Key Points
AhR activation leads to induction of MDSCs with immunosuppressive activity.
AhR-mediated induction of MDSCs is regulated by microRNA that target CXCR2.
Abstract
The compound 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD), an environmental contaminant, is a potent ligand for aryl hydrocarbon receptor (AhR). In the current study, we made an exciting observation that naive C57BL/6 mice that were exposed i.p. to TCDD showed massive mobilization of myeloid-derived suppressor cells (MDSCs) in the peritoneal cavity. These MDSCs were highly immunosuppressive and attenuated Con A–induced hepatitis upon adoptive transfer. TCDD administration in naive mice also led to induction of several chemokines and cytokines in the peritoneal cavity and serum (CCL2, CCL3, CCL4, CCL11, CXCL1, CXCL2, CXCL5, CXCL9, G-CSF, GM-CSF, VEGF, and M-CSF) and chemokine receptors on MDSCs (CCR1, CCR5, and CXCR2). Treatment with CXCR2 or AhR antagonist in mice led to marked reduction in TCDD-induced MDSCs. TCDD-induced MDSCs had high mitochondrial respiration and glycolytic rate and exhibited differential microRNA (miRNA) expression profile. Specifically, there was significant downregulation of miR-150-5p and miR-543-3p. These two miRNAs targeted and enhanced anti-inflammatory and MDSC-regulatory genes, including IL-10, PIM1, ARG2, STAT3, CCL11 and its receptors CCR3 and CCR5 as well as CXCR2. The role of miRs in MDSC activation was confirmed by transfection studies. Together, the current study demonstrates that activation of AhR in naive mice triggers robust mobilization of MDSCs through induction of chemokines and their receptors and MDSC activation through regulation of miRNA expression. AhR ligands include diverse compounds from environmental toxicants, such as TCDD, that are carcinogenic to dietary indoles that are anti-inflammatory. Our studies provide new insights on how such ligands may regulate health and disease through induction of MDSCs.
Introduction
The compound 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) is a halogenated aromatic hydrocarbon found in the environment as a contaminant with immunotoxic and carcinogenic properties. It is well characterized for its ability to act as a high-affinity ligand for aryl hydrocarbon receptor (AhR), a member of the basic helix-loop-helix/Per–AhR nuclear translocator protein (Arnt)–Sim (bHLH/PAS) family of transcription factors. In fact, AhR is required for induction of toxicity inasmuch as mice deficient in AhR are mostly resistant to TCDD-mediated toxicity (1). More recent studies have demonstrated that AhR may also play a crucial role in regulating various physiological and developmental processes, including the functions of the immune system (2).
AhR is localized in the cytoplasm and maintained in an inactive form by chaperone proteins. When TCDD binds to AhR, the chaperone proteins are released and the AhR–TCDD complex binds to the transcription factor Arnt. This complex (AhR–TCDD–Arnt) migrates to the nucleus and binds to specific DNA sequences called dioxin response elements (DREs), found on the promoters of various genes, thereby regulating the expression of these genes, such as CYP1A1 (3).
Extensive studies have shown that the immune system is one of most sensitive targets of TCDD. Multiple mechanistic pathways have been identified to delineate how AhR activation leads to regulation of the immune system. These include but are not limited to the following: activation of Fas, which expresses DREs, leading to induction of apoptosis of activated T cells (2, 4–6); induction of Foxp3 and regulatory T cells (Tregs) by virtue of the fact that Foxp3 expresses DREs on its promoter (3, 7), promoting Tregs while suppressing Th17 cells through decreased methylation of CpG islands of Foxp3 and increased methylation of IL-17 promoter (4, 8); and differential microRNA (miRNA) induction, such as decreased expression of miR-31, miR-219, and miR-490 that targeted Foxp3 and increased expression of miR-495 and miR-1192 that were specific to IL-17 (9).
AhR ligands also include dietary compounds, such as indole-3-carbinol (I3C), 3,3′-diindolylmethane (DIM), and resveratrol (10–14). The essential amino acid tryptophan, acquired from the diet, also serves as a source of AhR ligands (15). It is interesting to note that whereas some AhR ligands are considered to be highly toxic and carcinogenic, others constitute ligands that are endogenously produced or found in the diet that regulate immune response in health and disease. Also, whereas some AhR ligands, such as TCDD, are known to trigger Tregs, others such as 6-formylindolo[3,2-b]carbazole (FICZ) induce Th17 cells (7, 9). Thus, the precise mechanisms through which AhR ligands regulate the immune response needs further investigation.
Myeloid-derived suppressor cells (MDSCs) are heterogeneous populations derived from bone marrow (BM) and comprised of myeloid progenitors that under normal conditions differentiate into dendritic cells, granulocytes, and macrophages (16). In situations involving chronic infection, inflammation, trauma, or malignancy, the associated chemokines and cytokines induce abnormal accumulation of such immature myeloid cells that are highly immunosuppressive (17). In mice, these cells express CD11b+ and Gr-1+ surface markers (18, 19). MDSCs have contradictory roles in infection and immunity. They may act as a double-edged sword during the early and late stages of infection and inflammation, from promoting innate immunity in early stages to attenuating the immune system through inducing immunosuppressive conditions in late stages of infection (20). In some cases, increasing MDSCs during infection helps to reduce inflammatory extension and limit undesirable tissue damages (21). In cancers, MDSC accumulation can prevent anticancer immunity, thereby facilitating the tumor growth. MDSCs have also been shown to induce Tregs (22).
Because AhR activation by TCDD in naive mice can lead to immunosuppression and promote cancer. In the current study, we investigated whether TCDD can induce MDSCs in naive mice. We found that AhR activation by TCDD led to massive induction of MDSCs that were highly immunosuppressive. Furthermore, such an induction and activation of MDSCs resulted from production of chemokines and cytokines as well as regulation of miRNA. The current study demonstrates how AhR activation may regulate immune response through induction of MDSCs.
Materials and Methods
Experimental animals
Female C57BL/6 mice 8–10 wk old were purchased from The Jackson Laboratory. All mice were housed in specific pathogen–free conditions at the Association for Assessment and Accreditation of Laboratory Animal Care International–accredited University of South Carolina, School of Medicine, Animal Resource Facility. All experiments performed using mice in this manuscript were approved by the Institutional Animal Care and Use Committee, University of South Carolina.
Chemicals and reagents
TCDD was a kind gift from Dr. Steve Safe (Institute of Biosciences & Technology, Texas A&M Health Sciences Center, College Station, TX). 3-Methylcholanthrene (3-MC) was from Sigma-Aldrich. Con A, AhR antagonist (CH223191), and CXR2 antagonist (Sch527123) were purchased from Sigma-Aldrich. Culture medium reagents (RPMI 1640, penicillin–streptomycin, HEPES, l-Glutamine, FBS, and PBS) were purchased from Invitrogen Life Technologies (Carlsbad, CA). The following Abs were used for surface markers and/or intranuclear staining and were purchased from BioLegend (San Diego, CA): FITC or Alexa Fluor 700–conjugated anti-CD11b, PE- or BV510-conjugated-GR-1, Alexa Fluor 488–conjugated anti–Ly-6C, BV785-conjugated anti–Ly-6G, PE- or BV785-conjugated anti-CD4, PE-conjugated anti-CD3, and BV510-conjugated anti-NrP1. PE-conjugated anti–IL-17, BV605-conjugated anti–IL-10, BV650-conjugated anti–INF-γ, PerCP-Cy5.5–conjugated anti–TGF-β, Alexa Fluor 488–conjugated anti–IL-4, Alexa Fluor 488–conjugated anti-FOXP3, PE–Dazzle–conjugated anti-Helios, FITC-conjugated BV421-conjugated anti-MHC class II (MHC-II), BV605 or PE-conjugated anti-CD11c, PE-conjugated anti-Arg1, and allophycocyanin-conjugated anti-iNOS. Fc Blocker reagent was procured from BD Biosciences (San Diego). A Cytofix/Cytoperm Fixation/Permeabilization Kit was purchased from BD Biosciences. The True-Nuclear Transcription Factor Buffer Set was from BioLegend. EasySep PE Positive Selection Kits were purchased from STEMCELL Technologies (Vancouver, BC, Canada). RNeasy and miRNeasy Mini Kits, miScript Primer Assay Kit, and miScript SYBR Green PCR Kit were obtained from QIAGEN (Valencia, CA). The following reagents were purchased from Bio-Rad Laboratories (Madison, WI): iScript and miScript cDNA Synthesis Kits. Epicentre’s PCR PreMix F and Platinum Taq DNA Polymerase Kits were from Invitrogen Life Technologies. ELISA kits for IL-4, IL-10, and TGF-b (ELISA MAX Standard Set Mouse) were bought from BioLegend. XFp Glycolytic Rate Assay Kit and XFP Cell Mito Stress Test Kit were purchased from Agilent Technologies.
Induction of MDSCs in mice by TCDD
C57BL/6 mice were injected i.p. with TCDD (1–10 μg/kg) or the vehicle, corn oil. At various days postexposure, mice were euthanized and peritoneal cells were collected and washed twice with PBS. The cells were counted and stained for MDSC markers (CD11b and Gr-1), Granulocytic MDSC (G-MDSC: CD11b and Ly-6G), and monocytic MDSC (M-MDSC: CD11b and Ly-6C) populations were analyzed by flow cytometry (BD FACSCelesta), as described previously (23).
Purification of MDSCs and their subsets M-MDSCs and G-MDSCs
TCDD-induced MDSCs were purified from exudates of the peritoneal cavity, as described previously (24). In brief, peritoneal exudates were collected from TCDD-exposed mice and labeled with PE-conjugated Gr-1 Ab. A PE Selection Kit from STEMCELL Technologies was used for selection, and we followed the protocol from the company. After purification, flow cytometry (BD FACSCelesta) was used to assess the purity of MDSCs. We purified M-MDSC and G-MDSC subsets of MDSCs using FACSAria II cell sorter (BD FACSAria II). Peritoneal exudate cells were stained with CD11b and Ly-6C for M-MDSCs or CD11b and Ly-6G for G-MDSCs.
AhR and CXCR2 antagonists
Mice were injected i.p. with 10 or 50 mg/kg AhR antagonist (CH223191) or CXCR2 antagonist (Sch527123), respectively, 1 d before TCDD injection. Peritoneal exudates were collected on day 3 and stained with CD11b, GR-1, Ly-6G and Ly-6C to detect MDSCs and subsets of MDSCs.
Effect of TCDD-induced MDSCs and MDSC subsets (M-MDSCs and G-MDSCs) on T cell proliferation in vitro
To examine the suppressive effect of MDSCs on T cell proliferation, splenocytes (5 × 105) from C57BL/6 naive mice were cultured in the presence of Con A (2 μg/ml) together with different ratios of TCDD-generated MDSCs, M-MDSCs, and G-MDSCs for 24 h, as described (25). [3H]thymidine (1 μCi per well) was added to the cell cultures, and, after 18 h, radioactivity was measured using a liquid-scintillation counter (MicroBeta TriLux; PerkinElmer).
Mitochondrial respiration, glycolytic rate, and ATP rate
Oxygen consumption rates (OCR), proton efflux rate (PER), and ATP rate were measured in a total of 2 × 105 purified MDSCs from peritoneum of vehicle or TCDD-treated mice using an XF Extracellular Flux Analyzer (Seahorse Bioscience). For OCR, MDSCs were plated in an XF cell culture plate coated with 15 μg of Cell-Tak (BD Biosciences) in XF assay medium supplied with 1 mM pyruvate, 2 mM glutamine, and 10 mM glucose. MDSCs were analyzed under stressed conditions and in response to 1 μM oligomycin, 1 μM fluorocarbonyl-cyanide-phenylhydrazone (FCCP), and 0.5 μM rotenone and antimycin A. For PER, MDSCs were plated in an XF cell culture plate coated with 15 μg Cell-Tak in Phenol Red–free Base Medium enriched with 2 mM glutamine, 10 mM glucose, 1 mM pyruvate, and 5 mM HEPES as initial conditions. Cells were monitored under stressed conditions and in response to 0.5 μM rotenone plus antimycin A (Rot/AA) and 50 mM 2-deoxy-d-glucose (2-DG). For ATP rate, MDSCs were plated in an XF cell culture plate with 15 μg of Cell-Tak in DMEM enriched with 10 mM of glucose, 1 mM of pyruvate, and 2 mM of glutamine. ATP rate production was measured under stressed conditions in response to 1.5 μM of oligomycin and 0.5 μM rotenone plus antimycin A. All three tests were quantified by a Seahorse Bioscience XFp Extracellular Flux Analyzer (Agilent Technologies).
