Abstract
Endothelial-to-mesenchymal transition (EndMT) is the process wherein endothelial cells lose their typical endothelial cell markers and functions and adopt a mesenchymal-like phenotype. EndMT is required for development of the cardiac valves, the pulmonary and dorsal aorta, and arterial maturation, but activation of the EndMT programme during adulthood is believed to contribute to several pathologies including organ fibrosis, cardiovascular disease, and cancer. Non-coding RNAs, including microRNAs, long non-coding RNAs, and circular RNAs, modulate EndMT during development and disease. Here, we review the mechanisms by which non-coding RNAs facilitate or inhibit EndMT during development and disease and provide a perspective on the therapeutic application of non-coding RNAs to treat fibroproliferative cardiovascular disease.
Keywords: Non-coding RNA, Endothelial-mesenchymal transition (EndMT), Cardiac development, Cardiovascular disease, Plasticity
1. EndMT during development and pathology
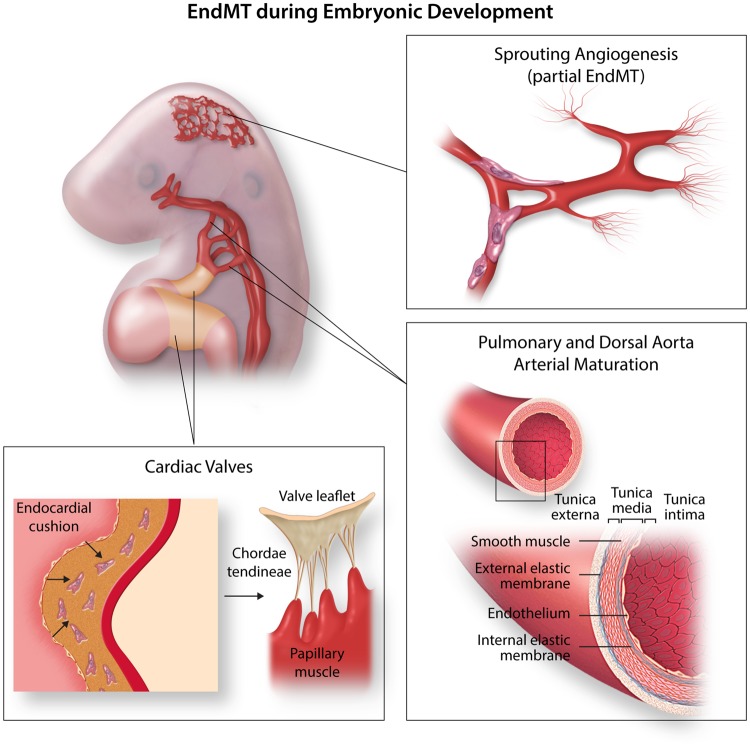
Endothelial cells form the inner layer of every single vessel in the body acting as a barrier between the blood or lymph and the rest of the tissues. Endothelial cells are highly plastic cells, that are able to differentiate into arterial, venous, lymphatic, and endocardial cell fates during development, but also into haematopoietic linages,1,2 or mesenchymal lineages through a process coined endothelial-to-mesenchymal transition (EndMT). During heart development, EndMT is essential for the formation of the valve mesenchyme.3,4 These valvulogenic regions are located in the atrioventricular canal (AVC) and the outflow tract (OFT) of the developing heart and undergo differential specification programmes compared to the cardiac chambers.5,6 Besides the involvement of EndMT during valve development, EndMT is involved in embryonic pulmonary artery development7 and in the formation of the smooth muscle component of the dorsal aorta (Figure 1).8 Furthermore, just recently, a partial EndMT process has been suggested pivotal to physiological angiogenic sprouting (Figure 1).9 This new concept may open new interpretations of vessel development, but also for heart development, where recent studies suggested a process similar to sprouting angiogenesis in the endocardium during ventricular trabeculation.10
Figure 1.
EndMT in development. During embryonic development, EndMT contributes to the cardiac valves, pulmonary and dorsal aorta arterial maturation, and sprouting angiogenesis (partial EndMT).
EndMT is a specific form of epithelial-to-mesenchymal transition (EMT) and the term was coined to refer specifically to the EMT affecting the endothelium, a squamous type of epithelium. The regulation of EndMT during normal developmental conditions shares many aspects with the general process of EMT; however, the molecular defects promoting pathologic EndMT are less understood. EndMT is characterized by loss of endothelial markers such as VE-Cadherin and CD31, and upregulation of the EndMT transcription factors SNAIL, SLUG, TWIST, ZEB1, and ZEB2 as well as mesenchymal markers such as α-SMA (ACTA2) and S100A4. EndMT-derived mesenchymal cells acquire a highly migratory and invasive potential which is accompanied by morphological changes from a cobble-stone endothelial morphology into a spindle-shaped myofibroblast-like morphology. In recent years, the reactivation of EndMT during adult life has gained increasing