Abstract
Cellular non-membranous RNA-granules, P-bodies (RNA processing bodies, PB) and stress granules (SG), are important components of the innate immune response to virus invasion. Mechanisms governing how a virus modulates PB formation remain elusive. Here, we report the important roles of GW182 and DDX6, but not Dicer, Ago2 and DCP1A, in PB formation, and that Kaposi’s sarcoma-associated herpesvirus (KSHV) lytic infection reduces PB formation through several specific interactions with viral RNA-binding protein ORF57. The wild-type ORF57, but not its N-terminal dysfunctional mutant, inhibits PB formation by interacting with the N-terminal GW-domain of GW182 and the N-terminal domain of Ago2, two major components of PB. KSHV ORF57 also induces nuclear Ago2 speckles. Homologous HSV-1 ICP27, but not EBV EB2, shares this conserved inhibitory function with KSHV ORF57. By using time-lapse confocal microscopy of HeLa cells co-expressing GFP-tagged GW182, we demonstrated that viral ORF57 inhibits primarily the scaffolding of GW182 at the initial stage of PB formation. Consistently, KSHV-infected iSLK/Bac16 cells with reduced GW182 expression produced far fewer PB and SG, but 100-fold higher titer of infectious KSHV virions when compared to cells with normal GW182 expression. Altogether, our data provide the first evidence that a DNA virus evades host innate immunity by encoding an RNA-binding protein that promotes its replication by blocking PB formation.
INTRODUCTION
In mammalian systems, mRNA-regulating RNA granules in somatic cells, which are comprised of both RNA processing bodies (PB) and stress granules (SG), have emerged as important players in the post-transcriptional regulation of gene expression (1–3). GW182 and de-capping/de-adenylating enzymes (DCP1/Ccr4-Caf1) are specific components of PB (4–7) where siRNA- or miRNA-guided mRNAs are translationally arrested and degraded (8). RNA-binding proteins, TIA-1 and poly(A)-binding protein (PABP), are two fundamental components of SG appearing during stress. In such physically and mechanically distinct, but spatially and functionally linked cytoplasmic non-membranous compartments (4), mRNA transcripts are packaged along with a discrete set of RNA-binding and scaffold proteins responsible for mRNA turnover and regulated translation under different cellular conditions. Notably, the two types of granules share several proteins such as XRN1, Argonaute 2 (Ago2), Fas-activated serine/threonine phosphoprotein (FAST), tristetraproline (TTP) and serve to regulate cellular homeostasis and RNA metabolism (4,9–11). Interestingly, a recent report observed single mRNAs interacting with SG and PB, with mRNAs moving bidirectionally between them (12).
Under normal physiological conditions, cytoplasmic PB are comprised primarily of mRNAs in complex with proteins involved with repressing translation and supporting 5′-to-3′ mRNA decay. Initially discovered over 20 years ago (13) as the cytoplasmic foci containing the 5′→3′ exoribonuclease XRN1, PB were rediscovered in 2002 as discrete GW182 bodies by immunostaining of cells with an autoimmune serum from a patient with neuropathy (14). Three GW182 paralogs exist in vertebrates (TNRC6A, TNRC6B and TNRC6C) (15,16) and are crucial for PB formation and miRNA-mediated gene silencing (17–21). In addition, various proteins from each phase of the general mRNA decay pathway, including mRNA de-adenylating (CCR4-Calf1 complex, PAN2 and PAN3) and RNA de-capping enzymes (including DCP1/2, HedIs, hEdc3 and RCK/p54) (5,7) were found in PB along with exonuclease XRN1. Besides GW182, PB also contain miRNAs and miRNA repression factors, including components of the RNA-induced silencing complex (RISC) such as Ago2, dead box helicases (MOV10 and DDX6) (18,22,23), RNA-binding protein CPEB (cytoplasmic polyadenylation element-binding protein) (24), AU-rich element binding protein TTP (25) and mRNA 5′ cap-binding protein eIF4E (26,27). The steps in mRNA degradation in PB are thought to involve decapping of deadenylated mRNA that renders the mRNA susceptible to 5′-3′ mRNA exonuclease activity or directs it to non-sense mediated decay (28). Given the list of PB constituents, PB are regarded as major sites of miRNA-mediated translational suppression and mRNA degradation (29). However, recent reports using single-molecule imaging indicated that mRNA decay might not be enriched within the PB (30) but rather occur predominantly outside of PB (31).
Viral manipulation of cellular gene expression is a key aspect of virus–host interactions that maintain conditions favorable for efficient viral replication. Mammalian PB and SG are important components of the host cell antiviral response. Several RNA and DNA viruses have been shown to evade or co-opt SG for their replication through different mechanisms (32–34). However, emerging evidence indicates that PB formation may be disrupted during virus infection, but the mechanism(s) of PB inhibition by viral machinery remains unknown. In this study, we demonstrated that B cells with lytic KSHV infection do not exhibit visible PB, but B cells with latent KSHV infection display many PB. A similar result was seen for epithelial Bac36 cells carrying a wild-type (wt) KSHV genome, but not for Bac36 cells carrying an ORF57-null KSHV genome (Δ57), suggesting that viral lytic ORF57, an RNA-binding protein and post-transcriptional regulator (35), might prevent the PB formation during KSHV lytic infection. KSHV ORF57 has an intrinsically disordered region (IDR) at its N-terminus for interactions with RNA and other RNA-binding proteins and a C-terminal region for its homodimerization (36,37). Subsequently, we demonstrated that ORF57 inhibits the PB formation by binding both GW182 and Ago2 and prevents the interaction of GW182 with Ago2. Thus, we provide the first experimental evidence of a viral protein directly interacting with host Ago2 and GW182 to disrupt PB formation and enhance virus production.
MATERIALS AND METHODS
Cell cultures and virus reactivation
Human HEK293T and HeLa cells were cultured in DMEM, Dicer wt (Dicer+/+) and Dicer knockout (Dicer-/-) HCT116 cells (38) in McCoy’s 5A, primary effusion lymphoma BCBL-1 cells (KSHV+) (39) and JSC-1 (KSHV+/EBV+) B-cell line (40) in RPMI-1640. All media were obtained from Thermo Fisher Scientific and supplemented with 10% fetal bovine serum (FBS, cat. # SH30070.03, HyClone, GE Healthcare) and 1× Penicillin-Streptomycin-Glutamine (cat. # 1037816, Thermo Fisher Scientific). HEK293T-derived Bac36 cell lines stably harboring a wt KSHV genome (Bac36-wt) or an ORF57-null KSHV genome (Bac36-Δ57) were established in our laboratory as described (41). KSHV-infected iSLK/Bac16 cells were cultivated as reported (42). All cultures were grown at 37°C in a 5% CO2 humidified atmosphere.
KSHV lytic infection in BCBL-1 cells or JSC-1 cells and Bac36 cells was reactivated by 1 mM sodium valproate (VA, cat. # P4543, Millipore Sigma) or 3 mM sodium butyrate (Bu, cat. # B5887, Millipore Sigma) treatment, respectively, for 24 h. KSHV lytic replication in iSLK/Bac16 was induced by simultaneous treatment with 1 mM sodium butyrate and 1 μg/ml doxycycline (DOX, cat. # NC0424034, Fisher Scientific).
Induction of cellular stress
A sodium arsenite (cat. # 38150, Sigma-Aldrich) solution (0.83 M) was prepared in water to serve as a 1660 × stock solution. To induce oxidative stress, the cells were cultivated in fresh culture medium containing 0.5 mM sodium arsenite for 30 min (43).
Antibodies and peptides
The antibodies used for this work and their respective working dilutions in Western blotting (WB) or indirect immunofluorescence assay (IFA) are as following. Rabbit polyclonal (1:2000, WB and 1:250, IFA) and mouse monoclonal anti-ORF57 (1:1000, WB and 1:250, IFA) antibodies were described earlier (41). Mouse monoclonal anti-RTA antibody (1:250, IFA) was obtained as a gift from Dr Koichi Yamanishi (Osaka University, Osaka, Japan). GW182-specific antiserum (1:1000, WB; 1:50, IFA) was an autoimmune serum from a patient with motor and sensory neuropathy (14). Mouse monoclonal anti-GW182 antibody (4B6) (https://www.kerafast.com/product/2282/anti-gw182tnrc6a-4b6-antibody) was purified and concentrated to 5–10 times more than the hybridoma supernatant. Anti-Flag M2 (1:1000, WB and 1:250, IFA; cat. # F1804), rabbit polyclonal anti-Flag antibody (1: 1000, WB; cat. # F7425) and rabbit polyclonal anti-HA (1:1000, WB, cat. # H6908) were obtained from Millipore Sigma. Mouse monoclonal anti-β-actin (1:2000, WB; cat. # sc-69879), rabbit polyclonal anti-Dicer (1:100, WB; cat. # sc-30226) and goat polyclonal anti-TIA-1 (1:1000, WB; 1:50, IFA; cat. # sc-1751) were purchased from Santa Cruz Biotechnology. Other antibodies used were rabbit polyclonal anti-Ago2 (1:1000, cat. # 07-590, EMD Millipore), anti-DDX6 (1:3000, WB; cat. # NB200-191, Novus Biologicals), anti-RHA (1:500, WB; cat. # ab26271, Abcam) and anti-PABP-C1 (1:1000, WB; cat. #ab21060, Abcam); rabbit monoclonal anti-DCP1A (1:2000, WB, 1:250, IFA; cat. # ab183709, Abcam), anti-CTCF (1:500, WB; clone D31H2, cat. #3418S, Cell Signaling) and anti-GAPDH (1:2000, WB; clone 14C10, cat # 2118, Cell Signaling); mouse monoclonal anti-Ago2/eIF2C2 (1:50, IFA; cat. # ab57113, Abcam), anti-eIF2AK2 (PKR) (1:1000, WB; cat. # H000005610-M01, Abnova) and hnRNP C1+C2 (1:250, WB; clone 4B4, cat. # ab10294, Abcam). All peroxidase-conjugated secondary antibodies used in WB (1:10000) were obtained from Millipore Sigma and all AlexaFluor-conjugated secondary antibodies (1:500) used in IFA were purchased from Thermo Fisher Scientific. TNRC6B-1-derived peptides (44) were synthesized by Peptide 2.0 (Chantilly, VA). Six peptides used in the study were E11 (DLDPRVLSNTGWGQTQIKQD), E12 (VLSNTGWGQTQIKQDTVWDI), G2 (KPVSGWGEGGQNEIGTWGNG), G3 (WGEGGQNEIGTWGNGGNASL), D3 (SWDNNNRSTGGSWNFGPQDS), A1(SNYANSTWGSGASSNNGTSP).
Plasmids
The following expression vectors were used in our studies: KSHV ORF57-Flag (wt, pVM7; mt NLS 2+3, pVM89), KSHV ORF59-Flag (pVM18) were described (41,45,46). Plasmid pCMV-FLAG 5.1 (Millipore Sigma) served as a negative control vector. The vector expressing FLAG/hemagglutinin (HA)-human Ago2 (hAgo2) (pIRESneo-Flag/HA Ago2, cat. # 10822) and the GFP-hAgo2 (pGFP-hAgo2, cat. # 11590) were obtained from Addgene. PCR-based mutagenesis approaches were used to create the Ago2 domain deleted mutants from the parent plasmid pIRESneo-Flag/HA Ago2: Ago2-ΔNL1 (removal of aa 1-226), Ago2-ΔN (removal of aa 53-135), Ago2-ΔPAZ (removal of aa 227-353) and Ago2-ΔPIWI (removal of aa 581-859). Plasmids for expression of GW182 (cat. # 41999) and its deletion mutants GW1Δ5 (cat. # 36440) or GW1Δ10 (cat. # 21517) were obtained from Addgene. To subclone ORF57 in pcDNA3-mRuby2, ORF57 was digested from pVM8 (46) using EcoRI and BamHI and first swapped to pVM24 (46) after its digestion with same set of enzymes. The resulting plasmid was again digested with NotI and BamHI to take out ORF57 and this fragment was ligated to pCDNA3-mRuby2 after its digestion with NotI and BamHI to obtain pcDNA3 ORF57-mRuby2. The accuracy of all plasmid was verified by sequencing. Plasmid expressing ORF57 homologues Herpes Simplex Virus Type 1 (HSV-1) ICP27-Flag (pKY15 derived from p320, gift from Dr Rozanne Sandri-Goldin) and Epstein Barr Virus (EBV) myc-EBV EB2 (pGS113, gift of Dr S. Swaminathan) were described (46).
