Abstract
Gastric cancer (GC) is one of the most common malignancies worldwide; however, understanding of its development and carcinogenesis is currently limited. Centromere protein O (CENPO), is a newly discovered constitutive centromeric protein, associated with cell death. The expression of CENPO in human cancers, including GC, is currently unknown. The aim of the present study was to investigate the clinical association between CENPO and GC, and to elucidate the potential mechanisms of CENPO in the process of GC progression. The results demonstrated that CENPO was expressed at high levels in GC and was correlated with p-TNM stage. In addition, CENPO was observed to be a marker of poor prognosis in patients with GC. Knockdown of CENPO contributed to GC cell growth inhibition and apoptosis induction. In addition, downregulation of CENPO expression suppressed GC cell growth in vivo. Furthermore, CENPO knockdown decreased ataxia telangiectasia mutated (ATM), cyclin D1 and c-Jun expression, indicating that the ATM signaling pathway may be involved in CENPO-mediated regulation of GC cell growth. In conclusion, increased CENPO expression may be associated with the aggressive tumor biology of GC and CENPO may present a novel therapeutic target and prognostic biomarker for patients with GC.
Keywords: gastric cancer, centromere protein O, cell proliferation, cell apoptosis, ataxia telangiectasia mutated signaling pathway
Introduction
Gastric cancer (GC) is one of the most common human cancers and is the second leading cause of cancer-associated death worldwide (1–3). Due to the progression of GC and high recurrence rates following surgical resection, GC remains a major public health issue. Therefore, elucidating the molecular mechanisms underlying GC tumorigenesis is required in order to identify prognostic markers and novel effective treatments.
Previous studies have demonstrated that the normal expression of centromere proteins is essential for mitosis (4) and abnormal core centromere proteins may increase the incidence of chromosomal instability, aneuploidy, and lead to oncogenesis (5,6). In a previous study, >40 proteins were identified in the interphase centromere complex (ICEN) by proteomic analysis (7); however, the functional role of a number of these centromeric proteins is currently unclear. Centromere protein O (CENPO; also known as ICEN 36), is a newly discovered constitutive centromeric protein that localizes at the centromere (8). CENPO facilitates the prevention of premature sister chromatid separation during recovery from spindle damage and is associated with cell death (9). Depletion of the CENPO protein by small interfering (si)RNA transfection leads to aneuploidy and an increase in aneuploid chromosomes, which may subsequently lead to disease or cancer development (8). However, the direct role and detailed molecular mechanisms of CENPO in cancer are currently unclear.
The results of the present study demonstrated that the expression of CENPO is markedly increased in GC and the effect of CENPO in the proliferation and tumorigenesis of GC cells was therefore investigated. Further analysis revealed that the ataxia telangiectasia mutated (ATM) signaling pathway was involved in the CENPO-mediated regulation of GC pathogenesis. These results suggest that CENPO may present a useful biomarker to assess GC progression and may also be a novel therapeutic target for patients with GC.
Materials and methods
Cell lines and culture conditions
Human GC cell lines, AGS and MGC-803, were purchased from the American Type Culture Collection and the human normal gastric epithelial cell line, GES-1, was purchased from Type Culture Collection of the Chinese Academy of Science. All cell lines were cultured in Dulbecco's Modified Eagle's Medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum (Thermo Fisher Scientific, Inc.) at 37°C in an atmosphere of 5% CO2.
Patients and tissue samples
Matched GC and adjacent normal tissue samples were obtained from 24 patients that underwent primary surgical treatment at The First Affiliated Hospital, Nanchang University (Nanchang, China), between March and November 2016. Patient characteristics are shown in Table I.
Table I.
Relationship between CENPO protein expression and clinicopathological characteristics in gastric cancer patients.
| CENPO protein expression | ||||
|---|---|---|---|---|
| Characteristic | Number of patients | Low (≤8) | High (>8) | P-value |
| All patients | 94 | 45 | 49 | |
| Sex | 0.393 | |||
| Male | 60 | 31 | 29 | |
| Female | 34 | 14 | 20 | |
| Age | >0.999 | |||
| <60 y.o. | 50 | 24 | 26 | |
| ≥60 y.o. | 44 | 21 | 23 | |
| Size of tumor | <0.0001 | |||
| ≤5 cm | 55 | 39 | 16 | |
| >5 cm | 39 | 6 | 33 | |
| T stages | 0.249 | |||
| T1+T2 | 14 | 9 | 5 | |
| T3+T4 | 80 | 36 | 44 | |
| Lymph node metastasis (pN) | 0.014 | |||
| N0 | 15 | 12 | 3 | |
| N1 | 11 | 6 | 5 | |
| N2 | 25 | 13 | 12 | |
| N3 | 43 | 14 | 29 | |
| p-TNM stages | 0.023 | |||
| I | 9 | 7 | 2 | |
| II | 16 | 11 | 5 | |
| III | 64 | 26 | 38 | |
| IV | 5 | 1 | 4 | |
| T stages | 0.219 | |||
| T1 | 6 | 4 | 2 | |
| T2 | 8 | 5 | 3 | |
| T3 | 75 | 32 | 43 | |
| T4 | 5 | 4 | 1 | |
TNM, tumor node metastasis; y.o., years old; CENPO, centromere protein O.