attention in the cardiovascular field due to its implication in numerous adult pathologies including pulmonary hypertension,11 atherosclerosis,12,13 brain vascular malformations,14 tissue fibrosis,15–17 and cancer progression18,19 (Figure 2). Under pathological conditions, several environmental factors induce EndMT, including high glucose, hypoxia, oxidative stress, pro-inflammatory cytokines, and disturbed shear stress (the frictional force of the blood flow on the endothelial cells).20–24 These environmental factors trigger the activation of signalling pathways which induce EndMT such as the canonical Transforming Growth Factor Beta (TGF-β) pathway or the non-canonical pathways such as Notch and Wnt.13 However, fibroblast growth factor (FGF) and mitogen-activated protein kinase (MAPK) modulate EndMT as inhibitory pathways.25 These signalling pathways are described in more detail below. The environmental factors and, thereby-induced signalling pathways, can also lead to the expression of non-coding RNAs, functional RNA molecules which are not translated into proteins. Non-coding RNAs include microRNAs (miRNAs), long non-coding RNAs (lncRNAs), and circular RNAs (circRNAs). These non-coding RNAs influence EndMT pathways during development and pathology.26 In this review, we summarize the current knowledge on the mechanisms by which non-coding RNAs regulate the EndMT process and outline their potential as therapeutic molecules to alleviate fibroproliferative diseases. Readers interested in further detail about the overall EndMT process, or its involvement during development or adult cardiovascular diseases, are referred to other recent articles covering these aspects in depth.13,27–29
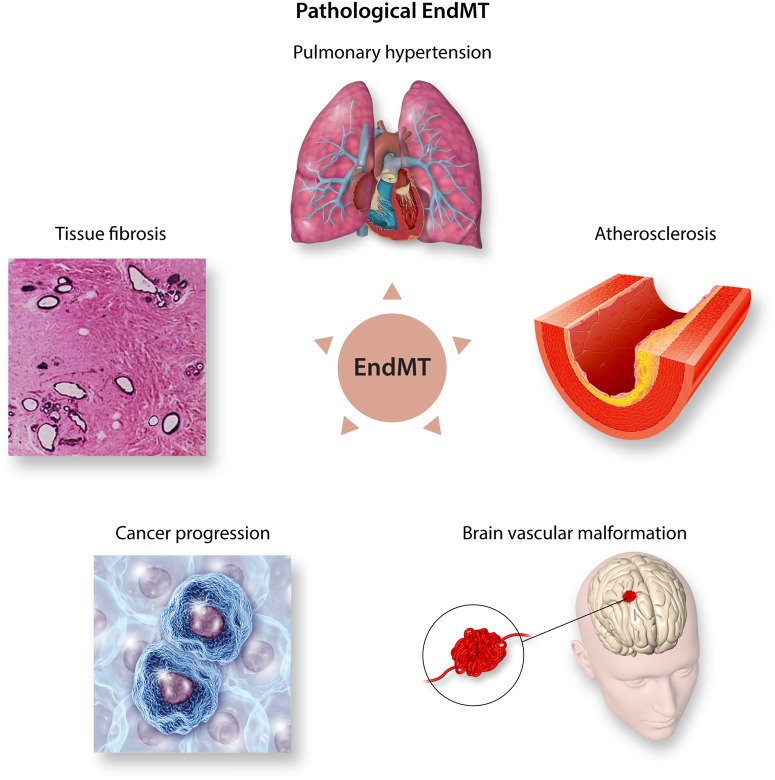
Figure 2.
Pathological EndMT. During adult life, reactivation of EndMT contributes to several pathologies such as pulmonary hypertension, atherosclerosis, brain vascular malformation, cancer progression, and tissue fibrosis.
2. Canonical EndMT pathways
2.1 The TGF-β signalling pathway
When considering the molecular mechanisms controlling EndMT, the most simplistic overview is a finely regulated network integrating TGF-β, bone morphogenetic proteins (BMPs), and the Notch signalling pathway (Figure 3).30 There are three ligands in the TGF-β family involved in EndMT; TGF-β1, -2, and -3.31 TGF-β ligands signal through tetrameric receptors formed by two type II TGF-β receptors (TGFβR2) and two type I TGF-β receptors (ALK5 and ALK2). Once activated, they induce an intracellular signalling pathway mediated by SMAD2 and SMAD3, which then interact with SMAD4, and the entire SMAD complex translocates to the nucleus and binds DNA-associated proteins to promote the expression of mesenchymal genes. In endothelial cells, TGF-β ligands also induce the activation of the type I TGF-β receptor ALK1 which antagonizes EndMT by signalling through SMAD1, 5, and 8, allowing certain crosstalk between the TGF-β and BMP pathways. TGF-β signalling can also occur in a SMAD-independent manner by activating intracellular signalling pathways such as MAP kinase (MAPK), Rho-like GTPase, ERK1/2, or phosphatidylinositol-3-kinase (PI3K)/AKT pathways,32,33 all of which are inducers of EndMT.
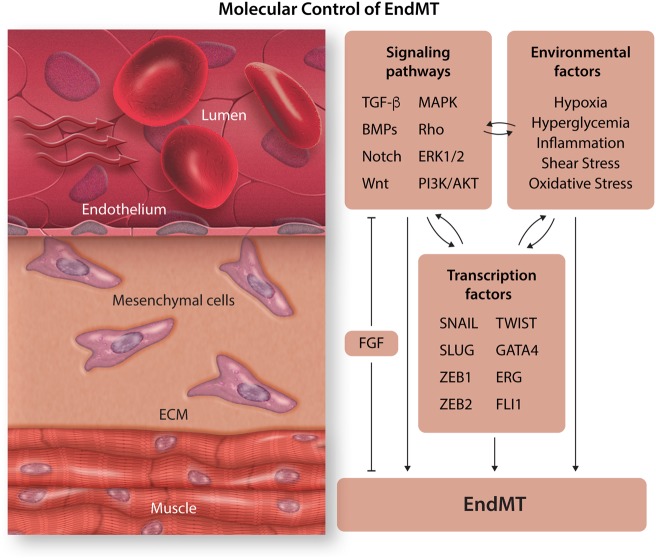
Figure 3.
Molecular control of EndMT. EndMT is induced by several environmental factors as well as signalling pathways which result in transcription of EndMT transcription factors which facilitate EndMT.