Plasmid transfections
All plasmid transfections were performed using LipoD293 In vitro DNA Transfection Reagent (cat. # SL100668, SignaGen Laboratories) according to the manufacturer’s instruction. Unless indicated, for IFA and Western blot analysis, HeLa (2.5 × 105) and HEK293T (5 × 105) cells were plated in a six-well plate a day prior DNA transfection with 1 μg of plasmid DNA/well. For IP, HeLa (2 × 106) and HEK293T (5 × 106) cells were plated in a 10-cm Petri dish a day prior transfection with 5 μg plasmid DNA/dish. The cells were harvested either by direct lysis in 2× SDS protein sample buffer containing 5% 2-mercaptoethanol (2-ME) (350 μl/six-well plate) or used for IP.
Immunoprecipitation (IP)
IP was performed as described earlier (47). Briefly, the individual proteins were ectopically expressed in HeLa or HEK293T cells grown in a 10-cm dish. Twenty-four hours after transfection, the cells were washed with 1× PBS (phosphate-buffered saline, [pH 7.4]) and lysed in 500 μl 1× RSB-200 lysis buffer (10 mM Tris-HCl [pH 7.5], 200 mM NaCl, 2.5 mM MgCl2, 0.1% NP-40 and (Nonidet P-40, a mild non-ionic detergent), 1 × cOmplete EDTA-free protease inhibitor cocktail [cat. # 11873580001, Roche]), briefly sonicated and cleared by centrifugation at 11500 × g for 10 min at 4°C. In the assays designed to detect the interaction of multiple independently expressed proteins, equal amount of each individual cell extract was mixed on ice. Before IP, all cell lysates were incubated with RNase A/T1 mixture (1.25 U RNase A/50U RNase T1 at final concentration, Thermo Fisher Scientific) for 10 min at RT followed by pre-cleaning with 80 μl pre-washed Sepharose CL-4B beads (cat. # CL4B200, Millipore Sigma). The pre-cleaned cell lysates were mixed with 80 μl (50% slurry) of antibody-coated protein A beads ([cat. # 16-125, Millipore Sigma], coated either with EZview anti-FLAG M2 [cat. # F2426, Millipore Sigma] or EZview anti-HA [cat. #. E6779, Millipore Sigma]) in 1 ml of IP buffer (50 mM HEPES [pH 7.5], 200 mM NaCl, 1 mM EDTA, 2.5 mM EGTA, 10% glycerol and 0.1% NP-40, 1 × EDTA-free cOmplete protease inhibitor cocktail [Roche]) and incubated overnight at 4°C followed by extensive wash with IP buffer. IP complexes on the beads were dissolved in 70 μl of 2× SDS protein sample buffer containing 50 mM dithiothreitol (DTT). Alternatively, the IP complexes were eluted with 50 μl of 100 μg/ml 3 × Flag (cat. # F4799, Millipore Sigma) or 100 μg/ml HA peptides (cat. # I2179, Millipore Sigma) in IP buffer for 2 h at 4°C. The eluant was mixed with 20 μl of 5× SDS protein buffer containing 50 mM DTT. Finally, 3–5% input lysates and 30–50% immunoprecipitated proteins were used for Western blot.
Electrophoresis and Western blot (WB)
For WB, all samples were heat-denatured at 95°C for 5–10 min before loading on NuPAGE Bis-Tris 4–12% gels (Thermo Fisher Scientific) followed by the electrophoresis in 1× MOPS SDS buffer (cat. # NP0001, Thermo Fisher Scientific). Separated proteins were transferred onto a nitrocellulose membrane and blocked for 1 h with 5% skimmed milk in 1× TBS (Tris-buffered saline) containing 0.05% Tween (TTBS). The incubation with primary antibodies diluted in TTBS was performed overnight at 4°C followed by 1 h at room temperature incubation with corresponding secondary peroxidase-conjugated antibodies diluted in 2% milk/TTBS. The specific signal generated with SuperSignal West Pico (cat. # 34080) or Femto Chemiluminescent Substrate (cat. # 34096, Thermo Fisher Scientific). Densitometric quantification of the individual protein band intensity was performed using ImageJ software (NIH).
Nuclear and cytoplasmic fractionation of HeLa cells were obtained using a nuclei isolation kit from Millipore Sigma (cat. # NUC101) according to the protocol provided by the company, with slight modification. Briefly, cells growing in a 60-mm dish were scraped in 1× PBS and lysed using 400 μl of Nuclei EZ lysis buffer. Nuclei were pelleted by centrifugation (500 × g) for 5 min at 4°C. The supernatant was collected as the cytoplasmic fraction and the obtained nuclear pellet was resuspended/washed gently once with 1× PBS and then resuspended in 400 μl of lysis buffer. Equal volumes of nuclear and cytoplasmic extracts or total protein samples were examined by a standard WB protocol after solubilizing in SDS protein sample buffer supplemented with 5% (vol/vol) 2-ME.
Indirect immunofluorescent assay (IFA) and confocal microscopy
Adherent HeLa and HEK293T cells were grown directly on glass coverslips. The non-adherent BCBL-1 cells were immobilized by spotting onto poly-d-lysine-treated (cat. # P4707, Millipore Sigma) glass coverslips. IFA was performed as described (46,47). Briefly, cells washed with PBS, fixed with 4% paraformaldehyde (cat. # 50-980-487, Fisher Scientific), permeabilized with 0.5% Triton X-100 (cat. # H5142, Promega) and blocked with 2% bovine serum albumin (cat. # W384A, Promega) dissolved in PBS containing 0.05% of Tween-20 (TPBS). Primary antibodies were diluted in blocking buffer and incubated with slides overnight at 4°C. AlexaFluor-conjugated secondary antibodies (1: 500, ThermoFisher Scientific) were diluted in blocking solution and incubated with slides at 37°C for 1 h in a humidified chamber. The slides were washed with TPBS and nuclei were counterstained with Hoechst 33342 dye (cat. # B2261, Millipore Sigma) or DAPI (cat. # D1306, Thermo Fisher Scientific) for 5 min before mounting in Prolong Gold Antifade mounting medium (cat. # P36934, Thermo Fisher Scientific). Confocal images were collected with a Zeiss LSM780 laser-scanning microscope (Carl Zeiss) equipped with 20 × Plan-Apochromat (N.A. 0.8) and 63 × Plan-Apochromat (N.A. 1.4) objective lenses. The x–y pixel sizes of 0.4 and 0.07 μm and optical slice thicknesses of 1.5 and 0.9 μm were used to acquire confocal images with the 20× and 63× objectives lenses, respectively. Confocal z-stacks were generated using the Imaris (version 8.0.2) image processing software (Bitplane).
Time-lapse microscopy
HeLa cells were growing in phenol red-free DMEM medium containing 10% FBS in a glass bottom dish and co-transfected with a vector expressing GFP/myc-GW182 along with a vector expressing either mRuby (negative control) or ORF57-mRuby. After transfection and 4 h incubation in a CO2 incubator, the glass bottom dish was transferred to a cultivation chamber attached to a confocal microscope. Live cell images of subcellular expression and localization of GFP-GW182 and mRuby/ORF57-mRuby were collected using a Zeiss LSM780 confocal microscope equipped with a 20× plan-apochromat objective lens (0.8 N.A.). Z-stack images covering 10 μm in the z-axis were collected every 45 min over 22 h.
GW182-derived peptide inhibition assay
ORF57-Flag and HA-Ago2 were ectopically expressed in HEK293T cells by transfection of 5 μg of plasmid DNA/10-cm dish. Twenty-four hours after transfection, the cells were lysed in 500 μl of ice-cold 1× RSB-200 lysis buffer/dish, briefly sonicated and cleared by centrifugation as described in IP section. The individual cell lysates of Ago2 (1000 μl) and ORF57 (500 μl) were separately incubated with RNase A/T1 mixture (containing 1.25 U RNase A/50U RNase T1 at final concentration, Ambion) for 10 min at room temperature and were used for two independent IP pulldown experiments. In the first IP pulldown, HA-Ago2 cell extract (1000 μl) was precleaned with 200 μl Sepharose CL-4B beads (cat. # CL4B200, Millipore Sigma) and incubated with 200 μl of EZview anti-HA-coated beads (cat. # E6779, Millipore Sigma) overnight at 4°C. After extensive washes with IP buffer, the HA-Ago2-coated beads were resuspended in 700 μl IP buffer. In parallel, 35 μl cell lysate containing ORF57 diluted in 500 μl of IP buffer was mixed with 5 or 10 nM of individual GW182 peptides and incubated at 4°C for 2 h. The peptide-ORF57 lysate mixture was then added to 50 μl of HA-Ago2-coated beads and incubated overnight at 4°C. After extensive washes with IP buffer, the HA-Ago2-associated protein complexes were eluted by incubation with 50 μl of IP buffer containing 100 μg/ml HA peptide (cat. # I2149, Millipore Sigma) for 2 h at 4°C. The eluant was mixed with 20 μl of 5× SDS protein buffer containing 5% 2-ME and used for WB. In the second IP pulldown, ORF57-containing extract was pre-cleaned with Sepharose CL-4B beads and then incubated with 350 μl of polyclonal anti-ORF57-coated protein G-beads overnight at 4°C. After the extensive wash with IP buffer, the remaining beads were resuspended in 700 μl IP buffer. In parallel, 70 μl of HA-Ago2 lysate diluted in 500 μl of IP buffer was mixed with 5 or 10 nM of individual GW182 peptides and incubated overnight at 4°C. The peptide-HA-Ago2 mixture was then incubated with 50 μl of ORF57 coated beads and followed by overnight incubation at 4°C. After extensive washes with IP buffer, the ORF57-associated protein complexes on the beads were eluted directly in 70 μl of 2× SDS protein sample buffer containing 5% 2-ME for WB.
Knockdown of host factor expression by siRNA and KSHV virus production
The siRNA knockdown of host factors was performed as described (48). Briefly, HeLa or KSHV-infected iSLK/Bac16 cells (42) growing in a six-well plate (2.5 × 105 cells/well) were transfected twice at a 24-h interval with 40 nM of ON-TARGETplus SMART-pools (Dharmacon) targeting human PKR/EIF2AK2 (L-003527-00-0005), TNRC6A/GW182 (L-014107-00-0005), CTCF (L-020165-00-0005), EIF2C2/Ago2 (L-004639-00-0005), DCP1A (L021242-00-0005), DDX6 (L-006371-00-0005) or Dicer duplex (P002010-01-05). ON-TARGETplus Non-targeting siRNA #1 was used as a negative control (D-001810-01). All siRNA transfections were carried out using LipoJet In Vitro Transfection Reagent (cat. # SL100468, SignaGen Laboratories). The knockdown efficiency was confirmed by WB using total cell extract that was collected 24 h after the second siRNA transfection. In the same time, the HeLa cells were fixed and co-stained with anti-GW182 and -DCP1A antibodies for the presence of PB. The iSLK/Bac16 were treated with a combination of 1 mM sodium butyrate and 1 μg/ml doxycycline to induce KSHV lytic replication as described (48). The amount of cell-free KSHV-GFP viruses in the culture media from siRNA-treated cells was determined by inoculation of freshly plated HEK293T cells. The amount of KSHV-infected GFP+ HEK293T cells were determined 48 h after infection by a direct fluorescent microscopy and analyzed by flow cytometry. The iSLK/Bac16 cells with or without gene-specific siRNA knockdown or lytic KSHV induction, but under arsenite-induced stress condition, were also stained for the presence of PB (GW182 staining) and SG (TIA-1 staining).