The Cancer Genome Atlas (TGCA) GC cohorts
A total of 443 GC samples with detailed clinicopathological data were obtained from the TGCA Stomach Adenocarcinoma database (http://www.cancergenome.nih.gov). Out of the 443 patients, 59 cases were diagnosed with stage I disease, 130 cases with stage II, 183 with stage III and 44 cases with stage IV. Staging information was not available for 27 cases. A total of 26 paired GC and normal tissue samples data and pathological information were collected and analyzed for the purposes of this study. The TCGA datasets were downloaded from the Broad Institute Firehose Pipeline8 (accessed, 12/03/2017).
Immunohistochemical staining (IHC) and scoring of tissue microarrays (TMAs)
GC TMAs were obtained from Shanghai Outdo Biotech Co., Ltd. Rabbit polyclonal anti-human CENPO antibodies (1:200; Origene Technologies, Inc.; cat no. TA349791) were used for IHC staining analysis according to a two-step protocol. The protein expression patterns of CENPO were analyzed by IHC examination in 94 GC tissue samples and 54 paired adjacent normal gastric tissues. The tissues were fixed in 10% formalin, for 16–24 h at room temperature, then embedded in paraffin and cut into 4 µm thick sections. The images of the specimens for IHC assay were captured by Motic Digital Slide Assistant System (Motic China Group Co., Ltd.). Then, 2 trained pathologists were independently blinded to evaluate CENPO staining intensity with Motic DSAssistant Lite version 1.0 (Motic Medical Diagnosis System Co. Ltd.). According to the staining intensity, scores were assigned as follows: 0, no staining; 1, pale yellow staining; 2, yellow staining; and 3, brown staining. The percentage of positive CENPO staining was defined as follows: 0, no positive cells; 1, ≤25% positive tumor cells; 2, 26–50% positive tumor cells; 3, 51–75% positive tumor cells; and 4, ≥76% positive tumor cells. The CENPO final score was calculated as the sum of the staining intensity and percentage positivity scores.
Plasmids, lentiviral vector construction and pathway inhibitor
Human CENPO (NM_024322) was subcloned into the GV-143 vector and the EGFP-CENPO plasmid, and the control empty vector were generated. All plasmids were constructed by Shanghai GeneChem Co., Ltd. The lentiviral vector system used in the present study was composed of the GV115-EGFP vector, which stably expresses siRNA, a marker enhanced green fluorescent protein (EGFP) fusion protein], pHelper 1.0 (gag/pol element) and Helper 2.0 (vesicularstomatitis virus G; VSVG element). The vectors, pHelper 1.0 and pHelper 2.0, contain imperative elements for virus packaging. The most effective double-stranded CENPO-targeted siRNA sequence, 5′-CAGCGTGAAAGCATGTATT-3′, was synthesized and cloned into the GV115-EGFP vector by Shanghai GeneChem Co., Ltd. Psc-NC (5′-TTCTCCGAACGTGTCACGT-3′) was used to generate the negative control lentiviral vectors (NC or control), which demonstrated no homology to any known human genes. Cancer cells were seeded in six-well plates (5×104 cells/well) until cell confluency reached 60% and then appropriate volumes of lentivirus (MOI=20) were added to the cells according to the multiplicity of infection, as recommended by the manufacturer. Following ≥72 h of lentivirus infection, the fluorescence intensity was observed using a fluorescence microscope (Olympus Corporation), the infection efficiency was calculated and the stable expression cell line was screened using the corresponding antibiotic. ATM pathway inhibitor, (KU-55933; MCE) was used in this study as in previous studies (10,11).
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was isolated using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. cDNA was synthesized by reverse transcription (RT) using the MX3000p (Agilent Technologies, Inc.) according to the manufacturer's protocol (Promega Corporation). The RT reaction conditions were 10 min at 70°C, 60 min at 42°C and 10 min at 70°C. The cDNA product was amplified using a SYBR Green Master Mix kit (Takara Bio, Inc.) and the MX3000p (Agilent Technologies, Inc.). GAPDH was used as the internal control. The primers used were as follows: CENPO, forward, 5′-CCAGCCGAGGAGTTTGTGTC-3′ and reverse, 5′-CCGGAGTGGTTTCTGTATGAC-3′; GAPDH, forward, 5′-TGACTTCAACAGCGACACCCA-3′, and reverse, 5′-CACCCTGTTGCTGTAGCCAAA-3′. The thermocycling conditions were as follows: 95°C for 4 min, followed by 35 cycles of 95°C for 15 sec, 60°C for 30 sec and 72°C for 30 sec. The relative mRNA expression (CENPO/GAPDH) was determined using the 2−ΔΔCq method (12).
Western blotting analysis
Human specimens and cancer cells were lysed in RIPA lysis buffer (Invitrogen; Thermo Fisher Scientific, Inc.) supplemented with protease inhibitor cocktail (Roche Applied Science), and the protein concentration was measured using the BCA Protein Assay kit (Beyotime Institute of Biotechnology). Protein lysates (20 µg) were separated by 12% SDS-PAGE and transferred to PVDF membranes. The membranes were subsequently blocked with 5% non-fat milk for 1 h at room temperature before they were incubated with primary antibodies overnight at 4°C. The membranes were then washed in TBST (0.1% Tween-20) three times for 5 min each time and then incubated with horseradish peroxidase-conjugated secondary antibodies, which including anti-mouse (1:2,000, cat. no. ZB2301, OriGene Technologies, Inc.) and anti-rabbit (1:2,000, cat. no. ZB2305, OriGene Technologies, Inc.) for 2 h at room temperature. This was followed by washing with TBST three times for 5 min each time. The bands were visualized using BeyoECL Plus reagent (Beyotime Institute of Biotechnology). The membranes were stripped and re-probed with an anti-GAPDH antibody (1:5,000; cat. no. sc47724; Santa Cruz Biotechnology, Inc.) as the loading control. The primary antibody against CENPO was purchased from OriGene Technologies, Inc. (1:500; cat. no. TA349791) and the antibodies against ATM (1:1,000; cat. no. ab32420), c-JUN (1:1,000; cat. no. ab40766), and cyclin D1 (CCND1; 1:800; cat. no. ab40754) were obtained from Abcam.