2.2 Transcription factors regulating EndMT
TGF-β ligands have been described as master regulators of EndMT by promoting the expression of the transcription factors SNAIL, SLUG, TWIST, ZEB1, and ZEB2.34 Although all of them repress the transcription of VE-Cadherin, they perform overlapping but non-redundant roles during EndMT. The transcription factors SNAIL and SLUG are pivotal to EndMT induction, whereas TWIST, ZEB1, and ZEB2 maintain the migratory phenotype.35 Furthermore, these transcription factors cross-regulate each other and their own expression, highlighting the fine molecular regulation of EndMT.36,39 Other transcription factors involved in the EndMT process include GATA4 and the ETS factors ERG and FLI1. Endothelial specific deletion of GATA4 culminates in reduced EndMT whereas the suppression of ERG or FLI1 expression enhances EndMT via activation of the TGF-β pathway, in particular SMAD2/3 transcription factors.40,41
2.3 The BMP signalling pathway
BMPs signal through four possible type I receptors (ALK1, ALK2, ALK3, and ALK6) and three type II receptors (BMPRII, ActRII, or ActRIIB). During valve development, BMP2, 4, 5, 6, and 7 ligands are expressed in the valvulogenic regions. Receptor complex activation promotes intracellular signalling mediated by the SMAD1, 5, and 8, that together with SMAD4 promote the nuclear translocation of the SMAD complex and the activation of BMP-responsive genes. During development, BMPs play critical roles in the patterning the AVC/OFT myocardium to provide a pro-EndMT environment and promote the transition of endothelial cells into invasive mesenchymal cells. In particular, BMP2 and BMP4 are the two ligands described to be responsible for EndMT in the AVC and OFT.5,42,43 It has to be noted that in contrast to its inducing role on EndMT during development, BMPs (in particular BMP7) have been described to inhibit pathological EndMT during adulthood.17,44
3. Non-canonical EndMT pathways
3.1 The notch signalling pathway
Notch is a cell-to-cell signalling pathway formed by four different receptors (Notch1–4). Notch is a key signal in the induction of EndMT during heart valve development. Notch promotes EndMT by inducing the expression of SNAIL and SLUG.45,46 Notch1 and 2 mutant embryos show heart valve defects that arise due to defective EndMT.45 The Notch pathway acts synergistically with the TGF-β2 and BMP2 pathways in the control of EndMT.46,47 TGF-β2 and BMP2 are able to induce SNAIL expression and to a weaker extent SLUG expression, whereas Notch activates SLUG and synergistically induces SNAIL in concert with TGF-β2 and BMP2.46,47 Furthermore, Notch controls the BMP2 expression pattern in the AVC myocardium.47
3.2 The Wnt signalling pathway
Canonical Wnt signalling involves signal transduction through stabilization of β-catenin. Stabilized β-catenin translocates to the nucleus where it interacts with DNA-associated transcription factors to induce target gene expression. EndMT is strongly inhibited in endothelial-specific β-catenin mutants.48,49 Wnt/β-catenin acts in the AVC endocardium downstream from TGF-β to promote the acquisition of a mesenchymal phenotype during EndMT.50 Canonical Wnt signalling induces EndMT by the induction of SNAIL and SLUG expression.51,52 In contrast, Wnt signalling through Wnt7a inhibits EndMT,53 highlighting the fine regulation of the EndMT process by the Wnt pathway.
3.3 Inflammatory EndMT
In general, the pathologies in which EndMT is recognized are associated with inflammatory activation in response to tissue damage. During the inflammatory response, pro-inflammatory cytokines including interleukin (IL)-1β and tumour necrosis factor alpha (TNF-α) activate the transcription factor nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) involved in the activation of other pro-inflammatory cytokines including IL-1, IL-6, TNF-α, or interferon gamma (IFNγ), hence creating a positive feedback regulatory loop. Activation of NF-κB promotes the induction of TGF-β1 and TGF-β2 and promotes EndMT.54 But NF-κB can also induce EndMT in a TGF-β-independent manner by directly activating SNAIL expression.55,56
4. Inhibitory EndMT pathways
4.1 The FGF signalling pathway
The FGF signalling pathway is formed by 22 different ligands that mediate their biological activity by binding to cell surface FGF receptors (FGFR1–4).57 Once activated, FGF receptors recruit similar intracellular signal transduction factors to other tyrosine kinase receptors including PI3K and MKK3/6, and activate similar intracellular signalling pathways including AKT-ERK1/2, Ras, or MAPK (Raf, MEK). The FGF pathway is an important regulator of endothelial TGF-β signalling mainly through FGFR1. Loss of FGFR1 signalling activates the EndMT programme in a TGF-β-dependent manner culminating in intimal hyperplasia and stenosis.58
5. Other factors regulating EndMT
Low oscillatory shear stress induces the expression of SNAIL, TWIST, and GATA4 in the endothelium at atheroprone sites,59,60 activates TGF-β signalling and decreases FGFR1 expression.12 Moreover, during hypoxia, HIF-1α accumulates in the nucleus61 where it activates hypoxia-induced genes including SNAIL, TWIST, and ALK5.62,63 Oxidative stress also promotes EndMT and has additive effects to TGF-β.64 Finally, the exposure to long-term high glucose levels, such as in diabetic patients, also promotes EndMT through PI3K/Akt/NF-κB pathways leading to cardiac fibrosis in diabetic patients.65 Altogether, this shows the diversity of factors and signalling pathways that modulate EndMT (Figure 3).
6. Non-coding RNAs
miRNAs are small fragments of RNA (typically 20–25 nucleotides). miRNAs can bind to complementary sequences within mRNA targets and then degrade the mRNA via cleavage or destabilization or inhibit the translation of mRNAs into protein.66 This represses gene and protein expression of direct targets of miRNAs but could also indirectly result in reactivation of gene and protein expression (because their repressors are downregulated by the miRNA). Increasing evidence describes a non-canonical role for miRNAs in transcriptional regulation as well, but the underlying mechanisms remain largely unknown. Non-coding RNAs are classified as lncRNAs when they contain more than 200 nucleotides. Their mechanism of action varies not only between different lncRNAs but also one particular lncRNA can act using different mechanisms. They can act as recruiters, tethers, and scaffolds for other regulatory factors involved in epigenetic modifications, or regulate gene transcription by acting as decoys, coregulators, or polymerase-II inhibitors. They are also involved in the organization of the different components of the transcription and splicing machinery, as well as the organization of subnuclear structures. lncRNAs are also involved in post-transcriptional regulation, controlling processes such as mRNA processing, stability, or translation, acting as sponges for miRNAs blocking their effects, or even can be the source of other small RNAs (for a review see reference 67). lncRNA can be also classified according to their structural conformation. On this regard, in this review we will separate the circRNAs from the rest of the lncRNA described. circRNAs are another subclass of non-coding RNAs generally formed by backsplicing that, similarly to other lncRNAs, function as miRNA sponges, RNA-binding protein sequestering factors, as well as regulators of gene expression by controlling mRNA transcription.68,69 Furthermore, circRNAs can also control gene transcription by interactions with phosphorylated Pol II or by competition with the pre-mRNA splicing machinery.70,71 circRNAs are highly stable molecules that form a covalently closed continuous loop making them resistant to RNA exonuclease digestion.72,73 The mechanisms of action of non-coding RNAs are summarized in Figure 4. Similar to lncRNAs, circRNAs can control EndMT by acting as sponges for miRNAs or by regulating the transcription of genes which can result in EndMT promotion or inhibition. In summary, miRNAs are important modulators as well which can affect gene and protein expression and thus modulate EndMT. lncRNAs and circRNAs, together with miRNAs, are therefore promising targets for therapeutic applications which aim to inhibit EndMT.