Proliferation assessment by BrdU and Ki67 labeling in vivo
To characterize MDSCs proliferation in vivo, mice were injected with BrdU (100 mg/kg) 2 h before TCDD treatment, as described previously (26, 27). Cells from the peritoneal exudate were harvested and were first stained using anti-mouse Abs against CD11b and Gr-1. After fixation and permeabilization, the cells were stained with PerCP-Cy5.5–conjugated anti-BrdU (BD Biosciences) and PE-conjugated anti-Ki67 Abs (BioLegend) using an intranuclear staining protocol (BD Biosciences). Quad-stained cells were analyzed by flow cytometry (BD FACSCelesta).
Con A–induced hepatitis and adoptive transfer of MDSCs
To generate hepatic inflammation, C57BL/6 mice were injected i.v. with Con A (12.5 mg/kg) as described previously (23). For adoptive transfer, Con A–injected mice received 5 million purified MDSCs from the peritoneal cavity (PC-MDSCs) or MDSCs from the BM (BM-MDSCs) of TCDD-treated mice 1 h before Con A injection. In experiments in which TCDD was tested for its ability to attenuate Con A–induced hepatitis, mice received TCDD (10 μg/kg) by the i.p. route 1 h before Con A injection. Mice were sacrificed 48 h after treatment. Spleens and livers were harvested, and blood was collected. Single-cell suspensions of splenocytes were prepared, and infiltrating mononuclear cells in the liver were isolated using a Percoll gradient, as described previously (12). Harvested cells from spleens and livers were stained with Abs against CD4 and INF-γ, IL-17, IL-4, or IL-10 to determine Th1, Th-17, Th-2, and induced Treg populations by flow cytometry (Beckman Coulter). IL-4 and TGF-β cytokine levels were detected in sera by ELISA.
ELISA
Serum from individual mice was collected and the concentration of various cytokines, including IL-4 and TGF-β was measured using ELISA MAXTM Standard SET Mouse Kit for respective cytokines. ELISAs were purchased from BioLegend.
Chemokine analysis
Serum and peritoneal exudates were collected from individual mice 3 d after TCDD or vehicle injection. G-CSF, GM-CSF, M-CSF, eotaxin (CCL11), LIF, LIX (CXCL5), KC (CXCL1), MCP-1 (CCL2), MIP-1A (CCL3), MIP-1B (CCL4), MIP-2 (CXCL2), MIG (CXCL9), RANTES (CCL5), and VEGF concentrations were determined by using a MILLIPLEX MAP Kit (MilliporeSigma) according to the manufacturer protocol and analyzed by the Bio-Plex chemiluminescence assay system (Bio-Rad Laboratories).
Histopathology of liver
Livers were harvested from mice and fixed in formaldehyde (4%) overnight, then embedded in paraffin and cut to∼6-μm thickness. The liver sections were then stained with H&E and analyzed by the Cytation 5 microscopic system (BioTek).
miRNA arrays and analysis
miRNA analysis was carried out as described previously (28). Total RNA, including miRNA, was isolated from PC-MDSCs after TCDD administration using miRNANeasy kit from QIAGEN and following the protocol of the company. miRNAs arrays were performed using Affymetrix miRNA Array (version 4). Raw files generated from the miRNA microarray were uploaded to Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession number GSE134337. Signal expression (fold change) of more than 3000 miRNAs was detected from the raw array data, and only those miRNAs that were altered more than 2-fold were considered for further analysis. The selected miRNAs were further analyzed for their targets and alignments using TargetScan, microRNA.org, and the miRWalk database. Furthermore, selected miRNAs were analyzed for their role in various diseases and pathways using Ingenuity Pathway Analysis (IPA) software. miRNAs from various groups were also analyzed for their relationship using LucidChart or a Venn diagram.
Real-time quantitative PCR to validate miRNAs and associated genes expression
Quantitative PCRs (Q-PCRs) were performed to determine the expression of selected miRNAs (miR-150-5p, and miR-543-3p) on cDNA synthesized from total RNAs, including miRNAs, isolated from PC-MDSCs post-TCDD or vehicle exposure. miScript Primer Assay Kits (QIAGEN) and SssAdvanced SYBR Green PCR kits from Bio-Rad Laboratories were used, and Q-PCR was performed following the protocol of the company. For Q-PCR, the run conditions to detect miRNA were as follows: 15 min at 95°C (initial activation step), followed by 40 cycles of 15 s at 94°C (denaturing temperature), 30 s at 60°C (annealing temperature), and 30 s at 70°C (extension temperature and fluorescence data collection). Fold changes of miRNA were calculated using ΔΔCt method, where Ct is the threshold cycle to detect fluorescence. The data were normalized to miRNAs against internal control miRNA (SNORD96A; QIAGEN), and fold change of miRNAs was calculated against control miRNA (SNORD96A), and the treatment group (TCDD) was compared with the vehicle group.
RT-PCR to determine the expression of IL-10, ARG2, STAT3, INOS1, and PIM1 in MDSCs post-TCDD or vehicle treatment
RT-PCR was performed to detect IL-10, ARG2, STAT3, PIM1, CCL11, CCR1, CCR3, CCR5, and CXCR2 expression. Primers of these genes were specifically designed using Integrated DNA Technologies tools (details of primers presented in Supplemental Table I). RT-PCR was performed for 40 cycles using the following conditions: 98°C (denaturing temperature) for 30 s, 98°C (annealing temperature) for 10 s, and 60°C (extension temperature) for 30 s. The PCR products, generated from mouse gene-specific primer pairs, were visualized with UV light, performing electrophoresis (1.2% agarose gel). The band intensity of PCR products was determined using the ChemiDoc image analysis system from Bio-Rad Laboratories (Hercules, CA). The expression of the above genes was normalized against PCR products generated from the mouse housekeeping gene GAPDH (internal control).
Statistical analysis
GraphPad Prism software version 6.01 (GraphPad Software, San Diego, CA) was used for statistical analysis. A standard t test or multiple t test with Holm–Sidak multiple comparisons corrections was used when comparing two groups for significance. A one-way or two-way ANOVA with post hoc Tukey multiple comparisons test was used to compare between the means of more than two groups. Error bars were expressed as mean ± SE of mean (±SEM), and significance was determined as having a p value <0.05. Each experiment was repeated at least twice with consistent results.
Results
TCDD induces MDSCs in mice
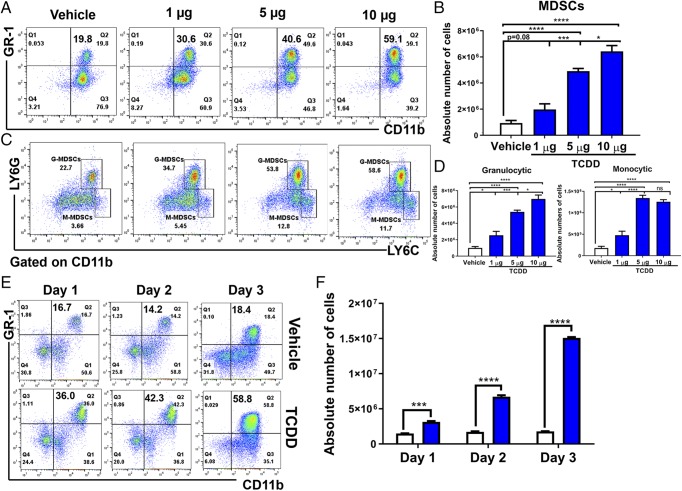
TCDD is a potent agonist of AhR, and thus we used TCDD to test whether activation of AhR leads to induction of MDSCs. To this end, three doses of TCDD (1, 5, and 10 μg/kg body weight) or vehicle were injected (i.p.) into C57BL/6 mice, and peritoneal exudates were collected on day 3 posttreatment. The presence of MDSCs in the peritoneal cavity was analyzed by staining the cells with fluorophore-labeled anti-mouse CD11b and GR1 Abs and using flow cytometry. There was a dose-dependent increase in both the percentages and numbers of CD11b+GR1+ cells in the peritoneal cavity (Fig. 1A, 1B) of mice that received TCDD, when compared with mice that received vehicle. Upon analysis of subsets of MDSCs (monocytic and granulocytic) by staining the cells with Abs against Ly-6C and Ly-6G, we observed a significant increase in both M-MDSCS (CD11b+Ly6G−Ly-6Chi) and G-MDSCs (CD11b+Ly-6G+Ly-6Clo) following TCDD treatment when compared with vehicle controls (Fig. 1C, 1D). However, there were significantly more G-MDSCs in both the percentages and total numbers when compared with M-MDSCs. To study the time course, we injected mice with TCDD and collected peritoneal exudates on days 1, 2, and 3 posttreatment. We observed significant induction of MDSCs on day 1 and their numbers continued to rise until day 3 (Fig. 1E, 1F).
FIGURE 1.
TCDD induces MDSCs in naive mice. Naive C57BL/6 mice were injected with TCDD (n = 6) or vehicle (n = 6) i.p. and at various days posttreatment, and cells from the peritoneal cavity were harvested and analyzed for MDSCs. (A) Representative plots from FlowJo software analysis of flow cytometry data showing induced MDSC in percentages following 1, 5, or 10 μg/kg TCDD administration when compared with vehicle. Cells were harvested on day 3. (B) Total number of MDSCs per mouse expressed as mean ± SEM, based on (A) description. (C) Representative dot plots showing M-MDSCs (CD11b+Ly-6G−Ly-6Chi) and G-MDSCs (CD11b+Ly-6G+Ly-6Clow) percentages following administration of 1, 5, or 10 μg/kg TCDD when compared with vehicle. (D) Total number of M-MDSCs and G-MDSCs per mouse expressed as mean ± SEM. (E) Time course of induction of MDSCs by 5 μg/kg TCDD. (F) Based on data in (E), total number of MDSCs per mouse at different days expressed as mean ± SEM. Statistical analysis was performed using one-way ANOVA with Tukey post hoc multiple comparisons test for (B) and (D). For (F), a multiple t test was performed using Holm–Sidak method multiple comparisons test. Data are representative of at least three independent experiments with reproducible results. Significance was designated as follows: *p < 0.05, ***p < 0.001, ****p < 0.0001.
