Abstract
Dysregulation of collagen type XII α1 chain (COL12A1) has been found in several cancer types and could be involved in tumor progression. However, its clinical significance in gastric cancer (GC) remains under exploration. Online databases (Gene Expression Omnibus and UALCAN), reverse transcription-quantitative PCR and immunohistochemistry were utilized in the present study to evaluate the expression of COL12A1 in GC tissues and cell lines. It was found that COL12A1 expression was notably upregulated in GC. Clinicopathological analysis showed that elevated COL12A1 expression was positively correlated with tumor invasiveness, metastasis and advanced clinical stage. The prognostic analysis suggested that high COL12A1 expression contributed to poor overall survival. Multivariate Cox analysis indicated that COL12A1 overexpression was a powerful independent prognostic indicator in patients with GC (hazard ratio, 1.896; 95% CI, 1.267–2.837; P=0.002). The results highlighted the importance of COL12A1 in GC and suggested its potential role as a candidate for clinical outcome prediction and targeted therapy in patients with GC.
Keywords: bioinformatics analysis, immunohistochemistry, collagen type XII α1 chain, extracellular matrix, prognosis
Introduction
Gastric cancer (GC), as one of the most common malignancies, is frequently accompanied by an unfavorable prognosis and low survival rate (1). Although progress has been made in therapeutic strategies, and early detection and clinical management have improved its prognosis, GC remains a complex clinical challenge, with the majority of patients being diagnosed during the later stages of the disease (2,3). Previously, it was reported that the aberrant expression of specific biomarkers was related to cancer development and a worse prognosis, and that some biomarkers affected the sensitivity of cancer to chemotherapeutic drugs (4). Therefore, it is of great benefit to identify novel biomarkers characterized by high sensitivity as well as high specificity that can be used as predictors for GC diagnosis and precise treatment.
Collagen type XII α1 chain (COL12A1), encoded by a gene whose chromosomal location is 6q12-q13, is a member of the fibril-associated collagen family harboring interrupted triple-helical collagen domains (5). COL12A1 acts as a bridge between fibrils and its mutations are reported to be related to myopathy (6). COL12A1 has received increasing attention due to its essential roles in human cancer, given that its overexpression has been identified in several different cancer types. A comparison of differentially expressed genes between 279 metastatic samples from multiple adenocarcinoma types and unmatched primary adenocarcinomas revealed elevated expression of collagen genes, including COL12A1, closely related to poor prognosis in several cancer types (7–9). Notably, COL12A1 was identified as a desmoplastic marker during myofibroblast differentiation in colorectal cancer (CRC), suggesting that COL12A1 upregulation is related to the malignant potential of the tumor (10). Besides, differential gene expression from microarray analyses showed that COL12A1 was highly expressed in renal cancer (8). Moreover, in ovarian cancer, COL12A1 expression exhibited a significantly increased level of expression in cisplatin-and doxorubicin-resistant cell lines relative to parental cell lines, and was associated with poor overall survival (OS) (7).
Although these results suggest a relationship between COL12A1 and cancer development and progression, its significance in GC remains an underexplored issue. Therefore, the present study evaluated COL12A1 expression using online databases, followed by quantification of COL12A1 mRNA and protein levels in clinical samples via reverse transcription-quantitative PCR (RT-qPCR) and immunohistochemistry (IHC), respectively. Furthermore, associations between COL12A1 expression and clinical features, as well as the clinical outcomes of patients with GC, were assessed.
Materials and methods
Patients and specimens
To measure COL12A1 expression, 64 paired fresh GC and matched control tissues were obtained between October 2011 and April 2014 at the First Affiliated Hospital of Zhejiang University. Table I shows the clinical features of the 64 GC patients. Paired formalin fixed paraffin-embedded (FFPE) samples (107 GC and 107 matched normal gastric tissues) were collected between January 2012 and December 2014. All 171 samples were obtained from 130 male and 41 female patients with a mean age of 64 years (range from 28–82 years) diagnosed with GC based on histopathology. Additionally, a tissue microarray containing 93 GC and 77 matched normal gastric tissues was purchased from Shanghai Outdo Biotech Co., Ltd. Patient clinicopathological information was collected; this included age, gender, tumor size, differentiation and the tumor-node-metastasis (TNM) stage pathological classification, according to the 8th edition of the American Joint Committee on Cancer guidelines (11). The study protocol was approved by the ethics committee of the First Affiliated Hospital of Zhejiang University.
Table I.
Clinicopathological parameters in 64 patients with gastric cancer.
| Variable | No. cases |
|---|---|
| Age (<60 years/≥60 years) | 22/42 |
| Sex (male/female) | 45/19 |
| Tumor size (≤5 cm/>5 cm) | 38/26 |
| Lauren's classification (intestinal type/diffuse type) | 26/38 |
| Differentiation (well to moderate/poor) | 28/36 |
| Tumor invasion (T1/T2/T3/T4) | 8/16/0/40 |
| Lymph node metastasis (N0/N1/N2/N3) | 16/10/11/27 |
Cell lines and culture conditions
Human GC cell lines (AGS, HGC-27, MKN45 and MGC-803) were obtained from the Institute of Cellular Biology (Chinese Academy of Science, Shanghai, China) and cultured in RPMI 1640 medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.). All cells were maintained at 37°C in a 5% CO2 atmosphere.