RESULTS
KSHV lytic infection inhibits PB formation in correlation with ORF57 expression
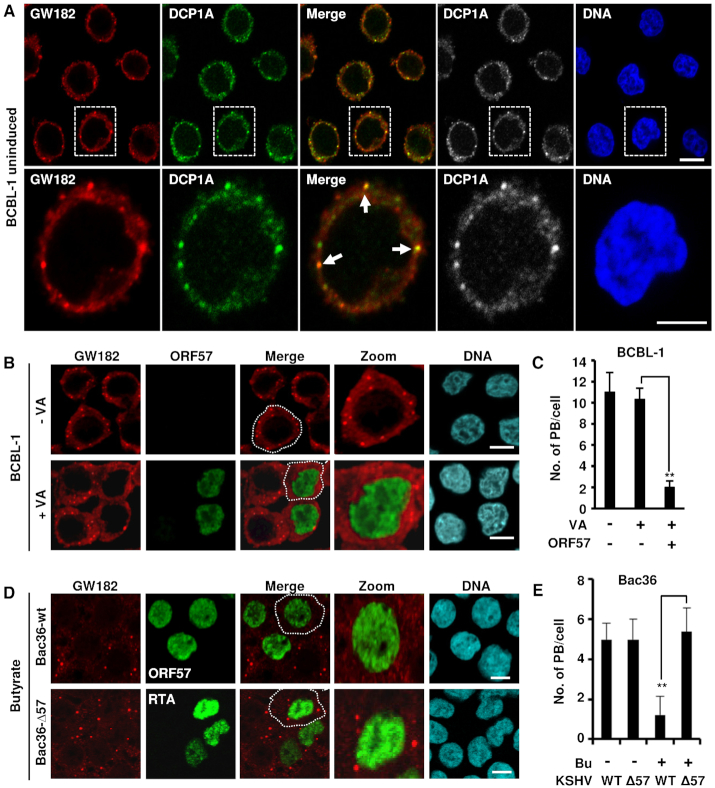
Previously, KSHV ORF57 was reported to prevent RNA-induced silencing complex (RISC) loading and miRNA-mediated inhibition of IL-6 mRNA to enhance IL-6 protein expression (45,49). Given that both miRNA-mediated RNA translation inhibition and RNA degradation occur in the PB (17–21), we used GW182-specific and DCP1A-specific antibodies to probe for PB by immunofluorescence in KSHV-infected BCBL-1 cells (39). The uninduced, latently infected BCBL-1 (Figure 1A) showed the presence of typical cytoplasmic GW182+ puncta that also contain DCP1A, typical for PB. During KSHV lytic infection and when ORF57 expression is induced in BCBL-1 cells by valproic acid (VA) (41), a dramatic reduction in the number of PB (∼2 PB/cell) occurs when compared to BCBL-1 cells with latent KSHV infection (no VA treatment) (10–12 PB/cell) (Figure 1B and C; Supplementary Figure S1). Similarly, the reduced number of PB was observed upon butyrate-treatment of HEK293T-derived Bac36 cells containing a wt KSHV genome (Bac36-wt) with ORF57 expression, but not in butyrate-treated Bac36 cells containing an ORF57-null KSHV genome (Bac36-Δ57) that only has viral RTA expression (Figure 1D and E). These data highlight the importance of viral ORF57 or lytic gene expression in suppressing PB formation during viral lytic infection.
Figure 1.
KSHV lytic infection inhibits PB formation by expression of viral ORF57. (A) GW182 and DCP1A are co-stained in the PB in BCBL-1 cells with no KSHV reactivation. BCBL-1 cells with latent KSHV infection were co-stained for GW182 (red) and DCP1A (green) using the corresponding antibodies and imaged by confocal microscopy. The nuclei were counterstained with Hoechst DNA stain. Arrows indicate the PB positively co-stained for both GW182 and DCP1A; bar = 5 μm. (B and C) Detection of PB during KSHV latent and lytic infection. (B) KSHV-infected BCBL-1 cells were induced with 1 mM valproic acid (VA, 1 mM) for lytic infection. Twenty-four hours after induction, BCBL-1 cells with (+VA) or without (-VA) virus lytic reactivation were co-immunostained for PB formation using a specific PB marker GW182 and for viral lytic protein ORF57 expression. The nuclei were counterstained with Hoechst dye. Images were captured by confocal microscopy; bar = 10 μm. (C) Number of PB in BCBL-1 cells during latent and lytic KSHV infection in the presence or absence of viral ORF57. Total of 50 cells in each group were counted in each experiment. The error bars represent SD from three independent experiments; **P < 0.01 in Student’s t-test. (D and E) ORF57, but not KSHV replication and transcription activator (RTA), suppresses PB formation during KSHV lytic infection in Bac36 cells. (D) Bac36 cells harboring a wild-type (wt) or ORF57-null KSHV genome (Δ57) (41) were induced by sodium butyrate (Bu, 3 mM). The cells with Bu induction for 24 h were then co-immunostained for the PB-specific marker GW182 and viral lytic protein ORF57 (Bac36-wt cells) or RTA (Bac36-Δ57 cells). The nuclei were counterstained with Hoechst dye. Images were obtained by confocal microscopy; bar = 10 μm. (E) Number of PB in Bac36-wt cells or Bac36-Δ57 cells during latent and lytic KSHV infection. Total of 50 cells in each group, including ORF57-positive Bac36-wt cells versus RTA-positive Bac36-Δ57 cells after lytic reactivation, were counted in each experiment. The error bars represent SD from three independent experiments; **P < 0.01 in Student’s t-test.
Viral ORF57 alone is enough to inhibit miRISC-independent PB formation
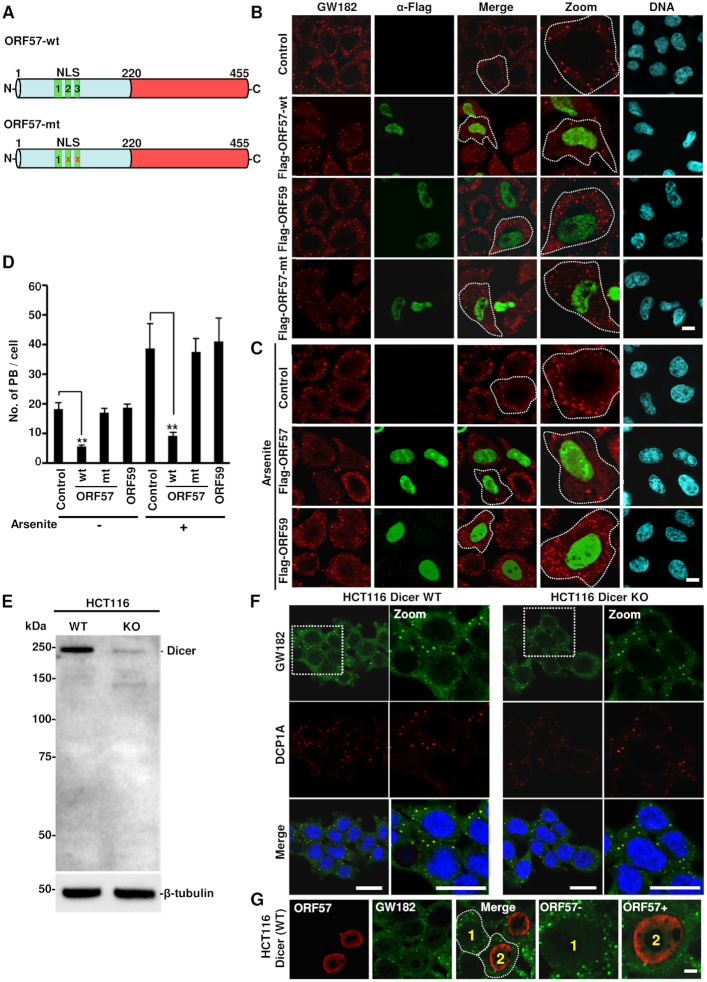
ORF57 has three nuclear localization signals (NLS) in its N-terminal IDR region that are involved in numerous ORF57 functions both in the nucleus and the cytoplasm (Figure 2A). Mutation in any two of these NLSs disrupts protein–protein interactions and renders ORF57 dysfunctional (46) (Figure 2A). To determine whether ORF57 alone is enough to block PB formation in the absence of other viral proteins, we transfected HeLa cells with a vector expressing a Flag-tagged wt ORF57, NLS2+3 mutant ORF57 (ORF57 mt), viral ORF59 (a viral nuclear DNA polymerase processivity factor) or an empty vector (control). Immunostaining with GW182 revealed a dramatic reduction in the number of PB (∼2–3 PB/cell) in ∼80% of ORF57-expressing cells over the cells expressing ORF57-mt, ORF59 or no ORF57 (∼15–20 PB/cell) (Figure 2B and D), indicating that only wt ORF57 can block the formation of PB. We also demonstrated that ORF57, but not ORF59, inhibited PB formation in arsenite-treated HeLa cells (Figure 2C and D) where more PB were induced by arsenite treatment (Figure 2D) (4). Together, these data demonstrate that ORF57 alone acts as a robust inhibitor of PB formation.
Figure 2.
ORF57 alone is enough to suppress PB formation. (A) Schematic diagrams of wild-type (wt) and mutant (mt) ORF57 protein structures. ORF57 protein composes of 455 amino acid residues and can be divided into a highly disordered N-terminal domain (light blue) and a highly structured C-terminal domain (red). The N-terminal ORF57 contains three nuclear localization signal motifs (NLS) diagrammed as NLS1, 2 and 3. The mt ORF57 contains the disrupted (x) NLS2 and NLS3 by introduction of point mutations and acts as a dysfunctional nuclear protein (46). (B) Inhibition of PB formation by ORF57 in HeLa cells. HeLa cells expressing ORF57-Flag (ORF57 wt or ORF57 mt) or ORF59-Flag for 24 h were co-immunostained with anti-human GW182 (red) and anti-Flag antibodies (green) to detect PB and viral proteins, respectively. HeLa cells transfected with an empty vector for 24 h served as control cells. The nuclei were counterstained with Hoechst; bar = 10 μm. (C) Inhibition of PB formation by ORF57 during stress. HeLa cells expressing ORF57-Flag or ORF59-Flag or an empty vector (control) for 24 h were treated with arsenite for 30 min and co-immunostained with anti-human GW182 (red) and anti-Flag (green) antibodies. The nuclei were stained with Hoechst; bar = 10 μm. (D) Quantification of PB/cell in untreated or arsenite-treated HeLa cells with or without ORF57-wt, ORF57-mt or ORF59. Total of 50 cells in each experiment were counted in three independent experiments. The error bar indicates mean ± SD. **P < 0.01 in Student’s t-test. (E andF) PB formation is independent on mature miRNAs. HCT116 cells with Dicer (wt) or without Dicer (KO) (E) as shown by Western blot were visualized by immunofluorescent staining using human GW182-specific antiserum and DCP1A antibody (F). The nuclei were stained with Hoechst; bar = 20 μm. (G) ORF57 expressed in wt HCT116 cells inhibits PB formation. HCT116 Dicer wt cells were transfected with an ORF57-Flag expression vector and 24 h after transfection co-stained with anti-GW182 antibody for the presence of PB and anti-Flag for ORF57 expression.
PB have been proposed to be the sites of mRNA decay and miRNA-mediated translational repression (8,28,29,50). Subsequently, we performed indirect immunofluorescence assay (IFA) for GW182 and DCP1A staining and examined the formation of PB in HCT116 cells with Dicer (for mature miRNA production) or without Dicer expression (no mature miRNA production) (38) (Figure 2E). As expected, many GW182-positive granules were found in Dicer-wt HCT116 cells. Surprisingly, HCT116 cells without Dicer (Dicer-KO) also displayed almost equally well in PB formation (Figure 2F). Consistently, both Dicer-wt and Dicer-KO cells showed no difference in arsenite-induced SG formation, as shown by positive TIA-1 staining (Supplementary Figure S2A). In addition, ectopic ORF57 was found to suppress PB formation in Dicer-wt HCT116 cells (Figure 2G) as seen in suppression of SG formation in both cell types (Supplementary Figure S2B and C). These data suggest that miRISC does not play an essential role in the PB formation, and therefore is not a target of ORF57 suppressive function.
