Abstract
Homoharringtonine (HHT) and imatinib have a synergistic effect in the clinical treatment of chronic myeloid leukemia (CML). The purpose of the present study was to explore the underlying mechanisms by which HHT enhanced imatinib sensitivity. K562 CML cells were treated with HHT and imatinib separately or in combination. Cell viability was detected by Cell Counting Kit-8 assay; apoptotic rates and protein expression levels of phosphorylated-tyrosine (p-Tyr) and p-CRK like proto-oncogene, adaptor protein (p-Crkl) were analyzed by flow cytometry; zinc-finger protein, X-linked (ZFX) overexpression plasmid was transfected to cells using electroporation; western blotting was used to detect the protein expression levels of PI3K, AKT, p-AKT and ZFX; and reverse transcription-quantitative PCR was used to measure ZFX mRNA expression levels. The results demonstrated that HHT and imatinib co-treatment had significant effects of proliferation inhibition and apoptosis induction on K562 CML cells compared with imatinib alone. Co-treatment also significantly downregulated the expression levels of p-Tyr, p-Crkl, PI3K and p-Akt compared with imatinib or HHT treatment. In addition, HHT downregulated ZFX mRNA and protein expression. ZFX overexpression reversed cell sensitivity to imatinib and HHT and also reduced the HHT-induced imatinib sensitization by increasing p-Akt expression. In conclusion, HHT may enhance the effect of imatinib on CML cells by downregulating ZFX.
Keywords: homoharringtonine, imatinib, apoptosis, tyrosine kinase, chronic myelogenous leukemia, zinc-finger protein, X-linked
Introduction
Chronic myeloid leukemia (CML) is characterized by the formation of the Philadelphia (Ph) chromosome, which occurs in pluripotent hematopoietic stem cells (1). This translocation generates the BCR activator of RhoGEF and GTPase (BCR)-ABL fusion gene that encodes p210BCR-ABL protein (2). The oncoprotein exhibits constitutive tyrosine kinase activity and serves a fundamental role in the formation of CML (3). Imatinib, a tyrosine kinase inhibitor (TKI), is the upfront treatment for Ph+ CML (4,5). However, drug resistance is a major reason for relapsed and refractory CML following the termination of imatinib treatment (6), and a number of mechanisms of resistance are independent of p210BCR-ABL upregulation and mutation status (7,8). Therefore, it is important to develop a technique to improve the therapeutic effects of imatinib.
Homoharringtonine (HHT) is a plant alkaloid with antitumor properties that is derived from trees of the genus Cephalotaxus; it has been widely used in China for the treatment of hematological malignancies since the 1970s (9,10). HHT has served an important role in the treatment of CML, both prior to the widespread use of TKIs, and at present following the development of TKI resistance (11–13). The FDA has approved HHT for CML refractory to TKIs (1). The anti-leukemic mechanism of HHT is based on the inhibition of protein synthesis (10). HHT reduces p210BCR-ABL protein expression level in BCR-ABL+ cells independently of BCR-ABL mutational status (13–15). HHT also has a synergistic relationship with imatinib in clinical therapy (16), but the working mechanism is poorly understood.
The zinc-finger protein, X-linked (ZFX) gene is on the mammalian X chromosome and is a transcriptional regulator involved in the maintenance of embryonic and hematopoietic stem cells (17,18). Previous studies suggest that ZFX serves a pivotal role in tumorigenesis of multiple types of cancer, including lung cancer, gastric cancer, breast cancer, malignant glioma and leukemia (19–23). Additionally, ZFX participates in drug resistance in hepatocellular carcinoma (24,25). The results of our previous study also demonstrated that ZFX may be involve in the regulation of cell proliferation and imatinib resistance in CML (26). Thus, the present study investigated the effects and mechanisms of HHT facilitating imatinib sensitivity in K562 human CML cells. The results indicated that HHT may enhance the effects of imatinib on CML cells by downregulating ZFX expression.
Materials and methods
Cell culture
K562 human CML cells were purchased from The Cell Bank of Type Culture Collection of The Chinese Academy of Sciences. Cells were cultivated in RPMI-1640 medium supplemented with 10% FBS (both Gibco; Thermo Fisher Scientific, Inc.) in a humidified atmosphere with 5% CO2 at 37°C. A range of concentrations of imatinib (Selleck Chemicals) or HHT (Bio-Techne) were added to the cells during experiments.
Transfection
To overexpress ZFX, the human ZFX sequence was amplified and cloned into the pEGFP-C1 expression plasmid by Shanghai GeneChem Co., Ltd. Cell transfection was performed by electroporation. Typically, 7×106 cells and pEGFP-C1-ZFX or empty vector were electroporated using a Bio-Rad Gene Pulser II (Bio-Rad Laboratories, Inc.) with 250 V voltage and 950 µFd electric capacity. Cells were subsequently resuspended in RPMI-1640 medium and cultured for 24–72 h.