RNA extraction and RT-qPCR
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) was used for total RNA extraction from fresh tissue samples and GC cell lines. Total RNA (1 µg) was reverse transcribed using the PrimeScript RT-PCR kit (Takara Bio, Inc.) with the reaction conditions: 15 min at 37°C and stored at 4°C. Then, RT-qPCR was conducted to evaluate COL12A1 expression on a StepOnePlus system (Applied Biosystems; Thermo Fisher Scientific, Inc.) and SYBR Green (Takara Bio, Inc.) with a two-step reaction condition: 30 sec at 95°C followed by 5 sec at 95°C and 60 sec at 60°C repeated for 40 cycles. The primers were as follows: COL12A1, forward, 5′-CCACAGGTTCAAGAGGTCCC-3′ and reverse, 5′-TGTGTTAGCCGGAACCTGGA-3′; and GAPDH forward, 5′-TGGGTGTGAACCATGAGAAGT-3′ and reverse. 5′-TGAGTCCTTCCACGATACCAA-3′. COL12A1 expression levels were quantified relative to those of GAPDH, which was used as the endogenous control, using the 2−ΔΔCq method (12).
IHC analysis
FFPE tissues were cut into 4-µm sections and the IHC staining against COL12A1 (cat. no. ab123104; monoclonal; raised in rabbit; 1:40 dilution; Abcam) was performed as previously described (13).
IHC staining evaluation
Each stained tissue section was reviewed and scored by two independent observers. Each section was examined in five fields at high magnification (×200) under a light microscope. The percentage of positive staining and the intensity of staining were scored based on previously published standards (14). The final immunoreactivity score (0–12) was equivalent to the score of the percentage of positive staining multiplied by the staining intensity score. Based on the analysis, the final score were defined as ‘high’ if the score was >4, and ‘low’ if the score was ≤4.
Bioinformatics analysis
A total of three gastric cancer gene expression profile datasets [GSE2701 (15) GSE63089 (16) and GSE79973 (17)] containing 95 GC tissues and 55 normal gastric tissues were downloaded from the Gene Expression Omnibus (GEO) database (18). The data were analyzed using the online tool GEO2R (https://www.ncbi.nlm.nih.gov/geo/geo2r) to analyze the mRNA expression of COL12A1 expression in GC and normal gastric tissues. UALCAN (19) was used to identify the up-or downregulation of COL12A1 in The Cancer Genome Atlas (TCGA) gastric cancer portal. The Human Protein Atlas (HPA) website (http://www.proteinatlas.org/) was used to explore the basic expression of COL12A1 in different organs. Three datasets (15,20,21) from Oncomine database (22) were used to analysis the relationship between COL12A1 mRNA expression and Lauren classification. The prognostic significance of COL12A1 was evaluated in an Affymetrix array (225,664_at; Thermo Fisher Scientific, Inc.) using Kaplan Meier-plotter (KM-plotter) (http://kmplot.com/analysis/index.php?p=service&cancer=gastric). The autoselected best threshold value was used to identify the high and low groups.
Statistical analysis
SPSS 19.0 (IBM Corp.) and GraphPad Prism 5.01 (GraphPad Software, Inc.) software were used to perform all statistical analyses. COL12A1 mRNA expression in GC relative to normal gastric tissues was analyzed by paired t-test. The χ2 test was used to analyze the relationships between COL12A1 expression and clinicopathological variables, and survival curves between high and low COL12A1 expression were plotted using the Kaplan-Meier method. Univariate and further multivariate analyses were conducted using the Cox proportional hazards model in order to identify prognostic indicators. P<0.05 was considered to indicate a statistically significant difference.
Results
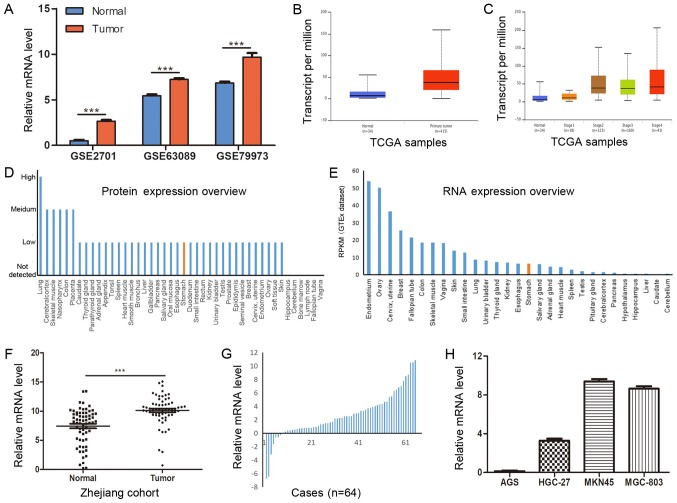
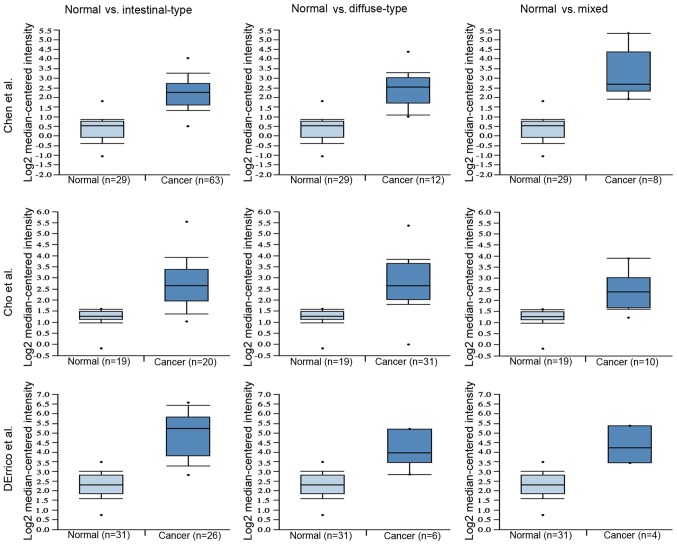
mRNA expression of COL12A1 is significantly elevated in GC tissues and cell lines. To evaluate COL12A1 mRNA expression in GC, three microarray datasets from GEO were analyzed and the results revealed that COL12A1 was highly expressed in GC (Fig. 1A; P<0.001). The results from the TCGA dataset also indicated that COL12A1 mRNA was markedly increased in GC compared with normal controls, and this was more apparent at advanced clinical stages (Fig. 1B and C). Additionally, the basic expression of COL12A1 in different organs was explored using HPA. As shown in Fig. 1D and E, the basic expression level of COL12A1 in the stomach was moderate compared to that in other organs. Validation of COL12A1 expression in 64 paired frozen GC and matched controls using RT-qPCR showed a significant elevation of COL12A1 mRNA levels in cancer samples (P<0.0001; Fig. 1F and G) relative to those in adjacent normal tissues. This was consistent with expectations. In addition, as shown in Fig. 1H, COL12A1 mRNA was highly expressed in three (HGC-27, MKN45 and MGC-803) of the four GC cell lines. Furthermore, the relationship between COL12A1 mRNA expression and Lauren classification was evaluated using the Oncomine database. As shown in Fig. 2, COL12A1 was markedly elevated in all the tumor tissues, regardless of Lauren classification.
