Abstract
The present study examined the involvement of autophagy as a mechanism in the protective effect of quercetin (QUE) on atherosclerosis (AS) in ApoE−/− mice. An AS model was established by feeding ApoE−/− mice a high-fat diet (HFD). Mice were divided into four experimental groups: The model, QUE, 3-methyladenine (3-MA) and QUE + 3-MA groups. Additionally, age-matched wild-type C57BL/6 mice were used as a Control group. Autophagosomes in the aorta were examined using a transmission electron microscope. Aorta pathology, serum lipid accumulation and collagen deposition were determined by hematoxylin and eosin, Oil Red O and Masson staining, respectively. The levels of cytokines, including tumor necrosis factor-α (TNF-α), interleukin-1β (IL-1β) and interleukin-18 (IL-18) were measured using ELISA assays. Protein levels of mTOR, microtubule associated protein 1 light chain 3a (LC3), P53 and cyclin dependent kinase inhibitor 1A (P21) in the aorta were analyzed using western blotting. ApoE−/− mice which were fed HFD exhibited substantial AS pathology, no autophagosomes, higher levels of TNF-α, IL-1β, IL-18 and mTOR and lower ratios of LC3 II/I. All these alterations were ameliorated and aggravated by QUE and 3-MA treatment, respectively. The inhibition of AS by QUE may be associated with the enhancement of autophagy and upregulation of P21 and P53 expression.
Keywords: atherosclerosis, autophagy, inflammation, quercetin
Introduction
Atherosclerosis (AS) is a common pathological basis of cardiovascular and cerebrovascular diseases, and inflammatory reactions and lipid metabolism disorders serve an important role (1). AS is prevalent in the elderly, and its incidence increases with age. Therefore, aging is a key causal factor of AS (2). In particular, a study has previously reported that cell senescence may be observed in atherosclerotic plaques in patients with AS (3). Oxidative stress damage and vascular inflammation caused by senescence can cause arterial dysfunction (4). Cyclin dependent kinase inhibitor 1A (P21) and P53 are considered to be marker proteins for senescence (5).
Autophagy protects the body from stress damage and delays the development of aging-associated diseases such as AS (6). Autophagy is an effective response against inflammation and oxidative stress in AS plaque cells, and thus it is considered to serve an important role in the initiation and development of AS (7). Autophagosomes observed under a transmission electron microscope (TEM) are regarded as the gold standard for autophagy detection (8). mTOR is a member of the PI3K-related family of protein kinases. mTOR integrates multiple upstream signaling pathways, interacts with regulator proteins and is a key regulatory molecule in the induction of autophagy (9). Microtubule associated protein 1 light chain 3α (LC3) is a cytosolic ubiquitin-like protein that binds to phosphatidylethanolamine to form autophagosomes (10). 3-methyladenine (3-MA) is a widely used autophagy inhibitor, which inhibits class III PI3K and blocks the early stage of autophagy (8).
The Traditional Chinese Medicine (TCM) monomer quercetin (QUE), a flavonoid compound with anti-inflammatory, antioxidative and lipid metabolism-modulating properties, has been demonstrated to have therapeutic effects in AS treatment (11). Previous studies have revealed that QUE can effectively interfere with AS development by regulating inflammatory mediator production and promoting cholesterol efflux (12,13). A previous study our laboratory revealed that QUE can effectively reduce oxidized-low density lipoprotein (ox-LDL)-induced RAW264.7 cell damage, reduce lipid accumulation and delay cell senescence (14). Additionally, it has been demonstrated that QUE serves an important role in the prevention and treatment of AS by upregulating autophagy in endothelial cells to protect cells from stress damage (15). However, it is unclear whether QUE exerts anti-atherosclerotic effects by regulating autophagy in ApoE−/− mouse models of AS induced by a high-fat diet (HFD).
In the present study, using an AS model in ApoE−/− mice, aorta autophagosomes were analyzed using TEM, protein levels of mTOR, LC3 II/I ratio, P53 and P21 in the aorta were examined using western blotting analysis, levels of the cytokines tumor necrosis factor-α (TNF-α), interleukin-1β (IL-1β) and interleukin-18 (IL-18) were assessed using ELISA assays, and aorta pathology, lipid accumulation and collagen deposition were investigated using hematoxylin and eosin (H&E), Oil Red O and Masson staining, respectively. Based on the results of these analyses, deeper mechanistic insights into QUE-mediated inhibition of AS development were gained.
Materials and methods
Experimental animals
A total of 96 specific pathogen-free (SPF) male ApoE−/− mice [age, 12 weeks; weight, 20±5 g; animal license no. SCXK (Jing) 2016-0006] and 24 age-matched male wild-type C57BL/6 mice [animal license no. SCXK (Jing) 2016-0006] were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd. The mice were kept in an SPF grade animal facility at the Animal Center of the Shanghai University of Traditional Chinese Medicine (Shanghai, China). The room temperature was kept constant at 24±1°C, the relative humidity was 50–70% with a 12 h light:dark cycle. All mice had free access to food and water. After 12 weeks, all mice were treated with cervical dislocation following anesthesia to collect samples. All animal experiments strictly followed the Guide for the Care and Use of Medical Laboratory Animals (16). The present study was approved by the Laboratory Animal Welfare and Ethics Committee of Shanghai University of Traditional Chinese Medicine (approval no. PZSHUTCM18113002).