Cell viability assay
Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc.) assay was used to examine cell viability following different treatments. K562 cells were seeded in a 96-well plate (8×103 cells/well), cultured for 24 h and treated with different concentrations of imatinib or HHT for 24 or 48 h at 37°C. Subsequently, 10 µl CCK-8 solution was added to the plate and the cells were incubated for 2 h at 37°C. Absorbance at 450 nm was measured using a microplate reader (BioTek Instruments, Inc.). HHT treatment or ZFX overexpression may affect cell viability; therefore, the relative viability of drug-treated cells was normalized to DMSO-treated cells to eliminate a false-positive effect.
Colony formation assay
Drug-treated K562 cells were cultured in a two-layer soft agar system as previously described (27). Single-cell suspensions were washed with RPMI-1640 medium, enumerated and plated into a 12-well plate (1 ml/well; 1×103 cells/ml). The feeder layer was prepared with agar equilibrated at 42°C. Following incubation for 10 days at 37°C, the colonies (≥40 cells for each) were counted under an inverted microscope (magnification, ×100; Olympus Corporation).
Apoptotic assay
Apoptosis was detected using the Annexin V-FITC/propidium iodide (PI) Apoptosis Detection kit (BD Biosciences) according to the manufacturer's protocol. Stained cells were analyzed using a flow cytometer (BD Biosciences), and cells were separated into normal, early apoptotic, late apoptotic and dead cells. The relative ratios of early apoptotic cells were analyzed by FlowJo software (version 10; FlowJo LLC).
Phosphorylated-tyrosine (p-Tyr) protein assay
Drug-treated K562 cells were fixed with 1% paraformaldehyde (BD Biosciences) for 30 min and permeabilized with 3% saponin (BD Biosciences) for 1.5 h (both at room temperature). Subsequently, the cells were stained using phycoerythrin-conjugated p-Tyr (1:1,000; cat. no. 558008) or p-CRK like proto-oncogene, adaptor protein (p-Crkl; 1:1,000; cat. no. 560788) antibodies (both BD Biosciences). Following washing with PBS, cells were recovered in 3% saponin and submitted to flow cytometric analysis (BD Biosciences), and mean fluorescence intensity (MFI) was recorded by FlowJo software (version 10) to observe the levels of p-Tyr and p-Crkl proteins.
Western blot analysis
Cells were lysed using RIPA buffer supplemented with a protease inhibitor cocktail (Sigma-Aldrich; Merck KGaA). The protein concentration was determined using a bicinchoninic acid assay, and equal amounts (20–30 µg) of total protein were separated by 6–10% SDS-PAGE and transferred onto PVDF membranes, which were then blocked with 5% skim milk for 2 h at room temperature. The primary antibodies against ZFX (1:1,000; cat. no. ab115998; Abcam), β-Actin (1:1,000; cat. no. ab8224; Abcam), PI3K (1:1,000; cat. no. 05–212; Merck KGaA), AKT (1:1,000; cat. no. 07-383; Merck KGaA) and p-AKT (1:1,000; cat. no. 04-736; Merck KGaA) were used to incubate the membranes overnight at 4°C. Following incubation with secondary goat anti-mouse (1:2,000; cat. no. ab6789; Abcam) or goat anti-rabbit (1:2,000; cat. no. ab6721; Abcam) antibody for 2 h at room temperature, blots were visualized using ECL Detection Reagent (cat. no. P0018; Beyotime Institute of Biotechnology) and analyzed using Image Lab software (Bio-Rad Laboratories, Inc.).
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was isolated from cells using TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.) and reverse transcribed with FastQuant RT kit (Tiangen Biotech Co., Ltd.). PCR was performed in triplicate with SuperReal PreMix Plus (Tiangen Biotech Co., Ltd.) using the Real-Time PCR Detection System (Roche Molecular Systems, Inc.). qPCR was conducted at 95°C for 5 min, followed by 40 cycles at 95°C for 10 sec, 65°C for 20 sec and 72°C for 30 sec. The primer sequences for ZFX and β-Actin were as follows: ZFX, forward 5′-GGCAGTCCACAGCAAGAAC-3′, reverse 5′-TTGGTATCCGAGAAAGTCAGAAG-3′; β-Actin, forward 5′-CTCCATCCTGGCCTCGCTGT-3′ and reverse 5′-GCTGTCACCTTCACCGTTCC-3′. Relative mRNA levels were normalized to β-Actin expression using the 2−ΔΔCq method (28).
Statistical analysis
SPSS 19.0 software (IBM Corp.) was used for statistical analysis. Data are expressed as the mean ± SD. Differences between two groups were assessed by Student's t-test. Statistical differences among multiple groups were analyze by one-way analysis of variance followed by a Bonferroni post hoc test. Each experiment was repeated for at least three times. P<0.05 was considered to indicate a statistically significant difference.
