Abstract
Some viruses package dsDNA together with large amounts of positively charged proteins, thought to help condense the genome inside the capsid with no evidence. Further, this role is not clear because these viruses have typically lower packing fractions than viruses encapsidating naked dsDNA. In addition, it has recently been shown that the major adenovirus condensing protein (polypeptide VII) is dispensable for genome encapsidation. Here, we study the morphology and mechanics of adenovirus particles with (Ad5-wt) and without (Ad5-VII-) protein VII. Ad5-VII- particles are stiffer than Ad5-wt, but DNA-counterions revert this difference, indicating that VII screens repulsive DNA-DNA interactions. Consequently, its absence results in increased internal pressure. The core is slightly more ordered in the absence of VII and diffuses faster out of Ad5-VII– than Ad5-wt fractured particles. In Ad5-wt unpacked cores, dsDNA associates in bundles interspersed with VII-DNA clusters. These results indicate that protein VII condenses the adenovirus genome by combining direct clustering and promotion of bridging by other core proteins. This condensation modulates the virion internal pressure and DNA release from disrupted particles, which could be crucial to keep the genome protected inside the semi-disrupted capsid while traveling to the nuclear pore.
INTRODUCTION
Regulation of dsDNA condensation is crucial for the management of genetic information. Major players in this regulation are specialized proteins which bend, wrap DNA, or form bridges between dsDNA strands (1). In eukaryotes, wrapping by H2A, H2B, H3 and H4 histone octamers provides the first organization level of chromatin, with ∼10 nm diameter nucleosomes forming a beads-on-string structure. Another histone, H1, is a bridger that clasps linker DNA at the nucleosome exit, stabilizing the structure (2). In bacteria, nucleoid-associated proteins have mainly bending and bridging roles (3). The highest known condensation of nucleic acids is the packing of viral genomes (4). In tailed bacteriophages and herpesviruses, naked dsDNA is stored inside the capsid at very high volume fractions approximately in the form of a spool, with polyamines and cations helping to relieve the electrostatic repulsion between dsDNA strands (4,5). In polyomaviruses, dsDNA is packed in complex with cell histones, forming a chromatin-like structure (6). Adenoviruses and some of their structural relatives (asfarviruses, chloroviruses, mimiviruses, marseilliviruses) encode and package positively charged proteins with similarities to cell histones (7–10). It has traditionally been thought that the main role of these proteins is to screen DNA-DNA electrostatic repulsion so that the viral genomes can be condensed to fit the limited space available inside the capsid. However, since these viruses have lower packing fractions than tailed phages (∼0.3 versus ∼0.5), the actual need for genome condensation agents is not that clear (11). Furthermore, knowledge about the organization and properties of nucleoprotein complexes inside virus particles is scarce.
Human adenovirus type 5 (Ad5) is the best characterized system encapsidating virus encoded proteins The Ad5 icosahedral, pseudoT = 25 capsid is composed of 240 hexon trimers, 12 penton-base pentamers, 12 penton base-associated fiber trimers and four minor coat proteins (IIIa, VI, VIII and IX) (12). This capsid encloses the non-icosahedral core, formed by a linear, 35 kbp dsDNA molecule bound to positively charged viral proteins that constitute ∼50% of the core molecular weight: 150 copies of protein V (41 kDa), 500–800 copies of protein VII (22 kDa), and 100–300 copies of protein μ (9 kDa) (13–15). Previous studies have shown that cores released from Ad5 particles disrupted with pyridine contain polypeptides V, VII, and μ, and have a thick (15–30 nm) fibrous appearance when observed at the electron microscope after negative staining or metal shadowing (16). After high ionic strength treatment of these cores, essentially only protein VII remains attached to the DNA, forming tangled structures with ∼200 beads of 9.5 nm diameter located at highly variable distances (∼10–130 nm) (16). These observations suggested a chromatin-like organization for the adenovirus genome, with protein VII acting as a wrapper, while protein μ could have a bridging function to form the thicker fibers (17,18). Currently, the only model for the core architecture in situ (in the crowded conditions within the intact virion) is based on cryo-electron tomography combined with molecular dynamics simulations, and proposes an organization of the DNA-protein complex as a fluid of soft repulsive particles (17).
The Ad5 particle becomes fully infectious only after maturation, which involves cleavage of several capsid and core proteins by the viral protease (19). Cleavage of proteins VII and μ results in decreased core condensation and increased internal pressure, priming the virion for sequential uncoating and facilitating genome release (20–23). However, it has recently been shown that the hypothesized condensing role of protein VII, the most abundant core protein in Ad5 (60%∼70% of the core protein mass), is not required for genome encapsidation. An Ad5 construct where the VII gene was excised from the genome using the Cre-lox recombination system has an assembly efficiency similar to the wild type virus (Ad5-wt), although the particles produced (Ad5-VII-) are not infectious due to a defect in entry (24). The actual cause for this entry defect is not clear. It has been hypothesized that, since the absence of VII represents a large loss of mass inside the virion, the entry defect may result from lack of internal pressurization, thereby hindering the start of the uncoating cascade. An alternative hypothesis is that the absence of VII would allow the genome to interact with positively charged proteins on the inner capsid surface, obstructing genome release (24). Therefore, more work is required to ascertain the actual role of core protein VII in adenovirus assembly and disassembly.
Theoretical models simulating the interactions of dsDNA and proteins within the capsid confinement predict that the presence of condensing proteins decreases internal capsid pressure, despite the decrease in free volume caused by their presence. These models also envisage that the natural tendency for the capsid contents to organize in layers near the confining capsid wall will increase when the strength of protein-DNA interactions is decreased (11). Experimentally, atomic force microscopy (AFM) of single virus particles in liquid (25) has proved the existence of internal pressure in tailed phages (26,27), where cryo-electron microscopy (cryo-EM) shows the naked genome ordered in concentric shells (5). Tailed phages release this pressure to translocate their genome into the host (28). No concentric shells are observed in Ad5 cryo-EM maps, and a change in internal pressure related to particle maturation has also been detected by AFM (18). Here we use AFM and cryo-EM to compare the mechanics and core organization of Ad5-wt and Ad5-VII– particles (Figure 1A), to understand how protein VII influences the architecture and physical properties of adenovirus particles.
Figure 1.
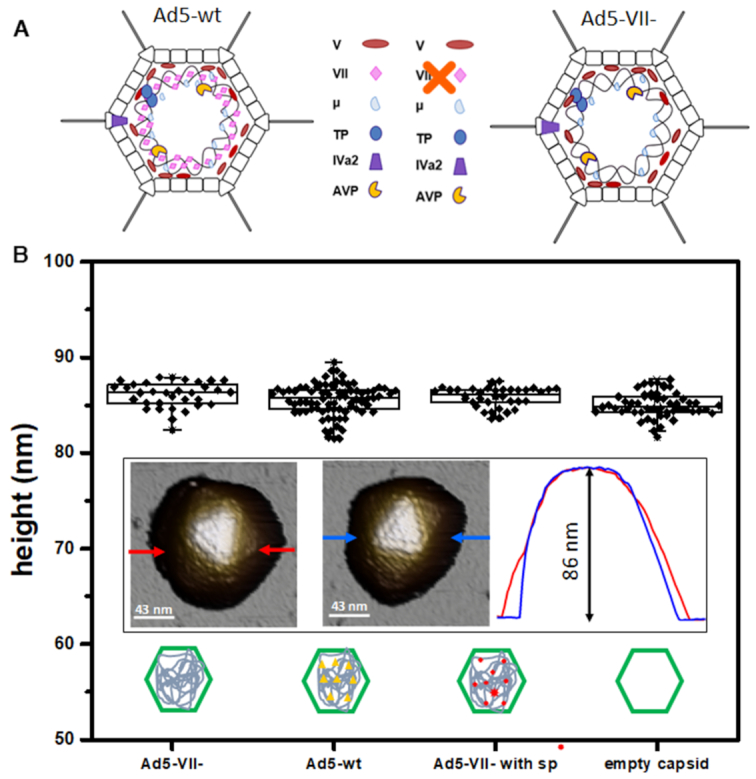
Ad5-wt and Ad5-VII– viral particles. (A) Schematic representation showing the core composition of the two specimens studied (modified from (15)). (B) Height distribution of all the viral particles imaged with AFM. The inset shows AFM topography images of Ad5-wt (left), Ad5-VII– (center) particles, and the topographical profiles (right) corresponding to the line between the red (Ad5-wt) and blue (Ad5-VII-) arrows.
