ABSTRACT
Microbial conversion of biomass to fatty acids (FA) and products derived thereof is an attractive alternative to the traditional oleochemical production route from animal and plant lipids. This study examined if NADPH-costly FA biosynthesis could be enhanced by overexpressing the transcription factor Stb5 in Saccharomyces cerevisiae. Stb5 activates expression of multiple genes encoding enzymes within the pentose phosphate pathway (PPP) and other NADPH-producing reactions. Overexpression of STB5 led to a decreased growth rate and an increased free fatty acid (FFA) production during growth on glucose. The improved FFA synthetic ability in the glucose phase was shown to be independent of flux through the oxidative PPP. RNAseq analysis revealed that STB5 overexpression had wide-ranging effects on the transcriptome in the batch phase, and appeared to cause a counterintuitive phenotype with reduced flux through the oxidative PPP. During glucose limitation, when an increased NADPH supply is likely less harmful, an overall induction of the proposed target genes of Stb5 (eg. GND1/2, TAL1, ALD6, YEF1) was observed. Taken together, the strategy of utilizing STB5 overexpression to increase NADPH supply for reductive biosynthesis is suggested to have potential in strains engineered to have strong ability to consume excess NADPH, alleviating a potential redox imbalance.
Keywords: Saccharomyces cerevisiae, STB5, transcription factor, NADPH, fatty acids, metabolic engineering
STB5 —encoding a transcriptional activator of genes related to NADPH biosynthesis — was overexpressed in an attempt to enhance FA biosynthesis and the resulting effects were studied at different growth conditions.
INTRODUCTION
The transformation of the current economy towards a biobased one is dependent on versatile technologies able to convert renewable feedstocks into chemical products and fuels. Within the field of metabolic engineering advances have been made to positively stimulate microbial biosynthesis of fatty acids (FA), the building blocks of oleochemicals (Pfleger, Gossing and Nielsen 2015; Marella et al. 2018). Oleochemicals represent a group of compounds with a diverse application range, for example in cosmetics and detergents as well as in industrial lubricants and transportation fuels. Currently, oleochemicals are predominantly produced from the FA molecules present in lipids such as vegetable oils — a global market which grew 4.5% annually the past decade due to the increased demand for biodiesel (OECD/FAO 2018). Microbial production of oleochemicals and biodiesel has distinct advantages compared to the traditional route based on plant or animal fats. A great variety of feedstocks can be utilized, including biomass with a high crop yield, hence resulting in a lower overall carbon footprint (Fargione et al. 2008). Furthermore, a microbe can be engineered to catalyze multiple steps of the conversion processes required to produce the sought-after product, from substrate degradation pathways to control of FA chain length and/or degree of saturation.
One of the most attractive hosts for metabolic engineering and industrial applications is the yeast Saccharomyces cerevisiae. Naturally, S. cerevisiae produces low quantities of lipids, but during the last decade it has been widely explored as a cell factory for FA and oleochemical synthesis (Zhang, Nielsen and Liu 2018). De novo FA synthesis is a cyclic cytosolic process, where acetyl-CoA is used as substrate to produce malonyl-CoA, which subsequently elongates FA chains by two carbons at a time, ultimately releasing fatty acyl-CoA chains. Engineering strategies to improve FA biosynthesis have generally focused on increasing the acetyl-CoA supply (de Jong et al. 2014; Zhou et al. 2016), de-regulating key-enzymes in the pathway (Shi et al. 2014; Ferreira et al. 2018a), preventing acyl-CoA mediated inhibition and/or FA degradation (Leber et al. 2014; Ferreira et al. 2018a) and overexpressing downstream pathways further converting the produced acyl-CoA or free fatty acid (FFA) (Ferreira et al. 2018a). FA biosynthesis consumes 2 NADPH per molecule of malonyl-CoA entering the FA elongation cycle after an initial acetyl-CoA priming reaction (Tehlivets, Scheuringer and Kohlwein 2007), resulting in a cost of 16 NADPH for one molecule of octadecanoic acid (C18). A recent study presented a rigorous reprogramming effort to turn S. cerevisiae into a lipogenic yeast, which partly included to re-route flux through the oxidative pentose phosphate pathway (PPP) for an increased NADPH generation (Yu et al. 2018). Besides the mentioned study, increasing NADPH supply remains a largely unexplored avenue to influence FA synthesis in S. cerevisiae.
The oxidative PPP is a major route for cytosolic NADPH generation in S. cerevisiae. It generates two NADPH per molecule of glucose consumed through the action of the two dehydrogenases, glucose-6-phosphate (G6P) dehydrogenase (Zwf1) and 6-phosphogluconate dehydrogenase (Gnd1/Gnd2) (Stincone et al. 2015). Other cytosolic routes leading to NADPH formation include the acetaldehyde dehydrogenase (Ald6), important for growth on glucose, and cytosolic isocitrate dehydrogenase (Idp2), which is essential for growth on acetate and oleate if ZWF1 is deleted (Minard and McAlister-Henn 2005). Mitochondrial NADPH supply stems from malic enzyme (Mae1), a NAD(H) kinase (Pos5) and mitochondrial isoforms of acetaldehyde dehydrogenase (Ald4/Ald5) and isocitrate dehydrogenase (Idp1) (Miyagi, Kawai and Murata 2009). Even though the mitochondrial membrane is not directly permeable for the cofactor, mitochondrial export of citrate and uptake of oxoglutarate catalyzed by the transporter Yhm2 is suggested to cause a net export of reducing equivalents to the cytosol, demonstrated by the reduced cytosolic NADPH/NADP+ ratio observed in a yhm2∆ mutant (Castegna et al. 2010). The NADPH/NADP+ ratio is believed to be more tightly regulated than the NADH/NAD+ ratio. For example, overexpression of cytosolic Pyc2 and NADPH-producing Mae1, either targeted to the mitochondria or the cytosol, did not generate a measurable difference in NADPH/NADP+ ratio while NADH/NAD+ levels decreased and the flux through the oxidative PPP was reduced, indicating a lower cytosolic need for NADPH (Moreira dos Santos et al. 2004).
As FA synthesis is carried out in the cytoplasm, the obvious native targets to increase NADPH supply are the cytosolic dehydrogenases Zwf1, Gnd1/2, Ald6 and Idp2. Of these, Ald6 is a part of the native enzymatic route generating FA from glucose, and overexpression has been attempted in combination with other modifications, mainly to increase flux towards acetyl-CoA and metabolites derived thereof (Shiba et al. 2007; Chen et al. 2013; Krivoruchko et al. 2013; de Jong et al. 2014). Idp2 is mainly responsible for NADPH generation during growth on non-fermentable carbon sources and was, together with Yhm2, one of the chosen targets to improve NADPH-supply by Yu et al. for FFA production (Yu et al. 2018). Redirection of flux through the oxidative PPP should in theory have a stronger effect on NADPH generation, as two molecules of NADPH are formed per G6P entering the pathway. However, flux through the G6P node is mainly believed to be determined by the concentrations of G6P and NADPH, suggesting that an increase in Zwf1 level may not have a great influence on the flux distribution (Stephanopoulos and Vallino 1991; Gombert et al. 2001). NADPH inhibits Zwf1 activity in crude cell free extracts (Llobell et al. 1988), which indicates that the NADPH concentration or the NADPH/NADP+ ratio dictates the flux entering the oxidative PPP. This was recently shown to be the case in Escherichia coli (Christodoulou et al. 2018). In yeast, overexpression of YNO1 encoding an NADPH oxidase caused a 10-fold increase in superoxide levels, while no such increase was present in a zwf1∆ mutant, indicating that flux through the oxidative PPP flux dynamically increases in response to NADPH demand (Rinnerthaler et al. 2012). If simultaneously considering the strong thermodynamic forces driving flux through glycolysis, it is apparent that a major intervention is required to efficiently redirect flux through the oxidative PPP.
Even though metabolite levels and post-transcriptional effects are likely to influence oxidative PPP activity (Wang et al. 2014), it has also been suggested to be regulated on a transcriptional level. The gene STB5 encodes a transcription factor (TF) reported to regulate multidrug resistance and oxidative stress response (Larochelle et al. 2006; Ouyang et al. 2018). In response to diamide treatment, Stb5 was shown to act as transcriptional activator of multiple genes within the PPP, including ZWF1, SOL3, GND1, GND2, RKI1, TAL1 and TKL1, while it is believed to repress the glycolytic gene PGI1, whose gene product competes with Zwf1 for the substrate G6P (Larochelle et al. 2006). In addition, STB5 was found to positively influence the expression of several other genes related to NADPH generation, such as ALD6, IDP2, ILV5, ALD4, GOR1 and YEF1 (Larochelle et al. 2006).This suggests that an increased flux through the PPP as well an increased pool of NADPH potentially could be achieved simply by enhancing the activity of STB5. Indeed, overexpression of STB5 from a low-copy plasmid increased NADPH levels 2.5 times in BY4727 background (Hector et al. 2009). Strong overexpression of STB5 prevented strain growth on glucose, but this growth defect was partly relieved when overexpressing a NADPH-consuming butanediol dehydrogenase, indicating that STB5 overexpression led to a redox imbalance (Cadiere, Galeote and Dequin 2010).
In this study, we evaluated if promoter replacement of STB5 could be used as a tool to positively influence FA synthesis, with the hypothesis that it potentially could activate several NADPH-producing reactions simultaneously while downregulating flux through upper glycolysis. In Fig. 1, an overview of central carbon metabolism and some of the key targets of Stb5 anticipated to be perturbed are shown. If successful this strategy could replace the requirement for promoter replacement of multiple genes, e.g. fine-tuned downregulation of PGI1 and overexpression of ZWF1, GND1, TAL1 and TKL1 that resulted in a 28% improvement in FA production (Yu et al. 2018).
Figure 1.
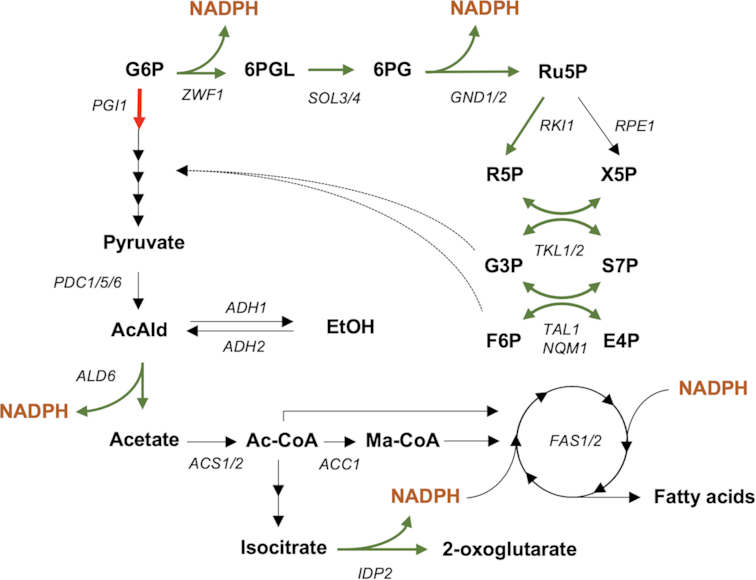
Overview of central carbon metabolism of S. cerevisiae and key targets of the TF Stb5. Positively and negatively regulated reactions are highlighted in green and red, respectively.
MATERIALS AND METHODS
Strains and plasmids
Investigated yeast strains were constructed from the laboratory yeast strain CEN.PK 113–5D. Table 1 shows name, genotype and origin of all strains used and constructed under the course of the study.
Table 1.