Results
HHT facilitates imatinib sensitivity in CML cells
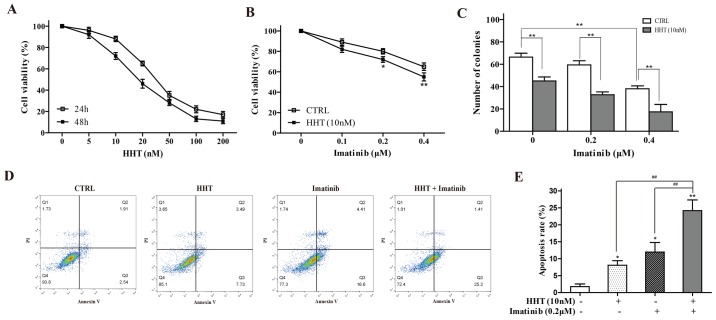
The effects of HHT on K562 cell viability were tested by CCK-8 assay. Following treatment for 24 or 48 h, HHT reduced cell viability in a dose- and a time-dependent manner (Fig. 1A). A HTT concentration of 10 nM was selected for subsequent experiments, as this concentration produced a small inhibitory effect at 24 h that would still enable the detection of further sensitization effects. 24-h co-treatment with 10 nM HHT and a range of concentrations of imatinib resulted in significantly greater inhibition of cell viability compared with imatinib alone (Fig. 1B). The results of a cloning experiment demonstrated the additive effect of imatinib and HHT on the reduced ability of K562 cells to form colonies compared with either drug alone (Fig. 1C). In addition, compared with HHT-alone or imatinib-alone groups, 24-h co-treatment with HHT and imatinib significantly increased the early apoptotic rate of K562 cells (Fig. 1D). An imatinib concentration of 0.2 µM imatinib was selected, as this concentration produced a small inhibitory effect upon which further sensitization could be observed, like for HTT.
Figure 1.
HHT enhances imatinib sensitivity in chronic myeloid leukemia cells. (A) Cell Counting Kit-8 assay was performed to measure the viability of K562 cells following treatment with a range of concentrations of HHT for 24 and 48 h. (B) Viability of K562 cells co-treated with 10 nM HHT and various concentrations of imatinib for 24 h. *P<0.05, **P<0.01 vs. CTRL. (C) K562 cell colony formation following co-treatment with 10 nM HHT and various concentrations of imatinib for 10 days. (D and E) Apoptosis of K562 cells co-treated with HHT or imatinib for 24 h was examined by flow cytometry. *P<0.05, **P<0.01 vs. CTRL. ##P<0.01 vs. HHT + imatinib. CTRL, control; HHT, homoharringtonine.
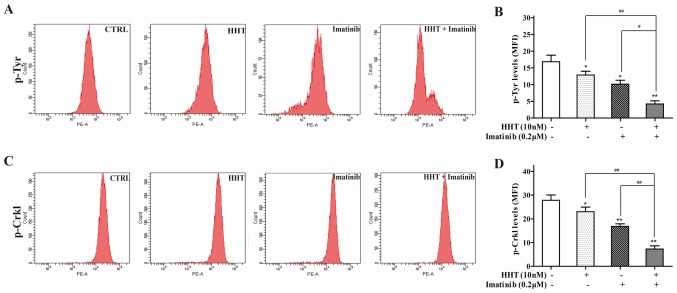
Effects of HHT combined with imatinib on tyrosine kinase activity in CML cells
Tyrosine kinase activity may be reflected by p-Try protein expression levels in BCR-ABL+ cells (29). Co-treatment of cells with HHT and imatinib for 24 h significantly decreased the MFI of p-Tyr staining compared with either treatment alone (Fig. 2A). p-Crkl, which has been reported as an appropriate measure of tyrosine kinase activity in CML (30), was also detected; the MFI of p-Crkl staining in co-treated cells was significantly deceased compared with the individual drug groups (Fig. 2B).
Figure 2.
HHT and imatinib inhibit tyrosine kinase activity in chronic myeloid leukemia cells. Flow cytometry assay was performed to analyze (A and B) p-Tyr and (C and D) p-Crkl levels in K562 cells treated with HHT or imatinib or co-treated with HHT and imatinib. *P<0.05, **P<0.01 vs. CTRL; #P<0.05, ##P<0.01 vs. HHT + imatinib. CTRL, control; HHT, homoharringtonine; MFI, mean fluorescence intensity; Crkl, CRK like proto-oncogene, adaptor protein; p-, phosphorylated; Tyr, tyrosine.
HHT regulates imatinib sensitivity through suppression of PI3K/Akt pathway in CML cells
The abnormal activation of PI3K/Akt pathway results in imatinib resistance in CML (31). Therefore, the expression levels of PI3K, Akt and p-Akt were assessed. Compared with imatinib treatment alone, co-treatment with HHT and imatinib for 24 h downregulated the expression levels of PI3K and p-Akt in K562 cells (Fig. 3). However, no difference in total Akt was detected. No notable difference in PI3K expression was observed between the HHT and co-treatment groups.