Characterization of TCDD-induced MDSCs
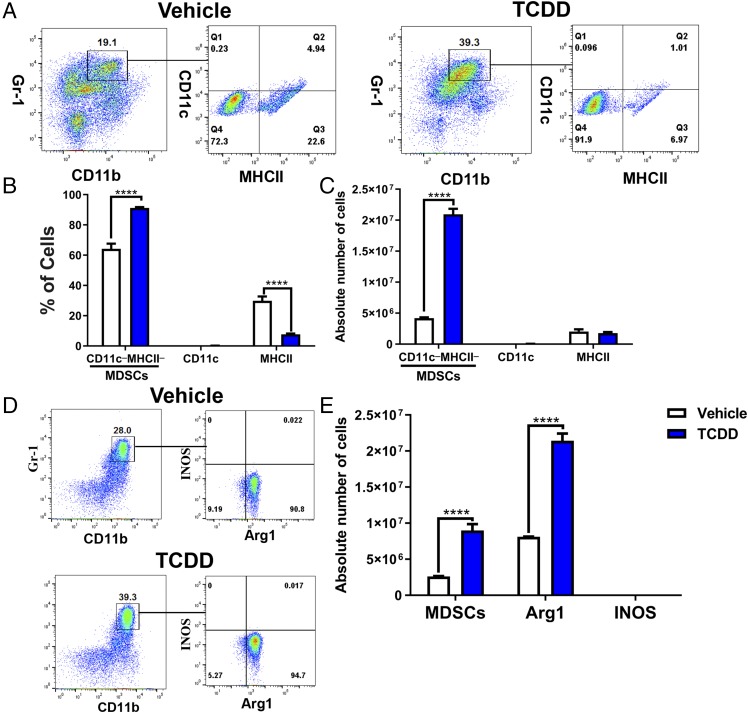
Because MDSCs are phenotypically similar to neutrophils and monocytes, we performed additional studies that help distinguish them from these cells (28, 29). M-MDSCs can be distinguished from the monocytes by the fact that M-MDSCs lack surface markers of monocytes, such as CD11c and MHC-II. Thus, we looked at the expression of these markers on TCDD-induced MDSCs and found that most of the TCDD-induced MDSCs (91.9%) did not express CD11c and MHC-II (Fig. 2A). Also, the proportion of such cells and their numbers significantly increased when compared with the vehicle controls (Fig. 2A–C). There are no precise markers that distinguish G-MDSCs from polymorphonuclear leukocytes, but murine polymorphonuclear leukocytes do not express Arginase-1 (Arg1), whereas G-MDSCs do, which gives them the immunosuppressive property. Thus, when we stained the TCDD-induced MDSCs for Arg1 and iNOS, we found that 94.7% of the MDSCs were Arg1+, whereas they failed to express iNOS (Fig. 2D, 2E). Functionally, MDSCs were also highly immunosuppressive, as detailed later in the article. Together, these data suggested that TCDD-induced cells were indeed MDSCs and lacked the properties of neutrophils or monocytes.
FIGURE 2.
MDSCs and MDSC subset characterization. Peritoneal exudate cells were harvested from vehicle- (n = 6) and TCDD-treated (n = 6) mice and stained for various markers. (A) Cells from vehicle- or TCDD-treated mice were gated on those that expressed CD11b+Gr-1+, and this population was further tested for MHC-II and CD11c. (B) Percentage of cells as shown in (A) that are negative or positive for MHC-II and CD11c markers. (C) Absolute number of CD11b+Gr-1+ cells per mouse that are negative or positive for MHC-II and CD11c markers. (D) Percentage of Arg1 and INOS expression in CD11b+Gr-1+ cells in vehicle and TCDD treatment groups. (E) Absolute number of CD11b+Gr-1+ cells and CD11b+Gr-1+ cells that express Arg1 and INOS. Vertical bars represent mean ± SEM. For bar graphs depicted, the multiple t test was performed using Holm–Sidak method multiple comparisons test. Data are representative of at least three independent experiments with reproducible results. Significance was designated as follows: ****p > 0.0001.
TCDD induces MDSCs through activation of AhR
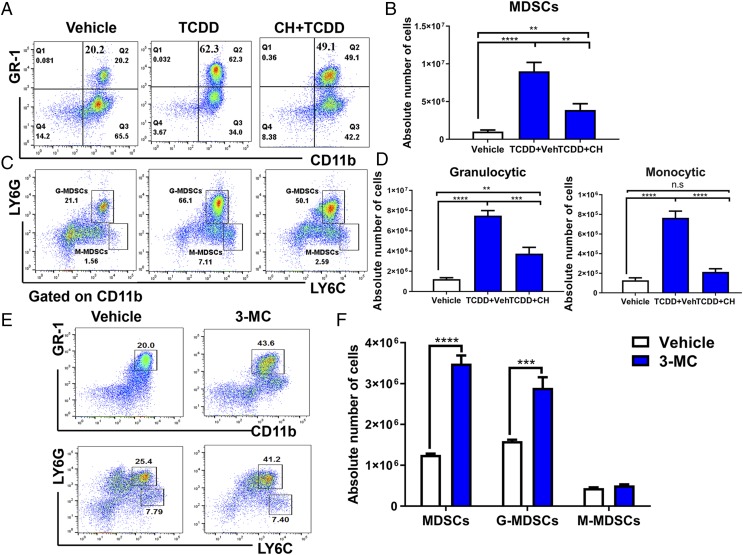
Because TCDD is a potent ligand for AhR, we tested the effect of AhR antagonist (CH223191) on the induction of MDSCs by TCDD. We found that administration of AhR antagonist significantly reduced the number of MDSCs (Fig. 3A, 3B) and MDSC subsets (Fig. 3C, 3D), thereby suggesting that TCDD was inducing the MDSCs, at least in part, through activation of AhR. To test whether the ability to induce MDSCs was restricted only to TCDD or whether it could also be seen using other AhR ligands, we injected 3-MC, a well-characterized AhR ligand (30) similar to TCDD, into mice i.p. and studied the PC-MDSCs. The data demonstrated that 3-MC was also able to induce high levels of MDSCs that belonged to both G-MDSCs and M-MDSCs, with the former being produced in larger numbers (Fig. 3E, 3F), similar to TCDD.
FIGURE 3.
AhR antagonist CH223191 treatment decreases TCDD-mediated MDSC induction. Naive C57BL/6 mice were injected with TCDD (10 μg/kg) i.p. as described in Fig. 1 legend. These mice were injected i.p with 10 mg/kg of AhR antagonist (CH223191, CH) 1 d before TCDD injection. Peritoneal exudates were collected on day 3 and stained for MDSCs. (A) Representative flow cytometric analysis showing MDSC percentages after treatment with AhR antagonist. (B) Total number of MDSCs per mouse expressed as mean ± SEM following treatment with AhR antagonist, based on description in (A). (C) Representative flow cytometric analysis showing percentages of MDSC subsets after treatment with AhR antagonist (CH). (D) Total number of MDSC subsets per mouse expressed as mean ± SEM following treatment with AhR antagonist. (E) Representative flow cytometric analysis showing percentages of MDSC (top panel) and MDSC subsets (bottom panel) after treatment with AhR agonist (3-MC). (F) Total number of MDSC subsets per mouse expressed as mean ± SEM following treatment with AhR agonist (3-MC). Statistical analysis was performed using one-way ANOVA with Tukey post hoc multiple comparisons test to determine significance for (B) and (D) between vehicle (n = 7) and TCDD (n = 7). For (F), significance between vehicle (n = 6) and 3-MC (n = 6) was determined using a multiple t test with Holm–Sidak method multiple comparisons test. Data are representative of at least two independent experiments with reproducible results. Significance was designated as follows: **p < 0.01, ***p < 0.001, ****p < 0.0001.
TCDD promotes the migration of MDSCs from BM to peritoneal cavity through chemokine induction
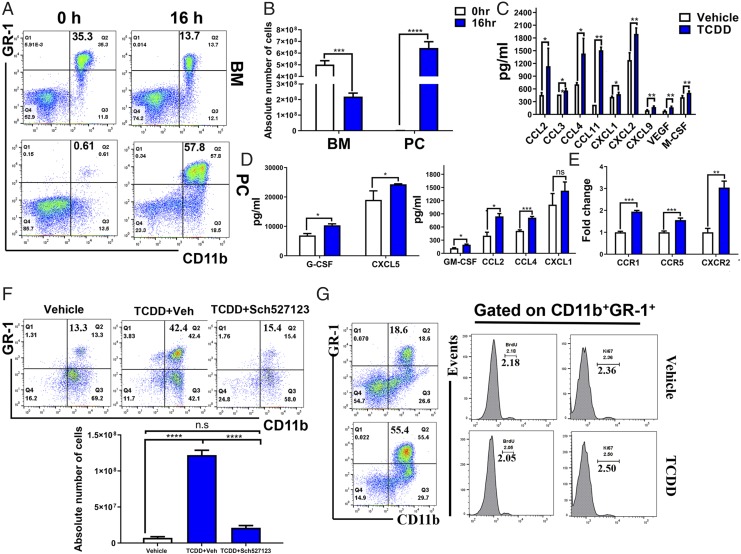
Normal mouse BM contains 20–30% CD11b+ GR-1+, whereas their proportion is much smaller in the spleens (2–4%) and they are absent in the lymph nodes (16). Thus, we next determined whether TCDD was promoting the migration of MDSCs from the BM. To this end, we enumerated CD11b+ GR-1+ cells in BM and peritoneal exudate at 0 and 16 h post-TCDD treatment by flow cytometry. Interestingly there was a significant decrease in the percentage of CD11b+ GR-1+ cells in the BM 16 h following TCDD treatment, whereas there was a significant increase in PC-MDSCs (Fig. 4A, 4B). These data suggested that CD11b+ GR-1+ cells from the BM may be migrating to the peritoneal cavity, the site of TCDD administration. It should be noted that these experiments were terminated early (16 h) because we wished to detect chemokines and cytokines that are normally induced at early time points. We were able to detect MDSCs as early as 16 h because of the high dose of TCDD used (10 μg/kg). Next, we investigated whether specific chemokines have a role in recruiting MDSCs from BM to peritoneal cavity. To that end, we detected some chemokines and cytokines that regulate MDSCs and found that the peritoneal fluid of TCDD-injected mice had significantly higher levels of MCP-1 (CCL2), MIP-1A (CCL3), MIP-1B (CCL4), eotaxin (CCL11), KC (CXCL1), MIP-2 (CXCL2), MIG (CXCL9), VEGF, and M-CSF when compared with controls (Fig. 4C). Also, we saw an increase in CCL2, CCL4, CXCL1, CXCL5, G-CSF, and GM-CSF in the serum of TCDD-treated mice (Fig. 4D). We also assessed chemokine receptor expression in MDSCs post-TCDD treatment. We found that three of the chemokine receptors increased in TCDD-induced MDSCs group (CCR1, CCR5, and CXCR2) when compared with the vehicle-induced MDSCs group (Fig. 4E). Next, we investigated whether blocking CXCR2 would have an effect on MDSCs recruitment to the peritoneal cavity. To this end, we injected CXCR2 antagonist Sch527123 in mice 1 d before TCDD treatment. After 3 d, MDSCs were assessed in the peritoneal cavity, and we observed that MDSC percentage and numbers were dramatically decreased following CXCR2 antagonist treatment when compared with controls (Fig. 4F). We also examined whether TCDD induces only the migration of MDSCs from BM to peritoneal cavity or whether it also induces MDSC proliferation in the peritoneal cavity. To that end, we injected BrdU into mice 2 h before TCDD treatment. After 48 h, we stained the peritoneal cells using fluorophore-labeled anti-mouse CD11b and GR-1 Abs as well as intranuclear BrdU and intracellular Ki67. We observed no significant difference in BrdU+ and Ki67+ cells between vehicle and TCDD groups, demonstrating that TCDD does not induce proliferation of MDSCs in the periphery (Fig. 4G).
FIGURE 4.