MATERIALS AND METHODS
Virus specimens
For Ad5-wt particles we used the E1-E3 deleted, GFP expressing Ad5/attP vector which is structurally wildtype (29). For empty capsids, we used light density particles produced by the Ad5 delayed packaging mutant FC31 (FC31-L3), which lack genome and core proteins but have a complete icosahedral shell (30). Both were propagated at 37°C in HEK293 cells and harvested at 36 and 56 h post-infection, respectively. The parent virus Ad5-VII-loxP contains loxP sites flanking the protein VII open reading frame, and was generated and propagated as described (24). Ad5-VII– particles were produced using the Cre66 cell line, a Cre recombinase expressing cell line derived from HEK-293 cells (a gift from Dr Stefan Kochaneck, University Ulm, Germany; unpublished), maintained in Dulbecco's modified Eagle's medium supplemented with 10% HyClone Fetalclone III serum, penicillin, streptomycin, and 0.25 mg/ml of Geneticin (Life Technologies). Cre66 cells (6 × 108 total cells) were infected at 200 CsCl-purified Ad5-VII-loxP virus particles/cell for 1 h at 37°C. Infected cells were harvested 3 days after infection when full cytopathic effect was evident. The cells were pelleted by centrifugation, medium removed, and cell pellets flash frozen in liquid nitrogen for storage. In all cases, particles were purified by equilibrium centrifugation in two consecutive CsCl gradients, desalted on a Bio-Rad 10 DC column, and stored in HBS buffer (20 mM HEPES, pH 7.8, 150 mM NaCl) supplemented with 10% glycerol for long term storage at –80°C.
Atomic force microscopy
Virus samples to be used for AFM assays were dialyzed against HBS at 4°C to remove glycerol, frozen in single-use, 5 μl aliquots and stored at -20°C. Stock particle concentrations were 2.3 × 1011 virus particles (vp)/ml for Ad5-wt, 2.7 × 1011 vp/ml for empty particles, and 3.1 × 1011 vp/ml for Ad5-VII-. For AFM measurements, single-use aliquots were thawed, diluted in 20 μl of HBS with 5 mM NiCl2 (Ni-HBS), and adsorbed onto freshly cleaved mica for 20 min at 4°C. After five washes with Ni-HBS, 200 μl of buffer were deposited on the mica surface to avoid desiccation. The AFM tip was prewetted with 30 μl of Ni-HBS so the total volume was about 260 μl. For experiments in the presence of spermidine, 2.3 μl of a 100 mM solution of spermidine (Sigma Aldrich S.L) in water were added to the sample drop, to obtain a final concentration of 1 mM spermidine. The AFM (Nanotec Electrónica S.L, Madrid, Spain) was operated in jumping mode plus (31) in liquid, with RC800PSA cantilevers (Olympus, Tokyo, Japan) with nominal spring constant of 0.05 N/m. The cantilever spring constants were calibrated with the Sader method (32). AFM exploration at low force (∼60 pN) was carried out to ensure a homogeneously dispersed population of particles with a 3-fold icosahedral symmetry orientation on the mica surface before starting measurements (Figure 1B, inset). This orientation maximizes the stable attachment of particles to the mica substrate for the mechanical characterization, because the icosahedral particles are resting on a triangular facet (21). For single indentation assays, viral particles were deformed with the AFM tip until breakage by means of a force versus z-piezo displacement experiment that is transformed into a force-indentation curve (FIC) (33). From these data the spring constant was obtained (Figure 2, inset left). In multiple indentation assays the particles were probed with consecutive indentations that reach lower forces that those of single indentation assays, to avoid virus collapse at once. While the first indentation just removes a few capsomers to crack open the capsid, subsequent indentations probe and modify the core in a controlled fashion as explained elsewhere (18). A typical multiple indentation assay is described in Supplementary Figure S1.
Figure 2.
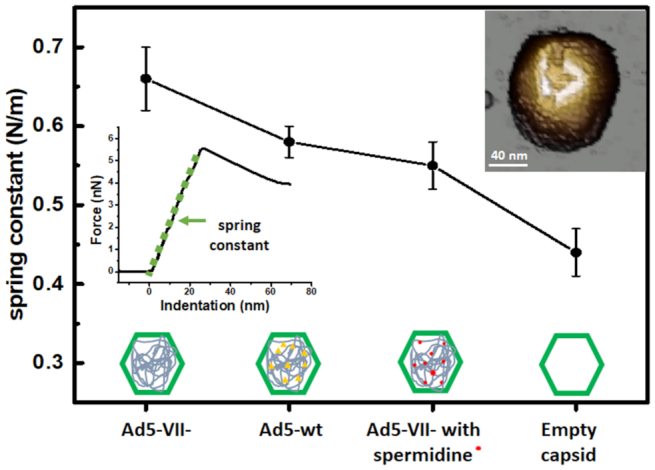
Mechanical properties of intact capsids. Stiffness (spring constants) measured for each specimen used in this work. Top right inset: Topography image of an adenovirus particle after performing a single indentation. Spring constant values are obtained from the force-indentation curve (FIC) (center left inset).
To analyze the structure of the core contents, we performed mechanical fatigue assays (21) consisting in the continuous imaging of a single virus particle at low force (∼70 pN). Imaging in jumping mode plus (31) implies that the virus is probed once at every pixel, inducing low energy impacts between the tip apex and the virus particle (34). This fatigue induces a gradual disassembly of the capsid, including capsomer disruption and core exposure (18). Obtaining consecutive images of the same particle allows us to monitor these morphological changes until the virion collapses, expelling a mixture of fibered and blob structures. We devised a procedure to estimate the size of the fibers and blobs laying on the substrate after particle disruption, based on statistical measures of height in specific regions of the AFM image (Supplementary Figures S4 and S5).
Cryo-electron microscopy and image processing
Purified virus particles were dialyzed for 1 h at 4°C against phosphate buffered saline (PBS, 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4 at pH 7.4) to remove the storage glycerol. Stock particle concentrations were 8.6 × 1011 vp/ml for Ad5-wt and 1.5 × 1012 vp/ml for Ad5-VII-. The Ad5-VII- sample was concentrated 11 times by spinning at 4°C in a 100 000 MWCO Amicon Ultra centrifugal filter (Millipore) before application to glow discharged Quantifoil R2/4 300 mesh Cu/Rh grids and vitrification in liquid ethane using a Leica CPC plunger. For the Ad5-wt control, particle concentration was increased by consecutively incubating the grid on 10 drops of the sample before the final blotting and plunging in liquid ethane (35). Cryo-EM images (4395 for wt, 1640 for VII-) were recorded in a 200 kV Talos Arctica microscope (FEI) equipped with a Falcon II detector, with a total dose of 50 e–/Å2 distributed over 32 frames, at a nominal pixel size of 1.4 Å and a defocus range between –0.5 and –3 μm. All image processing and 3D reconstruction tasks were performed within the Scipion framework (36). Frames were aligned using Motioncor2 and weighted according to the electron dose received before averaging (37). The contrast transfer function (CTF) was estimated using CTFFIND4 (38). Particles (26 661 for wt, 27 909 for VII-) were semi-automatically picked from micrographs corrected for the phase oscillations of the CTF (phase-flipped), extracted into 752 × 752 pixel boxes, normalized and resized to a sampling rate of 2.8 Å/px (376 px box size), using Xmipp (39). All 2D and 3D classifications and refinements were performed using RELION (40). 2D classification was used to discard low quality particles, and run for 25 iterations, with 50 classes, angular sampling 5 and regularization parameter T = 2. Classification in 3D was run for 40 iterations, with three classes, starting with an angular sampling of 3.7° and sequentially decreasing to 0.5, and regularization parameter T = 4. Icosahedral symmetry was imposed throughout the refinement process. The initial reference for 3D classification was a non-human adenovirus cryo-EM map (unpublished), low-pass filtered to 60 Å resolution. The class yielding the best resolution (23 060 particles for wt, 21 762 particles for VII-) was individually refined using the original 752 px boxed particles and the map obtained during the 3D classification as a reference, yielding a final map at 5.5 Å resolution for Ad5-wt and 5.0 Å for Ad5-VII-. Resolution was estimated according to the gold-standard FSC = 0.143 criterion as implemented in RELION auto-refine and postprocess routines (41). The actual sampling for both maps was estimated by comparison with a human adenovirus near atomic resolution model (PDB ID: 6B1T) (42) in UCSF Chimera (43), yielding a value of 1.37 Å/px. Xmipp was used to calculate histograms and radial average profiles, after both 3D maps were low pass filtered to 5.5 Å resolution and normalized to gray values with average 0 and standard deviation 1 within a region comprising the hexon shell (radii 322–427 Å). Radial average profiles were further rescaled to a common value range in the [−1, 1] interval.
