Abstract
A number of microRNAs (miRs) have been revealed to be involved in the development of osteoporosis, including postmenopausal osteoporosis. The aim of the present study was to investigate miR-135a-5p expression and the cellular function of miR-135a-5p and its underlying mechanism in postmenopausal osteoporosis. miR-135a-5p expression levels in the femoral neck trabecular bone tissue fragments from postmenopausal women with or without osteoporosis were detected by reverse transcription-quantitative polymerase chain reaction and western blot analysis. The role of miR-135a-5p during osteogenic differentiation was examined by performing gain- and loss-of-function experiments using miR-135a-5p mimic or inhibitor. TargetScan bioinformatics analysis was sued to predict targets of miR-135a-5p, which were confirmed using luciferase reporter assays. miR-135a-5p expression was significantly upregulated in femoral neck trabecular bone tissue fragments from postmenopausal women with osteoporosis compared with postmenopausal women without osteoporosis. In addition, miR-135a-5p expression levels significantly decreased during osteogenic differentiation in the C2C12 cell model. miR-135a-5p overexpression decreased the osteogenic potential of C2C12 cells, as miR-135a-5p overexpression significantly reduced the expression levels of several key osteoblast markers, whilst miR-135a-5p knockdown had the opposite effect. Furthermore, the current study demonstrated that RUNX2 was a direct target of miR-135a-5p. Rescue experiments indicated that RUNX2 overexpression significantly reversed the effect of miR-135a-5p mimic on the osteogenic potential of C2C12 cells, indicating that miR-135a-5p mediates osteogenic differentiation via direct targeting of RUNX2. Taken together, these results suggest that miR-135a-5p may serve a role in osteoporosis progression by regulating osteogenic differentiation via RUNX2.
Keywords: osteoporosis, microRNA-135a-5p, runt related transcription factor 2, osteoblast markers
Introduction
Osteoporosis is a common chronic metabolic bone disease characterized by low bone mineral density and low-traumata fractures (1,2). As a common disease among postmenopausal women, the main mechanism for the development of postmenopausal osteoporosis is the imbalance between bone formation and bone resorption resulting from a decreased estrogen level (3,4). China is the country with the largest number of elderly people worldwide (5). In China, it is estimated that at least 90 million people suffer from osteoporosis and by 2050 this will increase to 221 million (6,7). Osteoporosis is a chromic disease that predominantly affects the elderly worldwide, especially women (8), and therefore improvements in the prevention and treatment of osteoporosis are required.
Restoring and maintaining the balance between bone formation and bone resorption is an effective way to treat postmenopausal osteoporosis (9,10). However, due to the complex molecular mechanisms underlying osteogenic bone formation, and the lack of osteogenic drug targets, the current treatment of postmenopausal osteoporosis is focused on the inhibition of osteoclast activity and bone resorption capacity. Therefore, a greater understanding of the molecular mechanism underlying osteogenic bone formation, as well as identifying therapeutic targets with potential osteogenic effects, will provide novel strategies for the treatment of postmenopausal osteoporosis.
MicroRNAs (miRNAs or miRs) are a family of small non-coding single stranded RNAs, which can negatively regulate the expression of target genes during various cellular events, including proliferation, apoptosis and differentiation by binding to the 3′untranslated region (UTR) of target genes (11–14). Increasing evidence suggests that miRNAs are involved in the development of osteoporosis, including postmenopausal osteoporosis (15–18). However, the cellular function of miR-135a-5p in postmenopausal osteoporosis remains unknown.
The aim of the present study was to investigate miR-135a-5p expression and the cellular function of miR-135a-5p and its underlying mechanism in postmenopausal osteoporosis.
Materials and methods
Clinical samples
Bone fragments were obtained from 20 postmenopausal female patients (54–71 years old) with osteoporosis who underwent hip replacement for osteoporotic fractures (OP) at Ningbo First Hospital between April 2015 and April 2017. The control group were recruited at the same time and consisted of 20 postmenopausal female patients (52–74 years old) with osteoarthritis only and not osteoporosis, according to BMD and T-score measurements (0.791±0.081 and −0.26±0.831, respectively). Bone fragments, which were extracted from the transcervical region of the femoral neck, were dissected into smaller fragments, washed three times in PBS and stored at −80°C until further use. The present study was approved by the Ethics Committee of Ningbo First Hospital (Ningbo, China) and written informed consent was obtained from each patient.
To extract total RNA, bone fragments in TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) were homogenized using a tissue homogenizer (Omni International, Kennesaw, GA, USA) and total RNA was extracted using the RNA RNeasy kit (Qiagen GmbH, Hilden, Germany) according to the manufacturer's protocol.
Cell culture and treatment
Mouse myoblast cell line C2C12 (ATCC® CRL-1772™) was purchased from the American Type Culture Collection (Manassas, VA, USA). Cells were cultured in Dulbecco's modified Eagle medium (Invitrogen; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum (Invitrogen; Thermo Fisher Scientific, Inc.) and 1% streptomycin and penicillin mix solution, and maintained at 37°C in a 5% CO2-humidified incubator.
The C2C12 cell line is a typical pluripotent mesenchymal precursor cell line that possesses the potential to differentiate into myoblasts, chondroblasts and osteoblasts (19,20). In the current study, the C2C12 cell line was used as a cellular model of osteogenic differentiation. Osteogenic differentiation was induced following treatment with 2 nM bone morphogenetic protein 2 (BMP2; Invitrogen; Thermo Fisher Scientific, Inc.) for 24 h, as previously described (21).