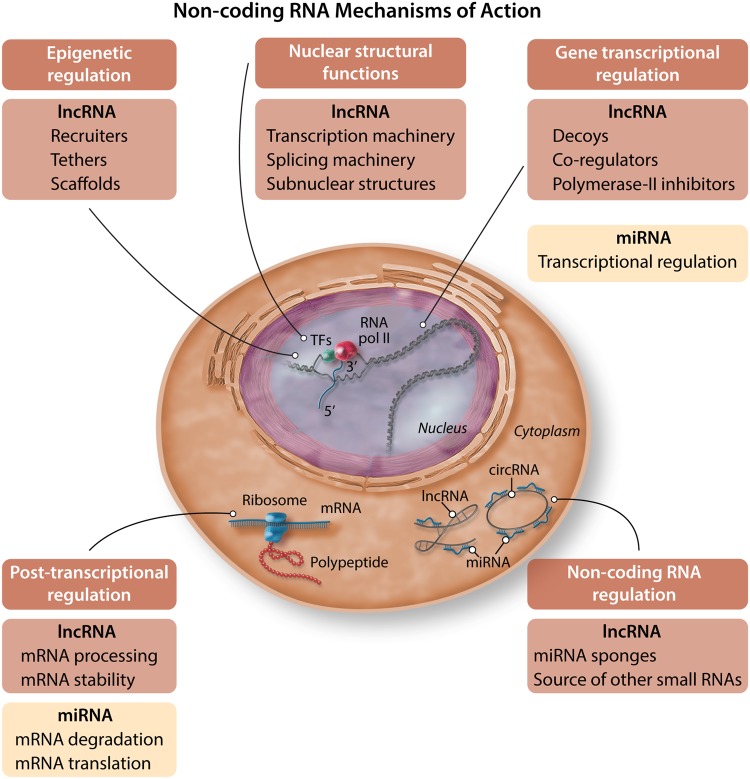
Figure 4.
Non-coding RNA mechanisms of action. Examples of how non-coding RNAs interfere with biological processes in cells (e.g. transcriptional regulation, post-transcriptional regulation, and sponging of miRNAs).
7. MicroRNAs inducing/facilitating EndMT
7.1 TGF-β2-induced EndMT is associated with differential expression of several miRNAs
TGF-β2-induced EndMT is associated with increased expression of miR-21, miR-27b, miR-30b, miR-125b, miR-195, Let-7c, Let-7g and decreased expression of miR-122a, miR-127, miR-196, and miR-375.74,75 This suggests a role for these miRNAs in modulating EndMT. Indeed, miR-21 and miR-27b facilitate EndMT.76,77 Inhibition of miR-21 reverts the TGF-β2-induced repression of VE-Cadherin and induction of S100A4.77 Consistently, kallistatin, an endogenous plasma protein, can reverse the TGF-β1-induced upregulation of miR-21 and thereby inhibit EndMT.76 Kallistatin can also reverse the expression of the EndMT transcription factor SNAIL, Akt phosphorylation, and activation of NF-κB, emphasizing that the inhibiting effect of kallistatin on EndMT is not only due to the inhibition of miR-21 expression.76 It has to be noted that inhibition of miR-21 did not affect ionizing radiation-induced EndMT.78 Another miRNA that is a positive regulator of EndMT is miR-27b. Inhibition of miR-27b suppresses EndMT by preclusion of ACTA2 and TAGLN (SM22α) expression, through an unidentified mechanism.75 The expression of another miRNA, miR-31, is not altered upon TGF-β2 treatment,79 yet inhibition of miR-31 suppresses both ACTA2 and TAGLN expression suggesting a role for miR-31 in facilitating EndMT.79 However, overexpression of miR-31 has limited effects on TGF-β2-induced EndMT suggesting that miR-31 does not induce EndMT directly but rather controls the magnitude of EndMT.79 Let-7c and Let-7g, two other miRNAs which are upregulated upon TGF-β2 treatment, are part of the Let-7 family which represses EndMT and will be discussed later in this review. The role of the other differentially expressed miRNAs in the regulation of EndMT remain unknown to date.
7.2 The role of miR-130 in modulating EndMT/EMT
miR-130b is upregulated in high proliferative and angiogenic-prone colorectal cancer, suggesting a role of miR-130b in proliferation, angiogenesis, EndMT, and/or EMT.80 Indeed, miR-130b overexpression facilitates tumour growth accompanied by enhanced proliferation, angiogenesis, and EMT.80 Overexpression of miR-130b results in decreased expression of E-Cadherin whereas EndMT transcription factors SNAIL and ZEB1 are increased.80 miR-130a expression is increased in pulmonary hypertension as well as in TGF-β1-treated endothelial cells, suggestive of a role for miR-130a in modulating both EndMT and EMT.81 Indeed, miR-130a enhances the TGF-β1-induced expression of ACTA2 and decreases CD31 expression.81 Interestingly, NF-κB can induce miR-130a expression and vice versa, illustrating the interplay between NF-κB and miRNAs.81
7.3 miR-374b and miR-449a modulate EndMT during atherosclerosis
Both miR-374b and miR-449a associate with EndMT in the context of atherosclerosis.82,83 Indeed, miR-449a expression culminates in reduced E-Cadherin expression and an increase in the expression of α-SMA and SMAD3.82 Importantly, antagonizing miR-449a expression inhibits the development of atherosclerosis in diabetic mice.82 Similarly to miR-449a, the expression of miR-374b is elevated in atheroprone regions in vivo, as well as TGF-β1-treated endothelial cells.83 Overexpression of miR-374b results in decreased expression of the endothelial markers VE-Cadherin and eNOS whereas the expression of mesenchymal markers TAGLN and Calponin increases.83 Interestingly, combined overexpression of miR-374b and MAPK7 (a known antagonist of EndMT) precludes the EndMT induction by miR-374b.83 Glucose treatment induces the expression of miR-328,84 and induces EndMT evidenced by an increase in the expression of mesenchymal markers Collagen-1 and Collagen-3 accompanied by activation of MEK1/2 and MAPK1/2.84 These data highlight the interplay between miRNAs and MAPK signalling, and identify MAPK signalling as a crucial regulator of EndMT.
