysis of intracellular pH reveals quantitative control of cell division rate by pHc in Saccharomyces cerevisiae, Genome Biol 13 (2012) R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [222].Xu X, Okamoto K, The Nem1-Spo7 protein phosphatase complex is required for efficient mitophagy in yeast, Biochem. Biophys. Res. Commun 496 (2018) 51–57. [DOI] [PubMed] [Google Scholar]
- [223].Holt LJ, Tuch BB, Villen J, Johnson AD, Gygi SP, Morgan DO, Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution, Science 325 (2009) 1682–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [224].Swaney DL, Beltrao P, Starita L, Guo A, Rush J, Fields S, Krogan NJ, Villen J, Global analysis of phosphorylation and ubiquitylation cross-talk in protein degradation, Nat. Methods 10 (2013) 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [225].Su W-M, Han GS, Dey P, Carman GM, Protein kinase A phosphorylates the Nem1-Spo7 protein phosphatase complex that regulates the phosphorylation state of the phosphatidate phosphatase Pah1 in yeast, J. Biol. Chem 293 (2018) 15801–15814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [226].Dubots E, Cottier S, Peli-Gulli MP, Jaquenoud M, Bontron S, Schneiter R, De Virgilio C, TORC1 regulates Pah1 phosphatidate phosphatase activity via the Nem1/Spo7 protein phosphatase complex, PLoS. One 9 (2014) e104194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [227].Soulard A, Cohen A, Hall MN, TOR signaling in invertebrates, Curr. Opin. Cell Biol 21 (2009) 825–836. [DOI] [PubMed] [Google Scholar]
- [228].Qiu Y, Hassaninasab A, Han G-S, Carman GM, Phosphorylation of Dgk1 diacylglycerol kinase by casein kinase II regulates phosphatidic acid production in Saccharomyces cerevisiae, J. Biol. Chem 291 (2016) 26455–26467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [229].Yang W-L, Carman GM, Phosphorylation and regulation of CTP synthetase from Saccharomyces cerevisiae by protein kinase A, J. Biol. Chem 271 (1996) 28777–28783. [DOI] [PubMed] [Google Scholar]
- [230].Choi H-S, Han G-S, Carman GM, Phosphorylation of yeast phosphatidylserine synthase by protein kinase A: identification of Ser46 and Ser47 as major sites of phosphorylation, J. Biol. Chem 285 (2010) 11526–11536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [231].Kim K-H, Carman GM, Phosphorylation and regulation of choline kinase from Saccharomyces cerevisiae by protein kinase A, J. Biol. Chem 274 (1999) 9531–9538. [DOI] [PubMed] [Google Scholar]
- [232].Broach JR, Deschenes RJ, The function of RAS genes in Saccharomyces cerevisiae, Adv. Cancer Res 54 (1990) 79–139. [DOI] [PubMed] [Google Scholar]
- [233].Thevelein JM, Signal Transduction in Yeast, Yeast 10 (1994) 1753–1790. [DOI] [PubMed] [Google Scholar]
- [234].Rajakumari S, Grillitsch K, Daum G, Synthesis and turnover of non-polar lipids in yeast, Prog. Lipid Res 47 (2008) 157–171. [DOI] [PubMed] [Google Scholar]