Cell transfection
C2C12 cells were seeded in 6-well plates at a density of 1×106 cells/well and cultured at 37°C for 24 h. miRNA mimic and inhibitor were obtained from Shanghai GenePharma Co., Ltd. (Shanghai, China). Cells were subsequently transfected with 100 nM miR-135a-5p mimic (5′-UAUGGCUUUUUAUUCCUAUGUGA-3′), 100 nM mimic control (5′-UCUCCAAACGUGUCACCUTT-3′), 100 nM miR-135a-5p inhibitor (5′-UCACAUAGGAAUAAAAAGCCAUA-3′), 100 nM inhibitor control (5′-CAGUACUUUUGUGUAGUACAA-3′), 2 µl control-plasmid (cat. no. sc-108083), 2 µl RUNX2-plasmid (cat. no. sc-400183-ACT; both Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA), miR-135a-5p mimic+control-plasmid or miR-135a-5p mimic+RUNX2-plasmid using Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. Cells without any treatment were considered as the control group. Transfection efficiency was detected following 24-h transfection using RT-qPCR.
Alkaline phosphatase (ALP) activity
Following a 24-h transfection with miR-135a-5p mimic, mimic control, miR-135a-5p inhibitor, inhibitor control, miR-135a-5p inhibitor+control-siRNA or miR-135a-5p inhibitor+RUNX2- siRNA, the ALP activity of C2C12 cells was detected. As previously described (19), the Alkaline Phosphatase Assay kit (cat. no. P0321; Beyotime Institute of Biotechnology, Shanghai, China) was used to detect the ALP activity according to the manufacturer's protocol. ALP activity was determined by measuring the absorbance at a wavelength of 405 nm using a microplate reader (BD Biosciences, Franklin Lakes, NJ, USA).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from C2C12 cells using TRIzol reagent, according to the manufacturer's protocol. Total RNA was reverse transcribed into cDNA using the High-Capacity cDNA Reverse Transcription kit (Applied Biosystems; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. qPCR was subsequently performed using the 2X SYBR Green PCR Master mix (Applied Biosystems; Thermo Fisher Scientific, Inc.). Primer sequences used for the qPCR were as follows: GAPDH forward, 5′CTTTGGTATCGTGGAAGGACTC3′ and reverse, 5′GTAGAGGCAGGGATGATGTTCT3′; U6 forward, 5′GCTTCGGCAGCACATATACTAAAAT3′ and reverse, 5′CGCTTCACGAATTTGCGTGTCAT3′; miR-135a-5p forward, 5′TTGGTCTTGTTTCCCGGTCC3′ and reverse, 5′TCACAGCTCCACAGGCTAAC3′; osteocalcin (OC) forward, 5′CTGACCTCACAGATCCCAAGC3′ and reverse, 5′TGGTCTGATAGCTCGTCACAAG3′; Osterix forward, 5′ACCAGGTCCAGGCAACAC3′ and reverse, 5′-GCAAAGTCAGATGGGTAAGTAG-3′; ALP forward, 5′CTTGACTGTGGTTACTGCTGATCA3′ and reverse, 5′GTATCCACCGAATGTGAAAACGT3′; and RUNX2 forward, 5′AGTCCCAACTTCCTGTGCTCC3′ and reverse, 5′CGGTAACCACAGTCCCATCTG3′. The thermocycling conditions were as follows: Initial denaturation at 95°C for 10 min; 35 cycles of 95°C for 15 sec and 55°C for 40 sec. Relative mRNA expression was quantified using the 2−ΔΔCq method and normalized to the internal reference gene U6 or GAPDH, respectively (22).
Western blot analysis
Following treatment with BMP2, total protein was extracted from C2C12 cells using RIPA buffer (cat. no. P0013E; Beyotime Institute of Biotechnology). Total protein was quantified using a bicinchoninic acid assay kit (cat. no. BCA1-1KT; Sigma-Aldrich; Merck KGaA) and 30 µg protein/lane was separated via SDS-PAGE on a 10% gel. The separated proteins were subsequently transferred onto polyvinylidene difluoride membranes (EMD Millipore, Billerica, MA, USA) and blocked for 1.5 h at room temperature with 5% non-fat milk. The membranes were incubated with primary antibodies ALP (cat. no. sc-365765), Osterix (cat. no. sc-393060; both Santa Cruz Biotechnology, Inc.), OC (cat. no. Ab93876; Abcam), Runx2 (cat. no. 12556) and β-actin (cat. no. 4970; both Cell Signaling Technology, Inc., Danvers, MA, USA; all 1:1,000) overnight at 4°C. Subsequently, membranes were incubated with a horseradish peroxidase-conjugated anti-rabbit immunoglobulin G secondary antibodies (1:2,000; cat. no. 7074; Cell Signaling Technology, Inc.) for 3 h at room temperature. Protein bands were visualized using an enhanced chemiluminescence reagent (Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol.
Dual-luciferase reporter assay
TargetScan bioinformatics software (www.targetscan.org/vert_72) was used to search for potential targets of miR-135a-5p, which identified RUNX2 as a potential target gene of miR-135a-5p. To confirm the direct binding between miR-135a-5p and RUNX2, the wild type (WT-RUNX2, 3′-GCAAUACAUUAUUAUAGCCAUAA-5′) and mutant (MUT-RUNX2, 3′-GCAAUACAUUAUUAUCCGGCAAA-5′) 3′UTR of RUNX2 was cloned into the pmiR-RB-Report™ luciferase reporter vector (Guangzhou RiboBio Co., Ltd., Guangzhou, China). Point mutations in the binding site for miR-135a-5p in the 3′UTR of RUNX2 were generated using the QuikChange Site-Directed Mutagenesis kit (Stratagene; Agilent Technologies, Inc., Santa Clara, CA, USA), according to the manufacturer's protocol. C2C12 cells were co-transfected with WT-RUNX2 or MUT-RUNX2 and miR-135a-5p or mimic control using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. Following 24-h transfection, luciferase activity was detected using the Dual-Luciferase® Assay system (Promega Corporation, Madison, WI, USA), according to the manufacturer's protocol. Firefly luciferase activity was normalized to Renilla luciferase activity.
