Abstract
With the increasing rate of chemoresistance in colorectal cancer (CRC) patients with advanced tumor stages, it is a matter of urgent importance to delineate the factors involved in the drug resistance process. In this study, gene expression profiles were downloaded from the Gene Expression Omnibus database and an integrated analysis with the aim of detecting hub long non-coding RNAs (lncRNAs) and their regulated, differentially expressed genes (DEGs) during treatment with oxaliplatin (OxPt) or irinotecan was conducted. A total of seven differentially expressed lncRNAs were correlated with OxPt resistance and 21 were correlated with resistance to SN-38, the active metabolite of irinotecan. Gene Ontology annotation and Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis confirmed that drug resistance was strongly associated with an imbalance between cell proliferation and apoptosis, cell energetic metabolism under hypoxic conditions, and angiogenesis. Moreover, a large number of lncRNA-targeted DEGs were located in extracellular exosomes. Further analyses identified four hub lncRNAs involved in the process of drug resistance, including CRNDE, H19, UCA1 and HOTAIR, which are predictive factors for treatment sensitivity. Among them, HOTAIR stands out as a strong factor, the elevated expression of which is also associated with advanced tumor node and metastasis stage and poor CRC disease prognosis.
Keywords: long noncoding RNA, colorectal cancer, chemoresistance, oxaliplatin, irinotecan
Introduction
Colorectal cancer (CRC) is a frequent malignant tumor in developed countries, with high morbidity and a high death rate. According to statistics published in 2015, CRC is the third most frequently diagnosed cancer in males and ranks second in females (1). According to the American Joint Committee on Cancer, the 5-year overall survival (OS) rate of CRC is 65.2% (2). Common risk factors for CRC include age, obesity, smoking and an unhealthy diet (3). Previous studies have found that several patients present with unresectable stage disease at initial diagnosis (4,5). Standard treatments for CRC include surgical resection, modern chemotherapy and radiation therapy. Although chemotherapy plays an important role in CRC treatment, the chemoresistance-reduced effectiveness of anti-neoplastic agents causes a lower survival rate of advanced-stage CRC patients.
Oxaliplatin (OxPt) is a third-generation platinum-based chemotherapeutic drug that inhibits the DNA replication and transcription of tumor cells (6). When used in combination with fluorouracil (5-FU), leucovorin and folinic acid, it is recognized as the first-line chemotherapy strategy for CRC (7). In recent years, the extensive use of OxPt has caused increased chemoresistance in clinical practice. Previous studies have found that the nucleotide excision repair pathway plays a key role in OxPt resistance (8,9). Moreover, the overexpression of chemoresistance-associated proteins (e.g., transforming growth factor-β and WBSCR22) and microRNAs, and increased messenger RNA levels of XPAC and ERCC1, are believed to predict chemoresistance to OxPt (10–12).
Irinotecan, an analogue of camptothecin isolated from the Chinese tree Camptotheca acuminata, was first approved by the Food and Drug Administration for the treatment of CRC in 1996 and is now used as the second choice for CRC chemotherapy (13). Irinotecan selectively inhibits topoisomerase I and induces the degradation of double-stranded DNA via intracellular modifications. Ultimately, irinotecan is activated by carboxylesterases to its active metabolite SN-38, which combines with topoisomerase and induces the breakage of DNA strands, DNA replication failure and cellular apoptosis (14,15). The therapeutic efficacy and toxicity of irinotecan are related to the expression of tumor-specific genes and the intra-tumor accumulation of SN-38 (16). Irinotecan treatment may also cause severe chemoresistance via variable levels of metabolic enzymes, reduced cellular accumulation from active drug efflux and reduced levels of Topo I expression, as well as different mutations and/or the activation of nuclear factor κB (NF-κB) (17).
Long non-coding RNAs (lncRNAs), defined as mRNA-like transcripts with lengths up to 200 nucleotides, are important members of the non-coding RNA family (18). lncRNA expression is regulated by both transcriptional and epigenetic factors. Several studies have revealed that lncRNAs are expressed in various tumor tissues and are involved in a number of cellular functions and developmental processes, such as cell growth, development, invasion, and apoptosis (19–21). Increasing lines of evidence suggest that lncRNAs are valuable bio-targets in the diagnosis and treatment of CRC (22). Recent studies have suggested that lncRNAs play a key role in drug function regulation and chemoresistance through various mechanisms in multiple cancers (23,24). At present, at least 70 CRC-related lncRNAs have been recognized, including HOTAIR, CCAT1, CCAT2, MALAT-1 and H19 (25). Changes in the expression of these lncRNAs could lead to chemotherapy and radiotherapy resistance. Therefore, further research and efforts are needed to clarify the chemoresistance mechanism of each lncRNA. Then a simple and reliable screening program based on the expression levels of lncRNAs can be developed to guide the selection of chemotherapy drugs.
In the current study, previously published RNA expression datasets on the chemoresistance of CRC in the Gene Expression Omnibus (GEO) database were searched. A comprehensive bioinformatics analysis was performed by defining the differentially expressed lncRNAs and other RNAs separately according to their chemoresistance to irinotecan or OxPt. The differentially expressed lncRNAs were correlated with differentially expressed genes (DEGs) and potential diseases based on the RNA-associated interactions database (RAID) and the mammalian lncRNA-disease repository (MNDR) database. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were also performed and a protein-protein interaction (PPI) network was established to screen for crucial genes and lncRNAs, whose hazard degrees were further demonstrated by evaluating their expression in tumors and according to tumor, node and metastasis (TNM) stage, as well as survival.
Materials and methods
Datasets and DEG identification
Gene expression profiles for chemoresistance to two chemotherapeutic drugs, OxPt and irinotecan (active metabolite SN-38), in three CRC cell lines (HCT116, HT29, and LoVo) were obtained from GSE42387 in the GEO database (26). Gene expression data were explored and visualized in the ggplot2 package of R software (version 3.2.0, http://ggplot2.tidyverse.org) for each sample in the nine groups (normal HCT116, OxPt-resistant HCT116, SN-38-resistant HCT116, normal HT29, OxPt-resistant HT29, SN-38-resistant HT29, normal LoVo, OxPt-resistant LoVo and SN-38-resistant LoVo). A detailed workflow of the data analysis is shown in Fig. S1. The limma package, which includes lmFit, eBayes and top Table functions, was used for the pairwise comparison of DEGs among the nine groups (27). The cutoff criteria were P<0.05 and abs(log2FC)>1, where FC indicates fold-change. The screened DEGs were divided into two groups, differentially expressed mRNAs and lncRNAs, which were further analyzed.
GO annotation and KEGG pathway enrichment analyses of DEGs
As mentioned above, two groups of DEGs: mRNAs and lncRNAs were analyzed. Based on RAID version 2.0 (28), the potential targets (confidence score >0.5) of the differentially expressed lncRNAs were obtained and intersected with DEGs found in the same group. Next, the intersected DEGs were divided into two groups: OxPt resistance and irinotecan resistance. GO annotation is a classic method used to identify the biological attributes of DEGs. This analysis comprises three parts: Biological process (BP), cell component (CC) and molecular function (MF). KEGG, a collection of genome, biological pathway, disease, drug and chemical substance databases, was used to identify the functional attributes of the DEGs. The Database for Annotation, Visualization and Integrated Discovery (DAVID; ver. 6.8) (29,30) was also applied for the functional interpretation of the two large lists of genes derived from previous analyses. Statistical significance was set at P<0.05.
PPI network construction and hub gene identification
A PPI network was constructed for the DEGs using STRING (31), an online functional protein association network tool, to detect potential relationships among the DEGs with confidence scores ≥0.4 and a maximum number of interactors of 1. The generated PPI network data were downloaded and imported into Cytoscape software (ver. 3.5.1), a bioinformatics tool used to create networks of protein interactions and reassess and integrate gene information from numerous embedded applications. The CentiScaPe plugin was used to determine the characteristics of each node in the PPI network, which gave each gene a degree score, the simplest topological index, allowing for immediate evaluation of the average number of edges (interactions) incident to the node (32). Genes with a degree ≥5 in significantly perturbed networks were defined as hub genes (mRNA). The differentially expressed lncRNA-mRNA interaction network was constructed and displayed by Cytoscape (version 3.5, http://www.cytoscape.org/). The lncRNA holding interactions with the greatest number of hub genes (mRNA) in the network were defined as hub lncRNAs.