Drugs and major reagents
The following drugs and reagents were used in the current study: QUE (cat. no. B20527; Shanghai Yuanye Bio-Technology Co., Ltd.), anti-rabbit mTOR (cat. no. 2983S; Cell Signaling Technology, Inc.), anti-rabbit LC3B (cat. no. 3868S; Cell Signaling Technology, Inc.), anti-rabbit P53 (cat. no. ab31333; Abcam), anti-rabbit P21 (cat. no. ab188224; Abcam), anti-rabbit GAPDH (cat. no. 5174S; Cell Signaling Technology, Inc.), IRDye® 800CW-conjugated goat anti-rabbit secondary antibody (cat. no. 926-32211; LI-COR Biosciences), IRDye® 800CW-conjugated goat anti-mouse secondary antibody (cat. no. 926-32210; LI-COR Biosciences), RIPA buffer (Beyotime Institute of Biotechnology), PMSF (Beyotime Institute of Biotechnology), bicinchoninic acid (BCA) protein assay kit (Beyotime Institute of Biotechnology), protein ladder (Thermo Fisher Scientific, Inc.), SDS-PAGE Gel Preparation kit (Beyotime Institute of Biotechnology). All ELISA kits were purchased from EK-Bioscience: Mouse total cholesterol (TC) ELISA kit (cat. no. Ek-M20591; Ek-Bioscience), mouse triglyceride (TG) ELISA kit (cat. no. Ek-M20590; Ek-Bioscience), mouse high density lipoprotein (HDL-C) ELISA kit (cat. no. Ek-M20589; Ek-Bioscience), mouse low density lipoprotein (LDL-C) ELISA kit (cat. no. Ek-M20588; Ek-Bioscience), mouse TNF-a ELISA kit (cat. no. EK-M21159; Ek-Bioscience), mouse IL-1β ELISA kit (cat. no. Ek-M20166; Ek-Bioscience) and mouse IL-18 ELISA kit (cat. no. Ek-M20162; Ek-Bioscience).
Modeling and grouping
After 1 week of acclimatization on a standard mouse diet, 96 male, 12-week-old ApoE−/− mice were randomly divided into four experimental groups (n=24 mice/group) using a random number table method as described previously (17): i) ApoE−/− mice + HFD (Model group); ii) ApoE−/− mice + HFD + QUE (QUE group); iii) apoE−/− mice + HFD + 3-MA (3-MA group); and iv) apoE−/− mice + HFD + 3-MA + QUE (QUE + 3-MA group). In addition, 24 age-matched wild-type C57BL/6 mice were used as normal controls (Control group). The HFD was established by adding 21% fat and 0.5% cholesterol to the standard mouse basal diet. The number of samples per group for each experiment was 6. The health and behavioral status of mice were monitored every 2 weeks. The QUE group was treated by daily oral gavage of a QUE solution (12.5 mg/kg), and the 3-MA group was treated by peritoneal injection of a water solution containing 3-MA (15 mg/kg) every other day, as previously described (13,18). Mice in the Model and Control groups were given an equal volume of distilled water by oral gavage on a daily basis. HFD feeding to establish the AS model and drug regimens started at the same time and lasted for 12 weeks.
Detection of AS indicators
Aorta pathology
Mice were sacrificed by cervical dislocation following anesthesia. Then, the heart and aorta from the thoracic to abdominal sections were removed and fixed in 10% formalin (room temperature; 1 day) solution for later paraffinization (six mice per group; thickness of sections, 5 µm; for H&E and Masson staining) or frozen sectioning (six mice per group; thickness of sections, 10–15 µm; for Oil Red O staining). H&E, Oil Red O and Masson staining (room temperature) were performed on the aorta section samples to determine aorta lipid plaque areas, lipid accumulation and collagen fiber content, respectively. Images were acquired using a digital scanning system (Pannoramic MIDI; 3DHISTECH, Ltd.). Semi-quantitative analysis was conducted as follows: Plaque area (PA; mm2)=aorta vessel area (VA; mm2)-aorta lumen area (LA; mm2), with normalized plaque area=PA/VA-LA; collagen fiber=collagen fiber area (mm2)/aorta lumen area (mm2); lipid content=Oil Red O positive area (mm2)/aorta lumen area (mm2).
Analysis of autophagosomes by TEM
Following anesthesia with pentobarbital sodium (50 mg/kg; i.p.), mice were sacrificed by cervical dislocation and the aortic arch was quickly isolated. Six mice per group were used for these experiments. A small piece of tissue was cut out and fixed in 2% glutaraldehyde-OsO4 for 2 h. Subsequent to ethanol series dehydration, epoxy resin embedding (37°C; 3 h), ultra-thin sectioning (70 nm), uranyl acetate (30 min) and citric acid staining (10 min), autophagosomes were observed using a TEM (JEOL JEM-1230, JEOL, Ltd.). All aforementioned procedures were performed at room temperature.
Serum lipid analysis by ELISA
Following anesthesia with pentobarbital sodium (50 mg/kg; i.p.), mice were sacrificed by cervical dislocation. Blood was collected using an orbital sinus blood collection method for ELISA analyses, and fresh tissue samples were collected for western blot analysis described below. Six mice per group were used for these experiments. Blood was allowed to set for 12 h prior to being centrifuged at 13,523 × g at 4°C for 30 min to obtain serum. TC, TG, HDL-C and LDL-C levels were measured using ELISA kits following the manufacturer's protocols. Optical density (OD)450 values of samples were acquired using a microplate reader (PowerWave™ XS; BioTek Instruments, Inc.) and used to calculate concentrations based on standard curves generated using serially diluted standards.