Statistical analysis
Data are presented as the mean ± standard deviation. All statistical analyses were performed using SPSS statistical software (version 18.0; SPSS, Inc., Chicago, IL, USA). The statistical significance of differences between two groups was analyzed using Student's t-test. One-way analysis of variance followed by Tukey's post hoc test was used to analyze differences among multiple groups. All experiments were repeated three times. P<0.05 was considered to indicate a statistically significant difference.
Results
Enhanced miR-135a-5p expression in postmenopausal women with osteoporosis
To determine whether miR-135a-5p was involved in the development of osteoporosis, the expression level of miR-135a-5p was detected using RT-qPCR in the bone tissue fragments from postmenopausal women with and without osteoporosis. The results revealed that the expression level of miR-135a-5p was significantly increased in postmenopausal women with osteoporosis compared with the postmenopausal women without osteoporosis (Fig. 1).
Figure 1.
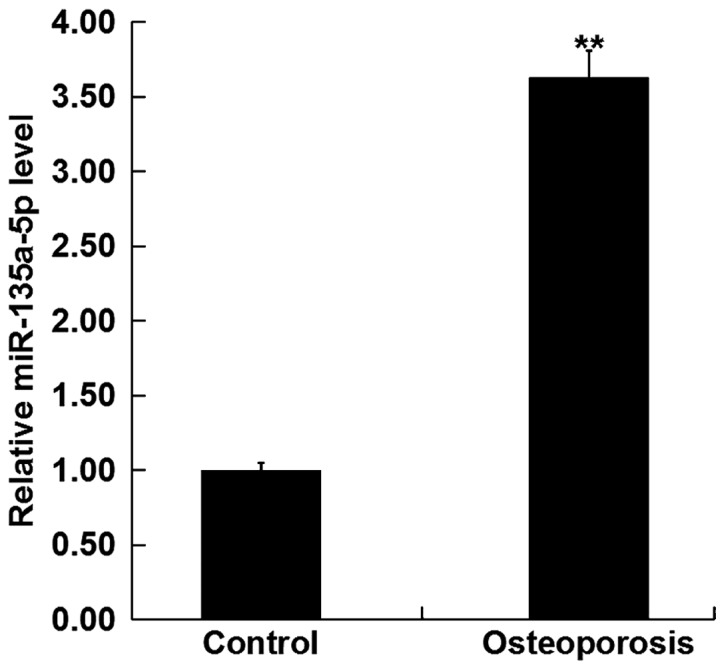
Relative miR-135a-5p expression in postmenopausal women with osteoporosis. The relative miR-135a-5p expression level was determined by, reverse transcription-quantitative polymerase chain reaction in bone tissue fragments from postmenopausal women with osteoporosis and without osteoporosis (Control). Data are presented as the mean ± standard deviation from three independent experiments. **P<0.01 vs. Control. miR, microRNA.
miR-135a-5p is downregulated during osteogenic differentiation induced by BMP2
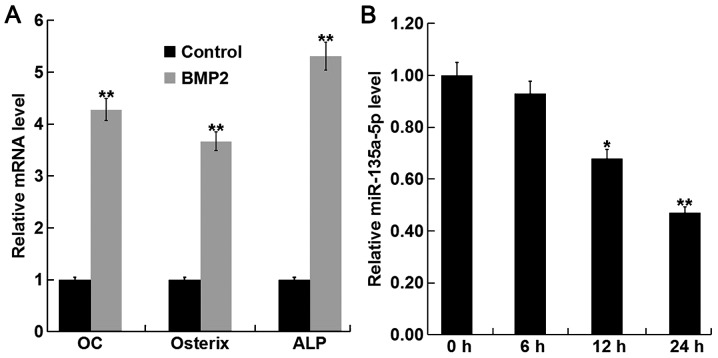
To investigate the function of miR-135a-5p in osteoporosis, the expression level of miR-135a-5p was detected during osteogenic differentiation induced by BMP2 in C2C12 cells. To confirm the successful induction of osteogenic differentiation, the relative mRNA expression level of key osteoblast markers OC, osterix, and ALP were detected using RT-qPCR. The relative mRNA expression levels of OC, osterix and ALP were significantly increased following treatment with BMP compared with the control group (Fig. 2A), indicating the successful induction of osteogenic differentiation. In addition, BMP2 treatment significantly decreased the expression level of miR-135a-5p in a time-dependent manner (Fig. 2B). Taken together, these results suggest that miR-135a-5p was downregulated during osteogenic differentiation.
Figure 2.
Relative miR-135a-5p expression during osteogenic differentiation. C2C12 cells were treated with 2 nM BMP2 for 24 h to induce osteogenic differentiation. (A) The relative mRNA expression level of OC, osterix and ALP were determined by RT-qPCR. **P<0.01 vs. Control. (B) The relative miR-135a-5p expression level was determined by RT-qPCR in C2C12 cells following treatment with BMP2 for 0, 6, 12 and 24 h. Data are presented as the mean ± standard deviation from three independent experiments. *P<0.05 and **P<0.01 vs. 0 h. miR, microRNA; BMP2, bone morphogenetic protein 2; OC, osteocalcin; ALP, alkaline phosphatase; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
Effects of miR-135a-5p in osteogenic differentiation in C2C12 cells
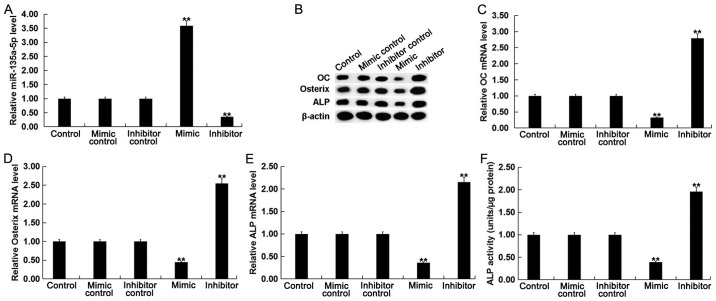
To further investigate the function of miR-135a-5p in osteogenic differentiation, C2C12 cells were treated with 2 nM BMP2 following transfection with miR-135a-5p mimic, mimic control, miR-135a-5p inhibitor or inhibitor control. Transfection efficiency was detected using RT-qPCR following 24-h transfection. The expression level of miR-135a-5p was significantly increased in BMP-treated C2C12 cells following transfection with miR-135a-5p mimic compared with the control (Fig. 3A).