Constructing the differentially expressed lncRNA-related disease network
Each lncRNA has a disease association profile. According to MNDR version 2.0 (33), a global view of the lncRNA-mediated disease network, which contains all of the differentially expressed lncRNA-related disease data (confidence score >0.5), was downloaded and integrated by R software. The disease network was constructed by Cytoscape. The disease label size was normalized to the number of lncRNAs related to each disease.
Hub lncRNA analysis
The hub lncRNAs were further investigated using Gene Expression Profiling Interactive Analysis, a newly developed interactive web server for analyzing the RNA sequencing expression data of 9,736 tumors and 8,587 normal samples from The Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression projects (34). The expression of each hub lncRNA in colon adenocarcinoma and rectum adenocarcinoma was compared with in normal tissue and to different tumor stages and plotted. Survival analysis was performed based on information from TCGA database. Relapse-free survival and OS were calculated. Hazard ratios with 95% confidence intervals and P-values calculated based on the Mantel-Cox log-rank test are displayed in Kaplan-Meier plots.
Results
Data quality control and DEG screening
A total of 32,705 genes were detected in each sample; their expression values and data distributions were similar between groups (Fig. S2A) and among samples (Fig. S2B). Principal component analysis showed that samples were easily grouped in different cell lines but not into certain drug resistance patterns (Fig. S2C). Therefore, further bioinformatics analysis should be performed separately based on different cell lines and chemoresistance. This dataset qualified for the following analysis.
Following data processing using limma, 255 DEGs in HCT116-OxPt vs. HCT116, 672 DEGs in HT29-OxPt vs. HT29, 491 DEGs in LoVo-OxPt vs. LoVo, 283 DEGs in HCT116-SN-38 vs. HCT116, 779 DEGs in HT29-SN-38 vs. HT29 and 1,713 DEGs in LoVo-SN-38 vs. LoVo were identified (Fig. S2D and E). The detailed DEGs information were shown in Tables SI–SVI.
Intersection between the predicted targets of lncRNAs and DEGs
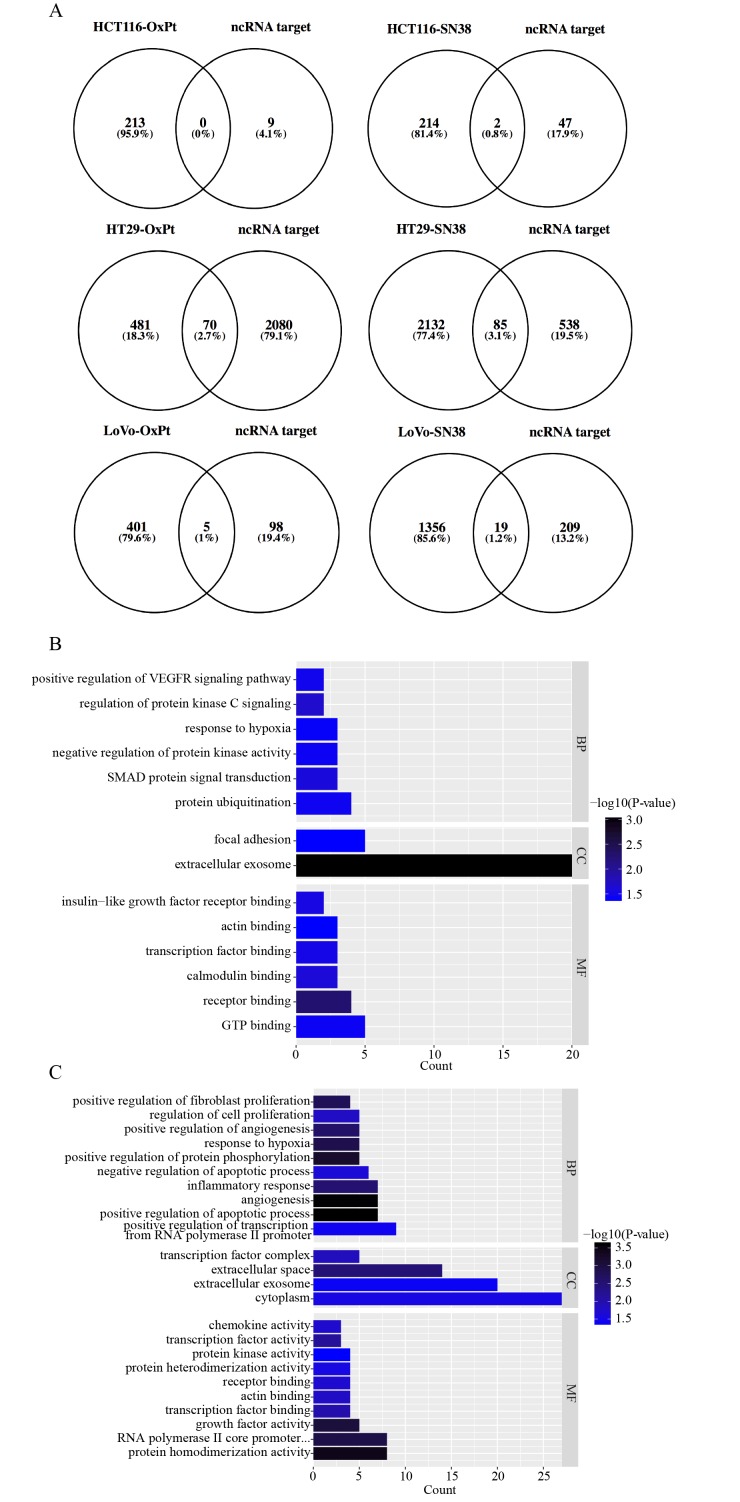
Among all of the DEGs identified in each comparison, the differentially expressed lncRNAs were isolated and are listed in Table I. The target prediction of each lncRNA was searched and obtained using RAID version 2.0. Next, the target genes were intersected with the identified DEGs in each comparison and are displayed as Venn diagrams (Fig. 1A). The intersected DEGs in the same drug management were united to perform GO annotation and KEGG pathway enrichment analyses based on DAVID. All significant (P<0.05) enriched entries in OxPt resistance are shown in a histogram (Fig. 1B), with the top three being protein ubiquitination, SMAD protein signal transduction and negative regulation of protein kinase activity in the BP GO annotation category; extracellular exosome and focal adhesion in the CC category; and GTP binding, receptor binding, and calmodulin binding in the MF category. All significant (P<0.05) enriched entries in irinotecan resistance are also shown in a histogram (Fig. 1C and D), with the top three being positive regulation of transcription from RNA polymerase II promoter, positive regulation of apoptotic process, angiogenesis in the BP GO annotation category; cytoplasm, extracellular exosome, and extracellular space in the CC category; protein homodimerization activity, RNA polymerase II core promoter proximal region sequence-specific DNA binding, and growth factor activity in the MF category; and pathways in cancer, microRNAs in cancer, and TNF signaling pathway in the KEGG pathway. Detailed information on the DEGs involved in the BP GO category and KEGG pathway are listed in Fig. 1E.
Table I.