Figure 1.
COL12A1 mRNA expression is upregulated in GC tissues and cell lines. (A) The expression of COL12A1 in three datasets. GSE7901 contains 45 GC and 5 normal gastric tissues. GSE63089 and GSE79973 contain 45 and 10 paired GC and normal gastric tissues, respectively. (B) The data-mining analysis showed that COL12A1 was upregulated in the TCGA gastric cancer portal (34 normal gastric samples and 415 primary tumor samples). (C) The data-mining analysis showed the expression status of COL12A1 in the TCGA gastric cancer portal based on different TNM stages (normal gastric samples and tumor samples classified to stage1, stage2, stage3 and stage4). (D) Basic protein expression levels of COL12A1 in normal organs. (E) Basic mRNA expression levels of COL12A1 in normal organs. (F) COL12A1 expression levels in 64 pairs of fresh normal gastric and GC tissues (COL12A1/GAPDH). (G) COL12A1 expression levels in 64 patients with GC (normalized to normal samples). (H) COL12A1 mRNA expression was examined by reverse transcription-quantitative PCR and normalized to GAPDH expression in four GC cell lines. ***P<0.001. COL12A1, collagen type XII α1 chain; TCGA, The Cancer Genome Atlas; GC, gastric cancer.
Figure 2.
COL12A1 protein expression is upregulated in all Lauren types. Three datasets (15,20,21) from Oncomine were used to evaluate the association between COL12A1 expression and Lauren classification. COL12A1, collagen type XII α1 chain.
COL12A1 protein expression status in GC tissues
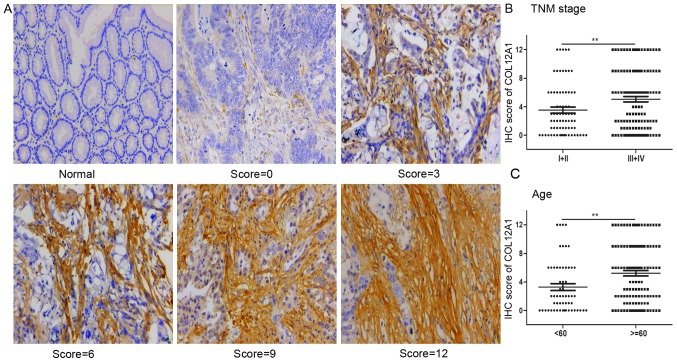
The elevated COL12A1 mRNA expression in GC suggested its carcinogenic role in this disease. To investigate its protein expression profiles in GC, IHC staining of COL12A1 protein was performed in a total of 200 GC and 184 corresponding normal tissues. IHC staining showed that COL12A1 protein expression was not observed in normal control tissues in the present cohort, while it was present in the stroma surrounding the GC cells. Representative COL12A1 protein expression images in normal or GC tissues are shown in Fig. 3A. These results suggested that COL12A1 is exclusively expressed in GC and not in matched normal tissues, and thus it may be a highly specific biomarker of GC.
Figure 3.
COL12A1 protein expression is upregulated in GC. (A) Representative images of COL12A1 IHC staining in GC samples. The IHC score is equal to the percentage of positive staining multiples staining intensity. Magnification, ×200. The IHC scores were significantly higher in (B) patients of advanced stage (III and IV) and (C) in patients >60 years old. **P<0.01. COL12A1, collagen type XII α1 chain; IHC, immunohistochemistry; GC, gastric cancer; TNM, tumor-node-metastasis.
Correlation between COL12A1 protein expression and clinicopathological features
Subsequently, the association between COL12A1 expression and clinical parameters was analyzed. Based on the median value of 4 for final immunoreactivity scoring for COL12A1 staining, elevated COL12A1 protein expression was present in 86 of 200 (43%) GC samples, with the other 114 (67%) samples exhibiting low expression profiles relative to normal controls. Additionally, in GC samples, the IHC scores of COL12A1 were increased in patients with more advanced tumor stages (stage III and IV; P<0.01; Fig. 3B) and in patients >60 years old (P<0.01; Fig. 3C). Furthermore, as shown in Table II, COL12A1 protein overexpression was positively correlated with sex (P=0.0027), tumor invasion (P=0.0001), lymph node metastasis (P=0.0121), distant metastasis (P=0.0366) and advanced TNM stage (P=0.0048); however, no significant associations with other clinical factors (such as age, tumor size and differentiation degree) were observed.
Table II.