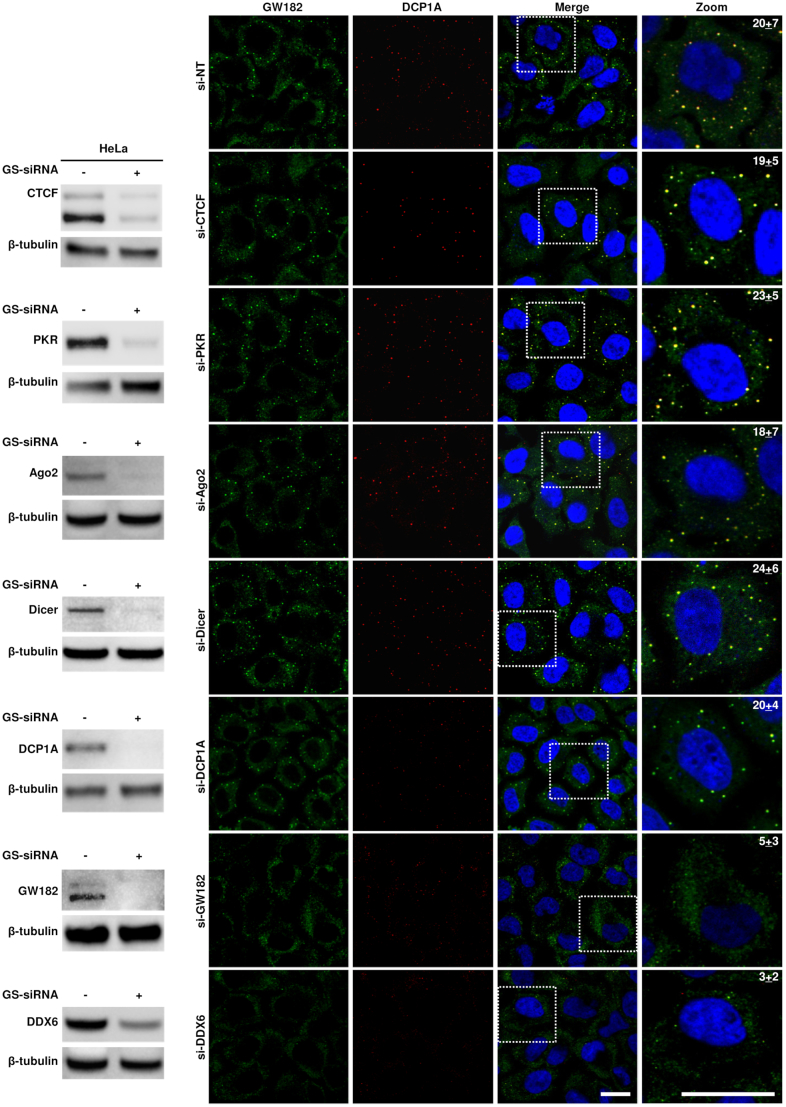
Although distinct combinations of proteins are found in PB and SG, they are dynamically linked RNA granules (1,4) and contain a subset of common protein components including Ago2, CPEB, eIF4E, TTP and other related RNA-binding proteins (51). To identify roles of the known proteins in PB formation, we further performed IFA analyses using HeLa cells with or without gene-specific siRNA knockdown of DCP1A, Dicer, Ago2, GW182 and DDX6, with CTCF and PKR as two unrelated protein controls, and examined the formation of PB by GW182 and DCP1A staining. As shown in Figure 3, only the knockdown of GW182 or DDX6 was found to block PB formation in HeLa cells and as expected, knockdown of CTCF and PKR had no effect on PB formation. Interestingly, we found that knockdown of DCP1A also did not show any effect on PB formation as seen in the knockdown of Dicer and Ago2, two major components of the miRNA pathway.
Figure 3.
Contribution of individual cellular factors to the formation of PB in HeLa cells that were treated with 40 nM of a gene-specific (GS-siRNA) or non-targeting (si-NT) siRNA. Twenty-four hours after transfection, the cells in each well were split into two wells and treated with the second round of 40 nM siRNA. The cells at 24 h after the second transfection were harvested in either 2× SDS/2-ME protein sample buffer or fixed with 4% paraformaldehyde. The knockdown efficiency of GS-siRNA (+) over si-NT (-) was determined by Western blot using corresponding antibodies with cellular β-tubulin served as a loading control. The standard IFA was carried out on fixed cells on coverslips to visualize the presence of PB using GW182 and DCP1A as the specific markers. The cells treated with si-NT served as control. The cell nuclei were counterstained with Hoechst dye. The number in a zoomed cell on the right is the number of PB per cell in a single plane image, averaged from 10 cells; bar = 20 μm.
GW182 inhibits infectious KSHV production
Mammalian PB have been viewed as an important component of host innate immunity to prevent virus replication. Several RNA viruses counteract PB formation for their productive infection (33,34). KSHV ORF57 is essential for KSHV replication and virus production (41,52). Therefore, we hypothesized that ORF57 functions to promote KSHV production by blocking PB formation in addition to blocking SG formation (48). To test this hypothesis, we first examined whether ORF57 could inhibit both PB and SG formation in KSHV-carrying iSLK/Bac16 cells (42) (Figure 4A). Subsequently, we knocked down the expression of GW182 in iSLK/Bac16 cells to disrupt host PB formation and then examined KSHV virion production from KSHV-reactivated iSLK/Bac16 cells with or without siRNA-mediated GW182 knockdown. The effect of GW182 knockdown in iSLK/Bac16 cells on infectious KSHV virion production was also compared with the effect of knocking down CTCF (a transcriptional repressor) and PKR (a protein kinase phosphorylating eIF2α to trigger SG formation), two reported regulatory host factors of KSHV replication (48,53) but not for PB formation (Figure 3). As shown in Figure 4B–D, we found that knocking down the expression of GW182, CTCF and PKR in iSLK/Bac16 cells (Figure 4B) significantly increased production of KSHV virions and led to iSLK/Bac16 culture supernatants to be highly infectious for HEK293T cells (Figure 4C). Quantitative analyses by flow cytometry indicated that siRNA knockdown of GW182 expression in the KSHV-reactivated iSLK/Bac16 cells led to a ∼100-fold increase in KSHV virion production compared to control (si-NT control), CTCF-knockdown (20-fold increase) and PKR-knockdown (64-fold increase) cells, all that have normal levels of GW182 (Figure 4D).
Figure 4.
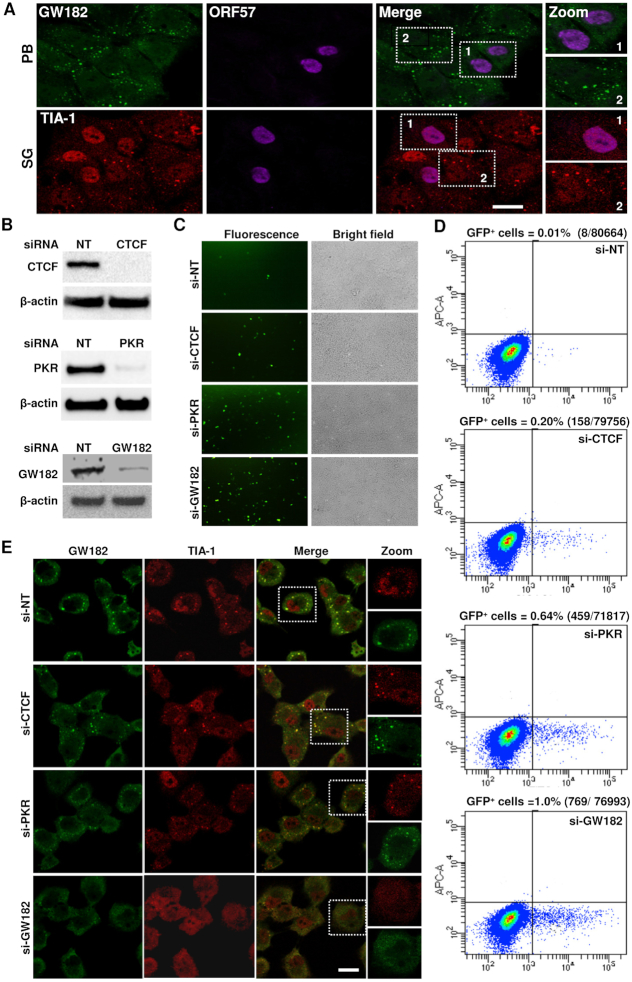
GW182 inhibits the production of infectious KSHV virions. (A) Inhibition of PB and SG formation by KSHV ORF57 in iSLK/Bac16 cells. The cells were cultivated 24 h in the presence of 1 mM sodium butyrate and 1 μg/ml doxycycline to reactivate KSHV lytic cycle before treated with 0.5 mM sodium arsenate for 30 min. The cells were co-stained for ORF57 together with the markers of PB (GW182) or SG (TIA-1); bar = 20 μm. (B) KSHV-infected iSLK/Bac16 cells were transfected twice with 40 nM of either a non-targeting siRNA (si-NT) or a siRNA specific for a transcriptional repressor CTCF (si-CTCF), SG regulator PKR (si-PKR) or PB co-factor GW182 (si-GW182) at an interval of 24 h. A fraction of cells from respective siRNA knockdown were compared for protein levels of CTCF, PKR or GW182 by immunoblotting with rabbit monoclonal anti-CTCF antibody (upper panel), mouse anti-PKR antibody (middle panel) or human anti-GW182 antiserum (bottom panel). Blots were stripped and re-probed with an anti-β-actin antibody as a loading control in each panel. (C and D) Increase of infectious KSHV virus production after siRNA knockdown of CTCF, PKR and GW182 expression in iSLK/Bac16 cells. KSHV lytic replication in iSLK/Bac16 cells with the specific siRNA knockdown (B) was induced by treatment with 1 mM sodium butyrate and 1 μg/ml doxycycline for 5 days. Supernatants containing the GFP-KSHV virus obtained from the induced cells were used to infect HEK293T cells. The infected cells after 48 h were observed for GFP expression as an indication of KSHV infection. Representative images of GFP+ cells are shown (C). Virus infected, GFP-positive HEK293T cells were quantitated by flow cytometry (D). Each transfection/induction was performed in triplicate and three replicate infections were performed with each supernatant. (E) iSLK/Bac16 cells were treated twice with 40 nM of a gene-specific or non-targeting (NT) siRNA at an interval of 24 h. The cells at 24 h after the second siRNA transfection were treated with 0.5 mM sodium arsenite for 30 min to induce stress granule formation. The fixed cells were then co-stained for GW182 and TIA-1 as markers of PB and SG, respectively, and imaged with confocal microscopy; bar = 20 μm.
To determine whether the increased production of infectious KSHV resulted from the gene-specific knockdown effect on PB formation in iSLK/Bac16 cells, we examined the cells with knockdown of individual genes by standard IFA staining for the formation of GW182-specific PB and TIA-1-specific SG. To our surprise, the majority of GW182-specific PB also contained SG-specific TIA-1 and knockdown of GW182 greatly reduced the formation of both GW182+ PB and TIA-1+ SG (Figure 4E). In contrast, knockdown of CTCF, PKR and Ago2 had no effect on PB formation as seen in control si-NT treated cells (Figure 4E and Supplementary Figure S3). Together, these data provide direct evidence that blockade of host RNA granule formation by the interaction of KSHV ORF57 with GW182 is an important viral molecular strategy to inhibit host innate immunity and allow progression of KSHV replication and productive infection.
ORF57 affects the subcellular distribution of Ago2 and induces nuclear Ago2 speckles
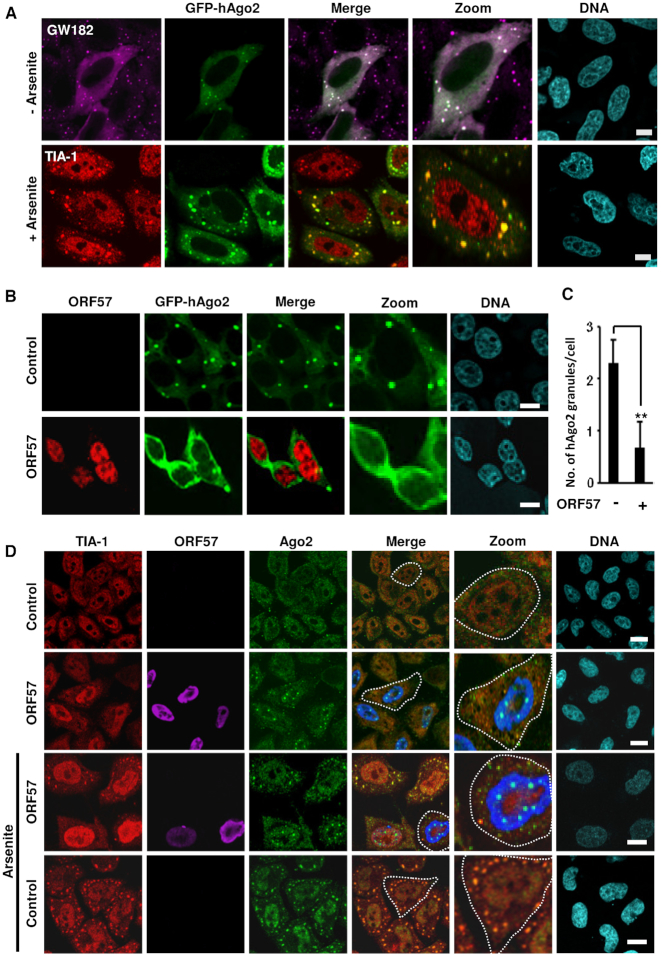
Given that ORF57 inhibits the formation of both PB and SG, two major cellular repositories for RNA granules, we next investigated whether ORF57 affects the recruitment of components common to both PB and SG. Notably, Ago2 is a common constituent of both PB and SG and has been proposed to be involved in miRNA-mediated RNA degradation in PB and RNA translation control in SG (4,9,54). First, we confirmed that transiently expressed GFP-hAgo2 colocalized in GFP-Ago2 granules with both GW182 (specific for PB) and TIA-1 (specific for SG) (Figure 5A). To address whether ORF57 directly influences Ago2 recruitment, we overexpressed GFP-hAgo2 in two cell lines, HEK293T (Figure 5B) and HeLa cells (Supplementary Figure S4A), in the absence or presence of ORF57. We found that, regardless of cell type, the GFP-hAgo2-positive granules were dramatically reduced in the presence of ORF57 expression, with GFP-hAgo2 exhibiting a homogenous cytoplasmic distribution (Figure 5B and C; Supplementary Figure S4A). Interestingly, ORF57 expression was found to induce the formation of endogenous Ago2 nuclear speckles (Figure 5D and Supplementary Figure S4B), whereas a viral DNA-binding protein ORF59 did not (Supplementary Figure S4B). The same result was observed for ORF57 in HeLa cells under arsenite-induced stress condition where ORF57 blocks SG formation (Figure 5D). Of note, ORF57 did not induce the nuclear speckles with GFP-hAgo2 (Figure 5B and Supplementary Figure S4A). Altogether, these data indicate that KSHV ORF57 interferes with distribution of both endogenous Ago2 and exogenous GFP-hAgo2 in mammalian cells. However, the distribution of the chimeric GFP-hAgo2 ectopically expressed differs from endogenous Ago2 in response to viral ORF57.