Drug resistance-related differentially expressed non-coding RNAs.
| Group | ID | Symbol | logFC | AveExpr | P-value | Change | Gene name |
|---|---|---|---|---|---|---|---|
| HCT116-OxPt | 285735 | LINC00326 | 1.46 | 7.31 | 2.1×10−05 | UP | Long intergenic non-protein coding RNA 326 |
| 284739 | LINC00176 | 1.11 | 8.93 | 1.3×10−02 | UP | Long intergenic non-protein coding RNA 176 | |
| HT29-OxPt | 283460 | HNF1A-AS1 | −1.72 | 7.65 | 5.0×10−05 | DOWN | HNF1A antisense RNA 1 |
| 643911 | CRNDEa | −1.97 | 10.82 | 1.4×10−06 | DOWN | Colorectal neoplasia differentially expressed | |
| LoVo-OxPt | 283120 | H19a | 1.00 | 7.06 | 5.4×10−04 | UP | H19, imprinted maternally expressed transcript |
| 64150 | DIO3OS | −1.79 | 8.51 | 2.5×10−06 | DOWN | DIO3 opposite strand/antisense RNA | |
| 196047 | EMX2OS | −1.46 | 7.87 | 2.5×10−03 | DOWN | EMX2 opposite strand/antisense RNA | |
| HCT116-SN38 | 652995 | UCA1a | −1.40 | 12.29 | 4.4×10−06 | DOWN | Urothelial cancer associated 1 |
| HT29-SN38 | 441951 | ZNFX1-AS1 | 1.35 | 10.22 | 9.2×10−05 | UP | ZNFX1 antisense RNA 1 |
| 727701 | LINC00165 | −1.03 | 7.43 | 5.5×10−05 | DOWN | Long intergenic non-protein coding RNA 165 | |
| 100169750 | PRINS | −1.50 | 9.81 | 6.8×10−06 | DOWN | Psoriasis associated RNA induced by stress | |
| 283460 | HNF1A-AS1 | −1.62 | 7.70 | 6.6×10−05 | DOWN | HNF1A antisense RNA 1 | |
| 554202 | MIR31HG | −1.49 | 8.07 | 1.3×10−05 | DOWN | MIR31 host gene | |
| 283131 | NEAT1 | 1.32 | 11.22 | 9.3×10−04 | UP | Nuclear paraspeckle assembly transcript 1 | |
| 100289410 | MCF2L-AS1 | −1.30 | 9.95 | 2.8×10−06 | DOWN | MCF2L antisense RNA 1 | |
| 643911 | CRNDEa | −1.17 | 11.21 | 2.9×10−05 | DOWN | Colorectal neoplasia differentially expressed | |
| LoVo-SN38 | 100093630 | SNHG8 | −1.49 | 9.80 | 1.3×10−03 | DOWN | Small nucleolar RNA host gene 8 |
| 253264 | ZNF503-AS1 | −2.28 | 7.90 | 8.6×10−07 | DOWN | ZNF503 antisense RNA 1 | |
| 60674 | GAS5 | −1.21 | 15.21 | 2.1×10−02 | DOWN | Growth arrest-specific 5 | |
| 64150 | DIO3OS | 1.07 | 9.94 | 4.0×10−05 | UP | DIO3 opposite strand/antisense RNA | |
| 196047 | EMX2OS | −1.10 | 8.05 | 8.5×10−03 | DOWN | EMX2 opposite strand/antisense RNA | |
| 128439 | SNHG11 | −1.39 | 10.12 | 4.7×10−04 | DOWN | Small nucleolar RNA host gene 11 | |
| 100124700 | HOTAIRa | 1.58 | 7.21 | 3.1×10−06 | UP | HOX transcript antisense RNA | |
| 114915 | EPB41L4A-AS1 | −1.42 | 15.27 | 3.0×10−02 | DOWN | EPB41L4A antisense RNA 1 | |
| 652995 | UCA1a | −1.45 | 7.58 | 1.6×10−04 | DOWN | Urothelial cancer associated 1 | |
| 401237 | LINC00340 | 1.99 | 7.58 | 1.7×10−06 | UP | Long intergenic non-protein coding RNA 340 | |
| 29092 | LINC00339 | −1.55 | 9.20 | 4.0×10−03 | DOWN | Long intergenic non-protein coding RNA 339 | |
| 79992 | AGPAT4-IT1 | 1.50 | 8.13 | 2.5×10−04 | UP | AGPAT4 intronic transcript 1 | |
| 388815 | LINC00478 | 3.03 | 8.11 | 2.7×10−08 | UP | Long intergenic non-protein coding RNA 478 |
Hub ncRNA. OxPt, oxaliplatin; nc, noncoding.
Figure 1.
Functional analyses of differentially expressed long non-coding RNA lncRNA-targeted DEGs. (A) A Venn diagram between lncRNA target genes in the database and screened DEGs for the six comparison groups. Histograms of GO functional classification of DEGs for the (B) OxPt resistance group and the (C) SN-38 resistance group. The x-axis represents the number of DEGs and the y-axis represents the GO terms. All GO terms were grouped into three ontologies: Biological process, cellular component and molecular function. The graph displays only significantly enriched GO terms (P<0.05); darker blue indicates greater significance. (D) A histogram of the KEGG pathway enrichment of DEGs screened in the SN-38 resistance group, where the x-axis represents the number of DEGs annotated in a pathway term; enriched pathways are shown on the y-axis. The graph displays only significantly enriched KEGG terms (P<0.05); darker red indicates greater significance. (E) A table listing biological process GO and KEGG terms in each group. lncRNA, long non-coding RNA; GO, gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; DEG, differentially expressed genes.
