Abstract
Prostate cancer is a common malignancy with a high mortality rate. Long non-coding RNA metastasis associated with lung adenocarcinoma transcript 1 (MALAT1) has been reported to serve tumor-promoting roles. However, the underlying mechanism requires further examination. In the present study, it was demonstrated that MALAT1 was increased while microRNA (miR/miRNA)-1-3p was decreased in prostate cancer cell lines. The silencing of MALAT1 inhibited migration, invasion and epithelial-mesenchymal transition, when epithelial (E)-cadherin expression level was increased, and neural (N)-cadherin, vimentin, Slug and Snail expression levels were decreased. Dual-luciferase reporter assay results demonstrated that miR-1-3p bound to MALAT1 and coronin 1C (CORO1C) 3′ untranslated region, and MALAT1 competed with CORO1C for the binding sites of miR-1-3p. MALAT1 inhibited the expression of miR-1-3p and vice versa. MALAT1 knockdown induced the decline of CORO1C, which was subsequently recovered by the miR-1-3p inhibitor. In addition, by inhibiting miR-1-3p or overexpressing CORO1C, the silencing of MALAT1-induced phenotypic alterations were restored. In conclusion, MALAT1 serving as a degradable miRNA sponge, may sequester miR-1-3p from CORO1C and by silencing MALAT1, migration, invasion and epithelial-mesenchymal transition may be inhibited in prostate cancer cells. MALAT1 and CORO1C may serve as novel clinical therapeutic targets for prostate cancer.
Keywords: prostate cancer, long noncoding RNA, miRNA sponge, metastasis associated lung adenocarnoma transcript 1, coronin 1C
Introduction
Prostate cancer is the most common reported malignancy of men in developed countries (1). A total of 307,500 men worldwide succumb to prostate cancer annually (1). The pathogenesis of prostate cancer involves multiple factors, including age, race, familial heredity and androgen levels (2). Androgen deprivation therapy is the most common clinical approach to treat prostate cancer. However, in the majority of patients castration-resistant prostate cancer (CRPC) develops, for which current anticancer treatments have yet to report positive clinical outcomes (3,4). Therefore, examining possible novel therapies is of great importance for the clinical treatment of prostate cancer.
In the recent years, the role of long noncoding RNAs (lncRNAs) in cancer pathogenesis and progression has gained increasing attention. LncRNAs are a class of endogenous noncoding RNAs with >200 nucleotides (5). LncRNA metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), also known as nuclear-enriched abundant transcript 2, was first indicated to be highly expressed in non-small cell lung cancer with a potential metastatic promoting role (6). Subsequently, MALAT1 was reported to be a nuclear-enriched transcript and widely conserved among 33 mammalian species (7). MALAT1 has been demonstrated to promote proliferation, migration, invasion, cell cycle transition and tumorigenesis in multiple types of malignant tumors, including ovarian cancer, cervical cancer and gastric cancer (8–11). MALAT1 is also indicated to be overexpressed in prostate cancer and silencing of MALAT1 inhibits proliferation, migration and invasion, and induces cell cycle arrest in prostate cancer cells (12).
MALAT1 has been reported to serve a binding role to microRNAs (miRs/miRNAs) by acting as a miRNA sponge or a competitive endogenous RNA (ceRNA). For instance, MALAT1 sponges miR-22 and upregulates the miR-22 targets matrix metallopeptidase 14 and Snail, and consequently enhances malignant melanoma growth and metastasis (13). In breast cancer cells, MALAT1 sponges miR-1-3p in order to promote malignancies (14,15). In addition, miR-1-3p is downregulated in prostate cancer specimens and its expression is negatively associated with prognosis. The overexpression of miR-1-3p inhibits proliferation and invasion in prostate cancer cells (16). Therefore, the aim of the present study is to examine whether the function of MALAT1 in prostate cancer is associated with miR-1-3p. This study investigated the association between MALAT1 and miR-1-3p in prostate cancer, and examined the underlying mechanism of MALAT1.
Materials and methods
Plasmid construction
To construct the lncRNA MALAT1 knockdown plasmid, two primers containing a sequence targeting the 5′-GGAAGATAGAAACAAGATA-3′ in the MALAT1 transcript were synthesized. The two primers were annealed and inserted into the pRNAH1.1 vector (Genscript, Nanjing, Jiangsu, China) at BamHI and HindIII sites. The sequence containing a negative control (NC) segment was inserted into the pRNAH1.1 vector and served as the control. The sequence information of the primers is presented in Table I.
Table I.
The information of knockdown, amplification PCR and qPCR primers used in this study.
| Name | Sequence (5′-3′) |
|---|---|
| shMALAT1 S | GATCCCCGGAAGATAGAAACAAGATATTCAAGAGATATCTTGTTTCTATCTTCCTTTTT |
| shMALAT1 AS | AGCTAAAAAGGAAGATAGAAACAAGATATCTCTTGAATATCTTGTTTCTATCTTCCGGG |
| shRNA NC S | GATCCCCTTCTCCGAACGTGTCACGTTTCAAGAGAACGTGACACGTTCGGAGAATTTTT |
| shRNA NC AS | AGCTAAAAATTCTCCGAACGTGTCACGTTCTCTTGAAACGTGACACGTTCGGAGAAGGG |
| CORO1C CDS F | CAAGGATCCATGAGGCGAGTGGTACGACAG |
| CORO1C CDS R | CGCCTCGAGTCAGGCTGCTATCTTTGCCAT |
| MALAT1 amplification PCR F | CACGGATCCGATAAAGGCTGAGTGTTGAG |
| MALAT1 amplification PCR F | CAACTCGAGCTTCGCATACGTGTGTCTGC |
| MALAT1 qPCR F | GACTTCAGGTCTGTCTGTTCT |
| MALAT1 qPCR R | CAACAATCACTACTCCAAGC |
| GAPDH qPCR F | GAAGGTCGGAGTCAACGGAT |
| GAPDH qPCR R | CCTGGAAGATGGTGATGGGAT |
| miR-1-3p RT | GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACATACAT |
| miR-1-3p qPCR F | GCGGGCTGGAATGTAAAG |
| U6 RT | GTTGGCTCTG GTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACAAAATATGG |
| U6 qPCR F | GCTTCGGCAGCACATATACT |
| qPCR Reverse | GTGCAGGGTCCGAGGTATTC |
MALAT1, metastasis-associated lung adenocarcinoma transcript 1; RT, reverse transcription; S, sense; AS, NC, negative control; antisense; F, forward; R, reverse; qPCR, quantitative polymerase chain reaction; miR, microRNA; sh, short hairpin; the coding sequence.