Serum TNF-α, IL-1β and IL-18 analysis by ELISA
Serum samples were prepared as described above. TNF-α, IL-1β and IL-18 levels were measured using ELISA kits according to the manufacturer's protocols. OD450 values of samples were acquired using a microplate reader (PowerWave™ XS; BioTek Instruments, Inc.) and used for calculating concentrations based on standard curves generated using serially diluted standards.
Measurement of Aorta mTOR, LC3, P53 and P21 protein levels by western blotting
Proteins were extracted from aorta tissues with RIPA lysis buffer containing PMSF. Following centrifugation at 13,523 × g at 4°C for 30 min, the supernatant was collected for protein quantification using the BCA method. Samples were mixed with 5X loading buffer (cat. no. P0015; Beyotime Institute of Biotechnology) and heated in boiling water for 10 min to denature proteins. These treated samples (amount of protein, 30 µg) were separated on SDS-PAGE gels (percentage of the gel, 12%) and then transferred to PVDF membranes (cat. no. IPVH00010; Millipore). The membranes were blocked with 5% skimmed milk (cat. no. C500625; Sangon Biotech) for 2 h at room temperature and subsequently incubated in the aforementioned primary antibody solutions (1:1,000 dilutions; 4°C) overnight. After washing, membranes were incubated with the aforementioned secondary antibodies (1:1,000) for 1 h at room temperature. An Odyssey far-infrared luminescence scanner (cat. no. 9120; Li-COR Biosciences) was used to capture the image. Protein band images were acquired and analyzed as integrated absorbance (IA; IA=mean OD × area) using Image J (version 1.8.0; National Institutes of Health), and the relative levels of target proteins were normalized to GAPDH (target protein IA/GAPDH IA).
Statistical analysis
Statistical analysis was conducted using SPSS software (version 23.0; IBM Corp.), and figures were generated using GraphPad Prism 5 software (GraphPad Software, Inc.). Results are presented as the mean ± SD. Each experiment was replicated ≥3 times. Differences between two groups were determined by unpaired t-tests and differences among three or more groups were assessed using one-way ANOVA followed by Student-Newman-Keuls test was then utilized. P<0.05 was considered to indicate a statistically significant difference.
Results
Serum lipid profiles
The Model group exhibited higher serum levels of TC and LDL-C, and lower levels of HDL-C compared with the Control group (P<0.01; Table I). Compared with the Model group, the QUE group had lower serum levels of TC and LDL-C (P<0.01; Table I). Compared with the 3-MA group, the QUE + 3-MA group had lower serum levels of TC and LDL-C (P<0.01; Table I). No statistically significant differences in TG content between the groups were observed.
Table I.
Comparison of serum lipid levels among groups.
| Group | Number of mice used | TC (mmol/l) | TG (mmol/l) | LDL-C (mmol/l) | HDL-C (mmol/l) |
|---|---|---|---|---|---|
| Control | 6 | 2.10±0.09 | 2.14±0.03 | 1.92±0.07 | 2.04±0.03 |
| Model | 6 | 6.86±0.12a | 2.21±0.06 | 5.28±0.04a | 1.30±0.01a |
| QUE | 6 | 5.81±0.05b | 2.15±0.04 | 4.48±0.06b | 1.32±0.02 |
| 3-MA | 6 | 7.23±0.07b | 2.26±0.05 | 5.74±0.07b | 1.29±0.05 |
| QUE + 3-MA | 6 | 6.74±0.07c | 2.20±0.03 | 4.69±0.04c | 1.31±0.02 |
Data are presented as the mean ± SD.
P<0.01 vs. Control
P<0.01 vs. Model
P<0.01 vs. 3-MA. 3-MA, 3-methyladenine; HDL-C, high-density lipoprotein; LDL-C, low-density lipoprotein; QUE, quercetin; TC, total cholesterol; TG, triglyceride.
Serum TNF-α, IL-1β and IL-18 levels
The Model group exhibited higher serum levels of TNF-α, IL-1β and IL-18 compared with the Control group (P<0.01; Table II). Compared with the Model group, serum levels of TNF-α, IL-1β and IL-18 were lower in the QUE group (P<0.01; Table II) but higher in the 3-MA group (P<0.01; Table II). The serum levels of all examined cytokines were lower in the QUE + 3-MA group compared with the 3-MA group (P<0.01; Table II).
Table II.
Comparison of serum TNF-α, IL-1β and IL-18 levels among groups.
| Group | Number of mice used | TNF-α (pg/ml) | IL-1β (pg/ml) | IL-18 (pg/ml) |
|---|---|---|---|---|
| Control | 6 | 333.51±7.57 | 58.97±1.77 | 92.17±1.17 |
| Model | 6 | 528.62±7.15a | 110.44±0.88a | 138.78±0.93a |
| QUE | 6 | 458.95±29.51b | 97.32±1.02b | 121.51±0.44b |
| 3-MA | 6 | 611.89±12.02b | 117.57±1.38b | 141.79±0.86b |
| QUE + 3-MA | 6 | 486.27±7.41c | 102.95±1.46c | 126.90±1.82c |
Data are presented as the mean ± SD.
P<0.01 vs. Control
P<0.01 vs. Model
P<0.01 vs. 3-MA. 3-MA, 3-methyladenine; IL-1β, interleukin 1β; IL-18, interleukin 18; QUE, quercetin; TNF-α, tumor necrosis factor α.
