Abstract
In the present study, the expression of microRNA (miR)-671-3p in non-small-cell lung cancer (NSCLC) was detected via reverse transcription-quantitative polymerase chain reaction analysis, and its role in cell proliferation, apoptosis, migration and invasion was investigated via Cell Counting Kit-8, colony formation, flow cytometry, Transwell and scratch assays, respectively. It was observed that the expression of miR-671-3p was upregulated in NSCLC tissues and cell lines (A549 and H1975). Treatment with miR-671-3p inhibitors suppressed cell proliferation, migration and invasion, and increased apoptosis in vitro, suggesting that miR-671-3p functions as an oncogene in NSCLC. In addition, forkhead box P2 (FOXP2) has been reported to be a tumor suppressor that is downregulated in several types of cancer, and its low expression was confirmed in NSCLC tissues and cell lines in the current study via western blotting. The results of the luciferase reporter assay also demonstrated that miR-671-3p targeted directly the 3′-untranslated region of FOXP2. Furthermore, overexpression of FOXP2 in A549 and H1975 cell lines suppressed the growth, migration and invasion, and promoted apoptosis, whereas these effects were reversed by transfection with miR-671-3p mimics, suggesting that miR-671-3p promoted tumor progression via regulating FOXP2. Taken together, the results reported in the present study implied that miR-671-3p may be a potential therapeutic target in NSCLC.
Keywords: microRNA-671-3p, forkhead box P2, proliferation, apoptosis, migration, invasion
Introduction
MicroRNAs (miRNAs) are short noncoding RNAs that regulate gene expression via hybridization to complementary sequences of mRNAs (1). miRNAs serve critical roles in biological processes and function as either tumor suppressors or oncogenes (2,3). An expanding body of evidence has identified a number of miRNA functions in cancer development, including invasion and metastasis (4).
As one of the most common malignancies worldwide, lung cancer is known to have high morbidity and mortality rates (5,6). The main types of lung cancer are small cell lung cancer and non-small-cell lung cancer (NSCLC), with the later accounting for ~85% of lung cancer cases (7). miRNAs have been reported to serve an important role in cancer development and are recently considered as key factors in lung cancer (2,8,9). For instance, miRNAs were observed to be epigenetic modulators in lung cancer, regulating the tumor microenvironment and the immune system (10). The members of let-7 miRNA family are highly expressed in normal lung tissue and have been found to negatively control multiple oncogenes (11).
A previous study has identified miR-671-3p as a potential osteoarthritis biomarker involved in metabolic processes (12). In breast cancer, miR-671-3p was observed to be downregulated and potentially function as a tumor suppressor by influencing the Wnt signaling cascade (13). However, the effects and mechanisms of miR-671-3p on the lung cancer cells remain unknown.
Forkhead box protein P2 (FOXP2) is a member of the forkhead box transcription factor family, and is expressed in various tissues (14). FOXP2 is involved in the pathogenesis and development of numerous cancers, including gastric, breast and hepatocellular cancers (15–17).
In this study, the aim was to determine the role of miR-671-3p in NSCLC progression and the potential underlying mechanisms. It was demonstrated that the expression of miR-671-3p was upregulated in NSCLC tissues and cell lines (A549 and H1975), and that miR-671-3p regulated NSCLC cell proliferation, apoptosis, migration and invasion via directly targeting FOXP2.
Materials and methods
Human tissue samples
The study was reviewed and approved by the Ethics Committee of the First People's Hospital of Changzhou (Changzhou, China). Clinical samples (NSCLC tissues and adjacent normal tissues) were collected from 40 NSCLC patients [23 males and 17 females; 11 patients aged <60 years old (45–59 years old) and 29 patients aged ≥60 years old (60–75 years old)] who received surgery at The First People's Hospital of Changzhou (Changzhou, China) between July 2015 and November 2017; and informed consent was obtained from all patients. All diagnoses of NSCLC were confirmed using pathological assays, including computed tomography, nuclear magnetic resonance imaging and immunohistochemistry; none of the patients included in the study had previously received any cancer treatment. All tissues were immediately frozen in liquid nitrogen and stored at −80°C for future experiments.
Cell culture and cell transfection
Human normal lung 16HBE and human NSCLC (A549 and H1975) cell lines were obtained from the Chinese Academy of Science Cell Bank (Shanghai, China). Cells were cultured in RPMI-1640 medium (HyClone; GE Healthcare Life Sciences, Logan, UT, USA) supplied with 10% fetal bovine serum (FBS; Biowest, Barcelona, Spain) at 37°C with 5% CO2. miR-671-3p mimics, inhibitors, mimics NC and inhibitors NC were purchased from Shanghai GenePharma Co., Ltd. (Shanghai, China). pcDNA.3.1-FOXP2 and pcDNA3.1 empty vector (2 µg) were inserted into a pCDF-CMV-MCS-EF1-Peru lentiviral vector using 293T cells (5×104; Chinese Academy of Science Cell Bank) and Entranster™-H (Engreen Biosystem Co., Ltd., Beijing, China); all the vectors were synthesized by Shanghai GenePharma Co., Ltd. (Shanghai, China). The lentiviral supernatant was collected 48 h after the transfection and used to infect cancer cells. The empty pcDNA3.1 vector was used as negative control. The following primers for FOXP2 were used: Sense, 5′-GGATCCATGATGCAGGAATCTGCG-3′, and antisense, 5′-CTCGAGTCATTCCAGATCTTCAGAT-3′. Cells (5×104) were transfected with 200 nM miR-671-3p mimic, 50 nM miR-671-3p inhibitor and 8×108 TU/ml pcDNA3.1-FOXP2 lentivirus by using Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA). After 24 or 48 h, transfected cells were collected and used in RNA or protein assays.