Identification of hub lncRNAs and their associated disease network
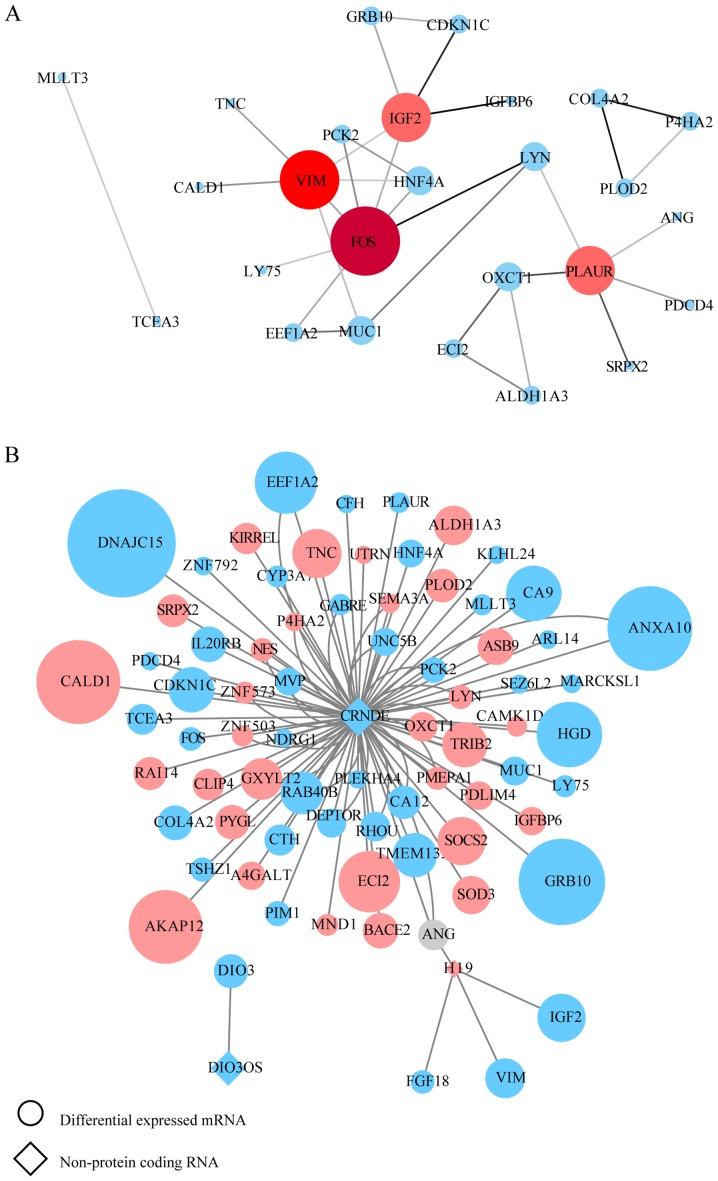
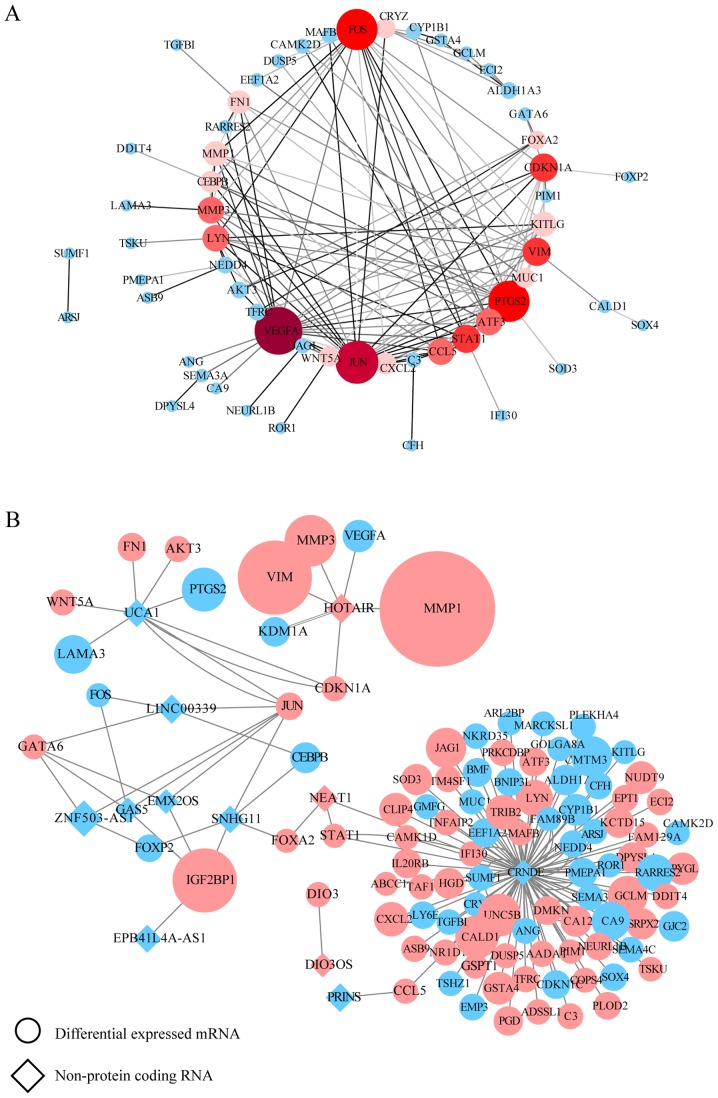
There was a positive association between mRNA and protein expression changes; therefore, putative PPI network maps were constructed for the united datasets of chemoresistance using the STRING database and Cytoscape. Excluding the DEGs distributed on the edge of the PPI network, the remaining 26 and 57 central DEGs in the OxPt and SN-38 groups, respectively, were evaluated using the CentiScaPe plugin. The degree index of each central DEG was calculated and hub genes were defined using the criterion that the degree value must be ≥5. In the OxPt group, the hub genes were FOS, VIM, PLAUR and IGF2. In the SN-38 group, the hub genes were VEGFA, JUN, PTGS2, FOS, VIM, STAT1, CDKN1A, MMP3, CCL5, LYN, ATF3, KITLG, MMP1, FN1, CEBPB, CXCL2, MUC1, CRYZ, WNT5A and FOXA2 (Tables SVII and SVIII). These hub genes are marked in red in the PPI network (Figs. 2A and 3A). In a similar manner, the interaction network between each specific lncRNA and its potential targets in DEGs were visualized using Cytoscape in Figs. 2B and 3B. Up- and downregulated DEGs are colored in red and blue, respectively. The log2FC value is also indicated by the size of the node. Each hub lncRNA was identified by calculating its downstream hub genes whose count should be ranked in the top three and larger than 2. During OxPt screening, the lncRNAs CRNDE (target DEGs: PLAUR and FOS) and H19 (target DEGs: IGF2 and VIM) stood out. SN-38 screening revealed the following lncRNAs: CRNDE (target DEGs: STAT1, MUC1, LYN, KITLG, CXCL2, CRYZ, CCL5 and ATF3), HOTAIR (target DEGs: VEGFA, CDKN1A, VIM, MMP3, and MMP1), and UCA1 (target DEGs: CDKN1A, JUN, WNT5A, PTGS2, and FN1).
Figure 2.
PPI network construction for the OxPt resistance group. (A) PPI network for screened lncRNA-targeted DEGs. Genes marked in red represent hub genes with a degree ≥5 (color and size are increased along with degree value). The darker the connection line, the greater the confidence score. (B) lncRNA-mRNA association network. Circles indicate mRNAs and diamonds indicate non-protein-coding RNAs. A larger node indicates a larger log2FC value. Upregulated genes are shown in pink and downregulated genes are shown in blue. PPI, protein-protein interaction; lncRNA, long noncoding RNA; OxPt, oxaliplatin; DEG, differentially expressed genes; FC, fold-change.
Figure 3.
PPI network construction for the SN-38 resistance group. (A) PPI network for screened lncRNA-targeted DEGs. Genes marked in red represent hub genes with a degree ≥5 (color and size are increased along with degree value). The darker the connection line, the greater the confidence score. (B) lncRNA-mRNA association network. Circles indicate mRNAs and diamonds indicate non-protein-coding RNAs. A larger node indicates a larger log2fold-change value. Upregulated genes are shown in pink and downregulated genes are shown in blue. PPI, protein-protein interaction; lncRNA, long noncoding RNA; DEG, differentially expressed genes.
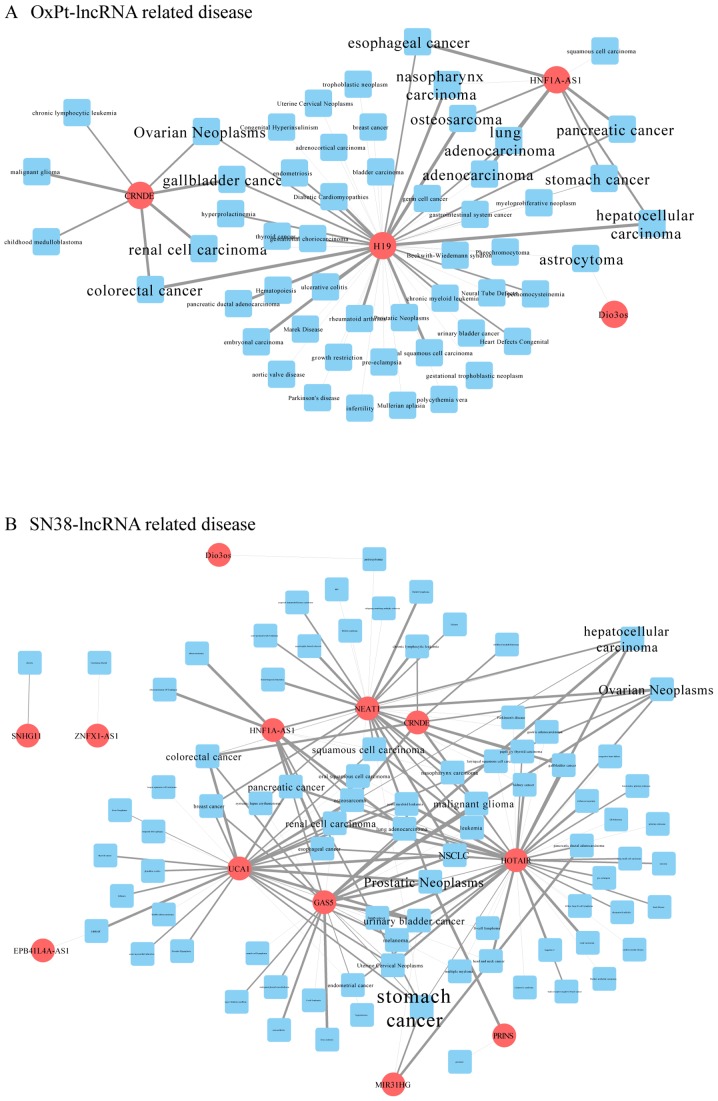
Information on lncRNA-associated diseases was obtained in full scale according to the MNDR database. By integrating this information in Cytoscape, the major related diseases associated with the differentially expressed lncRNAs were clearly sorted out. The more lncRNAs a disease is connected to, the larger its label. As shown in Fig. 4, cancer is the word that most frequently appears. Among them, CRC is obvious in the OxPt-lncRNA-related disease network, which is strongly connected to CRNDE and H19 (Fig. 4A), whereas the SN-38-lncRNA-related disease network is strongly connected to GAS5, UCA1, NEAT1, and CRNDE (Fig. 4B).