Aorta pathology
H&E staining
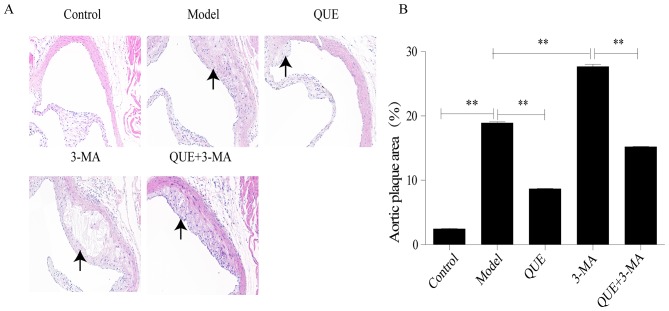
Following HFD feeding for 12 weeks, mouse aortic roots exhibited significant formation of AS plaque compared with the Control group (P<0.01; Fig. 1A and B). Compared with the Model group, areas of AS plaque were significantly smaller in the QUE group (P<0.01; Fig. 1A and B), and significantly larger in the 3-MA group (P<0.01; Fig. 1A and B). Compared with the 3-MA group, the QUE + 3MA group exhibited significantly smaller areas of AS plaque (P<0.01; Fig. 1A and B).
Figure 1.
Effect of QUE on aortic plaque areas in mice. (A) Hematoxylin and eosin staining of mice aortas. Magnification, ×200. (B) Percentage of aortic plaque area. Data are presented as the mean ± SD, n=6. **P<0.01. 3-MA, 3-methyladenine; QUE, quercetin.
Oil red O staining
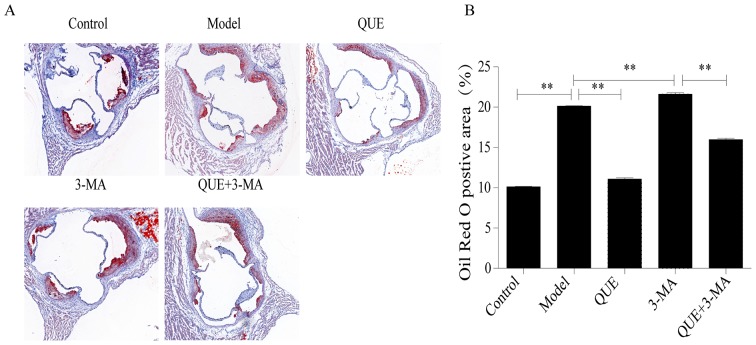
Aortic roots of mice in the Model group had larger amounts of red-stained lipid accumulation compared with the Control group. Compared with the Model group, lipid content was significantly less abundant in the QUE group (P<0.01; Fig. 2A and B), and more abundant in the 3-MA group (P<0.01; Fig. 2A and B). Lipid content in the QUE + 3-MA group were significantly less abundant than those in the 3-MA group (P<0.01; Fig. 2A and B).
Figure 2.
Effect of QUE on the accumulation of lipid in mice aortas. (A) Oil Red O staining. Magnification, ×40. (B) Percentage of lipid accumulation. Data are presented as the mean ± SD, n=6. **P<0.01. 3-MA, 3-methyladenine; QUE, quercetin.
Masson staining
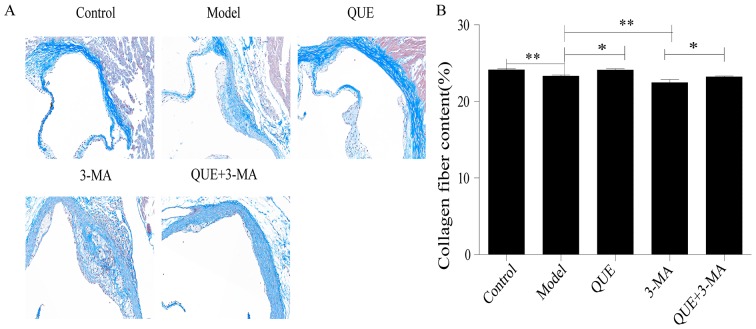
Aortic walls in the Control group exhibited large amounts of collagen and smooth muscle fibers, while the AS plaques in the Model group contained thin layers of fibrous caps and few matrix fibers (P<0.01; Fig. 3A and B). Compared with the Model group, the QUE group exhibited more collagen fibers (P<0.05; Fig. 3A and B), whilst the fibrous caps of plaques in the 3-MA group were thinner and contained less collagen (P<0.01; Fig. 3A and B). The collagen fibers in aortas from the QUE + 3-MA group were more abundant compared with the 3-MA group (P<0.05; Fig. 3A and B).
Figure 3.
Effect of QUE on the area of plaque collagen in ApoE−/− mice. (A) Masson staining. Magnification, ×200. (B) Percentage of collagen content. Data are presented as the mean ± SD, n=6. *P<0.05 and **P<0.01. 3-MA, 3-methyladenine; QUE, quercetin.