RESULTS AND DISCUSSION
Absence of protein VII stiffens the Ad5 intact particle
To investigate the role of protein VII in the physical properties of Ad5, we first analyzed the mechanics of Ad5-wt and Ad5-VII– intact particles by single nanoindentation assays (Materials and Methods). AFM topography images that resolve individual protein capsomers revealed no substantial differences between Ad5-wt and Ad5-VII– particles (Figure 1B, inset). Height measurements of all particles included in this study (Ad5-wt, Ad5-VII– and empty shells) provided values ∼86 nm (Figure 1B) consistent with the expected capsid diameter and indicating that none of them suffered measurable deformations induced by the interaction with the substrate (21,44,45).
The spring constants of Ad5-wt and empty particles, as measured by single indentation assays (Figure 2), were kwt = 0.58 ± 0.02 N/m (mean ± SEM; N = 53) and kE = 0.44 ± 0.03 N/m (mean ± SEM; N = 54) in agreement with previously published values showing that the presence of the core stiffens Ad5 particles (18). For Ad5-VII-, the elastic constant was higher, kVII– = 0.66 ± 0.04 N/m (mean ± SEM; N = 33) (Figure 2). Therefore, Ad5-VII– particles are stiffer than Ad5-wt.
Two possible reasons for stiffening in Ad5 have been described. First, upon maturation cleavage of proteins VII and μ, the screening of DNA-DNA electrostatic repulsion by the core proteins is decreased, resulting in an increase of internal pressure and a stiffer particle (18). Second, a point mutation in capsid protein VI that partially hinders its maturation cleavage also produced an increase in particle stiffness (46). Since it has been observed that the Ad5-VII– particles also present deficient proteolytic cleavage of protein VI (24), we next investigated the mechanical properties of the Ad5-VII– core, to clarify the cause of the stiffening observed.
Absence of protein VII softens the Ad5 core
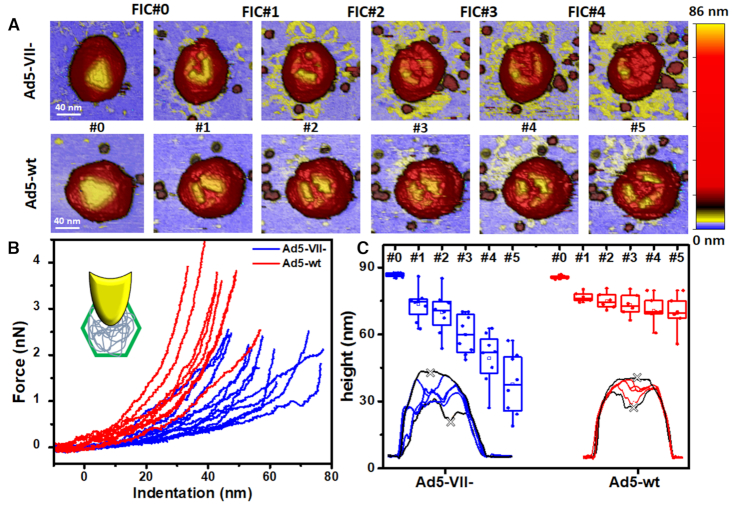
After the first deformation induced by the AFM tip has fractured the capsid shell (Figure 2, top right inset), successive indentations probe directly the viral core through the opened crack (18). Figure 3A presents the typical evolution of Ad5-wt and Ad5-VII– particles in a multiple indentation experiment. The first indentation (FIC#0) on the intact shell (Figure 3A#0) evidences a linear behavior according to the thin shell theory (33). This initial indentation opens a crack at the icosahedral facet with a similar size to the apex of the AFM tip (∼20 nm) (Figure 3A#1), allowing direct access of the tip to the core. As previously observed for Ad5 mature and immature particles (18), subsequent FICs present a non-linear Hertzian behaviour (Supplementary Figure S1) in contrast with the linear deformation found in intact particles (Figure 2, left chart inset). During the first indentation after capsid cracking (Figure 3A, #1), Ad5-wt cores are not deformed beyond 40 nm at 3 nN, while Ad5-VII– cores undergo larger indentations, up to ∼60 nm, at forces as low as 2 nN (Figure 3B). This difference indicates that it is easier for the AFM tip to penetrate/deform Ad5-VII– than Ad5-wt cores. That is, in the absence of protein VII the Ad5 core is softer and less condensed.
Figure 3.
Mechanical properties of the core. (A) Images showing the evolution Ad5-wt and Ad5-VII– particles during multiple indentation assays. Images are colored by height, as indicated by the color bar at the right hand side. FICs can be found in Supplementary Figure S1. (B) FIC curves corresponding to the first indentation performed on the core of 8 Ad5-wt (red) and 12 Ad5-VII– (blue) particles. (C) Evolution of the core (as indicated by the minimum height in the crater, grey crosses) for eight Ad5-VII– and six Ad5-wt particles. Insets: Examples of topographical profiles obtained through the crater opened by the indentation.
Absence of protein VII increases the Ad5 internal pressure
It is generally accepted that viral genomes modulate the mechanics of virus particles by increasing their stiffness (47–50). If the genome is assumed to be a solid material filling the cavity of the capsid, the deformation of the capsid-genome ensemble can be simplified by considering genome and capsid as individual springs associated in parallel. The AFM tip roughly compress the shell (kE) and the core by the same amount d. Thus the total force on the AFM tip becomes –(kEd + kcored) = –(kE + kcore) d, yielding and equivalent spring constant kE + kcore as in the case of springs in parallel. In this approximation, the spring constants of the Ad5 core in the presence or absence of protein VII would be 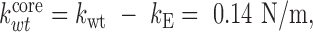 and
and 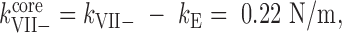 predicting that the core in Ad5-wt would be softer than in Ad5-VII– when confined within the icosahedral shell. In contrast, one could expect that in the absence of VII, the core would be softer because it has less molecular mass. In fact, we found this tendency when probing directly the mechanics of the opened cores (Figure 3B). This behavior is reminiscent of our previous observation on the mechanics of Ad5 mature and immature particles, where we found that upon maturation the core becomes softer, but the capsid becomes stiffer due to an increase in internal pressure (18).
predicting that the core in Ad5-wt would be softer than in Ad5-VII– when confined within the icosahedral shell. In contrast, one could expect that in the absence of VII, the core would be softer because it has less molecular mass. In fact, we found this tendency when probing directly the mechanics of the opened cores (Figure 3B). This behavior is reminiscent of our previous observation on the mechanics of Ad5 mature and immature particles, where we found that upon maturation the core becomes softer, but the capsid becomes stiffer due to an increase in internal pressure (18).
To investigate whether particle stiffening in Ad5-VII– was also related to changes in genome condensation and internal pressurization, we modified the factors governing the energetics of confined DNA (DNA bending and DNA-DNA electrostatic repulsion) by incubation with the condensing agent spermidine (18,27,51). Spermidine is a trivalent polyamine that screens the phosphate negative charges, induces dsDNA condensation in solution and acts inside virus capsids changing the DNA conformation inside phages (52,53). We have previously shown that spermidine decreases the Ad5-wt elastic constant to 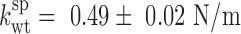 (mean ± SEM, N = 36), but does not change the elasticity of empty particles, indicating that this polyamine can diffuse through the capsid shell to modify the electrostatic interaction between DNA strands (18). We now find that upon incubation with spermidine, the Ad5-VII– particle spring constant decreased to
(mean ± SEM, N = 36), but does not change the elasticity of empty particles, indicating that this polyamine can diffuse through the capsid shell to modify the electrostatic interaction between DNA strands (18). We now find that upon incubation with spermidine, the Ad5-VII– particle spring constant decreased to 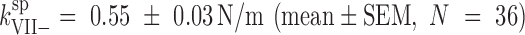 , which is similar to that of Ad5-wt (Figure 2). This result indicates that genome condensation induced by spermidine softens the Ad5-VII– particle, and therefore informs about the role of protein VII in screening DNA negative charges.