Association between COL12A1 protein expression and clinicopathological characteristics in 200 patients with gastric cancer.
| COL12A1 expression | ||||
|---|---|---|---|---|
| Characteristics | No. patients | High | Low | P-value |
| All cases | 200 | 86 | 114 | |
| Age, years | 0.764 | |||
| <60 | 68 | 28 | 40 | |
| ≥60 | 132 | 58 | 74 | |
| Sex | 0.0027 | |||
| Male | 143 | 71 | 72 | |
| Female | 57 | 15 | 42 | |
| Tumor size, cm | ||||
| ≤5 | 98 | 41 | 57 | 0.7764 |
| >5 | 102 | 45 | 57 | |
| Tumor invasion (T) | 0.0001 | |||
| T1+T2 | 31 | 3 | 25 | |
| T3+T4 | 169 | 83 | 89 | |
| Lymph node metastasis (N) | 0.0121 | |||
| N0 | 48 | 13 | 35 | |
| N1+N2+N3 | 152 | 73 | 79 | |
| Distant metastasis (M) | 0.0366 | |||
| M0 | 184 | 75 | 109 | |
| M1 | 16 | 11 | 5 | |
| Differentiation | 0.7751 | |||
| Well to moderate | 86 | 38 | 48 | |
| Low | 114 | 48 | 66 | |
| TNM stage | 0.0048 | |||
| I+II | 74 | 22 | 52 | |
| III+IV | 126 | 64 | 62 | |
| Sample type | <0.0001 | |||
| Tumor | 200 | 86 | 115 | |
| Normal | 184 | 0 | 184 | |
TNM, tumor-node-metastasis; COL12A1, collagen type XII α1 chain.
Survival analysis of GC patients
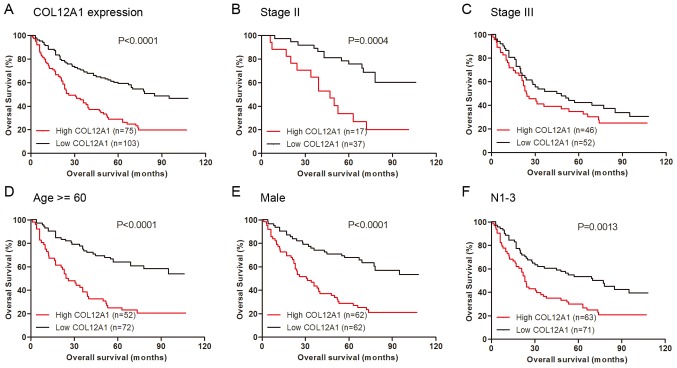
As described, elevated COL12A1 expression was positively related to advanced TNM stage. Therefore, the question of whether high COL12A1 expression influenced GC prognosis was investigated. Of the 200 patients included in the study, 22 lacked survival information. Therefore, 178 patients were used to explore the correlations between COL12A1 expression level and survival status. The results revealed that patients with low expression levels of COL12A1 protein exhibited more extended periods of OS than those with high COL12A1 protein levels (Fig. 4A; P<0.0001). Similar results were acquired in patients at intermediate TNM stage II (Fig. 4B; P=0.0004), although not in those at stage III (Fig. 4C; P>0.05). When stratified by age, sex or lymph node metastasis, the results indicated that high COL12A1 expression was positively associated with worse OS in patients >60 years old (P<0.0001), in male patients (P<0.0001), and in patients with lymph node metastasis (P=0.0013) (Fig. 4D-F).
Figure 4.
Kaplan-Meier survival analysis. (A) COL12A1 overexpression was positively associated with worse OS. The association between COL12A1 expression and OS in patients of (B) stage II and (C) stage III is presented. Stratified analysis indicated that COL12A1 was associated with a worse OS in (D) patients >60 years old, (E) male patients and (F) patients with lymph node metastasis. OS, overall survival; COL12A1, collagen type XII α1 chain.
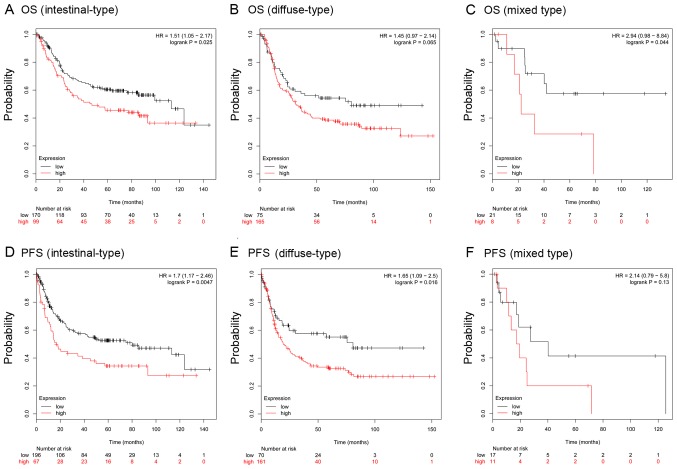
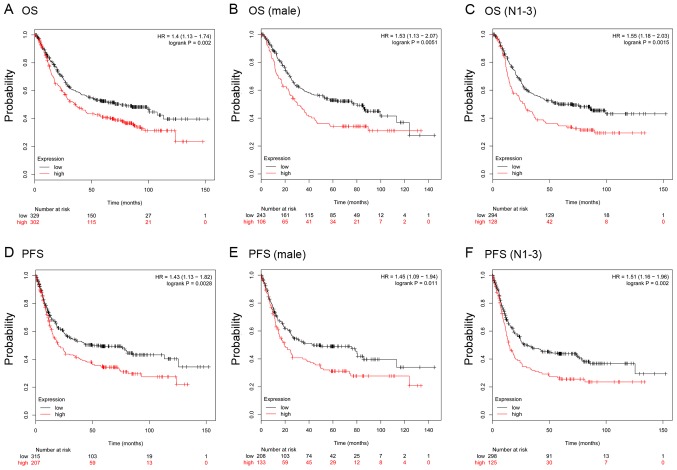
To further verify the prognostic role of COL12A1, the KM-plotter database, which contains a total of 875 GC samples, was used. For COL12A1, 631 GC samples from one Affymetrix array (225664_at; Thermo Fisher Scientific, Inc.) were analyzed in the present study. It was predicted that in this cohort, COL12A1 overexpression was positively related to worse OS [Fig. 5A; hazard ratio (HR), 1.4; 95% CI, 1.13–1.72; log-rank P=0.002] and progression free survival (PFS) (Fig. 5D; HR, 1.43; 95% CI, 1.13–1.82; log-rank P=0.0028). Further analysis in male patients and patients with lymph node metastasis revealed similar results in OS (Fig. 5B and C) and PFS (Fig. 5E and F). Stratification analysis showed that COL12A1 was an indicator of inferior OS for intestinal-and mixed-type GC and an indicator of inferior PFS for intestinal-and diffuse-type GC (Fig. 6). Moreover, the univariate analysis suggested tumor invasiveness, lymph node invasion, distant metastasis, advanced clinical stage and COL12A1 high expression were significantly associated with OS, and further multivariate Cox analysis confirmed that distant metastasis and COL12A1 overexpression could be regarded as powerful independent prognostic factors for OS in GC (P<0.05; Table III); large-scale studies are warranted to verify these results further.