7.4 The role of miR-9 and miR-342-5p in modulating EndMT and angiogenesis
Contrasting effects of miR-9 and miR-342-5p highlight differential effects of miRNAs on certain biologic processes. miR-9 induces EndMT with decreased VE-Cadherin expression and a corresponding increase in N-Cadherin.85 miR-9 also represses NF-κB expression and inflammation whereas it promotes tube formation.85 On the other hand, miR-342-5p also induces EndMT, but in contrast to miR-9, this inhibits lumen formation and angiogenic sprouting.86 These data show that although there is redundancy in miRNAs that induce EndMT, there are also distinct outcomes.85,86 Indirectly, this also suggests that pro-angiogenic conditions may or may not be pro-EndMT, and vice versa.
8. miRNAs inhibiting EndMT
8.1 miR-15a, miR-23b, and miR-199a as inhibitors of EMT/EndMT during development
While the abovementioned miRNAs function to promote EndMT, others exhibit an inhibitory effect. For example, the levels of miR-15a, miR-23b, and miR-199a are elevated during AVC development in the embryonic chick heart, suggesting a role for these miRNAs in modulating both EMT and EndMT.87 Indeed, miR-15a, miR-23b, or miR-199a inhibit EMT/EndMT in AVC explants.87 In line with this, miR-23 also inhibits TGF-β-induced EMT/EndMT by inhibiting the TGF-β-induced expression ACTA2 and SNAIL.88 Overexpression of miR-199a-5p in human umbilical vein endothelial cells (HUVECs) as well as irradiated HUVECs undergoing EndMT induced the expression of α-SMA and Collagen-1 in co-cultured human foetal lung fibroblasts.89 This suggests that miR-199a-5p as well as EndMT in itself are important for myofibroblast activation of neighbouring fibroblasts.89
8.2 The role of miR-483 in modulating EndMT
miR-483 reduces oscillatory flow-induced EndMT by decreasing the expression of SNAIL, SLUG, TWIST, and TAGLN,90 which coincides with a reduction in inflammatory activation of IL-6, ICAM-1, and VCAM-1.90 Concurrently, endothelial cells expressing miR-483 decrease the expression of mesenchymal markers induced by serum of Kawasaki patients, while increasing the expression of endothelial markers.91 Interestingly, statin (atorvastatin) treatment in combination with serum of Kawasaki patients induces the expression of miR-483 and similarly represses EndMT.91 This highlights the potential of pharmacological agents to interfere with the EndMT programme through non-coding RNAs. Moreover, it is tempting to speculate that repression of EndMT is among the so-called ‘pleiotropic beneficial effects’ attributed to statins.
8.3 The role of miR-148 in inhibiting EndMT
Fibrodysplasia ossificans progressiva is a congenital disorder associated with skeletal malformations and heterotopic calcification in which EndMT is involved. While gain of function mutations in ALK2 are associated with this disease, the activin A receptor type I (ACVR1) gene is also implicated. Notably, both are associated with the TGF-β pathway signalling. ACVR1 is a target of miR-148a, and the constitutive activation of ACVR1 is known to induce EndMT. Indeed, expression of miR-148a represses BMP signalling in endothelial cells,92 suggesting that miR-148a can modulate EndMT. To our knowledge, this hypothesis has not been directly examined. In contrast, the expression of miR-148b does not decrease ACVR1 expression, implying that miR-148a and miR-148b have different gene targets.93 Overexpression of miR-148b increases migration, proliferation and angiogenesis in HUVECs.93 On the other hand, inhibition of miR-148b induces EndMT both in vitro and in a mouse model of skin wound healing.93 In line with this, TNF-α and IL-1β treatment in HUVECs decreased miR-148b levels and induced EndMT, which is precluded by the overexpression of miR-148b.93
8.4 Let-7, miR-424, and miR-503 inhibit EndMT
The inflammation-induced loss of FGF signalling decreases the expression levels of Let-7, culminating in the activation of TGF-β signalling and thus EndMT.94 Let-7b inhibits EndMT in a murine transplant arteriopathy model.94 Moreover, the major plasma metabolite HT-3O sulfate, with antioxidant and anti-inflammatory properties, protects against IL-1β-induced EndMT by restoring Let-7 expression.95 IL-13 treatment induces EndMT accompanied by decreased levels of miR-424 and miR-503, suggesting a role for these miRNAs in modulating EndMT.96 Indeed, inhibition of miR-424 increases the expression of the mesenchymal markers α-SMA and N-Cadherin.96 Furthermore, miR-424 or miR-503 inhibit the migration of endothelial cells.96
8.5 miR-18a-5p and miR-532 inhibit EndMT during cardiac fibrosis and myocardial infarction
miR-18a-5p inhibits glucose-induced EndMT by decreasing the expression of the mesenchymal markers S100A4, Vimentin, and Fibronectin and increasing the expression of CD31.97 miR-18a-5p attenuates both cardiac fibrosis and EndMT in diabetic mice.97 Since miR-18a-5p targets Notch2, this might explain the underlying mechanism of how miR-18a-5p inhibits EndMT.97 Knockdown of another miRNA, miR-532, in a mouse model of myocardial infarction elevates the abundance of Collagen-1/CD31 and α-SMA/CD31 double-positive cells, indicative of active EndMT.98 Indeed, knockdown of miR-532 enhanced TGF-β2-induced EndMT by increasing the expression of Collagen-3, SNAIL, and ACTA2 while decreasing the expression of CD31 and vWF.98
8.6 The role of miR-218, miR-221, miR-302c, and miR-494 in inhibiting EndMT
miR-302c inhibits EndMT in vitro.99 Interestingly, co-implantation of a human hepatocellular carcinoma cell line and endothelial cells with loss or gain of 302c drastically differed hepatocellular carcinoma growth in mice, implying that miR-302c in endothelial cells may suppress endothelial cell-mediated tumour growth.99 Reprogramming by the RhoA-Rock-canonical BMP signalling pathways is associated with increased expression of miR-302b and miR-302c.100 This demonstrates the essential role of miR-302b/c in modulating endothelial cell behaviour. Another miRNA, miR-218, decreases CTGF expression, thereby increasing the expression of E-Cadherin while reducing Vimentin and Fibronectin expression in a human colon cancer cell line (HCT116 cells).101 When HUVECs were treated with conditioned medium from miR-218 overexpressing HCT116 cells, this suppressed angiogenesis.101 These data suggest that miR-218 not only inhibits EndMT/EMT but also angiogenesis.101 miR-221 also suppresses angiogenesis by downregulating ZEB2 expression in HUVECs.102 Treatment of HUVECs with conditioned medium from miR-494 overexpressing decidua-derived mesenchymal stem cells also impairs capillary formation.103 However, the role of miR-221 and miR-494 in regulating EndMT needs further elucidation.