To establish the overexpression plasmid of coronin 1C (CORO1C), the coding sequence (CDS) was amplified with human cDNA as the template, in the presence of CORO1C CDS primers. Following sequence analysis, the amplification segment was inserted into pcDNA3.1 vector (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) with BamHI and XhoI sites. The MALAT1 sequence targeted by miR-1-3p was also cloned into the pcDNA3.1 vector with BamHI and XhoI sites, in the presence of amplification PCR primers. The information regarding the CORO1C CDS and MALAT1 amplification PCR primers is presented in Table I.
Cell culture and transfection
Prostatic epithelial cell line RWPE2 was purchased from Shanghai Zhongqiaoxinzhou Biotech (Shanghai, China) and cultured with prostatic epithelial cell medium (Shanghai Zhongqiaoxinzhou Biotech) at 37°C in 5% CO2. Prostate cancer cell lines DU145 and PC-3 were purchased from Procell Life Science & Technology Co., Ltd., (Wuhan, China), and cultured with minimum essential medium (MEM) or Ham's F-12K medium (Procell Life Science & Technology Co., Ltd.) supplemented with 10% fetal bovine serum (FBS; Biological Industries, Kibbutz Beit Haemek, Israel). Prostate cancer cell lines LNCaP and 22RV1 were purchased from Shanghai Zhongqiaoxinzhou Biotech and cultured with RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% FBS. The cells were seeded in 6-well plates and transfected with short hairpin RNA (sh)MALAT1 plasmid (~6.8 kb) with Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher Scientific Inc.). Following 24 h, the cells were treated with 100 µg/ml G418 for 2 weeks and returned to normal medium. Subsequent to culturing for ~2 weeks, the monoclonal cell masses were selected. The MALAT1-knocked down cells were obtained.
After culturing for 24 h, the LNCaP and 22RV1 cells were transfected with miR-1-3p mimics or inhibitor in serum-free medium, in the presence of Lipofectamine 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.). A total of 4 h later, the transfection medium was replaced with normal medium and the cells were cultured for other experiments.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
The total RNA was extracted using a TRIpure RNA extraction kit (BioTek China, Beijing, China) and the concentration was measured with an ultraviolet spectrophotometer (Thermo Fisher Scientific Inc.). The RNA was reverse transcribed into cDNA with M-MLV reverse transcriptase (BioTek China) and 2×Power Taq PCR Master Mix buffer (BioTeke), in the presence of oligo(dT) and random primers, or specific miRNA RT primers (Sangon Biotech Co., Ltd., Shanghai, China): Denaturation at 70°C for 2 min, annealing at 25°C for 10 min, and extension at 42°C for 50 min. Subsequently, the cDNA was used for RT-qPCR with SYBR Green (Beijing Solarbio Science & Technology Co., Ltd., Beijing, China) to detect the expression levels of MALAT1 and miR-1-3p, with GAPDH or U6 as the respective internal control. The procedure was as follows: 94°C for 5 min 10 sec, 60°C for 20 sec, 72°C for 30 sec, and 40 cycles of 72°C for 2 min 30 sec, 40°C for 1 min 30 sec, melting from 60–94°C each 1°C for 1 sec, and finally incubation at 25°C for several minutes. The data were calculated using the 2−ΔΔCq method (17). The information on primers is presented in Table I.
Western blot analysis
The protein was extracted with radioimmunoprecipitation assay lysis buffer (Beyotime Institute of Biotechnology, Haimen, China) and the concentration was determined with a bicinchoninic acid protein assay kit (Beyotime Institute of Biotechnology). Following protein denaturation by boiling, the proteins were separated with SDS-PAGE (5, 8, 10 and 14% separation gel for different sized proteins) and transferred onto a polyvinylidene difluoride (PVDF) membrane (EMD Millipore, Billerica, MA, USA). Following blocking with 5% skimmed milk at room temperature for 1 h, the PVDF membrane was incubated with one of the following antibodies at 4°C overnight: Rabbit anti-CORO1C (1:500; cat. no. 14749-1-AP; Wuhan Sanying Biotechnology, Wuhan, China), mouse anti-epithelial (E)-cadherin (1:1,000; cat. no. 14472), rabbit anti-neural (N)-cadherin (1:1,000; cat. no. 4061), rabbit anti-vimentin (1:500; cat. no. 5741; all Cell Signaling Technology, Inc., Danvers, MA, USA), rabbit anti-Slug (1:500; cat. no. ab27568; Abcam, Cambridge, UK), rabbit anti-Snail (1:1,000; cat. no. 3879; Cell Signaling Technology, Inc.). Following rinsing with TBST (TBS buffer with 0.15% Tween-20), the PVDF membrane was incubated with the corresponding secondary antibody labeled with IgG at 37°C for 45 min. The PVDF membrane was reacted with enhanced chemiluminescent reagent (Beyotime Institute of Biotechnology) for 5 min, followed by signal exposure. The immunoblotting bands were analyzed with Gel-Pro-Analyzer 4 software (Media Cybernetics, Rockville, MD, USA). GAPDH served as the internal control. The GAPDH level was detected via incubation with rabbit anti-GAPDH (1:500; cat. no. bs-2188R; Bioss, Beijing, China) and corresponding secondary antibody as described above.