Figure 4.
lncRNA-related disease network construction. (A) The OxPt resistance group and (B) the SN-38 resistance group. lncRNAs are indicated in red circles. Diseases are indicated in blue square. The disease label size is normalized to the number of lncRNAs related to each disease. Lnc, long noncoding; OxPt, oxaliplatin.
lncRNA expression validation and survival analysis in TCGA
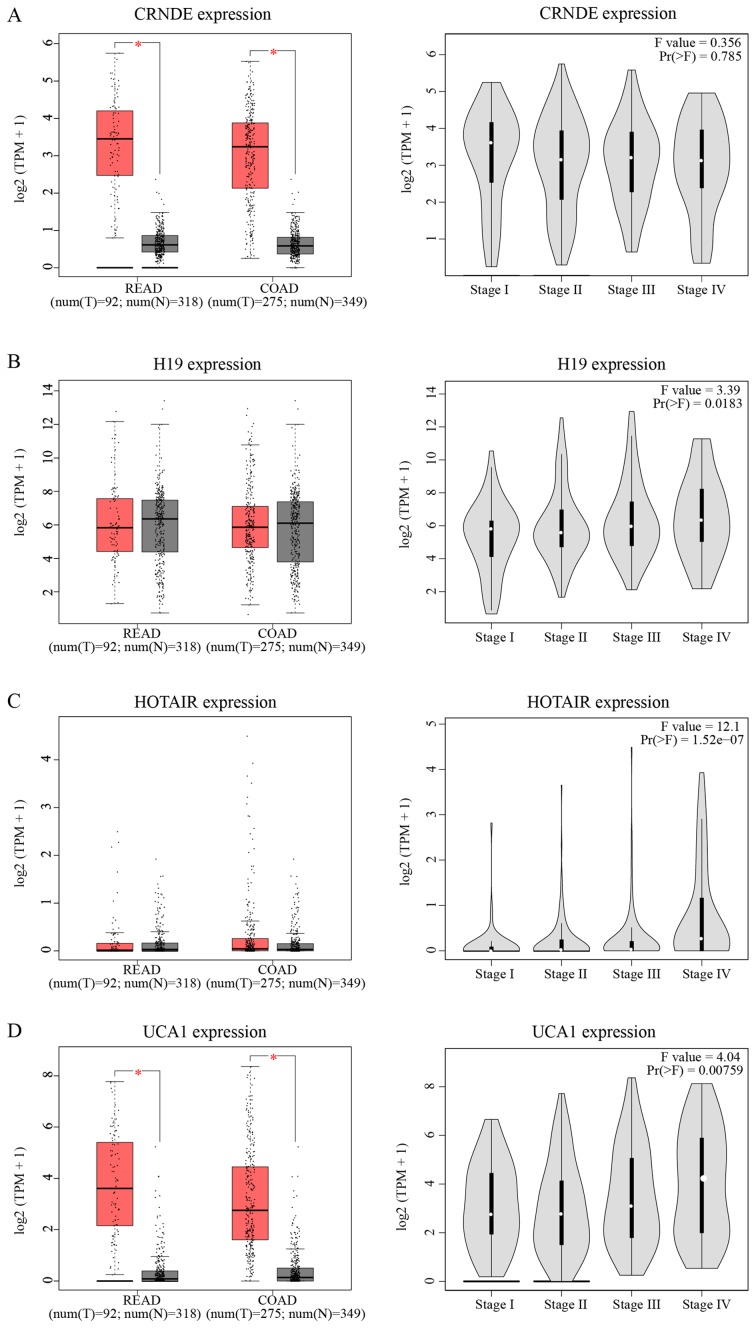
TCGA database was utilized to validate and compare hub lncRNA expression in tumor and normal tissues. Although the results cannot relate the expression levels of these lncRNAs to chemoresistance, it did, to a certain extent, partially confirm the hazard degree of each lncRNA. As indicated in Fig. 5A-D, the expression of CRNDE and UCA1 was significantly increased in tumor tissue compared with in normal tissue (P<0.05). Except for CRNDE, the expression of three other lncRNAs increased as the TNM stage increased. In Fig. 5E-H, only HOTAIR showed a significant relationship with OS and disease-free survival (DFS; P<0.05). Patients with increased expression of HOTAIR suffered a shorter OS (HR=1.9, P=0.0066) and DFS (HR=1.8, P=0.012) compared to those with weaker expression.
Figure 5.
Clinical significances of hub lncRNAs. Analyses of hub lncRNAs expression in tumor vs. normal tissues and in different tumor stages for (A) CRNDE, (B) H19, (C) HOTAIR, and (D) UCA1. The red box stands for tumor tissue; grey box is for normal tissue; and dots presents each sample value in left panel. *P<0.05; other P-values are shown on the diagrams. Overall and disease-free survival analyses of hub lncRNAs (E) CRNDE, (F) H19, (G) HOTAIR, and (H) UCA1. P-values are shown on the diagrams. READ, rectum adenocarcinoma; COAD, colon adenocarcinoma
Discussion
In recent years, the prevalence of CRC has increased worldwide. Due to the popularity of prophylactic screening, CRC can be diagnosed and treated increasingly early (3). However, many patients are in late stages when diagnosed (≥stage III or high-risk stage II) and require neoadjuvant or adjuvant chemotherapy (7). Some patients must face a tough reality-chemoresistance-which results in a shorter survival owing to limited chemotherapy choices. Although a previous bioinformatics analysis has revealed differences in gene expression patterns in chemoresistant cells, such as original study of GSE42387 done by Jensen et al (26), few have determined pivotal lncRNAs involved in this process. Therefore, a comprehensive analysis at the whole-genome level is necessary to identify key lncRNAs and provide detailed gene signatures for effective chemotherapy. The present study aimed at exploring the relationship between aberrant expressed lncRNAs and mRNAs. The GEO dataset was re-analyzed by relaxing DEG screening condition to gain more information. Only DEGs predicted to be the targets of differential expressed lncRNAs were involved in the following analysis.
To gain further insight into the biological pathways and identify key factors involved in chemoresistance, functional analyses of lncRNA-targeted DEGs, including GO annotation and KEGG pathway enrichment analyses were performed. The present study found that several DEGs are components of the extracellular exosome. Interestingly, the exosome is believed to confer chemoresistance to pancreatic cancer (35), breast cancer (36), gastric cancer (37) and CRC (38). It is believed that due to their cell-to-cell communication, exosomes protect tumor cells from the cytotoxic effects of chemotherapy drugs and transfer chemoresistance properties to nearby cells (39). The items enriched in GO biological processes indicate that chemoresistance occurring during both OxPt and irinotecan therapy is associated with an imbalance between cell proliferation and apoptosis regulation, cell energetic metabolism under hypoxic conditions, and angiogenesis. However, it seems that irinotecan treatment yields more DEGs than OxPt treatment. The present study hypothesized that this is why OxPt is the first choice for the clinical therapy of CRC.
In the present study, four hub lncRNAs were identified: CRNDE, H19, UCA1 and HOTAIR. CRNDE is a 1,910-nt cancer-secreted lncRNA transcribed by human chromosome 16 (16q12.2). Decades of research have shown that the high expression of CRNDE is involved in the progression of several cancers, such as renal cell carcinoma (40), hepatocellular carcinoma (41) and glioma tumors (42). Liu et al (43) found exosomal CRNDE-h in the sera of CRC patients. They also found a positive correlation between serum levels of CRNDE-h and regional lymph node or distant metastasis. Therefore, they believe that exosomal CRNDE can be used as a serum biochemical marker in the diagnosis and prognosis of CRC. Another recent study revealed that the overexpression of CRNDE promoted the development of CRC by suppressing the expression of dual-specificity phosphatase 5 (DUSP5) and CDKN1A. Moreover, the knockdown of CRNDE significantly blocked cell proliferation and induced apoptosis (44). In glioma, CRNDE promoted cell growth and invasion via mTOR signaling (42). The involvement of CRNDE in the chemoresistance of CRC treatment has also been assessed by a number of researchers (45,46). After treating CRC cells with different concentrations of 5-FU, Han et al (45) found that CRNDE-knockdown CRC cells exhibited increased sensitivity to 5-FU and OxPt treatment. Together, these results suggest that CRNDE is closely related to the development and chemoresistance of CRC.