Strains and plasmids used and constructed within this study.
| Strain | Genotype | Origin |
|---|---|---|
| CEN.PK 113–5D | MATa MAL2–8c SUC2 ura3–52 | P. Kötter, University of Frankfurt, Germany |
| AB11 | MATa MAL2–8c SUC2 ura3–52 faa1∆ faa4∆ | This study |
| AB12 | CEN.PK 113–5D PSTB5Δ::PCUP1 | This study |
| AB13 | CEN.PK 113–5D PSTB5Δ::PCYC1 | This study |
| AB14 | CEN.PK 113–5D PSTB5Δ::PADH1 | This study |
| AB15 | CEN.PK 113–5D PSTB5Δ::PTPI1 | This study |
| AB16 | CEN.PK 113–5D PSTB5Δ::PPGK1 | This study |
| AB17 | AB11 PSTB5Δ::PCYC1 | This study |
| AB18 | AB11 PSTB5Δ::PADH1 | This study |
| AB19 | AB11PSTB5Δ::PTPI1 | This study |
| AB20 | AB11 PSTB5Δ::PPGK1 | This study |
| AB21 | AB11 zwf1∆ | This study |
| AB22 | AB19 zwf1∆ | This study |
| AB23 | AB11 gpp1∆ | This study |
| AB24 | AB19 gpp1∆ | This study |
| AB25 (see Table 2) | AB11 pSP-GM1 | This study |
| AB26 (see Table 2) | AB11 pAB10 | This study |
| AB27 (see Table 2) | AB23 pSP-GM1 | This study |
| AB28 (see Table 2) | AB23 pAB10 | This study |
| AB29 (see Table 2) | AB24 pSP-GM1 | This study |
| AB30 (see Table 2) | AB24 pAB10 | This study |
| AB31 | AB15 zwf1∆ | This study |
Culture media
Strains were routinely grown in yeast extract peptone dextrose (YPD) medium or plates (10 g L−1 yeast extract, 20 g L−1 peptone and 20 g L−1 glucose, additional 20 g L−1 agar for plates), and when appropriate YPD plates containing 200 mg L−1 of the antibiotic G418 sulphate. When transformed with plasmids, strains were grown on SD-URA dropout plates (6.9 g L−1 yeast nitrogen base w/o amino acids (Formedium), 0.77 g L−1 complete supplement mixture w/o uracil (Formedium), 20 g L−1 glucose and 20 g L−1 agar).
Shake flask and BioLector cultivations were performed in minimal medium containing 7.5 g L−1 (NH4)2SO4, 14.4 g L−1 KH2PO4, 0.5 g L−1 MgSO4·7H2O and 20 g L−1 glucose, with the pH adjusted to 6.5. Sterile solutions of glucose, 1 mL L−1 vitamins and 2 mL L−1 trace metal solution (described previously (Verduyn et al. 1992)) were added after autoclavation. 60 mg L−1 uracil was added to the media if the strains did not carry a plasmid. In BioLector cultivations, a 100 mM CuSO4 solution was used to obtain cultivation media with final concentrations of 100, 200 or 400 µM Cu2+.
Bioreactor cultivations were conducted in minimal medium containing 5 g L−1 (NH4)2SO4, 3 g L−1 KH2PO4, 0.5 g L−1 MgSO4·7H2O, 20 g L−1 glucose, 1 mL L−1 vitamins, 1 mL L−1 trace metal solution, 60 mg L−1 uracil and 50 µL L−1 antifoam (Sigma-Aldrich).
Strain and plasmid construction
Chromosomal modifications were introduced into CEN.PK 113–5D using a plasmid based CRISPR/Cas9 system, which required the construction of plasmids targeting each desired site of modification and promoter-replacement cassettes. Furthermore, a plasmid containing a phosphoketolase gene from Bifidobacterium breve and a phosphotransacetylase gene from Clostridium kluyveri was constructed. See a complete list of plasmids used and constructed in this study in Table 2.
Table 2.
Description and origin of plasmids used and constructed in this study.
| Plasmid | Description | Origin |
|---|---|---|
| pCfB2311 | 2µ origin; PSNR52-gRNA-TSUP4; natMX marker | (Stovicek et. al. 2015) |
| pCfB2312 | CEN/ARS origin; PTEF1-cas9-TCYC1; kanMX marker | (Stovicek et. al. 2015) |
| pgRNA.STB5 | pCfB2312 PSNR52-gRNA(STB5)-TSUP4 | This study |
| pgRNA.FAA1/FAA4 | pCfB2312 PSNR52-gRNA(FAA1)-TSUP4 PSNR52-gRNA(FAA4)-TSUP4 | This study |
| pgRNA.ZWF1 | pCfB2312 PSNR52-gRNA(ZWF1)-TSUP4 | This study |
| pgRNA.GPP1 | pCfB2312 PSNR52-gRNA(GPP1)-TSUP4 | This study |
| pSP-GM1 | PTEF1-PPGK1 bidirectional promoter (2μ, URA3) | (Chen et. al. 2012) |
| pAB3 | pSP-GM1 PTEF1-xfpk(B. breve) | (Bergman et. al. 2016) |
| pAB10 | pSP-GM1 PTEF1-xfpk(B. breve) PPGK1-pta(C. kluyveri) | This study |
gRNA selections were made using the CRISPy tool (Jakounas et al. 2015) for gene deletions, and Deskgen's online Knockin tool (www.deskgen.com) for STB5 promoter exchange, as it allows for guide selection within the promoter region with accompanying activity and off-target scores (Doench et al. 2016). The selected 20 bp guide sequences are listed in Table S1 (Supporting Information).
The plasmid CfB2311 (Stovicek et al. 2015) containing a gRNA cassette with the SNR52 promoter and SUP4 terminator, was used as an initial template for gRNA cassette amplification. The gRNA cassettes were amplified as two individual fragments, where the 20 bp targeting sequences were added as complementary overhangs on the reverse primer for the first fragment, which comprises PSNR52, and on the forward primer for the second fragment, which comprises the structural region of the gRNA and TSUP4. The resulting polymerase chain reaction (PCR) products were fused in a second PCR, generating the final gRNA cassette. All primers and oligonucleotides used in this study are listed in Table S2 (Supporting Information), and details about the gRNA-fragment assembly strategy are described in Section A. PCR were performed using Phusion high fidelity DNA polymerase (Thermo Fisher Scientific). Resulting PCR products were purified using either a Gene Jet PCR or Gel Extraction purification Kit (Thermo Fisher Scientific).
The Cas9 and gRNA were co-expressed from the centromeric plasmid pCfB2312 (Stovicek et al. 2015). Plasmids were constructed by linearizing the vector with Pfl23II (Thermo Fisher Scientific) and cloning either a single (PSTB5, ZWF1, GPP1) or two gRNA-cassettes (FAA1/FAA4) into it by Gibson cloning (New England Biolabs). The Gibson reactions were used to transform E. coli DH5alpha according to the method of Inoue et al. (Inoue, Nojima and Okayama 1990). Transformants were used to inoculate 5 mL LB-medium with 80 mg L−1 ampicillin and grown at 37°C overnight. Plasmids were extracted using a GeneJET Plasmid Miniprep Kit (Thermo Fisher Scientific). The obtained plasmids were confirmed by sequencing (Eurofins Genomics), and named pgRNA.STB5, pgRNA.FAA1/FAA4, pgRNA.ZWF1 and pgRNA.GPP1.
The yeast promoters of CUP1, CYC1, ADH1, TPI1 and PGK1 were amplified from genomic DNA and flanked with 237 and 300 bp long STB5 recognition sequences. The upstream sequence was located 662 bp away from the start codon, and the downstream recognition sequence was the 300 first base pairs of the gene (see Table S2, Section B, Supporting Information, for primer sequences and details about fragment assembly). The obtained fusion constructs were used as double stranded donors in the CRISPR/Cas9 transformations, and thus deleted 662 bp of the native promoter when integrated upstream of STB5.
Yeast transformations of freshly prepared competent cells were performed using the LiAc/SS carrier DNA/PEG method (Gietz and Schiestl 2007). One nmol of (each) 120 bp DS-repair oligo (Table S2, Section C, Supporting Information) and 1 µg of pgRNA-plasmid were used in transformations to achieve gene deletions. For promoter replacements, 1 µg of repair fragment (i.e. promoter constructs) and pgRNA plasmid were used. As a control, each pgRNA-plasmid was transformed without DS-oligo to verify its functionality. After a heat chock, cells were resuspended in YPD and incubated at 30°C for 2–3 h to allow expression of the kanMX marker gene. Transformants were selected on YPD + G418 plates, incubated for at least 2 days at 30°C, and restreaked twice before confirming the deletions and integrations with colony PCR and sequencing. The pgRNA plasmids were looped out by cultivating strains in 5 mL YPD overnight, streaking 10 µL on a YPD plate. As soon as individual colonies were formed, 10 of these were replica plated on YPD and YPD + G418 plates to find isolates that had lost the plasmid. Sequential modifications were performed according to Table 1.
Plasmid pAB10 was constructed based on plasmid pAB3 (Bergman et al. 2016). The phosphotransacetylase gene from C. kluyveri (pta(CK)) was codon optimized for use in S. cerevisiae (Genscript, see Table S3, Supporting Information, for sequence). PPGK1 and TCYC1 from pSP-GM1 (Chen et al. 2012) and pta(CK) were amplified using the primers listed in Table S1, Section D (Supporting Information), after which the three fragments were fused. Plasmid pAB3 was linearized using SmaI/XhoI, purified from gel, and the PPGK1-pta(CK)-TCYC1 fusion fragment was inserted into the pAB3-backbone using homologous recombination in yeast as described previously (Bergman et al. 2016). The resulting plasmid pAB10 or pSP-GM1 were used to transform strains AB11, AB23 and AB24, generating strains AB25-AB30.
Cultivation procedures
BioLector
Growth behavior of promoter-exchanged strains or strains carrying plasmids were evaluated in a BioLector (m2p-labs, Baesweiler, Germany). Cultures were prepared in biological triplicates. Single colonies were cultivated in 3 mL minimal medium overnight at 30°C and 200 rpm. The precultures were used to inoculate 1 mL of medium to an initial OD600 of 0.1 using minimal medium containing either 0, 100, 200 or 400 µM Cu2+. Cultivations were conducted at 30°C with 1200 rpm shaking in a 48-well microtiter plate (MTP-48-B FlowerPlate, m2p-labs). The optical density was measured with 15 min intervals until strains reached stationary phase at a filter gain of 30. Maximum specific growth rates (µmax) were calculated from the slope of the ln(OD600) plotted against time during exponential phase.
Shake flasks
Cultivations of strains for NADPH assay and FFA quantification were performed in biological triplicates, and strains to be evaluated in the survival assay were cultivated in biological duplicates. Single colonies were used to inoculate 3 mL of medium cultivated overnight at 30°C and 200 rpm. The precultures were used to inoculate 20 mL of medium in 100 mL unbaffled shake flasks. The initial OD600 was set to 0.005 to 0.1 depending on their growth rate in order to be able to synchronize time of harvest. Flasks were incubated at 30°C with 200 rpm orbital shaking. Samples were harvested around an OD600 0.6–1.0 to be used in NADPH and survival assay (see sections ‘NADPH assay’ and ‘Survival assay’ for processing). For FA quantification, samples were taken at the end of exponential phase and after 72 h of cultivation (i.e. stationary phase) by withdrawing cells and media, chilling on ice while quickly measuring OD600 and thereafter storing the samples at -20°C until time of sample preparation (see section ‘FAME analysis’ for processing).
Bioreactor cultivations
Bioreactor cultivations were conducted in biological quadruplicates. Precultures were prepared in 5 mL of minimal medium in 15 mL cultivation tubes using single colonies as inoculum for each replicate, and grown overnight at 30°C and 200 rpm orbital shaking. The precultures were used to inoculate 50 mL minimal medium in 250 mL unbaffled shake flasks to an OD600 of 0.05–0.1 in order to scale up the preculture. The shake flasks were once again incubated at 30°C and 200 rpm, until they reached an OD600 of 0.8–1.0, at which a defined volume of each cell culture was centrifuged, the cell pellet was re-suspended in 50 mL fresh minimal medium for bioreactor cultivation, and used to inoculate the bioreactors (DasGip Parallel Bioreactor Systems for Microbiology (Eppendorf)) to an initial OD600 of 0.1 in a volume of 600 mL. During the course of the cultivation, the temperature was set to 30°C, stirring to 600 rpm and aeration to 36 L h−1 (0.1 VVM). The pH was adjusted to 5.0 using 2 M solutions of HCl and KOH. Composition of the exhaust gas was examined using a DasGip GA4 gas analyzer (Eppendorf). The strains were allowed to grow until early ethanol phase, after which feeding and outflow was turned on corresponding to a dilution rate of 0.1 h−1. Samples for OD600 and high-performance liquid chromatography (HPLC) were taken continuously during exponential growth phase and at a couple of times during chemostat to verify entry into steady state conditions. Physiological parameters and fluxes (q) for the batch phase were calculated using a cell dry weight/OD600 conversion factor obtained during mid exponential phase (OD600 ∼1.0–1.5). A biomass composition of CH1.8O0.5N0.2 was assumed (Villadsen, Nielsen and Lidén 2011).