Bioinformatic analysis
StarBase v2.0 (http://starbase.sysu.edu.cn/) was used to predict the candidate miRNAs of MALAT1. TargetScan 7.2 (http://www.targetscan.org/vert_72/) was subsequently used to predict the targeted genes of miR-1-3p.
Dual-luciferase reporter assay
To demonstrate the binding of MALAT1 to miR-1-3p, the DNA segment containing the sequence miR-1-3p, was cloned into the dual-luciferase reporter, pmirGLO (Promega Cooperation, Madison, WI, USA). The 293T cells (Procell Life Science & Technology Co., Ltd.) were transfected with the dual-luciferase reporter containing the MALAT1 segment or its mutant sequence and miR-1-3p mimics, in presence of Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher Scientific Inc.). Following 48 h, the cells were treated with dual-luciferase reporter assay system (Promega Cooperation, Madison, WI, USA) and the firefly and the Renilla luciferase activities were detected.
Similarly, the DNA segment containing CORO1C 3′UTR sequence targeted by miR-1-3p, was cloned into dual-luciferase reporter constructs. The firefly and Renilla luciferase activities were detected.
Wound healing assay
The LNCaP and 22RV1 cells were cultured until they reached 80% confluence, and treated with 1 µg/ml mitomycin C (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) for 1 h. The wound was made with a 200 µl pipette tip and the cells were subsequently cultured. At 0 and 24 h, the wound size was viewed under an inverted light microscope (magnification ×100; Olympus Corporation, Tokyo, Japan).
Transwell assay
The Transwell assay was performed and membrane precoated with Matrigel to detect the invasive ability of LNCaP and 22RV1 cells. The Transwell chamber with an 8.0 µm pore polycarbonate membrane (Corning Inc., Corning, NY, USA) was pre-coated with Matrigel (Becton, Dickinson and Company; BD Biosciences, Franklin Lakers, NJ, USA) at 37°C. A total of 200 µl cell suspension containing 2×104 cells was added into the upper chamber and 800 µl medium containing 30% FBS was added into the lower chamber. After 24 h culture at 37°C, the cells on the reverse surface of the polycarbonate membrane were fixed with 4% paraformaldehyde at room temperature for 20 min, stained with 0.5% cresol violet at room temperature for 5 min and images were captured under an inverted light microscope (×200 magnification).
Statistical analysis
The data in the present study were presented as the mean ± standard deviation replicated in triplicate. Results were analyzed with one-way or two-way analysis of variance with Bonferroni's post hoc multiple comparisons. GraphPad Prism 5.01 (GraphPad Software, Inc., La Jolla, CA, USA) was used for analysis of data in this study. P<0.05 was considered to indicate a statistically significant difference.
Results
MALAT1 is increased in prostate cancer cells and inhibits miR-1-3p expression
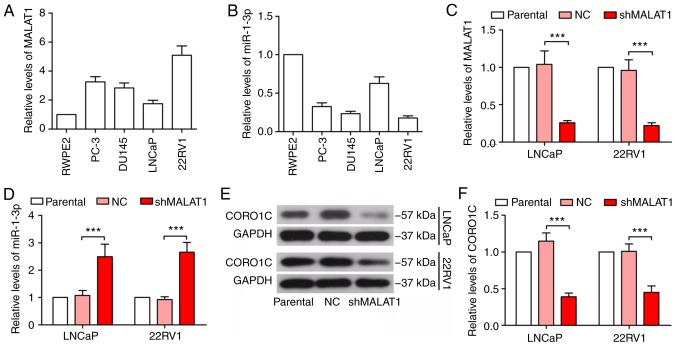
The expression levels of MALAT1 and miR-1-3p were detected in the normal prostatic epithelial cell line, RWPE2 and prostate cancer cell lines, PC-3, DU145, LNCaP and 22RV1. As indicated in Fig. 1A and B, the expression of MALAT1 was variably upregulated in prostate cancer cells, compared with RWPE2 cells, while miR-1-3p was downregulated in prostate cancer cells. LNCaP and 22RV1 cells were selected to investigate the role of MALAT1, since the expression levels of MALAT1 and miR-1-3p differed between LNCaP and 22RV1 cells. In addition, LNCaP cells are androgen-dependent, whereas 22RV1 cells are androgen-independent (18). MALAT1 was significantly silenced in LNCaP and 22RV1 cells, and its effectiveness was verified by RT-qPCR (P<0.001; Fig. 1C). The expression level of miR-1-3p was significantly increased following MALAT1 knockdown (P<0.001; Fig. 1D). At the same time, CORO1C was decreased in MALAT1-silenced cells (Fig. 1E and F), suggesting that MALAT1 served a regulatory role in miR-1-3p and CORO1C expression levels.
Figure 1.
MALAT1 is increased in prostate cancer cells and inhibits miR-1-3p expression. The relative expression levels of (A) MALAT1 and (B) miR-1-3p in human normal prostatic epithelial cell line, RWPE2, and prostate cancer cell lines PC-3, DU145, LNCaP and 22RV1 were detected by quantitative polymerase chain reaction. The relative levels of (C) MALAT1 and (D) miR-1-3p in prostate cancer cells following MALAT1 knockdown. (E) Western blotting and (F) quantitative analysis of the protein level of CORO1C in MALAT1-silenced prostate cancer cells was detected with GAPDH as the internal control. ***P<0.001. MALAT1, metastasis-associated lung adenocarcinoma transcript 1; CORO1C, coronin 1C; miR, microRNA; NC, negative control; sh, short hairpin.