Identifying the source of TCDD-induced MDSCs. Naive C57BL/6 mice were injected with TCDD (10 μg/kg) i.p. as described in Fig. 1 legend. (A) Representative flow cytometric analysis showing the percentage of MDSCs in BM 16 h after TCDD treatment when compared with 0 h (top panel). MDSCs percentage in peritoneal cavity (PC) after 16 h compared with 0 h (bottom panel). (B) Total number of MDSCs per mouse expressed as mean ± SEM, based on description in (A), where n = 5 for each experimental condition. (C) Measurement of chemokines from the peritoneal exudates of vehicle (n = 4) and TCDD (n = 4) mice with data expressed as mean ± SEM. (D) Detection of chemokines in the serum from vehicle- (n = 4) and TCDD-treated (n = 4) mice with data expressed as mean ± SEM. (E) Q-PCR validation of CCR1, CCR5, and CXCR2 expression in vehicle- (n = 4) and TCDD-treated (n = 4) mice, with data expressed as mean ± SEM. (F) Flow cytometric analysis of MDSC percentage (top panel) and absolute numbers (bottom panel) following treatment with vehicle (n = 5), TCDD (n = 5), or TCDD and CXCR2 antagonist (n = 3). (G) Representative plots of BrdU labeling and anti-Ki67 staining at 48 h after TCDD treatment. The left panel shows staining for MDSCs and right panel shows staining for BrdU and Ki67 on gated MDSCs. Vertical bars represent mean ± SEM. For (B)–(D), significance was determined using a multiple t test with Holm–Sidak method multiple comparisons test. For (F), significance was determined using one-way ANOVA and Tukey multiple comparisons test. Data are representative of at least three independent experiments with reproducible results. Significance was designated as follows: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
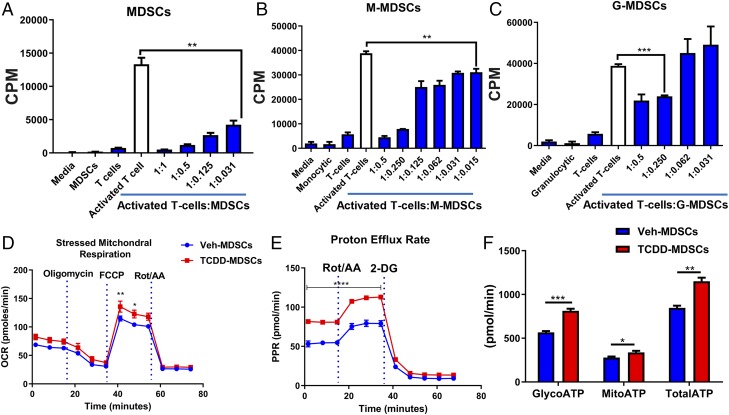
TCDD-induced PC-MDSCs, M-MDSCs, and G-MDSCs mediate suppression of T cell activation in vitro
Immune suppression is the hallmark feature of MDSCs; thus, to determine whether TCDD-induced MDSCs can suppress T cell activation, we performed T cell proliferation assays using Con A (2 μg/ml) in the presence or absence of TCDD-induced MDSCs collected from the peritoneal cavity. We observed dose-dependent suppression of T cell activation in the presence of MDSCs, and, furthermore, whereas both G-MDSCs and M-MDSCs were suppressive, the latter were found to be more effective (Fig. 5A–C). It should be noted that, whereas TCDD induced more G-MDSCs than M-MDSCs (Fig. 1C, 1D), on a per cell basis, M-MDSCs were more immunosuppressive in T cell proliferation assays than G-MDSCs (Fig. 5B, 5C), thereby suggesting that, overall, both subpopulations may contribute significantly toward suppressing T cell proliferation.
FIGURE 5.
TCDD-induced MDSCs suppress T cell proliferation and exhibit different metabolic profile. (A–C) TCDD-induced–purified PC-MDSCs or M-MDSCs and G-MDSCs were incubated with spleen cells activated with Con A at different ratios, and T cell proliferation was assessed by [3H]-thymidine incorporation assay. Data are depicted as mean ± SEM of triplicate cultures (n = 3) shown as counts per minute (CPM). (D and E) OCR and glycolytic PER in TCDD-induced PC-MDSCs (n = 3) compared with vehicle-induced PC-MDSCs (n = 3). (F) ATP production rate in the experimental groups including MDSCs from vehicle- (n = 3) or TCDD-treated mice (n = 3). For (A)–(C), one-way ANOVA with Tukey multiple comparisons test was used to determine significance. For (D) and (E), two-way ANOVA with Tukey multiple comparisons test was used to determine significance. For (E), significance, ****p < 0.0001, was found from 0 to 40 min time points and is illustrated with a bar across these six data points. For (F), a multiple t test with Holm–Sidak method of multiple comparison was used to determine significance. Data are representative of at least two independent experiments with reproducible results. Significance was defined as follows: p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
TCDD-induced PC-MDSCs have a high mitochondrial respiration and glycolytic rate
Tumor-infiltrating MDSCs have been shown to have an increased OCR and extracellular acidification rate when compared with splenic-MDSCs (31). To explore the nature of TCDD-induced MDSCs, we tested the OCR and PER as well as the ATP rate using a Seahorse Bioscience XFp Extracellular Flux Analyzer. We observed that TCDD-induced MDSCs from the peritoneal cavity had higher OCR, PER, and ATP production rates in comparison with vehicle-induced PC-MDSCs (Fig. 5D–F). We also observed that all PER parameters, including basal glycolysis, percentage of PER from glycolysis, and compensatory glycolysis, were higher in TCDD-induced PC-MDSCs when compared with vehicle-induced MDSCs (Fig. 5E). Also, TCDD-induced MDSCs showed higher rates of mitoATP, glycoATP, and total ATP in comparison with vehicle-induced MDSCs (Fig. 5F). Because TCDD-induced MDSCs had increased mitochondrial respiration (OCR) and glycolysis (PER), and overall energy (ATP) compared with vehicle controls, these data suggested that TCDD-induced MDSCs had increased capacity to respond to energy demands (e.g., immunosuppressive function) because they had more available energy.
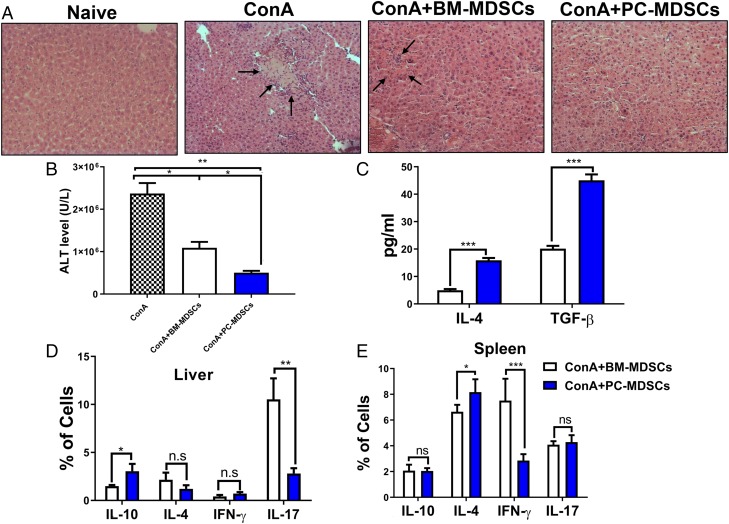
TCDD-induced MDSCs protect Con A–induced liver damage in vivo
Next, we investigated whether TCDD-induced MDSCs were functional in vivo in suppressing T cell response. We had shown earlier that MDSCs can protect liver from Con A–mediated hepatitis in vivo (32). To that end, we i.v. injected naive mice with purified TCDD-induced PC-MDSCs or BM, followed 1 h later with Con A. Spleens and livers of mice treated with PC-MDSCs or BM-MDSCs were harvested 48 h after treatment. Upon histological analysis of liver, there was reduced liver inflammation in mice that received TCDD-induced PC-MDSCs when compared with mice that received MDSCs from BM (Fig. 6A). Analysis of alanine transaminase (ALT) in sera showed significantly reduced levels in mice that received TCDD-induced PC-MDSCs when compared with mice that received MDSCs from BM (Fig. 6B). Moreover, levels of IL-4 and TGF-β (anti-inflammatory cytokines) increased in the sera of mice that received TCDD-induced PC-MDSCs when compared with mice that received BM-MDSCs (Fig. 6C). These data suggested that TCDD-induced PC-MDSCs are more immunosuppressive in function in vivo when compared with BM-derived MDSCs from the same mice.
FIGURE 6.
TCDD-induced MDSCs protect from Con A–induced liver damage and inflammation in vivo following adoptive transfer. C57BL/6 mice were injected i.v. with Con A (12.5 mg/kg), and these mice received the injection 1 h before an adoptive transfer of 5 million purified PC-MDSCs or BM-MDSCs from TCDD-treated mice. Mice were sacrificed after 48 h posttreatment for further analysis to include the following groups: naive (n = 3), Con A (n = 3), Con A plus BM-MDSCs (n = 3), Con A plus PC-MDSCs (n = 3). (A) H&E stain of liver tissue at original magnification ×20. (B) Measurement of ALT in sera with significance determined by using ANOVA and Tukey multiple comparisons test. Vertical error bars represent mean ± SEM. (C) Measurement of TGF-β and IL-4 level in sera. (D and E) Percentages of cells expressing various cytokines determined by flow cytometry in spleen (D) and liver (E). For (C)–(E), one-way ANOVA with Tukey multiple comparisons test was used to determine significance, with vertical error bars representing mean ± SEM. Data are representative of at least three independent experiments with reproducible results. Significance was defined as follows: *p < 0.05, **p < 0.01, ***p < 0.001.
TCDD-induced MDSCs reduce inflammation in liver and spleen of Con A–induced hepatitis mice
To better understand the suppressive effect of TCDD-induced PC-MDSCs, we purified mononuclear cells from livers as well as spleens from mice treated with PC-MDSCs and BM-MDSCs as described in Materials and Methods. The mononuclear cells and splenocytes were stained to determine Th1, Th17, Th2, and induced Tregs. There was significant reduction in CD4+IL-17+ in the liver and CD4+IFN-γ+ in the spleens of mice treated with TCDD-induced PC-MDSCs when compared with cells treated with BM-MDSCs (Fig. 6D, 6E). In contrast, there was significant upregulation of CD4+IL-10+ in the liver and CD4+IL-4+ in the spleens of mice treated with PC-MDSCs when compared with cells from mice treated with BM-MDSCs (Fig. 6D, 6E).
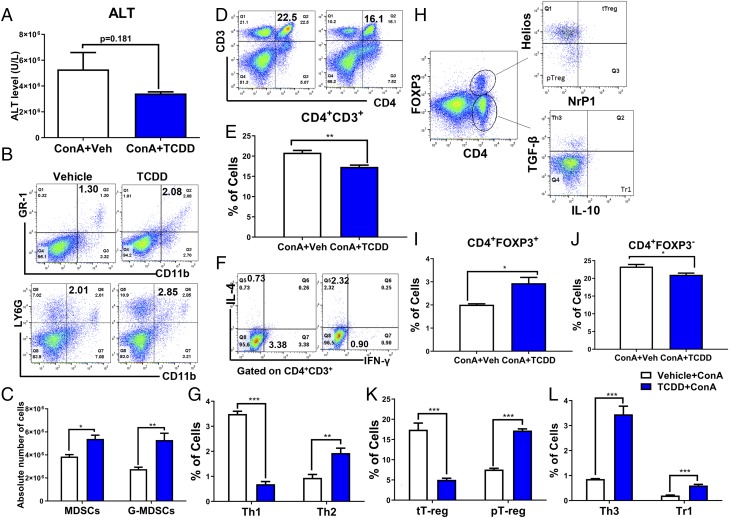
TCDD reduces inflammation in Con A–induced hepatitis in mice by generating MDSCs and Tregs
To test whether TCDD would directly suppress Con A–induced hepatitis and whether this is regulated by MDSCs, we injected mice with TCDD (10 μg/kg) or vehicle 1 h before Con A (10 μg/kg) injection (i.v.). We noted that TCDD was able to decrease ALT levels thereby showing protection of liver damage (Fig. 7A). Next, we stained the spleen cells and liver mononuclear cells for MDSCs, various T helper cells (Th1, Th2, and Th3), and Treg subsets (Treg, peripheral Treg, and Tr1). We observed significant increase in MDSCs and G-MDSCs in spleens of mice exposed to TCDD, when compared with mice treated with vehicle (Fig. 7B, 7C). Furthermore, there was a significant reduction in the percentages of CD3+CD4+ cells and Th1 cells and an increase in the percentages of Th2 cells in TCDD-treated mice when compared with vehicle-treated mice (Fig. 7G). For Treg and its subsets (Fig. 7H), there was a significant increase in the percentages of CD4+Foxp3+ populations (Fig. 7I) and in the percentages of peripheral Tregs, Tr1, and Th3 cells in spleen of TCDD-treated mice in comparison with vehicle-treated mice (Fig. 7K, 7L). However, the percentages of CD4+Foxp3− (Fig. 7J) and thymus Tregs (Fig. 6K) were significantly decreased in mice were treated with TCDD when compared with mice treated with vehicle.