Figure 5.
Ago2 granules are positive for GW182 and TIA-1 co-staining and ORF57 interferes with Ago2 distribution. (A) Ago2 granules can be co-stained with GW182 and TIA-1. GFP-hAgo2-transfected HeLa cells were stained for PB using GW182-specific antiserum (top row) or treated with arsenite (0.5 mM for 30 min) to induce SG before staining with anti-TIA-1 (a SG-specific marker) antibody (lower row). The nuclei were stained with Hoechst dye. Images were acquired by confocal microscopy; bar = 10 μM. (B and C) ORF57 prevents the formation of GFP-hAgo2 granules in HEK293T cells. HEK293T cells were co-transfected by a vector expressing GFP-hAgo2 with an empty vector (control) or an ORF57-Flag vector. The subcellular localization of GFP-hAgo2 was examined by fluorescence microscopy, and ORF57-Flag was immunostained using anti-ORF57 antibodies (red) (B). Images were captured using confocal microscopy; bar = 10 μm. (C) Quantitative analysis of number of GFP-hAgo2 granules per cell. Total of 50 cells in (B) were counted from each experiment and the average number of GFP-hAgo2 granules per cell is shown in bar graph as a mean ± SD from three independent experiments. **P < 0.01 in Student’s t-test. (D) ORF57 induces the formation of nuclear Ago2 granules in HeLa cells with or without stress. HeLa cells transfected with a vector expressing ORF57 or an empty vector (control) were left untreated or treated with 0.5 mM sodium arsenite for 30 min. Fixed cells were co-immunostained with anti-TIA-1 (for SG), anti-Ago2 and anti-ORF57 antibodies. The nuclei were stained with Hoechst dye. Images were captured by confocal microscopy. Each image is representative of three independent experiments; bar = 10 μm.
ORF57 interacts with Ago2 and GW182, two major components of PB
To further understand how ORF57 interferes with Ago2 distribution and PB formation, we first determined whether the presence of ORF57 influenced the expression of Ago2, GW182 and DCP1A in HeLa, BCBL-1 and iSLK/Bac16 cells, and found that it did not (Supplementary Figure S5A–C). Then, we examined the interaction of ORF57 with Ago2 to determine how it might influence the required Ago2–GW182 binding for reported Ago2 functions (17,18,55). An immunoprecipitation (IP) pulldown of ORF57 in JSC-1 cells with KSHV lytic infection followed by Western blot analyses of ORF57-associated proteins showed the interaction of ORF57 with Ago2 and RNA helicase A (RHA), another PB component (56,57) (Supplementary Figure S5D). An anti-RHA IP also showed RHA interaction with Ago2 and viral ORF57 in the KSHV-infected cells (Supplementary Figure S5D). Further co-IP studies revealed that the interaction of ORF57 and Ago2 was independent of RNA and not affected by RNase digestion (Supplementary Figure S5E).
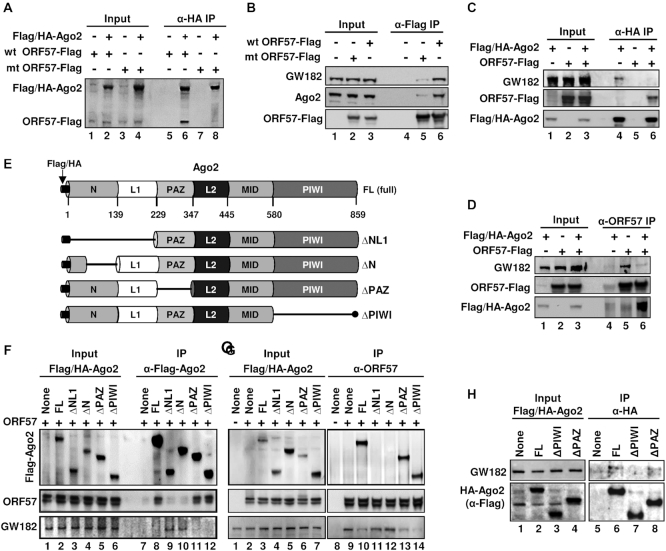
Most importantly, we found that only the wild-type (wt) ORF57, but not its N-terminal dysfunctional mutant (mt), was highly interactive with both ectopic Ago2 (Figure 6A, compare lanes 6 to 8) and endogenous Ago2 and GW182 (Figure 6B, compare lanes 6 to 5). Given that the N-terminal IDR of ORF57 interacts with both Ago2 and GW182, we then examined whether wt ORF57 might interfere with the Ago2–GW182 interaction as reported (17,18,55). As shown in Figure 6C and D, further co-IP experiments demonstrated that the Ago2–GW182 interaction is sensitive to viral ORF57. We showed that ectopic HA-Ago2 and viral ORF57 separately bind endogenous GW182 as shown by co-IP with anti-HA antibody (Figure 6C, lane 4) and anti-ORF57 antibody (Figure 6D, lane 5), but this HA-Ago2–GW182 interaction could be easily blocked in the presence of ORF57 as shown in anti-HA antibody (Figure 6C, lane 6) and anti-ORF57 co-IP experiments (Figure 6D, lane 6).
Figure 6.
Viral ORF57 interacts with GW182 and Ago2 and affects their binding activity. (A and B) ORF57-wt, but not its dysfunctional mutant (ORF57-mt), interacts with Ago2 and GW182. HEK293T cells were transfected separately with an empty vector (-) or a vector expressing Flag-tagged ORF57-wt or ORF57-mt or Flag/HA-tagged Ago2. Total cell lysate at 24 h after each transfection was mixed as indicated and digested with RNase A/T1 before co-immunoprecipitation for ORF57-associated proteins using an anti-HA antibody (A) or the cell lysates from transfection of an empty vector, Flag-ORF57-wt or Flag-ORF57-mt were digested with RNase A/T1 and co-immunoprecipitated for ORF57-associated endogenous GW182 and Ago2 using an anti-Flag antibody (B). The proteins in the co-IP were blotted for Ago2 and ORF57 by an anti-Flag antibody (A) or for GW182, Ago2 and ORF57 by the corresponding antibodies (B). (C and D) ORF57 disrupts Ago2–GW182 interaction. Total cell extract from HEK293T cells transfected by a vector expressing Flag/HA-Ago2, ORF57-Flag or an empty vector was mixed each other as indicated and then digested with RNase A/T1. The resulting protein complexes were co-immunoprecipitated by anti-HA (C) or anti-ORF57 (D) antibody-immobilized beads. The proteins in the co-IP were blotted for GW182, Flag/HA-Ago2 and ORF57 by the corresponding antibodies.(E–G) ORF57 interacts with the N-terminal domain of Ago2 and prevents Ago2 interaction with GW182. (E) Schematic diagrams (not in scale) of full-length (FL) Ago2 and its deletion mutants with the deleted region indicated by a solid line shows six characteristic domains of Ago2 (N, L1, PAZ, L2, MID and PIWI). (F and G) Mapping of ORF57-Ago2 interacting domains and their effect on Ago2–GW182 binding. HEK293T cell extract containing HA/Flag-Ago2 or its deletion mutant ΔNL1, ΔN, ΔPAZ or ΔPIWI was separately mixed with the cell extract containing untagged ORF57. The individual mixture was then digested with RNase A/T1 and immunoprecipitated using anti-Flag antibody for Ago2 (F) or anti-ORF57 antibody (G). The proteins in the co-IP were blotted for Ago2-associated ORF57 and endogenous GW182 (F) or ORF57-associated Flag/HA-Ago2 and endogenous GW182 (G) with the corresponding antibodies. A smaller band of ORF57 is the caspase-7 cleaved ORF57 (82). (H) The PIWI domain, not PAZ, of Ago2 is required to interact with GW182. HEK293T cell extract containing Flag/HA-Ago2 or its deletion mutant ΔPAZ or ΔPIWI were digested with RNase A/T1 and immunoprecipitated overnight using anti-HA-coated beads. The immunoprecipitated complexes were eluted by HA peptides and analyzed by Western blot using an anti-Flag antibody to detect Ago2 and anti-GW182 human serum to detect the endogenous GW182.
ORF57 interacts with the N-terminal domain of Ago2 to prevent Ago2–GW182 interaction
Ago2 has four major functional domains (N, PAZ, MID and PIWI) and two structural linker domains (L1 and L2) (58,59) (Figure 6E). The N-terminal domain mediates the unwinding of RNA inside Ago2 in RISC, PAZ domain binds to the miRNA 3′ overhang nucleotides, the MID domain binds to the 5′ overhang and the PIWI domain exerts endonuclease activity in addition to its important role of engaging GW182 (58,60,61). To determine the specific domain of Ago2 that interacts with ORF57, four domain-specific deletion mutants of Ago2 ΔNL1, ΔN, ΔPAZ or ΔPIWI (Figure 6E) as well as the full-length (FL) Ago2 each bearing a chimeric Flag-HA tag were individually expressed separately from untagged ORF57.
By mixing the individual Ago2 cell extract with the ORF57-containing cell extract, followed by RNase A/T1 treatment, anti-Flag (Figure 6F) or anti-ORF57 (Figure 6G) co-IP experiments showed that the full-length Ago2, ΔPAZ and ΔPIWI interacted with ORF57 (lanes 8, 11 and 12 in Figure 6F and lanes 10, 13 and 14 in Figure 6G), but the ΔNL1 and ΔN did not (lanes 9 and 10 in Figure 6F and lanes 11 and 12 in Figure 6G). Further studies showed that this ORF57–Ago2 interaction led to reduced Ago2–GW182 binding and could be confirmed by reblotting with an anti-GW182 antibody (see the corresponding lanes in Figure 6F and G). Although full-length Ago2 (Figure 6C, lane 4 and Figure 6H, lane 6), ΔNL1 and ΔN (Figure 6F, lanes 9 and 10), and ΔPAZ (Figure 6H, lane 8) interact with endogenous GW182, the ΔPIWI, as expected, was unable to interact Ago2 in a separate co-IP (Figure 6H, compare lane 7 to lanes 6 and 8). Thus, these data provide the evidence that ORF57 binding to the N-terminal domain of Ago2 prevents the Ago2–GW182 interaction, a hypothetical prerequisite for PB formation (19,55).
ORF57 interacts with the N-terminal GW-rich domain of GW182
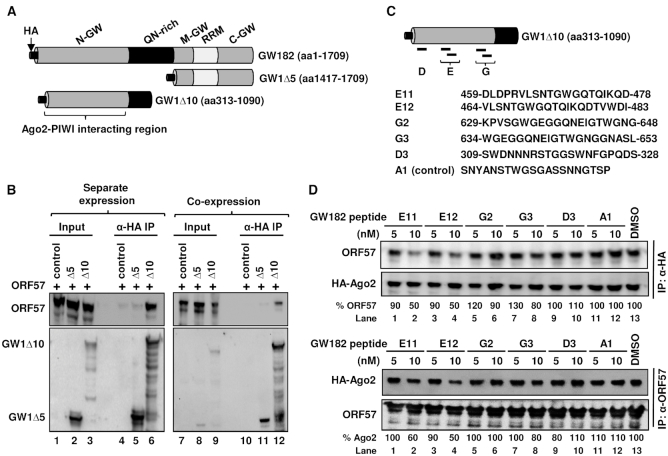
Like Ago2, human GW182 is also primarily a cytoplasmic RNA-binding protein in many continuous cell lines (54) (Supplementary Figure S5F). GW182 (TNRC6A-2) protein is a 182-kDa autoantigen composed of 1709 amino acids (16) (Supplementary Figure S6). GW182 (TNRC6A-2), TNRC6A-1 and TNRC6B-1 (Supplementary Figure S6) have also been shown to be essential for microRNA-mediated gene silencing and PB formation and contain three non-overlapping glycine and tryptophan (GW)-rich domains at the N-terminal (N), Middle (M) and C-terminal (C) regions, a glutamine/asparagine (QN)-rich domain and a RNA-recognition motif (RRM) (14,44,55,62) (Figure 7A). It is known that the PIWI domain of Ago2 (Figure 6H) interacts with GW182 at multiple GW-rich domains (19,55).