MALAT1 knockdown inhibits migration, invasion and epithelial-mesenchymal transition (EMT) in prostate cancer cells
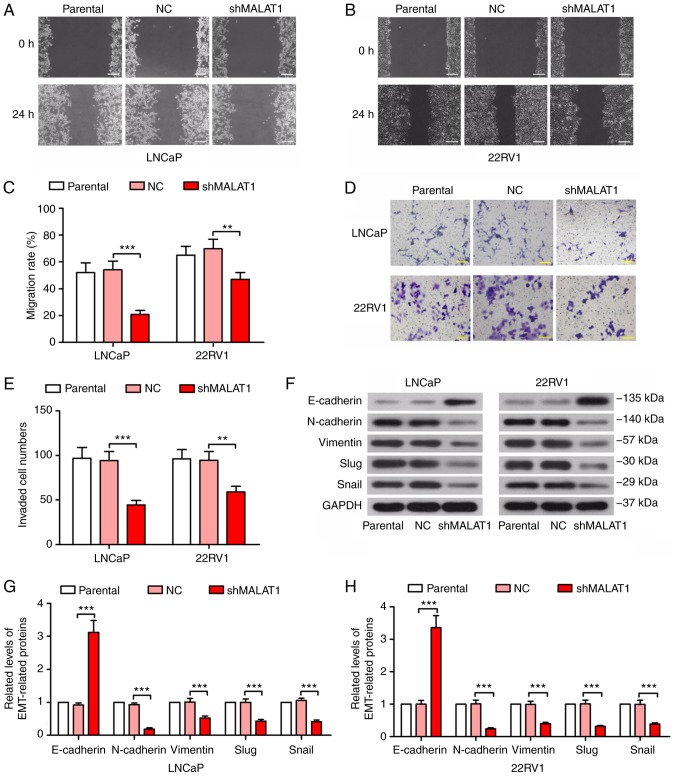
Phenotypic experiments were performed to detect the effect of MALAT1 on prostate cancer cell malignancy. As indicated in Fig. 2, the knockdown of MALAT1 significantly decreased the migratory ability of prostate cancer cells (P<0.01; Fig. 2A-C). In addition, the MALAT1-silenced LNCaP and 22RV1 cells indicated a significant decrease in invasive ability (P<0.01; Fig. 2D and E). Taking into consideration the important role of EMT on cell migration and invasion in cancer cells, a number of EMT-associated proteins were detected. The results on the expression levels of E-cadherin indicated a >3-fold increase and the expression levels of N-cadherin, vimentin, Slug and Snail were decreased by ≥50% in MALAT1-silenced prostate cancer cells compared with NC (Fig. 2F and H).
Figure 2.
MALAT1 knockdown inhibited migration, invasion and EMT in prostate cancer cells. A wound healing assay was performed to measure the migratory ability of (A) LNCaP and (B) 22RV1 cells following MALAT1 knockdown (Scale bar, 200 µm). (C) The migration rate in A and B was quantified. A Transwell assay with Matrigel was performed to detect the invasive ability of MALAT1-silenced (D) LNCaP and (E) 22RV1 cells (Scale bar, 100 µm). (F) The expression levels of several EMT-associated proteins, E-cadherin, N-cadherin, vimentin, Slug and Snail in (G) LNCaP and (H) 22RV1 cells. **P<0.01 and ***P<0.001. MALAT1, metastasis-associated lung adenocarcinoma transcript 1; EMT, epithelial-mesenchymal transition; NC, negative control; n, neural; E, epithelial; sh, short hairpin.
MALAT1is bound to miR-1-3p and regulates CORO1C expression
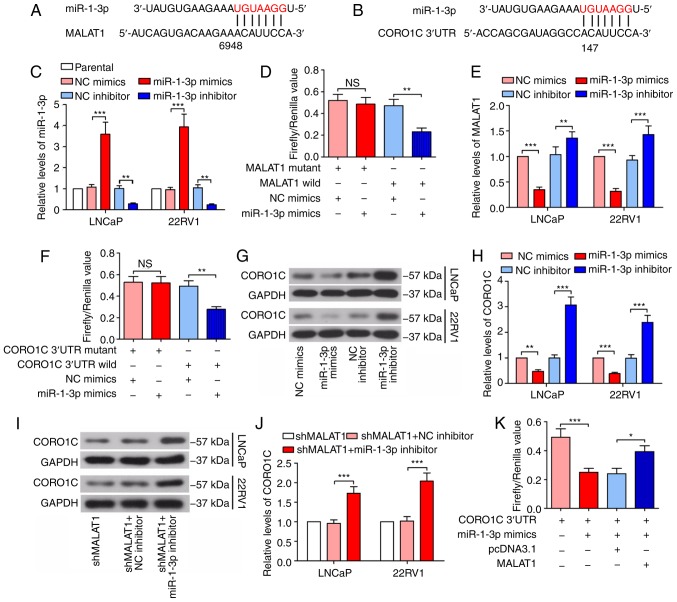
Bioinformatics analysis was conducted using starBase v2.0 (http://starbase.sysu.edu.cn/) to predict the candidate miRNAs of MALAT1 and miR-1-3p was identified (Fig. 3A). TargetScan 7.2 (http://www.targetscan.org/vert_72/) was subsequently used and it was indicated that miR-1-3p seed sequence directly binds to 3′UTR of CORO1C mRNA (Fig. 3B). Following confirmation of the efficacy of the miR-1-3p mimic and inhibitor (Fig. 3C), a dual-luciferase reporter assay results demonstrated that the firefly/Renilla value of the wild type MALAT1 was decreased by 51% by the miR-1-3p mimic, and the firefly/Renilla value of CORO1C 3′UTR was decreased by 43% by the miR-1-3p mimic in 293T cells (Fig. 3D and F), demonstrating the binding of miR-1-3p to MALAT1 and 3′UTR of the CORO1C mRNA. As indicated in Fig. 3E, the expression levels of MALAT1 were decreased following transfection of with the miR-1-3p mimic, while they decreased following miR-1-3p inhibition, suggesting that MALAT1 may be a degradable sponge. The CORO1C expression level decreased following miR-1-3p overexpression and increased following miR-1-3p inhibition (Fig. 3G and H), in addition, the MALAT1 silencing-induced decrease of the CORO1C expression level was rescued by the inhibition of miR-1-3p (Fig. 3I and J). In addition, the dual-luciferase reporter assay demonstrated that MALAT1 attenuated the binding of miR-1-3p to the CORO1C 3′UTR (the luciferase activity of CORO1C 3′UTR + miR-1-3p mimics + MALAT1 group was compared with that of CORO1C 3′UTR + miR-1-3p mimics + pcDNA3.1 group; Fig. 3K), suggesting that MALAT1 additionally competes with CORO1C for the binding sites of miR-1-3p.