Samples for RNAseq, OD600, cell dry weight and HPLC analysis were simultaneously taken at three times points: in mid exponential phase, early ethanol phase and in chemostat conditions. For RNAseq, cell suspension from the bioreactor was directly cooled on crushed ice and centrifuged for 5 min at 5000 g. The cell pellet was washed with ice-cold phosphate-buffered saline (PBS), transferred to Eppendorf tubes and re-centrifuged at 0°C and 10 000 g. PBS was removed and pellets were snap-frozen in liquid N2. Cell pellets for RNAseq analysis were stored at -80°C until being processed.
NADPH assay
NADPH was quantified using an NADP/NADPH Quantification Kit (Sigma-Aldrich). Strains to be evaluated for NADPH content were cultivated in biological triplicates as described above. For each sample harvested, 5 mL of cell culture of an OD600 between 0.5–1.0 was immediately quenched in 20 mL -40°C methanol, centrifuged at 4000 g and -10°C for 3 min. The collected cell pellets were re-dissolved in 1 mL -40°C methanol, transferred to pre-cooled 1.5 mL tubes, centrifuged at 10 000 g and -10°C for 1 min, after which the supernatant was removed and the sample was snap-frozen in liquid N2 and thereafter stored at −80°C.
The same day samples were to be used for NADPH quantification, the samples were freeze dried at 0.1 mbar using a Christ alpha 2–4 LSC (Christ Gefriertrocknungsanlagen) for 4 h. Thereafter, samples were dissolved in 1 mL NADP/NADPH Extraction Buffer, transferred to a pre-cooled 2 mL tubes with 0.5 mm glass beads and homogenized using a FastPrep -24 (MP Biomedicals) (4 cycles of 5.5 m/s for 30 s, 5 min resting on ice in between runs). The homogenized mixtures were transferred to pre-cooled 1.5 mL tubes, centrifuged at maximal speed in a table top centrifuge for 5 min at 0°C, and the supernatant was collected and kept on ice. The majority of the volume of the collected samples was run through 10 kDa cut-off spin filters to deprotonize the samples and the filtrates were used to quantify the NADPH according to the manufacturer's protocol. The remaining crude cell free extract was used for protein concentration determination with a Pierce BCA Protein Assay Kit (Thermo Fisher Scientific) according to the manufacturer's instructions. NADPH readings were normalized to protein content.
Survival assay
Strains to be evaluated for survival in response to H2O2 were cultivated in biological duplicates as described above. For each sample harvested, 4 mL of cell culture was immediately added to 4 mL of freshly prepared minimal medium containing H2O2 at concentrations of 0, 2, 4 and 8 mM, resulting in active hydrogen peroxide concentrations of 0, 1, 2 and 4 mM. The tubes were incubated at 30°C and 200 rpm shaking for 30 min, after which the tubes were centrifuged at 4000 g for 5 min. Cells were washed in 10 mL deionized H2O, recentrifuged and once again resuspended in 10 mL deionized H2O. The OD600 was measured in duplicates, dilutions of each sample were made and 10 µL corresponding to approximately 50 cells (based on the assumption that 1 mL of cell suspension of OD600 of 1.0 contains 3 × 107 cells) were spotted in duplicates for each sample. The average colony counts were calculated based on two biological (shake flasks) and technical (plate streaks) replicates (n = 4).
Extracellular metabolite analysis
In order to quantify extracellular metabolites, fermentation samples were filtered through a 0.45 μm nylon filter (VWR International AB) and analysed with HPLC. The HPLC system UltiMate® 3000 (Dionex) was utilized equipped with an Aminex® HPX-87H ion exclusion column (Bio-Rad). The system was operated using 5 mM H2SO4 as eluent at a flow rate of 0.6 mL/min and a column temperature of 45°C. Quantification of glucose, glycerol, acetate and ethanol was performed using a refractive index detector (512 μRIU), while pyruvate and succinate were quantified using an UV detector.
FAME analysis
FFA produced by strains with mutations faa1∆ faa4∆ were simultaneously extracted and derivatized to methyl esters using a protocol modified from Haushalter et al (Haushalter et al. 2014). For each sample, 250 µL of cell culture (cells + medium) was transferred to a glass vial; 15 µL of tetrabutylammonium hydroxide was added, immediately followed by 250 µL of 200 mM methyl iodide and 100 mg L−1 pentadecanoic acid (internal standard) in dichloromethane (DCM). The tube was capped and samples placed in a vortex mixer for 30 min at 1400 rpm. To promote phase separation, the samples were centrifuged after which 150 µL of the DCM layer was transferred to a GC vial with glass insert. The samples were evaporated for 30 min using a miVac sample concentrator (SP Scientific), and as a last step resuspended in 150 µL hexane. Using a gas chromatograph (Focus GC, Thermo Fisher Scientific) equipped with a Zebron ZB-5MS GUARDIAN capillary column (30 m × 0.25 mm × 0.25 μm, Phenomenex) and a Flame Ionization Detector (FID, Thermo Fisher Scientific), the derivatized FAME samples were analyzed. The injection volume was 1 µL and the inlet temperature was set to 280°C. The GC program was as follows: initial temperature 50°C; hold for 2 min; ramp to 140°C during 3 min; ramp to 280°C during 14 min; hold for 3 min. The flow rate of the carrier gas (helium) was 1.0 mL min−1. The produced FAMEs were quantified by the co-analysis of FAME standards of physiologically relevant chain length (C8-C18) and pentadecanoic acid (C15). The obtained data were processed with Xcalibur software.
RNAseq sample preparation, profiling and analysis
Cell pellets were thawed on ice, and RNA was extracted using an RNeasy Mini Kit (Qiagen). Samples were treated with TURBO DNase (Thermo Fischer Scientific) to remove genomic DNA contamination. RNA integrity was evaluated with a 2100 Bioanalyzer (Agilent Technologies). RNA samples were processed with a TruSeq Stranded mRNA HT Sample Prep Kit (Illumina) to generate a poly-A enriched cDNA library, and samples were sequenced using a NextSeq 500 (Illumina), paired end, 2 × 76 bp. Read pairs ranged between 3.0 to 4.1 million. The raw data can be retrieved from the European Nucleotide Archive with access number ERP112694. The raw reads were processed with the NGI RNAseq Pipeline (https://github.com/SciLifeLab/NGI-RNAseq) version 1.4, see Table S4 (Supporting Information) for more information. The S288c reference genome was used to map the reads (https://www.ensembl.org/Saccharomyces_cerevisiae/Info/Index), and the percentage of uniquely mapped reads was between 86.5–93.6%. Raw read counts were calculated with feature Counts (Liao, Smyth and Shi 2014) and FPKM values calculated with StringTie (Pertea et al. 2015). Differential gene expression analysis was performed with DESeq2 (Love, Huber and Anders 2014), adjusted P-values were calculated according to the method of Benjamini–Hochberg. The Platform for Integrative Analysis of Omics (PIANO) R package (Varemo, Nielsen and Nookaew 2013) was utilized to analyse enriched GO terms and perform reporter TF analysis, where the adjusted P-values and log2 fold change from the differential gene expression analysis were used as input, except for the GSA methods maxmean, gsea and page, which used Wald statistics from DESeq2 as input. GO term annotations were collected from Ensembl (www.ensembl.org), and the interaction between TFs and genes, based on both binding and expression evidence, from Yeastract (http://www.yeastract.com). From the reporter TF analysis, the top five scoring TFs from the distinct directional up and distinct directional down regulation for each condition were selected and reported with their significance calculated from a null-distribution and adjusted with the Benjamini–Hochberg method.
RESULTS
Overexpression of STB5 reduces maximum specific growth rate and final biomass concentration on glucose
As a previous study had reported that strong overexpression of STB5 disabled a wine yeast derivative to grow on glucose (Cadiere, Galeote and Dequin 2010), we initially decided to replace the native promoter of STB5 with the Cu2+-responsive promoter of CUP1 (PCUP1) to investigate if and at what level of induction STB5 overexpression would repress growth on glucose in CEN.PK113–5D. The maximum specific growth rates of the control and the PCUP1-STB5 strain treated with Cu2+ concentrations ranging from 0–400 µM are listed in Table 3A.
Table 3.
Maximum specific growth rates (µ(max) [h−1]) of STB5-promoter exchanged strains in CEN.PK113–5D background.
| A)STB5overexpression mediated by copper induction | ||||
|---|---|---|---|---|
| Cu2+ (µM) | Control (CEN.PK 113–5D) | PCUP1-STB5 (AB12) | ||
| ‘0’ | 0.317 ± 0.034 | 0.344 ± 0.018 | ||
| 100 | 0.254 ± 0.014 | 0.259 ± 0.003 | ||
| 200 | 0.284 ± 0.014 | 0.227 ± 0.008 * | ||
| 400 | 0.181 ± 0.004 | 0.107 ± 0.021 * | ||
| B)STB5overexpression mediated by constitutive promoters | ||||
| Control (CEN.PK 113–5D) | PCYC1-STB5 (AB13) | PADH1-STB5 (AB14) | PTPI1-STB5 (AB15) | PPGK1-STB5 (AB16) |
| 0.326 ± 0.012 | 0.265 ± 0.006 * | 0.239 ± 0.002 * | 0.246 ± 0.009 * | 0.219 ± 0.014 * |
Asterisks (*) indicate a significant difference (P < 0.05; Student´s t-test, two-sided) compared to the control at the same conditions. Strains were grown in minimal media with 2% glucose and 60 mg/L uracil. ‘Zero µM’ Cu2+ concentration in reality corresponds to approximately 6 µM due to the presence of Cu2+ in the trace metal solution added to the media. Values correspond to the average of biological triplicates ± standard deviation.
Cu2+ concentrations of 100, 200 and 400 µM increasingly reduced the µmax of the control, which is to be expected due to the toxicity of copper (Avery, Howlett and Radice 1996). At zero and 100 µM, there was no difference in µmax between the two strains, while a greater decrease was observed for PCUP1-STB5 at 200 and 400 µM, corresponding to approximately 80 and 60% of the growth rate observed for the control at the same Cu2+ concentration. The maximum cell density was nevertheless exclusively reduced in PCUP1-STB5 at all tested concentrations of Cu2+ exceeding zero, by 13–20% of what was observed for the control (Table S5, Supporting Information).
It cannot be excluded that STB5 expression was induced to a certain extent in the control strain as well, as Cu2+ toxicity is believed to be mediated through the formation of reactive oxygen species (Valko, Morris and Cronin 2005), which possibly could activate a redox stress response mediated by Stb5 (Larochelle et al. 2006). If so, the replacement of the native STB5 promoter with PCUP1 shows that the effects of Stb5 binding can be intensified beyond the natural level of induction. Irrespective of such effects, we show that an elevated level of STB5 expression reduces but does not entirely prevent growth on glucose in CEN.PK113–5D, as previously observed by Cadiere, Galeote and Dequin (2010).
In order to avoid the toxic effect of copper, we decided to evaluate a set of yeast constitutive promoters of varying strength. The four endogenous promoters of CYC1, ADH1, TPI1 and PGK1 were selected to increase STB5 expression, where PCYC1 represent a weaker promoter whereas PADH1, PTPI1 and PPGK1 are considered stronger with varying relative activities (Partow et al. 2010; Peng et al. 2015). Consistent with the Cu2+ induction experiments, the constitutive promoter exchanges resulted in a significantly decreased µmax in all constructed strains (Table 3B). Replacement of the native promoter with PCYC1 reduced growth rate to approximately 80% of the control. The strongest effect was observed for the glycolytic promoter PPGK1, for which µmax was close to 0.22 h−1, corresponding to 67% of the control. Similar to when utilizing the inducible PCUP1 promoter, final biomass concentrations were also reduced in the constitutive STB5 overexpressing strains, ranging from 13% for PCYC1 to 21–22% for PADH1, PTPI1 and PPGK1 compared to the control (Table S5, Supporting Information).