Figure 7.
ORF57 interacts with the N-terminal GW-rich domain (N-GW) of GW182 to disrupt GW182 interaction with Ago2. (A) Schematic diagram (not in scale) of GW182 (TNRC6A-2, 1709-aa) domains and two GW182 deletion mutants. (B) Mapping of ORF57–GW182 interacting domains. Total cell extract from HEK293T cells transfected with a vector expressing the HA-tagged N-terminal half of GW182 (GW1Δ10) or the C-terminal half of GW182 (GW1Δ5) or an empty vector (control) was mixed with the cell extract containing Flag-tagged ORF57 (B, left panel). Alternatively, Flag-tagged ORF57 was co-expressed in HEK293T cells with HA-tagged GW1Δ10, GW1Δ5, or an empty vector as a control (B, right panel). The cell extracts were then digested with RNase A/T1 before immunoprecipitation with an anti-HA antibody. The proteins pulled down by the co-IP were blotted for GW182-associated ORF57 by an anti-ORF57 antibody or by an anti-HA antibody for GW182. (C) Schematic illustration (not in scale) of ORF57-interacting GW1Δ10 (aa 313-1090) and its derived peptides previously shown to bind Ago2. The sequence and deduced position of each peptide (E11, E12, G2, G3 and D3) in GW182 are originated from a reported study on TNRC6B-1 (1723-aa) (44). A control peptide (A1) not binding to Ago2 (44) served as a control. (D) GW182-derived peptide competitive binding assays. HA-tagged Ago2 from HEK293T cells was immobilized on anti-HA antibody-coated beads (top panel). The HEK293T cell extract containing ORF57 was pre-mixed with individual GW182 peptides for 2 h at room temperature. This mixture was then added to the Ago2-immobilized beads and was incubated overnight at 4°C. The ORF57 protein associated with HA-ago2 beads in the pulldown assays was blotted by using an anti-ORF57 antibody and an anti-HA antibody for the constant Ago2 level (top panel). The binding of ORF57 (%) to HA-Ago2 in the presence of a GW182 peptide was measured based on the band intensity after normalized to the ORF57 level bound to HA-Ago2 in the presence of DMSO, which set as 100%. Alternatively, ORF57 from HEK293T cells was immobilized on polyclonal anti-ORF57 antibody-coated beads (lower panel). HA-Ago2 expressed in HEK293T cells was pre-mixed with the individual GW182 peptides for 2 h at room temperature. The mixture was then added to the ORF57-immobilized beads and incubated overnight at 4°C. The HA-Ago2 protein associated with ORF57 in the presence of GW182 peptides in the pulldown assays was blotted using an anti-HA antibody and the constant ORF57 level in each sample was blotted with anti-ORF57 antibody for the pulldown efficiency (lower panel). The binding of HA-Ago2 (%) to ORF57 in the presence of a GW182 peptide was measured as described above.
Knowing that ORF57 interacts with endogenous GW182 (Figure 6B, lane 6; Figure 6D, lane 5; and Figure 6G, lane 9), we set out to map the interacting domain of GW182. Flag-tagged ORF57 was expressed in HEK293T cells either separately from or co-expressed with a truncated HA-tagged GW182 containing the middle and C-terminal GW-rich domains (GW1Δ5, aa 1417-1709) or a N-terminal GW-rich (N-GW) domain plus part of the QN-rich domain (GW1Δ10, aa 313-1090). The HA-GW182 containing cell extracts were mixed with the Flag-ORF57 containing extracts in different combinations (ORF57 plus GW1Δ5 or GW1Δ10) and then immunoprecipitated by using anti-HA coated beads. As shown in Figure 7B, ORF57 was found to interact with GW1Δ10, but not with GW1Δ5 either in the mixture from the separate expression (left panel, compare lanes 6 to 5) or co-expression (right panel, compare lanes 12 to 11) conditions. These results provide the first evidence that ORF57 interacts with the N-GW domain of GW182, the same domain responsible for interacting with the Ago2 PIWI domain.
A 25-aa region in the N-GW domain of GW182 is responsible for competitive interaction with ORF57 and Ago2
Given the fact that the GW1Δ10 bearing more than 700 aa residues contains an Ago2-PIWI interacting domain (44) and the N-terminal IDR of ORF57 interaction with GW182 (Figure 6B) could be reduced in the presence of Flag/HA-Ago2 (Figure 6D, compare lanes 5 to 6; Figure 6G, compare lanes 9 to 10), we further determined that part of the N-GW domain of GW182 was responsible for ORF57 interaction by using TNRC6B-derived peptides that interact with Ago2 with high affinity (44) (Figure 7C, Supplementary Figure S6) in a co-IP based competitive binding assay. Here, HA-tagged Ago2 on the anti-HA-coated beads was incubated with a cell extract containing ORF57 already pre-mixed with individual GW182 peptides or DMSO as a control. A peptide A1 derived from TNRC6B (Figure 7C and Supplementary Figure S6) and known to not interact with Ago2 (44) served as a peptide control. We expected reduced ORF57–Ago2 interaction if a competitive GW182 peptide bound ORF57 in the pre-mixed reaction. The Ago2-associated ORF57 in the pulldowns was then blotted with anti-ORF57 antibody for ORF57 or anti-HA antibody for Ago2.
As expected, the carrier solvent (1%) DMSO and control peptide A1 had no effect on the interaction of Ago2 with ORF57 (Figure 7D top panel, lanes 11–13). However, peptides E11 and E12, when pre-mixed with ORF57, decreased the ORF57–Ago2 interaction by ∼10% at 5 nM to almost 50% at 10 nM (Figure 7D top panel, compare lanes 1–4 to lane 13). Other peptides G2, G3 and D3 that exhibit high affinity for Ago2 binding (44) did not show any obvious influence on the ORF57–Ago2 interaction at 5 nM (Figure 7D top panel, lanes 5, 7 and 9) and only weak inhibition (10–20%) at 10 nM (Figure 7D top panel, compare lanes 6 and 8 to lane 13) in this competitive binding assay.
Convincingly, in experiments where the GW182 peptides were pre-mixed with Ago2 in a cell extract before incubation with ORF57 immobilized on the anti-ORF57 beads, showed a similar 40–50% inhibitory effect of the peptides E11 and E12 at 10 nM on ORF57-Ago2 interaction (Figure 7D lower panel, compare lanes 2 and 4 to lane 13), whereas the peptide G3 at 10 nM or D3 at 5 nM displayed only ∼20% reduction (Figure 7D lower panel, compare lanes 8 and 9 to lane 13) and D3 at 10 nM exhibited no effect (Figure 7D lower panel, lane 10). These results clearly indicate that ORF57 interacts with the N-GW domain of GW182 through an aa 459–483 domain, the same GW182 region that has high binding affinity to Ago2.
ORF57 alters the scaffolding activity of GW182 at the initial stage of PB formation
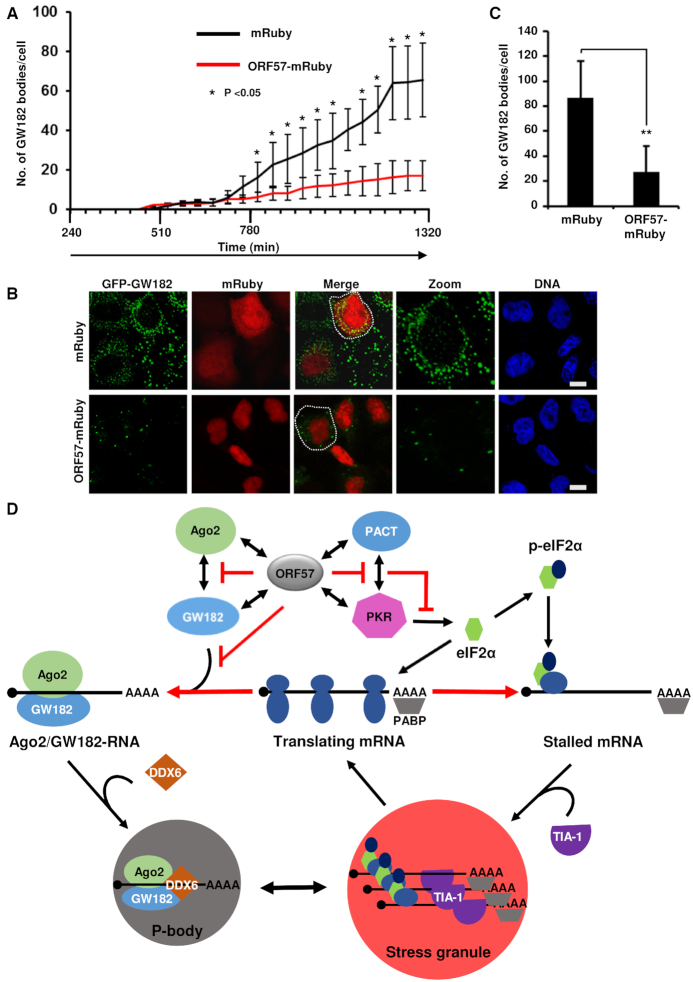
GW182 is a scaffolding protein and this activity is essential for PB formation (63). Therefore, we set out to determine the dynamics of PB formation by observing the scaffolding activity of GW182 in the presence or absence of ORF57. Time-lapse confocal microscopy of the HeLa cells co-expressing GFP-tagged GW182 (16) with an empty control vector expressing mRuby only showed that GW182 expression is localized in small puncta that can aggregate to form bigger puncta (21). As shown in Figure 8A and Supplementary Figure S7A and B, the formation of these small puncta increased over the time (60–80/per cell) and their aggregation resulted in a heterogeneous mixture of small to larger GW182-positive bodies. Interestingly, the number of these bodies was dramatically reduced to ∼10–15/cell in the presence of ORF57. A careful examination of these bodies both in living cells at 22 h post-transfection (Figure 8B) and in fixed cells at 24 h post-transfection (Figure 8C) showed that ORF57 did block the formation of the smaller puncta, but had little influence on the larger puncta that had already formed (Supplementary Videos S1 and 2). Altogether, these results indicate that, by interacting with Ago2 and GW182 and blocking Ago2–GW182 interaction (Figure 8D), ORF57 inhibits primarily the scaffolding of GW182 at the initial stage of PB formation.
Figure 8.
ORF57 prevents the early stages of PB formation by disturbing the scaffolding activity of GW182. (A) ORF57 prevents the early stages of dynamic PB formation. HeLa cells were co-transfected with a vector expressing GFP-myc-GW182 along with a vector expressing either mRuby or ORF57-mRuby. The subcellular expression of GFP-myc-GW182 (for PB), mRuby or ORF57-mRuby in alive cells was monitored every 45-min starting from 4 h to 22 h after transfection for appearance of GFP (green)- or mRuby (red)-mediated fluorescence (Supplementary Figure S7). Images were captured at each time point using a confocal time-lapse microscope attached to a CO2 incubator (Supplementary Figure S7). GW182 bodies per cell in five cells at each time point in each experiment were counted and plotted in bar graphs. The error bar indicates mean ± SD from a total of 10 cells from two successful experiments. *P < 0.05 in Student’s t-test. (B) A representative image from mRuby or ORF57-mRuby co-expression with GFP-myc-GW182 in HeLa cells was selected at 22 h after transfection; bar = 10 μm. (C) Quantification of GFP-myc-GW182-positive PB per cell in the presence of mRuby or ORF57-mRuby 24 h after co-transfection. The error bar in each group indicates mean ± SD in 50 fixed cells double positive for both mRuby and GFP from three independent experiments. **P < 0.01 in Student’s t-test. (D) A schematic model depicting the inhibitory steps of KSHV ORF57 toward the formation of PB and SG. ORF57 interacts with Ago2 and GW182 and blocks Ago2 and GW182 interaction and GW182 scaffolding activity, or interacts with PACT and PKR and blocks PACT and PKR interaction and PKR activation (48), thus suppressing the formation of both PB and SG that are interchangeable in mammalian cells. The stalled mRNAs in SG could be released for protein translation after stress relief.