Figure 3.
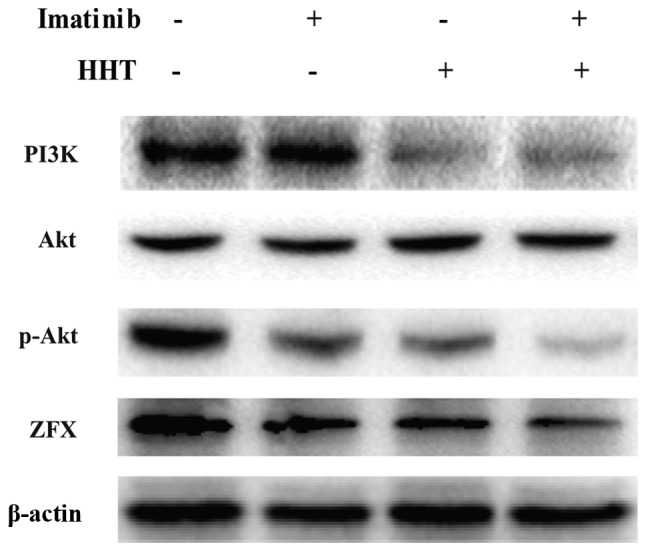
Effects of HHT and imatinib on the PI3K/AKT pathway in chronic myeloid leukemia cells. Western blotting was performed to detect expression levels of PI3K, AKT, and p-Akt in K562 cells treated with HHT or imatinib for 48 h. CTRL, control; HHT, homoharringtonine; p-, phosphorylated; ZFX, zinc-finger protein, X-linked.
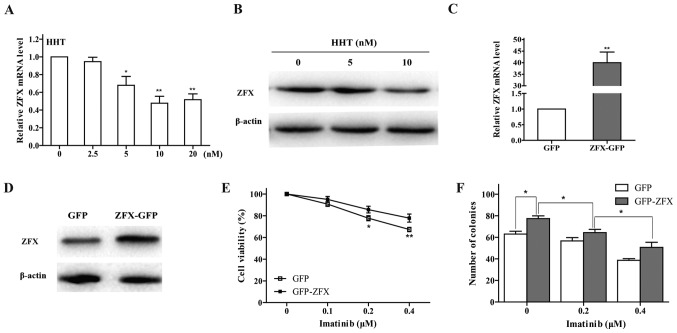
HHT downregulates ZFX expression levels in CML cells
The results of our previous study demonstrated that ZFX silencing may inhibit CML cell growth through the PI3K/AKT pathway (26). To test whether the effect of HHT on CML cells was associated with ZFX expression, RT-qPCR and western blot analyses were performed; the results revealed that the mRNA and protein expression levels of ZFX were dose-dependently decreased by HHT treatment for 24 h in K562 cells (Fig. 4A and B). In addition, HHT and imatinib co-treatment downregulated ZFX protein expression levels (Fig. 3).
Figure 4.
Effects of ZFX on imatinib resistance in chronic myeloid leukemia cells. (A) mRNA and (B) protein expression levels of ZFX in K562 cells were determined by RT-qPCR and western blotting, respectively, following treatment with HHT. *P<0.05 and **P<0.01 vs. CTRL. (C) RT-qPCR and (D) western blotting were used to examine ZFX mRNA and protein expression levels, respectively, in K562 cells following transfection with ZFX-GFP and empty GFP plasmid. **P<0.01 vs. GFP. (E) Viability of K562 cells transfected with ZFX-GFP treated with imatinib for 24 h. *P<0.05, **P<0.01 vs. GFP. (F) K562 cell colony formation following transfection with ZFX-GFP and treatment with imatinib for 10 days. *P<0.05. GFP, green fluorescent protein; HHT, homoharringtonine; RT-qPCR, reverse transcription-quantitative PCR; ZFX, zinc-finger protein, X-linked.
Overexpression of ZFX induces imatinib resistance in CML cells
To validate the effects of ZFX on CML cell response to imatinib, K562 cells transfected with either ZFX-GFP or empty GFP plasmids were incubated with imatinib for 24 h. ZFX mRNA and protein expression levels were successfully increased following transfection with the ZFX-GFP plasmid (Fig. 4C and D). The results of the CCK-8 assay indicated that overexpression of ZFX decreased the sensitivity of K562 cells to imatinib (Fig. 4E). In addition, overexpression of ZFX reversed the inhibitory effects of imatinib on colony formation in K562 cells (Fig. 4F).