FIGURE 7.
TCDD treatment attenuates Con A–induced hepatitis and associated inflammation. Con A was used to induce hepatitis as described in Fig. 6 legend. These mice received TCDD (10 μg/kg) by i.p. route 1 h before Con A injection followed by analysis of spleens and liver for inflammation. (A) ALT level in serum of hepatitis-induced mice treated with vehicle (n = 5) or TCDD (n = 5). (B and C) Percentage and total numbers of MDSCs and G-MDSCs in the spleens of two groups, respectively. (D and E) Percentage of CD3+CD4+ cells in the spleens, respectively. (F and G) Percentages of Th1 (CD3+CD4+IFNγ+) and Th2 (CD3+CD4+IL-4+) cells in splenocytes. (H–L) Percentage of Tregs and Treg subsets from spleens of experimental groups. Vertical error bars represent mean ± SEM. For (A), (I), and (J), significance was determined using a parametric unpaired two-tailed t test with Welch corrections. For (C), (G), (K), and (L), significance was determined using a multiple t test with Holm–Sidak method multiple comparisons test. Data are representative of at least three independent experiments with reproducible results. Significance was defined as follows: *p < 0.05, **p < 0.01, ***p < 0.001.
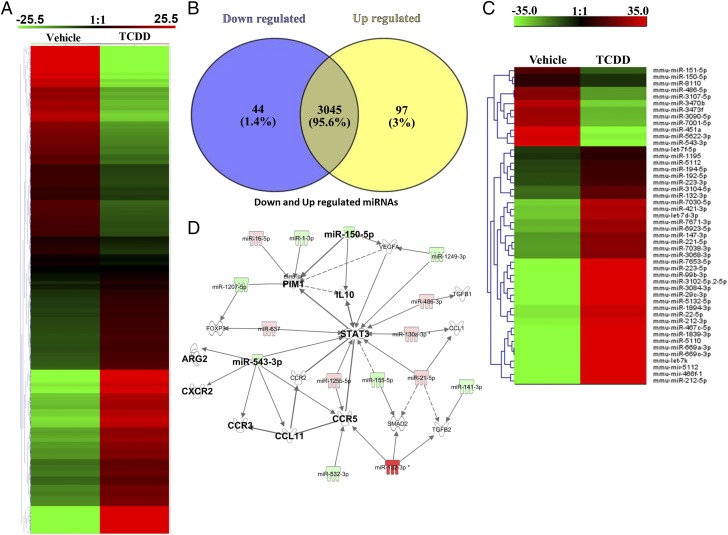
TCDD altered miRNAs expression in PC-MDSCs
Our previous studies demonstrated that miRNA play a critical role in MDSC induction (33). To that end, we investigated the role of miRNA in the induction of MDSCs by TCDD by performing miRNA microarray using MDSCs derived from vehicle or TCDD-treated groups. There were more than 3195 miRNAs that were analyzed by the microarray (Fig. 8A), of which the expression of only 141 miRNAs were altered by a >2-fold change in TCDD-treated groups when compared with vehicle controls (Fig. 8A). As shown in the Venn diagram, there were 3054 miRNAs that showed no change, whereas 97 miRNAs were upregulated, and 44 miRNAs were downregulated (Fig. 8B). The corresponding heatmap depicts upregulated (n = 97) and downregulated (n = 44) miRNAs in TCDD-induced MDSCs when compared with vehicle-induced MDSCs (Fig. 8C). Furthermore, upon analysis of the 141 dysregulated miRNAs using IPA software, we observed a direct relationship between various miRNAs and the target genes, including miR-150-5p and the target genes IL-10 and PIM1 and miR-543-3p and the target genes ARG2 and STAT3 as well as CCL11, CCR3, CCR5, and CXCR2 (Fig. 8D). These data suggested that TCDD-mediated alterations in the miRNAs may regulate the expression of various target molecules, such as IL-10, PIMI, ARG2, STAT3, CCL11, CCR3, CCR5, and CXCR2, which are prominently involved in the induction and functions of MDSCs.
FIGURE 8.
TCDD-mediated alterations in miRNA expression in MDSCs. Naive mice were treated with TCDD as described in Fig. 1 legend. The PC-MDSCs were analyzed for miRNA expression between vehicle- and TCDD-treated mice, with MDSCs pooled from five different mice per experimental group. (A) Heat map of miRNAs expression in MDSCs from vehicle and TCDD groups showing more than 3000 miRNAs. (B) Venn diagram showing miRNA that are upregulated or downregulated in TCDD group when compared with vehicle controls. (C) Heat map of upregulated and downregulated (>2-fold) miRNAs in vehicle versus TCDD groups. (D) IPA was used to determine interaction between upregulated and downregulated miRNAs and targeted genes. Data are representative of one independent experiment.
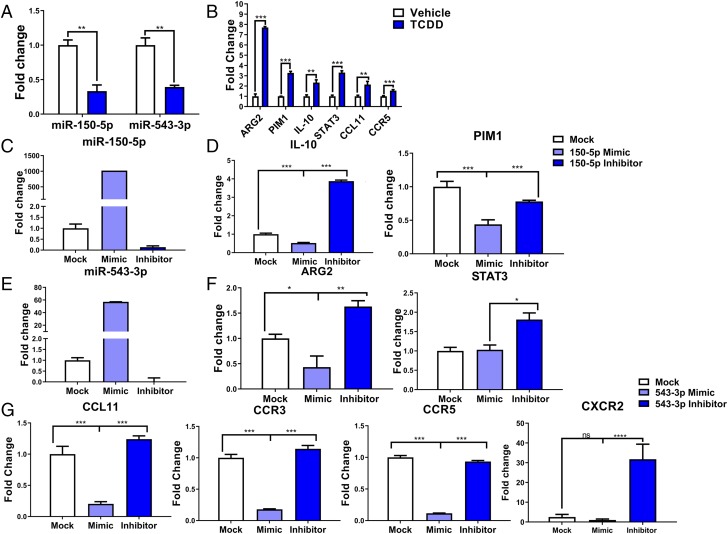
Real-time Q-PCR to validate selective miRNAs and the target genes
Based on the complementary binding affinity of miR-150-5p with IL-10 and PIM1 genes and miR-543-3p with ARG2 and STAT3 genes, as well as with CCL11, CCR3, CCR5, and CXCR2 (Supplemental Fig. 1), we selected miR-150-5p and miR-543-3p to validate their expression. PCR data showed significant downregulation of both miR-150-5p and miR-543-3p in TCDD-induced PC-MDSCs when compared with vehicle-induced MDSCs (Fig. 9A). Next, we examined the expression of target genes IL-10 and PIM1 (miR-150-5p target genes), ARG2, STAT3, CCL11, and CCR5 (miR-154-3p target genes) by performing PCR. We observed significant upregulation of all these genes in TCDD-induced PC-MDSCs when compared with vehicle-induced MDSCs (Fig. 9B). These data demonstrated that TCDD-alters the expression of miRNA in MDSCs, which may lead to their induction and functions.
FIGURE 9.
Q-PCR analysis of miRNA-150-5p and miRNA-543-3p and specific targeted genes. MDSCs were isolated as described in Fig. 1 legend in vehicle (n = 5) or TCDD (n = 5) mice. (A) Expression of mir-150-5p and mir-543-3p in TCDD-induced MDSCs when compared with vehicle. (B) Expression of targeted genes IL-10, PIM1, ARG2, STAT3, CCL11, CCR3, and CCR5 in TCDD-induced MDSCs when compared with vehicle. (C) Induction of mir-150-5p expression with mimic compared with mock and inhibitor. (D) Induction of IL-10 and PIM1 expression with inhibitor of mir-150-5p compared with mimic of mir-150-5p. (E) Expression of mir-543-3p with mimic compared with mock and inhibitor. (F and G) Expression of ARG2, STAT3, CCL11, CCR3, CCR5, and CXCR2 in the presence of inhibitor of mir-543-3p compared with mimic of mir-543-3p. For (A) and (B), significance was determined using a multiple t test with Holm–Sidak method multiple comparisons test. For (C)–(G), using triplicate (n = 3) wells for each experimental condition, one-way ANOVA with Tukey multiple comparisons test was used to determine significance. Data are representative of at least three independent experiments with reproducible results. Significance was defined as follows: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Analysis of miR-150-5p and miR-543-3p and specific targeted gene expression
To further understand the role of miR-150-5p in IL-10 and PIM1 expression and miR-543-3p in ARG2, STAT3, CCL11, CCR3, CCR5, and CXCR2 expression, we performed a series of transfection assays. To that end, MDSCs were mock-transfected or transfected with mature miR-150-5p, mature miR-543-3p, anti–miR-150-5p inhibitor or anti–miR-543-3p inhibitor, and cultured for 24hrs. Next, the expression of various miRNAs and their target molecules were studied. Mock-transfected MDSCs showed basal level expression of both miR-150-5p and miR-543-3p (Fig. 9C, 9E) but MDSCs transfected with mature miR-150-5p and miR-543-3p showed significantly upregulated expression of both miRNAs in transfected MDSCs (Fig. 9C, 9E). However, transfection of MDSCs with anti–miR-150-5p and anti–miR-543-3p inhibitors showed downregulated expression of both miRNAs in MDSCs (Fig. 9C, 9E). These data demonstrated that transfection of MDSCs with mature miRNAs or anti-miRNAs inhibitors showed expected results. To further understand the role of these two miRNAs, we performed PCR to determine the expression of IL-10, PIM1, STAT3, ARG2, CC11, CR3, CR5, and CXCR2 genes. The expression of IL-10 and PIM1 was significantly reduced in MDSCs in the presence of miR-150-5p; however, the expression of these genes was significantly increased in MDSCs in the presence of anti-miRNA inhibitor (Fig. 9D). Fig. 9F, 9G and showed significant reduction in the expression of ARG2, CCL11, CCR3, CCR5, and CXCR2 genes in MDSCs in the presence of miR-543-3p mimic. However, there was a significant increase in the expression of these genes in the presence of anti-miRNA inhibitor (Fig. 9F, 9G). Together, these data demonstrated that TCDD-mediated alterations in the expression of miRNA might play a critical role in MDSC induction and functions.
Discussion
In the current study, we made an exciting observation that TCDD potently induces MDSCs at the site of administration, even in naive animals. Also, TCDD was able to induce MDSCs in an inflammatory model of Con A–induced hepatitis. We found that TCDD-induced MDSCs were highly immunosuppressive, as demonstrated using in vitro and adoptive transfer experiments. TCDD induced both the subsets of MDSCs: G-MDSCs (CD11b+Ly-6G+Ly-6Clo) and M-MDSCs (CD11b+Ly-6G−Ly-6Chi). Additionally, we found that TCDD caused alterations in the expression of miRNA in MDSCs that were likely to promote the immunosuppressive functions of MDSCs. G-MDSCs and M-MDSCs are morphologically and phenotypically similar to neutrophils and monocytes (28), respectively. However, murine G-MDSCs differ from neutrophils in that the former cells express Arg1 and are highly immunosuppressive (28), which was observed in the current study. Also, M-MDSCs differ from monocytes in that the latter cells express MHC-II and CD11c (28), and we found that TCDD-induced M-MDSCs did not express these markers.