RNA isolation and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
For miR-671-3p detection, total RNA was extracted from cells and tissues with the miRcute miRNA Isolation kit (Tiangen, Shanghai, China) and reverse-transcribed using One Step PrimeScript miRNA cDNA Synthesis kit (Takara Biotechnology Co., Ltd., Dalian, China) according to the manufcaturer's protocols. For FOXP2 mRNA expression detection, total RNA was extracted using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and then reverse-transcribed using M-MLV reverse transcriptase (Promega Corporation, Madison, WI, USA) according to the manufacturer's protocols. Subsequently, qPCR was performed using the SYBR ExScript RT-qPCR kit (Takara Biotechnology Co., Ltd.) in an ABI 7500 Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.). U6 and GAPDH served as the endogenous control for miRNA and mRNA, respectively and each sample was detected in triplicate. The reaction was conducted in a total volume of 20 µl, containing 1 µl cDNA, 10 µl SYBR Premix EX Taq, 1 µl of each primer (10 µM), and 7 µl ddH2O. The PCR program was conducted at 95°C for 3 min, followed by 40 cycles of 95°C for 10 sec, and 60°C for 30 sec. The primers used were as follows: GAPDH, forward 5′-TGAACTGAAAGCTCTCCACC-3′, reverse, 5′-CTGATGTACCAGTTGGGGAA-3′; U6, forward 5′-CTCGCTTCGGCAGCACA-3′, reverse, 5′-AACGCTTCACGAATTTGCGT-3′; FOXP2, forward 5′-CCACGAAGACCTCAATGGTT-3′, reverse, 5′-TCACGCTGAGGTTTCACAAG-3′; and miR-671-3p, forward 5′-CCGGUUCUCAGGGCUCCACC-3′ and reverse, 5′-GTGCAGGGTCCGAGGT-3′ (Sangon Biotech Co., Ltd., Shanghai, China). All fold changes were calculated using the comparative Cq method (2−ΔΔCq), using GAPDH or U6 for normalization (18).
Western blotting
Total proteins from cells and tissues were lysed with radioimmunoprecipitation assay lysis buffer (Beyotime Institute of Biotechnology, Haimen, China), and the concentration of protein was measured using a bicinchoninic acid assay kit (Beyotime Institute of Biotechnology). A total of 50 µg total protein per lane was separated by 10% SDS-PAGE and blotted onto polyvinylidene fluoride membranes. Next, the membrane was blocked using 5% non-fat milk in PBS-0.5% Tween 20 at room temperature for 3 h and probed with primary antibodies against FOXP2 (1:500; ab16046; Abcam, Cambridge, MA, USA), B-cell lymphoma 2 (Bcl-2; 1:1,000; ab32124; Abcam), Bcl-2-associated X protein (Bax; 1:1,000; ab32503; Abcam), caspase-3 (1:1,000; ab13847; Abcam), caspase-9 (1:1,000, ab13847; Abcam), matrix metalloproteinase-2 (MMP-2; 1:1,000; ab37150; Abcam), MMP-9 (1:1,000; ab73734; Abcam) and GAPDH (1:5,000; G8795; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) at 4°C overnight. The membranes were then incubated with goat anti-mouse secondary antibody conjugated with horseradish peroxidase (1:5,000; ab97040; Abcam) for 1 h at room temperature. Finally, the membranes were visualized using an enhanced chemiluminescence kit (Beyotime Institute of Biotechnology). The bands were scanned using a densitometer, and the gray values of the bands were calculated automatically by the ImageJ software (version 1.45k; National Institutes of Health, Bethesda, MD, USA).
Cell proliferation and colony formation assays
Cell proliferation was detected by Cell Counting Kit (CCK)-8 and 5-ethynyl-2′-deoxyuridine (EdU) assays according to the manufacturers' protocol. Briefly, cells were seeded at a density of 1×104 cells/well in 96-well plates and cultured for 24 h before transfection. After transfection for 48 h, 10 µl of CCK-8 solution (Beyotime Institute of Biotechnology) was added and incubated for 1 h at 37°C. Finally, the optical density value at 450 nm was determined using a microplate reader (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
For colony formation assay, cells were seeded in a 6-well plate at a density of 1,000 cells/well at 37°C for 2 weeks. Subsequently, the cells were fixed with 95% methanol for 30 min at room temperature and stained with 0.1% crystal violet for 15 min at room temperature, and the colonies were examined.
Cell apoptosis assay
Cell apoptosis was detected by flow cytometry analysis using a fluorescein isothiocyanate (FITC)-Annexin V/propidium iodide (PI) Apoptosis Detection kit (BD Pharmingen; BD Biosciences, San Jose, CA, USA). Briefly, the cells were collected and adjusted to the concentration of 1×105 cells/ml. A cell suspension of 200 µl was prepared for each sample, washed twice with cold PBS and resuspended in 100 µl binding buffer (Beyotime Institute of Biotechnology). Then, 5 µl Annexin V-FITC (20 µg/ml) and 5 µl PI (50 µg/ml) were added and the cells were incubated on ice in the dark for 15 min. Finally, 400 µl binding buffer was added and fluorescence determination of apoptotic cells was performed was performed using a FC500 flow cytometer equipped with Cell Quest 3.0 software (BD Biosciences). All analyses were performed in triplicate.
Scratch assay
Cells were seeded in 6-well plates at a density of 5×105 cells/well until 80% of the cells were attached to the plate wall, and then rinsed in PBS twice to remove any floating cells. Next, wounds were made in each well with sterile pipette tips. Subsequently, the cells were washed twice with PBS, and 2 ml RPMI-1640 culture medium containing 10% FBS was added to the culture plate. Images of the cells were captured at 24 and 48 h after wound generation to assess the cell migration ability.
Transwell invasion assay
Transwell assays were conducted to analyze the migration and invasion abilities of cells. Briefly, 4×104 cells in 100 µl RPMI-1640 culture medium supplemented with 10% FBS were placed in the upper chamber of Transwell plates (Corning Inc., Corning, NY, USA), which were pre-coated with Matrigel (growth factor reduced; BD Biosciences). Dulbecco's modified Eagle's medium (Beyotime Institute of Biotechnology) with 20% FBS was added to the lower chambers. After 24-h incubation, cells that had invaded across the membrane were fixed in 95% methanol at room temperature and stained with 0.1% crystal violet for 10 min at room temperature. Cells in nine randomly selected fields were counted under an inverted microscope (Carl Zeiss AG, Jena, Germany) at ×200 magnification, and the number of invading cells was expressed as the mean cell number in these fields.