Figure 3.
Effect of miR-135a-5p in osteogenic differentiation. To explore the function of miR-135a-5p in osteoblast differentiation, C2C12 cells were treated with 2 nM BMP2 for 24 h following transfection with miR-135a-5p mimic, mimic control, miR-135a-5p inhibitor or inhibitor control, respectively, for 2 h. (A) The relative miR-135a-5p expression level was determined by RT-qPCR in C2C12 cells following transfection with and treatment with BMP2. (B) The relative protein expression level of OC, Osterix and ALP were determined by western blot analysis in C2C12 cells following transfection and treatment with BMP2. The relative mRNA expression level of (C) OC, (D) Osterix and (E) ALP were determined by RT-qPCR in C2C12 cells following transfection and treatment with BMP2. (F) ALP activity was examined in C2C12 cells following transfection and treatment with BMP2. Data are presented as the mean ± standard deviation from three independent experiments. **P<0.01 vs. Control. miR, microRNA; BMP2, bone morphogenetic protein 2; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; OC, osteocalcin; ALP, alkaline phosphatase.
To determine the effect of miR-135a-5p in osteogenic differentiation, changes in mRNA and protein expression levels of specific osteoblast markers were examined. The mRNA and protein expression levels of OC, osterix and ALP were significantly decreased in BMP-treated C2C12 cells following transfection with miR-135a-5p mimic compared with the control, whilst the opposite effect was observed following transfection with miR-135a-5p (Fig. 3B-E). In addition, it was observed that compared with the control group, the ALP activity in BMP2-treated cells was significantly suppressed following miR-135a-5p overexpression, whilst ALP activity was significantly enhanced following miR-135a-5p knockdown (Fig. 3F). Taken together, these results suggest that miR-135a-5p can inhibit osteogenic differentiation.
RUNX2 is a direct target of miR-135a-5p
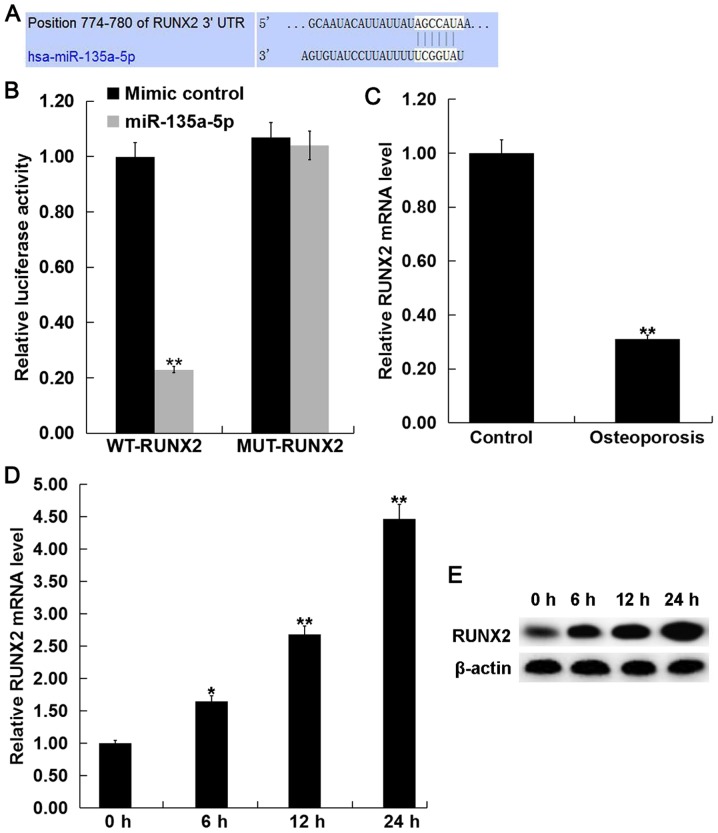
To investigate the molecular mechanism of miR-135a-5p in osteogenic differentiation, TargetScan (http://www.targetscan.org) was used to predict the potential target genes of miR-135a-5p. RUNX2 was identified as a potential target of miR-135a-5p (Fig. 4A). To confirm the prediction, luciferase reporter assays were performed to validate the direct interaction between miR-135a-5p and RUNX2. Luciferase activity was significantly decreased following co-transfection with miR-135a-5p mimic and WT-RUNX2 compared with MUT-RUNX2, which had no significant effect on luciferase activity and miR-135a-5p mimic (Fig. 4B), indicating that miR-135a-5p directly targets RUNX2.
Figure 4.