8.7 miR-192, miR-194, miR-497, miR-29, and Let-7 as inhibitors of EndMT in kidney disease
miR-192 or miR-194 increase E-cadherin and decrease the expression of N-Cadherin and ZEB2,104 suggesting that a decrease in miR-192 or miR-194 during autosomal dominant polycystic kidney disease contributes to EMT/EndMT.104 Another miRNA which might be involved in inhibiting EndMT is miR-497.105 Melatonin inhibits TGF-β2-induced EndMT by attenuating the TGF-β2-induced reduction in miR-497 expression, thereby suppressing the expression of ROCK1 and ROCK2.105 Melatonin also reverses the loss of miR-497 and increase in EndMT in glomeruli of diabetic rats, showing the overall importance of miR-497 in the inhibition of EndMT in the context of kidney disease.105 Linagliptin, a new dipeptidyl peptidase-4 (DPP-4) inhibitor that is used to treat diabetes, is another pharmacological agent that inhibits EndMT and ameliorates kidney fibrosis in diabetic mice.106,107 Importantly, linagliptin modulates miR-29 expression, and the miR-29 family suppresses EndMT, suggesting that linagliptin inhibits EndMT by restoring miR-29 expression levels in chronic kidney disease.106 Diabetic mice also have decreased expression of N-acetyl-seryl-aspartyl-lysyl-proline (AcSDKP), an endogenous anti-fibrotic peptide, which is associated with insufficient levels of anti-fibrotic miRNAs in the kidney such as the miR-29 and Let-7 families.108 Importantly, administration of AcSDKP to diabetic mice, and also to TGF-β2-treated endothelial cells, decreases the expression of TGF-βR1 and SMAD3 phosphorylation.108,109 AcSDKP restores the expression of both the miR-29 and Let-7 miRNA families,108 while Linagliptin reverses the decrease in expression of the Let-7 family via restoration of the FGF signalling.109 This demonstrates that anti-fibrotic interventions induce a similar anti-fibrotic miRNA profile both in vivo and in vitro.106,108 Interestingly, overexpression of Let-7 in combination with TGF-β2 treatment induces the expression of the miR-29 family, and vice versa.108 This suggests a crosstalk between the miR-29 and Let-7 miRNA families in facilitating anti-fibrotic and EndMT inhibitory effects.108
9. miRNAs which have different effects on EndMT in development and pathology
9.1 The role of the miR-200 family in modulating EndMT
miR-200a inhibits TGF-β-induced EndMT.110 Similarly, another miR-200 family member, miR-200b, prevents both glucose- and TGF-β1-induced EndMT.111 Endothelial cell-specific overexpression of miR-200b in diabetic mice prevents glucose-induced EndMT in the heart as well as in retinal tissues.111,112 In addition, overexpression of miR-200b associates with angiogenesis suppression.113 Downregulation of the miR-200 family results in the upregulation of the EndMT transcription factors SNAIL and ZEB1.114 This underlines the role of the miR-200 family in inhibiting EndMT. Individuals with bicuspid aortic valves (and therefore a higher risk of developing aortic aneurysm) have lower expression of miR-200c suggesting potential activation of both EMT and EndMT.114 Individuals with bicuspid aortic valves are also associated with a non-coding variant, called rs6601627, which is suggested to interact with GATA4.115 Importantly, CRISPR/Cas9-mediated disruption of GATA4 impairs TGF-β2 and BMP2-induced EndMT in endothelial cells derived from human induced pluripotent stem cells.115 This demonstrates that non-coding RNAs are important in regulating both EMT and EndMT in aortic valves. In contrast to the above, during development, SNAIL-induced repression of the miR-200 family promotes the generation of Flk1-positive endothelial cells, suggesting that during development the miR-200 family has an opposite role and supports the maintenance of the endothelial character.116 Indeed, lower levels of the miR-200 family in human embryonic stem cells is also associated with differentiation into vascular endothelial cells.117 Furthermore, miR-200a did not affect EMT/EndMT during development of the AVC in the developing chicken heart.87 This suggests that the miR-200 family has different roles during development and pathological conditions in different species.