Bioinformatics analysis
The TargetScan database 7.1 (http://www.targetscan.org/) was used to predict putative targets of miR-671-3p.
Luciferase reporter assay
The wild-type 3′-untranslated region (3′UTR) of FOXP2 gene was amplified using the following primers: Sense, 5′-GGTACCCTTTACAAACAGTTTTGACAG-3′, and antisense, 5′-AAGCTTTGGTGTGAATGATGACTGG-3′. The PCR products were cloned into the pGL3-basic vector using KpnI/HindIII sites to obtain the pGL3-FOXP2-3′UTR vector. A mutant version of this construct (pGL3-FOXP2-mut-3′UTR) carrying 4-bp substitutions in the miR-671-3p target sites was also obtained by site-directed mutagenesis using the QuikChange Site-Directed Mutagenesis kit (Agilent Technologies, Inc., Santa Clara, CA, USA). The following primers were used for amplifying the mutant 3′UTR: Fragment 1, forward 5′-CAGCGATCGCGAACTGACTTGTGAAACCTCAGCG-3′, reverse, 5′-CTCGCAGTTACTTCCAGTCCCTCAAAGCC-3′; and fragment 2, forward 5′-GTCTTTGGGTCATGATCAACGAACCGG-3′ and reverse, 5′-TATGTTTAAACTTTATAAATGGGTCAAAAAGAATTAGA-3′. A549 and H1975 cells were seeded at a density of 2×105 cells/well onto 24-well plates 12 h prior to the transfection, and then co-transfected with miR-671-3p mimics or negative control (NC) mimics along with the pGL3-FOXP2-3′UTR or pGL3-FOXP2-mut-3′UTR using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). After 36 h of transfection, the luciferase activity was measured with a Dual-Glo Luciferase Assay System (Promega Corporation, Madison, WI, USA) and normalized to Renilla luciferase.
Statistical analysis
All results are reported as the mean ± standard deviation, and were obtained using at least three independent replicates. All statistical analyses were performed using GraphPad software (version 5.0; GraphPad Software, Inc., La Jolla, CA, USA). Differences between two groups were calculated by Student's t-test. Differences among three or more groups were calculated by one-way analysis of variance test, followed by Tukey's multiple comparison test. Pearson correlation analysis was used to investigate the association between FOXP2 and miRNA-671-3p. A P-value of <0.05 was considered to denote a difference that was statistically significant.
Results
High expression of miR-671-3p in NSCLC tissues and cell lines
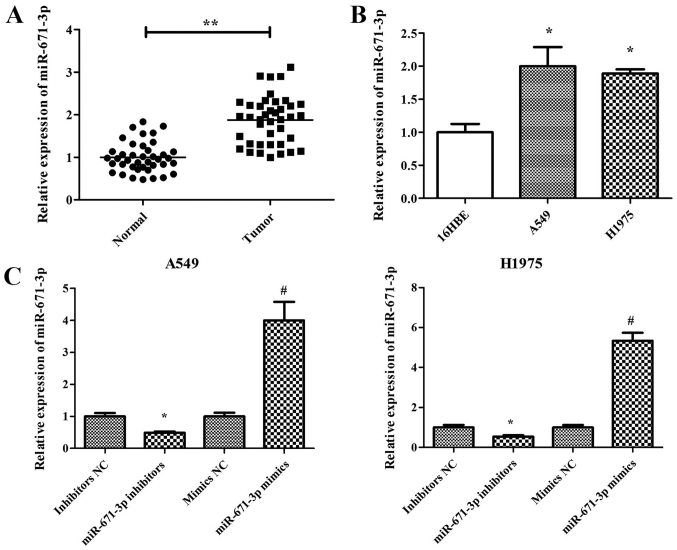
miR-671-3p has been reported to be a tumor suppressor in breast cancer (19). However, the effects and underlying mechanisms of miR-671-3p in lung cancer remain unknown. Therefore, the current study examined the expression pattern of miR-671-3p in NSCLC. miR-671-3p expression in 40 human NSCLC and adjacent normal lung tissues was measured by RT-qPCR. The results demonstrated that miR-671-3p expression in NSCLC tissues was significantly higher compared with that in adjacent normal tissues (P<0.05; Fig. 1A). Furthermore, miR-671-3p expression in two NSCLC cell lines (A549 and H1975) was measured, and the normal lung 16HBE cell line was used as the control. Similar to the results in NSCLC tissues, the expression level of miR-671-3p was significantly upregulated in A549 and H1975 cells compared with that in 16HBE cells (Fig. 1B). These findings suggested that miR-671-3p is upregulated in NSCLC tissues and cells.
Figure 1.
High expression of miR-671-3p in NSCLC tissues and cell lines. (A) Expression levels of miR-671-3p in NSCLC tissues and adjacent normal tissues (**P<0.01). (B) Expression levels of miR-671-3p in NSCLC and normal cell lines (*P<0.05 vs. normal 16HBE cells). (C) Expression levels of miR-671-3p in NSCLC cell lines transfected with miR-671-3p inhibitors, miR-671-3p mimics or the corresponding NC (*P<0.05 vs. inhibitors NC; #P<0.05 vs. mimics NC). Data are presented as the mean ± standard deviation of three independent experiments. miR, microRNA; NSCLC, non-small-cell lung cancer; NC, negative control.