RUNX2 is a direct target of miR-135a-5p. (A) TargetScan bioinformatics analysis was used to predict the miR-135a-5p binding site in the 3′UTR of RUNX2. (B) Luciferase reporter assays were performed in C2C12 cells following co-transfection with luciferase reporter plasmids containing wild-type RUNX2 3′UTR or mutant RUNX2 3′UTR and miR-135a-5 mimic or mimic control. Data are presented as the mean ± standard deviation from three independent experiments. **P<0.01 vs. Mimic control. (C) The relative RUNX2 mRNA expression level was determined by RT-qPCR in bone tissue fragments from postmenopausal women with osteoporosis and without osteoporosis (Control). The relative RUNX2 (D) mRNA and (E) protein expression levels were detected by RT-qPCR and western blot analysis, respectively, in C2C12 cells following treatment with BMP2 for 0, 6, 12 and 24 h. Data are presented as the mean ± standard deviation from three independent experiments. *P<0.05 and **P<0.01 vs. Control. RUNX2, runt related transcription factor 2; miR, microRNA; UTR, untranslated region; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; BMP2, bone morphogenetic protein 2.
The mRNA expression level of RUNX2 was detected using RT-qPCR in the bone tissue fragments from postmenopausal women with and without osteoporosis. The current study demonstrated that the mRNA expression level of RUNX2 was significantly decreased in postmenopausal women with osteoporosis compared with the postmenopausal women without osteoporosis (Fig. 4C). Furthermore, BMP2 treatment significantly increased the mRNA and protein expression levels of RUNX2 in C2C12 cells in a time-dependent manner (Fig. 4D and E).
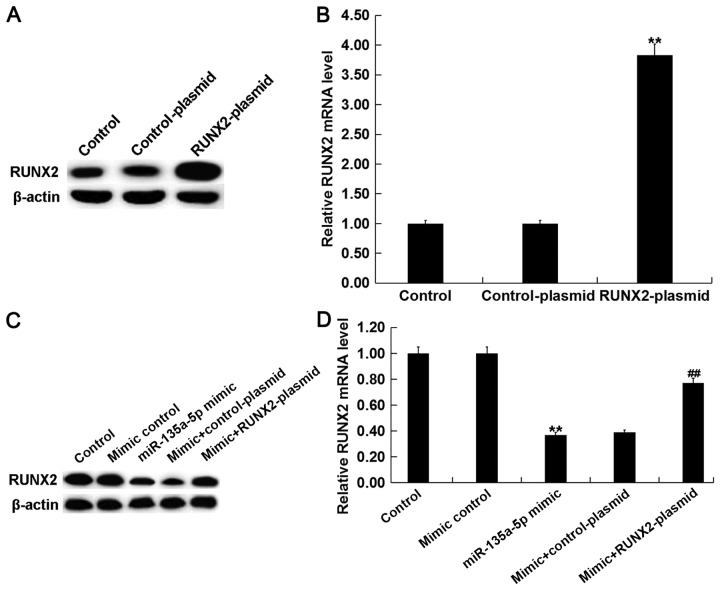
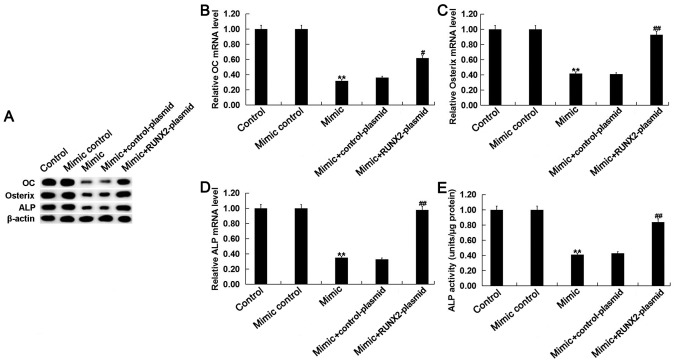
miR-135a-5p overexpression inhibits osteogenic differentiation in C2C12 cells by targeting RUNX2. To investigate whether miR-135a-5p serves a role in osteogenic differentiation by direct targeting of RUNX2, C2C12 cells were treated with 2 nM BMP2 following transfection with miR-135a-5p mimic, mimic control, control-plasmid, RUNX2-plasmid, miR-135a-5p mimic+control-plasmid or miR-135a-5p mimic+RUNX2-plasmid. Transfection efficiency was detected using RT-qPCR following 24-h transfection. The expression level of RUNX2 was significantly increased in BMP-treated C2C12 cells following transfection with RUNX2-plasmid compared with the control (Fig. 5A and B). The relative protein and mRNA expression levels of RUNX2 were decreased in BMP2-treated C2C12 cells following transfection with miR-135a-5p mimic, however the miR-135a-5p-induced effects were reversed following transfection with RUNX2-plasmid (Fig. 5C and D).
Figure 5.
miR-135a-5p regulates RUNX2 expression in BMP2 treated C2C12 cells. C2C12 cells were treated with 2 nM BMP2 for 24 h following transfection with miR-135a-5p mimic, mimic control, control-plasmid, RUNX2-plasmid, miR-135a-5p mimic+control-plasmid or miR-135a-5p mimic+RUNX2-plasmid, respectively, for 2 h. The relative RUNX2 (A) protein and (B) mRNA expression levels were detected by western blot and RT-qPCR analysis, respectively, in C2C12 cells following transfection with control-plasmid and RUNX2-plasmid. The relative RUNX2 (C) protein and (D) mRNA expression level was detected by western blot and RT-qPCR analysis, respectively, in C2C12 cells following transfection with mimic control, miR-135a-5p mimic, miR-135a-5p mimic+control-plasmid or miR-135a-5p mimic+RUNX2-plasmid. Data are presented as the mean ± standard deviation from three independent experiments. **P<0.01 vs. Control; ##P<0.01 vs. Mimic group. miR, microRNA; RUNX2, runt related transcription factor 2; BMP2, bone morphogenetic protein 2; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
The mRNA and protein expression levels of OC, osterix and ALP were decreased in BMP-treated C2C12 cells following transfection with miR-135a-5p mimic compared with the control, however these effects were reversed following transfection with RUNX2-plasmid (Fig. 6A-D). In addition, ALP activity was suppressed by miR-135a-5p overexpression however the addition of RUNX2-plasmid reversed the effect and significantly enhanced ALP activity (Fig. 6E).