Overexpression of STB5 fails to generate a measurable surplus of NADPH and reduces survival after H2O2 shock
After establishing that overexpression of STB5 did not prevent growth on glucose, we wanted to investigate if its overexpression generated an increase in NADPH in CEN.PK113–5D. A previous report, which investigated the function of the STB5 regulated gene YMR315W, showed that STB5 overexpression from a plasmid led to a 2.5-fold increase in NADPH levels in a BY4727 background. Thus, we quantified the NADPH content of the control CEN.PK113–5D and the STB5 promoter exchanged strains (AB13-AB16) in exponentially growing cells, but failed to measure a significant difference in NADPH levels (Figure S1, Supporting Information).
Instead, we turned to explore if the promoter exchanged strains would be more suited to handle treatment with an oxidizing agent, as NADPH supplies the reducing potential of most cellular antioxidant systems, including the glutathione system (Morano, Grant and Moye-Rowley 2012). In order to circumvent a beneficial effect of STB5 upregulation in the wild type, which likely would occur if strains were plated on oxidative media, we instead decided to shock exponentially growing cells with different concentrations of H2O2. The experiment included the control (CEN.PK113–5D), the negative control zwf1∆ (AB31), which is not able to catalyze the first reaction in the oxidative PPP, and two STB5-promoter exchanged strains: PCYC1-STB5 (AB13) and PTPI1-STB5 (AB15). The proportion of H2O2-shocked cells that survived the treatment are presented in Fig. 2.
Figure 2.
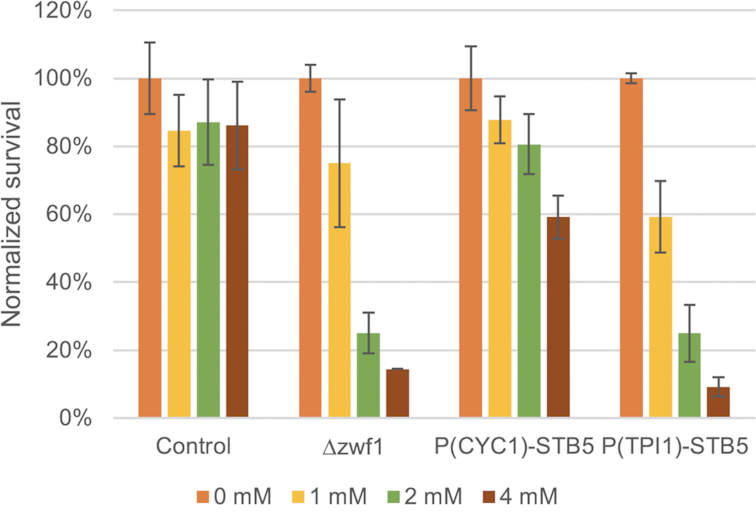
Survival of the strains CEN.PK 113–5D, zwf1∆ and PCYC1-STB5 and PTPI1-STB5 after liquid H2O2-shock. The values indicate the proportion of surviving cells at each concentration compared to the value observed for each strain at 0 mM H2O2. Values are averages of two biological and two technical replicates; error bars represent ± standard deviation.
The control only showed a slightly reduced viability after H2O2 shock at all tested concentrations (1, 2 and 4 mM), indicating an efficient oxidative stress response. As expected, the zwf1∆ mutant showed a sharp decrease in survival at higher concentration of H2O2. The majority of the colonies were also small indicating that they indeed had been severely damaged by the exposure. Contrary to the hypothesis that overexpression of STB5 could boost oxidative stress resistance via increased NADPH levels, survival of PCYC1-STB5 was significantly reduced at 4 mM H2O2 compared to the control. Similarly, survival of the PTPI1-STB5 strain was reduced to levels close to that observed for the zwf1∆ mutant. This shows that overexpression of STB5 increases sensitivity to oxidative stress, possibly through a mechanism which limits flux through the oxidative PPP.
Overexpression of STB5 increases FA production in glucose phase in an oxidative PPP independent manner
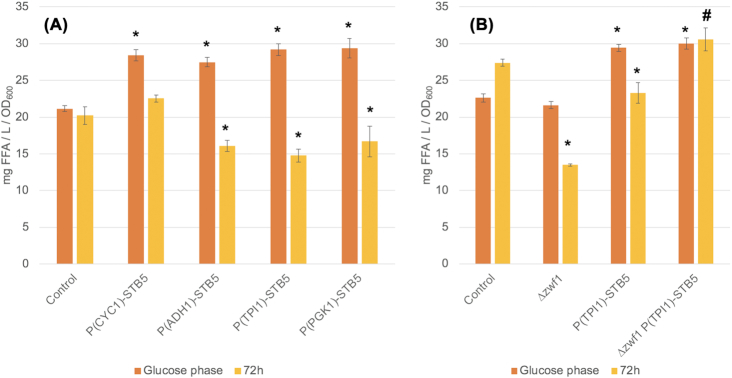
Even though we were unable to directly demonstrate that STB5 overexpression was linked to an increased NADPH supply, we still decided to investigate the initial question of this study: Can STB5 overexpression be utilized as a strategy to improve NADPH dependent FA synthesis? In order to have a more efficient FA production (and drain of NADPH) the promoter replacements were integrated into the double deletion strain CEN.PK 113–5D faa1∆faa4∆. This strain cannot activate FFA to acyl-CoA and likely experiences less acyl-CoA mediated inhibition of FA synthesis (Faergeman and Knudsen 1997). It also excretes visual levels of FFAs into the cultivation medium which can easily be quantified (Scharnewski et al. 2008; Leber et al. 2014). Interestingly, while in faa1∆faa4∆ strains with a strong promoter controlling STB5, the growth rate was reduced in a similar manner as in CEN.PK113–5D (62–72% of that observed for the control), the growth rate of PCYC1-STB5 was not significantly altered compared to the control (Table S6, Supporting Information).
Figure 3A shows the results of the FFA quantification of STB5-overexpression strains (AB17-AB20) compared with the control (AB11). The titer values (mg FFA/L) were normalized to the OD600 at sampling to minimize influence of sampling time point and/or biomass yield. Samples were taken at the end of the glucose phase when cells had stopped growing exponentially as well as at 72 h when cells had entered stationary phase. The quantification showed that all levels of overexpression of STB5 had a significant beneficial effect on FFA formation in the glucose phase, resulting in a level between 130–139% of that observed for the control, while no significant difference was observed between the STB5 overexpression strains. At 72 h, there was a small albeit non-significant increase in final FFA titers observed for the weaker promoter exchange (PCYC1), while all of the stronger promoter exchanges significantly reduced the production of FFAs to 73–83% of that observed for the control. This suggests that strong overexpression of STB5 specifically has a negative effect on FFA synthesis in the ethanol phase.
Figure 3.
FAA quantification of CEN.PK 113–5D faa1∆faa4∆ strains after glucose was depleted (Glucose phase) and at stationary phase (72 h). A) Influence of different levels of STB5 overexpression on FA synthetic ability. B) Influence of ZWF1 deletion on FA synthetic ability in the control and PTPI1-STB5 strain (# indicates that the strain failed to consume the produced ethanol). Strains were grown in minimal medium with 2% glucose with the addition of 60 mg/L uracil. Values are averages of three biological replicates, error bars ± standard deviation. Asterisks (*) indicate a significant change from the control (P < 0.05; Student´s t-test, two-sided) at the corresponding sampling point.
We wondered if the specific increase in FFA synthesis during the glucose phase could be linked to an increased redirection of carbon flux through the oxidative PPP, as indicated by the regulatory targets reported previously (Larochelle et al. 2006). Thus, we deleted ZWF1 in strain AB11 and AB19 and quantified the produced FFAs in the control (AB11), zwf1∆ (AB21), PTPI1-STB5 (AB19) and zwf1∆ PTPI1-STB5 (AB22) at the end of the glucose phase and at 72 h, the results of which are shown in Fig. 3B. Compared to the first experiment, FFA production in the ethanol phase was higher for the control and PTPI1-STB5. However, the observed trends between the control and PTPI1-STB5 were similar to those previously observed. Interestingly, the FFA production of zwf1∆ was not affected in the glucose phase, while a substantial decrease was observed at stationary phase, where FFA production was measured to be merely 50% of that observed for the control. Neither did the deletion of ZWF1 influence FFA synthesis of the PTPI1-STB5 strain in the glucose phase, suggesting that the beneficial effect of STB5 overexpression on FFA production was independent of oxidative PPP flux. The zwf1∆ PTPI1-STB5 strain failed to resume growth after glucose was depleted, which is why the FFA production remained at the same level for the glucose phase and at 72 h, i.e. the presumed negative influence of STB5 overexpression on FFA synthesis during growth on ethanol was masked.
Synergistic effects of an increased acetyl-CoA supply and STB5 overexpression on FA production in glucose phase
Based on the observation that overexpression of STB5 using weak (PCYC1) as well as strong promoters (PADH1, PTPI1 and PPGK1) produced a very similar beneficial effect on FA production in the glucose phase (Fig. 3A), even though the maximum specific growth rates of the respective strains differed substantially (Table S6, Supporting Information), we decided to investigate if FFA production in the glucose phase could be limited by other factors than NADPH—such as the precursor acetyl-CoA. We utilized a pathway consisting of two heterologous proteins: a phosphoketolase (Xfpk), which can convert fructose-6-phosphate or xylulose-5-phosphate to acetyl phosphate (AcP) and erythrose-4-phosphate or glyceraldehyde-3-phosphate, respectively, and a phosphotransacetylase (Pta), which catalyzes the reversible conversion of AcP to acetyl-CoA. The constructed plasmid pAB10 encodes an efficient Xfpk from B. breve (Bergman et al. 2016) and a Pta from C. kluyveri (Meadows et al. 2016). pAB10 and the empty plasmid pSP-GM1, respectively, were used to transform the strains AB11, AB23 and AB24, generating the six strains AB25-AB30. The deletion of glycerol-3-phosphatase gene GPP1 in strain AB27-AB30 blocks the major route of endogenous hydrolysis of AcP to acetate (Bergman et al. 2016), which otherwise would cause energy loss, and increase CO2 production and respiratory demand of a strain expressing a phosphoketolase (Bergman et al. 2019). Fig. 4 shows the FFA titers obtained at the end of glucose phase normalized to sampling OD600.
Figure 4.
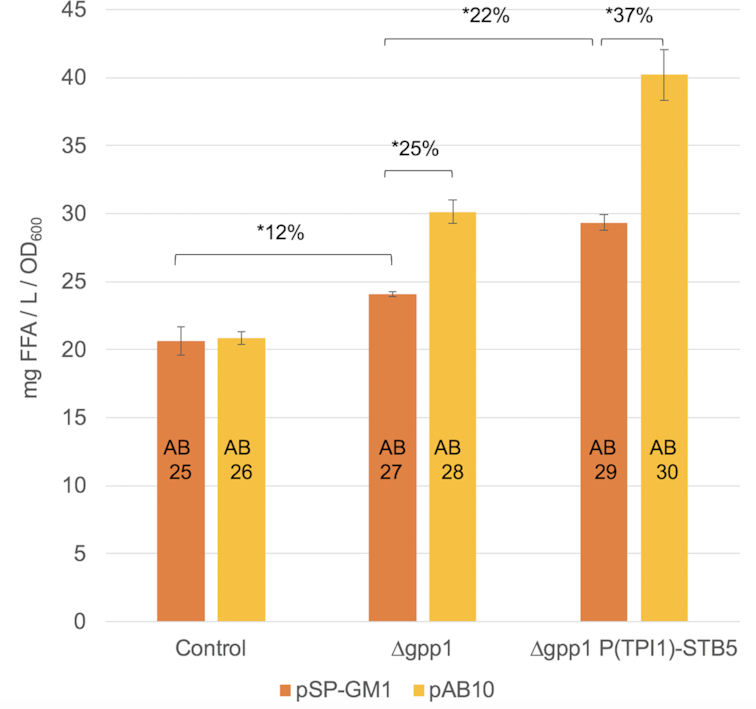
Influence of additional acetyl-CoA supply on FFA production of CEN.PK 113–5D faa1∆faa4∆ strains on glucose. The indicated host strains were either transformed with the empty plasmid pSP-GM1 or pAB10, expressing a phosphoketolase from B. breve and and phosphotransacetylase from C. kluyveri. Strains were grown in minimal medium with 2% glucose and harvested at or close to glucose depletion. Values are averages of three biological replicates, error bars ± standard deviation. Asterisks (*) indicate a significant change (P < 0.05; Student´s t-test, two-sided).