, which is similar to that of Ad5-wt (Figure 2). This result indicates that genome condensation induced by spermidine softens the Ad5-VII– particle, and therefore informs about the role of protein VII in screening DNA negative charges.
Together, our data on core and particle stiffness show that the absence of protein VII loosens the core compaction, decreasing the rigidity of the virus core (Figure 3B) and increasing the pressure within intact particles (Figure 2). We can estimate the magnitude of the change in internal pressure using the continuous elasticity prediction for the spring constant of a pressurized thin spherical shell indented by a point force (54):
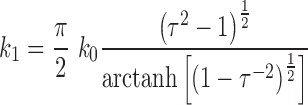 |
(1) |
Here, 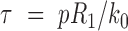 is a dimensionless parameter comparing the relative strength of pressure
is a dimensionless parameter comparing the relative strength of pressure 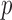 against the elastic constant of the unpressurized shell
against the elastic constant of the unpressurized shell 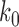 , and
, and 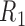 is the middle radius of the adenovirus capsid shell (
is the middle radius of the adenovirus capsid shell ( 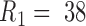 nm). Taking
nm). Taking  , and
, and 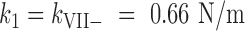 , and solving Equation (1) for
, and solving Equation (1) for  , we obtain an estimate of 3.4 ± 0.7 MPa for the increase of pressure due to the lack of protein VII. This variation of pressure is similar to that generated during maturation (3 ± 1 MPa) (18), consistent with both effects resulting from changes in genome compaction by the core proteins. An increase in internal pressure is also consistent with the behavior predicted by theoretical models for weaker protein-DNA interaction regimes (11).
, we obtain an estimate of 3.4 ± 0.7 MPa for the increase of pressure due to the lack of protein VII. This variation of pressure is similar to that generated during maturation (3 ± 1 MPa) (18), consistent with both effects resulting from changes in genome compaction by the core proteins. An increase in internal pressure is also consistent with the behavior predicted by theoretical models for weaker protein-DNA interaction regimes (11).
Absence of protein VII facilitates genome diffusion out of the Ad5 capsid
In multiple indentation experiments, elongated structures with a height compatible with dsDNA strands (55) (Figure 3A, yellow) appeared on the substrate surrounding Ad5-VII– particles after the first fracture of the shell (Figure 3A#1, top), but not in Ad5-wt (Figure 3A#1, bottom). Material consistent with unpacked DNA seemed to be more abundant in Ad5-VII– particles that in Ad5-wt for the equivalent indentation (Figure 3A). In a previous work we showed that combining AFM with fluorescence microscopy in a similar unpacking experiment resulted in light emission in the presence of the dsDNA accesibility reporter YOYO1 (20). This qualitative observation suggests that it is easier to exit the disrupted shell for the VII-free genome than for the VII-bound one. An alternative possibility would be that the VII-free genome has a larger tendency to adsorb to the mica substrate than the VII-bound DNA. Measurements of the topographical profile across the crater produced by the successive indentations support the first possibility. Because topographical profiles include both the crater and its rims, we used the lowest height inside the crack as an indicator of the remaining core contents (Figure 3C, insets). Plotting the evolution of this parameter along six consecutive indentations for eight Ad5-VII– and six Ad5-wt particles (Figure 3C) showed that, indeed, the core components are leaving faster the Ad5-VII– than the Ad5-wt cracked particles.
To further explore this aspect, we also compared how soon unpacked DNA was observed when particle disruption was induced by mechanical fatigue assays (M&M). Mechanical fatigue caused by repeated AFM imaging of the same particle at very low forces (∼100 pN) promotes gradual disruption of the capsid, while allowing real time monitoring of the disassembly pathway from the initially intact to the final collapsed state (Figure 4A and videos SM1 and SM2) (21). Complete particle disruption by mechanical fatigue requires a different number of images for each individual particle. Thus, we quantify the time required to first observe dsDNA expelled from the capsid during a fatigue experiment as a percentage of the total particle life time. That is, 0% experiment time refers to the initial image when the particle is intact, and 100% to the final image when the particle is collapsed. These experiments indicated that dsDNA exits Ad5-VII– particles earlier (30% experiment time) than Ad5-wt (70% experiment time) (Figure 4B).
Figure 4.
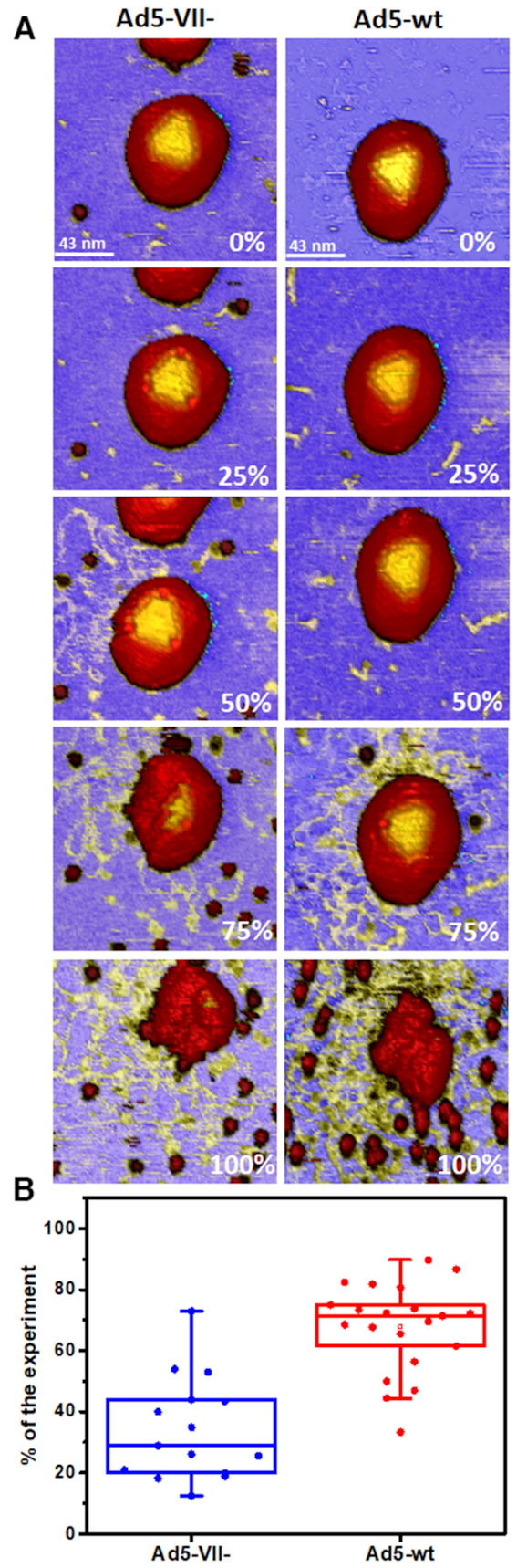
Genome exposure in mechanical fatigue experiments. (A) Topographical images of one Ad5-VII– (left) and one Ad5-wt (right) particle at different times in the experiment (indicated as percentage of the total experiment time). (B) Boxplot showing the percentage of experiment time when the genome was observed on the mica surface.
Because of the experimental design, at the end of mechanical fatigue or multiple indentation assays most of the analyzed particles have released their genome. However, in single indentation assays, only one image is taken after breakage of the capsid, and released DNA is not always observed, indicating that the genome can remain inside the cracked capsid shell. Unpacked DNA was observed in only 3% of Ad5-wt particles after a single indentation, in contrast to 42% of the Ad5-VII– particles (Supplementary Table S1). All these results are compatible with a less condensed genome in Ad5-VII– that leaves the capsid faster than in Ad5-wt.