Figure 6.
miR-135a-5p inhibits osteogenic differentiation by targeting RUNX2. C2C12 cells were treated with 2 nM BMP2 for 24 h following transfection with miR-135a-5p mimic, mimic control, control-plasmid, RUNX2-plasmid, miR-135a-5p mimic+control-plasmid or miR-135a-5p mimic+RUNX2-plasmid, respectively, for 2 h. (A) The relative protein expression level of OC, Osterix and ALP were determined by western blot analysis in C2C12 cells following transfection and treatment with BMP2. The relative mRNA expression level of (B) OC, (C) Osterix and (D) ALP were determined by reverse transcription-quantitative polymerase chain reaction in C2C12 cells following transfection and treatment with BMP2. (E) ALP activity was examined in C2C12 cells following transfection and treatment with BMP2. Data are presented as the mean ± standard deviation from three independent experiments. **P<0.01 vs. Control; #P<0.05 vs. Mimic; ##P<0.01 vs. Mimic; miR, microRNA; RUNX2, runt related transcription factor 2; BMP2, bone morphogenetic protein 2; OC, osteoclacin; ALP, alkaline phosphatase.
Discussion
In the present study, the miR-135a-5p expression level was significantly increased in postmenopausal women with osteoporosis. However, during osteogenic differentiation in vitro, the miR-135a-5p expression level was significantly decreased. miR-135a-5p overexpression decreased the expression of osteoblast makers, OC, osterix and ALP, as well as suppressing ALP activity. By contrast, miR-135a-5p knockdown increased the expression of osteoblast makers, and enhanced ALP activity. Therefore, the current study demonstrated the inhibitory effect of miR-135a-5p on osteogenic differentiation. In addition, bioinformatics analysis was used to predict RUNX2 as a direct target of miR-135a-5p. The RUNX2 expression level was significantly decreased in postmenopausal women with osteoporosis. However, during osteogenic differentiation in vitro, the RUNX2 expression level was significantly increased. Taken together, the results suggest that miR-135a-5p inhibited osteogenic differentiation by targeting RUNX2 and therefore miR-135a-5p may be a promising therapeutic target for the treatment of osteoporosis.
In China, osteoporosis is a common bone-related disease (23). The incidence of osteoporosis is high in postmenopausal women due to several independent risk factors which include, estrogen deficiency, persistent calcium loss and aging (24). Osteoporosis occurs due to an imbalance between osteoclastic bone resorption and osteoblastic bone formation (25). Restoring and maintaining the balance between bone formation and bone resorption is an effective way to treat postmenopausal osteoporosis (9,10). Although some progress has been made in the treatment of postmenopausal osteoporosis, there are currently no effective therapies (26). Therefore, understanding of the molecular mechanism underlying osteogenic bone formation, as well as identifying therapeutic targets with potential osteogenic effects, may provide novel strategies for the treatment of postmenopausal osteoporosis.
Several studies have indicated that miRNAs serve an important role in the regulation of osteoblastic differentiation or bone formation (27,28). The study of miRNAs in postmenopausal osteoporosis presents a novel direction for the diagnosis and treatment of postmenopausal osteoporosis (15–18). There have been relatively few studies investigating the cellular function of miR-135a-5p. Chen et al (29) reported that miR-135a-5p could inhibit 3T3-L1 adipogenesis by activating the Wnt/β-catenin signaling pathway. Wang et al (30) demonstrated that serum miR-135a-5p expression was upregulated in colorectal cancer and suggested that miR-135a-5p expression may be used as a diagnostic biomarker for colorectal cancer. Yao et al (31) demonstrated that miR-135a-5p promoted proliferation and metastasis in hepatocellular carcinoma cells via direct targeting of Kruppel-like factor 4. In addition, miR-135a-5p is thought to serve an inhibitory role in several types of cancer including thyroid carcinoma, head and neck squamous cell carcinoma and glioblastoma (32–34). Furthermore, miR-135a-5p is involved in the neuroprotective effects of hydrogen sulfide against Parkinson's disease (35). TargetScan was used to predict potential target genes of miR-135a-5p. TargetScan revealed hundreds of target genes of miR-135a-5p, including RUNX2. RUNX2, a transcription factor that belongs to the runt homology domain protein family, is the predominant transcription factor for osteoblast differentiation and bone formation and it may therefore be a potential target for the treatment of osteoporosis (36,37). These findings suggest that miR-135a-5p may serve a role in osteogenic differentiation and osteoporosis. However, the role of miR-135a-5p in postmenopausal osteoporosis remain unknown. Therefore, the aim of the present study was to investigate miR-135a-5p expression in postmenopausal women with osteoporosis as well as the cellular function of miR-135a-5p and its underlying mechanism in osteoblast differentiation.
The relative miR-135a-5p expression level in the bone tissue fragments of postmenopausal women with and without osteoporosis was examined. The current study demonstrated that the expression level of miR-135a-5p was significantly upregulated in postmenopausal women with osteoporosis. Further analysis indicated that miR-135a-5p expression was downregulated during osteogenic differentiation. Overexpression of miR-135a-5p inhibited osteogenic differentiation, whilst miR-135a-5p knockdown enhanced osteogenic differentiation. To further investigate the underlying molecular mechanism of miR-135a-5p in osteogenic differentiation, the direct interaction between miR-135a-5p and RUNX2 was predicted using TargetScan and confirmed using dual-luciferase reporter assay. To investigate whether miR-135a-5p serves a role in osteogenic differentiation by direct targeting of RUNX2, rescue experiments were performed. The inhibitory effect of miR-135a-5p on osteogenic differentiation was reversed by RUNX2 overexpression, indicating that miR-135a-5p can inhibit osteogenic differentiation via RUNX2.