TEM observations
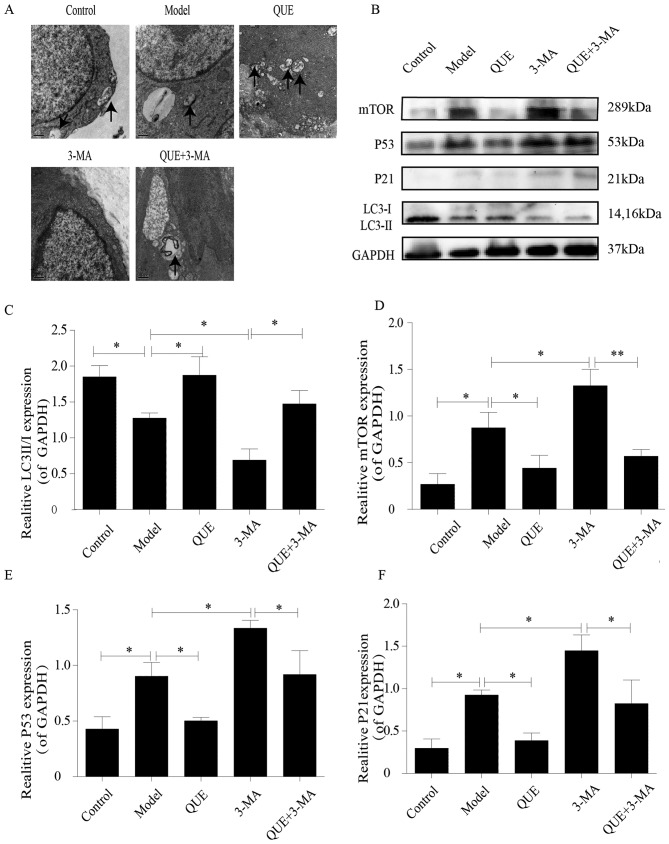
Macrophages in aortas of the Control group exhibited intact structures and contained small numbers of autophagosomes, which were less abundant in the Model group. Aortic macrophages in the QUE group exhibited more autophagosomes Compared with the Model group, whilst the 3-MA group exhibited no autophagosome formation. However, unlike the 3-MA group, autophagosomes were found in the aortic macrophages of the QUE + 3-MA group (Fig. 4A).
Figure 4.
Effects of QUE on autophagosomes and protein expression levels of P53, P21 mTOR and LC3 II/I. (A) Effect of QUE on autophagosomes of mouse aortic macrophages. Magnification, ×20,000. n=3. (B) Western blotting results for LC3 II/I, mTOR, P53 and P21 expression in the mouse aorta. (C-F) Expression levels of (C) LC3 II/I, (D) mTOR, (E) P53 and (F) P21. Data are presented as the means ± SD, n=3. *P<0.05 and **P<0.01. 3-MA, 3-methyladenine; LC3, microtubule associated protein 1 light chain 3α; P21, cyclin dependent kinase inhibitor 1A; QUE, quercetin.
Aortic expression of the autophagy-associated proteins mTOR, LC3, P53 and P21
The aortas of mice in the Control group exhibited higher of LC3 II/I ratios and lower expression levels of mTOR, P53 and P21 compared with the Model group, which had lower LC3 II/I ratio (P<0.05; Fig. 4B and C) and higher expression levels of mTOR (P<0.05; Fig. 4B and D), P53 (P<0.05; Fig. 4B and E) and P21 (P<0.05; Fig. 4B and F). Compared with the Model group, the QUE group exhibited higher ratio of LC3 II/I and lower expression of mTOR, P53, and P21 (P<0.05; Fig. 4B-F); and the 3-MA group exhibited lower ratios of LC3 II/I and higher expression levels of mTOR, P53 and P21 (P<0.05; Fig. 4B-F). Compared with the 3-MA group, the QUE + 3-MA group exhibited higher ratios of LC3 II/I (P<0.05; Fig. 4B and C) and lower expression levels of mTOR (P<0.01; Fig. 4B and D), P53 and P21 (P<0.05; Fig. 4B, E and F).
Discussion
AS is a potential pathological basis of cardiovascular and cerebrovascular events, and inflammation and lipid metabolism serve an important role in its occurrence and development (19). Autophagy is a lysosomal degradation process that inhibits cellular senescence by removing damaged organelles and long-lived proteins (20). Autophagy is a metabolic process that degrades damaged cells and proteins, and defects in autophagy are closely associated with senescence. Studies have indicated that enhancing autophagy can prevent senescence and reduce age-associated pathological alterations in the heart and kidney, leading to improved health status in mice (21,22). Autophagy, a process known to be protective in age-associated cardiovascular disease, may serve an important role in the initiation and development of AS (23). The pathology of AS is characterized by vascular smooth muscle cell (VSMC) apoptosis, vascular endothelial cell (VEC) remodeling and macrophage-mediated inflammation (2). Weakened autophagy takes place in all major cell types in AS plaques, resulting in impaired macrophage apoptosis and increased senescence of blood vessel endothelial and smooth muscle cells (2). Since VMSCs are the only cells in the plaque fibrous cap that generate collagen fibers, promoting autophagy of VSMCs helps stabilize the plaque and prevents its rupture (24). It has been demonstrated that the presence of senescent VSMCs in plaques is associated with AS development, and impaired autophagy may increase the senescence process of VSMCs to drive the formation of plaque and the development AS (25). Similarly, inhibiting autophagy promotes the senescence of VECs to increase AS development (23).