Cell proliferation assay
Cells in the logarithmic growth phase were resuspended in standard medium and seeded in 96-well plates at a density of 2×103 cells/well. The number of EGFP-positive cells was counted using a Celigo Imaging Cytometer (Nexcelom Bioscience LLC) on 5 consecutive days.
Apoptosis assay
Cells transfected with siRNA or NC were harvested at 5 days following transfection and apoptosis levels were assessed using an Annexin V-based flow cytometer (Guava easyCyte HT, EMD Millipore). Harvested cells (200 µl) at a concentration of 1×106 cells/ml, were added to fluorescence-activated cell sorting (FACS) tubes and stained with Annexin V-APC (cat. no. 88-8007, eBioscience, Thermo Fisher Scientific, Inc.) in the dark at room temperature for 15 min.
Colony forming assay
Transfected and control GC cells were seeded in 6-well plates at a density of 800 cells/well. The medium was refreshed every 2–3 days. Following incubation for 14 days at 37°C, the cells were washed twice with PBS, fixed in 4% paraformaldehyde for 20 min at room temperature, stained with crystal violet for 10 min at room temperature and then washed three times with double-distilled H2O. Images of the colonies were captured using a Leica DM IL light microscope (Leica Microsystems, Inc.).
Gene expression profiling and data analysis
MGC-803 cells that have been transfected with si-CENPO and control MGC-803 cells were used to compare gene expression patterns by microarray analysis. Following transfection of MGC-803 cells with CENPO siRNA, total RNA was extracted and 50–500 ng of RNA was used to generate biotin-modified amplified RNA (aRNA) using a GeneChip 3′IVT Express kit (Affymetrix; Thermo Fisher Scientific, Inc.). Reverse transcription was performed using a T7 oligo (dT) primer and a first-strand IVT Labeling Master Mix was used to produce multiple copies of biotin-modified aRNA. The aRNA was then purified and quantified. Following fragmentation, the aRNA was hybridized to the GeneChip PrimeView™ human array (Affymetrix; Thermo Fisher Scientific, Inc.). The chips were subsequently stained with streptavidin-phycoerythrin (MoBiTec, Goettingen, Germany) for 10 min at 25°C and washed in a GeneChip Fluidics Station 450. The microarray signals were scanned and analyzed using the GeneChip Array Scanner 3000 (Thermo Fisher Scientific, Inc.). Differentially expressed genes (DEGs) between the two groups were defined as those exhibiting a >1.5-fold or a <0.5-fold-change in gene expression and where the P-value was <0.05 following correction. Ingenuity Pathway Analysis (IPA; http://www.ingenuity.com) (13,14) was performed for significant DEGs and enrichment pathway analysis.
Xenograft tumor model
All animal experiments were approved by the Ethics Board of The Institute of The First Affiliated Hospital, Nanchang University. A total of 4×106 MGC-803 GC cells (100 µl) with CENPO overexpression or knockdown were injected subcutaneously into 4-week-old female BALB/c nude mice (10 mice/group, 2 group (n=20), weight ~15 g), which were purchased from Shanghai Lingchang Biological Technology Co., Ltd. The mice had free access to clean food and water, and maintained at 18–29°C, with relative humidity of 50–60%, 10–14 h light/dark cycle (lights turned on at 8:00 and turned off at 18:00 every day). Every 3 days, tumor sizes were recorded and the volumes were calculated using the following equation: Tumor volume=length × width2 × π/6. The mice were sacrificed at day 24, at which point the tumors were collected and images were captured.
Statistical analysis
Each experiment was performed in triplicate. The SPSS software package (version, 22.0; IBM Corp.) was used to perform statistical analysis of the results. Data are expressed as the mean ± standard deviation. Significant differences between two groups were analyzed using Student's t-test. Analysis of variance analysis was used for the comparison of multiple groups and a Dunnett test was used as a post hoc test. Pearson's chi-squared test was used to compare the clinicopathologic features. Overall survival curves were generated using Kaplan-Meier plots and assessed using the log-rank test. P<0.05 was used to indicate a statistically significant difference.