Another hub lncRNA identified in the present study, lncRNA H19, is a key oncogenic mediator coded by chromosome 11. According to previous studies, H19 is overexpressed in various cancers, including breast cancer (47), thyroid carcinoma (48) and lung cancer (49). H19 also plays an oncogenic role in the development of CRC. The expression of H19 is increased in CRC tissues from advanced TNM stage patients (50). Enhanced H19 expression was also found in a CRC cell line and treatment with H19 significantly induced the proliferation of CRC cells, suggesting an underlying relationship between H19 and colorectal carcinogenesis (51). H19 also plays an important role in the migration and invasion of CRC cells (52). Increasing research attention has been given to the chemoresistance of H19 in the treatment of cancer (53,54). Emerging evidence also indicates that H19 knockout in the HT-29 cell line (MTX-resistant colorectal cells) reduces the risk of drug resistance. Further investigations showed that this H19-related chemoresistance is mediated by the Wnt/β-catenin signaling pathway (55).
HOTAIR is a novel oncogenic lncRNA transcribed by the HoxC gene. As with the abovementioned lncRNAs, HOTAIR has also been identified in several cancers, including osteosarcoma (56), cervical cancer (57) and gastric cancer (58). Similarly, HOTAIR exerts its oncogenic role by regulating CRC cell proliferation, invasion, and migration by mediating p21 (59). Elevated HOTAIR expression has also been detected in CRC cells and tissues relative to adjacent control tissues (60,61). The precise role of HOTAIR in chemoresistance in the chemotherapy of CRC was also recently reported. Li et al (62) indicated that HOTAIR promotes the chemoresistance of CRC cells to 5-FU by suppressing miR-218 and activating NF-κB signaling. Another study performed by Xiao et al (63) found that HOTAIR knockout improved the sensitivity of CRC cells to cisplatin and paclitaxel and that this mechanism may, at least in part, be related to the expression of miR-203a-3p and activation of the Wnt/β-catenin signaling pathway.
The regulatory role of UCA1 in chemotherapy drug resistance in CRC was also investigated. The upregulated lncRNA UCA1 is correlated with the progression of lung cancer, esophageal squamous cell carcinoma and CRC (64–66) and predicts a poor prognosis. Compared with control tissues, an increased level of UCA1 was found in CRC tissues and cells and the level of UCA1 was positively correlated with tumor size and depth (67). Further in vitro experiments suggested that UCA1 induces CRC cell proliferation and inhibits apoptosis. Recent studies have also revealed the potential role of UCA1 in chemoresistance in CRC. CRC cells with UCA1 knockdown exhibit sensitivity to 5-FU by increasing tumor cell apoptosis. Conversely, UCA1 overexpression may result in increased resistance to 5-FU treatment, suggesting that UCA1 may predict the response to CRC chemotherapy (68). Furthermore, overexpression of UCA1 has a consanguineous connection with radioresistance in CRC treatment (69).
Collectively, the current study identified four pivotal lncRNAs, CRNDE, H19, UCA1 and HOTAIR, which are strongly associated with CRC chemoresistance and can be used as predictive factors for treatment sensitivity and tumor prognosis. Functional enrichment analysis indicated that an imbalance between cell proliferation and apoptosis, cell energetic metabolism under hypoxic conditions, and angiogenesis were the key biological processes whereby lncRNAs perturbed gene expression and altered sensitivity of tumor cells to chemotherapy. However, the results of the present study are based entirely on bioinformatics analyses and lack in vivo and in vitro experimental evidence. Further research is required to delineate their potential roles in chemoresistance.
Supplementary Material
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used during the present study are downloaded from TCGA and GEO datasets and analyzed data is available from the corresponding author on reasonable request.
Authors' contributions
FFS was involved in the conception and design of the study, project administration and drafted the manuscript; FFS and WWL collected data from the internet, performed bioinformatics analyses and organized the figures and tables; WWL, performed the literature search. JQ was involved in the conception and design of the study, supervised the study and critically reviewed the manuscript. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.O'Connell JB, Maggard MA, Ko CY. Colon cancer survival rates with the new American Joint Committee on cancer sixth edition staging. J Natl Cancer Inst. 2004;96:1420–1425. doi: 10.1093/jnci/djh275. [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Fedewa SA, Ahnen DJ, Meester RGS, Barzi A, Jemal A. Colorectal cancer statistics, 2017. CA Cancer J Clin. 2017;67:177–193. doi: 10.3322/caac.21395. [DOI] [PubMed] [Google Scholar]
- 4.Moriarity A, O'Sullivan J, Kennedy J, Mehigan B, McCormick P. Current targeted therapies in the treatment of advanced colorectal cancer: A review. Ther Adv Med Oncol. 2016;8:276–293. doi: 10.1177/1758834016646734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Mestier L, Manceau G, Neuzillet C, Bachet JB, Spano JP, Kianmanesh R, Vaillant JC, Bouché O, Hannoun L, Karoui M. Primary tumor resection in colorectal cancer with unresectable synchronous metastases: A review. World J Gastrointest Oncol. 2014;6:156–169. doi: 10.4251/wjgo.v6.i6.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raymond E, Faivre S, Woynarowski JM, Chaney SG. Oxaliplatin: Mechanism of action and antineoplastic activity. Semin Oncol. 1998;25:4–12. [PubMed] [Google Scholar]
- 7.Benson AB, Venook AP, Al-Hawary MM, Cederquist L, Chen YJ, Ciombor KK, Cohen S, Cooper HS, Deming D, Engstrom PF, et al. NCCN guidelines insights: Colon cancer, version 2.2018. J Natl Compr Canc Netw. 2018;16:359–369. doi: 10.6004/jnccn.2018.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bahrami A, Amerizadeh F, Hassanian SM, ShahidSales S, Khazaei M, Maftouh M, Ghayour-Mobarhan M, Ferns GA, Avan A. Genetic variants as potential predictive biomarkers in advanced colorectal cancer patients treated with oxaliplatin-based chemotherapy. J Cell Physiol. 2018;233:2193–2201. doi: 10.1002/jcp.25966. [DOI] [PubMed] [Google Scholar]
- 9.Hammond WA, Swaika A, Mody K. Pharmacologic resistance in colorectal cancer: A review. Ther Adv Med Oncol. 2016;8:57–84. doi: 10.1177/1758834015614530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yan D, Tu L, Yuan H, Fang J, Cheng L, Zheng X, Wang X. WBSCR22 confers oxaliplatin resistance in human colorectal cancer. Sci Rep. 2017;7:15443. doi: 10.1038/s41598-017-15749-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mao L, Li Y, Zhao J, Li Q, Yang B, Wang Y, Zhu Z, Sun H, Zhai Z. Transforming growth factor-β1 contributes to oxaliplatin resistance in colorectal cancer via epithelial to mesenchymal transition. Oncol Lett. 2017;14:647–654. doi: 10.3892/ol.2017.6209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gnoni A, Russo A, Silvestris N, Maiello E, Vacca A, Marech I, Numico G, Paradiso A, Lorusso V, Azzariti A. Pharmacokinetic and metabolism determinants of fluoropyrimidines and oxaliplatin activity in treatment of colorectal patients. Curr Drug Metab. 2011;12:918–931. doi: 10.2174/138920011798062300. [DOI] [PubMed] [Google Scholar]