Homologous HSV-1 ICP27, but not EBV EB2, exerts a similar function as KSHV ORF57 in the inhibition of PB formation
Given that HSV ICP27 and EBV EB2 (SM) proteins are homologues to KSHV ORF57, we reasoned that the ability to block PB formation by KSHV ORF57 might be a conserved function in other herpesviruses. To investigate this possibility, we expressed HSV-1 ICP27 and EBV EB2 in HeLa cells and examined their influence on PB formation. As shown in Supplementary Figure S8A and B, both ICP27 and KSHV ORF57, but not EB2 protein, inhibited PB formation. Quantitative analysis of PB counts in 50 cells with each viral protein expression indicated that both ORF57 and ICP27 expressing cells appeared an average of only ∼2 GW182-positive granules per cell, whereas the cells with EB2 expression or no viral protein expression displayed ∼20 GW182-positive granules per cell (Supplementary Figure S8B). These data indicate that ORF57 and ICP27 expression leads to at least a ∼90% reduction of PB formation and that HSV-1 ICP27, but not EBV EB2, shares this conserved function with KSHV ORF57 in blocking PB formation.
DISCUSSION
PB are constitutively present in cells and increase in sizes and numbers when translational arrest occurs. Various studies indicate that PB play important roles in RNA surveillance, RNA decay, miRNA-mediated silencing and translational control (3,64). In this report, the miRISC-dependent PB formation could not be further confirmed either by using Dicer-KO HCT116 cells or siRNA knockdown of Dicer and Ago2 expression in HeLa and iSLK/Bac16 cells. We showed that HCT116 Dicer-KO cells deficient in production of mature miRNAs (38) exhibit no difference from the wt Dicer cells in PB formation. Knockdown of Dicer, Ago2 and DCP1A expression in HeLa cells and Ago2 expression in iSLK/Bac16 cells also exhibited no effect on PB formation. In contrast, we found that GW182 and DDX6 are critical and knockdown of GW182 and DDX6 expression led to severe reduction in PB formation. Our time-lapse confocal microscopy showed that GW182 is essential for PB formation by acting as a scaffolding protein, as reported (63).
Despite that Ago2 and DCP1A appear non-essential in PB formation, the interaction of GW182 with Ago2 is considered to dock miRNA-bound Ago2, stabilize Ago2-miRNA interactions and thereby maintain the presence of Ago2 in the cytoplasmic PB (63). This takes place through the N-GW domain of GW182 binding to the three evenly spaced tryptophan-binding pockets in the PIWI domain of Ago2 (44,65). We confirmed that deletion of the PIWI domain from Ago2 prevents Ago2 interaction with GW182. Moreover, we found that the N-terminal IDR region of KSHV ORF57 binds to both the N-terminal domain of Ago2 and the N-GW region (aa 459–483) of GW182 in the context of other domains, thereby preventing the GW182–Ago2 interaction and the scaffold activity of GW182 (Figure 8D). This viral strategy to disrupt PB formation is similar to KSHV inhibition of SG formation by viral ORF57 interacting with PKR and PACT (48) (Figure 8D).
Host PB have been viewed as macromolecular machinery used by the innate immune system to protect against virus infection (66). Disruption of PB formation by RNA viruses has been reported for poliovirus through viral 3C proteinase-mediated degradation of host DCP1A (67), hantavirus by viral N protein binding to DCP1A (68), rotavirus by its NSP1-mediated degradation of PAN3 (69), West Nile virus through recruitment of GW182 and DCP1A to viral replication site (70), and hepatitis C virus by relocation of PB components to lipid droplet (71–74). The finding that DCP1A appears non-essential for PB formation in our report indicates that more studies are needed on how virus-induced degradation of DCP1A could inhibit PB formation. Moreover, PB formation was also found to be unrelated to DCP2 protein level (75).
In this report, KSHV was discovered to inhibit PB formation for its replication by encoding a viral ORF57 to block GW182’s scaffolding activities. By analogy, disruption of PB formation by knocking down GW182 expression in KSHV-infected cells was found to increase KSHV production by ∼100-fold, further signifying the important roles of host RNA granules restricting productive KSHV infection (48). We noticed that knockdown of GW182 expression affects not only the formation of PB, but also SG in iSLK/Bac16 cells expressing only a minimal amount of GW182. Interestingly, we found that ORF57 homolog HSV-1 ICP27, but not EBV EB2, exerts a similar function to ORF57 in the inhibition of PB formation. The molecular mechanism by which three homologs in the herpesvirus family exhibit a functional disparity on PB inhibition remains to be determined.
Both GW182 and Ago2 are nuclear-cytoplasmic shuttling proteins (54,76). Ago2 could be primarily nuclear or cytoplasmic, depending on types of cells and tissues (43). The subcellular localization of GW182 is executed by a nuclear export signal (NES) and a nuclear localization signal (NLS) (54), whereas Ago2 lacks a NLS and can be also transported into the nucleus by importin 8 when loaded with miRNA (76,77). In this study, we found that ORF57 interacts with Ago2 to promote accumulation of endogenous Ago2, but not the ectopic GFP-hAgo2 or Flag/HA-Ago2, in the nucleoplasm as speckles in HeLa cells. A similar finding to Ago2 was reported for the TNRC6A, by which TNRC6A transports Ago2 protein into the nucleus through its Ago2-interacting motif(s) (21). The induction and functions of these nuclear Ago2 speckles by KSHV ORF57 remain to be investigated.
By following the liquid–liquid phase separation model for RNP granule assembly (78), a recent report (65) indicated that miRNA-bound Ago2 and GW182 could be condensed into phase-separated droplets. The condensation of miRISC droplets led to RNP granule assembly (79,80). However, our results that both Ago2 and Dicer are non-essential for PB formation support a miRISC-independent model of PB formation. KSHV ORF57 bound to its RNA targets and interacts with RNA-binding proteins through its N-terminal IDR domain (35,36). By extrapolation, ORF57 inhibits the early stage of GW182 granule formation primarily by its IDR domain interacting with GW182 and thus probably by preventing droplet formation in the absence of miRNA. Once the droplet is formed or at the late stage of PB assembly, it may escape from ORF57’s inhibitory activity. Whether these escaped GW182-granules are a separate class of RNA granules as described (23,81) remain to be determined.
In conclusion, this study shows that PB assembly in mammalian cells is miRISC-independent. KSHV inhibits PB formation by viral ORF57 interacting with Ago2 and GW182 in lytic infection, leading to virus multiplication.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Bert Vogelstein for Dicer wt and knockout HCT116 cell lines, Dr Jae Jung for iSLK2/Bac16 cells, Rozanne Sandri-Goldin for HSV-1 ICP27 and Sankar Swaminathan for the EBV EB2 expression vectors.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Cancer Institute, National Institutes of Health [ZIA SC 010357 to Z.M.Z.]. Funding for open access charge: Intramural Research Budget [ZIA SC 010357].
Conflict of interest statement. None declared.
REFERENCES
- 1. Buchan J.R., Parker R.. Eukaryotic stress granules: the ins and outs of translation. Mol. Cell. 2009; 36:932–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Stoecklin G., Kedersha N.. Relationship of GW/P-bodies with stress granules. Adv. Exp. Med. Biol. 2013; 768:197–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Luo Y., Na Z., Slavoff S.A.. P-bodies: composition, properties, and functions. Biochemistry. 2018; 57:2424–2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kedersha N., Stoecklin G., Ayodele M., Yacono P., Lykke-Andersen J., Fritzler M.J., Scheuner D., Kaufman R.J., Golan D.E., Anderson P.. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 2005; 169:871–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zheng D., Ezzeddine N., Chen C.Y., Zhu W., He X., Shyu A.B.. Deadenylation is prerequisite for P-body formation and mRNA decay in mammalian cells. J. Cell Biol. 2008; 182:89–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Aizer A., Brody Y., Ler L.W., Sonenberg N., Singer R.H., Shav-Tal Y.. The dynamics of mammalian P body transport, assembly, and disassembly in vivo. Mol. Biol. Cell. 2008; 19:4154–4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen C.Y., Shyu A.B.. Deadenylation and P-bodies. Adv. Exp. Med. Biol. 2013; 768:183–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liu J., Valencia-Sanchez M.A., Hannon G.J., Parker R.. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat. Cell Biol. 2005; 7:719–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Leung A.K., Calabrese J.M., Sharp P.A.. Quantitative analysis of Argonaute protein reveals microRNA-dependent localization to stress granules. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:18125–18130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mollet S., Cougot N., Wilczynska A., Dautry F., Kress M., Bertrand E., Weil D.. Translationally repressed mRNA transiently cycles through stress granules during stress. Mol. Biol. Cell. 2008; 19:4469–4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Anderson P., Kedersha N.. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat. Rev. Mol. Cell Biol. 2009; 10:430–436. [DOI] [PubMed] [Google Scholar]
- 12. Moon S.L., Morisaki T., Khong A., Lyon K., Parker R., Stasevich T.J.. Multicolour single-molecule tracking of mRNA interactions with RNP granules. Nat. Cell Biol. 2019; 21:162–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bashkirov V.I., Scherthan H., Solinger J.A., Buerstedde J.M., Heyer W.D.. A mouse cytoplasmic exoribonuclease (mXRN1p) with preference for G4 tetraplex substrates. J. Cell Biol. 1997; 136:761–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Eystathioy T., Chan E.K., Tenenbaum S.A., Keene J.D., Griffith K., Fritzler M.J.. A phosphorylated cytoplasmic autoantigen, GW182, associates with a unique population of human mRNAs within novel cytoplasmic speckles. Mol. Biol. Cell. 2002; 13:1338–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ding L., Han M.. GW182 family proteins are crucial for microRNA-mediated gene silencing. Trends Cell Biol. 2007; 17:411–416. [DOI] [PubMed] [Google Scholar]
- 16. Eulalio A., Tritschler F., Izaurralde E.. The GW182 protein family in animal cells: new insights into domains required for miRNA-mediated gene silencing. RNA. 2009; 15:1433–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu J., Rivas F.V., Wohlschlegel J., Yates J.R. III, Parker R., Hannon G.J.. A role for the P-body component GW182 in microRNA function. Nat. Cell Biol. 2005; 7:1261–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jakymiw A., Lian S., Eystathioy T., Li S., Satoh M., Hamel J.C., Fritzler M.J., Chan E.K.. Disruption of GW bodies impairs mammalian RNA interference. Nat. Cell Biol. 2005; 7:1267–1274. [DOI] [PubMed] [Google Scholar]
- 19. Lian S.L., Li S., Abadal G.X., Pauley B.A., Fritzler M.J., Chan E.K.. The C-terminal half of human Ago2 binds to multiple GW-rich regions of GW182 and requires GW182 to mediate silencing. RNA. 2009; 15:804–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Eulalio A., Helms S., Fritzsch C., Fauser M., Izaurralde E.. A C-terminal silencing domain in GW182 is essential for miRNA function. RNA. 2009; 15:1067–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nishi K., Nishi A., Nagasawa T., Ui-Tei K.. Human TNRC6A is an Argonaute-navigator protein for microRNA-mediated gene silencing in the nucleus. RNA. 2013; 19:17–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Meister G., Landthaler M., Peters L., Chen P.Y., Urlaub H., Luhrmann R., Tuschl T.. Identification of novel argonaute-associated proteins. Curr. Biol. 2005; 15:2149–2155. [DOI] [PubMed] [Google Scholar]
- 23. Ayache J., Benard M., Ernoult-Lange M., Minshall N., Standart N., Kress M., Weil D.. P-body assembly requires DDX6 repression complexes rather than decay or Ataxin2/2L complexes. Mol. Biol. Cell. 2015; 26:2579–2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wilczynska A., Aigueperse C., Kress M., Dautry F., Weil D.. The translational regulator CPEB1 provides a link between dcp1 bodies and stress granules. J. Cell Sci. 2005; 118:981–992. [DOI] [PubMed] [Google Scholar]
- 25. Fenger-Gron M., Fillman C., Norrild B., Lykke-Andersen J.. Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol. Cell. 2005; 20:905–915. [DOI] [PubMed] [Google Scholar]