Results
CENPO is upregulated in GC and associated with a poor prognosis
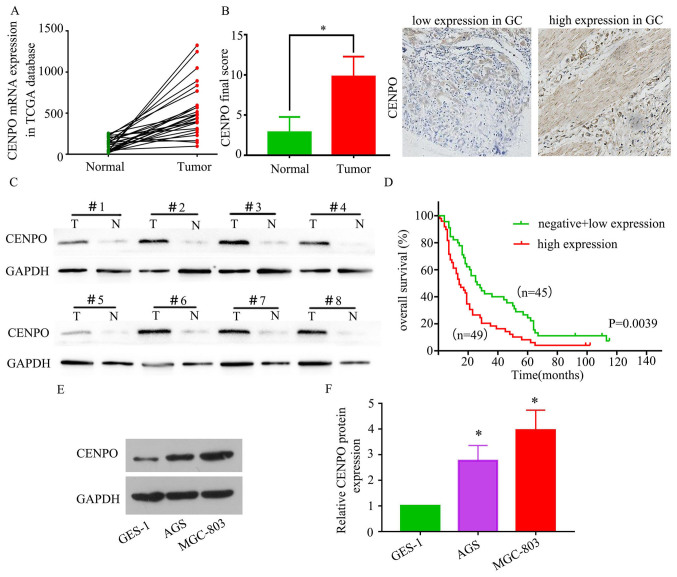
To evaluate the significance of CENPO in GC, the expression of CENPO in TCGA datasets was first examined. As demonstrated in Fig. 1A, significantly increased CENPO mRNA levels were observed in cancer tissues when compared with paired normal tissues (P<0.0001). To confirm these results, CENPO expression in clinical specimens was assessed by TMA analysis of 94 GC tissues and paired normal controls. The results indicated that the CENPO protein expression score was significantly increased in GC tissues compared with paired normal tissues (P<0.05; Fig. 1B). Patients were divided into high (final score of 9–12) and low (final score ≤8) expression groups (Fig. 1B). To evaluate the clinical significance of CENPO in GC, the association between high CENPO expression and the clinicopathological features of patients with GC was assessed. Statistical analysis demonstrated that high CENPO expression was significantly associated with tumor size (P<0.0001), lymph node metastasis (P=0.014) and p-TNM stage (P=0.023; Table I). CENPO expression was then detected in 24 paired GC specimens using western blotting analysis. As demonstrated in Fig. 1C, high CENPO expression was observed in GC specimens when compared with paired normal tissues. Patients with higher levels of CENPO protein expression exhibited shorter survival times (P=0.0039; Fig. 1D). In addition, CENPO expression in GES-1 and two different GC cell lines (AGS and MGC-803) were examined by western blot analysis. As demonstrated in Fig. 1E and F, CENPO protein expression in GES-1 cells was significantly decreased compared with in GC cell lines (P<0.01); AGS and MGC-803 GC cells exhibited high levels of CENPO expression (AGS vs. GES-1; P<0.0001; MGC-803 vs. GES-1; P<0.0001). Taken together, these results revealed that CENPO expression is consistently elevated in GC and may be associated with aggressive tumor behavior.
Figure 1.
CENPO is upregulated in GC and its expression is associated with poor patient prognosis. (A) CENPO mRNA expression levels in 26 primary GC tissues and paired normal tissues according to the TCGA dataset (P<0.0001). (B) Immunohistochemical staining of CENPO in normal and GC tissues (Image magnification, ×200). *P<0.05. (C) Protein expression levels of CENPO in 24 paired primary GC tissues as determined by western blot analysis. (D) Overall survival rates between CENPO high and low expression groups (P=0.0039). (E) Western blotting and (F) graphical analysis of CENPO expression was detected in GES-1 and GC cell lines, AGS and MGC-803. *P<0.05 vs. GES-1. CENPO, centromere protein O; GC, gastric cancer; TCGA, The Cancer Genome Atlas.
CENPO regulates GC cell growth and apoptosis in vitro
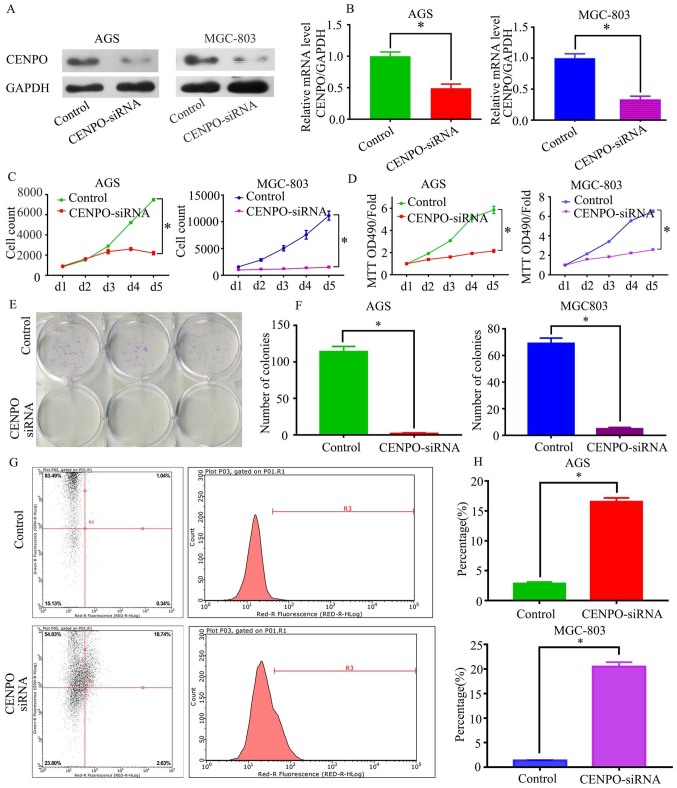
To investigate the biological effects of CENPO in GC, AGS and MGC-803 cell lines were selected for further analysis. Following transfection of cells with CENPO siRNA, CENPO protein and mRNA levels were significantly reduced in the CENPO siRNA group when compared with the controls (P<0.05; Fig. 2A and B). Celigo and MTT assay analyses were used to investigate the effect of CENPO on cell growth. Cell counts and optical density (OD) values were determined at 1–5 days following transfection. The results demonstrated that the cell counts and OD values in the CENPO siRNA group were significantly decreased when compared with the control group, indicating that cell proliferation was significantly reduced (P<0.0001; Fig. 2C and D). In addition, colony formation assays demonstrated that knockdown of CENPO expression inhibited colony formation of GC cells, which is consistent with the cell proliferation observations (P=0.0009 and P<0.0001; Fig. 2E and F). Changes in cell proliferation are often associated with changes in the cell cycle or apoptosis. Therefore, apoptosis analysis using Annexin V-FACS was performed. The results indicated that the percentage of apoptotic cells in the CENPO siRNA group was significantly increased when compared with the controls (P<0.0001; Fig. 2G and H). Together, these results demonstrate that increased CENPO expression may promote the growth of GC cells and inhibit apoptosis.