TCDD is well characterized for its immunosuppressive properties that are mediated through activation of AhR (34–37). TCDD-mediated immunosuppression may involve multiple pathways because AhR ligation leads to activation of DREs found on a significant number of genes (38–40), including those involved in the regulation of immune response. Some mechanistic pathways that have been previously identified include the following: induction of Fas that expresses DRE on its promoter and upregulation of FasL leading to enhanced activation-induced cell death/apoptosis (2, 5, 6, 39, 41–44), upregulation of Foxp3, which also expresses DREs, thereby leading to the increased induction of Tregs (3, 7) while suppressing Th17 cells via epigenetic regulation (8). In addition to Tregs, it is becoming increasingly clear that MDSCs also play a critical role as regulators of inflammation (23, 45, 46). Whether AhR activation can lead to induction of MDSCs in naive mice has not been investigated thus far, and therefore formed the rationale for the pursuit of the current study. Interestingly, we found that TCDD treatment led to robust induction of MDSCs in naive animals in a dose-dependent manner, and such an induction was AhR dependent.
It has been reported that there are two groups of interconnected signals in MDSC accumulation and activation. The first group of signals is responsible for immature myeloid cell expansion, whereas the second group of signals is responsible for their activation and promotion of pathologic activity (47). The first group of signals includes GM-CSF, M-CSF, G-CSF, IL-6, VEGF, and polyunsaturated fatty acids, and these signals primarily act via STAT3 and STAT5. However, this signaling alone is not sufficient without activating a second set of signals, which is mainly provided by proinflammatory molecules, such as IFN-γ, IL-1β, IL-4, IL-6, IL-13, TNF, and TLR ligand, which use NF-κB, STAT1, and STAT6 transcription factors for their activation (47–49). Chemokines and their receptors, such as CCL5, CXCL17, CXCL2, CCR5, and CXCR2 play a critical role in MDSCs migration from BM to the tumor environment (50–52). Furthermore, we observed increased expression of chemokine receptors post-TCDD administration on MDSCs, such as CCR1 (receptor for CCL3 and CCL4), CCR5 (receptor for CCL4 and CCL11), and CXCR2 (receptor for CXCL1, CXCL2 and CXCL5). One of the chemokine receptors that is expressed on MDSCs that has effective role in MDSC migration is CXCR2. CXCR2-deficient mice showed significantly decreased MDSC induction in an inflammatory model of endometriosis when compared with wild-type mice (53). Also, we found in the current study that blocking CXCR2 using an antagonist Sch527123 almost completely inhibited TCDD-mediated MDSC induction. These data suggested that CXCR2 plays a critical role in TCDD-mediated induction of MDSCs. Additionally, we found that downregulation of miR-543-3p by TCDD, which directly targets CXCR2 (which we confirmed with transfection experiments), may be the key mechanism through which CXCR2 is induced in MDSCs. Importantly, another signal that is involved in MDSC accumulation includes GM-CSF, G-CSF, M-CSF, and VEGF (50, 54), and we found an increase in these factors in both serum and peritoneal exudate of the TCDD-treated group when compared with vehicle. These data together suggested that TCDD-induced upregulation of chemokines and their receptors were responsible for the migration and accumulation of PC-MDSCs. We were surprised to see that TCDD induced many chemokines; thus, we checked to see whether such chemokines might be induced following AhR activation involving DREs. We performed in silico analysis to identify the promoters of various chemokines and chemokine receptors through CpG island detection to find out whether any of these promoters expressed DREs. This analysis demonstrated that several chemokines and chemokine receptor promoters had numerous DREs in their promoter regions (Supplemental Table II). Also, the observation that TCDD caused a decrease in MDSCs in the BM, whereas they caused an increase in PC-MDSCs suggested that TCDD-induced chemokines in the peritoneal cavity may have caused MDSC mobilization from BM to peritoneal exudate. This was also supported by the observation that TCDD treatment led to a significant decrease in BM-MDSCs with a consequent increase in PC-MDSCs.
Additionally, we found that the PC-MDSCs induced by TCDD were not actively dividing, thereby supporting the idea that migration rather than proliferation was occurring in the TCDD-induced MDSCs. We noted that TCDD treatment also caused activation of MDSCs and affected their function. Energy metabolic pathways used by MDSCs may play a critical role on the immunosuppressive functions of MDSCs, and an increase in this pathway is a sign of increased immunosuppressive activity (31). In a recent report, tumor-bearing MDSCs were shown to have higher glycolytic rates that reduced reactive oxygen species–associated apoptosis, and in these same studies, blocking glycolysis in vivo decreased accumulation of MDSCs as well as reducing their immunosuppressive capabilities (51). We observed that TCDD-induced MDSCs had higher OCR, PER, and ATP production rates. These results, in addition to recent findings concerning the significant role energy pathways play in MDSC survival and function, suggest that AhR-activated MDSCs have increased energy available to suppress the immune response. TCDD-induced MDSCs also exhibited a significant increase in cell lineage-specific transcription factors and cytokines, including IL-10, PIM1, STAT3, and ARG2.
MDSCs constitute a heterogeneous population of cells representing a pathological state of activation of myeloid cells that have acquired a highly immunosuppressive phenotype (28). These cells are of great interest in cancer as well as in inflammation, as they potently suppress the cytotoxic activities of NK and NK T cells as well as the immune responses mediated by CD4 and CD8 T cells (52, 55). Under normal conditions, precursor myeloid cells from BM differentiate into mature granulocytes, macrophages, or dendritic cells as they home to peripheral organs. However, enhanced mediator production during pathological conditions, such as cancer, infections, trauma, inflammation, and autoimmunity, as well as sometimes in response to certain natural compounds, leads to increased proliferation of immature myeloid cells, whereas they block their terminal differentiation, resulting in the accumulation of immunosuppressive MDSCs (56). Recent studies from our laboratory have demonstrated the induction of MDSCs is an important mechanism through which several natural compounds exert immunosuppressive or anti-inflammatory properties, including marijuana, cannabinoids, and resveratrol (23, 32, 57, 58).
Functional analysis of TCDD-induced MDSCs showed potent immunosuppressive effects on T cell proliferation in vitro in the current study. We noted that, whereas both G-MDSC and M-MDSCs were immunosuppressive against T cell proliferation in vitro, the latter cells were more potent. We also demonstrated that TCDD-induced MDSCs were functionally immunosuppressive in vivo as well. Upon adoptive transfer of TCDD-induced MDSCs into Con A–injected mice, there was significant protection of the liver from acute inflammation. This was found to be associated with increased polarization of Th2 cells and Tregs and decreased induction of Th1 and Th17 cells, along with increased production of the anti-inflammatory cytokines TGF-β and IL-4 levels in these mice. These data are consistent with our previous studies demonstrating that MDSCs induced by cannabinoids can suppress Con A–induced hepatitis in vivo upon adoptive transfer (23). Importantly, we also noted that administration of TCDD into Con A–injected mice also led to protection of the liver, and we also saw similar cell phenotypes that include increased presence of MDSCs, Th2 cells, Tregs, and subsets of Tregs, but a decrease in the induction of Th1 cells. MDSCs can induce Tregs through disruption of the Th17/Treg balance because MDSCs have the potential to convert Th17 cells into Foxp3+ Tregs, enhancing the shift of the immune response from inflammation to tolerance (59). This is also achieved by increasing IL-10 or TGF-β production (22, 60). In the current study, we noted that, upon adoptive transfer of TCDD-induced MDSCs, there was significant reduction in CD4+IL-17+ and CD4+IFN-γ+ cells, whereas there was an increase in CD4+IL-10+. MDSCs have previously been shown to regulate inflammation by suppressing T cells that produce IFN-γ and IL-17 (61). In addition, MDSC depletion in vivo leads to increased production of IL-6, IFN-γ, and a Th17 response (61). MDSCs can also promote the expansion of Tregs via TGF-β–dependent (62) and TGF-β–independent pathways (63). In a recent study, it was shown that MDSC-derived IL-10 plays a critical role in attenuating inflammatory arthritis by blocking Th17 cells while promoting Tregs (22).
Several studies have shown that miRNA play an influential role in the toxicity of TCDD in animal models (41, 64–67). Studies have also shown that miRNA play a critical role in the regulation of MDSCs in different diseases models (25, 33, 68–70). For example, inhibition of miR-9 has been shown to promote the differentiation of MDSCs, whereas overexpression of miR-9 markedly enhanced the function of MDSCs (71). Also, miR-155 and miR-21 were the two most upregulated miRNAs during the induction of MDSCs from the BM cells by GM-CSF, IL-6, and TGF-B (72). Chen and his colleagues (73) found that miR-17-5p, miR-20a, miR-223, miR-21, miR-155, miR-494, miR-690, and miR-101 are of particular interest for tumor MDSC accumulation and function. In this study, we also observed that TCDD-induced MDSCs exhibited significant downregulation in the expression of miRNA, including miR-150-5p and miR-543-3p. The analysis in silico demonstrated that both miR-150-5p and miR-543-3p expressed DREs on their promoters (Supplemental Table II) and thus may help regulate their expression. Upon further characterization of these two miRNAs, we observed that these two miRNAs targeted several anti-inflammatory genes, including IL-10, PIM1, ARG2, and STAT3 as well as CCL11 and its receptors CCR3 and CCR5. Transfection studies confirmed that these miRNAs caused upregulation of these anti-inflammatory genes. IL-10, ARG2, and STAT3 have been well characterized for their ability to downregulate the production of proinflammatory cytokines, such as INF-γ, IL-2, IL-3, and TNF-α (52–56). Additionally, the STAT3 transcription factor is a hallmark of MDSCs, as STAT3 is directly involved in the accumulation and expansion of MDSCs in humans and mice. Arg1 found in MDSCs is directly involved in depleting l-arginine availability for T cells in the inflamed microenvironment and thereby inhibit T cell proliferation. MDSCs not only suppress T cell activation by IL-10 production but also by interacting with macrophages to increase IL-10 production and decrease IL-12 secretion (74–76). PIM1 is a member in the serine/threonine kinases family and has been implicated in the regulation of apoptosis, metabolism, cell cycle, and migration. PIM1 was observed to be overexpressed in numerous solid tumors and was accompanied by MDSCs accumulation (77). From the current study, we noted that CCL11 and its receptors CCR3 and CCR5 were specifically connected to MDSC migration from the BM to the peritoneal exudate. IPA shed light on the relationship between the genes STAT3, IL-10, PIM1, ARG2, CCL11, CCR3, and CCR5 with two downregulated miRNAs, miR-150-5p and miR-543-3p. miR-150-5p targets IL-10 and PIM1, whereas miR-543-3p targets ARG2, STAT3, CCL11, CCR3, and CCR5 genes. Although we have explained how various cytokines, chemokines, and transcription factors induced by TCDD are linked to MDSC induction and function, our results also demonstrated that CXCR2 signaling may play a dominant role in MDSC induction.
In summary, the current study demonstrates that activation of AhR triggers massive accumulation of MDSCs, even in naive mice without any inflammatory signal, resulting from induction of chemokines and their receptors. Furthermore, we observed that TCDD also causes changes in the expression of miRNA within the MDSCs, which can alter gene expression to promote their anti-inflammatory functions. AhR activation is a double-edged sword. On one hand, AhR plays a key role in intestinal homeostasis, as deficits in AhR signaling have been linked to experimental and human intestinal bowel disease (78). In contrast, AhR ligands such as the environmental contaminants TCDD or methylcholanthrene (MC), are highly toxic and are considered to be carcinogens. Interestingly, this dual action of AhR activation correlates with MDSCs, which are known to suppress inflammation and autoimmune disease (79) as well as promote tumor development and progression (80). Further studies aimed at addressing the nature of AhR-induced MDSCs in the regulation of health and disease should provide useful clues to regulate disease pathogenesis.
Supplementary Material
This work was supported by funding sources from the National Institutes of Health (R01ES019313, R01MH094755, R01AI123947, R01AI129788, P01AT003961, P20GM103641, and R01AT006888) and the Ministry of Higher Education and Scientific Research, Iraq.