- 13.Conti JA, Kemeny NE, Saltz LB, Huang Y, Tong WP, Chou TC, Sun M, Pulliam S, Gonzalez C. Irinotecan is an active agent in untreated patients with metastatic colorectal cancer. J Clin Oncol. 1996;14:709–715. doi: 10.1200/JCO.1996.14.3.709. [DOI] [PubMed] [Google Scholar]
- 14.Cecchin E, Corona G, Masier S, Biason P, Cattarossi G, Frustaci S, Buonadonna A, Colussi A, Toffoli G. Carboxylesterase isoform 2 mRNA expression in peripheral blood mononuclear cells is a predictive marker of the irinotecan to SN38 activation step in colorectal cancer patients. Clin Cancer Res. 2005;11:6901–6907. doi: 10.1158/1078-0432.CCR-05-0602. [DOI] [PubMed] [Google Scholar]
- 15.Chabot GG, Robert J, Lokiec F, Canal P. Irinotecan pharmacokinetics. Bull Cancer. 1998:11–20. (In French) [PubMed] [Google Scholar]
- 16.Hecht JR. Gastrointestinal toxicity of irinotecan. Oncology (Williston Park) 1998;12:72–78. [PubMed] [Google Scholar]
- 17.Xu Y, Villalona-Calero M. Irinotecan: Mechanisms of tumor resistance and novel strategies for modulating its activity. Ann Oncol. 2002;13:1841–1851. doi: 10.1093/annonc/mdf337. [DOI] [PubMed] [Google Scholar]
- 18.Ransohoff JD, Wei Y, Khavari PA. The functions and unique features of long intergenic non-coding RNA. Nat Rev Mol Cell Biol. 2018;19:143–157. doi: 10.1038/nrm.2017.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21:1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 20.Kim T, Croce CM. Long noncoding RNAs: Undeciphered cellular codes encrypting keys of colorectal cancer pathogenesis. Cancer Lett. 2018;417:89–95. doi: 10.1016/j.canlet.2017.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H, Ma SQ, Huang J, Chen XP, Zhou HH. Roles of long noncoding RNAs in colorectal cancer metastasis. Oncotarget. 2017;8:39859–39876. doi: 10.18632/oncotarget.16339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han D, Wang M, Ma N, Xu Y, Jiang Y, Gao X. Long noncoding RNAs: Novel players in colorectal cancer. Cancer Lett. 2015;361:13–21. doi: 10.1016/j.canlet.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 23.Wang M, Han D, Yuan Z, Hu H, Zhao Z, Yang R, Jin Y, Zou C, Chen Y, Wang G, et al. Long non-coding RNA H19 confers 5-Fu resistance in colorectal cancer by promoting SIRT1-mediated autophagy. Cell Death Dis. 2018;9:1149. doi: 10.1038/s41419-018-1187-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li L, Shang J, Zhang Y, Liu S, Peng Y, Zhou Z, Pan H, Wang X, Chen L, Zhao Q. MEG3 is a prognostic factor for CRC and promotes chemosensitivity by enhancing oxaliplatin-induced cell apoptosis. Oncol Rep. 2017;38:1383–1392. doi: 10.3892/or.2017.5828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo J, Qu J, Wu DK, Lu ZL, Sun YS, Qu Q. Long non-coding RNAs: A rising biotarget in colorectal cancer. Oncotarget. 2017;8:22187–22202. doi: 10.18632/oncotarget.14728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jensen NF, Stenvang J, Beck MK, Hanáková B, Belling KC, Do KN, Viuff B, Nygård SB, Gupta R, Rasmussen MH, et al. Establishment and characterization of models of chemotherapy resistance in colorectal cancer: Towards a predictive signature of chemoresistance. Mol Oncol. 2015;9:1169–1185. doi: 10.1016/j.molonc.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yi Y, Zhao Y, Li C, Zhang L, Huang H, Li Y, Liu L, Hou P, Cui T, Tan P, et al. RAID v2. 0: An updated resource of RNA-associated interactions across organisms. Nucleic Acids Res. 2017;45:D115–D118. doi: 10.1093/nar/gkw1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 31.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2018;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scardoni G, Tosadori G, Faizaan M, Spoto F, Fabbri F, Laudanna C. Biological network analysis with CentiScaPe: Centralities and experimental dataset integration. F1000Res. 2014;3:e139. doi: 10.12688/f1000research.4477.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cui T, Zhang L, Huang Y, Yi Y, Tan P, Zhao Y, Hu Y, Xu L, Li E, Wang D. MNDR v2.0: An updated resource of ncRNA-disease associations in mammals. Nucleic Acids Res. 2018;46:D371–D374. doi: 10.1093/nar/gkx1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Patel GK, Khan MA, Bhardwaj A, Srivastava SK, Zubair H, Patton MC, Singh S, Khushman M, Singh AP. Exosomes confer chemoresistance to pancreatic cancer cells by promoting ROS detoxification and miR-155-mediated suppression of key gemcitabine-metabolising enzyme, DCK. Br J Cancer. 2017;116:609–619. doi: 10.1038/bjc.2017.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen WX, Liu XM, Lv MM, Chen L, Zhao JH, Zhong SL, Ji MH, Hu Q, Luo Z, Wu JZ, Tang JH. Exosomes from drug-resistant breast cancer cells transmit chemoresistance by a horizontal transfer of microRNAs. PLoS One. 2014;9:e95240. doi: 10.1371/journal.pone.0095240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ji R, Zhang B, Zhang X, Xue J, Yuan X, Yan Y, Wang M, Zhu W, Qian H, Xu W. Exosomes derived from human mesenchymal stem cells confer drug resistance in gastric cancer. Cell Cycle. 2015;14:2473–2483. doi: 10.1080/15384101.2015.1005530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu Y, Yan C, Mu L, Huang K, Li X, Tao D, Wu Y, Qin J. Fibroblast-derived exosomes contribute to chemoresistance through priming cancer stem cells in colorectal cancer. PLoS One. 2015;10:e0125625. doi: 10.1371/journal.pone.0125625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brinton LT, Sloane HS, Kester M, Kelly KA. Formation and role of exosomes in cancer. Cell Mol Life Sci. 2015;72:659–671. doi: 10.1007/s00018-014-1764-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ding C, Han F, Xiang H, Xia X, Wang Y, Dou M, Zheng J, Li Y, Xue W, Ding X, Tian P. LncRNA CRNDE is a biomarker for clinical progression and poor prognosis in clear cell renal cell carcinoma. J Cell Biochem. 2018;119:10406–10414. doi: 10.1002/jcb.27389. [DOI] [PubMed] [Google Scholar]
- 41.Wang H, Ke J, Guo Q, Barnabo Nampoukime KP, Yang P, Ma K. Long non-coding RNA CRNDE promotes the proliferation, migration and invasion of hepatocellular carcinoma cells through miR-217/MAPK1 axis. J Cell Mol Med. 2018;22:5862–5876. doi: 10.1111/jcmm.13856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Y, Wang Y, Li J, Zhang Y, Yin H, Han B. CRNDE, a long-noncoding RNA, promotes glioma cell growth and invasion through mTOR signaling. Cancer Lett. 2015;367:122–128. doi: 10.1016/j.canlet.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 43.Liu T, Zhang X, Gao S, Jing F, Yang Y, Du L, Zheng G, Li P, Li C, Wang C. Exosomal long noncoding RNA CRNDE-h as a novel serum-based biomarker for diagnosis and prognosis of colorectal cancer. Oncotarget. 2016;7:85551–85563. doi: 10.18632/oncotarget.13465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ding J, Li J, Wang H, Tian Y, Xie M, He X, Ji H, Ma Z, Hui B, Wang K, Ji G. Long noncoding RNA CRNDE promotes colorectal cancer cell proliferation via epigenetically silencing DUSP5/CDKN1A expression. Cell Death Dis. 2017;8:e2997. doi: 10.1038/cddis.2017.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Han P, Li JW, Zhang BM, Lv JC, Li YM, Gu XY, Yu ZW, Jia YH, Bai XF, Li L, et al. The lncRNA CRNDE promotes colorectal cancer cell proliferation and chemoresistance via miR-181a-5p-mediated regulation of Wnt/β-catenin signaling. Mol Cancer. 2017;16:9. doi: 10.1186/s12943-017-0583-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gao H, Song X, Kang T, Yan B, Feng L, Gao L, Ai L, Liu X, Yu J, Li H. Long noncoding RNA CRNDE functions as a competing endogenous RNA to promote metastasis and oxaliplatin resistance by sponging miR-136 in colorectal cancer. Onco Targets Ther. 2017;10:205–216. doi: 10.2147/OTT.S116178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Han J, Han B, Wu X, Hao J, Dong X, Shen Q, Pang H. Knockdown of lncRNA H19 restores chemo-sensitivity in paclitaxel-resistant triple-negative breast cancer through triggering apoptosis and regulating Akt signaling pathway. Toxicol Appl Pharmacol. 2018;359:55–61. doi: 10.1016/j.taap.2018.09.018. [DOI] [PubMed] [Google Scholar]