TEM can provide an accurate and detailed view of autophagosome structures, and it is thus taken as the gold standard for determining autophagy levels (8). mTOR is a key molecule in the process of autophagy induction. When energy is sufficient, mTORC1 is activated, and autophagy is inhibited by highly phosphorylated unc-51 like autophagy activating kinase 1 (ULK1) complex and autophagy related 13 (Atg13) (26). When energy is low, mTORC1 activity is inhibited, the dephosphorylated Atg13 and ULK1 are inhibited, and subsequently form a complex with RB1 inducible coiled-coil 1 to induce nucleation and elongation of autophagosomes (26). LC3 is a molecular marker of autophagy, and LC3-I is modified and transformed by ubiquitin to become lipid-soluble LC3-II, which is involved in the formation of autophagosomes (27). As a transcription factor, P53 accumulates in senescent cells and directly leads to cell senescence (28). P21 is a negative regulator of the cell cycle, and serves a key role in cellular senescence (29). In the present study, an AS animal model was induced by feeding ApoE−/− mice a HFD for 12 weeks. Compared with the Control group, the aortas of model mice exhibited typical AS plaques. Oil Red O staining and serum lipid analysis suggested a disorder in lipid metabolism; TEM results revealed fewer autophagosomes and altered autophagy marker proteins. Western blotting revealed increased expression in senescence factors P53 and P21. Following treatment with 3-MA, the area of aortic plaques increased compared with Model group. In addition, the lipid metabolism disorder was further aggravated and protein expression levels of mTOR, P53 or P21 were increased further whereas the LC3 II/I ratio was decreased, no autophagosomes were observed in the cells from the 3-MA group.
AS is a chronic inflammatory disease mediated by a network of pro-inflammatory cytokines, including the interleukin and TNF families (30). TNF-α is a subclinical indicator for predicting AS. As a major pro-inflammatory mediator found in AS plaques, TNF-α activates the secretion of other pro-inflammatory mediators, prevents the synthesis of the extracellular matrix, and thus negatively impacts plaque stability (31). The proinflammatory cytokine IL-1β serves a key role in the initiation and development of AS. IL-1β promotes the release of metalloproteinases which destroy the connective tissue matrix in plaques to render them frail (32). IL-18 is a novel member of the IL-1 family that has been demonstrated to stimulate the expression of the inflammatory cytokines TNF-α and interferon-γ and promote the pathogenesis of AS (33). Higher expression levels of IL-18 in AS plaques have been reported and are considered to contribute to weakened stability of plaques (33). Autophagy, an important protective response to pathological stress, serves a role in regulating inflammation. Defects in autophagy cause the production and release of pro-inflammatory mediators, and conversely, the induction of autophagy helps eliminate inflammatory aggregation (34). A previous study revealed that autophagy has an anti-AS effect by inhibiting AS-associated inflammation, the lipid-lowering drug atorvastatin stabilizes AS plaques and inhibits AS development by reducing the production of the pro-inflammatory cytokines TNF-α, IL-1β and IL-18, a mechanism which involves the induction of autophagy (35). In agreement with these findings, the present study revealed that the mice in the Model group exhibited higher serum levels of TNF-α, IL-1β and IL-18 compared with the Control group, and serum levels of these cytokines were further increased following treatment with the autophagy inhibitor 3-MA.
QUE is a flavonoid compound with multiple biological activities, including anti-inflammatory, anti-senescence and cardiovascular protective properties (36). TCM herbs South Dodder Seed Chinese Dodder Seed and Taxillus sutchuenensis (Lecomte) Danser contain QUE (37). A previous study in our laboratory reported that QUE reduced ox-LDL-induced damage in RAW264.7 cells, reduced lipid precipitation and delayed cellular senescence (14). In a peroxide-induced oxidative stress model in human umbilical vein endothelial cells, QUE has been demonstrated to inhibit vascular cell adhesion molecule 1 and CD80 expression, resulting in an anti-AS effect (38). In a study using HFD-induced AS in ApoE−/− mice, QUE was found to exert its anti-AS effects by suppressing scavenger receptor expression and reducing ox-LDL absorption (39). It has previously been reported that QUE protects endothelial cells and effectively prevents AS by upregulating autophagy via the ERK signaling pathway (15). In an in vitro study using RSC96 cells incubated in high-glucose medium, QUE reduced cell damage, which was associated with upregulated expression levels of LC3 and increased numbers of autophagosomes (40).
The present study demonstrated that QUE could alleviate AS lesions induced by a HFD in ApoE−/− mice, reduce lipid accumulation in aortic roots and reduce serum levels of TC and LDL-C, as well as the expression levels of TNF-α, IL-1β and IL-18. TEM revealed an increased quantity of autophagosomes in the aortas of mice in the QUE group compared with the Model group. In addition, the ratio of LC3 II/I in mouse aortas was significantly increased, while mTOR, P53 and P21 protein expression levels were downregulated compared with the Model group. The aforementioned results suggested that the mechanism of QUE in the inhibition of AS may be associated with the enhancement of autophagy and the delay of senescence.
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the National Natural Science Foundation of China (grant no. 81873348), the Shanghai Nature Science Fund (grant no. 16ZR1433900) and the Shanghai Health and Family Planning Commission Fund (grant no. 201640217).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
DS, HC, CC and SX conceived the experiments and experimental plan. HC, QJ and LY performed the experiments and collected and analyzed the data. HC and QJ wrote the manuscript, DS, HC and QJ reviewed and edited the manuscript. All authors approved the final version of these manuscript and figures.