Figure 3.
MALAT1 binds to miR-1-3p and regulates CORO1C expression. (A) The seed sequence of miR-1-3p bound to MALAT1. (B) The seed sequence of miR-1-3p bound to the 3′UTR of CORO1C mRNA. (C) The expression levels of miR-1-3p in LNCaP and 22RV1 cells were detected following overexpression or inhibition of miR-1-3p. (D) Dual-luciferase reporter assay was performed in 293T cells to confirm the direct binding between miR-1-3p and MALAT1. (E) Reverse transcription-quantitative polymerase chain reaction was used for detection of MALAT1 expression levels following transfection with miR-1-3p mimics or inhibitor in LNCaP and 22RV1 cells. (F) The binding of miR-1-3p to CORO1C 3′UTR was confirmed by dual-luciferase reporter assay. (G) Western blotting and (H) quantitative analysis of the CORO1C expression levels in LNCaP and 22RV1 cells following transfection with the miR-1-3p mimic or inhibitor were examined. (I) Western blotting and (J) quantitative analysis of the protein levels of CORO1C in prostate cancer cells were determined following interference of MALAT1 and miR-1-3p. (K) The firefly/Renilla value of CORO1C 3′UTR was determined following overexpression of miR-1-3p and MALAT1 in 293T cells. *P<0.05, **P<0.01 and ***P<0.001. MALAT1, metastasis-associated lung adenocarcinoma transcript 1; CORO1C, coronin 1C; 3′UTR, 3′ untranslated region; NS, no significance; microRNA, miR.
The MALAT1 knockdown-induced malignancy changes are recovered by miR-1-3p inhibition
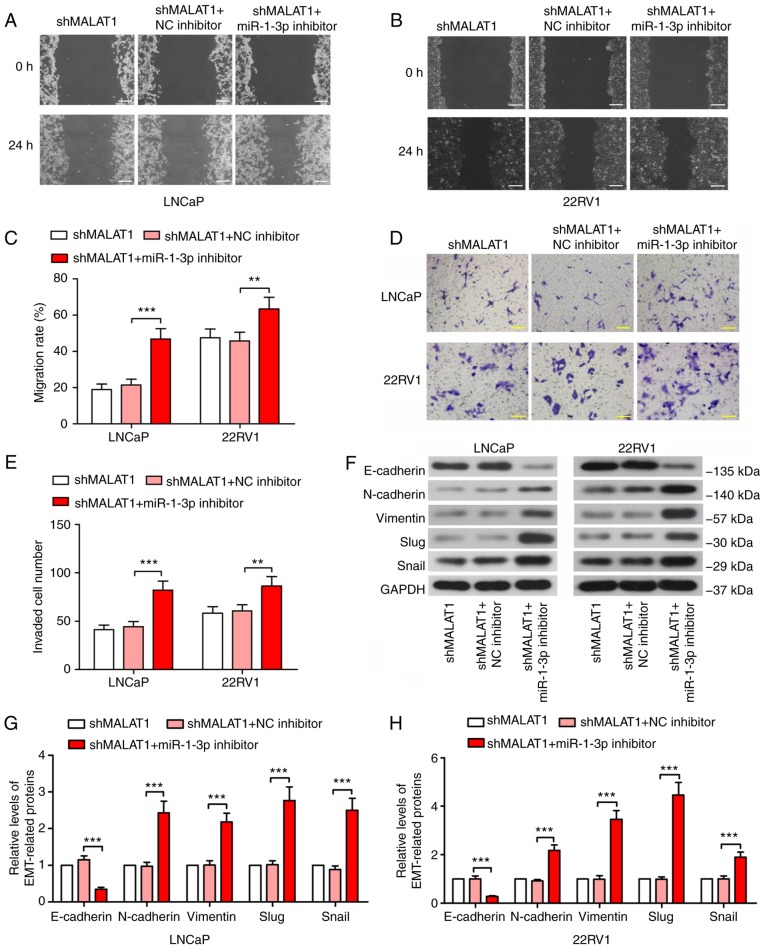
To confirm the underlying mechanism of action of MALAT1, phenotypic experiments were performed in MALAT1-silenced cells following miR-1-3p inhibition. The wound healing assay results revealed that the suppression of the migratory ability of MALAT1 knockdown-induced cells was significantly recovered by miR-1-3p in prostate cancer cells (P<0.01; Fig. 4A-C). Similar results were observed in the Transwell assay (Fig. 4D and E). Additionally, the expression level of E-cadherin was decreased by ≥70% and the expression levels of N-cadherin, vimentin, Slug and Snail indicated a 2-fold increase following transfection with the miR-1-3p inhibitor (Fig. 4F and H), suggesting enhanced EMT.
Figure 4.
MALAT1-knockdown-induced malignancy changes are recovered by inhibition of miR-1-3p. A wound healing assay was performed to measure the migratory ability of (A) LNCaP and (B) 22RV1 cells following inhibition of MALAT1 and miR-1-3p (Scale bar, 200 µm). (C) The migration rate in LNCaP and 22RV1 cells was quantified. (D) A Transwell assay with Matrigel and (E) quantitative analysis to detect the invasive ability of prostate cancer cells following silencing of MALAT1 and miR-1-3p (Scale bar, 100 µm). (F) Western blotting of the expression levels of EMT-associated proteins, E-cadherin, N-cadherin, vimentin, Slug and Snail in (G) LNCaP and (H) 22RV1. **P<0.01 and ***P<0.001. MALAT1, metastasis-associated lung adenocarcinoma transcript 1; EMT, epithelial-mesenchymal transition; N, neural; e, epithelial; sh, short hairpin; NC, negative control; miR, microRNA.