- 48.Li M, Chai HF, Peng F, Meng YT, Zhang LZ, Zhang L, Zou H, Liang QL, Li MM, Mao KG, et al. Estrogen receptor β upregulated by lncRNA-H19 to promote cancer stem-like properties in papillary thyroid carcinoma. Cell Death Dis. 2018;9:1120. doi: 10.1038/s41419-018-1077-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang Z, Lei W, Hu HB, Zhang H, Zhu Y. H19 promotes non-small-cell lung cancer (NSCLC) development through STAT3 signaling via sponging miR-17. J Cell Physiol. 2018;233:6768–6776. doi: 10.1002/jcp.26530. [DOI] [PubMed] [Google Scholar]
- 50.Tsang WP, Ng EK, Ng SS, Jin H, Yu J, Sung JJ, Kwok TT. Oncofetal H19-derived miR-675 regulates tumor suppressor RB in human colorectal cancer. Carcinogenesis. 2010;31:350–358. doi: 10.1093/carcin/bgp181. [DOI] [PubMed] [Google Scholar]
- 51.Han D, Gao X, Wang M, Qiao Y, Xu Y, Yang J, Dong N, He J, Sun Q, Lv G, et al. Long noncoding RNA H19 indicates a poor prognosis of colorectal cancer and promotes tumor growth by recruiting and binding to eIF4A3. Oncotarget. 2016;7:22159–22173. doi: 10.18632/oncotarget.8063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang W, Redpath RE, Zhang C, Ning N. Long non-coding RNA H19 promotes the migration and invasion of colon cancer cells via MAPK signaling pathway. Oncol Lett. 2018;16:3365–3372. doi: 10.3892/ol.2018.9052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ma H, Yuan L, Li W, Xu K, Yang L. The LncRNA H19/miR-193a-3p axis modifies the radio-resistance and chemotherapeutic tolerance of hepatocellular carcinoma cells by targeting PSEN1. J Cell Biochem. 2018;119:8325–8335. doi: 10.1002/jcb.26883. [DOI] [PubMed] [Google Scholar]
- 54.Si X, Zang R, Zhang E, Liu Y, Shi X, Zhang E, Shao L, Li A, Yang N, Han X, et al. LncRNA H19 confers chemoresistance in ERα-positive breast cancer through epigenetic silencing of the pro-apoptotic gene BIK. Oncotarget. 2016;7:81452–81462. doi: 10.18632/oncotarget.13263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu KF, Liang WC, Feng L, Pang JX, Waye MM, Zhang JF, Fu WM. H19 mediates methotrexate resistance in colorectal cancer through activating Wnt/β-catenin pathway. Exp Cell Res. 2017;350:312–317. doi: 10.1016/j.yexcr.2016.12.003. [DOI] [PubMed] [Google Scholar]
- 56.Wang N, Meng X, Liu Y, Chen Y, Liang Q. LPS promote Osteosarcoma invasion and migration through TLR4/HOTAIR. Gene. 2019;680:1–8. doi: 10.1016/j.gene.2018.09.031. [DOI] [PubMed] [Google Scholar]
- 57.Kim HJ, Lee DW, YIm GW, Nam EJ, Kim S, Kim SW, Kim YT. Long non-coding RNA HOTAIR is associated with human cervical cancer progression. Int J Oncol. 2015;46:521–530. doi: 10.3892/ijo.2014.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xian HP, Zhuo ZL, Sun YJ, Liang B, Zhao XT. Circulating long non-coding RNAs HULC and ZNFX1-AS1 are potential biomarkers in patients with gastric cancer. Oncol Lett. 2018;16:4689–4698. doi: 10.3892/ol.2018.9199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lin K, Jiang H, Zhang LL, Jiang Y, Yang YX, Qiu GD, She YQ, Zheng JT, Chen C, Fang L, Zhang SY. Down-regulated LncRNA-HOTAIR suppressed colorectal cancer cell proliferation, invasion, and migration by mediating p21. Dig Dis Sci. 2018;63:2320–2331. doi: 10.1007/s10620-018-5127-z. [DOI] [PubMed] [Google Scholar]
- 60.Lu X, Liu Z, Ning X, Huang L, Jiang B. The long noncoding RNA HOTAIR promotes colorectal cancer progression by sponging miR-197. Oncol Res. 2018;26:473–481. doi: 10.3727/096504017X15105708598531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Luo ZF, Zhao D, Li XQ, Cui YX, Ma N, Lu CX, Liu MY, Zhou Y. Clinical significance of HOTAIR expression in colon cancer. World J Gastroenterol. 2016;22:5254–5259. doi: 10.3748/wjg.v22.i22.5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li P, Zhang X, Wang L, Du L, Yang Y, Liu T, Li C, Wang C. lncRNA HOTAIR contributes to 5FU resistance through suppressing miR-218 and activating NF-κB/TS signaling in colorectal cancer. Mol Ther Nucleic Acids. 2017;8:356–369. doi: 10.1016/j.omtn.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xiao Z, Qu Z, Chen Z, Fang Z, Zhou K, Huang Z, Guo X, Zhang Y. LncRNA HOTAIR is a prognostic biomarker for the proliferation and chemoresistance of colorectal cancer via MiR-203a-3p-mediated Wnt/ss-catenin signaling pathway. Cell Physiol Biochem. 2018;46:1275–1285. doi: 10.1159/000489110. [DOI] [PubMed] [Google Scholar]
- 64.Wang HM, Lu JH, Chen WY, Gu AQ. Upregulated lncRNA-UCA1 contributes to progression of lung cancer and is closely related to clinical diagnosis as a predictive biomarker in plasma. Int J Clin Exp Med. 2015;8:11824–11830. [PMC free article] [PubMed] [Google Scholar]
- 65.Shalini S, Dorstyn L, Dawar S, Kumar S. Old, new and emerging functions of caspases. Cell Death Differ. 2015;22:526–539. doi: 10.1038/cdd.2014.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li JY, Ma X, Zhang CB. Overexpression of long non-coding RNA UCA1 predicts a poor prognosis in patients with esophageal squamous cell carcinoma. Int J Clin Exp Pathol. 2014;7:7938–7944. [PMC free article] [PubMed] [Google Scholar]
- 67.Han Y, Yang YN, Yuan HH, Zhang TT, Sui H, Wei XL, Liu L, Huang P, Zhang WJ, Bai YX. UCA1, a long non-coding RNA up-regulated in colorectal cancer influences cell proliferation, apoptosis and cell cycle distribution. Pathology. 2014;46:396–401. doi: 10.1097/PAT.0000000000000125. [DOI] [PubMed] [Google Scholar]
- 68.Bian Z, Jin L, Zhang J, Yin Y, Quan C, Hu Y, Feng Y, Liu H, Fei B, Mao Y, et al. LncRNA-UCA1 enhances cell proliferation and 5-fluorouracil resistance in colorectal cancer by inhibiting miR-204-5p. Sci Rep. 2016;6:23892. doi: 10.1038/srep23892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang X, Liu W, Xu X, Zhu J, Wu Y, Zhao K, He S, Li M, Wu Y, Zhang S, et al. Downregulation of long noncoding RNA UCA1 enhances the radiosensitivity and inhibits migration via suppression of epithelialmesenchymal transition in colorectal cancer cells. Oncol Rep. 2018;40:1554–1564. doi: 10.3892/or.2018.6573. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used during the present study are downloaded from TCGA and GEO datasets and analyzed data is available from the corresponding author on reasonable request.