9.2 The role of miR-126 in regulating EndMT
Combined knockdown of ERG and FLI1 induces EndMT which is accompanied by low miR-126 expression levels, suggesting a role for miR-126 in modulating EndMT.118 Indeed, miR-126 limits the expression of ACTA2, TAGLN, Collagen-1, and SLUG and partially counteracts the reduction in VE-Cadherin and CD31 expression.118 Interestingly, treatment of HUVECs with conditioned medium from tumours also resulted in decreased expression of both ERG and FLI1, suggesting that the decrease of these transcription factors by soluble mediators from the tumour microenvironment can promote EndMT and therewith tumour progression.118 In line with this, miR-126 suppresses the expression of the mesenchymal genes ACTA2, TAGLN, and myocardin while maintaining the expression of progenitor markers.119 Also, miR-126 reverses the TGF-β1-induced activation of FoxO3 and SMAD4 and decrease in PI3K and Akt, suggesting novel pathways involved in the modulation of EndMT.119 In contrast to the above data, knockdown of miR-126a-5p reverses the hypoxia-induced decrease in CD31 and increase in α-SMA.120 Altogether, while the majority of studies suggest that miR-126 inhibits EndMT, it appears that in specific contexts miR-126 may exert pro-EndMT effects.
9.3 The role of miR-155 and miR-20a in modulating EndMT
Inhibition of miR-155 reverses the TGF-β-induced expression of SNAIL, SLUG, TWIST, and Vimentin, suggesting that miR-155 inhibits EndMT.121 In contrast, inhibition of miR-155 does not affect TGF-β3-induced EndMT in mouse embryonic endothelial cells.122 Moreover, overexpression of miR-155 inhibits TGF-β3-induced EndMT, suggesting that induction of miR-155 expression represses EndMT.122 This demonstrates that the mechanism by which miR-155 regulates EndMT is different during developmental and pathological contexts, which might be explained by the maturity of the cells or the difference in TGF-β isoform. miR-20a also has differential effect on EndMT in development and adult pathology. During the development of the OFT in the mouse embryos, deletion of BMP4/7 inhibits EMT/EndMT which could be rescued by the expression of miR-17/20a, indicative of an EndMT inductor role for this miRNA.123 In contrast, FGF2-induced expression of miR-20a limits EndMT by reducing the expression of TGF-β receptor 1/2 and SARA (which recruits SMAD2 and SMAD3 to the TGF-β receptor complex) in adult endothelial cells.124
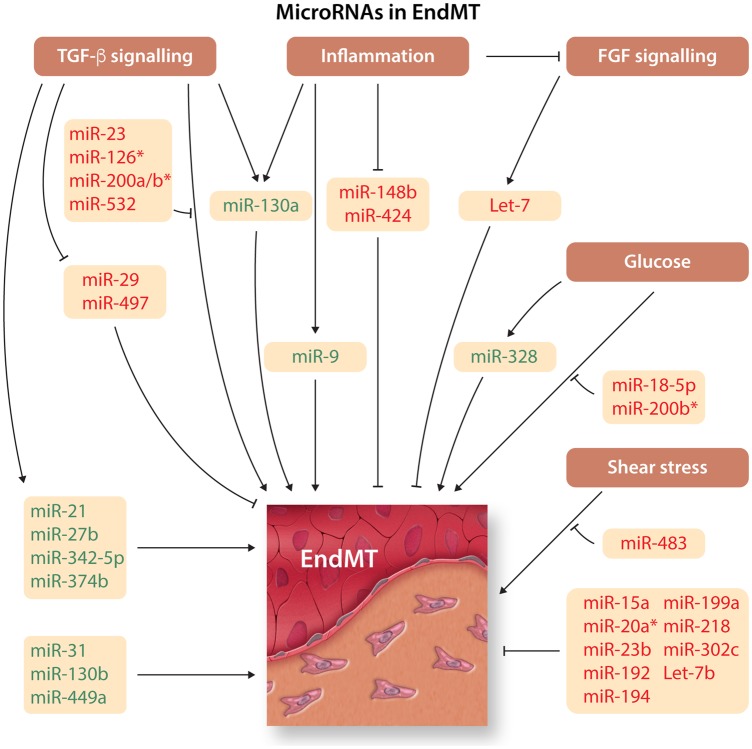
In all, a remarkable number of miRNAs are associated to the EndMT regulatory programme. These are summarized in Figure 5 and Table 1.
Figure 5.
MicroRNA in EndMTOverview of microRNAs (miRNAs) that induce or inhibit EndMT. miRNA inducers and inhibitors of EndMT are depicted in green and red, respectively. *A different role during pathology and development.
Table 1.
MicroRNAs in EndMT
| miRNAs | Biological context | Targets | Experimental model |
|---|---|---|---|
| Inducing EndMT | |||
| miR-9 | Development/Pathology— Lymphangiogenesis/Inflammation | NF-κB1 | In vitro—Rat mesenteric lymphatic endothelial cells85 |
| miR-17 | Development—OFT formation | Vegfa |
|
| miR-21 | Pathology—Fibrosis | ? | |
| miR-27b | Molecular characterization | Elk1, Nrp2, PlxnA2, PlxnD1 | In vitro—Mouse pancreatic microvascular endothelial cells75 |
| miR-31 | Pathology—Inflammation | VAV3 | In vitro—Mouse pancreatic microvascular endothelial cells79 |
| miR-130a | Pathology—Pulmonary hypertension | BMPR2 |
|
| miR-130b | Pathology—Colorectal cancer | PPARγ |
|
| miR-143 | Pathology—Ischaemic stroke | HECTD1 | In vivo—Mouse brain136 |
| miR-328 | Pathology—Diabetes | ? | In vitro—HUVECs84 |
| miR-342-5p | Development—Angiogenesis | Endoglin |
|
| miR-374b | Pathology—Neointimal hyperplasia | RAC1, MAP3K3, MAP3K7, MAPK7, MEF2D, KLF4 |
|
| miR-449a | Pathology—Atherosclerosis | AdipoR2 |
|
| Inhibiting EndMT | |||
| miR-15a | Development—AVC formation | ? | Ex vivo—Chicken AVC explants87 |
| miR-18a-5p | Pathology—Diabetes | Notch2 |
|
| miR-23b | Development—AVC formation | ? | Ex vivo—Chicken AVC explants87 |
| Development—Cardiac valve formation | Has2, Icat, Tmem2 |
|
|
| miR-29a | Pathology—Bladder carcinoma | VEGFA |
|
| miR-29 family | Pathology—Diabetes-related kidney fibrosis | DPP-4 | |
| miR-30d | Pathology—Neuroinflammatory disorders | ATG5 |
|
| miR-145 | Pathology—Neointimal hyperplasia | TGFBR2, SMAD3 | In vitro—Endothelial progenitor cells125 |
| miR-148b | Physiology—Skin wound healing | TGFB2, SMAD2 |
|
| miR-186-5p | Pathology—Prostate cancer | Twist1 | In vitro—Prostate cancer cells127 |
| miR-192/194 | Pathology—Kidney disease | ZEB2, CDH2 |
|