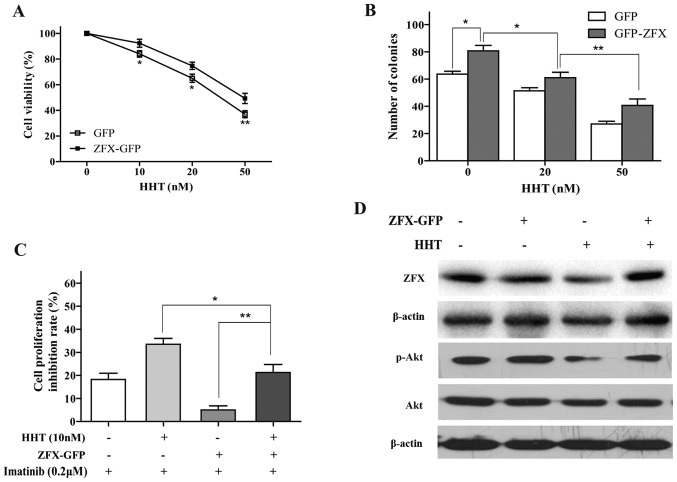
Overexpression of ZFX attenuates the effects of HHT on CML cells
Overexpression of ZFX reversed the HHT-induced inhibition of proliferation and clone formation in K562 cells (Fig. 5A and B). Additionally, the effects of HHT on imatinib sensitivity were attenuated by overexpression of ZFX; the cell inhibition rate in the HHT + ZFX group was significantly increased compared with that in the ZFX group (Fig. 5C). ZFX overexpression also reversed HHT-induced decrease of p-AKT expression (Fig. 5D). Thus, the data suggested that ZFX may participate in HHT-induced imatinib sensitivity in CML cells.
Figure 5.
Effects of ZFX overexpression on HHT-treated chronic myeloid leukemia cells. (A) Viability of K562 cells transfected with ZFX-GFP and treated with HHT for 24 h. *P<0.05, **P<0.01 vs. GFP. (B) Cell colony formation following transfection with ZFX-GFP overexpression vector transfection and treatment with various concentrations of HHT for 10 days. (C) Imatinib sensitivity was determined using Cell Counting Kit-8 assay in K562 cells transfected with ZFX-GFP following treatment with HHT for 24 h. (D) Protein expression levels of ZFX and p-AKT were determined by western blotting following transfection with ZFX-GFP and treatment with HHT. *P<0.05, **P<0.01. CTRL, control; GFP, green fluorescent protein; HHT, homoharringtonine; p-, phosphorylated; ZFX, zinc-finger protein, X-linked.
Discussion
HHT treatment is effective for patients with CML and may provide an effective treatment for patients with TKI-resistant CML with BRC-ABL mutations (32,33). Thus, co-treatment with HHT and imatinib may provide a novel approach to improve the efficiency of CML treatment. The results of the present study have demonstrated a potential mechanism of HHT in enhancing the effect of imatinib on CML cells.
In the present study, HHT treatment increased CML cell sensitivity to imatinib. Co-treatment with HHT and imatinib resulted in decreases in K562 cell viability and in the number of colonies formed compared with either treatment alone. In addition, co-treatment induced apoptosis in K562 cells more effectively compared with individual treatments. These results were consistence with our previous study in imatinib-resistant CML cells (34). Therefore, HHT may facilitate imatinib sensitivity by inducing apoptosis in CML.
HHT is a broad-spectrum protein TKI that inhibits signaling protein phosphorylation by oncogenic proteins, such as Janus kinase 2 V617F and p210BCR-ABL, thus blocking the survival signaling pathways of leukemia cells (10,35). Constitutive Try phosphorylation is the major characteristic of BCR-ABL+ cells (1). In the present study, flow cytometry was used to measure the p-Tyr and p-Crkl expression levels, which were previously demonstrated to be abnormally high in CML cells (30,36). The results of the present study demonstrated that co-treatment with HHT and imatinib decreased p-Tyr and p-Crkl expression levels compared with individual drug treatment. HHT reduces p210BCR-ABL expression in BCR-ABL+ cells (37), which may partially explain the additive interaction between HHT and imatinib.
The PI3K/AKT signaling pathway is essential for CML cell viability, and may be an effective target for therapeutic intervention in imatinib-resistant CML (3,31). Previous studies have reported that HHT mediates myeloid cell apoptosis by inhibiting the PI3K/AKT signaling pathway (13,38). In the present study, PI3K and p-AKT expression levels were decreased by co-treatment with HHT and imatinib compared with either treatment alone. These data indicated that HHT may enhance the effects of imatinib through synergistic inhibition of the PI3K/AKT pathway.
To further examine the crucial molecules except for p210BCR-ABL underlying the mechanisms via HHT enhances imatinib sensitivity through the PI3K/AKT pathway, the role of ZFX, which is a known mediator of biological function in cancer cells (39–41), was investigated. ZFX mRNA and protein expression levels were downregulated by HHT. Overexpression of ZFX reversed imatinib- or HHT-induced inhibition of cell viability. In addition, ZFX overexpression significantly weakened HHT-induced imatinib sensitization and reversed the inhibitory effect of HHT on p-AKT expression. Thus, ZFX may be responsible for abolishing the sensitization of imatinib mediated by HHT.