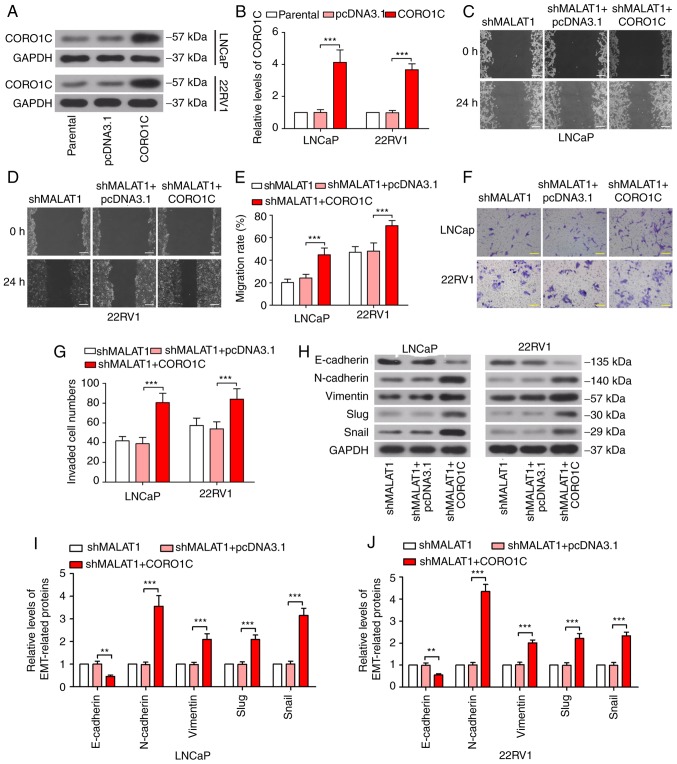
The phenotypic alterations induced by silencing MALAT1 are abolished by forced expression of CORO1C
In order to confirm the competitive association between MALAT1 and CORO1C, CORO1C was overexpressed in MALAT1-silenced cells. CORO1C expression was significantly increased following transfection with a CORO1C overexpression plasmid (Fig. 5A and B), which proved the effectiveness of the CORO1C ectopic expression plasmid. The wound healing assay revealed that the shMALAT1-induced decline in migratory ability was rescued by forced expression of CORO1C (Fig. 5C-E). The Transwell assay demonstrated similar results for the invasion of prostate cancer cells (Fig. 5F and G). As presented in Fig. 5H-J, E-cadherin expression level was significantly decreased (P<0.01) and N-cadherin, vimentin, Slug and Snail was significantly increased following CORO1C overexpression, compared with the MALAT1-silenced cells (P<0.001), suggesting that shMALAT1-induced EMT inhibition was ceased by forced expression of CORO1C.
Figure 5.
Phenotypic alterations induced by silencing of MALAT1 is abolished by forced expression of CORO1C. (A) Western blotting and (B) quantitative analysis of the CORO1C expression levels were detected following CORO1C overexpression. Wound healing assay was performed to measure the migratory rate in (C) LNCaP and (D) 22RV1 cells following MALAT1 knockdown or/and CORO1C overexpression (Scale bar, 200 µm) and (E) quantitative analysis. (F) Transwell assay and (G) quantitative analysis to investigate the invasive ability of LNCaP and 22RV1 cells (Scale bar, 100 µm). (H) Western blotting of the expression levels of EMT-associated proteins, E-cadherin, N-cadherin, vimentin, Slug and Snail in (I) LNCaP and (J) 22RV1. ***P<0.001. MALAT1, metastasis-associated lung adenocarcinoma transcript 1; EMT, epithelial-mesenchymal transition; CORO1C, coronin 1C; N, neural; e, epithelial; sh, short hairpin; NC, negative control; miR, microRNA.
Discussion
MALAT1 is located at the nuclear speckles that are enriched with pre-mRNA splicing factors (7,19). MALAT1 has been demonstrated to interact with a set of serine/arginine-rich (SR) family splicing regulators (SRSF1), including SRSF1/2/3 (20,21). The depletion of MALAT1 alters the phosphorylation status of SR proteins and causes mislocalization of splicing factors in nuclear speckles. The repression of MALAT1 in HeLa cells alters the pattern of alternative splicing of particular pre-mRNAs (20). These results suggest that MALAT1 indirectly controls alternative splicing by changing the distribution of splicing regulators in the nuclear speckles. However, the Malat1 knockout mouse does not exhibit similar phenotypes. In the embryo fibroblasts isolated from the Malat1 knockout mouse, the formation/structure of nuclear speckles and the phosphorylation status/localization of SR proteins are presented correctly (22). The discrepancy between MALAT1 function in vitro and in vivo could be explained as follows: MALAT1 only serves certain roles under specific conditions (23).
MALAT1 also can post-transcriptionally control gene expression through sponging miRNAs as a ceRNA (24). Through this mechanism, MALAT1 sequesters miRNAs through miRNA responsive elements located in its sequence, therefore, relieving the inhibitory effects of tumor suppressor miRNAs on oncogenic targets and leading to phenotypic alterations, including proliferation and invasion (7). In the present study, it was demonstrated that MALAT1 served as a miRNA sponge, bound to the seed sequence of miR-1-3p and attenuated the binding of miR-1-3p to CORO1C 3′UTR. Additionally, MALAT1 expression levels were affected by the miR-1-3p mimic or inhibitor, suggesting that MALAT1 may be a degradable sponge. Nevertheless, the effect of MALAT1 on miR-1-3p and CORO1C in vivo has not been investigated. The authors plan to next establish a Malat1 knockout rodent model and detect the expression levels of miR-1-3p and CORO1C, to confirm whether the effect of MALAT1 in vivo and in vitro is consistent.