Figure 5.
KM plotter predicts the prognostic role of COL12A1 in patients with different Lauren classifications. OS in (A) intestinal-, (B) diffuse- and (C) mixed-type GC patients; PFS in (D) intestinal-, (E) diffuse- and (F) mixed-type GC patients. OS, overall survival; PFS, progression-free survival; COL12A1, collagen type XII α1 chain; GC, gastric cancer.
Figure 6.
KM plotter predicts a poorer prognosis for gastric cancer patients with high expression of COL12A1. (A) OS in all patients (n=631); (B) OS in male patients; and (C) OS in patients with lymph node metastasis. (D) PFS (n=522) in all patients; (E) PFS in male patients; and (F) PFS in patients with lymph node metastasis. OS, overall survival; PFS, progression-free survival; COL12A1, collagen type XII α1 chain.
Table III.
Univariate and multivariate analyses of COL12A1 expression and patient survival.
| Overall survival | ||||
|---|---|---|---|---|
| Characteristics | HR | (95% CI) | P-value | |
| Univariate analysis (n=178) | ||||
| Age (≥60 years vs. <60 years) | 1.436 | 0.943 | 2.18 | 0.091 |
| Sex (male vs. female) | 0.835 | 0.56 | 1.24 | 0.377 |
| Tumor size (>5 cm vs. ≤5 cm) | 1.6 | 0.15 | 1.097 | 2.33 |
| Tumor invasion (T3+T4 vs. T1+T2) | 2.018 | 1.12 | 3.606 | 0.018 |
| Lymph node metastasis (N1-3 vs. N0) | 1.85 | 1.148 | 2.981 | 0.011 |
| Distant metastasis (M1 vs. M0) | 4.232 | 2.277 | 7.864 | <0.001 |
| Differentiation (poor vs. well to moderate) | 1.352 | 0.922 | 1.983 | 0.123 |
| TNM stage (III+IV vs. I+II) | 2.348 | 1.546 | 3.566 | <0.001 |
| COL12A1 expression (high vs. low) | 2.361 | 1.614 | 3.453 | <0.001 |
| Multivariate analysis | ||||
| Tumor invasion (T3+T4 vs. T1+T2) | 1.516 | 0.833 | 2.76 | 0.174 |
| Lymph node metastasis (N1-3 vs. N0) | 1.474 | 0.903 | 2.406 | 0.12 |
| Distant metastasis (M1 vs. M0) | 2.919 | 1.547 | 5.51 | 0.001 |
| COL12A1 expression (high vs. low) | 1.896 | 1.267 | 2.837 | 0.002 |
Patients with P<0.05 in univariate analysis were selected for multivariate analysis of survival. HR, hazard ratio; COL12A1, collagen type XII α1 chain; TNM, tumor-node-metastasis.
Discussion
Early diagnosis of GC is a long-standing problem; however, early diagnosis can significantly benefit the clinical outcomes of GC patients by facilitating the implementation of prompt and more effective treatment. Therefore, extensive efforts have been expended to better understand the potential mechanisms associated with cancer development. Among these mechanisms, dysregulation of the extracellular matrix (ECM), a network of macromolecules between cells that is mainly composed of collagens, has attracted substantial attention (23). The steady state of ECM remodeling, in which damaged or old molecules are degraded and simultaneously replaced by new versions, is a key process involved in maintaining tissue homeostasis (24). Recently, dysregulation of ECM proteins was revealed in numerous cancer types (23,25). Alterations in ECM composition and quantity strongly influence tumor and stromal cell properties, which activate downstream signaling that modulates cell behavior and contributes to tumor progression (26). A review noted that the ECM is involved in neoplastic progression by modulating cancer hallmarks, and that its influence might vary in different cancer types (27). Together, these data indicate that ECM proteins are critical in the development of human malignancies and may represent promising candidates for early diagnosis and the personalization of clinical treatments.
COL12A1 is an ECM protein and, to date, its known disorders are mainly restricted to connective tissue pathology. Specifically, COL12A1 mutations are involved in blended muscle and connective tissue phenotypes, often resulting in poorer prognoses compared with patients without COL12A1 mutations (28). Additionally, elevated COL12A1 expression is related to anterior cruciate ligament (ACL) tears, with higher expression levels in acute compared with chronic ACL damage (29).
The protein encoded by COL12A1, collagen type XII, interacts specifically with fibers, especially those containing collagen I (30). Recently, dysregulation of the ECM has been observed in a large spectrum of human cancer types, including CRC, and ovarian, breast and renal cell carcinomas. In CRC, COL12A1 expression is increased and COL12A1 levels are upregulated in tumors with microsatellite stability relative to tumors exhibiting high degrees of microsatellite instability (31). Notably, proteomics analyses have been conducted to search for markers of CRC-invasion fronts, with the results suggesting that type XII collagen is a potent marker of myofibroblast differentiation, implying that the COL12A1 protein is closely related to tumor metastasis in CRC (32). In ovarian cancer cell lines, the COL12A1 expression level is significantly increased in cells resistant to cisplatin and/or doxorubicin (33), and high COL12A1 expression levels are positively correlated with worse OS in patients with ovarian cancer or renal cell carcinoma (8,34).