In conclusion, the results of the present study demonstrated that HHT may increase imatinib sensitivity of CML cells through downregulation of ZFX expression, which leads to the inhibition of PI3K/AKT pathway, thereby providing new insight into the therapeutic strategy of CML treatment.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
JW and BW drafted the manuscript. JW, BW, YS, XL and YD collected, analyzed and interpreted the data. YL and CW conceived and designed the present study. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Apperley JF. Chronic myeloid leukaemia. Lancet. 2015;385:1447–1459. doi: 10.1016/S0140-6736(13)62120-0. [DOI] [PubMed] [Google Scholar]
- 2.Ren R. Mechanisms of BCR-ABL in the pathogenesis of chronic myelogenous leukaemia. Nat Rev Cancer. 2005;5:172–183. doi: 10.1038/nrc1567. [DOI] [PubMed] [Google Scholar]
- 3.Steelman LS, Pohnert SC, Shelton JG, Franklin RA, Bertrand FE, McCubrey JA. JAK/STAT, Raf/MEK/ERK, PI3K/Akt and BCR-ABL in cell cycle progression and leukemogenesis. Leukemia. 2004;18:189–218. doi: 10.1038/sj.leu.2403241. [DOI] [PubMed] [Google Scholar]
- 4.Druker BJ, Talpaz M, Resta DJ, Peng B, Buchdunger E, Ford JM, Lydon NB, Kantarjian H, Capdeville R, Ohno-Jones S, Sawyers CL. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N Engl J Med. 2001;344:1031–1037. doi: 10.1056/NEJM200104053441402. [DOI] [PubMed] [Google Scholar]
- 5.Kantarjian HM, Cortes JE, O'Brien S, Giles F, Garcia-Manero G, Faderl S, Thomas D, Jeha S, Rios MB, Letvak L, et al. Imatinib mesylate therapy in newly diagnosed patients with Philadelphia chromosome-positive chronic myelogenous leukemia: High incidence of early complete and major cytogenetic responses. Blood. 2003;101:97–100. doi: 10.1182/blood-2002-02-0545. [DOI] [PubMed] [Google Scholar]
- 6.Hochhaus A, Kreil S, Corbin AS, La Rosée P, Müller MC, Lahaye T, Hanfstein B, Schoch C, Cross NC, Berger U, et al. Molecular and chromosomal mechanisms of resistance to imatinib (STI571) therapy. Leukemia. 2002;16:2190–2196. doi: 10.1038/sj.leu.2402741. [DOI] [PubMed] [Google Scholar]
- 7.Jabbour EJ, Cortes JE, Kantarjian HM. Resistance to tyrosine kinase inhibition therapy for chronic myelogenous leukemia: A clinical perspective and emerging treatment options. Clin Lymphoma Myeloma Leuk. 2013;13:515–529. doi: 10.1016/j.clml.2013.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Binato R, Mencalha A, Pizzatti L, Scholl V, Zalcberg I, Abdelhay E. RUNX1T1 is overexpressed in imatinib mesylate-resistant cells. Mol Med Rep. 2009;2:657–661. doi: 10.3892/mmr_00000153. [DOI] [PubMed] [Google Scholar]
- 9.Powell RG, Weisleder D, Smith CR., Jr Antitumor alkaloids for Cephalataxus harringtonia: Structure and activity. J Pharm Sci. 1972;61:1227–1230. doi: 10.1002/jps.2600610812. [DOI] [PubMed] [Google Scholar]
- 10.Lü S, Wang J. Homoharringtonine and omacetaxine for myeloid hematological malignancies. J Hematol Oncol. 2014;7:2. doi: 10.1186/1756-8722-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kantarjian HM, Talpaz M, Smith TL, Cortes J, Giles FJ, Rios MB, Mallard S, Gajewski J, Murgo A, Cheson B, O'Brien S. Homoharringtonine and low-dose cytarabine in the management of late chronic-phase chronic myelogenous leukemia. J Clin Oncol. 2000;18:3513–3521. doi: 10.1200/JCO.2000.18.20.3513. [DOI] [PubMed] [Google Scholar]
- 12.Legros L, Hayette S, Nicolini FE, Raynaud S, Chabane K, Magaud JP, Cassuto JP, Michallet M. BCR-ABL(T315I) transcript disappearance in an imatinib-resistant CML patient treated with homoharringtonine: A new therapeutic challenge. Leukemia. 2007;21:2204–2206. doi: 10.1038/sj.leu.2404772. [DOI] [PubMed] [Google Scholar]
- 13.Wang L, You LS, Ni WM, Ma QL, Tong Y, Mao LP, Qian JJ, Jin J. β-Catenin and AKT are promising targets for combination therapy in acute myeloid leukemia. Leuk Res. 2013;37:1329–1340. doi: 10.1016/j.leukres.2013.06.023. [DOI] [PubMed] [Google Scholar]