Figure 2.
Functional role of CENPO in regulating GC cell growth and apoptosis in vitro. CENPO (A) protein and (B) mRNA expression were decreased following transfection with CENPO siRNA. (C) Celigo and (D) MTT assays were performed to determine the proliferation of GC cells transduced with the CENPO siRNA lentivirus. (E) A colony formation assay was performed using GC cells transduced with the CENPO siRNA lentivirus. (F) Number of AGS and MGC-803 cell colonies that formed following transduction with the CENPO siRNA lentivirus. The percentage of apoptotic (G) AGS and (H) MGC-803 cells was significantly increased in the CENPO siRNA group P<0.0001. *P<0.05. CENPO, centromere protein O; GC, gastric cancer; siRNA, small-interfering RNA.
Downregulation of CENPO reduces tumorigenicity in MGC-803-xenograft nude mice
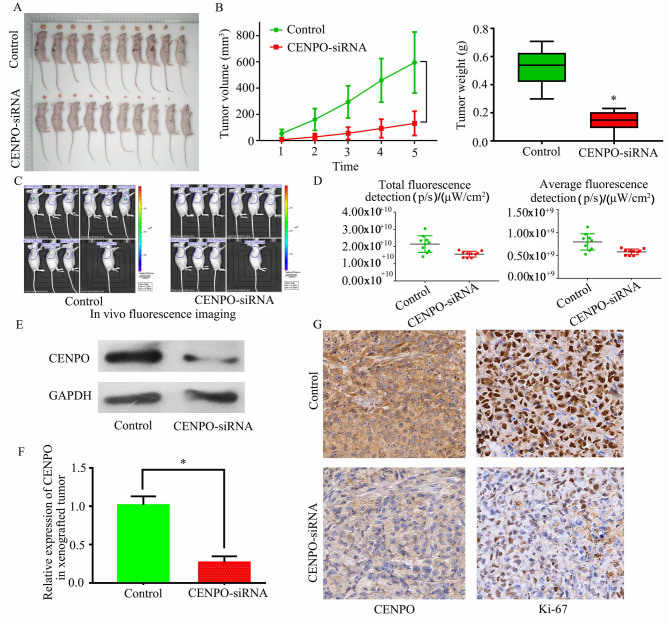
Using a nude mouse model, tumors that formed in mice injected with CENPO siRNA-transfected MGC-803 cells exhibited reduced proliferative capacity than tumors that formed from control cells (Fig. 3A). The tumor volumes and weights were significantly reduced in the CENPO siRNA group at the endpoint (P<0.0001; Fig. 3B). In addition, total fluorescence (P=0.0059) and average fluorescence (P=0.0019) were decreased in the CENPO siRNA group (Fig. 3C and D). Furthermore, CENPO expression in tumors was decreased in the CENPO siRNA group according to western blotting and RT-qPCR analyses (P=0.0007; Fig. 3E and F). The expression of CENPO and the Ki-67 percentage IHC scores in tumor cells from the CENPO siRNA group were also relatively decreased (Fig. 3G). Collectively, these results suggest that CENPO may be involved in mediating GC cell proliferation in vivo.
Figure 3.
Downregulation of CENPO expression reduces tumorigenicity in MGC-803 ×enograft nude mice. (A) Sacrificed nude mice from the control and CENPO siRNA groups and their corresponding tumors. (B) Tumor volumes and weights between the control and the CENPO siRNA groups were compared. *P<0.05 vs. control. (C) CENPO downregulation reduced tumorigenicity, as determined by bioluminescence detection and (D) graphical analysis. (E) Western blotting and (F) reverse transcription-quantitative PCR analyses were performed to compare CENPO expression in tumors from the control and CENPO siRNA groups. (G) CENPO and Ki-67 expression were detected by immunohistochemical staining (Image magnification, ×400). *P<0.05. CENPO, centromere protein O; siRNA, small-interfering RNA.