Ethics approval and consent to participate
The present study was approved by the Laboratory Animal Welfare and Ethics Committee of Shanghai University of Traditional Chinese Medicine (approval no. PZSHUTCM18113002; Shanghai, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Ross R. Atherosclerosis-an inflammatory disease. N Engl J Med. 1999;340:115–126. doi: 10.1056/NEJM199901143400207. [DOI] [PubMed] [Google Scholar]
- 2.Grootaert MOJ, Lynn R, Schrijvers DM, De Meyer GRY, Martinet W. Defective autophagy in atherosclerosis: To die or to senesce? Oxid Med Cell Longe 2018. 2018:7687083. doi: 10.1155/2018/7687083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang JC, Bennett M. Aging and atherosclerosis: Mechanisms, functional consequences, and potential therapeutics for cellular senescence. Cir Res. 2012;111:245–259. doi: 10.1161/CIRCRESAHA.111.261388. [DOI] [PubMed] [Google Scholar]
- 4.Sun L, Dou F, Chen J, Chi H, Xing S, Liu T, Sun S, Chen C. Salidroside slows the progression of EA.hy926 cell senescence by regulating the cell cycle in an atherosclerosis model. Mol Med Rep. 2018;17:257–263. doi: 10.3892/mmr.2017.7872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim YY, Jee HJ, Um JH, Kim YM, Bae SS, Yun J. Cooperation between p21 and Akt is required for p53-dependent cellular senescence. Aging Cell. 2017;16:1094–1103. doi: 10.1111/acel.12639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kitada M, Ogura Y, Koya D. The protective role of Sirt1 in vascular tissue: Its relationship to vascular aging and atherosclerosis. Aging (Albany NY) 2016;8:2290–2307. doi: 10.18632/aging.101068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Netea-Maier RT, Plantinga TS, van de Veerdonk FL, Smit JW, Netea MG. Modulation of inflammation by autophagy: Consequences for human disease. Autophagy. 2016;12:245–260. doi: 10.1080/15548627.2015.1071759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, Adachi H, Adams CM, Adams PD, Adeli K, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition) Autophagy. 2016;12:1–222. doi: 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gurumurthy S, Xie SZ, Alagesan B, Kim J, Yusuf RZ, Saez B, Tzatsos A, Ozsolak F, Milos P, Ferrari F, et al. The Lkb1 metabolic sensor maintains haematopoietic stem cell survival. Nature. 2010;468:659–663. doi: 10.1038/nature09572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jacquin E, Leclerc-Mercier S, Judon C, Blanchard E, Fraitag S, Florey O. Pharmacological modulators of autophagy activate a parallel noncanonical pathway driving unconventional LC3 lipidation. Autophagy. 2017;13:854–867. doi: 10.1080/15548627.2017.1287653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li C, Zhang WJ, Frei B. Quercetin inhibits LPS-induced adhesion molecule expression and oxidant production in human aortic endothelial cells by p38-mediated Nrf2 activation and antioxidant enzyme induction. Redox Biol. 2016;9:104–113. doi: 10.1016/j.redox.2016.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhaskar S, Sudhakaran PR, Helen A. Quercetin attenuates atherosclerotic inflammation and adhesion molecule expression by modulating TLR-NF-κB signaling pathway. Cell Immunol. 2016;310:131–140. doi: 10.1016/j.cellimm.2016.08.011. [DOI] [PubMed] [Google Scholar]
- 13.Cui Y, Hou P, Li F, Liu Q, Qin S, Zhou G, Xu X, Si Y, Guo S. Quercetin improves macrophage reverse cholesterol transport in apolipoprotein E-deficient mice fed a high-fat diet. Lipids Health Dis. 2017;16:9. doi: 10.1186/s12944-016-0393-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li S, Cao H, Shen D, Jia Q, Chen C, Xing SL. Quercetin protects against ox-LDL-induced injury via regulation of ABCAl, LXR-α and PCSK9 in RAW264.7 macrophages. Mol Med Rep. 2018;18:799–806. doi: 10.3892/mmr.2018.9048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhi K, Li M, Bai J, Wu Y, Zhou S, Zhang X, Qu L. Quercitrin treatment protects endothelial progenitor cells from oxidative damage via inducing autophagy through extracellular signal-regulated kinase. Angiogenesis. 2016;19:311–324. doi: 10.1007/s10456-016-9504-y. [DOI] [PubMed] [Google Scholar]
- 16.Wang J. Shanghai Sci Tech Publishers; 2012. Guide for the care and use of medical laboratory animals. [Google Scholar]
- 17.Xiye F. Medical laboratory zoology. People's Health Press. 1995;156 [Google Scholar]
- 18.Wu X, He L, Chen F, He X, Cai Y, Zhang G, Yi Q, He M, Luo J. Impaired autophagy contributes to adverse cardiac remodeling in acute myocardial infarction. PLoS One. 2014;9:e112891. doi: 10.1371/journal.pone.0112891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zimmer S, Grebe A, Bakke SS, Bode N, Halvorsen B, Ulas T, Skjelland M, De Nardo D, Labzin LI, Kerksiek A, et al. Cyclodextrin promotes atherosclerosis regression via macrophage reprogramming. Sci Transl Medi. 2016;8:333ra50. doi: 10.1126/scitranslmed.aad6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kwon Y, Kim JW, Jeoung JA, Kim MS, Kang C. Autophagy is pro-senescence when seen in close-up, but anti-senescence in long-shot. Mol Cells. 2017;40:607–612. doi: 10.14348/molcells.2017.0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fernández ÁF, Sebti S, Wei Y, Zou Z, Shi M, McMillan KL, He C, Ting T, Liu Y, Chiang WC, et al. Disruption of the beclin 1-BCL2 autophagy regulatory complex promotes longevity in mice. Nature. 2018;558:136–140. doi: 10.1038/s41586-018-0162-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Revuelta M, Matheu A. Autophagy in stem cell aging. Aging Cell. 2017;16:912–915. doi: 10.1111/acel.12655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiong Y, Yepuri G, Forbiteh M, Yu Y, Montani JP, Yang Z, Ming XF. ARG2 impairs endothelial autophagy through regulation of MTOR and PRKAA/AMPK signaling in advanced atherosclerosis. Autophagy. 2014;10:2223–2238. doi: 10.4161/15548627.2014.981789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tai S, Hu XQ, Peng DQ, Zhou SH, Zheng XL. The roles of autophagy in vascular smooth muscle cells. Int J Cardiol. 2016;211:1–6. doi: 10.1016/j.ijcard.2016.02.128. [DOI] [PubMed] [Google Scholar]