The expression of the heterologous Xfpk/Pta pathway using plasmid pAB10 did not have a beneficial effect on FA production in the control strain (AB26 vs AB25). The introduction of gpp1∆ (AB27) had a significantly beneficial effect on FFA synthesis, which increased 12% compared the control strain (AB25). Overexpression of the Xfpk/Pta pathway in the gpp1∆ strain (AB28) caused a significant increase (25%) in FFA production (AB28 vs AB27), likely due to the reduced degradation of AcP into acetate. This indicates that Xfpk/Pta constitutes a functional route for increasing the acetyl-CoA supply in the gpp1∆ host, also shown previously (Meadows et al. 2016).
The gpp1∆ PTPI1-STB5 strain (AB29) showed a similar level of FFA production (30 mg L−1 OD600−1) compared to what was observed for PTPI1-STB5 without deletion of GPP1 (Fig. 3A and B), and corresponded to a 22% increase compared to the gpp1∆ strain (AB 29 vs AB27). The introduction of the Xfpk/Pta pathway into the gpp1∆ PTPI1-STB5 background (AB30) increased FFA production further, corresponding to a 37% increase compared to the same strain harboring the empty plasmid pSP-GM1 (AB30 vs AB29), and a 67% increase compared to the gpp1∆ strain carrying pSP-GM1 (AB30 vs AB27). These results indicate that the effects of STB5-overexpression and an increased acetyl-CoA supply are not just additive, but act in synergy to increase FFA synthesis.
Overexpression of STB5 increases expression of a majority of suggested target genes in steady state conditions while the activation is limited in batch culture
In order to obtain a better understanding for the effects resulting from STB5 overexpression, we decided to conduct batch and continuous bioreactor cultivations of the non-modified strain (CEN.PK113–5D) and PTPI1-STB5 (AB15). The calculated parameters maximum specific growth rate (µmax), biomass yield (YS/X), flux-coefficients (q), carbon balance and respiratory quotient (RQ) are summarized in Table S7 (Supporting Information). As described previously, STB5 overexpression negatively influenced the maximum specific growth rate, which caused a general decrease in the calculated specific carbon flux values in the exponential phase. If the calculated fluxes were normalized to glucose uptake rate and carbon content, glycerol, ethanol and biomass production rates still remained significantly reduced. At the same time, oxygen consumption qO2 increased slightly, and the RQ was reduced, indicating that the STB5 overexpression mutant has a metabolism of more respiratory character. However, both of these parameters were altered at a non-significant level (p(qO2) = 0.24 and p(RQ) = 0.068).
During the course of the bioreactor cultivation, we took samples for RNAseq analysis in three different conditions: 1) in the glucose-consumption phase, when STB5 overexpression appeared to influence strain growth the most, 2) in the ethanol-consumption phase, where a negative effect on FA synthetic ability was observed and 3) in steady state conditions—which is the preferred sampling choice for RNAseq data analysis as a difference in growth between two strains will not influence gene expression, for example as it is the case with genes related to ribosome biogenesis and stress response (Regenberg et al. 2006).
The overall variation of the data is visualized in a principal component analysis plot (Fig. 5), which shows a clear separation between sample groups. The largest variation was observed for samples harvested at different conditions (i.e. glucose (G), ethanol (E) and steady state (S)), consistent with the major rearrangement in metabolism associated with a shift from fermentative to respiratory growth. The largest variance within control (ref) and STB5 overexpressing strain (s) was observed for samples harvested in the glucose phase (refG and sG for control and STB5-overexpression, respectively), while the variation appeared to be relatively small in steady state conditions (S). In the glucose phase, STB5 overexpression appears to shift the cellular state to one that to a higher degree resembles respiratory conditions, indicated by the lateral shift along the first principle component in Fig. 5.
Figure 5.
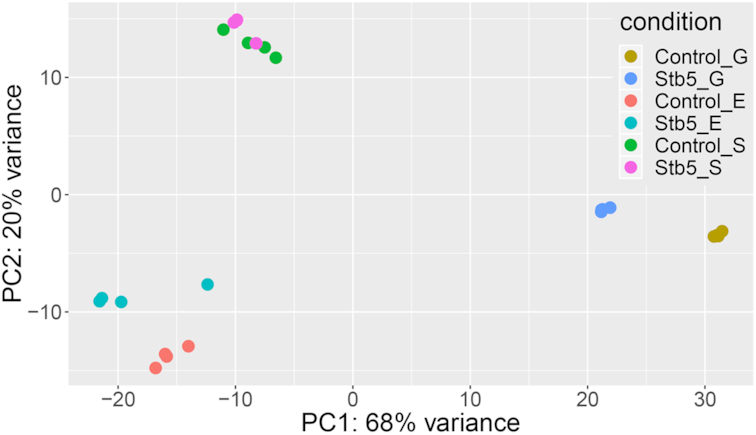
Principal component analysis plot of RNAseq data from control and STB5 overexpression strain. ’Control’ and ‘Stb5’ indicate the control CEN.PK-113 5D and STB5 overexpression strain AB15, respectively, while G, E and S correspond to the sampling point condition (Glucose, Ethanol and Steady state, respectively).
In Table 4, an overall outcome of the differential expressional analysis is shown. In the glucose phase, 3695 transcripts were significantly changed compared to the control (padj < 0.01). The number of significantly changed transcripts were lower in the ethanol phase, and decreased even further at steady state. In the glucose and ethanol phases, the majority of differentially expressed genes were positively influenced by STB5 overexpression, as indicated by the greater number of transcripts with a large positive log2-fold change (LFC), while this trend could not be observed during steady state.
Table 4.
Number of significantly changed genes and number of genes with |LFC| > 0.5 and 1.0 in PTPI1-STB5 compared to the control strain AB15 in glucose phase, ethanol phase and steady state when cultivated in bioreactors.
| Number of transcripts | |||||
|---|---|---|---|---|---|
| Sampling point | padj < 0.01 | LFC > 0.5 | LFC > 1.0 | LFC < −0.5 | LFC < −1.0 |
| Glucose phase | 3695 | 1085 | 536 | 703 | 130 |
| Ethanol phase | 1568 | 577 | 266 | 305 | 82 |
| Steady state | 879 | 187 | 61 | 176 | 63 |
STB5 expression was significantly enhanced compared to the control in all conditions, with a LFC of 4.83, 1.75 and 3.29 in glucose, ethanol and steady state, respectively. The normal expression levels of STB5 were low at all conditions, with FPKM-values between 16 and 27, which for example can be compared to that of the glycolytic gene PGK1 in the glucose phase, averaging close to 3000 (Table S8, Supporting Information).
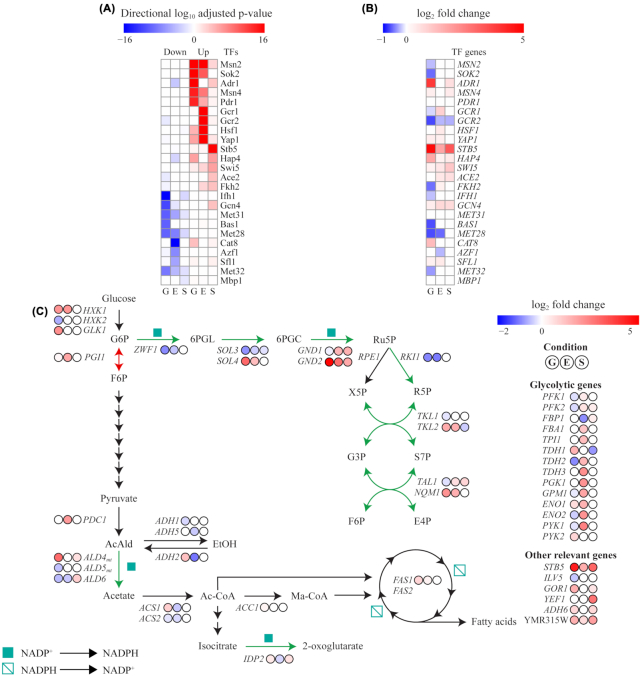
When conducting a gene set analysis, numerous GO terms that were indicated to be distinctly downregulated upon STB5 overexpression in the glucose phase were connected to ribosomal processes, such as rRNA processing and export, ribosomal subunit biogenesis and assembly, and amino acid synthesis (Fig. S2, Supporting Information). This was expected, as PTPI1-STB5 had an approximately 40 and 20% lower maximum specific growth rate and biomass yield, respectively, compared to the control (Table S7, Supporting Information), and thus should require less protein synthesis. Simultaneously, gene sets connected to respiratory processes were stimulated by overexpression of STB5 in the glucose phase (Fig. S2, Supporting Information), which correlated with the lower RQ value observed for PTPI1-STB5 (Table S7, Supporting Information).
We performed a reporter TF analysis to get a broader understanding if certain TFs were likely to be of importance for the overall transcriptional responses observed in the different growth stages. The result of the analysis highlights the top five reporter TFs related to a coordinated up- and downregulation of target genes in each condition, shown in Fig. 6A, and the LFC of each reported TF compared to the control in the same condition is presented in Fig. 6B. Adr1, essential for activation of glucose-repressed genes when cells enter the diauxic shift (Tachibana et al. 2005), stood out as a key TF in the glucose phase, together with Msn2, Msn4, Pdr1 and Sok2–transcriptional activators involved in stress response, pleiotropic drug resistance and cAMP-dependent protein kinase signal transduction. Of these, only ADR1 showed a prominent degree of upregulation (Fig. 6B). In the study by Ouyang et al, ADR1 and MSN4 was indicated as direct targets of Stb5, although, ADR1 was reported to be negatively regulated (Ouyang et al. 2018). Among other TFs which show a good correlation of target/TF expression is for example Ifh1, a ribosomal gene expression regulator, which is downregulated in the glucose phase as are its target genes. In the ethanol phase, GCR1 encoding a transcriptional activator of genes involved in glycolysis is upregulated, which correlates well with a strong upregulation of majority of the glycolytic genes in the ethanol phase (Fig. 6C). Interestingly, Stb5 was only highlighted by the TF reporter analysis to have a significant positive influence on its target genes during steady state conditions, even though it was more strongly expressed in the glucose phase. This indicates that mechanisms are present to counteract the effects of Stb5-mediated regulation in glucose and ethanol phase. Furthermore, the other reporter TFs in steady state have lower significance compared to that observed for Stb5, indicating that fewer secondary effects were influencing expression compared to the situation in glucose and ethanol phase.
Figure 6.
Transcriptional analysis of STB5 overexpression in different conditions, where G, E and S correspond to the sampling point ‘Glucose’, ‘Ethanol’ and ‘Steady State’, respectively. A) TF reporter analysis, revealing the top five scored TFs predicted to drive the down- and upregulated transcriptional changes at each of the tested conditions. B) Differential expression of the reporter TF genes in respective conditions (padj < 0.01). C) Differential expression analysis of genes involved in the PPP, glycolysis, FA synthesis and other previously suggested STB5 target genes (padj < 0.01).