CORO1C is an actin-binding protein that serves a key role in cell motility (25). Directional cell migration is essential for multiple physiological processes. For instance, the migration of leukocytes into damaged or infected tissues is crucial for an effective immune response and the migration of keratinocytes and dermal fibroblasts is a key aspect of wound healing (26). Additionally, migration is of significance for metastasis of malignant tumors and multiple reports have demonstrated the function of CORO1C in cancer (27–29). For instance, CORO1C is highly expressed in diffuse gliomas and CORO1C knockdown inhibits proliferation, motility and invasion in glioblastoma cells (29). The role of CORO1C in prostate cancer, to the best of our knowledge, has yet to be reported. The inhibition of miR-1-3p led to increased expression of CORO1C, accompanied by an increased migration, invasion and EMT rate in prostate cancer, which suggested the metastatic promoting role of CORO1C. The tumor suppressing role of miR-1-3p has been reported in multiple tumors (30,31), including prostate cancer (16). In the present study, it was demonstrated that silencing MALAT1 inhibited the expression of CORO1C by relieving the binding to miR-1-3p. The shMALAT1-induced phenotypic changes may be reversed by the miR-1-3p inhibitor or the CORO1C overexpression plasmid. Recently, another team reported similar results (32). They indicated that MALAT1 served as a sponge to bind to miR-1-3p and sequestered miR-1-3p away from its target KRAS, and the knockdown of MALAT1 accelerated KRAS-induced apoptosis in prostate cancer (32).
Prostate cancer is an androgen-dependent tumor, although it always develops to CRPC. In the present study, LNCaP and 22RV1 cells were selected to perform experiments. LNCaP cells were androgen-dependent, while 22RV1 cells were androgen-independent (18). However, the results of LNCaP and 22RV1 cells were similar, suggesting that the effect of MALAT1 on phenotypes of prostate cancer cells and expression of miR-1-3p and CORO1C, may be independent of androgens. The present study also demonstrated that the expression of MALAT1 was increased in several prostate cancer cell lines, including LNCaP and 22RV1, compared with normal prostatic epithelial cells, and the expression of miR-1-3p was decreased. It is well known that androgen and androgen receptor (AR) signaling are essential for the progression of prostate cancer, so the results suggested that the functions of MALAT1 may be associated with androgen and AR signaling. Therefore, further experiments with androgens are essential to deeply expound the mechanism of MALAT1 in prostate cancer.
In conclusion, the present study demonstrated that the silencing of MALAT1 inhibited migration, invasion and EMT by serving as a miRNA sponge and impairing the binding of miR-1-3p to CORO1C in prostate cancer cells. The present study identified the role of CORO1C in prostate cancer cells and demonstrated that MALAT1 regulated the CORO1C expression by competing with CORO1C for the binding sites of miR-1-3p. These findings may provide novel insight for clinical therapies for prostate cancer.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
HW and XD designed the research and conducted the experiments. ZL, LL, KG and SX performed experiments, and analyzed the data. XD drafted the manuscript. All authors read the manuscript and approved the submission.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Attard G, Parker C, Eeles RA, Schröder F, Tomlins SA, Tannock I, Drake CG, de Bono JS. Prostate cancer. Lancet. 2016;387:70–82. doi: 10.1016/S0140-6736(14)61947-4. [DOI] [PubMed] [Google Scholar]
- 3.Yuan X, Cai C, Chen S, Chen S, Yu Z, Balk SP. Androgen receptor functions in castration-resistant prostate cancer and mechanisms of resistance to new agents targeting the androgen axis. Oncogene. 2014;33:2815–2825. doi: 10.1038/onc.2013.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clarke NW. Landmarks in non-hormonal pharmacological therapies for castration-resistant prostate cancer. BJU Int. 2012;110(Suppl 1):S14–S22. doi: 10.1111/j.1464-410X.2012.11430.x. [DOI] [PubMed] [Google Scholar]
- 5.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ji P, Diederichs S, Wang W, Böing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 7.Amodio N, Raimondi L, Juli G, Stamato MA, Caracciolo D, Tagliaferri P, Tassone P. MALAT1: A druggable long non-coding RNA for targeted anti-cancer approaches. J Hematol Oncol. 2018;11:63. doi: 10.1186/s13045-018-0606-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu L, Wang X, Guo Y. Long non-coding RNA MALAT1 is upregulated and involved in cell proliferation, migration and apoptosis in ovarian cancer. Exp Ther Med. 2017;13:3055–3060. doi: 10.3892/etm.2017.4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang L, Bai HS, Deng Y, Fan L. High MALAT1 expression predicts a poor prognosis of cervical cancer and promotes cancer cell growth and invasion. Eur Rev Med Pharmacol Sci. 2015;19:3187–3193. [PubMed] [Google Scholar]
- 10.Sun R, Qin C, Jiang B, Fang S, Pan X, Peng L, Liu Z, Li W, Li Y, Li G. Down-regulation of MALAT1 inhibits cervical cancer cell invasion and metastasis by inhibition of epithelial-mesenchymal transition. Mol Biosyst. 2016;12:952–962. doi: 10.1039/C5MB00685F. [DOI] [PubMed] [Google Scholar]