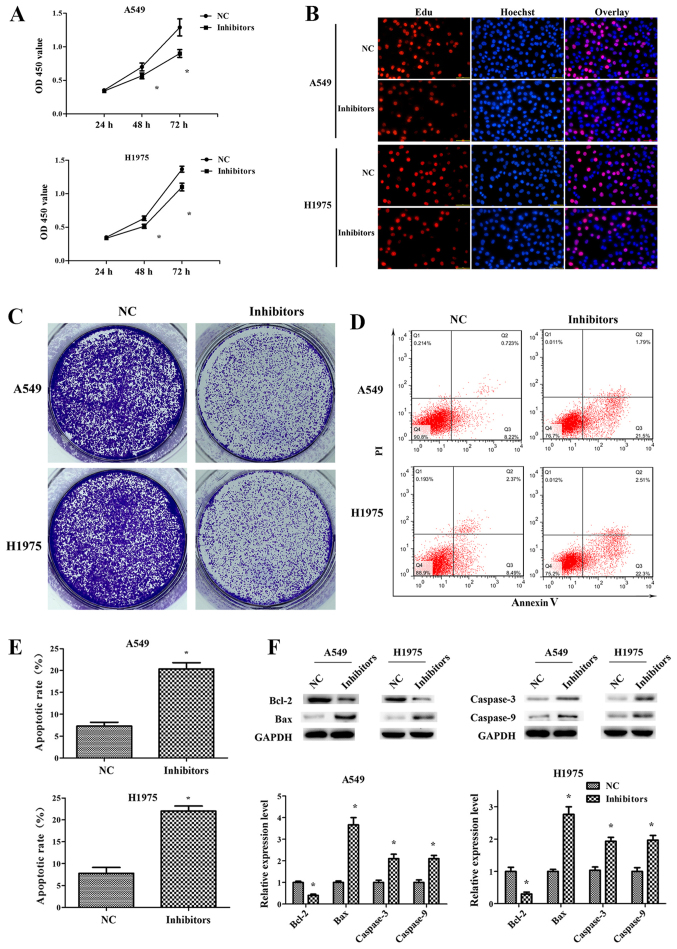
Downregulation of miR-671-3p inhibits NSCLC cell proliferation and induces apoptosis in vitro
Sustained proliferation is one of the hallmarks of cancer, and the fundamental basis of cancer dissemination and metastasis (20). In order to explore the role of miR-671-3p in NSCLC cell proliferation, miR-671-3p was knocked down in A549 and H1975 cells by transfection with miR-671-3p inhibitors. The RT-qPCR results confirmed that miR-671-3p was efficiently downregulated following transfection with miR-671-3p inhibitors, as compared with the NC group (Fig. 1C). Next, the CCK-8 assay results indicated that the proliferation capacity was decreased in A549 and H1975 cells transfected with miR-671-3p inhibitors (Fig. 2A). Similarly, EdU and colony formation assays were revealed reduced cell proliferation and colony formation in the inhibitor group compared with the NC group (Fig. 2B and C). In addition, fewer early and late apoptotic cells were detected in cells transfected with NC as compared with those transfected with miR-671-3p inhibitors, indicating that miR-671-3p downregulation promoted the apoptosis of A549 and H1975 cells (Fig. 2D and E). Furthermore, the levels of apoptosis-associated proteins were detected by western blotting. It was observed that the anti-apoptotic protein Bcl-2 was significantly downregulated in the miR-671-3p inhibitor group, whereas pro-apoptotic proteins Bax, caspase-3 and caspase-9 were markedly upregulated (Fig. 2F).
Figure 2.
Downregulation of miR-671-3p inhibits the proliferation and induces the apoptosis of non-small-cell lung cancer cells in vitro. A549 and H1975 cell lines were transfected with miR-671-3p inhibitors or NC, and then subjected to various assays. (A) Cell Counting Kit-8 and (B) EdU assays were performed to examine the cell proliferation (×100 magnification). (C) Colony formation assay. (D) Flow cytometry assay and (E) apoptosis rate. (F) Western blotting was used to determine the Bcl-2, Bax, caspase-3 and caspase-9 protein levels. All data are presented as the mean ± standard deviation, and each experiment was performed in triplicate. *P<0.05 vs. NC group. miR, microRNA; NC, negative control; Bcl-2, B-cell lymphoma 2; Bax, Bcl-2-associated X protein.
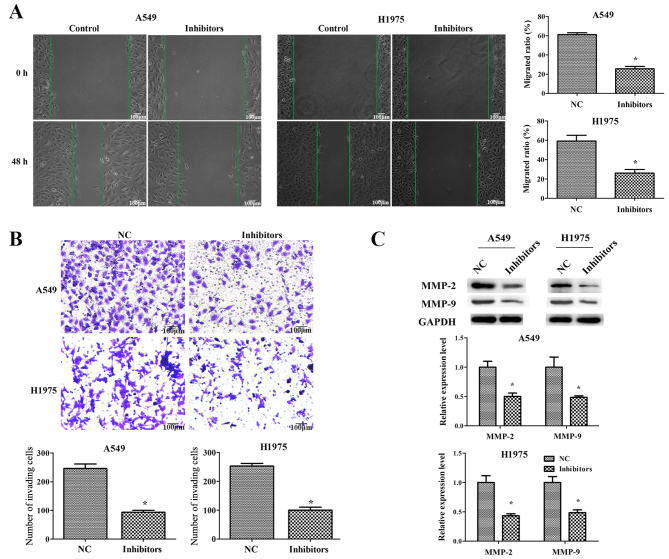
Downregulation of miR-671-3p suppresses NSCLC cell migration and invasion in vitro
To explore the function of miR-671-3p in NSCLC cell migration and invasion, scratch and transwell assays were performed. The results of the scratch assay demonstrated that there was significant difference in the migrated distance between the control and miR-671-3p inhibitor groups (P<0.05; Fig. 3A). As displayed in Fig. 3B, the transwell assay revealed that an increased number of cells had invaded to the lower chamber in the control group as compared with that in the miR-671-3p inhibitor group.
Figure 3.
Downregulation of miR-671-3p inhibits non-small-cell lung cancer cell migration and invasion in vitro. A549 and H1975 cells were transfected with miR-671-3p inhibitors or NC. (A) Cell migration was examined by scratch assay, scale bar 100 μm. (B) Cell invasion was examined by transwell assay (×100 magnification). (C) Western blotting for MMP-2 and MMP-9 protein level detection. All data are presented as the mean ± standard deviation, and each experiment was performed in triplicate. *P<0.05 vs. NC group. miR, microRNA; NC, negative control; MMP, matrix metalloproteinase.
Matrix metalloproteinases (MMPs) have an important role in cancer cell migration and invasion by degrading the extracellular matrix (21,22). Therefore, western blotting was then performed to detect the expression levels of MMP-2 and MMP-9. As shown in Fig. 3C, MMP-2 and MMP-9 were significantly decreased in A549 and H1975 cells transfected with miR-671-3p inhibitors. Taken together, these results indicated that miR-671-3p downregulation inhibited NSCLC cell migration and invasion in vitro.