Figure 6C shows a selection of genes related to central carbon metabolism and previously indicated target genes of Stb5 and having significant altered expression (padj < 0.01). With respect to the genes of the PPP, which in a BY4741 background have been suggested to be activated by Stb5 in response to oxidative stress (Larochelle et al. 2006), all genes but two were differentially regulated in the glucose phase. ZWF1, SOL3, RKI1, TAL1 and TKL1 were significantly downregulated, the expression level of GND1 and RPE1 remained unchanged, while SOL4, GND2, NQM1 and TKL2 were induced. It should however be noted that SOL4, GND2, NQM1 and TKL2 had lower FPKM values than their corresponding paralogs in the glucose phase (Table S8, Supporting Information). When the raw read counts for each gene pair were added together and the combined LFC for SOL, GND, TAL and TKL were calculated with DESeq2, only GND has a slightly positive value while the other PPP reactions were affected negatively (significant LFC-values: -0.69, 0.18, -0.26 and -0.24, respectively), suggesting a reduced capacity for flux through the PPP in the glucose phase. In addition, the proposed effect of Stb5 being able to act as a transcriptional silencer of PGI1 could not be seen in our study in any of the conditions.
Other NADPH-associated regulatory effects of Stb5 in response to oxidative stress in a BY4741 background included activation of ALD6, IDP2, ILV5, GOR1, YEF1, ALD4 and ADH6 (Larochelle et al. 2006). Among these, ALD6 and ILV5 were downregulated, YEF1 remained unchanged while IDP2, GOR1, ALD4 and ADH6 were upregulated in response to STB5 overexpression in the glucose phase. Even though IDP2 had a positive LFC in the DE analysis, it is unlikely that it has a large influence on metabolism in the glucose phase due to its low FPKM value (Table S8, Supporting Information). ALD4 was strongly upregulated, but the subcellular localization of the corresponding enzyme is within the mitochondria and the suggested redox shuttle gene YHM2 was significantly downregulated (LFC = −1.31).
During growth on glucose, FA synthesis requires flux from pyruvate through the so-called pyruvate dehydrogenase (PDH) bypass, comprised by Pdc, Ald and Acs enzymes. PDC-gene expression was not significantly changed in the glucose phase. As mentioned in the section above, ALD6, responsible for the catalysis of acetaldehyde to acetate, was significantly downregulated in the glucose phase in strain PTPI1-STB5. The alcohol dehydrogenase Adh1 is the primary enzyme to divert flux from the PDH bypass, catalyzing the conversion of acetaldehyde to ethanol, while alcohol dehydrogenase Adh2 catalyzes the reverse reaction. Ethanol formation was reduced in strain PTPI1-STB5 (Table S7, Supporting Information), also indicated by the reduced expression of ADH1 (LCF = −0.39) and upregulation of ADH2. When the raw read counts for ACS1 and ACS2 were combined, a small negative effect on expression was observed (LFC = −0.22). With regards to the genes directely related to FA biosynthesis, ACC1 and FAS1 were significantly upregulated (LFC = 0.21 and LFC = 0.90).
In the ethanol phase, STB5 overexpression led to an approximate 3-fold increase in STB5 transcripts compared to the control, which is in agreement with a lower activity of the TPI1 promoter under ethanol consumption (Peng et al. 2015). A large number of transcriptional processes were however affected, indicating that limited overexpression of STB5 is enough to influence the expression of its target genes. As previously mentioned, STB5 overexpression caused a coordinated upregulation of glycolytic genes (Fig. 6C) in the ethanol phase, which likely is mediated through an upregulation of the TF gene GCR1 highlighted in the TF analysis (Fig. 6A and B), which is instrumental in the expression of glycolytic genes (Uemura and Fraenkel 2000). Simultaneously, gluconeogenic genes (PDB1, PCK1 and PYC1) and the initial gene in the oxidative PPP (ZWF1) were downregulated compared to the control, indicating that flux through the oxidative PPP was reduced in the ethanol phase (Fig. 6C). GND1, GND2, TAL1, NQM1 and TKL2 were all significantly upregulated, suggesting that precursors for biosynthesis could be obtained from the non-oxidative PPP. Compared to growth on glucose, expression of NADPH-producing IDP2 increased markedly in the ethanol phase for both strains (Table S8, Supporting Information) which correlates well with previous reports that its expression is induced in the presence of non-fermentable carbon sources (Haselbeck and Mcalisterhenn 1993). Nevertheless, both IDP2 and ALD6 were in the ethanol phase downregulated in the PTPI1-STB5 strain compared to the control, while neither of ILV5, GOR1, YEF1, ALD4 and ADH6 were differentially expressed. Furthermore, a number of glyoxylate and TCA cycle genes were significantly downregulated (ICL1, MLS1, MDH2, SFC1, LDC1, SDH3 and FUM1) in the ethanol phase. Gene-set analysis of differentially expressed genes in the ethanol phase (Fig. S3, Supporting Information) also highlights a distinct downregulation of processes such as cristae formation and ATP synthesis coupled proton transport, suggesting that the strain had a reduced respiratory capacity in the ethanol phase.
At steady state conditions, the overexpression mutant had a higher consumption of oxygen and production of carbon dioxide while the biomass yield was about 91% of that observed for the control (Table S7, Supporting Information), indicating a higher degree of respiration. These observations were also reflected in the transcriptional data, as gene set analysis shows that ribosomal processes were downregulated while respiratory and mitochondrial processes were upregulated (Fig. S4, Supporting Information). Within the PPP, transcription of ZWF1, SOL4 and RKI1 were not significantly changed, SOL3 and TKL2 were downregulated (LFC = −0.27 and -0.49, respectively), while GND1, GND2, RPE1 and TAL1 were upregulated (LFC = 1.64, 1.81, 0.22 and 0.89, respectively). Of the other genes indicated to be activated by Stb5 in the study of Larochelle et al. (2006), almost all were upregulated: ALD6, IDP2, GOR1, YEF1, ALD4 and ADH6 (LFC = 1.0, 0.61, 0.53, 2.96, 0.63 and 0.82). This agrees with the reporter TF analysis, which highlighted that Stb5 target genes were considered significantly upregulated in steady state conditions but not in glucose nor ethanol phase (Fig. 6A).
DISCUSSION
The targeting of TFs has been attempted previously in metabolic engineering strategies in S. cerevisiae, mainly to abolish certain types of negative regulation, such as the deletion of OPI1 to boost phospholipid synthesis (Feng, Lian and Zhao 2015). Fewer studies have investigated the overexpression of transcriptional activators to influence production, a likely result of the typically broad target range of TF.
We decided to evaluate overexpression of the TF STB5 to influence FA biosynthesis, as previous reports had suggested it to have a primary role in the oxidative stress response via the transcriptional activation of genes related to NADPH generation (Larochelle et al. 2006; Hector et al. 2009; Ouyang et al. 2018). Previously, it has been shown that a rerouting of glycolytic flux through the oxidative PPP mediated by a coordinated upregulation of several PPP genes and downregulation of PGI1 had a beneficial effect on FFA synthesis (Yu et al. 2018). Larochelle et al. reported that STB5, in part, is likely to mediate that particular effect when the cell is exposed to oxidative stress (Larochelle et al. 2006), which is why we were interested to know if a similar result could be obtained by simply replacing the native STB5 promoter. If functional, this potentially could constitute an easy-to-implement strategy to boost NADPH supply for various applications.
Indeed, during the preparation of this manuscript, several recent reports were published exploring if overexpression of STB5 could increase efficiency of biosynthetic processes dependent on NADPH or a high flux through the PPP (Hong et al. 2019; Kim et al. 2018; Park et al. 2018). Hong et al. observed that STB5 and OLE1 co-overexpression significantly increased lycopene titers in an engineered strain (Hong et al. 2019). Park et al. overexpressed STB5 as a means to enhance the flux through the PPP, and found the strategy positively stimulated shinorine production (Park et al. 2018). However, even while Kim et al. observed a 1.2-fold increase in available NADPH upon STB5 overexpression, the titer of their target-product protopanaxadiol was negatively affected (Kim et al. 2018). The results of these studies highlight the broad potential of STB5 overexpression as a metabolic engineering strategy, but simultaneously raise questions about the systemic effects STB5 overexpression may have on the host—a gap which our analysis seeks to fill.
In a previous study by Cadiere et al., STB5 was overexpressed by replacement of the native promoter with PTDH3, which resulted in the strain not being able to grow on glucose, an effect that was attributed to the strain´s inability to re-oxidize a surplus of cellular NADPH (Cadiere, Galeote and Dequin 2010). In our study, we found that overexpression of STB5 only resulted in a reduction in growth rate (Table 3) and final biomass concentration (Table S5, Supporting Information). This could be attributed to the use of different yeast strains, as Cadiere et al. used a wine yeast derivative likely to be dissimilar to CEN.PK113–5D. Even though we observed that a stronger expression was linked to a greater decrease in µmax, the difference was relatively small when comparing the effect of utilizing either PCYC1 or PPGK1. PCYC1 is considered to be a relatively weak promoter, and reduced µmax to 80% of the value for the control, while the strong glycolytic promoter PPGK1 resulted in a µmax corresponding to 67% of the control. This could potentially be a result of target saturation—that even a limited overexpression of STB5 successfully allows the TF to bind most of its target promoters and mediate transcriptional changes, while a stronger expression has a less pronounced effect. Our obtained RNAseq data show that the average FPKM of STB5, CYC1 and PGK1 in the control strain equals 16, 595 and 3015 in the glucose phase (Table S8, Supporting Information), respectively, which if directly translated to induction capacity of STB5 supports that a saturation effect occurs. However, as the average FPKM of TPI1 in the glucose phase was 2459 while it equaled 494 for STB5 when it was placed under the control of PTPI1, a direct correlation between expression strength of the native genes and corresponding promoter-exchanged target cannot be made.
Our efforts to quantify cellular NADPH content in the exponential phase (Fig. S1, Supporting Information) did not deliver results resembling the outcome by Hector et al., who demonstrated a 2.5-fold increase in NADPH when STB5 was overexpressed from a plasmid (Hector et al. 2009). Also in that particular study, a different yeast background was used (BY4727) compared to the one utilized herein (CEN.PK113–5D). Previous reports indicated that the NADP+/NADPH ratio is difficult to perturb in a CEN.PK background via genetic modifications. In response to overexpression of the NADPH-producing malic enzyme, CEN.PK113–7D counteracted imposed changes by reducing flux through the oxidative PPP, which resulted in an unchanged NADPH/NADP+ ratio (Moreira dos Santos et al. 2004). It is likely that STB5-overexpressing strains in our study counteracted a potentially increased NADPH synthesis with an increased consumption, for example by producing more FAs, (Fig. 3) or by partly overriding the STB5-mediated regulation by other mechanisms, such as transcriptionally downregulating key components of the PPP (Fig. 6C).
We also indirectly assessed NADPH supply in living cells by evaluating how well cells handle oxidative stress. For example, strains devoid of ZWF1 and oxidative PPP flux have frequently been shown to be sensitive to oxidative agents, as was the case here (Fig. 2). Contradictory to our initial hypothesis, overexpression of STB5 reduced resistance against oxidative stress in a similar way to that observed for the zwf1∆ mutant. Partly, this sensitivity could be related to a reduced oxidative PPP flux, indicated by the RNAseq data (Fig. 6C). In response to oxidative perturbations, the cell requires an immediate stabilization of its redox state, which has been shown to be dependent on metabolic reconfiguration—namely a dynamic redirection of glycolytic flux towards the oxidative PPP (Ralser et al. 2009). This rerouting is initiated via the inhibition of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) in response to oxidative perturbation, which occurs within seconds of exposure and thus effectively blocks the glycolytic flux. In comparison, transcriptional adaptation of PPP-gene expression to oxidative stress occurs after about 4–7 min (Chechik et al. 2008), during which time a redox collapse would have detrimental effects on cell physiology and survival. Furthermore, the cellular oxidative stress response is dependent on parameters additional to NADPH supply, and overexpression of STB5 for example significantly reduced expression of GSH1, encoding the initial step in glutathione biosynthesis, (LFC = −0.75), which possibly could limit survival in response to oxidative stress.