The microarray data presented in this article have been submitted to the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE134337.
The online version of this article contains supplemental material.
- AhR
- aryl hydrocarbon receptor
- ALT
- alanine transaminase
- Arg1
- Arginase-1
- Arnt
- AhR nuclear translocator protein
- BM
- bone marrow
- BM-MDSC
- MDSC from the BM
- DRE
- dioxin response element
- G-MDSC
- granulocytic MDSC
- IPA
- Ingenuity Pathway Analysis
- 3-MC
- 3-methylcholanthrene
- MDSC
- myeloid-derived suppressor cell
- MHC-II
- MHC class II
- miRNA
- microRNA
- M-MDSC
- monocytic MDSC
- OCR
- oxygen consumption rate
- PC-MDSC
- MDSC from the peritoneal cavity
- PER
- proton efflux rate
- Q-PCR
- quantitative PCR
- TCDD
- 2,3,7,8-tetrachlorodibenzo-p-dioxin
- Treg
- regulatory T cell.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Fernandez-Salguero P. M., Hilbert D. M., Rudikoff S., Ward J. M., Gonzalez F. J. 1996. Aryl-hydrocarbon receptor-deficient mice are resistant to 2,3,7,8-tetrachlorodibenzo-p-dioxin-induced toxicity. Toxicol. Appl. Pharmacol. 140: 173–179. [DOI] [PubMed] [Google Scholar]
- 2.McIntosh B. E., Hogenesch J. B., Bradfield C. A. 2010. Mammalian Per-Arnt-Sim proteins in environmental adaptation. Annu. Rev. Physiol. 72: 625–645. [DOI] [PubMed] [Google Scholar]
- 3.Schmidt J. V., Bradfield C. A. 1996. Ah receptor signaling pathways. Annu. Rev. Cell Dev. Biol. 12: 55–89. [DOI] [PubMed] [Google Scholar]
- 4.Fisher M. T., Nagarkatti M., Nagarkatti P. S. 2004. Combined screening of thymocytes using apoptosis-specific cDNA array and promoter analysis yields novel gene targets mediating TCDD-induced toxicity. Toxicol. Sci. 78: 116–124. [DOI] [PubMed] [Google Scholar]
- 5.Camacho I. A., Singh N., Hegde V. L., Nagarkatti M., Nagarkatti P. S. 2005. Treatment of mice with 2,3,7,8-tetrachlorodibenzo-p-dioxin leads to aryl hydrocarbon receptor-dependent nuclear translocation of NF-kappaB and expression of Fas ligand in thymic stromal cells and consequent apoptosis in T cells. J. Immunol. 175: 90–103. [DOI] [PubMed] [Google Scholar]
- 6.Camacho I. A., Nagarkatti M., Nagarkatti P. S. 2002. 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD) induces Fas-dependent activation-induced cell death in superantigen-primed T cells. Arch. Toxicol. 76: 570–580. [DOI] [PubMed] [Google Scholar]
- 7.Quintana F. J., Basso A. S., Iglesias A. H., Korn T., Farez M. F., Bettelli E., Caccamo M., Oukka M., Weiner H. L. 2008. Control of T(reg) and T(H)17 cell differentiation by the aryl hydrocarbon receptor. Nature 453: 65–71. [DOI] [PubMed] [Google Scholar]
- 8.Singh N. P., Singh U. P., Singh B., Price R. L., Nagarkatti M., Nagarkatti P. S. 2011. Activation of aryl hydrocarbon receptor (AhR) leads to reciprocal epigenetic regulation of FoxP3 and IL-17 expression and amelioration of experimental colitis. PLoS One 6: e23522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singh N. P., Singh U. P., Rouse M., Zhang J., Chatterjee S., Nagarkatti P. S., Nagarkatti M. 2016. Dietary indoles suppress delayed-type hypersensitivity by inducing a switch from proinflammatory Th17 cells to anti-inflammatory regulatory T cells through regulation of microRNA. J. Immunol. 196: 1108–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loub W. D., Wattenberg L. W., Davis D. W. 1975. Aryl hydrocarbon hydroxylase induction in rat tissues by naturally occurring indoles of cruciferous plants. J. Natl. Cancer Inst. 54: 985–988. [PubMed] [Google Scholar]
- 11.Chitrala K. N., Yang X., Nagarkatti P., Nagarkatti M. 2018. Comparative analysis of interactions between aryl hydrocarbon receptor ligand binding domain with its ligands: a computational study. BMC Struct. Biol. 18: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Busbee P. B., Nagarkatti M., Nagarkatti P. S. 2015. Natural indoles, indole-3-carbinol (I3C) and 3,3′-diindolylmethane (DIM), attenuate staphylococcal enterotoxin B-mediated liver injury by downregulating miR-31 expression and promoting caspase-2-mediated apoptosis. PLoS One 10: e0118506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rouse M., Singh N. P., Nagarkatti P. S., Nagarkatti M. 2013. Indoles mitigate the development of experimental autoimmune encephalomyelitis by induction of reciprocal differentiation of regulatory T cells and Th17 cells. Br. J. Pharmacol. 169: 1305–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singh N. P., Hegde V. L., Hofseth L. J., Nagarkatti M., Nagarkatti P. 2007. Resveratrol (trans-3,5,4′-trihydroxystilbene) ameliorates experimental allergic encephalomyelitis, primarily via induction of apoptosis in T cells involving activation of aryl hydrocarbon receptor and estrogen receptor. Mol. Pharmacol. 72: 1508–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hubbard T. D., Murray I. A., Perdew G. H. 2015. Indole and tryptophan metabolism: endogenous and dietary routes to Ah receptor activation. Drug Metab. Dispos. 43: 1522–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gabrilovich D. I., Nagaraj S. 2009. Myeloid-derived suppressor cells as regulators of the immune system. Nat. Rev. Immunol. 9: 162–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gabrilovich D. I., Ostrand-Rosenberg S., Bronte V. 2012. Coordinated regulation of myeloid cells by tumours. Nat. Rev. Immunol. 12: 253–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Young M. R., Endicott R. A., Duffie G. P., Wepsic H. T. 1987. Suppressor alveolar macrophages in mice bearing metastatic Lewis lung carcinoma tumors. J. Leukoc. Biol. 42: 682–688. [DOI] [PubMed] [Google Scholar]
- 19.Seung L. P., Rowley D. A., Dubey P., Schreiber H. 1995. Synergy between T-cell immunity and inhibition of paracrine stimulation causes tumor rejection. Proc. Natl. Acad. Sci. USA 92: 6254–6258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dai J., El Gazzar M., Li G. Y., Moorman J. P., Yao Z. Q. 2015. Myeloid-derived suppressor cells: paradoxical roles in infection and immunity. J. Innate Immun. 7: 116–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goh C., Narayanan S., Hahn Y. S. 2013. Myeloid-derived suppressor cells: the dark knight or the joker in viral infections? Immunol. Rev. 255: 210–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park M. J., Lee S. H., Kim E. K., Lee E. J., Baek J. A., Park S. H., Kwok S. K., Cho M. L. 2018. Interleukin-10 produced by myeloid-derived suppressor cells is critical for the induction of Tregs and attenuation of rheumatoid inflammation in mice. Sci. Rep. 8: 3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hegde V. L., Nagarkatti M., Nagarkatti P. S. 2010. Cannabinoid receptor activation leads to massive mobilization of myeloid-derived suppressor cells with potent immunosuppressive properties. Eur. J. Immunol. 40: 3358–3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sido J. M., Yang X., Nagarkatti P. S., Nagarkatti M. 2015. Δ9-Tetrahydrocannabinol-mediated epigenetic modifications elicit myeloid-derived suppressor cell activation via STAT3/S100A8. J. Leukoc. Biol. 97: 677–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hegde V. L., Tomar S., Jackson A., Rao R., Yang X., Singh U. P., Singh N. P., Nagarkatti P. S., Nagarkatti M. 2013. Distinct microRNA expression profile and targeted biological pathways in functional myeloid-derived suppressor cells induced by Δ9-tetrahydrocannabinol in vivo: regulation of CCAAT/enhancer-binding protein α by microRNA-690. J. Biol. Chem. 288: 36810–36826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang X., Hegde V. L., Rao R., Zhang J., Nagarkatti P. S., Nagarkatti M. 2014. Histone modifications are associated with Δ9-tetrahydrocannabinol-mediated alterations in antigen-specific T cell responses. J. Biol. Chem. 289: 18707–18718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singh U. P., Singh N. P., Singh B., Price R. L., Nagarkatti M., Nagarkatti P. S. 2012. Cannabinoid receptor-2 (CB2) agonist ameliorates colitis in IL-10(-/-) mice by attenuating the activation of T cells and promoting their apoptosis. Toxicol. Appl. Pharmacol. 258: 256–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Veglia F., Perego M., Gabrilovich D. 2018. Myeloid-derived suppressor cells coming of age. Nat. Immunol. 19: 108–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bronte V., Brandau S., Chen S. H., Colombo M. P., Frey A. B., Greten T. F., Mandruzzato S., Murray P. J., Ochoa A., Ostrand-Rosenberg S., et al. 2016. Recommendations for myeloid-derived suppressor cell nomenclature and characterization standards. Nat. Commun. 7: 12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pansoy A., Ahmed S., Valen E., Sandelin A., Matthews J. 2010. 3-methylcholanthrene induces differential recruitment of aryl hydrocarbon receptor to human promoters. Toxicol. Sci. 117: 90–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hossain F., Al-Khami A. A., Wyczechowska D., Hernandez C., Zheng L., Reiss K., Valle L. D., Trillo-Tinoco J., Maj T., Zou W., et al. 2015. Inhibition of fatty acid oxidation modulates immunosuppressive functions of myeloid-derived suppressor cells and enhances cancer therapies. Cancer Immunol. Res. 3: 1236–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hegde V. L., Nagarkatti P. S., Nagarkatti M. 2011. Role of myeloid-derived suppressor cells in amelioration of experimental autoimmune hepatitis following activation of TRPV1 receptors by cannabidiol. PLoS One 6: e18281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kadhim S., Singh N. P., Zumbrun E. E., Cui T., Chatterjee S., Hofseth L., Abood A., Nagarkatti P., Nagarkatti M. 2018. Resveratrol-mediated attenuation of Staphylococcus aureus enterotoxin B-induced acute liver injury is associated with regulation of microRNA and induction of myeloid-derived suppressor cells. Front. Microbiol. 9: 2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Greenlee W. F., Dold K. M., Irons R. D., Osborne R. 1985. Evidence for direct action of 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) on thymic epithelium. Toxicol. Appl. Pharmacol. 79: 112–120. [DOI] [PubMed] [Google Scholar]
- 35.Holladay S. D., Lindstrom P., Blaylock B. L., Comment C. E., Germolec D. R., Heindell J. J., Luster M. I. 1991. Perinatal thymocyte antigen expression and postnatal immune development altered by gestational exposure to tetrachlorodibenzo-p-dioxin (TCDD). Teratology 44: 385–393. [DOI] [PubMed] [Google Scholar]
- 36.Esser C. 1994. Dioxins and the immune system: mechanisms of interference. A meeting report. Int. Arch. Allergy Immunol. 104: 126–130. [DOI] [PubMed] [Google Scholar]
- 37.Kerkvliet N. I., Shepherd D. M., Baecher-Steppan L. 2002. T lymphocytes are direct, aryl hydrocarbon receptor (AhR)-dependent targets of 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD): AhR expression in both CD4+ and CD8+ T cells is necessary for full suppression of a cytotoxic T lymphocyte response by TCDD. Toxicol. Appl. Pharmacol. 185: 146–152. [DOI] [PubMed] [Google Scholar]