- 11.Li Y, Wu Z, Yuan J, Sun L, Lin L, Huang N, Bin J, Liao Y, Liao W. Long non-coding RNA MALAT1 promotes gastric cancer tumorigenicity and metastasis by regulating vasculogenic mimicry and angiogenesis. Cancer Lett. 2017;395:31–44. doi: 10.1016/j.canlet.2017.02.035. [DOI] [PubMed] [Google Scholar]
- 12.Ren S, Liu Y, Xu W, Sun Y, Lu J, Wang F, Wei M, Shen J, Hou J, Gao X, et al. Long noncoding RNA MALAT-1 is a new potential therapeutic target for castration resistant prostate cancer. J Urol. 2013;190:2278–2287. doi: 10.1016/j.juro.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 13.Luan W, Li L, Shi Y, Bu X, Xia Y, Wang J, Djangmah HS, Liu X, You Y, Xu B. Long non-coding RNA MALAT1 acts as a competing endogenous RNA to promote malignant melanoma growth and metastasis by sponging miR-22. Oncotarget. 2016;7:63901–63912. doi: 10.18632/oncotarget.11564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chou J, Wang B, Zheng T, Li X, Zheng L, Hu J, Zhang Y, Xing Y, Xi T. MALAT1 induced migration and invasion of human breast cancer cells by competitively binding miR-1 with cdc42. Biochem Biophys Res Commun. 2016;472:262–269. doi: 10.1016/j.bbrc.2016.02.102. [DOI] [PubMed] [Google Scholar]
- 15.Jin C, Yan B, Lu Q, Lin Y, Ma L. Reciprocal regulation of Hsa-miR-1 and long noncoding RNA MALAT1 promotes triple-negative breast cancer development. Tumour Biol. 2016;37:7383–7394. doi: 10.1007/s13277-015-4605-6. [DOI] [PubMed] [Google Scholar]
- 16.Hudson RS, Yi M, Esposito D, Watkins SK, Hurwitz AA, Yfantis HG, Lee DH, Borin JF, Naslund MJ, Alexander RB, et al. MicroRNA-1 is a candidate tumor suppressor and prognostic marker in human prostate cancer. Nucleic Acids Res. 2012;40:3689–3703. doi: 10.1093/nar/gkr1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 18.van Bokhoven A, Varella-Garcia M, Korch C, Johannes WU, Smith EE, Miller HL, Nordeen SK, Miller GJ, Lucia MS. Molecular characterization of human prostate carcinoma cell lines. Prostate. 2003;57:205–225. doi: 10.1002/pros.10291. [DOI] [PubMed] [Google Scholar]
- 19.Yoshimoto R, Mayeda A, Yoshida M, Nakagawa S. MALAT1 long non-coding RNA in cancer. Biochim Biophys Acta. 2016;1859:192–199. doi: 10.1016/j.bbagrm.2015.09.012. [DOI] [PubMed] [Google Scholar]
- 20.Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schor IE, Llères D, Risso GJ, Pawellek A, Ule J, Lamond AI, Kornblihtt AR. Perturbation of chromatin structure globally affects localization and recruitment of splicing factors. PLoS One. 2012;7:e48084. doi: 10.1371/journal.pone.0048084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang B, Arun G, Mao YS, Lazar Z, Hung G, Bhattacharjee G, Xiao X, Booth CJ, Wu J, Zhang C, Spector DL. The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep. 2012;2:111–123. doi: 10.1016/j.celrep.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang X, Hamblin MH, Yin KJ. The long noncoding RNA Malat1: Its physiological and pathophysiological functions. RNA Biol. 2017;14:1705–1714. doi: 10.1080/15476286.2017.1358347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Humphries CL, Balcer HI, D'Agostino JL, Winsor B, Drubin DG, Barnes G, Andrews BJ, Goode BL. Direct regulation of Arp2/3 complex activity and function by the actin binding protein coronin. J Cell Biol. 2002;159:993–1004. doi: 10.1083/jcb.200206113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vicente-Manzanares M, Webb DJ, Horwitz AR. Cell migration at a glance. J Cell Sci. 2005;118:4917–4919. doi: 10.1242/jcs.02662. [DOI] [PubMed] [Google Scholar]
- 27.Mataki H, Enokida H, Chiyomaru T, Mizuno K, Matsushita R, Goto Y, Nishikawa R, Higashimoto I, Samukawa T, Nakagawa M, et al. Downregulation of the microRNA-1/133a cluster enhances cancer cell migration and invasion in lung-squamous cell carcinoma via regulation of Coronin 1C. J Hum Genet. 2015;60:53–61. doi: 10.1038/jhg.2014.111. [DOI] [PubMed] [Google Scholar]
- 28.Lim JP, Shyamasundar S, Gunaratne J, Scully OJ, Matsumoto K, Bay BH. YBX1 gene silencing inhibits migratory and invasive potential via CORO1C in breast cancer in vitro. BMC Cancer. 2017;17:201. doi: 10.1186/s12885-017-3187-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thal D, Xavier CP, Rosentreter A, Linder S, Friedrichs B, Waha A, Pietsch T, Stumpf M, Noegel A, Clemen C. Expression of coronin-3 (coronin-1C) in diffuse gliomas is related to malignancy. J Pathol. 2008;214:415–424. doi: 10.1002/path.2308. [DOI] [PubMed] [Google Scholar]
- 30.Shang A, Yang M, Shen F, Wang J, Wei J, Wang W, Lu W, Wang C, Wang C. MiR-1-3p suppresses the proliferation, invasion and migration of bladder cancer cells by up-regulating SFRP1 expression. Cell Physiol Biochem. 2017;41:1179–1188. doi: 10.1159/000464379. [DOI] [PubMed] [Google Scholar]
- 31.Wang Z, Wang J, Chen Z, Wang K, Shi L. MicroRNA-1-3p inhibits the proliferation and migration of oral squamous cell carcinoma cells by targeting DKK1. Biochem Cell Biol. 2018;96:355–364. doi: 10.1139/bcb-2017-0015. [DOI] [PubMed] [Google Scholar]
- 32.Chang J, Xu W, Du X, Hou J. MALAT1 silencing suppresses prostate cancer progression by upregulating miR-1 and downregulating KRAS. Onco Targets Ther. 2018;11:3461–3473. doi: 10.2147/OTT.S164131. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.