CENPO silencing suppresses ATM signaling pathways
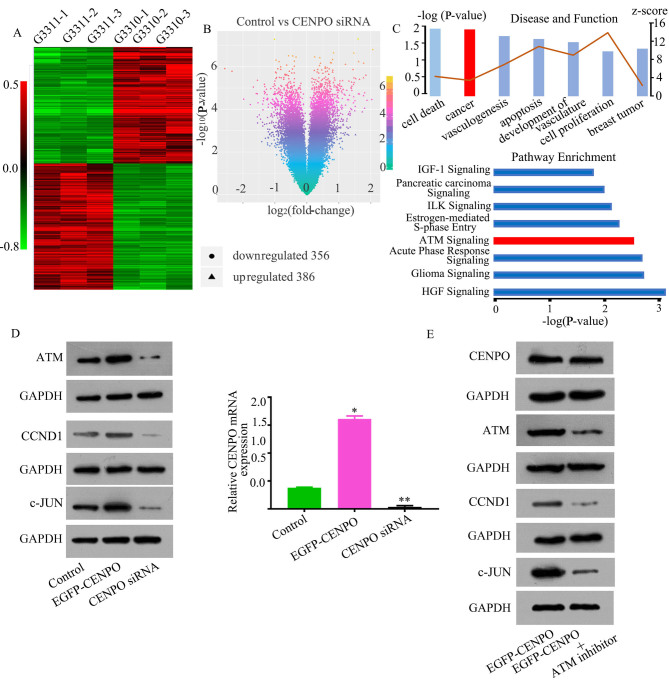
To investigate the molecular mechanisms underlying CENPO-mediated regulation of GC proliferation, a microarray platform was used to compare the gene expression profiles of CENPO-depleted MGC-803 cells with control cells. This analysis identified 356 downregulated genes and 386 upregulated genes (absolute fold-change >2; P<0.05; Fig. 4A and B). IPA disease and functional analysis demonstrated that cell death, cancer, vasculogenesis, apoptosis and cell proliferation were closely associated with CENPO dysregulation (Fig. 4C). IPA pathway enrichment analysis demonstrated that the ATM signaling pathway was a significantly enriched cancer-associated signaling pathway in response to CENPO depletion, which suggests that this pathway may be involved in affected functional processes in CENPO-depleted cells (Fig. 4C). The CENPO lentivirus, EGFP-CENPO, was then transfected into MGC-803 cells. Compared with the control group, the expression of ATM, CCND1 and c-Jun were significantly increased in the EGFP-CENPO group, and were significantly decreased in the CENPO siRNA group (P<0.0001; Fig. 4D). In order to verify these results, the ATM inhibitor, KU-55933 was added to the culture medium of cells in the EGFP-CENPO group. Western blotting results indicated that ATM, CCND1 and c-Jun expression levels were reduced (Fig. 4E). These results suggest that activation of ATM, CCND1 and c-Jun are involved in CENPO-mediated regulation of GC cell tumorigenicity.
Figure 4.
CENPO is associated with the ATM signaling pathway according to gene expression profiling and data analysis. (A) A microarray platform was used to analyze the gene expression profiles in MGC-803 cells following CENPO knockdown. A heatmap present all of the differently expressed genes, including 356 genes that were downregulated and 386 genes that were upregulated. (B) A volcano plot present all of the differently expressed genes between CENPO control group and CENPO siRNA group. (C) CENPO may be involved in different biological processes, molecular functions and diseases according to IPA analysis. (D) ATM, CCND1 and c-Jun protein expression in the control group, the EGFP-CENPO group and the CENPO siRNA group. *P<0.0001 EGFP-CENPO vs. Control. **P<0.0001 CENPO siRNA vs. Control. (E) ATM, CCND1 and c-Jun expression in EGFP-CENPO group, with the addition of the ATM inhibitor. CENPO, centromere protein O; ATM, ataxia telangiectasia mutated; IPA, Ingenuity Pathway Analysis; CCND1, cyclin D1; EGFP-CENPO, enhanced green fluorescent protein CENPO overexpression vector; siRNA, small-interfering RNA.
Discussion
Centromeres are essential elements that facilitate chromosome separation and normal cell division during meiosis and mitosis (15). Previously, >40 CENPs have been discovered (7). Abnormal localization or overexpression of these proteins leads to disordered cell division and chromosomal aneuploidy; these changes have been closely associated with cancer (16). Previous studies have demonstrated that numerous malignancies exhibit high CENPH or CENPF expression (17,18). However, the molecular function of CENPO in cancer is currently unclear.
In the present study, CENPO was observed to be frequently upregulated in GC cells and tissues, and high CENPO expression was associated with high TNM stage and poor prognosis. This suggests that high CENPO expression may serve important roles in the progression of GC. In chicken DT40 cells, knockdown of CENPO expression was associated with reduced cell proliferation and mitotic defects (19). The results of the present study are in accordance with these observations. Knockdown of CENPO expression in GC cells led to a reduction in cell proliferation and induction of apoptosis in the current study. In addition, overexpression of CENPO promoted GC tumor growth, which was further confirmed using a subcutaneous xenograft tumor model. These results suggest that CENPO may be involved in the pathogenesis and pathological progression of GC.
To further clarify the molecular mechanisms underlying the role of CENPO in GC tumors, microarray analysis was performed. The expression of 386 genes was upregulated and 356 genes was downregulated in CENPO siRNA GC cells group. Bioinformatics analysis of the identified DEGs demonstrated that these genes were implicated in different biological processes and molecular functions. The results suggest that CENPO may be an essential regulator of cancer cell death, vasculogenesis, cell proliferation, cell apoptosis and GC tumorigenesis. Pathway enrichment analysis indicated that the ATM signaling pathway was a dysregulated cancer-associated signaling pathway in response to CENPO depletion. The ATM protein functions as a critical regulator of the DNA damage response and coordinates DNA repair, checkpoint activation, and metabolic changes in response to oxidative stress and DNA double-strand breaks (20,21). However, previous reports suggest that it may also function as an oncogene and promote tumorigenicity in breast cancer (22–24). The present study demonstrated that overexpression of CENPO in GC cells increased ATM, CCND1 and c-Jun expression, while knockdown of CENPO had the opposite effect. Previous studies indicate that numerous tumor types exhibit high CCND1 expression, which has been associated with poor patient outcomes and which may promote cancer cell growth (25,26). It has been reported that the absence of functional ATM leads to CCND1-mediated genomic instability, which ultimately facilitates cell transformation and tumor formation (27). Previous studies have also demonstrated that CCND1 translation may be regulated by ATM to control cell cycle progression (28,29). c-Jun is a downstream molecule of the mitogen-activated protein kinase signaling cascade and contributes to numerous cellular functions, such as apoptosis, proliferation, survival and tumorigenesis (30,31). It has been reported that ATM serves an important role in oxidative stress-mediated signaling, which enhances c-Jun phosphorylation due to its role as an ionizing radiation-induced oxidative stress sensor and by modulating intracellular redox homeostasis (32). The results of the present study suggest that CENPO mediates ATM signaling to increase CCND1 and c-Jun expression, which may subsequently promote GC progression. The results therefore provide direction for future research that will aim to investigate the regulatory mechanism of CENPO.