Even though the experiment of strain survival in response to oxidative stress indicated a potential reduction in NADPH supply, we observed positive effects of STB5 overexpression on FA synthesis, specifically in the glucose phase (Fig. 3A). Based on the initial hypothesis that STB5 overexpression could redirect flux through the oxidative PPP, it initially appeared logical as the largest flux diversion could be achieved when there is a steady inflow of glucose. However, when weighing in the reduced survival of PTPI1-STB5 in response to oxidative stress (Fig. 2), that the beneficial effect on FFA synthesis was independent of flux through Zwf1 (Fig. 3B) and differential gene expression analysis results of PPP genes (Fig. 6C), the beneficial effect on FFA synthesis most likely has other causes, potentially mediated through an increased NADPH supply via other enzymatic routes. The RNAseq analysis showed that previously reported Stb5-target genes encoding NADPH-producing enzymes such as ALD4, ADH6, IDP2, GOR1 and YMR315W were significantly upregulated while ALD6 and ILV5 were downregulated (Fig. 6C). An increased net activity of corresponding enzymes, and/or reduced consumption of NADPH through other reactions, would not have been of help during an oxidative perturbation, as it would cause an upstream blockage of the carbon flux at GAPDH, but it could still represent a source of NADPH for FA synthesis.
We made an interesting observation with regards to the origin of NADPH supply for FA synthesis in the control strain. There was a 50% reduction in FFA synthesis in the zwf1∆ strain at the end of the cultivation (Fig. 3B), which indicates that the oxidative PPP has a significant role in FA synthesis in S. cerevisiae during ethanol consumption. This observation is consistent with the fact that the majority of NADPH for FA synthesis in the oleaginous yeast Yarrowia lipolytica stems from the oxidative PPP (Wasylenko, Ahn and Stephanopoulos 2015). Interestingly, no negative effect of zwf1∆ on FFA production was observed during the exponential phase. This could potentially be attributed to a low requirement of NADPH and biosynthetic precursors during fermentative growth when the biomass yield is low, exemplified by the low YSX in exponential phase (0.157) compared to that observed in complete respiratory conditions during steady state (0.517) (Table S7, Supporting Information). Strong overexpression of STB5 led to a negative effect on FA synthesis in the ethanol phase (Fig. 3A). Based on the observation made in the zwf1∆ strain, this could possibly be explained by the ethanol phase specific upregulation of genes related to glycolysis and reduced expression of gluconeogenic genes and ZWF1 observed in the PTP11-STB5 strain. Furthermore, a reduced expression of genes related to respiration was also observed, which should affect generation of ATP, also demanded in high quantities during FA synthesis.
Both mild (PCYC1) to strong (PADH1/PADH1/PPGK1) overexpression of STB5 resulted in a relatively equal improvement on FA production (Fig. 3A), while the growth rate between the strains differed significantly (Table S6, Supporting Information). Thus, we wondered if NADPH supply was increased to a larger extent in strains where STB5 expression was controlled by a strong promoter, but that the precursor acetyl-CoA instead could constitute a limitation for FA biosynthesis in the glucose phase. In order to study if this was the case, we employed a phosphoketolase pathway as shown by previous studies to enhance acetyl-CoA availability (Meadows et al. 2016). Initially, no beneficial effect on FA synthesis was observed when the Xfpk/Pta pathway was expressed in the control strain (Fig. 4, AB26 vs AB25). This is likely due to the efficient native conversion of acetyl phosphate to acetate via Gpp1 and Gpp2 (Bergman et al. 2016), also strengthening the hypothesis that the acetate to acetyl-CoA conversion is limited in glucose phase. Therefore, we decided to delete the gene encoding the phosphatase Gpp1 in the control and STB5-overexpression strain, and showed that this deletion alone had a positive effect on FA synthesis (AB27 vs AB25). Possibly, this could be related to the fact that glycerol-3-phosphate—which is likely to accumulate in the gpp1∆ strain—is a substrate for phospholipid and triacylglycerol (TAG) synthesis. Cellularly produced FFAs in yeast have been shown to stem from the hydrolysis of neutral lipids and phospholipids (Leber et al. 2014; Ferreira et al. 2018b). Expression of Xfpk/Pta in this background significantly increased FFA production by 25% (AB28 vs AB27), while its expression in the gpp1∆ PTPI1-STB5 background increased FFA production by 37% (AB30 vs AB29). The increased acetyl-CoA supply provided by the Xfpk/Pta-pathway and consequences of STB5 overexpression apparently had a synergistic effect on FFA synthesis, as the sum of the individual effects on FFA synthesis was lower than the observed dual effect. This would be an expected outcome if STB5 overexpression were able to increase the NADPH supply, as a surplus of NADPH only will be efficiently utilized for FA synthesis if acetyl-CoA is readily available. However, the RNAseq analysis pointed out that expression of ACC1 and FAS1 was upregulated in response to STB5 overexpression, which also could lead to a more efficient utilization of cytosolic acetyl-CoA. Unfortunately, the impact of these two separate effects cannot be resolved.
STB5 overexpression by itself does not appear to be able to provoke the same gene expression changes in the glucose phase as observed when cells are treated with oxidative stress (Larochelle et al. 2006). Ouyang et al. investigated Stb5-mediated regulation in different environmental conditions in absence of oxidative stress agents, and showed that Stb5 was bound to the promoter of ZWF1 in all instances, whereas deletion of STB5 did not affect ZWF1 transcription (Ouyang et al. 2018). However, when exposed to diamide, ZWF1 induction has been shown to be strongly reduced in a stb5∆ strain compared to the control (Larochelle et al. 2006). Similarly, Hector et al. observed a moderate induction of a reporter gene placed under the control of a Stb5-regulated promoter by overexpressing STB5, while when treating the cells with the oxidative agent diamide, they observed a 2-fold increase in activity (Hector et al. 2009). Possibly, the cellular consequences brought about by oxidative stress, such as a reduced NADPH/NADP+ ratio, are required for proper binding of Stb5 to its binding motif or subsequent induction.
Additionally, NADPH produced by the cell will efficiently be consumed when exposed to oxidative stress—a redox sink which was not present in strain PTPI1-STB5 (AB15). A perturbation of the redox balance would have global cellular effects and could potentially be a starting signal to the complex transcriptional reprogramming observed in PTPI1-STB5 in the glucose phase. The maintenance of redox homeostasis typically involves to counteract a drop in the NADPH/NADP+ ratio, a situation which for example is commonly encountered during aerobic respiration when ROS form due to leakage of electrons from the respiratory chain (Aung-Htut et al. 2012). The reverse situation is likely to be a rare event in nature, but as mentioned previously, overexpression of a malic enzyme targeted to the cytosol was shown to decrease flux through the oxidative PPP (Moreira dos Santos et al. 2004). Furthermore, an increased cytosolic NADPH availability mediated via a deletion of a NADP+-dependent glutamate dehydrogenase gene GDH1 was found to decrease transcription of ZWF1, GND1 and ALD6 (Bro, Regenberg and Nielsen 2004). This correlates well with the observed downregulation of ZWF1 and ALD6, and the anticipated but not actualized Stb5-mediated upregulation of GND1, in strain PTPI1-STB5 in exponential phase.
Furthermore, Ouyang et al. found, among others genes, GND1, TAL1, ALD6 and YEF1 to be bound by Stb5 in the control strain and simultaneously downregulated in a stb5∆ strain during glucose-limited conditions, indicating that Stb5 acts directly as a transcriptional activator of these genes in the absence of oxidative stress (Ouyang et al. 2018). Even though these genes were either not significantly changed or even downregulated during the exponential phase in our study, during chemostat conditions all of the mentioned genes were significantly upregulated in response to STB5 overexpression (LFC corresponding to 1.64, 0.89, 1.02 and 2.96). Furthermore, repression of ZWF1 transcription was not observed in steady state conditions, and all but one of the remaining target genes suggested by Larochelle et al. (IDP2, GOR1, ALD4 and ADH6) were also upregulated. The cellular NADPH/NADP+ ratio is generally held high in order to be able to drive biosynthetic reactions, and the cytosolic ratio in CEN.PK 113–7D has been reported to be 22.0 and 15.6 during exponential phase and steady state, respectively (Zhang et al. 2015). This supports that an increase in NADPH supply would be particularly damaging in the exponential phase, which could be an explanation for the larger reduction in biomass yield observed for strain PTPI1-STB5 in the exponential phase (81% of control) compared to steady state (91% of control) (Table S7, Supporting Information). Taken together, this strongly implies that the Stb5-mediated gene regulation partially is superseded by stronger regulatory mechanisms to counteract a redox imbalance in the batch phase.
We observed a mitigation of the negative influence of mild STB5 overexpression (promoter PCYC1) had on growth rate when a strain with a greater ability for FA synthesis was utilized (Table 3 vs Table S6, Supporting Information). With this in mind, optimally a mild to moderate overexpression of STB5 in a yeast strain with excellent ability to consume the NADPH during product formation is suggested for optimal performance. This would likely increase biosynthetic ability of the desired compound while at the same time reducing undesirable systemic effects. Furthermore, as negative effects on FFA synthesis were observed in the ethanol phase, the utilization of a glucose-specific promoter could be of interest to evaluate.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Eduard Kerkhoven for support during bioreactor cultivations, Kate Campbell for helpful discussions regarding PPP and NADPH metabolism, and staff at the Novo Nordisk Foundation Center for Biosustainability, DTU, Denmark, for conducting the RNA sequencing.
Authors’ contributions: A.B. conceived the study, performed majority of experiments, analyzed and interpreted the data and wrote manuscript. D.V. constructed yeast strains AB12-AB20 and conducted preliminary studies of these. J.H. conducted the RNAseq profiling and analysis. J.H., J.N., Y.C. and V.S. discussed the results and edited the manuscript. All authors read and approved the final manuscript.
FUNDING
This work was supported by the Knut and Alice Wallenberg Foundation, the Novo Nordisk Foundation [Grant number NNF10CC1016517], the Swedish Foundation for Strategic Research, the Swedish Research Council, FORMAS and the Carl Tryggers Foundation.
Conflict of interest. None declared.