Although increased attention has been focused on elucidating the significance of COL12A1 in human cancer, the detailed mechanism of its involvement remains unknown. The present study, to the best of our knowledge, is the first to analyze COL12A1 expression in GC at both the mRNA and protein levels. Specifically, the present data revealed that COL12A1 expression was markedly increased in GC tissues compared with corresponding normal controls, both at the mRNA and protein levels. Additionally, COL12A1 mRNA was highly expressed in several GC cell lines except AGS, which may be attributed to the fact that different cell lines have different backgrounds, and thus it is reasonable for a certain gene to be expressed differentially in different cell lines. The IHC assay of COL12A1 showed that COL12A1 was mainly expressed in the stromal cells, especially in fibroblasts as showed in Fig. 3. Fibroblasts are essential components of the ECM and are responsible for the production of ECM. Cancer-associated fibroblasts (CAFs) are a cluster of cells characterized by their morphology and the expression of protein markers, including α-smooth muscle actin, fibroblast activated protein, neuron-glial antigen-2 and fibroblast specific protein (35–37). CAFs mainly originate from local fibroblasts or fibroblast precursors. Studies have linked CAFs to tumor progression, metastasis and poor clinical outcomes in various cancer types (36–38). The present results suggested that COL12A1 may be used as a potential marker of CAFs.
Additionally, Lauren classification is widely used for the histological classification of gastric adenocarcinoma (39). Based on these criteria, the majority of GC cases can be divided into either intestinal or diffuse types, with the remaining cases classified as mixed or unclassified. The two GC types differ from each other in a number of ways (40–43). For example, compared to intestinal-type GC, diffuse-type GC is characterized by a lower response rate to chemotherapy, a worse prognosis, and higher peritoneal or distant recurrence rates (44,45). Therefore, in-depth elucidation of the underlying mechanisms of diffuse-type GC is required. In the present study, it was found that COL12A1 was highly expressed in intestinal-and diffuse-type GC, and was associated with worse clinical outcomes (OS and PFS).
Further analysis suggested that high COL12A1 protein expression was associated with tumor invasiveness, clinical metastasis and a worse TNM stage. Prognostic analysis demonstrated a positive association between high expression of COL12A1 and worse prognosis, and the results were in accordance with the prognosis prediction of KM-plotter. Therefore, the present findings revealed a tumor-promoting role of COL12A1 in GC. However, there are several limitations to the present study. For example, the study was retrospectively designed and the Lauren classification status of the samples was lacking. Large-scale prospective studies with more comprehensive information are required. In vitro and in vivo studies validating the tumor-promoting role of COL12A1 in the aggressiveness of GC and fibroblast cells by up-or downregulating its expression are required. Furthermore, explorations of the underlying molecular biological mechanisms of COL12A1 in GC are also warranted.
In conclusion, the present study revealed that COL12A1 expression was notably increased in GC tissues, and elevated COL12A1 protein levels were positively associated with aggressive clinical features. Importantly, COL12A1 protein expression levels represent a potential prognostic indicator for OS in patients with GC. These results indicated that COL12A1 plays a tumor-promoting role and might be a potential candidate for therapeutic targeting, based on its putative role in tumor progression and prognosis prediction in patients with GC.
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the National Natural Science Foundation of China (grant nos. 81201089 and 81272676) and the Natural Science Foundation of Zhejiang Province, China (grant nos. LY15H160026 and LY15H160012).
Availability of data and materials
The data used and/or analyzed in the present study are available from the corresponding author on reasonable request.
Authors' contributions
XJ, MW, XX and LT designed the study. XJ, MW, YH and LZ wrote the manuscript and conducted the immunohistochemical analyses. XJ, MW, XX, KH, YH and ZX collected the clinical samples and managed the clinical data. XJ, KH, HYW and HHW contributed to bioinformatics analysis. KH, HYW and ZX were involved in project management, and contributed to preparing and making figures and tables. HYW, HHW and LT supervised the study. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The study protocol was approved by the ethics committee of the First Affiliated Hospital of Zhejiang University and written informed consent was obtained from all participants in the study.