Protein VII changes the organization of the Ad5 genome in virio and ex virio
In virio
To investigate how the absence of protein VII reflects on the Ad5 core organization inside the intact particle, we compared cryo-EM icosahedrally averaged maps for the Ad5-wt and Ad5-VII– particles (Figure 5). Although the Ad5 core is not icosahedrally ordered, interesting differences could be observed. In the Ad5-VII– particle, the core density was weaker and more heterogeneous than in Ad5-wt, as can be appreciated from visual inspection of the map cross-sections (Figure 5A) and comparison of gray level statistics. When gray levels in the region corresponding to the hexon shell were normalized to the same value range in both maps, in the Ad5-VII– core the average gray level was lower, and the standard deviation larger (Supplementary Figure S2 and Table S2). This observation indicates that, when protein VII (which accounts for 26–30% of the core mass) is not present, the particle contents are more loosely distributed within the capsid cavity. This looser conformation is consistent with the changes in core condensation detected with AFM experiments.
Figure 5.
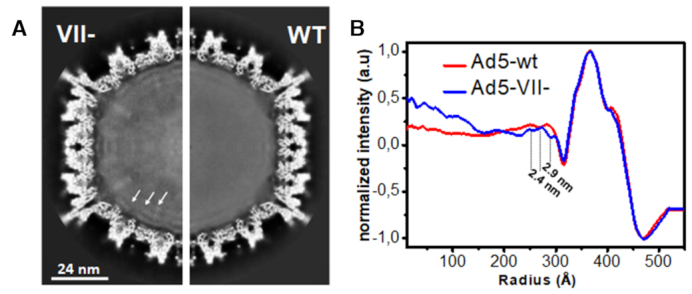
Organization of the core inside the virus particles. (A) Central section of 3D cryo-EM maps of Ad5-VII– and Ad5-wt. Higher density is shown in white. White arrows highlight the three concentric shells in Ad5-VII. (B) Radial average profiles of the Ad5-VII– and Ad5-wt maps.
In the Ad5-VII– map, three weak concentric shells appear near the inner wall of the capsid that are not present in Ad5-wt (Figure 5A). The distance between shells (2.4 and 2.9 nm, Figure 5B) is similar to the distances observed between the concentric layers of naked dsDNA in tailed bacteriophages (56,57). The presence of these weak layers points to an organization of the Ad5-VII– genome somewhat similar to that found in viruses not packaging proteins. This organization agrees with theoretical models predicting that weakening the interactions between condensing proteins and DNA would result in an increased tendency to form layers beneath the confining capsid surface (11).
Ex virio
More information on the conformation of the Ad5 core nucleoprotein complex can be derived from the observation of unpacked contents at the end of in situ AFM mechanical fatigue assays (videos SM1 and SM2, Supplementary Figure S3). Fibered shapes (yellow) and homogeneously dispersed formations (blobs, red) surround the major debris located at the center of the images (Figure 6A and Supplementary Figure S3). The characterization of structures with a size similar to the radius of the AFM tip apex is strongly influenced by dilation artefacts in the lateral dimension (58). However, vertical measurements are not affected by these dilation effects. Therefore, we used the height as a size indicator of these objects to find that the vertical thickness of the fibered shapes is compatible with the dsDNA diameter as measured by AFM (55). A close-up view of these areas reveals that some structures can be resolved as individual filaments (Figure 6B). However, topographical profiles show that in Ad5-wt the filament diameter is larger than in Ad5-VII– (Figure 6C, inset). A statistical analysis (Supplementary Figure S5, Materials and Methods)) confirmed this aspect, with average filament heights of ∼2 nm for Ad5-wt and 1.5 nm for Ad5-VII– (Figure 6C). Also, filament heights presented a broader dispersion (∼1 to ∼3 nm) in Ad5-wt. We interpret the larger thickness in Ad5-wt filaments as being caused by the association of different regions in the dsDNA genome. The thickness dispersion indicates that this association would include at most three dsDNA chains. These results suggest that the presence of protein VII promotes condensation of dsDNA by formation of bundles, perhaps via bridging.
Figure 6.
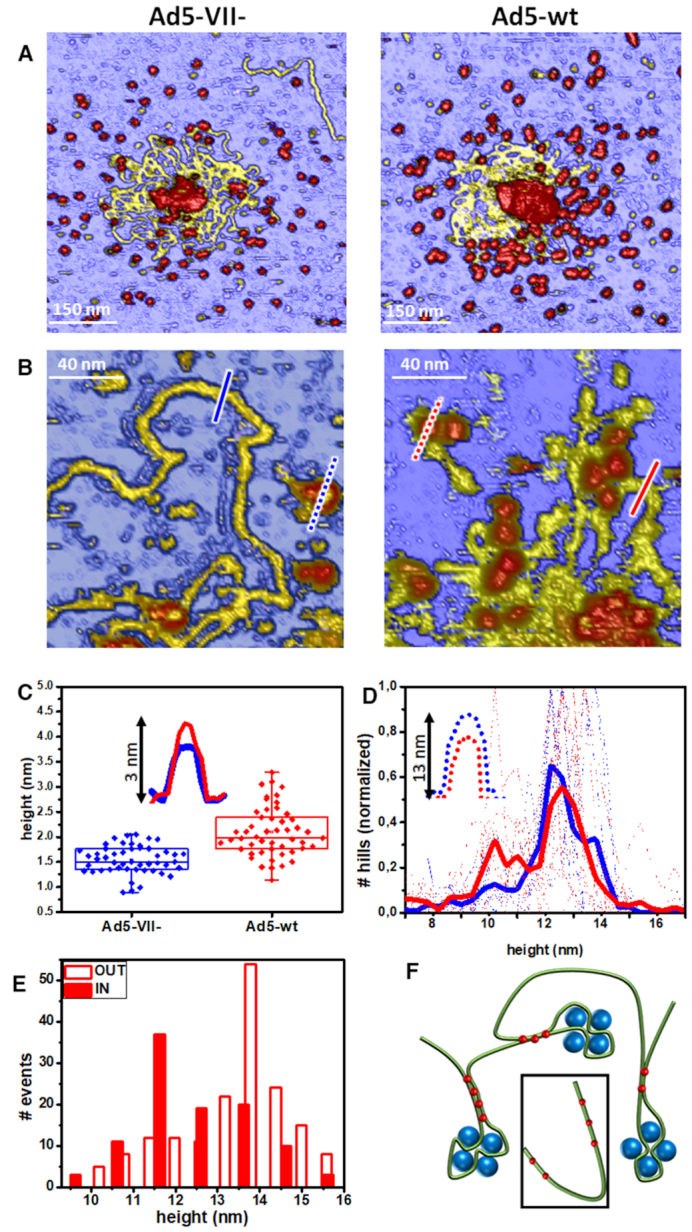
Analysis of unpacked core contents. (A) Example images showing the final stages of Ad5-VII– (left) and Ad5-wt (right) particles after mechanical fatigue disruption. (B) Close-up details of the released contents. (C) Statistics of fibered material heights calculated as explained in Supplementary Figure S4. The inset shows two example topographical profiles across the fibered material (blue and red solid lines in B). (D) Histograms of the blob height distribution obtained for 11 Ad5-wt (red) and 9 Ad5-VII– (blue) disrupted particles. Solid lines present the average of the dotted lines coming from individual particles (Supplementary Figure S5). Inset: example topographical profiles across the blobs (dashed lines in B). (E) Histograms showing the height of the Ad5-wt blobs in contact (red) and not in contact (white) with DNA. The equivalent histogram for Ad5-VII– in shown in Supplementary Figure S5C. (F) Cartoon showing a hypothetical model for the condensing role of protein VII. Green, DNA; blue, protein VII; red, protein μ. The inset illustrates the situation where the bridging action of μ is hindered by the absence of clusters formed by the VII-DNA complexes.