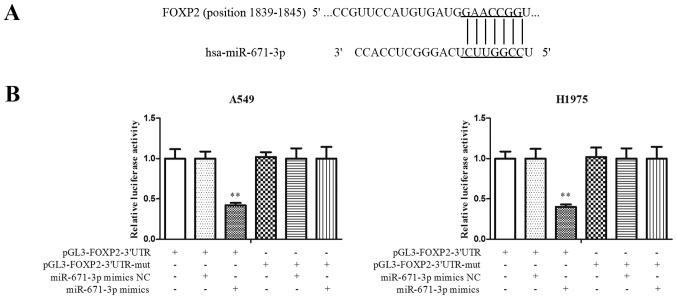
miR-671-3p directly targets FOXP2 in NSCLC
To explore the molecular mechanisms by which miR-671-3p promoted tumor cell progression, a search for putative miR-671-3p targets was conducted using TargetScan software. FOXP2 was predicted to be a potential candidate target of miR-671-3p (Fig. 4A). A luciferase reporter assay was then performed to detect the interaction between FOXP2 and miR-671-3p. As shown in Fig. 4B, overexpression of miR-671-3p decreased the luciferase signal in FOXP2-3′UTR cells by ~60% (P<0.05). However, no significant differences were identified between the miR-671-3p mimics and mimics NC groups in cells transfected with FOXP2-3′UTR-mut. These results indicated that miR-671-3p targeted the 3′UTR of FOXP2 directly in NSCLC.
Figure 4.
miR-671-3p directly targets FOXP2 in non-small-cell lung cancer. (A) Predicted binding sites in the 3′UTR of FOXP2 mRNA and seed sequence of miR-671-3p. (B) Dual luciferase report assay, conducted to confirm whether miR-671-3p targets FOXP2. All data were presented as the mean ± standard deviation and each was performed in triplicate. **P<0.01 vs. pGL3-FOXP2-3′UTR. miR, microRNA; FOXP2, forkhead box P2; 3′UTR, 3′-untranslated region; mut, mutant; NC, negative control.
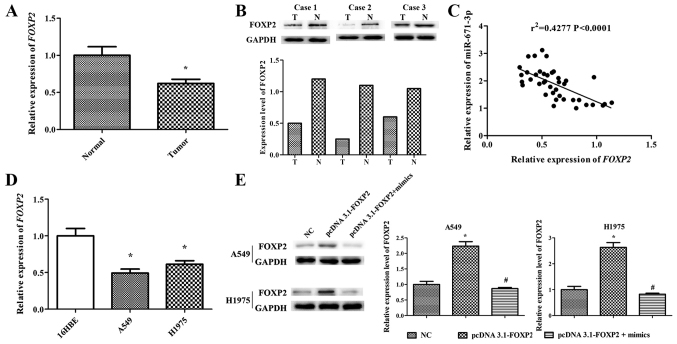
miR-671-3p expression is negatively correlated with FOXP2 in NSCLC tissues and cell lines
RT-qPCR and western blotting were performed to determine the mRNA and protein levels of FOXP2 in 40 human NSCLC lung tissues. The results indicated that FOXP2 expression in NSCLC lung tissues was markedly lower compared with that in adjacent normal lung tissues (P<0.05; Fig. 5A and B). In order to further explore the association between the FOXP2 and miR-671-3p expression levels, Pearson correlation analysis was conducted, and the results demonstrated that miR-671-3p expression was negatively correlated with FOXP2 expression in NSCLC tissues (r2=0.4277, P<0.0001; Fig. 5C). Furthermore, NSCLC cell lines (A549 and H1975) exhibited significantly lower FOXP2 expression in comparison with that in the normal lung 16HBE cell line (Fig. 5D).
Figure 5.
miR-671-3p expression is negatively correlated with FOXP2 expression in NSCLC tissues and cell lines. (A) FOXP2 mRNA expression level in NSCLC tissues. (B) Western blotting was used to determine FOXP2 protein expression in NSCLC tissues. (C) Pearson correlation analysis between miR-671-3p level and FOXP2 mRNA expression. (D) FOXP2 mRNA expression in A549 and H1975 cell lines. (E) Western blotting was applied to detect FOXP2 protein levels in A549 and H1975 cell lines transfected with pcDNA3.1-FOXP2 or NC. All data are presented as the mean ± standard deviation, and each experiment was performed in triplicate. *P<0.05 vs. corresponding control group; #P<0.05 vs. pcDNA3.1-FOXP2 group. miR, microRNA; FOXP2, forkhead box P2; NSCLC, non-small-cell lung cancer; T, tumor tissue; N, normal tissue; NC, negative control.