In conclusion, the current study demonstrated that miR-135a-5p expression was upregulated in postmenopausal women with osteoporosis, and miR-135a-5p inhibited osteogenic differentiation via direct targeting of RUNX2. Therefore, miR-135a-5p may be a promising therapeutic target for the treatment of postmenopausal osteoporosis.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Ningbo Natural Science Foundation Project (grant no. 2017A610278).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
XS contributed to study design, data collection, statistical analysis and data interpretation. ZZ contributed to data collection, manuscript preparation and the literature search.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of Ningbo First Hospital (Ningbo, China) and written informed consent was obtained from each patient.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Dalle Carbonare L, Valenti MT, Zanatta M, Donatelli L, Lo Cascio V. Circulating mesenchymal stem cells with abnormal osteogenic differentiation in patients with osteoporosis. Arthritis Rheum. 2009;60:3356–3365. doi: 10.1002/art.24884. [DOI] [PubMed] [Google Scholar]
- 2.Rachner TD, Khosla S, Hofbauer LC. Osteoporosis: Now and the future. Lancet. 2011;377:1276–1287. doi: 10.1016/S0140-6736(10)62349-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kikuta S, Tanaka N, Kazama T, Kazama M, Kano K, Ryu J, Tokuhashi Y, Matsumoto T. Osteogenic effects of dedifferentiated fat cell transplantation in rabbit models of bone defect and ovariectomy-induced osteoporosis. Tissue Eng Part A. 2013;19:1792–1802. doi: 10.1089/ten.tea.2012.0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lubkowska A, Dobek A, Mieszkowski J, Garczynski W, Chlubek D. Adiponectin as a biomarker of osteoporosis in postmenopausal women: Controversies. Dis Markers 2014. 2014:975178. doi: 10.1155/2014/975178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He W, Goodkind D, Kowal P. Census Bureau, International population reports, P95/16-1, An Aging World; 2015. Washington DC: U.S.: Government Publishing Office; 2016. U.S. [Google Scholar]
- 6.Wang Y, Tao Y, Hyman ME, Li J, Chen Y. Osteoporosis in china. Osteoporos Int. 2009;20:1651–1662. doi: 10.1007/s00198-009-0925-y. [DOI] [PubMed] [Google Scholar]
- 7.Li Y, Xuan M, Wang B, Yang J, Zhang H, Zhang XZ, Guo XH, Lü XF, Xue QY, Yang GY, et al. Comparison of parathyroid hormone (1–34) and elcatonin in postmenopausal women with osteoporosis: An 18-month randomized, multicenter controlled trial in China. Chin Med J (Engl) 2013;126:457–463. doi: 10.1166/jbn.2018.2579. [DOI] [PubMed] [Google Scholar]
- 8.Pouresmaeili F, Kamalidehghan B, Kamarehei M, Goh YM. A comprehensive overview on osteoporosis and its risk factors. Ther Clin Risk Manag. 2018;14:2029–2049. doi: 10.2147/TCRM.S138000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marie PJ, Kassem M. Osteoblasts in osteoporosis: Past, emerging, and future anabolic targets. Eur J Endocrinol. 2011;165:1–10. doi: 10.1530/EJE-11-0132. [DOI] [PubMed] [Google Scholar]
- 10.Ruiz-Gaspà S, Blanch-Rubió J, Ciria-Recasens M, Monfort J, Tío L, Garcia-Giralt N, Nogués X, Monllau JC, Carbonell-Abelló J, Pérez-Edo L. Reduced proliferation and osteocalcin expression in osteoblasts of male idiopathic osteoporosis. Calcif Tissue Int. 2010;86:220–226. doi: 10.1007/s00223-010-9336-5. [DOI] [PubMed] [Google Scholar]
- 11.Hammond SM. An overview of microRNAs. Adv Drug Deliv Rev. 2015;87:3–14. doi: 10.1016/j.addr.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soifer HS, Rossi JJ, Saetrom P. MicroRNAs in disease and potential therapeutic applications. Mol Ther. 2017;15:2070–2079. doi: 10.1038/sj.mt.6300311. [DOI] [PubMed] [Google Scholar]
- 13.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 14.O'Connell RM, Rao DS, Chaudhuri AA, Baltimore D. Physiological and pathological roles for microRNAs in the immune system. Nat Rev Immunol. 2010;10:111–122. doi: 10.1038/nri2708. [DOI] [PubMed] [Google Scholar]
- 15.Jin D, Wu X, Yu H, Jiang L, Zhou P, Yao X, Meng J, Wang L, Zhang M, Zhang Y. Systematic analysis of lncRNAs, mRNAs, circRNAs and miRNAs in patients with postmenopausal osteoporosis. Am J Transl Res. 2018;10:1498–1510. [PMC free article] [PubMed] [Google Scholar]
- 16.Mandourah AY, Ranganath L, Barraclough R, Vinjamuri S, Hof RV, Hamill S, Czanner G, Dera AA, Wang D, Barraclough DL. Circulating microRNAs as potential diagnostic biomarkers for osteoporosis. Sci Rep. 2018;8:8421. doi: 10.1038/s41598-018-26525-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feng Q, Zheng S, Zheng J. The emerging role of microRNAs in bone remodeling and its therapeutic implications for osteoporosis. Biosci Rep. 2018;38(pii):BSR20180453. doi: 10.1042/BSR20180453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li H, Wang Z, Fu Q, Zhang J. Plasma miRNA levels correlate with sensitivity to bone mineral density in postmenopausal osteoporosis patients. Biomarkers. 2014;19:553–556. doi: 10.3109/1354750X.2014.935957. [DOI] [PubMed] [Google Scholar]