Topographical profiles (Figure 6D, inset) also pointed out to different heights for the blobs in the Ad5-wt and Ad5-VII– images (Figure 6A and B). These particles likely correspond to coat and core proteins coming from the collapsed virus structures after fatigue. We carried out a statistical analysis of the blob heights for 11 Ad5-wt and 10 Ad5-VII– disrupted particles analyzed in ∼10 different experimental sessions. A histogram of normalized blob heights was calculated for each virus particle (Supplementary Figure S5). Then, histograms for all Ad5-wt or all Ad5-VII– particles were averaged (Figure 6D). These charts reveal a major population of objects between 12 and 13 nm for both Ad5-wt and Ad5-VII-, compatible with the size of adenovirus capsomers (15,59). An additional population at ∼10 nm was observed in Ad5-wt but not in Ad5-VII-. This size is close to the 9 nm beads previously observed in metal shadowed cores after high ionic strength treatments that removed most core proteins except VII (16). We classified both the small and large blobs in Ad5-wt particles in two categories, depending on whether they were in contact with the DNA or isolated on the mica surface (Figure 6A). Objects found isolated on the substrate corresponded mainly to the larger blobs (Figure 6E, empty red), while most of the smaller blobs appeared associated with filaments (Figure 6E, solid red), pointing to their interaction with dsDNA. That is, Ad5-wt collapsed particles release objects smaller than capsomers and located preferentially in association with DNA strands, that do not appear when Ad5-VII– particles are disrupted. Therefore, these objects must correspond to the presence of core protein VII.
We can estimate the number of protein VII monomers in a small blob by comparison with the known mass of the hexon trimer forming the larger blobs. The molecular mass of a hexon trimer is 324 kDa, while the molecular mass of mature protein VII is 19 kDa (19). If we consider the large and small blobs as spheres with radii rL ∼6.5 and rS∼ 5 nm, respectively, the number of protein VII monomers per small blob would be 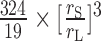 ∼9. However, several considerations from the previous literature would be reluctant with the formation of these VII nonamers. First, since in one Ad5 virion there are 252 capsomers (240 hexon trimers + 12 pentons) and between 500 and 800 copies of protein VII (13,14), a fully disrupted particle should present at most (800/9)/(252 + (800/9)) ∼26% of small objects. In our Ad5-wt images we observe ∼38% of small objects, above the highest expected value if the protein formed nonamers. This excess could be understood if a considerable number of capsomers remained bound forming capsid debris located at the center of the AFM images (Figure 6A and Supplementary Figure S3). Second, assuming that the small blobs contain 9 protein VII monomers, the estimated number of VII oligomers per viral particle (between 55 and 88) would be 2–4 times smaller than the number of beads previously observed for pyridine cores (60). Third, a high order multimer of VII would also be incompatible with previous cross-linking results on disrupted virions and extracted cores, where only dimers were detected (61).
∼9. However, several considerations from the previous literature would be reluctant with the formation of these VII nonamers. First, since in one Ad5 virion there are 252 capsomers (240 hexon trimers + 12 pentons) and between 500 and 800 copies of protein VII (13,14), a fully disrupted particle should present at most (800/9)/(252 + (800/9)) ∼26% of small objects. In our Ad5-wt images we observe ∼38% of small objects, above the highest expected value if the protein formed nonamers. This excess could be understood if a considerable number of capsomers remained bound forming capsid debris located at the center of the AFM images (Figure 6A and Supplementary Figure S3). Second, assuming that the small blobs contain 9 protein VII monomers, the estimated number of VII oligomers per viral particle (between 55 and 88) would be 2–4 times smaller than the number of beads previously observed for pyridine cores (60). Third, a high order multimer of VII would also be incompatible with previous cross-linking results on disrupted virions and extracted cores, where only dimers were detected (61).
It is also possible that we are overestimating the amount of VII monomers per blob, if part of the mass considered corresponds not only to protein VII, but also to associated DNA. In fact, an interesting model can be proposed if we consider each blob as a unit formed by one or several molecules of protein VII, wrapped or otherwise asymmetrically associated with a certain region of DNA. Protein VII appears to interact with DNA to a large extent electrostatically, as indicated by the experiments with spermidine (Figure 2). In each of these VII-DNA units, the repulsive monopole electrostatic interactions would be effectively screened. Remaining higher order (multipole) interactions in the protein-DNA complex combined with short range (steric) repulsion may act as effective sticky ‘patches’ and induce the formation of particularly stable clusters with conformations determined by the structural and interaction anisotropy of the individual complexes, such as cubes (8 effective particles), octahedra (6 effective particles) or tetrahedra (4 effective particles) (62). Depending on the interaction details, clusters may be more or less polydisperse, but it is worth noting that we observe a reasonably sharp peak in the AFM small blob size distribution (Figure 6D) which suggests a largely monodisperse cluster population. The fact that clusters do not form in Ad5-VII– cores suggests that protein VII is essential for their formation. The clustering model should be taken with some care, as it does not take explicitly into account the role of other core proteins and the fact that the Ad5 DNA is a single topological entity. If this model is correct, it would mean that the interaction between the protein-DNA complexes is more complicated than previously proposed in the full confinement (isotropic, weakly repulsive potential) (17). In any case, our data show that objects compatible with multimers of protein VII bound to DNA, and bundles of dsDNA, are present in Ad5-wt unpacked cores, but not in Ad5-VII-.
CONCLUSIONS
We have used AFM and cryo-EM to explore how the absence of core protein VII influences the physical properties and core organization of human adenovirus particles. In the absence of protein VII, the Ad5 particle becomes stiffer and the core becomes softer. Incubation with spermidine softens the Ad5-VII– particle. Thus, our results support the role of protein VII in screening the DNA negative charges, thereby modulating capsid pressure. A decrease in the core condensation when VII is not present accelerates genome diffusion through fractures of the capsid shell. Therefore, the defect in virus entry observed in Ad5-VII– particles (24) is not due to either lack of internal pressure or trapping of the genome inside the particle. Our images of the Ad5 core as it is progressively extracted from the capsid in a physiological environment do not show beads-on-a-string structures as previously shown in released cores dried on a surface for EM analysis where most proteins but VII had been chemically removed (16). Rather, we find beads that can be attributed to clusters of protein VII-DNA complexes, adjacent to bundles consisting of a few dsDNA chains that suggest condensing by a bridging interaction (Figure 6A and B, right). Since the bridging seems to happen away from the actual position of the VII-DNA cluster, we propose that protein VII promotes bundling of DNA by facilitating the bridging action of the small core protein μ (18). The 9 kDa precursor form of protein μ is cleaved in three 20–30 residue long fragments by the Ad5 protease (19), which would correspond to objects of radii ∼1.3 nm as estimated in a similar way as for the red blobs above. We suggest that protein VII could assist the action of μ by bending the DNA molecule so that two branches of it become closer, within the bridging range of μ (Figure 6F).
Although the condensing action of protein VII is not needed for DNA packaging during adenovirus assembly, it might play a role in other steps of the infectious cycle. For instance, it has been proposed that protein VII protects the Ad5 genome from the cellular DNA damage response once inside the nucleus (63). Adenovirus uncoating in the cell occurs in a stepwise manner, starting at the plasma membrane and continuing at the early endosome, where vertex capsomers (pentons) and peripheral core components are released (64,65). Afterward the partially disrupted particle escapes the endosome and travels to the nuclear pore by transport along the microtubular network (66). We hypothesize that the condensation induced by protein VII may be required to keep the genome protected inside the semi-disrupted capsid while trafficking across the cytosol, before reaching the nuclear pore.
DATA AVAILABILITY
The Ad5-wt and Ad5-VII– maps have been deposited at the Electron Microscopy Databank with ID EMD-4448 (Ad5-wt) and EMD-4424 (Ad5-VII-).
Supplementary Material
ACKNOWLEDGEMENTS
We acknowledge the CNB-CIB cryo-EM facility for data acquisition, and Mara I. Laguna for expert technical help.
Author contributions: N.M.G. performed AFM experiments. M.H.P., M.P.I. and C.S.M. carried out cryo-EM studies. P.O., P.H., M.P.I. and G.N.C. produced virus samples. D.R. performed theoretical calculations. N.M.G., M.H.P., C.S.M., A.S. and P.J.dP. analyzed data. C.S.M. and P.J.dP. coordinated the study and wrote the manuscript, with input from all other authors.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Spanish Ministry of Economy and Competitivity [FIS2017-89549-R; “María de Maeztu” Program for Units of Excellence in R&D MDM-2014-0377; and FIS2017-90701-REDT] and Human Frontiers Science Program [HFSPO RGP0012/2018] to P.J.P. Spanish State Research Agency and European Regional Development Fund [BFU2016-74868-P] and Spanish Ministry of Economy, Industry and Competitivity [BIO2015-68990-REDT, the Spanish Adenovirus Network, AdenoNet] to C.S.M. MINECO/FEDER, UE [FIS2015-67837-P] to D.R. National Institutes of Health [CA122677 and AI102577] to P.H. M.H.-P. is a recipient of a Juan de la Cierva postdoctoral contract funded by the Spanish Ministry of Economy and Competitivity. M.P.-I. holds a predoctoral fellowship funded by La Caixa Foundation. Funding for open access charge: Secretaría de Estado de Investigación, Desarrollo e Innovación [FIS2017-89549-R].