- 14.Dong Y, Gao X, Zhao Y, Wei M, Xu L, Yang G, Liu L. Semi-random mutagenesis profile of BCR-ABL during imatinib resistance acquirement in K562 cells. Mol Med Rep. 2017;16:9409–9414. doi: 10.3892/mmr.2017.7835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fresno M, Jiménez A, Vázquez D. Inhibition of translation in eukaryotic systems by harringtonine. Eur J Biochem. 1977;72:323–330. doi: 10.1111/j.1432-1033.1977.tb11256.x. [DOI] [PubMed] [Google Scholar]
- 16.Tipping AJ, Mahon FX, Zafirides G, Lagarde V, Goldman JM, Melo JV. Drug responses of imatinib mesylate-resistant cells: Synergism of imatinib with other chemotherapeutic drugs. Leukemia. 2002;16:2349–2357. doi: 10.1038/sj.leu.2402775. [DOI] [PubMed] [Google Scholar]
- 17.Galan-Caridad JM, Harel S, Arenzana TL, Hou ZE, Doetsch FK, Mirny LA, Reizis B. Zfx controls the self-renewal of embryonic and hematopoietic stem cells. Cell. 2007;129:345–357. doi: 10.1016/j.cell.2007.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harel S, Tu EY, Weisberg S, Esquilin M, Chambers SM, Liu B, Carson CT, Studer L, Reizis B, Tomishima MJ. ZFX controls the self-renewal of human embryonic stem cells. PLoS One. 2012;7:e42302. doi: 10.1371/journal.pone.0042302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang M, Xu S, Yue W, Zhao X, Zhang L, Zhang C, Wang Y. The role of ZFX in non-small cell lung cancer development. Oncol Res. 2012;20:171–178. doi: 10.3727/096504012X13548165987493. [DOI] [PubMed] [Google Scholar]
- 20.Wu S, Lao XY, Sun TT, Ren LL, Kong X, Wang JL, Wang YC, Du W, Yu YN, Weng YR, et al. Knockdown of ZFX inhibits gastric cancer cell growth in vitro and in vivo via downregulating the ERK-MAPK pathway. Cancer Lett. 2013;337:293–300. doi: 10.1016/j.canlet.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 21.Zhu Z, Li K, Xu D, Liu Y, Tang H, Xie Q, Xie L, Liu J, Wang H, Gong Y, et al. ZFX regulates glioma cell proliferation and survival in vitro and in vivo. J Neurooncol. 2013;112:17–25. doi: 10.1007/s11060-012-1032-z. [DOI] [PubMed] [Google Scholar]
- 22.Yang H, Lu Y, Zheng Y, Yu X, Xia X, He X, Feng W, Xing L, Ling Z. shRNA-mediated silencing of ZFX attenuated the proliferation of breast cancer cells. Cancer Chemother Pharmacol. 2014;73:569–576. doi: 10.1007/s00280-014-2379-y. [DOI] [PubMed] [Google Scholar]
- 23.Weisberg SP, Smith-Raska MR, Esquilin JM, Zhang J, Arenzana TL, Lau CM, Churchill M, Pan H, Klinakis A, Dixon JE, et al. ZFX controls propagation and prevents differentiation of acute T-lymphoblastic and myeloid leukemia. Cell Rep. 2014;6:528–540. doi: 10.1016/j.celrep.2014.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang S, Shu R, Yue M, Zhang S. Effect of over-expression of Zinc-finger protein (ZFX) on self-renewal and drug-resistance of hepatocellular carcinoma. Med Sci Monit. 2016;22:3025–3034. doi: 10.12659/MSM.897699. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Lai KP, Chen J, He M, Ching AK, Lau C, Lai PB, To KF, Wong N. Overexpression of ZFX confers self-renewal and chemoresistance properties in hepatocellular carcinoma. Int J Cancer. 2014;135:1790–1799. doi: 10.1002/ijc.28819. [DOI] [PubMed] [Google Scholar]
- 26.Wu J, Wei B, Wang Q, Ding Y, Deng Z, Lu X, Li Y. ZFX facilitates cell proliferation and imatinib resistance in chronic myeloid leukemia cells. Cell Biochem Biophys. 2016;74:277–283. doi: 10.1007/s12013-016-0725-x. [DOI] [PubMed] [Google Scholar]
- 27.Yang J, Ikezoe T, Nishioka C, Udaka K, Yokoyama A. Bcr-Abl activates AURKA and AURKB in chronic myeloid leukemia cells via AKT signaling. Int J Cancer. 2014;134:1183–1194. doi: 10.1002/ijc.28434. [DOI] [PubMed] [Google Scholar]