Patient consent for publication
Written informed consent was obtained from all participants.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Yang SY, Roh KH, Kim YN, Cho M, Lim SH, Son T, Hyung WJ, Kim HI. Surgical outcomes after open, laparoscopic and robotic gastrectomy for gastric cancer. Ann Surg Oncol. 2017;24:1770–1777. doi: 10.1245/s10434-016-5482-y. [DOI] [PubMed] [Google Scholar]
- 3.Miller KD, Siegel RL, Lin CC, Mariotto AB, Kramer JL, Rowland JH, Stein KD, Alteri R, Jemal A. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin. 2016;66:271–289. doi: 10.3322/caac.21349. [DOI] [PubMed] [Google Scholar]
- 4.Pu X, Ye Y, Wu X. Development and validation of risk models and molecular diagnostics to permit personalized management of cancer. Cancer. 2014;120:11–19. doi: 10.1002/cncr.28393. [DOI] [PubMed] [Google Scholar]
- 5.Gerecke DR, Olson PF, Koch M, Knoll JH, Taylor R, Hudson DL, Champliaud MF, Olsen BR, Burgeson RE. Complete primary structure of two splice variants of collagen XII and assignment of alpha 1 (XII) collagen (COL12A1), alpha 1 (IX) collagen (COL9A1) and alpha 1 (XIX) collagen (COL19A1) to human chromosome 6q12-q13. Genomics. 1997;41:236–242. doi: 10.1006/geno.1997.4638. [DOI] [PubMed] [Google Scholar]
- 6.Hicks D, Farsani GT, Laval S, Collins J, Sarkozy A, Martoni E, Shah A, Zou Y, Koch M, Bönnemann CG, et al. Mutations in the collagen XII gene define a new form of extracellular matrix-related myopathy. Hum Mol Genet. 2014;23:2353–2363. doi: 10.1093/hmg/ddt637. [DOI] [PubMed] [Google Scholar]
- 7.Sun F, Ding W, He JH, Wang XJ, Ma ZB, Li YF. Stomatin-like protein 2 is overexpressed in epithelial ovarian cancer and predicts poor patient survival. BMC Cancer. 2015;15:746. doi: 10.1186/s12885-015-1723-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao H, Ljungberg B, Grankvist K, Rasmuson T, Tibshirani R, Brooks JD. Gene expression profiling predicts survival in conventional renal cell carcinoma. PLoS Med. 2006;3:e13. doi: 10.1371/journal.pmed.0030013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramaswamy S, Ross KN, Lander ES, Golub TR. A molecular signature of metastasis in primary solid tumors. Nat Genet. 2003;33:49–54. doi: 10.1038/ng1060. [DOI] [PubMed] [Google Scholar]
- 10.Torres S, Bartolomé RA, Mendes M, Barderas R, Fernandez-Aceñero MJ, Peláez-García A, Peña C, Lopez-Lucendo M, Villar-Vázquez R, de Herreros AG, et al. Proteome profiling of cancer-associated fibroblasts identifies novel proinflammatory signatures and prognostic markers for colorectal cancer. Clin Cancer Res. 2013;19:6006–6019. doi: 10.1158/1078-0432.CCR-13-1130. [DOI] [PubMed] [Google Scholar]
- 11.Ji X, Bu ZD, Yan Y, Li ZY, Wu AW, Zhang LH, Zhang J, Wu XJ, Zong XL, Li SX, et al. The 8th edition of the american joint committee on cancer tumor-node-metastasis staging system for gastric cancer is superior to the 7th edition: Results from a Chinese mono-institutional study of 1663 patients. Gastric Cancer. 2018;21:643–652. doi: 10.1007/s10120-017-0779-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 13.Gao B, Lin J, Jiang Z, Yang Z, Yu H, Ding L, Yu M, Cui Q, Dunavin N, Zhang M, Li M. Upregulation of chemokine CXCL10 enhances chronic pulmonary inflammation in tree shrew collagen-induced arthritis. Sci Rep. 2018;8:9993. doi: 10.1038/s41598-018-28404-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baptista MZ, Sarian LO, Vassallo J, Pinto GA, Soares FA, de Souza GA. Prognostic significance of GRP78 expression patterns in breast cancer patients receiving adjuvant chemotherapy. Int J Biol Markers. 2011;26:188–196. doi: 10.5301/JBM.2011.8624. [DOI] [PubMed] [Google Scholar]
- 15.Chen X, Leung SY, Yuen ST, Chu KM, Ji J, Li R, Chan AS, Law S, Troyanskaya OG, Wong J, et al. Variation in gene expression patterns in human gastric cancers. Mol Biol Cell. 2003;14:3208–3215. doi: 10.1091/mbc.e02-12-0833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang X, Ni Z, Duan Z, Xin Z, Wang H, Tan J, Wang G, Li F. Overexpression of E2F mRNAs associated with gastric cancer progression identified by the transcription factor and miRNA co-regulatory network analysis. PLoS One. 2015;10:e0116979. doi: 10.1371/journal.pone.0116979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He J, Jin Y, Chen Y, Yao HB, Xia YJ, Ma YY, Wang W, Shao QS. Downregulation of ALDOB is associated with poor prognosis of patients with gastric cancer. Onco Targets Ther. 2016;9:6099–6109. doi: 10.2147/OTT.S110203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clough E, Barrett T. The gene expression omnibus database. Methods Mol Biol. 2016;1418:93–110. doi: 10.1007/978-1-4939-3578-9_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK, Varambally S. UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cho JY, Lim JY, Cheong JH, Park YY, Yoon SL, Kim SM, Kim SB, Kim H, Hong SW, Park YN, et al. Gene expression signature-based prognostic risk score in gastric cancer. Clin Cancer Res. 2011;17:1850–1857. doi: 10.1158/1078-0432.CCR-10-2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.D'Errico M, de Rinaldis E, Blasi MF, Viti V, Falchetti M, Calcagnile A, Sera F, Saieva C, Ottini L, Palli D, et al. Genome-wide expression profile of sporadic gastric cancers with microsatellite instability. Eur J Cancer. 2009;45:461–469. doi: 10.1016/j.ejca.2008.10.032. [DOI] [PubMed] [Google Scholar]
- 22.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: A cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/S1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karsdal MA, Nielsen SH, Leeming DJ, Langholm LL, Nielsen MJ, Manon-Jensen T, Siebuhr A, Gudmann NS, Rønnow S, Sand JM, et al. The good and the bad collagens of fibrosis-their role in signaling and organ function. Adv Drug Deliv Rev. 2017;121:43–56. doi: 10.1016/j.addr.2017.07.014. [DOI] [PubMed] [Google Scholar]