REFERENCES
- Aung-Htut MT, Ayer A, Breitenbach Met al.. Oxidative stresses and ageing. Subcell Biochem. 2012;57:13–54. [DOI] [PubMed] [Google Scholar]
- Avery SV, Howlett NG, Radice S. Copper toxicity towards Saccharomyces cerevisiae: dependence on plasma membrane fatty acid composition. Appl Environ Microbiol. 1996;62:3960–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman A, Hellgren J, Moritz Tet al.. Heterologous phosphoketolase expression redirects flux towards acetate, perturbs sugar phosphate pools and increases respiratory demand in Saccharomyces cerevisiae. Microb Cell Fact. 2019;18:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman A, Siewers V, Nielsen Jet al.. Functional expression and evaluation of heterologous phosphoketolases in Saccharomyces cerevisiae. Amb Express. 2016;6:115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bro C, Regenberg B, Nielsen J. Genome-wide transcriptional response of a Saccharomyces cerevisiae strain with an altered redox metabolism. Biotechnol Bioeng. 2004;85:269–76. [DOI] [PubMed] [Google Scholar]
- Cadiere A, Galeote V, Dequin S. The Saccharomyces cerevisiae zinc factor protein Stb5p is required as a basal regulator of the pentose phosphate pathway. FEMS Yeast Res. 2010;10:819–27. [DOI] [PubMed] [Google Scholar]
- Castegna A, Scarcia P, Agrimi Get al.. Identification and functional characterization of a novel mitochondrial carrier for citrate and oxoglutarate in Saccharomyces cerevisiae. J Biol Chem. 2010;285:17359–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chechik G, Oh E, Rando Oet al.. Activity motifs reveal principles of timing in transcriptional control of the yeast metabolic network. Nat Biotechnol. 2008;26:1251–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Daviet L, Schalk Met al.. Establishing a platform cell factory through engineering of yeast acetyl-CoA metabolism. Metab Eng. 2013;15:48–54. [DOI] [PubMed] [Google Scholar]
- Chen Y, Partow S, Scalcinati Get al.. Enhancing the copy number of episomal plasmids in Saccharomyces cerevisiae for improved protein production. FEMS Yeast Res. 2012;12:598–607. [DOI] [PubMed] [Google Scholar]
- Christodoulou D, Link H, Fuhrer Tet al.. Reserve flux capacity in the pentose phosphate pathway enables Escherichia coli’s rapid response to oxidative stress. Cell Syst. 2018;6:569–78 e7. [DOI] [PubMed] [Google Scholar]
- de Jong BW, Shi S, Siewers Vet al.. Improved production of fatty acid ethyl esters in Saccharomyces cerevisiae through up-regulation of the ethanol degradation pathway and expression of the heterologous phosphoketolase pathway. Microb Cell Fact. 2014;13:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench JG, Fusi N, Sullender Met al.. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol. 2016;34:184–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faergeman NJ, Knudsen J. Role of long-chain fatty acyl-CoA esters in the regulation of metabolism and in cell signalling. Biochem J. 1997;323:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fargione J, Hill J, Tilman Det al.. Land clearing and the biofuel carbon debt. Science. 2008;319:1235–38. [DOI] [PubMed] [Google Scholar]
- Feng XY, Lian JZ, Zhao HM. Metabolic engineering of Saccharomyces cerevisiae to improve 1-hexadecanol production. Metab Eng. 2015;27:10–19. [DOI] [PubMed] [Google Scholar]
- Ferreira R, Teixeira PG, Gossing Met al.. Metabolic engineering of Saccharomyces cerevisiae for overproduction of triacylglycerols. Metab Eng Commun. 2018a;6:22–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira R, Teixeiraa PG, Siewers Vet al.. Redirection of lipid flux toward phospholipids in yeast increases fatty acid turnover and secretion. P Natl Acad Sci USA. 2018b;115:1262–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Schiestl RH. Frozen competent yeast cells that can be transformed with high efficiency using the LiAc/SS carrier DNA/PEG method. Nat Protoc. 2007;2:1–4. [DOI] [PubMed] [Google Scholar]
- Gombert AK, dos Santos MM, Christensen Bet al.. Network identification and flux quantification in the central metabolism of Saccharomyces cerevisiae under different conditions of glucose repression. J Bacteriol. 2001;183:1441–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haselbeck RJ, McAlister-Henn L. Function and expression of yeast mitochondrial NAD- and NADPH-specific isocitrate dehydrogenases. J Biol Chem. 1993;268:12116–22. [PubMed] [Google Scholar]
- Haushalter RW, Kim W, Chavkin TAet al.. Production of anteiso-branched fatty acids in Escherichia coli; next generation biofuels with improved cold-flow properties. Metab Eng. 2014;26:111–8. [DOI] [PubMed] [Google Scholar]
- Hector RE, Bowman MJ, Skory CDet al.. The Saccharomyces cerevisiae YMR315W gene encodes an NADP(H)-specific oxidoreductase regulated by the transcription factor Stb5p in response to NADPH limitation. New Biotechnol. 2009;26:171–80. [DOI] [PubMed] [Google Scholar]
- Hong J, Park SH, Kim Set al.. Efficient production of lycopene in Saccharomyces cerevisiae by enzyme engineering and increasing membrane flexibility and NAPDH production. App Microbiol Biotechnol. 2019;103:211–223. [DOI] [PubMed] [Google Scholar]
- Inoue H, Nojima H, Okayama H. High efficiency transformation of Escherichia coli with plasmids. Gene. 1990;96:23–28. [DOI] [PubMed] [Google Scholar]
- Jakounas T, Sonde I, Herrgard Met al.. Multiplex metabolic pathway engineering using CRISPR/Cas9 in Saccharomyces cerevisiae. Metab Eng. 2015;28:213–22. [DOI] [PubMed] [Google Scholar]
- Kim JE, Jang IS, Sung BHet al.. Rerouting of NADPH synthetic pathways for increased protopanaxadiol production in Saccharomyces cerevisiae. Sci Rep-Uk. 2018;8:15820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krivoruchko A, Serrano-Amatriain C, Chen Yet al.. Improving biobutanol production in engineered Saccharomyces cerevisiae by manipulation of acetyl-CoA metabolism. J Ind Microbiol Biotechnol. 2013;40:1051–56. [DOI] [PubMed] [Google Scholar]
- Larochelle M, Drouin S, Robert Fet al.. Oxidative stress-activated zinc cluster protein Stb5 has dual activator/repressor functions required for pentose phosphate pathway regulation and NADPH production. Mol Cell Biol. 2006;26:6690–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leber C, Polson B, Fernandez-Moya Ret al.. Overproduction and secretion of free fatty acids through disrupted neutral lipid recycle in Saccharomyces cerevisiae. Metab Eng. 2014;28:54–62. [DOI] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–30. [DOI] [PubMed] [Google Scholar]
- Llobell A, Lopezruiz A, Peinado Jet al.. Glutathione-reductase directly mediates the stimulation of yeast glucose-6-phosphate-dehydrogenase by Gssg. Biochem J. 1988;249:293–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marella ER, Holkenbrink C, Siewers Vet al.. Engineering microbial fatty acid metabolism for biofuels and biochemicals. Curr Opin Biotech. 2018;50:39–46. [DOI] [PubMed] [Google Scholar]
- Meadows AL, Hawkins KM, Tsegaye Yet al.. Rewriting yeast central carbon metabolism for industrial isoprenoid production. Nature. 2016;537:694–97. [DOI] [PubMed] [Google Scholar]
- Minard KI, McAlister-Henn L. Sources of NADPH in yeast vary with carbon source. J Biol Chem. 2005;280:39890–96. [DOI] [PubMed] [Google Scholar]
- Miyagi H, Kawai S, Murata K. Two sources of mitochondrial NADPH in the yeast Saccharomyces cerevisiae. J Biological Chem. 2009;284:7553–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morano KA, Grant CM, Moye-Rowley WS. The response to heat shock and oxidative stress in Saccharomyces cerevisiae. Genetics. 2012;190:1157–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira dos Santos M, Raghevendran V, Kotter Pet al.. Manipulation of malic enzyme in Saccharomyces cerevisiae for increasing NADPH production capacity aerobically in different cellular compartments. Metab Eng. 2004;6:352–63. [DOI] [PubMed] [Google Scholar]
- OECD/FAO. OECD-FAO Agricultural Outlook 2018–2027. Paris: OECD Publishing, 2018. [Google Scholar]
- Ouyang L, Holland P, Lu Het al.. Integrated analysis of the yeast NADPH-regulator Stb5 reveals distinct differences in NADPH requirements and regulation in different states of yeast metabolism. FEMS Yeast Res. 2018;18,foy091. [DOI] [PubMed] [Google Scholar]
- Park SH, Lee K, Jang JWet al.. Metabolic engineering of Saccharomyces cerevisiae for production of shinorine, a sunscreen material, from xylose. Acs Synth Biol. 2019;8:346–357. [DOI] [PubMed] [Google Scholar]
- Partow S, Siewers V, Bjorn Set al.. Characterization of different promoters for designing a new expression vector in Saccharomyces cerevisiae. Yeast. 2010;27:955–64. [DOI] [PubMed] [Google Scholar]
- Peng BY, Williams TC, Henry Met al.. Controlling heterologous gene expression in yeast cell factories on different carbon substrates and across the diauxic shift: a comparison of yeast promoter activities. Microb Cell Fact. 2015;14:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Pertea GM, Antonescu CMet al.. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotech. 2015;33:290–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfleger BF, Gossing M, Nielsen J. Metabolic engineering strategies for microbial synthesis of oleochemicals. Metab Eng. 2015;29:1–11. [DOI] [PubMed] [Google Scholar]
- Ralser M, Wamelink MMC, Latkolik Set al.. Metabolic reconfiguration precedes transcriptional regulation in the antioxidant response. Nat Biotechnol. 2009;27:604–605. [DOI] [PubMed] [Google Scholar]
- Regenberg B, Grotkjaer T, Winther Oet al.. Growth-rate regulated genes have profound impact on interpretation of transcriptome profiling in Saccharomyces cerevisiae. Genome Biol. 2006;7:R107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinnerthaler M, Buttner S, Laun Pet al.. Yno1p/Aim14p, a NADPH-oxidase ortholog, controls extramitochondrial reactive oxygen species generation, apoptosis, and actin cable formation in yeast. P Natl Acad Sci USA. 2012;109:8658–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharnewski M, Pongdontri P, Mora Get al.. Mutants of Saccharomyces cerevisiae deficient in acyl-CoA synthetases secrete fatty acids due to interrupted fatty acid recycling. FEBS J. 2008;275:2765–78. [DOI] [PubMed] [Google Scholar]
- Shi S, Chen Y, Siewers Vet al.. Improving production of malonyl coenzyme A-derived metabolites by abolishing Snf1-dependent regulation of Acc1. mBio. 2014;5:e01130–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba Y, Paradise EM, Kirby Jet al.. Engineering of the pyruvate dehydrogenase bypass in Saccharomyces cerevisiae for high-level production of isoprenoids. Metab Eng. 2007;9:160–8. [DOI] [PubMed] [Google Scholar]
- Stephanopoulos G, Vallino JJ. Network rigidity and metabolic engineering in metabolite overproduction. Science. 1991;252:1675–81. [DOI] [PubMed] [Google Scholar]
- Stincone A, Prigione A, Cramer Tet al.. The return of metabolism: Biochemistry and physiology of the pentose phosphate pathway. Biol Rev. 2015;90:927–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stovicek V, Borodina I, Forster J. CRISPR–Cas system enables fast and simple genome editing of industrial Saccharomyces cerevisiae strains. Metab Eng Commun. 2015;2:13–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana C, Yoo JY, Tagne JBet al.. Combined global localization analysis and transcriptome data identify genes that are directly coregulated by Adr1 and Cat8. Mol Cell Biol. 2005;25:2138–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tehlivets O, Scheuringer K, Kohlwein SD. Fatty acid synthesis and elongation in yeast. Biochim Biophys Acta. 2007;1771:255–70. [DOI] [PubMed] [Google Scholar]
- Uemura H, Fraenkel DG. Glucose metabolism in gcr mutants of Saccharomyces cerevisiae. J Bacteriol. 2000;182:2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valko M, Morris H, Cronin MT. Metals, toxicity and oxidative stress. Curr Med Chem. 2005;12:1161–208. [DOI] [PubMed] [Google Scholar]
- Varemo L, Nielsen J, Nookaew I. Enriching the gene set analysis of genome-wide data by incorporating directionality of gene expression and combining statistical hypotheses and methods. Nucleic Acids Res. 2013;41:4378–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verduyn C, Postma E, Scheffers WAet al.. Effect of benzoic acid on metabolic fluxes in yeasts: a continuous-culture study on the regulation of respiration and alcoholic fermentation. Yeast. 1992;8:501–17. [DOI] [PubMed] [Google Scholar]
- Villadsen I, , Nielsen J, Lidén G, Bioreaction Engineering Principles. Third Edition. New York: Springer Science+Business Media, 2011. [Google Scholar]
- Wang YP, Zhou LS, Zhao YZet al.. Regulation of G6PD acetylation by SIRT2 and KAT9 modulates NADPH homeostasis and cell survival during oxidative stress. Embo J. 2014;33:1304–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasylenko TM, Ahn WS, Stephanopoulos G. The oxidative pentose phosphate pathway is the primary source of NADPH for lipid overproduction from glucose in Yarrowia lipolytica. Metab Eng. 2015;30:27–39. [DOI] [PubMed] [Google Scholar]
- Yu T, Zhou YJ, Huang Met al.. Reprogramming yeast metabolism from alcoholic fermentation to lipogenesis. Cell. 2018;174;1549–58. [DOI] [PubMed] [Google Scholar]
- Zhang JR, ten Pierick A, van Rossum HMet al.. Determination of the cytosolic NADPH/NADP ratio in Saccharomyces c erevisiae using shikimate dehydrogenase as sensor reaction. Sci Rep-Uk. 2015;5:12846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Nielsen J, Liu Z. Metabolic engineering of Saccharomyces cerevisiae for production of fatty acid-derived hydrocarbons. Biotechnol Bioeng. 2018;115:2139–47. [DOI] [PubMed] [Google Scholar]
- Zhou YJJ, Buijs NA, Zhu ZWet al.. Production of fatty acid-derived oleochemicals and biofuels by synthetic yeast cell factories. Nat Commun. 2016;7:11709. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.