- 38.Mimura J., Fujii-Kuriyama Y. 2003. Functional role of AhR in the expression of toxic effects by TCDD. Biochim. Biophys. Acta 1619: 263–268. [DOI] [PubMed] [Google Scholar]
- 39.Kerkvliet N. I. 2002. Recent advances in understanding the mechanisms of TCDD immunotoxicity. Int. Immunopharmacol. 2: 277–291. [DOI] [PubMed] [Google Scholar]
- 40.Sulentic C. E., Holsapple M. P., Kaminski N. E. 1998. Aryl hydrocarbon receptor-dependent suppression by 2,3,7, 8-tetrachlorodibenzo-p-dioxin of IgM secretion in activated B cells. Mol. Pharmacol. 53: 623–629. [PubMed] [Google Scholar]
- 41.Singh N. P., Singh U. P., Guan H., Nagarkatti P., Nagarkatti M. 2012. Prenatal exposure to TCDD triggers significant modulation of microRNA expression profile in the thymus that affects consequent gene expression. PLoS One 7: e45054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Singh N. P., Nagarkatti M., Nagarkatti P. 2008. Primary peripheral T cells become susceptible to 2,3,7,8-tetrachlorodibenzo-p-dioxin-mediated apoptosis in vitro upon activation and in the presence of dendritic cells. Mol. Pharmacol. 73: 1722–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Faulconer L., Camacho I., Nagarkatti M., Nagarkatti P. S. 2006. Superantigen-primed T cells exposed to 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) replicate poorly following recall encounter. Arch. Toxicol. 80: 134–145. [DOI] [PubMed] [Google Scholar]
- 44.Camacho I. A., Nagarkatti M., Nagarkatti P. S. 2004. Evidence for induction of apoptosis in T cells from murine fetal thymus following perinatal exposure to 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD). Toxicol. Sci. 78: 96–106. [DOI] [PubMed] [Google Scholar]
- 45.Elliott D. M., Singh N., Nagarkatti M., Nagarkatti P. S. 2018. Cannabidiol attenuates experimental autoimmune encephalomyelitis model of multiple sclerosis through induction of myeloid-derived suppressor cells. Front. Immunol. 9: 1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sido J. M., Nagarkatti P. S., Nagarkatti M. 2015. Δ9-Tetrahydrocannabinol attenuates allogeneic host-versus-graft response and delays skin graft rejection through activation of cannabinoid receptor 1 and induction of myeloid-derived suppressor cells. J. Leukoc. Biol. 98: 435–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Condamine T., Gabrilovich D. I. 2011. Molecular mechanisms regulating myeloid-derived suppressor cell differentiation and function. Trends Immunol. 32: 19–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Condamine T., Mastio J., Gabrilovich D. I. 2015. Transcriptional regulation of myeloid-derived suppressor cells. J. Leukoc. Biol. 98: 913–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yan D., Yang Q., Shi M., Zhong L., Wu C., Meng T., Yin H., Zhou J. 2013. Polyunsaturated fatty acids promote the expansion of myeloid-derived suppressor cells by activating the JAK/STAT3 pathway. Eur. J. Immunol. 43: 2943–2955. [DOI] [PubMed] [Google Scholar]
- 50.Umansky V., Sevko A. 2013. Tumor microenvironment and myeloid-derived suppressor cells. Cancer Microenviron. 6: 169–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jian S. L., Chen W. W., Su Y. C., Su Y. W., Chuang T. H., Hsu S. C., Huang L. R. 2017. Glycolysis regulates the expansion of myeloid-derived suppressor cells in tumor-bearing hosts through prevention of ROS-mediated apoptosis. Cell Death Dis. 8: e2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dolcetti L., Marigo I., Mantelli B., Peranzoni E., Zanovello P., Bronte V. 2008. Myeloid-derived suppressor cell role in tumor-related inflammation. Cancer Lett. 267: 216–225. [DOI] [PubMed] [Google Scholar]
- 53.Zhang T., Zhou J., Man G. C. W., Leung K. T., Liang B., Xiao B., Ma X., Huang S., Huang H., Hegde V. L., et al. 2018. MDSCs drive the process of endometriosis by enhancing angiogenesis and are a new potential therapeutic target. Eur. J. Immunol. 48: 1059–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dolcetti L., Peranzoni E., Ugel S., Marigo I., Fernandez Gomez A., Mesa C., Geilich M., Winkels G., Traggiai E., Casati A., et al. 2010. Hierarchy of immunosuppressive strength among myeloid-derived suppressor cell subsets is determined by GM-CSF. Eur. J. Immunol. 40: 22–35. [DOI] [PubMed] [Google Scholar]
- 55.Ostrand-Rosenberg S. 2010. Myeloid-derived suppressor cells: more mechanisms for inhibiting antitumor immunity. Cancer Immunol. Immunother. 59: 1593–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bronte V., Apolloni E., Cabrelle A., Ronca R., Serafini P., Zamboni P., Restifo N. P., Zanovello P. 2000. Identification of a CD11b(+)/Gr-1(+)/CD31(+) myeloid progenitor capable of activating or suppressing CD8(+) T cells. Blood 96: 3838–3846. [PMC free article] [PubMed] [Google Scholar]
- 57.Singh U. P., Singh N. P., Singh B., Hofseth L. J., Taub D. D., Price R. L., Nagarkatti M., Nagarkatti P. S. 2012. Role of resveratrol-induced CD11b(+) Gr-1(+) myeloid derived suppressor cells (MDSCs) in the reduction of CXCR3(+) T cells and amelioration of chronic colitis in IL-10(-/-) mice. Brain Behav. Immun. 26: 72–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guan H., Singh N. P., Singh U. P., Nagarkatti P. S., Nagarkatti M. 2012. Resveratrol prevents endothelial cells injury in high-dose interleukin-2 therapy against melanoma. PLoS One 7: e35650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hoechst B., Gamrekelashvili J., Manns M. P., Greten T. F., Korangy F. 2011. Plasticity of human Th17 cells and iTregs is orchestrated by different subsets of myeloid cells. Blood 117: 6532–6541. [DOI] [PubMed] [Google Scholar]
- 60.Lebrun A., Lo Re S., Chantry M., Izquierdo Carerra X., Uwambayinema F., Ricci D., Devosse R., Ibouraadaten S., Brombin L., Palmai-Pallag M., et al. 2017. CCR2+ monocytic myeloid-derived suppressor cells (M-MDSCs) inhibit collagen degradation and promote lung fibrosis by producing transforming growth factor-β1. J. Pathol. 243: 320–330. [DOI] [PubMed] [Google Scholar]
- 61.Arocena A. R., Onofrio L. I., Pellegrini A. V., Carrera Silva A. E., Paroli A., Cano R. C., Aoki M. P., Gea S. 2014. Myeloid-derived suppressor cells are key players in the resolution of inflammation during a model of acute infection. Eur. J. Immunol. 44: 184–194. [DOI] [PubMed] [Google Scholar]
- 62.Huang B., Pan P. Y., Li Q., Sato A. I., Levy D. E., Bromberg J., Divino C. M., Chen S. H. 2006. Gr-1+CD115+ immature myeloid suppressor cells mediate the development of tumor-induced T regulatory cells and T-cell anergy in tumor-bearing host. Cancer Res. 66: 1123–1131. [DOI] [PubMed] [Google Scholar]
- 63.Serafini P., Mgebroff S., Noonan K., Borrello I. 2008. Myeloid-derived suppressor cells promote cross-tolerance in B-cell lymphoma by expanding regulatory T cells. Cancer Res. 68: 5439–5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Moffat I. D., Boutros P. C., Celius T., Linden J., Pohjanvirta R., Okey A. B. 2007. microRNAs in adult rodent liver are refractory to dioxin treatment. Toxicol. Sci. 99: 470–487. [DOI] [PubMed] [Google Scholar]
- 65.Yoshioka W., Higashiyama W., Tohyama C. 2011. Involvement of microRNAs in dioxin-induced liver damage in the mouse. Toxicol. Sci. 122: 457–465. [DOI] [PubMed] [Google Scholar]
- 66.Hanieh H. 2015. Aryl hydrocarbon receptor-microRNA-212/132 axis in human breast cancer suppresses metastasis by targeting SOX4. Mol. Cancer 14: 172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sofo V., Götte M., Laganà A. S., Salmeri F. M., Triolo O., Sturlese E., Retto G., Alfa M., Granese R., Abrão M. S. 2015. Correlation between dioxin and endometriosis: an epigenetic route to unravel the pathogenesis of the disease. Arch. Gynecol. Obstet. 292: 973–986. [DOI] [PubMed] [Google Scholar]
- 68.Reithmair M., Buschmann D., Märte M., Kirchner B., Hagl D., Kaufmann I., Pfob M., Chouker A., Steinlein O. K., Pfaffl M. W., Schelling G. 2017. Cellular and extracellular miRNAs are blood-compartment-specific diagnostic targets in sepsis. J. Cell. Mol. Med. 21: 2403–2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang R., Liu C., Niu Y., Jing Y., Zhang H., Wang J., Yang J., Zen K., Zhang J., Zhang C. Y., Li D. 2016. MicroRNA-128-3p regulates mitomycin C-induced DNA damage response in lung cancer cells through repressing SPTAN1. Oncotarget 8: 58098–58107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lacedonia D., Palladino G. P., Foschino-Barbaro M. P., Scioscia G., Carpagnano G. E. 2017. Expression profiling of miRNA-145 and miRNA-338 in serum and sputum of patients with COPD, asthma, and asthma-COPD overlap syndrome phenotype. Int. J. Chron. Obstruct. Pulmon. Dis. 12: 1811–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tian J., Rui K., Tang X., Ma J., Wang Y., Tian X., Zhang Y., Xu H., Lu L., Wang S. 2015. MicroRNA-9 regulates the differentiation and function of myeloid-derived suppressor cells via targeting Runx1. J. Immunol. 195: 1301–1311. [DOI] [PubMed] [Google Scholar]
- 72.Li L., Zhang J., Diao W., Wang D., Wei Y., Zhang C. Y., Zen K. 2014. MicroRNA-155 and MicroRNA-21 promote the expansion of functional myeloid-derived suppressor cells. J. Immunol. 192: 1034–1043. [DOI] [PubMed] [Google Scholar]
- 73.Chen S., Zhang Y., Kuzel T. M., Zhang B. 2015. Regulating tumor myeloid-derived suppressor cells by MicroRNAs. Cancer Cell Microenviron. 2: e637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rébé C., Végran F., Berger H., Ghiringhelli F. 2013. STAT3 activation: a key factor in tumor immunoescape. JAK-STAT 2: e23010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Munder M., Schneider H., Luckner C., Giese T., Langhans C. D., Fuentes J. M., Kropf P., Mueller I., Kolb A., Modolell M., Ho A. D. 2006. Suppression of T-cell functions by human granulocyte arginase. Blood 108: 1627–1634. [DOI] [PubMed] [Google Scholar]
- 76.Sinha P., Clements V. K., Bunt S. K., Albelda S. M., Ostrand-Rosenberg S. 2007. Cross-talk between myeloid-derived suppressor cells and macrophages subverts tumor immunity toward a type 2 response. J. Immunol. 179: 977–983. [DOI] [PubMed] [Google Scholar]
- 77.Narlik-Grassow M., Blanco-Aparicio C., Carnero A. 2014. The PIM family of serine/threonine kinases in cancer. Med. Res. Rev. 34: 136–159. [DOI] [PubMed] [Google Scholar]
- 78.Lamas B., Richard M. L., Leducq V., Pham H. P., Michel M. L., Da Costa G., Bridonneau C., Jegou S., Hoffmann T. W., Natividad J. M., et al. 2016. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat. Med. 22: 598–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wheeler M. A., Rothhammer V., Quintana F. J. 2017. Control of immune-mediated pathology via the aryl hydrocarbon receptor. J. Biol. Chem. 292: 12383–12389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kumahara T. 1989. [Measurement of radiation energy and its application. II. 2. Electronics for energy measurement]. Radioisotopes 38: 537–544. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.