- 24.Hansen NU, Genovese F, Leeming DJ, Karsdal MA. The importance of extracellular matrix for cell function and in vivo likeness. Exp Mol Pathol. 2015;98:286–294. doi: 10.1016/j.yexmp.2015.01.006. [DOI] [PubMed] [Google Scholar]
- 25.Gilkes DM, Semenza GL, Wirtz D. Hypoxia and the extracellular matrix: Drivers of tumour metastasis. Nat Rev Cancer. 2014;14:430–439. doi: 10.1038/nrc3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Millar MW, Corson N, Xu L. The adhesion g-protein-coupled receptor, GPR56/ADGRG1, inhibits cell-extracellular matrix signaling to prevent metastatic melanoma growth. Front Oncol. 2018;8:8. doi: 10.3389/fonc.2018.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pickup MW, Mouw JK, Weaver VM. The extracellular matrix modulates the hallmarks of cancer. EMBO Rep. 2014;15:1243–1253. doi: 10.15252/embr.201439246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Punetha J, Kesari A, Hoffman EP, Gos M, Kamińska A, Kostera-Pruszczyk A, Hausmanowa-Petrusewicz I, Hu Y, Zou Y, Bönnemann CG, JȨdrzejowska M. Novel Col12A1 variant expands the clinical picture of congenital myopathies with extracellular matrix defects. Muscle Nerve. 2017;55:277–281. doi: 10.1002/mus.25232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brophy RH, Tycksen ED, Sandell LJ, Rai MF. Changes in transcriptome-wide gene expression of anterior cruciate ligament tears based on time from injury. Am J Sports Med. 2016;44:2064–2075. doi: 10.1177/0363546516643810. [DOI] [PubMed] [Google Scholar]
- 30.Bader HL, Keene DR, Charvet B, Veit G, Driever W, Koch M, Ruggiero F. Zebrafish collagen XII is present in embryonic connective tissue sheaths (fascia) and basement membranes. Matrix Biol. 2009;28:32–43. doi: 10.1016/j.matbio.2008.09.580. [DOI] [PubMed] [Google Scholar]
- 31.Ortega P, Moran A, Fernandez-Marcelo T, De Juan C, Frias C, Lopez-Asenjo JA, Sanchez-Pernaute A, Torres A, Diaz-Rubio E, Iniesta P, Benito M. MMP-7 and SGCE as distinctive molecular factors in sporadic colorectal cancers from the mutator phenotype pathway. Int J Oncol. 2010;36:1209–1215. doi: 10.3892/ijo_00000604. [DOI] [PubMed] [Google Scholar]
- 32.Karagiannis GS, Petraki C, Prassas I, Saraon P, Musrap N, Dimitromanolakis A, Diamandis EP. Proteomic signatures of the desmoplastic invasion front reveal collagen type XII as a marker of myofibroblastic differentiation during colorectal cancer metastasis. Oncotarget. 2012;3:267–285. doi: 10.18632/oncotarget.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Januchowski R, Świerczewska M, Sterzynska K, Wojtowicz K, Nowicki M, Zabel M. Increased expression of several collagen genes is associated with drug resistance in ovarian cancer cell lines. J Cancer. 2016;7:1295–1310. doi: 10.7150/jca.15371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chudasama D, Bo V, Hall M, Anikin V, Jeyaneethi J, Gregory J, Pados G, Tucker A, Harvey A, Pink R, Karteris E. Identification of novel cancer biomarkers of prognostic value using specific gene regulatory networks (GRN): A novel role of RAD51AP1 for ovarian and lung cancers. Carcinogenesis. 2018;39:407–417. doi: 10.1093/carcin/bgx122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang L, Xu AM, Liu S, Liu W, Li TJ. Cancer-associated fibroblasts in digestive tumors. World J Gastroenterol. 2014;20:17804–17818. doi: 10.3748/wjg.v20.i47.17804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ostman A, Augsten M. Cancer-associated fibroblasts and tumor growth-bystanders turning into key players. Curr Opin Genet Dev. 2009;19:67–73. doi: 10.1016/j.gde.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 37.Kalluri R, Zeisberg M. Fibroblasts in cancer. Nat Rev Cancer. 2006;6:392–401. doi: 10.1038/nrc1877. [DOI] [PubMed] [Google Scholar]
- 38.Luo H, Tu G, Liu Z, Liu M. Cancer-associated fibroblasts: A multifaceted driver of breast cancer progression. Cancer Lett. 2015;361:155–163. doi: 10.1016/j.canlet.2015.02.018. [DOI] [PubMed] [Google Scholar]
- 39.Lauren P. The two histological main types of gastric carcinoma: Diffuse and so-called intestinal-type carcinoma. An attempt at a histo-clinical classification. Acta Pathol Microbiol Scand. 1965;64:31–49. doi: 10.1111/apm.1965.64.1.31. [DOI] [PubMed] [Google Scholar]
- 40.Sunakawa Y, Lenz HJ. Molecular classification of gastric adenocarcinoma: Translating new insights from the cancer genome atlas research network. Curr Treat Options Oncol. 2015;16:17. doi: 10.1007/s11864-015-0331-y. [DOI] [PubMed] [Google Scholar]
- 41.Riquelme I, Saavedra K, Espinoza JA, Weber H, García P, Nervi B, Garrido M, Corvalán AH, Roa JC, Bizama C. Molecular classification of gastric cancer: Towards a pathway-driven targeted therapy. Oncotarget. 2015;6:24750–24779. doi: 10.18632/oncotarget.4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vauhkonen M, Vauhkonen H, Sajantila A, Sipponen P. Differences in genomic instability between intestinal-and diffuse-type gastric cancer. Gastric Cancer. 2005;8:238–244. doi: 10.1007/s10120-005-0346-3. [DOI] [PubMed] [Google Scholar]
- 43.Ma J, Shen H, Kapesa L, Zeng S. Lauren classification and individualized chemotherapy in gastric cancer. Oncol Lett. 2016;11:2959–2964. doi: 10.3892/ol.2016.4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marrelli D, Polom K, Neri A, Roviello F. Clinical impact of molecular classifications in gastric cancer. Updates Surg. 2018;70:225–232. doi: 10.1007/s13304-018-0546-0. [DOI] [PubMed] [Google Scholar]
- 45.Lee JH, Chang KK, Yoon C, Tang LH, Strong VE, Yoon SS. Lauren histologic type is the most important factor associated with pattern of recurrence following resection of gastric adenocarcinoma. Ann Surg. 2018;267:105–113. doi: 10.1097/SLA.0000000000002040. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used and/or analyzed in the present study are available from the corresponding author on reasonable request.