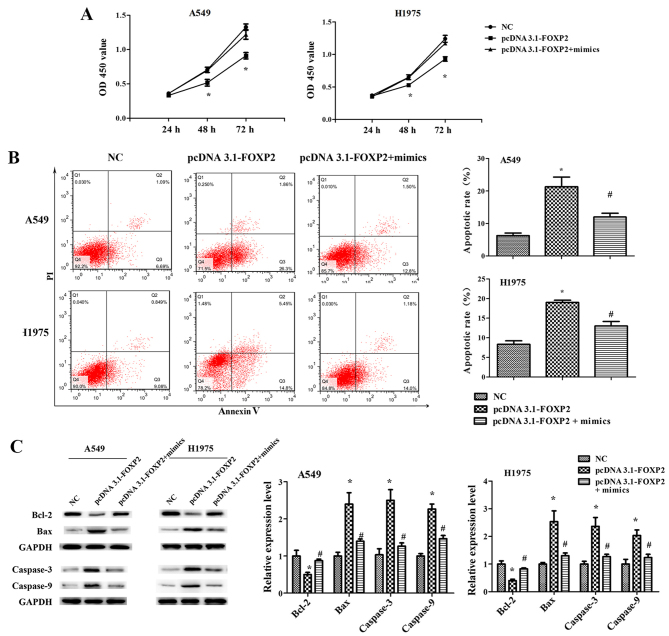
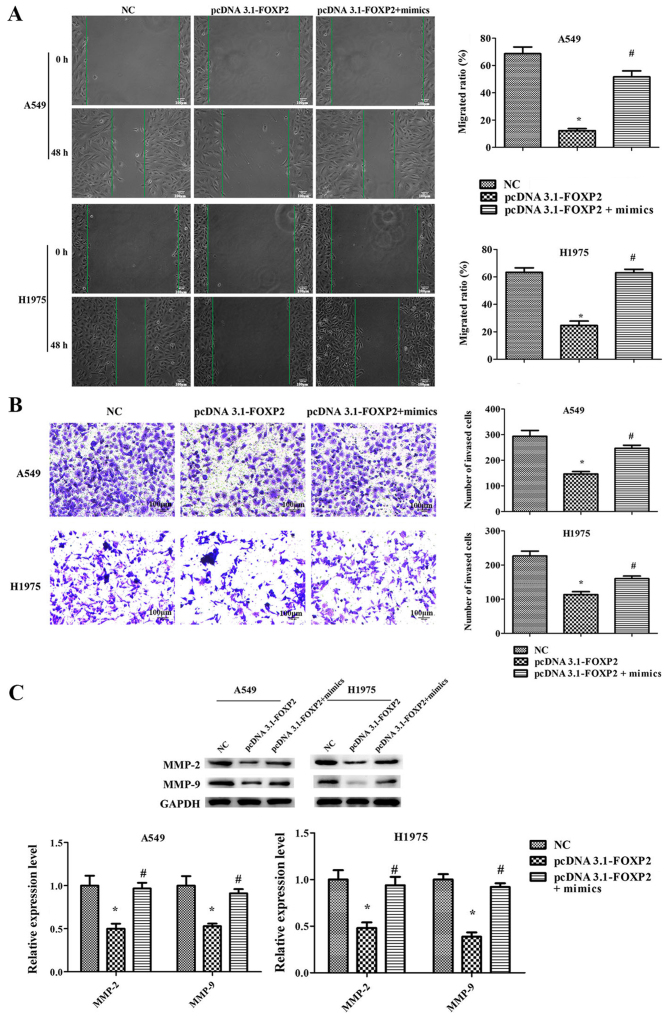
miR-671-3p promotes NSCLC cell proliferation, migration and invasion, and suppresses apoptosis by downregulating FOXP2 in vitro
To explore the role of FOXP2 in NSCLC cell progression, the pcDNA3.1-FOXP2 vector was constructed. FOXP2 expression level in the pcDNA3.1-FOXP2 cell group was ~2.5 times that in NC cells. However, transfection with miR-671-3p mimics significantly downregulated FOXP2 expression in pcDNA3.1-FOXP2 cells (Fig. 5E).
Furthermore, it was observed that FOXP2 overexpression significantly suppressed the proliferation, migration and invasion ability of A549 and H1975 cells, while it induced cell apoptosis (Figs. 6 and 7). However, the effects of pcDNA3.1-FOXP2 on cell proliferation, apoptosis, migration and invasion were reversed in the presence of miR-671-3p mimics (Figs. 6 and 7). These findings implied that miR-671-3p promotes NSCLC cell proliferation, migration and invasion, and suppresses apoptosis by downregulating FOXP2 in vitro.
Figure 6.
miR-671-3p promotes NSCLC cell proliferation and suppresses apoptosis by downregulating FOXP2 in vitro. A549 and H1975 cell lines transfected with NC or pcDNA3.1-FOXP2 or pcDNA3.1-FOXP2 and miR-671-3p mimics. (A) Cell Counting Kit-8 assay. (B) Flow cytometry apoptosis assay. (C) Western blotting for detection of Bcl-2, Bax, caspase-3 and caspase-9 protein levels. All data are presented as the mean ± standard deviation, and each experiment was performed in triplicate. *P<0.05 vs. NC group; #P<0.05 vs. pcDNA3.1-FOXP2 group. miR, microRNA; FOXP2, forkhead box P2; NSCLC, non-small-cell lung cancer; NC, negative control; Bcl-2, B-cell lymphoma 2; Bax, Bcl-2-associated X protein.
Figure 7.
miR-671-3p promotes NSCLC cell proliferation, migration and invasion, and suppresses apoptosis by downregulating FOXP2 in vitro. A549 and H1975 cell lines transfected with NC or pcDNA3.1-FOXP2 or pcDNA3.1-FOXP2 and miR-671-3p mimics. (A) Scratch migration assay, scale bar 100 μm. (B) Transwell invasion assay (×100 magnification). (C) Western blotting for detection of MMP-2 and MMP-9 protein levels. All data are presented as the mean ± standard deviation, and each experiment was performed in triplicate. *P<0.05 vs. NC group; #P<0.05 vs. pcDNA3.1-FOXP2 group. miR, microRNA; FOXP2, forkhead box P2; NSCLC, non-small-cell lung cancer; NC, negative control; MMP, matrix metalloproteinase.
Discussion
It is known that dysregulation of specific miRNAs can be involved in the initiation and progression of cancer (23). Various miRNAs have been reported to function as oncogenes or tumor suppressors in NSCLC by regulating the expression of downstream target genes (24,25). A previous study revealed that miR-671-5p served tumor suppressor roles in breast cancer; overexpression of miR-671-5p attenuated the proliferation and invasion of breast cancer cells by targeting FOXM1 (26). To the best of our knowledge, this is the first study to investigate the role of miR-671-3p in NSCLC. In the present study, the expression of miR-671-3p was increased in NSCLC tissues and cell lines (A549 and H1975), as compared with normal lung tissue and cells, respectively. To investigate the potential oncogenic roles of miR-671-3p in NSCLC cells, cells were transfected with miR-671-3p inhibitors. Inhibition of miR-671-3p was reported to suppress the proliferation, migration and invasion, and promote the apoptosis of A549 and H1975 cells.