- 25.Grootaert MO, da Costa Martins PA, Bitsch N, Pintelon I, De Meyer GR, Martinet W, Schrijvers DM. Defective autophagy in vascular smooth muscle cells accelerates senescence and promotes neointima formation and atherogenesis. Autophagy. 2015;11:2014–2032. doi: 10.1080/15548627.2015.1096485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yuan HX, Russell RC, Guan KL. Regulation of PIK3C3/VPS34 complexes by MTOR in nutrient stress-induced autophagy. Autophagy. 2013;9:1983–1995. doi: 10.4161/auto.26058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu J, Shen Y, Qian HY, Liu LJ, Zhou BC, Xiao Y, Mao JN, An GY, Rui MZ, Wang T, Zhu CL. Effects of mild hypothermia on the ROS and expression of caspase-3 mRNA and LC3 of hippocampus nerve cells in rats after cardiopulmonary resuscitation. World J Emerg Med. 2014;5:298–305. doi: 10.5847/wjem.j.issn.1920-8642.2014.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tran D, Bergholz J, Zhang H, He H, Wang Y, Zhang Y, Li Q, Kirkland JL, Xiao ZX. Insulin-like growth factor-1 regulates the SIRT1-p53 pathway in cellular senescence. Aging Cell. 2014;13:669–678. doi: 10.1111/acel.12219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Si X, Shao C, Li J, Jia S, Tang W, Zhang J, Yang J, Wu X, Luo Y. Loss of p21 promoted tumorigenesis in the background of telomere dysfunctions induced by TRF2 and Wrn deficiency. Int J Biol Sci. 2018;14:165–177. doi: 10.7150/ijbs.23477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.ten Kate GL, Sijbrands EJ, Staub D, Coll B, ten Cate FJ, Feinstein SB, Schinkel AF. Noninvasive imaging of the vulnerable atherosclerotic plaque. Curr Probl Cardiol. 2010;35:556–591. doi: 10.1016/j.cpcardiol.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 31.Lee AS, Kim JS, Lee YJ, Kang DG, Lee HS. Anti-TNF-α activity of Portulaca oleracea in vascular endothelial cells. Int J Mol Sci. 2012;13:5628–5644. doi: 10.3390/ijms13055628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Z, Lerman LO, Tang H, Barber C, Wan L, Hui MM, Furenlid LR, Woolfenden JM. Inflammation imaging of atherosclerosis in Apo-E-deficient mice using a (99m)Tc-labeled dual-domain cytokine ligand. Nucl Med Biol. 2014;41:785–792. doi: 10.1016/j.nucmedbio.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Şahin M, Ugan Y, Tunç ŞE, Akın Ş, Köroğlu B, Kutlucan A, Sütçü R, Yeşildağ A, Kılbaş A. Potential role of interleukin-18 in patients with rheumatoid arthritis-associated carotid intima-media thickness but not insulin resistance. Eur J Rheumatol. 2014;1:135–139. doi: 10.5152/eurjrheumatol.2014.140046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Z, Wang G, Feng D, Zu G, Li Y, Shi X, Zhao Y, Jing H, Ning S, Le W, et al. Targeting the miR-665-3p-ATG4B-autophagy axis relieves inflammation and apoptosis in intestinal ischemia/reperfusion. Cell Death Dis. 2018;9:483. doi: 10.1038/s41419-018-0518-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peng S, Xu LW, Che XY, Xiao QQ, Pu J, Shao Q, He B. Atorvastatin inhibits inflammatory response, attenuates lipid deposition, and improves the stability of vulnerable atherosclerotic plaques by modulating autophagy. Front Pharmacol. 2018;9:438. doi: 10.3389/fphar.2018.00438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang M, Xie Z, Gao W, Pu L, Wei J, Guo C. Quercetin regulates hepatic cholesterol metabolism by promoting cholesterol-to-bile acid conversion and cholesterol efflux in rats. Nutr Res. 2016;36:271–279. doi: 10.1016/j.nutres.2015.11.019. [DOI] [PubMed] [Google Scholar]
- 37.Sun X, Yamasaki M, Katsube T, Shiwaku K. Effects of quercetin derivatives from mulberry leaves: Improved gene expression related hepatic lipid and glucose metabolism in short-term high-fat fed mice. Nutr Res Pract. 2015;9:137–143. doi: 10.4162/nrp.2015.9.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang D, Liu X, Liu M, Chi H, Liu J, Han H. Protective effects of quercetin and taraxasterol against H2O2-induced human umbilical vein endothelial cell injury in vitro. Exp Ther Med. 2015;10:1253–1260. doi: 10.3892/etm.2015.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu L, Gao C, Yao P, Gong Z. Quercetin alleviates high-fat diet-induced oxidized low-density lipoprotein accumulation in the liver: Implication for autophagy regulation. Biomed Res Int 2015. 2015:607531. doi: 10.1155/2015/607531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qu L, Xiao-Chun L, Gu B, Zhang H, Dai W, SHI Y. Quercetin up-regulates autophagy in RSC96 cells culutured in high glucose via the pathway of Akt-Mtor. Basic Clin Med. 2015;5:596–602. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.