- 28.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 29.Desplat V, Lagarde V, Belloc F, Chollet C, Leguay T, Pasquet JM, Praloran V, Mahon FX. Rapid detection of phosphotyrosine proteins by flow cytometric analysis in Bcr-Abl-positive cells. Cytometry A. 2004;62:35–45. doi: 10.1002/cyto.a.20030. [DOI] [PubMed] [Google Scholar]
- 30.La Rosée P, Holm-Eriksen S, Konig H, Härtel N, Ernst T, Debatin J, Mueller MC, Erben P, Binckebanck A, Wunderle L, et al. Phospho-CRKL monitoring for the assessment of BCR-ABL activity in imatinib-resistant chronic myeloid leukemia or Ph+ acute lymphoblastic leukemia patients treated with nilotinib. Haematologica. 2008;93:765–769. doi: 10.3324/haematol.12186. [DOI] [PubMed] [Google Scholar]
- 31.Burchert A, Wang Y, Cai D, von Bubnoff N, Paschka P, Müller-Brüsselbach S, Ottmann OG, Duyster J, Hochhaus A, Neubauer A. Compensatory PI3-kinase/Akt/mTor activation regulates imatinib resistance development. Leukemia. 2005;19:1774–1782. doi: 10.1038/sj.leu.2403898. [DOI] [PubMed] [Google Scholar]
- 32.Visani G, Isidori A. Resistant chronic myeloid leukemia beyond tyrosine-kinase inhibitor therapy: Which role for omacetaxine. Expert Opin Pharmacother. 2014;15:1–3. doi: 10.1517/14656566.2014.850491. [DOI] [PubMed] [Google Scholar]
- 33.Cortes J, Lipton JH, Rea D, Digumarti R, Chuah C, Nanda N, Benichou AC, Craig AR, Michallet M, Nicolini FE, et al. Phase 2 study of subcutaneous omacetaxine mepesuccinate after TKI failure in patients with chronic-phase CML with T315I mutation. Blood. 2012;120:2573–2580. doi: 10.1182/blood-2012-03-415307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu JJ, Ding YH, Deng ZK, Shi YY, Lu XY, Li YF. Effect of homoharringtonine combined with Imatinib on the K562/G01 cells and its mechanism. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2017;25:80–84. doi: 10.7534/j.issn.1009-2137.2017.01.013. (In Chinese) [DOI] [PubMed] [Google Scholar]
- 35.Tong H, Ren Y, Zhang F, Jin J. Homoharringtonine affects the JAK2-STAT5 signal pathway through alteration of protein tyrosine kinase phosphorylation in acute myeloid leukemia cells. Eur J Haematol. 2008;81:259–266. doi: 10.1111/j.1600-0609.2008.01116.x. [DOI] [PubMed] [Google Scholar]
- 36.Hamilton A, Alhashimi F, Myssina S, Jorgensen HG, Holyoake TL. Optimization of methods for the detection of BCR-ABL activity in Philadelphia-positive cells. Exp Hematol. 2009;37:395–401. doi: 10.1016/j.exphem.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 37.Chen Y, Hu Y, Michaels S, Segal D, Brown D, Li S. Inhibitory effects of omacetaxine on leukemic stem cells and BCR-ABL-induced chronic myeloid leukemia and acute lymphoblastic leukemia in mice. Leukemia. 2009;23:1446–1454. doi: 10.1038/leu.2009.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meng H, Yang C, Jin J, Zhou Y, Qian W. Homoharringtonine inhibits the AKT pathway and induces in vitro and in vivo cytotoxicity in human multiple myeloma cells. Leuk Lymphoma. 2008;49:1954–1962. doi: 10.1080/10428190802320368. [DOI] [PubMed] [Google Scholar]
- 39.Jiang H, Zhang L, Liu J, Chen Z, Na R, Ding G, Zhang H, Ding Q. Knockdown of zinc finger protein X-linked inhibits prostate cancer cell proliferation and induces apoptosis by activating caspase-3 and caspase-9. Cancer Gene Ther. 2012;19:684–689. doi: 10.1038/cgt.2012.53. [DOI] [PubMed] [Google Scholar]
- 40.Li K, Zhu ZC, Liu YJ, Liu JW, Wang HT, Xiong ZQ, Shen X, Hu ZL, Zheng J. ZFX knockdown inhibits growth and migration of non-small cell lung carcinoma cell line H1299. Int J Clin Exp Pathol. 2013;6:2460–2467. [PMC free article] [PubMed] [Google Scholar]
- 41.Fang Q, Fu WH, Yang J, Li X, Zhou ZS, Chen ZW, Pan JH. Knockdown of ZFX suppresses renal carcinoma cell growth and induces apoptosis. Cancer Genet. 2014;207:461–466. doi: 10.1016/j.cancergen.2014.08.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.