- 19.Katagiri T, Yamaguchi A, Komaki M, Abe E, Takahashi N, Ikeda T, Rosen V, Wozney JM, Fujisawa-Sehara A, Suda T. Bone morphogenetic protein-2 converts the differentiation pathway of C2C12 myoblasts into the osteoblast lineage. J Cell Biol. 1994;127:1755–1766. doi: 10.1083/jcb.127.6.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shin CS, Lecanda F, Sheikh S, Weitzmann L, Cheng SL, Civitelli R. Relative abundance of different cadherins defines differentiation of mesenchymal precursors into osteogenic, myogenic, or adipogenic pathways. J Cell Biochem. 2000;78:566–577. doi: 10.1002/1097-4644(20000915)78:4<566::AID-JCB6>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Y, Gao Y, Cai L, Li F, Lou Y, Xu N, Kang Y, Yang H. MicroRNA-221 is involved in the regulation of osteoporosis through regulates RUNX2 protein expression and osteoblast differentiation. Am J Transl Res. 2017;9:126–135. [PMC free article] [PubMed] [Google Scholar]
- 22.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 23.Feng Z, Liu C, Guan X, Mor V. China's rapidly aging population creates policy challenges in shaping a viable long-term care system. Health Aff (Millwood) 2012;31:2764–2773. doi: 10.1377/hlthaff.2012.0535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Black DM, Rosen CJ. Postmenopausal osteoporosis. N Engl J Med. 2016;374:2096–2097. doi: 10.1056/NEJMcp1513724. [DOI] [PubMed] [Google Scholar]
- 25.Raisz LG. Pathogenesis of osteoporosis: Concepts, conflicts, and prospects. J Clin Invest. 2005;115:3318–3325. doi: 10.1172/JCI27071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tella SH, Gallagher JC. Prevention and treatment of postmenopausal osteoporosis. J Steroid Biochem Mol Biol. 2014;142:155–170. doi: 10.1016/j.jsbmb.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lian JB, Stein GS, van Wijnen AJ, Stein JL, Hassan MQ, Gaur T, Zhang Y. MicroRNA control of bone formation and homeostasis. Nat Rev Endocrinol. 2012;8:212–227. doi: 10.1038/nrendo.2011.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Valenti MT, Dalle Carbonare L, Mottes M. Role of microRNAs in progenitor cell commitment and osteogenic differentiation in health and disease (Review) Int J Mol Med. 2018;41:2441–2449. doi: 10.3892/ijmm.2018.3452. [DOI] [PubMed] [Google Scholar]
- 29.Chen C, Peng Y, Peng Y, Peng J, Jiang S. miR-135a-5p inhibits 3T3-L1 adipogenesis through activation of canonical Wnt/β-catenin signaling. J Mol Endocrinol. 2014;52:311–320. doi: 10.1530/JME-14-0013. [DOI] [PubMed] [Google Scholar]
- 30.Wang Q, Zhang H, Shen X, Ju S. Serum microRNA-135a-5p as an auxiliary diagnostic biomarker for colorectal cancer. Ann Clin Biochem. 2017;54:76–85. doi: 10.1177/0004563216638108. [DOI] [PubMed] [Google Scholar]
- 31.Yao S, Tian C, Ding Y, Ye Q, Gao Y, Yang N, Li Q. Down-regulation of Krüppel-like factor-4 by microRNA-135a-5p promotes proliferation and metastasis in hepatocellular carcinoma by transforming growth factor-β1. Oncotarget. 2016;7:42566–42578. doi: 10.18632/oncotarget.9934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao X, Sun Z, Li H, Jiang F, Zhou J, Zhang L. MiR-135a-5p modulates biological functions of thyroid carcinoma cells via targeting VCAN 3′-UTR. Cancer Biomark. 2017;20:207–216. doi: 10.3233/CBM-170566. [DOI] [PubMed] [Google Scholar]
- 33.Guo LM, Ding GF, Xu W, Ge H, Jiang Y, Chen XJ, Lu Y. MiR-135a-5p represses proliferation of HNSCC by targeting HOXA10. Cancer Biol Ther. 2018:1–28. doi: 10.1080/15384047.2018.1450112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin J, Wen X, Zhang X, Sun X, Yunzhi L, Peng R, Zhu M, Wang M, Zhang Y, Luo W, et al. miR-135a-5p and miR-124-3p inhibit malignancy of glioblastoma by downregulation of syndecan binding protein. J Biomed Nanotechnol. 2018;14:1317–1329. doi: 10.1166/jbn.2018.2579. [DOI] [PubMed] [Google Scholar]
- 35.Liu Y, Liao S, Quan H, Lin Y, Li J, Yang Q. Involvement of microRNA-135a-5p in the protective effects of hydrogen sulfide against parkinson's disease. Cell Physiol Biochem. 2016;40:18–26. doi: 10.1159/000452521. [DOI] [PubMed] [Google Scholar]
- 36.Chen D, Zhao M, Mundy GR. Bone morphogenetic proteins. Growth Factors. 2004;22:233–241. doi: 10.1080/08977190412331279890. [DOI] [PubMed] [Google Scholar]
- 37.Kim JY, Cheon YH, Kwak SC, Baek JM, Yoon KH, Lee MS, Oh J. Emodin regulates bone remodeling by inhibiting osteoclastogenesis and stimulating osteoblast formation. J Bone Miner Res. 2014;29:1541–1553. doi: 10.1002/jbmr.2183. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.