FOXP2 is a member of the FOX family of proteins, which are involved in a number of important biological processes including cell metabolism, development, differentiation, proliferation, apoptosis, migration and invasion (27,28). FOXP2 was the first characterized FOX family member that was relevant to the human ability to develop speech (29,30). In recent years, FOXP2 has been reported to serve an important role in cancer (31). FOXP2 was found to be overexpressed in lymphoma cells (32), while low expression of FOXP2 was correlated with poor survival in hepatocellular carcinoma (33). In addition, Cuiffo et al (16) reported that elevated miR-199a levels and suppressed FOXP2 expression levels are prominent features of malignant clinical breast cancer. Furthermore, downregulation of FOXP2 expression was observed in gastric cancer cells and tissues, while miR-190 was observed to regulate the pathological process of gastric cancer through the FOXP2 pathway (15). Another study indicated that miR-618 inhibited prostate cancer migration and invasion by targeting FOXP2 (34). The role of FOXP2 in cancer development has also been reported in several other studies (35–37). However, to the best of our knowledge, the current study is the first to report on the expression of FOXP2 and its direct targeting by miR-671-3p in NSCLC.
In the present study, downregulation of FOXP2 was observed in NSCLC tissues and cells. Bioinformatics and luciferase assays revealed that FOXP2 was direct target gene of miR-671-3p in NSCLC cells. RTq-PCR and western blotting demonstrated that overexpression of miR-671-3p reduced the expression of FOXP2 at the mRNA and protein levels. Furthermore, it was observed that the overexpression of FOXP2 inhibited the proliferation, migration and invasion, and promoted the apoptosis of A549 and H1975 cells. Conversely, co-transfection with miR-671-3p mimics attenuated the effects of pcDNA3.1-FOXP2. The results indicated that FOXP2 is a downstream target, and a functional mediator of the effects of miR-671-3p in NSCLC.
In conclusion, the present study revealed that miR-671-3p improved the proliferation, migration and invasion of NSCLC cells, as well as suppressed cell apoptosis, by regulating FOXP2 expression. These findings suggested that miR-671-3p functions as an oncogene and may be a potential therapeutic target in NSCLC.
Acknowledgements
Not applicable.
Funding
No funding was recieved.
Availability of data and materials
The datasets used during the current study are available from the corresponding author on reasonable request.
Authors' contributions
ZYL and ZZZ were major contributors in sample collection, the conduction of experiments, data interpretation and manuscript preparation. HB, QDZ, SJZ, LZ and XQZ participated in sample collection, conduction of experiments and data interpretation. JZ was responsible for experimental design and revised this manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Informed consent was obtained from each patient, and this study was approved by the Ethics Committee of the First People's Hospital of Changzhou, China.
Patient consent for publication
Not applicable.
Competing interests
The authors declare no conflict of interest.
References
- 1.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang Y, Yang Q, Wang S. MicroRNAs: A new key in lung cancer. Cancer Chemother Pharmacol. 2014;74:1105–1111. doi: 10.1007/s00280-014-2559-9. [DOI] [PubMed] [Google Scholar]
- 3.Yin R, Guo L, Zhang W, Zheng J. The pleiotropic effects of mirnas on tumor angiogenesis. J Cell Biochem. 2015;116:1807–1815. doi: 10.1002/jcb.24679. [DOI] [PubMed] [Google Scholar]
- 4.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 5.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 6.Hess KR, Varadhachary GR, Taylor SH, Wei W, Raber MN, Lenzi R, Abbruzzese JL. Metastatic patterns in adenocarcinoma. Cancer. 2006;106:1624–1633. doi: 10.1002/cncr.21778. [DOI] [PubMed] [Google Scholar]
- 7.McIntyre A, Ganti AK. Lung cancer-A global perspective. J Surg Oncol. 2017;115:550–554. doi: 10.1002/jso.24532. [DOI] [PubMed] [Google Scholar]
- 8.Cho WC. Role of miRNAs in lung cancer. Expert Rev Mol Diagn. 2009;9:773–776. doi: 10.1586/erm.09.57. [DOI] [PubMed] [Google Scholar]
- 9.Hashemi ZS, Khalili S, Forouzandeh Moghadam M, Sadroddiny E. Lung cancer and miRNAs: A possible remedy for anti-metastatic, therapeutic and diagnostic applications. Expert Rev Respir Med. 2017;11:147–157. doi: 10.1080/17476348.2017.1279403. [DOI] [PubMed] [Google Scholar]
- 10.Rusek AM, Abba M, Eljaszewicz A, Moniuszko M, Niklinski J, Allgayer H. MicroRNA modulators of epigenetic regulation, the tumor microenvironment and the immune system in lung cancer. Mol Cancer. 2015;14:34. doi: 10.1186/s12943-015-0302-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson CD, Esquela-Kerscher A, Stefani G, Byrom M, Kelnar K, Ovcharenko D, Wilson M, Wang X, Shelton J, Shingara J, et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007;67:7713–7722. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- 12.Ntoumou E, Tzetis M, Braoudaki M, Lambrou G, Poulou M, Malizos K, Stefanou N, Anastasopoulou L, Tsezou A. Serum microRNA array analysis identifies miR-140-3p, miR-33b-3p and miR-671-3p as potential osteoarthritis biomarkers involved in metabolic processes. Clin Epigenetics. 2017;9:127. doi: 10.1186/s13148-017-0428-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiong DD, Chen H, He RQ, Lan AH, Zhong JC, Chen G, Feng ZB, Wei KL. MicroRNA-671-3p inhibits the development of breast cancer: A study based on in vitro experiments, in-house quantitative polymerase chain reaction and bioinformatics analysis. Int J Oncol. 