Conflict of interest statement. None declared.
REFERENCES
- 1. Luijsterburg M.S., White M.F., Driel R., Dame R.T.. The major architects of chromatin: architectural proteins in bacteria, Archaea and Eukaryotes. Crit. Rev. Biochem. Mol. Biol. 2008; 43:393–418. [DOI] [PubMed] [Google Scholar]
- 2. Zhou B.-R., Feng H., Kato H., Dai L., Yang Y., Zhou Y., Bai Y.. Structural insights into the histone H1-nucleosome complex. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:19390–19395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gruber S. Multilayer chromosome organization through DNA bending, bridging and extrusion. Curr. Opin. Microbiol. 2014; 22:102–110. [DOI] [PubMed] [Google Scholar]
- 4. Teif V.B., Bohinc K.. Condensed DNA: condensing the concepts. Prog. Biophys. Mol. Biol. 2011; 105:208–222. [DOI] [PubMed] [Google Scholar]
- 5. Black L.W., Thomas J.A.. Condensed genome structure. Adv. Exp. Med. Biol. 2012; 726:469–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Germond J.E., Hirt B., Oudet P., Gross-Bellark M., Chambon P.. Folding of the DNA double helix in chromatin-like structures from simian virus 40. Proc. Natl. Acad. Sci. U.S.A. 1975; 72:1843–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Borca M.V., Irusta P.M., Kutish G.F., Carillo C., Afonso C.L., Burrage A.T., Neilan J.G., Rock D.L.. A structural DNA binding protein of African swine fever virus with similarity to bacterial histone-like proteins. Arch. Virol. 1996; 141:301–313. [DOI] [PubMed] [Google Scholar]
- 8. Okamoto K., Miyazaki N., Reddy H.K.N., Hantke M.F., Maia F.R.N.C., Larsson D.S.D., Abergel C., Claverie J.-M., Hajdu J., Murata K. et al.. Cryo-EM structure of a Marseilleviridae virus particle reveals a large internal microassembly. Virology. 2018; 516:239–245. [DOI] [PubMed] [Google Scholar]
- 9. Wulfmeyer T., Polzer C., Hiepler G., Hamacher K., Shoeman R., Dunigan D.D., Etten J.L.V., Lolicato M., Moroni A., Thiel G. et al.. Structural Organization of DNA in Chlorella Viruses. PLoS One. 2012; 7:e30133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Xiao C., Kuznetsov Y.G., Sun S., Hafenstein S.L., Kostyuchenko V.A., Chipman P.R., Suzan-Monti M., Raoult D., McPherson A., Rossmann M.G.. Structural studies of the giant mimivirus. PLoS Biol. 2009; 7:e92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Marion S., San Martín C., Šiber A.. Role of condensing particles in polymer confinement: a model for virus-packed ‘minichromosomes’. Biophys. J. 2017; 113:1643–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu H., Jin L., Koh S.B.S., Atanasov I., Schein S., Wu L., Zhou Z.H.. Atomic structure of human adenovirus by cryo-EM reveals interactions among protein networks. Science. 2010; 329:1038–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. van Oostrum J., Burnett R.M.. Molecular composition of the adenovirus type 2 virion. J. Virol. 1985; 56:439–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Benevento M., Di Palma S., Snijder J., Moyer C.L., Reddy V.S., Nemerow G.R., Heck A.J.R.. Adenovirus composition, proteolysis, and disassembly studied by in-depth qualitative and quantitative proteomics. J. Biol. Chem. 2014; 289:11421–11430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. San Martín C. Latest insights on adenovirus structure and assembly. Viruses. 2012; 4:847–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Vayda M.E., Rogers A.E., Flint S.J.. The structure of nucleoprotein cores released from adenovirions. Nucleic Acids Res. 1983; 11:441–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pérez-Berná A.J., Marion S., Chichón F.J., Fernández J.J., Winkler D.C., Carrascosa J.L., Steven A.C., Šiber A., San Martín C.. Distribution of DNA-condensing protein complexes in the adenovirus core. Nucleic Acids Res. 2015; 43:4274–4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ortega-Esteban A., Condezo G.N., Pérez-Berná A.J., Chillón M., Flint S.J., Reguera D., San Martín C., de Pablo P.J.. Mechanics of viral chromatin reveals the pressurization of human adenovirus. ACS Nano. 2015; 9:10826–10833. [DOI] [PubMed] [Google Scholar]
- 19. Mangel W.F., San Martín C.. Structure, function and dynamics in adenovirus maturation. Viruses. 2014; 6:4536–4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ortega-Esteban A., Bodensiek K., San Martín C., Suomalainen M., Greber U.F., de Pablo P.J., Schaap I.A.. Fluorescence tracking of genome release during mechanical unpacking of single viruses. ACS Nano. 2015; 9:10571–10579. [DOI] [PubMed] [Google Scholar]
- 21. Ortega-Esteban A., Pérez-Berná A.J., Menéndez-Conejero R., Flint S.J., San Martín C., de Pablo P.J.. Monitoring dynamics of human adenovirus disassembly induced by mechanical fatigue. Sci. Rep. 2013; 3:1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pérez-Berná A.J., Marabini R., Scheres S.H.W., Menéndez-Conejero R., Dmitriev I.P., Curiel D.T., Mangel W.F., Flint S.J., San Martín C.. Structure and uncoating of immature adenovirus. J. Mol. Biol. 2009; 392:547–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pérez-Berná A.J., Ortega-Esteban A., Menéndez-Conejero R., Winkler D.C., Menéndez M., Steven A.C., Flint S.J., de Pablo P.J., San Martín C.. The role of capsid maturation on adenovirus priming for sequential uncoating. J. Biol. Chem. 2012; 287:31582–31595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ostapchuk P., Suomalainen M., Zheng Y., Boucke K., Greber U.F., Hearing P.. The adenovirus major core protein VII is dispensable for virion assembly but is essential for lytic infection. PLOS Pathog. 2017; 13:e1006455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. de Pablo P.J. Atomic force microscopy of virus shells. Semin. Cell Dev. Biol. 2018; 73:199–208. [DOI] [PubMed] [Google Scholar]
- 26. Evilevitch A., Roos W.H., Ivanovska I.L., Jeembaeva M., Jonsson B., Wuite G.J.L.. Effects of salts on internal DNA pressure and mechanical properties of phage capsids. J. Mol. Biol. 2011; 405:18–23. [DOI] [PubMed] [Google Scholar]
- 27. Hernando-Pérez M., Miranda R., Aznar M., Carrascosa J.L., Schaap I.A.T., Reguera D., de Pablo P.J.. Direct measurement of phage phi29 stiffness provides evidence of internal pressure. Small. 2012; 8:2366–2370. [DOI] [PubMed] [Google Scholar]
- 28. Gonzalez-Huici V., Salas M., Hermoso J.M.. The push-pull mechanism of bacteriophage O29 DNA injection. Mol. Microbiol. 2004; 52:529–540. [DOI] [PubMed] [Google Scholar]
- 29. Alba R., Hearing P., Bosch A., Chillón M.. Differential amplification of adenovirus vectors by flanking the packaging signal with attB/attP-ΦC31 sequences: Implications for helper-dependent adenovirus production. Virology. 2007; 367:51–58. [DOI] [PubMed] [Google Scholar]
- 30. Condezo G.N., Marabini R., Ayora S., Carazo J.M., Alba R., Chillón M., San Martín C.. Structures of adenovirus incomplete particles clarify capsid architecture and show maturation changes of packaging protein L1 52/55k. J. Virol. 2015; 89:9653–9664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ortega-Esteban A., Horcas I., Hernando-Pérez M., Ares P., Pérez-Berná A.J., San Martín C., Carrascosa J.L., de Pablo P.J., Gómez-Herrero J.. Minimizing tip-sample forces in jumping mode atomic force microscopy in liquid. Ultramicroscopy. 2012; 114:56–61. [DOI] [PubMed] [Google Scholar]
- 32. Sader J.E., Chon J.W.M., Mulvaney P.. Calibration of rectangular atomic force microscope cantilevers. Rev. Sci. Instrum. 1999; 70:3967–3969. [Google Scholar]