In conclusion, the results of the current study are to the best of our knowledge the first to report increased expression of CENPO in GC and an association with poor patient prognosis. CENPO protein regulate GC cell growth and apoptosis via targeting of the ATM signaling pathway. However, future studies that aim to elucidate the detailed regulatory mechanisms of CENPO in GC tumorigenesis are required, in order to facilitate translation of these findings into clinical practice.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Science Foundation of China (grant nos. 81460373 and 81360362) and the ‘Talent 555 Project’ of Jiangxi Province.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
YT and ZJ curated the data. YC, JX, GZ, YT and LW conducted formal analysis and acquired funding. ZL and ZJ were involved in designing the methodology. YC, JX, GZ and LW were involved in project administration. ZL and ZJ composed the original draft of the manuscript. YC and JX wrote the manuscript. YT and ZJ edited the manuscript.
Ethics approval and consent to participate
The present study was approved by The Ethics Board of the Institute of the First Affiliated Hospital, Nanchang University (Nanchang, China). The Ethics Board also supervised and examined the whole process of the present study. All participants agreed to join the study and provided written informed consent.
Patient consent for publication
All participants provided written informed consent for publication.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Ferro A, Peleteiro B, Malvezzi M, Bosetti C, Bertuccio P, Levi F, Negri E, La Vecchia C, Lunet N. Worldwide trends in gastric cancer mortality (1980–2011), with predictions to 2015, and incidence by subtype. Eur J Cancer. 2014;50:1330–1344. doi: 10.1016/j.ejca.2014.01.029. [DOI] [PubMed] [Google Scholar]
- 2.Corso S, Giordano S. How can gastric cancer molecular profiling guide future therapies? Trends Mol Med. 2016;22:534–544. doi: 10.1016/j.molmed.2016.05.004. [DOI] [PubMed] [Google Scholar]
- 3.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Ca Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 4.Bakhoum SF, Compton DA. Kinetochores and disease. Keeping microtubule dynamics in check! Curr Opin Cell Biol. 2012;24:64–70. doi: 10.1016/j.ceb.2011.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duensing A, Duensing S. Centrosomes, polyploidy and cancer. Adv Exp Med Biol. 2010;676:93–103. doi: 10.1007/978-1-4419-6199-0_6. [DOI] [PubMed] [Google Scholar]
- 6.Kramer A, Neben K, Ho AD. Centrosome replication, genomic instability and cancer. Leukemia. 2002;16:767–775. doi: 10.1038/sj.leu.2402454. [DOI] [PubMed] [Google Scholar]
- 7.Obuse C, Yang H, Nozaki N, Goto S, Okazaki T, Yoda K. Proteomics analysis of the centromere complex from HeLa interphase cells: UV-damaged DNA binding protein 1 (DDB-1) is a component of the CEN-complex, while BMI-1 is transiently co-localized with the centromeric region in interphase. Genes Cells. 2004;9:105–120. doi: 10.1111/j.1365-2443.2004.00705.x. [DOI] [PubMed] [Google Scholar]
- 8.Izuta H, Ikeno M, Suzuki N, Tomonaga T, Nozaki N, Obuse C, Kisu Y, Goshima N, Nomura F, Nomura N, Yoda K. Comprehensive analysis of the ICEN (Interphase Centromere Complex) components enriched in the CENP-A chromatin of human cells. Genes to cells. 2006;11:673–684. doi: 10.1111/j.1365-2443.2006.00969.x. [DOI] [PubMed] [Google Scholar]
- 9.Hori T, Okada M, Maenaka K, Fukagawa T. CENP-O class proteins form a stable complex and are required for proper kinetochore function. Mol Biol Cell. 2008;19:843–854. doi: 10.1091/mbc.e07-06-0556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Denais CM, Gilbert RM, Isermann P, McGregor AL, te Lindert M, Weigelin B, Davidson PM, Friedl P, Wolf K, Lammerding J. Nuclear envelope rupture and repair during cancer cell migration. Science. 2016;352:353–358. doi: 10.1126/science.aad7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Orthwein A, Noordermeer SM, Wilson MD, Landry S, Enchev RI, Sherker A, Munro M, Pinder J, Salsman J, Dellaire G, et al. A mechanism for the suppression of homologous recombination in G1 cells. Nature. 2015;528:422–426. doi: 10.1038/nature16142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real time quantitative PCR and the 2(-Delta Delta C(T)) Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 13.Felciano RM, Bavari S, Richards DR, Billaud JN, Warren T, Panchal R, Kramer A. Predictive systems biology approach to broad-spectrum, host-directed drug target discovery in infectious diseases. Pac Symp Biocomput. 2013:17–28. [PubMed] [Google Scholar]