2018;52:1801–1814. doi: 10.3892/ijo.2018.4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen MT, Sun HF, Li LD, Zhao Y, Yang LP, Gao SP, Jin W. Downregulation of FOXP2 promotes breast cancer migration and invasion through TGFβ/SMAD signaling pathway. Oncol Lett. 2018;15:8582–8588. doi: 10.3892/ol.2018.8402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jia WZ, Yu T, An Q, Yang H, Zhang Z, Liu X, Xiao G. MicroRNA-190 regulates FOXP2 genes in human gastric cancer. Onco Targets Ther. 2016;9:3643–3651. doi: 10.2147/OTT.S103682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cuiffo BG, Campagne A, Bell GW, Lembo A, Orso F, Lien EC, Bhasin MK, Raimo M, Hanson SE, Marusyk A, et al. MSC-regulated microRNAs converge on the transcription factor FOXP2 and promote breast cancer metastasis. Cell Stem Cell. 2014;15:762–774. doi: 10.1016/j.stem.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 17.Yu Z, Lin X, Tian M, Chang W. microRNA-196b promotes cell migration and invasion by targeting FOXP2 in hepatocellular carcinoma. Oncol Rep. 2018;39:731–738. doi: 10.3892/or.2017.6130. [DOI] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Chen X, Lu P, Wang DD, Yang SJ, Wu Y, Shen HY, Zhong SL, Zhao JH, Tang JH. The role of miRNAs in drug resistance and prognosis of breast cancer formalin-fixed paraffin-embedded tissues. Gene. 2016;595:221–226. doi: 10.1016/j.gene.2016.10.015. [DOI] [PubMed] [Google Scholar]
- 20.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 21.Jiménez MJ, Balbin M, Alvarez J, Komori T, Bianco P, Holmbeck K, Birkedal-Hansen H, López JM, López-Otín C. A regulatory cascade involving retinoic acid, Cbfa1, and matrix metalloproteinases is coupled to the development of a process of perichondrial invasion and osteogenic differentiation during bone formation. J Cell Biol. 2001;155:1333–1344. doi: 10.1083/jcb.200106147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khokha R, Denhardt DT. Matrix metalloproteinases and tissue inhibitor of metalloproteinases: A review of their role in tumorigenesis and tissue invasion. Invasion Metastasis. 1989;9:391–405. [PubMed] [Google Scholar]
- 23.Tan W, Liu B, Qu S, Liang G, Luo W, Gong C. MicroRNAs and cancer: Key paradigms in molecular therapy. Oncol Lett. 2018;15:2735–2742. doi: 10.3892/ol.2017.7638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang Y, Ding L, Hu Q, Xia J, Sun J, Wang X, Xiong H, Gurbani D, Li L, Liu Y, Liu A. MicroRNA-218 functions as a tumor suppressor in lung cancer by targeting IL-6/STAT3 and negatively correlates with poor prognosis. Mol Cancer. 2017;16:141. doi: 10.1186/s12943-017-0710-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang Y, Li H, Liu Y, Chi C, Ni J, Lin X. miR-4319 hinders YAP expression to restrain non-small cell lung cancer growth through regulation of LIN28-mediated RFX5 stability. Biomed Pharmacother. 2019;115:108956. doi: 10.1016/j.biopha.2019.108956. [DOI] [PubMed] [Google Scholar]
- 26.Tan X, Fu Y, Chen L, Lee W, Lai Y Rezaei K, Tabbara S, Latham P, Teal CB, Man YG, et al. miR-671-5p inhibits epithelial-to-mesenchymal transition by downregulating FOXM1 expression in breast cancer. Oncotarget. 2016;7:293–307. doi: 10.18632/oncotarget.6344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chiu YC, Li MY, Liu YH, Ding JY, Yu JY, Wang TW. Foxp2 regulates neuronal differentiation and neuronal subtype specification. Dev Neurobiol. 2014;74:723–738. doi: 10.1002/dneu.22166. [DOI] [PubMed] [Google Scholar]
- 28.Katoh M, Katoh M. Human FOX gene family (Review) Int J Oncol. 2004;25:1495–1500. [PubMed] [Google Scholar]
- 29.Tsui D, Vessey JP, Tomita H, Kaplan DR, Miller FD. FoxP2 regulates neurogenesis during embryonic cortical development. J Neurosci. 2013;33:244–258. doi: 10.1523/JNEUROSCI.1665-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Enard W, Przeworski M, Fisher SE, Lai CS, Wiebe V, Kitano T, Monaco AP, Pääbo S. Molecular evolution of FOXP2, a gene involved in speech and language. Nature. 2002;418:869–872. doi: 10.1038/nature01025. [DOI] [PubMed] [Google Scholar]
- 31.Herrero MJ, Gitton Y. The untold stories of the speech gene, the FOXP2 cancer gene. Genes Cancer. 2018;9:11–38. doi: 10.18632/genesandcancer.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Campbell AJ, Lyne L, Brown PJ, Launchbury RJ, Bignone P, Chi J, Roncador G, Lawrie CH, Gatter KC, Kusec R, Banham AH. Aberrant expression of the neuronal transcription factor FOXP2 in neoplastic plasma cells. Br J Haematol. 2010;149:221–230. doi: 10.1111/j.1365-2141.2009.08070.x. [DOI] [PubMed] [Google Scholar]
- 33.Yan X, Zhou H, Zhang T, Xu P, Zhang S, Huang W, Yang L, Gu X, Ni R, Zhang T. Downregulation of FOXP2 promoter human hepatocellular carcinoma cell invasion. Tumour Biol. 2015;36:9611–9619. doi: 10.1007/s13277-015-3701-y. [DOI] [PubMed] [Google Scholar]
- 34.Song XL, Tang Y, Lei XH, Zhao SC, Wu ZQ. miR-618 inhibits prostate cancer migration and invasion by targeting FOXP2. J Cancer. 2017;8:2501–2510. doi: 10.7150/jca.17407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen MT, Sun HF, Li LD, Zhao Y, Yang LP, Gao SP, Jin W. Downregulation of FOXP2 promotes breast cancer migration and invasion through TGFβ/SMAD signaling pathway. Oncol Lett. 2018;15:8582–8588. doi: 10.3892/ol.2018.8402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cuiffo BG, Karnoub AE. Silencing FOXP2 in breast cancer cells promotes cancer stem cell traits and metastasis. Mol Cell Oncol. 2015;3:e1019022. doi: 10.1080/23723556.2015.1019022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu J, Liu P, Tang H, Shuang Z, Qiu Q, Zhang L, Song C, Liu L, Xie X, Xiao X. FOXP2 Promotes tumor proliferation and metastasis by targeting GRP78 in triple-negative breast cancer. Curr Cancer Drug Targets. 2018;18:382–389. doi: 10.2174/1568009618666180131115356. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used during the current study are available from the corresponding author on reasonable request.