- 33. Ivanovska I.L., de Pablo P.J., Ibarra B., Sgalari G., MacKintosh F.C., Carrascosa J.L., Schmidt C.F., Wuite G.J.L.. Bacteriophage capsids: Tough nanoshells with complex elastic properties. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:7600–7605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hernando-Pérez M., Lambert S., Nakatani-Webster E., Catalano C.E., de Pablo P.J.. Cementing proteins provide extra mechanical stabilization to viral cages. Nat. Commun. 2014; 5:4520. [DOI] [PubMed] [Google Scholar]
- 35. Snijder J., Borst A.J., Dosey A., Walls A.C., Burrell A., Reddy V.S., Kollman J.M., Veesler D.. Vitrification after multiple rounds of sample application and blotting improves particle density on cryo-electron microscopy grids. J. Struct. Biol. 2017; 198:38–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. de la Rosa-Trevín J.M., Quintana A., Del Cano L., Zaldívar A., Foche I., Gutiérrez J., Gómez-Blanco J., Burguet-Castell J., Cuenca-Alba J., Abrishami V. et al.. Scipion: A software framework toward integration, reproducibility and validation in 3D electron microscopy. J. Struct. Biol. 2016; 195:93–99. [DOI] [PubMed] [Google Scholar]
- 37. Zheng S.Q., Palovcak E., Armache J.-P., Verba K.A., Cheng Y., Agard D.A.. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 2017; 14:331–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rohou A., Grigorieff N.. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 2015; 192:216–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. de la Rosa-Trevín J.M., Otón J., Marabini R., Zaldívar A., Vargas J., Carazo J.M., Sorzano C.O.S.. Xmipp 3.0: an improved software suite for image processing in electron microscopy. J. Struct. Biol. 2013; 184:321–328. [DOI] [PubMed] [Google Scholar]
- 40. Scheres S.H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012; 180:519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Scheres S.H.W., Chen S.. Prevention of overfitting in cryo-EM structure determination. Nat. Methods. 2012; 9:853–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dai X., Wu L., Sun R., Zhou Z.H.. Atomic structures of minor proteins VI and VII in the human adenovirus. J. Virol. 2017; 91:e00850-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E.. UCSF chimera - a visualization system for exploratory research and analysis. J. Comput. Chem. 2004; 25:1605–1612. [DOI] [PubMed] [Google Scholar]
- 44. Llauró A., Schwarz B., Koliyatt R., de Pablo P.J., Douglas T.. Tuning viral capsid nanoparticle stability with symmetrical morphogenesis. ACS Nano. 2016; 10:8465–8473. [DOI] [PubMed] [Google Scholar]
- 45. Zeng C., Hernando-Pérez M., Dragnea B., Ma X., van der Schoot P., Zandi R.. Contact mechanics of a small icosahedral virus. Phys. Rev. Lett. 2017; 119:038102. [DOI] [PubMed] [Google Scholar]
- 46. Denning D., Bennett S., Mullen T., Moyer C., Vorselen D., Wuite G.J.L., Nemerow G., Roos W.H.. Maturation of adenovirus primes the protein nano-shell for successful endosomal escape. Nanoscale. 2019; 11:4015–4024. [DOI] [PubMed] [Google Scholar]
- 47. Carrasco C., Carreira A., Schaap IaT., Serena P.A., Gómez-Herrero J., Mateu M.G., de Pablo P.J.. DNA-mediated anisotropic mechanical reinforcement of a virus. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:13706–13711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sae-Ueng U., Li D., Zuo X., Huffman J.B., Homa F.L., Rau D., Evilevitch A.. Solid-to-fluid DNA transition inside HSV-1 capsid close to the temperature of infection. Nat. Chem. Biol. 2014; 10:861–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Snijder J., Uetrecht C., Rose R.J., Sanchez-Eugenia R., Marti G.A., Agirre J., Guérin D.M.A., Wuite G.J.L., Heck A.J.R., Roos W.H.. Probing the biophysical interplay between a viral genome and its capsid. Nat. Chem. 2013; 5:502–509. [DOI] [PubMed] [Google Scholar]
- 50. Carrasco C., Castellanos M., de Pablo P.J., Mateu M.G.. Manipulation of the mechanical properties of a virus by protein engineering. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:4150–4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Grønbech-Jensen N., Mashl R.J., Bruinsma R.F., Gelbart W.M.. Counterion-induced attraction between rigid polyelectrolytes. Phys. Rev. Lett. 1997; 78:2477–2480. [Google Scholar]
- 52. Bloomfield V.A., Crothers D.M., Tinoco I., Hearst J.E., Wemmer D.E., Killman P.A., Turner D.H.. Nucleic Acids: Structures, Properties, and Functions, 1st edn. 2000; Sausalito: University Science Books. [Google Scholar]
- 53. Leforestier A., Livolant F.. Structure of toroidal DNA collapsed inside the phage capsid. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:9157–9162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Vella D., Ajdari A., Vaziri A., Boudaoud A.. The indentation of pressurized elastic shells: from polymeric capsules to yeast cells. J. R. Soc. Interface. 2012; 9:448–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Moreno-Herrero F., Colchero J., Baró A.M.. DNA height in scanning force microscopy. Ultramicroscopy. 2003; 96:167–174. [DOI] [PubMed] [Google Scholar]
- 56. Lander G.C., Johnson J.E., Rau D.C., Potter C.S., Carragher B., Evilevitch A.. DNA bending-induced phase transition of encapsidated genome in phage λ. Nucleic Acids Res. 2013; 41:4518–4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Comolli L.R., Spakowitz A.J., Siegerist C.E., Jardine P.J., Grimes S., Anderson D.L., Bustamante C., Downing K.H.. Three-dimensional architecture of the bacteriophage phi29 packaged genome and elucidation of its packaging process. Virology. 2008; 371:267–277. [DOI] [PubMed] [Google Scholar]
- 58. Villarrubia J.S. Algorithms for Scanned Probe Microscope Image Simulation, Surface Reconstruction, and Tip Estimation. J. Res. Natl. Inst. Stand. Technol. 1997; 102:425–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Snijder J., Reddy V.S., May E.R., Roos W.H., Nemerow G.R., Wuite G.J.L.. Integrin and defensin modulate the mechanical properties of adenovirus. J. Virol. 2013; 87:2756–2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mirza M.A., Weber J.. Structure of adenovirus chromatin. Biochim. Biophys. Acta BBA - Gene Struct. Expr. 1982; 696:76–86. [DOI] [PubMed] [Google Scholar]
- 61. Chatterjee P.K., Vayda M.E., Flint S.J.. Interactions among the three adenovirus core proteins. J. Virol. 1985; 55:379–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zhang Z., Glotzer S.C.. Self-assembly of patchy particles. Nano Lett. 2004; 4:1407–1413. [DOI] [PubMed] [Google Scholar]
- 63. Karen K.A., Hearing P.. Adenovirus core protein VII protects the viral genome from a DNA damage response at early times after infection. J. Virol. 2011; 85:4135–4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Greber U.F., Willetts M., Webster P., Helenius A.. Stepwise dismantling of adenovirus 2 during entry into cells. Cell. 1993; 75:477–486. [DOI] [PubMed] [Google Scholar]
- 65. Puntener D., Engelke M.F., Ruzsics Z., Strunze S., Wilhelm C., Greber U.F.. Stepwise loss of fluorescent Core protein V from human adenovirus during entry into cells. J. Virol. 2011; 85:481–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Bremner K.H., Scherer J., Yi J., Vershinin M., Gross S.P., Vallee R.B.. Adenovirus transport via direct interaction of cytoplasmic dynein with the viral capsid hexon subunit. Cell Host Microbe. 2009; 6:523–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Ad5-wt and Ad5-VII– maps have been deposited at the Electron Microscopy Databank with ID EMD-4448 (Ad5-wt) and EMD-4424 (Ad5-VII-).