- 14.Krämer A, Green J, Pollard J, Jr, Tugendreich S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics. 2014;30:523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McAinsh AD, Meraldi P, Draviam VM, Toso A, Sorger PK. The human kinetochore proteins Nnf1R and Mcm21R are required for accurate chromosome segregation. EMBO J. 2006;25:4033–4049. doi: 10.1038/sj.emboj.7601293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gordon DJ, Resio B, Pellman D. Causes and consequences of aneuploidy in cancer. Nat Rev Genet. 2012;13:189–203. doi: 10.1038/nrg3123. [DOI] [PubMed] [Google Scholar]
- 17.Liao WT, Song LB, Zhang HZ, Zhang X, Zhang L, Liu WL, Feng Y, Guo BH, Mai HQ, Cao SM, et al. Centromere protein H is a novel prognostic marker for nasopharyngeal carcinoma progression and overall patient survival. Clin Cancer Res. 2007;13:508–514. doi: 10.1158/1078-0432.CCR-06-1512. [DOI] [PubMed] [Google Scholar]
- 18.Mi YJ, Gao J, Xie JD, Cao JY, Cui SX, Gao HJ, Yao SP, Liu T, Zhang YY, Guo CH, et al. Prognostic relevance and therapeutic implications of centromere protein F expression in patients with esophageal squamous cell carcinoma. Dis Esophagus. 2013;26:636–643. doi: 10.1111/dote.12002. [DOI] [PubMed] [Google Scholar]
- 19.Kagawa N, Hori T, Hoki Y, Hosoya O, Tsutsui K, Saga Y, Sado T, Fukagawa T. The CENP-O complex requirement varies among different cell types. Chromosome Res. 2014;22:293–303. doi: 10.1007/s10577-014-9404-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi M, Kipps T, Kurzrock R. ATM Mutations in Cancer: Therapeutic Implications. Mol Cancer Ther. 2016;15:1781–1791. doi: 10.1158/1535-7163.MCT-15-0945. [DOI] [PubMed] [Google Scholar]
- 21.Karnitz LM, Zou L. Molecular pathways: Targeting ATR in cancer therapy. Clin Cancer Res. 2015;21:4780–4785. doi: 10.1158/1078-0432.CCR-15-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stagni V, Manni I, Oropallo V, Mottolese M, Di Benedetto A, Piaggio G, Falcioni R, Giaccari D, Di Carlo S, Sperati F, et al. ATM kinase sustains HER2 tumorigenicity in breast cancer. Nat Commun. 2015;6:6886. doi: 10.1038/ncomms7886. [DOI] [PubMed] [Google Scholar]
- 23.Liu R, Tang J, Ding C, Liang W, Zhang L, Chen T, Xiong Y, Dai X, Li W, Xu Y, et al. The depletion of ATM inhibits colon cancer proliferation and migration via B56γ2-mediated Chk1/p53/CD44 cascades. Cancer Lett. 2017;390:48–57. doi: 10.1016/j.canlet.2016.12.040. [DOI] [PubMed] [Google Scholar]
- 24.Chen WT, Ebelt ND, Stracker TH, Xhemalce B, Van Den Berg CL, Miller KM. ATM regulation of IL-8 links oxidative stress to cancer cell migration and invasion. Elife. 2015;4:e07270. doi: 10.7554/eLife.07270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ortiz A, Garcia D, Vicente Y. Prognostic significance of cyclin D1 protein expression and gene amplification in invasive breast carcinoma. PLoS One. 2017;12:e0188068. doi: 10.1371/journal.pone.0188068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qie S, Diehl JA. Cyclin D1, cancer progression, and opportunities in cancer treatment. J Mol Med. 2016;94:1313–1326. doi: 10.1007/s00109-016-1475-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vaites LP, Lian Z, Lee EK, Yin B, DeMicco A, Bassing CH, Diehl JA. ATM deficiency augments constitutively nuclear cyclin D1-driven genomic instability and lymphomagenesis. Oncogene. 2014;33:129–133. doi: 10.1038/onc.2012.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hitomi M, Yang K, Stacey AW, Stacey DW. Phosphorylation of cyclin D1 regulated by ATM or ATR controls cell cycle progression. Mol Cell Biol. 2008;28:5478–5493. doi: 10.1128/MCB.02047-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Y, Yang DQ. The ATM Inhibitor KU-55933 suppresses cell proliferation and induces apoptosis by blocking Akt in cancer cells with overactivated Akt. Mol Cancer Ther. 2010;9:113–125. doi: 10.1158/1535-7163.MCT-08-1189. [DOI] [PubMed] [Google Scholar]
- 30.Meng Q, Xia Y. c-Jun, at the crossroad of the signaling network. Protein Cell. 2011;2:889–898. doi: 10.1007/s13238-011-1113-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peluso I, Yarla NS, Ambra R, Pastore G, Perry G. MAPK signalling pathway in cancers: Olive products as cancer preventive and therapeutic agents, Semin. Cancer Biol. 2019;56:185–195. doi: 10.1016/j.semcancer.2017.09.002. [DOI] [PubMed] [Google Scholar]
- 32.Lee SA, Dritschilo A, Jung M. Role of ATM in oxidative stress-mediated c-Jun phosphorylation in response to ionizing radiation and CdCl2. J Biol Chem. 2001;276:11783–11790. doi: 10.1074/jbc.M004517200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.