- 26. Andrei M.A., Ingelfinger D., Heintzmann R., Achsel T., Rivera-Pomar R., Luhrmann R.. A role for eIF4E and eIF4E-transporter in targeting mRNPs to mammalian processing bodies. RNA. 2005; 11:717–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ferraiuolo M.A., Basak S., Dostie J., Murray E.L., Schoenberg D.R., Sonenberg N.. A role for the eIF4E-binding protein 4E-T in P-body formation and mRNA decay. J. Cell Biol. 2005; 170:913–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Eulalio A., Behm-Ansmant I., Schweizer D., Izaurralde E.. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol. Cell Biol. 2007; 27:3970–3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Parker R., Sheth U.. P bodies and the control of mRNA translation and degradation. Mol. Cell. 2007; 25:635–646. [DOI] [PubMed] [Google Scholar]
- 30. Horvathova I., Voigt F., Kotrys A.V., Zhan Y., Artus-Revel C.G., Eglinger J., Stadler M.B., Giorgetti L., Chao J.A.. The dynamics of mRNA turnover revealed by single-molecule imaging in single cells. Mol. Cell. 2017; 68:615–625. [DOI] [PubMed] [Google Scholar]
- 31. Pitchiaya S., Mourao M.D.A., Jalihal A.P., Xiao L., Jiang X., Chinnaiyan A.M., Schnell S., Walter N.G.. Dynamic recruitment of single RNAs to processing bodies depends on RNA functionality. Mol. Cell. 2019; 74:521–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Reineke L.C., Lloyd R.E.. Diversion of stress granules and P-bodies during viral infection. Virology. 2013; 436:255–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lloyd R.E. Regulation of stress granules and P-bodies during RNA virus infection. Wiley. Interdiscip. Rev. RNA. 2013; 4:317–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang Q., Sharma N.R., Zheng Z.M., Chen M.. Viral regulation of RNA granules in infected cells. Virol. Sin. 2019; 34:175–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Majerciak V., Zheng Z.M.. KSHV ORF57, a protein of many faces. Viruses. 2015; 7:604–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Majerciak V., Pripuzova N., Chan C., Temkin N., Specht S.I., Zheng Z.M.. Stability of structured Kaposi's sarcoma-associated herpesvirus ORF57 protein is regulated by protein phosphorylation and homodimerization. J. Virol. 2015; 89:3256–3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yuan F., Gao Z.Q., Majerciak V., Bai L., Hu M.L., Lin X.X., Zheng Z.M., Dong Y.H., Lan K.. The crystal structure of KSHV ORF57 reveals dimeric active sites important for protein stability and function. PLoS Pathog. 2018; 14:e1007232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cummins J.M., He Y., Leary R.J., Pagliarini R., Diaz L.A. Jr, Sjoblom T., Barad O., Bentwich Z., Szafranska A.E., Labourier E. et al.. The colorectal microRNAome. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:3687–3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Renne R., Zhong W., Herndier B., McGrath M., Abbey N., Kedes D., Ganem D.. Lytic growth of Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat. Med. 1996; 2:342–346. [DOI] [PubMed] [Google Scholar]
- 40. Cannon J.S., Ciufo D., Hawkins A.L., Griffin C.A., Borowitz M.J., Hayward G.S., Ambinder R.F.. A new primary effusion lymphoma-derived cell line yields a highly infectious Kaposi’s sarcoma herpesvirus-containing supernatant. J. Virol. 2000; 74:10187–10193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Majerciak V., Pripuzova N., McCoy J.P., Gao S.J., Zheng Z.M.. Targeted disruption of Kaposi's sarcoma-associated herpesvirus ORF57 in the viral genome is detrimental for the expression of ORF59, K8alpha, and K8.1 and the production of infectious virus. J. Virol. 2007; 81:1062–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Brulois K.F., Chang H., Lee A.S., Ensser A., Wong L.Y., Toth Z., Lee S.H., Lee H.R., Myoung J., Ganem D. et al.. Construction and manipulation of a new Kaposi's sarcoma-associated herpesvirus bacterial artificial chromosome clone. J. Virol. 2012; 86:9708–9720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sharma N.R., Wang X., Majerciak V., Ajiro M., Kruhlak M., Meyers C., Zheng Z.M.. Cell type- and tissue context-dependent nuclear distribution of human Ago2. J. Biol. Chem. 2016; 291:2302–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pfaff J., Hennig J., Herzog F., Aebersold R., Sattler M., Niessing D., Meister G.. Structural features of Argonaute-GW182 protein interactions. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:E3770–E3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kang J.G., Pripuzova N., Majerciak V., Kruhlak M., Le S.Y., Zheng Z.M.. Kaposi’s sarcoma-associated herpesvirus ORF57 promotes escape of viral and human interleukin-6 from microRNA-mediated suppression. J. Virol. 2011; 85:2620–2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Majerciak V., Yamanegi K., Nie S.H., Zheng Z.M.. Structural and functional analyses of Kaposi sarcoma-associated herpesvirus ORF57 nuclear localization signals in living cells. J. Biol. Chem. 2006; 281:28365–28378. [DOI] [PubMed] [Google Scholar]
- 47. Massimelli M.J., Majerciak V., Kruhlak M., Zheng Z.M.. Interplay between polyadenylate-binding protein 1 and Kaposi's sarcoma-associated herpesvirus ORF57 in accumulation of polyadenylated nuclear RNA, a viral long noncoding RNA. J. Virol. 2013; 87:243–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sharma N.R., Majerciak V., Kruhlak M.J., Zheng Z.M.. KSHV inhibits stress granule formation by viral ORF57 blocking PKR activation. PLoS Pathog. 2017; 13:e1006677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kang J.G., Majerciak V., Uldrick T.S., Wang X., Kruhlak M., Yarchoan R., Zheng Z.M.. Kaposi’s sarcoma-associated herpesviral IL-6 and human IL-6 open reading frames contain miRNA binding sites and are subject to cellular miRNA regulation. J. Pathol. 2011; 225:378–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Aizer A., Kalo A., Kafri P., Shraga A., Ben-Yishay R., Jacob A., Kinor N., Shav-Tal Y.. Quantifying mRNA targeting to P-bodies in living human cells reveals their dual role in mRNA decay and storage. J. Cell Sci. 2014; 127:4443–4456. [DOI] [PubMed] [Google Scholar]
- 51. Kedersha N., Anderson P.. Mammalian stress granules and processing bodies. Methods Enzymol. 2007; 431:61–81. [DOI] [PubMed] [Google Scholar]
- 52. Han Z., Swaminathan S.. Kaposi’s sarcoma-associated herpesvirus lytic gene ORF57 is essential for infectious virion production. J. Virol. 2006; 80:5251–5260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Li D.J., Verma D., Mosbruger T., Swaminathan S.. CTCF and Rad21 act as host cell restriction factors for Kaposi's sarcoma-associated herpesvirus (KSHV) lytic replication by modulating viral gene transcription. PLoS Pathog. 2014; 10:e1003880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Nishi K., Takahashi T., Suzawa M., Miyakawa T., Nagasawa T., Ming Y., Tanokura M., Ui-Tei K.. Control of the localization and function of a miRNA silencing component TNRC6A by Argonaute protein. Nucleic Acids Res. 2015; 43:9856–9873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yao B., Li S., Lian S.L., Fritzler M.J., Chan E.K.. Mapping of Ago2-GW182 functional interactions. Methods Mol. Biol. 2011; 725:45–62. [DOI] [PubMed] [Google Scholar]
- 56. Robb G.B., Rana T.M.. RNA helicase A interacts with RISC in human cells and functions in RISC loading. Mol. Cell. 2007; 26:523–537. [DOI] [PubMed] [Google Scholar]
- 57. Carroll J.S., Munchel S.E., Weis K.. The DExD/H box ATPase Dhh1 functions in translational repression, mRNA decay, and processing body dynamics. J. Cell Biol. 2011; 194:527–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Schirle N.T., MacRae I.J.. The crystal structure of human Argonaute2. Science. 2012; 336:1037–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kidwell M.A., Doudna J.A.. Activating silent argonautes. Nat. Struct. Mol. Biol. 2013; 20:769–771. [DOI] [PubMed] [Google Scholar]
- 60. Meister G. Argonaute proteins: functional insights and emerging roles. Nat. Rev. Genet. 2013; 14:447–459. [DOI] [PubMed] [Google Scholar]
- 61. Ye Z., Jin H., Qian Q.. Argonaute 2: a novel rising star in cancer research. J. Cancer. 2015; 6:877–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yao B., Li S., Jung H.M., Lian S.L., Abadal G.X., Han F., Fritzler M.J., Chan E.K.. Divergent GW182 functional domains in the regulation of translational silencing. Nucleic Acids Res. 2011; 39:2534–2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Braun J.E., Huntzinger E., Izaurralde E.. The role of GW182 proteins in miRNA-mediated gene silencing. Adv. Exp. Med. Biol. 2013; 768:147–163. [DOI] [PubMed] [Google Scholar]
- 64. Chan E.K., Fritzler M.J.. Ten Years of Progress in GW/P Body Research. 2013; NY: Springer. [Google Scholar]
- 65. Sheu-Gruttadauria J., MacRae I.J.. Phase transitions in the assembly and function of human miRISC. Cell. 2018; 173:946–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Beckham C.J., Parker R.. P bodies, stress granules, and viral life cycles. Cell Host. Microbe. 2008; 3:206–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Dougherty J.D., White J.P., Lloyd R.E.. Poliovirus-mediated disruption of cytoplasmic processing bodies. J. Virol. 2011; 85:64–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Mir M.A., Duran W.A., Hjelle B.L., Ye C., Panganiban A.T.. Storage of cellular 5′ mRNA caps in P bodies for viral cap-snatching. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:19294–19299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Bhowmick R., Mukherjee A., Patra U., Chawla-Sarkar M.. Rotavirus disrupts cytoplasmic P bodies during infection. Virus Res. 2015; 210:344–354. [DOI] [PubMed] [Google Scholar]
- 70. Chahar H.S., Chen S., Manjunath N.. P-body components LSM1, GW182, DDX3, DDX6 and XRN1 are recruited to WNV replication sites and positively regulate viral replication. Virology. 2013; 436:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ariumi Y., Kuroki M., Kushima Y., Osugi K., Hijikata M., Maki M., Ikeda M., Kato N.. Hepatitis C virus hijacks P-body and stress granule components around lipid droplets. J. Virol. 2011; 85:6882–6892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Perez-Vilaro G., Scheller N., Saludes V., Diez J.. Hepatitis C virus infection alters P-body composition but is independent of P-body granules. J. Virol. 2012; 86:8740–8749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Perez-Vilaro G., Fernandez-Carrillo C., Mensa L., Miquel R., Sanjuan X., Forns X., Perez-del-Pulgar S., Diez J.. Hepatitis C virus infection inhibits P-body granule formation in human livers. J. Hepatol. 2015; 62:785–790. [DOI] [PubMed] [Google Scholar]
- 74. Biegel J.M., Pager C.T.. Hepatitis C virus exploitation of processing bodies. J. Virol. 2016; 90:4860–4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Jang G.J., Yang J.Y., Hsieh H.L., Wu S.H.. Processing bodies control the selective translation for optimal development of Arabidopsis young seedlings. Proc. Natl. Acad. Sci. U.S.A. 2019; 116:6451–6456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Weinmann L., Hock J., Ivacevic T., Ohrt T., Mutze J., Schwille P., Kremmer E., Benes V., Urlaub H., Meister G.. Importin 8 is a gene silencing factor that targets argonaute proteins to distinct mRNAs. Cell. 2009; 136:496–507. [DOI] [PubMed] [Google Scholar]
- 77. Wei Y., Li L., Wang D., Zhang C.Y., Zen K.. Importin 8 regulates the transport of mature microRNAs into the cell nucleus. J. Biol. Chem. 2014; 289:10270–10275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Lin Y., Protter D.S., Rosen M.K., Parker R.. Formation and maturation of Phase-Separated liquid droplets by RNA-Binding proteins. Mol. Cell. 2015; 60:208–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Jonas S., Izaurralde E.. The role of disordered protein regions in the assembly of decapping complexes and RNP granules. Genes Dev. 2013; 27:2628–2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Protter D.S.W., Rao B.S., Van T.B., Lin Y., Mizoue L., Rosen M.K., Parker R.. Intrinsically disordered regions can contribute promiscuous interactions to RNP granule assembly. Cell Rep. 2018; 22:1401–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Hubstenberger A., Courel M., Benard M., Souquere S., Ernoult-Lange M., Chouaib R., Yi Z., Morlot J.B., Munier A., Fradet M. et al.. P-Body purification reveals the condensation of repressed mRNA regulons. Mol. Cell. 2017; 68:144–157. [DOI] [PubMed] [Google Scholar]
- 82. Majerciak V., Kruhlak M., Dagur P.K., McCoy J.P. Jr, Zheng Z.M.. Caspase-7 cleavage of Kaposi sarcoma-associated herpesvirus ORF57 confers a cellular function against viral lytic gene expression. J. Biol. Chem. 2010; 285:11297–11307. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.