| miR-199a | Development—AVC formation | ? | Ex vivo—Chicken AVC explants87 |
| Pathology—Radiation-induced pulmonary fibrosis | ? | In vitro—HUVECs89 | |
| miR-218 | Pathology—Colorectal cancer | CTGF |
|
| miR-221 | Pathology—Tumour angiogenesis | ZEB2 | In vitro—HUVECs102 |
| miR-302c | Pathology—Hepatocellular carcinoma | MTDH |
|
| Pathology—Corneal blindness | ? | In vitro—Human corneal endothelial cells100 | |
| miR-424/503 | Pathology—Pulmonary hypertension | ? | In vitro—Human pulmonary artery endothelial cells96 |
| miR-483 | Pathology—Aortic valve calcification | ? |
|
| Pathology—Kawasaki disease | CTGF |
|
|
| miR-494 | Pathology—Preeclampsia | VEGF | In vitro—HUVECs103 |
| miR-497 | Pathology—Diabetic Nephropathy | ROCK1/2 | In vitro—Human renal glomerular endothelial cells105In vivo—Diabetic rats |
| miR-532 | Pathology—Myocardial Infarction | prss23 |
|
| Let-7 family | Pathology—Neointima formation | ? |
|
| Pathology—Inflammation | ? | In vitro—HUVECs and human retinal endothelial cells95 | |
| Pathology—Diabetes-related kidney fibrosis | ? | ||
| Differential effects on EndMT | |||
| miR-20a | Development—Outflow tract cushion development | Vegfa |
|
| Pathology—Molecular characterization | TGFBR1/2, SARA | In vitro—HUVECs124 | |
| miR-126 | Pathology—Tumour progression | ? |
|
| Pathology—Neointimal hyperplasia | PIK3R2 | In vitro—Rat bone marrow-derived endothelial progenitor cells119 | |
| Development/Pathology—Pulmonary vasculature remodelling | ? |
|
|
| miR-155 | Pathology—Molecular characterization | SKI | In vitro—Human coronary artery endothelial cells121 |
| Development—Molecular characterization | ? | In vitro—Mouse embryonic endothelial cells122 | |
| miR-200a | Pathology—Cardiac fibrosis | GRB2 | In vitro—Human aortic endothelial cells110 |
| miR-200b | Pathology—Diabetic retinopathy | ? |
|
| Pathology—Diabetic cardiomyopathy | ? |
|
|
| Pathology/Physiology—Angiogenesis | Ets1 | In vitro—Human microvascular endothelial cells113 | |
| miR-200c | Pathology—Aneurysm formation | ? |
|
| miR-200 family | Development—Vasculogenesis | Flk1, Ets1 | In vitro—Mouse embryonic stem cells116 |
| Development—Differentiation | ? | In vitro—Human embryonic stem cells117 | |
List of miRNAs regulating EndMT categorized into their inducing, inhibiting or differential role on EndMT. Question mark refers to unknown targets in this context.
AdipoR2, adiponectin receptor 2; ATG5, autophagy related 5; BMPR2, bone morphogenetic protein receptor 2; CDH2, cadherin-2; CTGF, connective tissue growth factor; DPP-4, dipeptidyl peptidase-4; Elk1, ETS transcription factor; Ets1, protein c-ets-1; Flk1, foetal liver kinase 1; GRB2, growth factor receptor-bound protein 2; Has2, hyaluronic acid synthase 2; Icat, beta-catenin-interacting protein; KLF4, kruppel-like factor 4; MAP3K3/7, mitogen-activated protein kinase kinase kinase 3/7; MAPK7, mitogen-activated protein kinase 7; MEF2D, myocyte-specific enhancer factor 2D; MTDH, metadherin; NF-κB, nuclear factor kappa-light-chain-enhancer of activated B cells; Nrp2, neurophilin 2; PIK3R2, PI3K regulatory subunit p85 beta; PlxnA2, plexin A2; PlxnD1, plexin D1; PPARγ, perixome proliferator-activated receptor γ; RAC1, ras-related C3 botulinum toxin substrate 1; ROCK, rho-associated, coiled-coil-containing protein kinase; SARA, Smad anchor for receptor activation; SKI, SKI proto-oncogene; SMAD, SMAD family member; TGFB, transforming growth factor beta; TGFBR, transforming growth factor beta receptor; Tmem2, transmembrane protein 2; Twist1, twist-related protein 1; VAV3, guanine nucleotide exchange factor VAV3; VEGF, vascular endothelial growth factor; ZEB2, zinc finger E-box-binding homeobox 2.
10. lncRNAs promoting EndMT
Among all the lncRNAs identified, only a few lncRNAs have been implicated in the regulation of EndMT hitherto. Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) is a lncRNA extensively associated with cancer metastasis and recently described as an EndMT inducer. MALAT1 competitively binds to miR-145, a known miRNA that inhibits TGF-β1-induced EndMT by directly targeting TGF-βR2 and SMAD3. Acting as a miRNA sponge, MALAT1 blocks the inhibitory activity of the miR-145 and promotes EndMT.125 Another lncRNA identified as an EndMT promoter is GATA6-AS. The long non-coding antisense transcript of GATA6 (GATA6-AS) interacts with the epigenetic regulator LOXL2 to regulate endothelial gene expression via changes in H3K4me3 methylation, including genes encoding for periostin and cyclooxygenase-2. GATA6-AS expression is induced in endothelial cells by hypoxia. Inhibition of GATA6-AS blocks TGF-β2-induced EndMT in vitro and promotes blood vessel formation in mice.126 A third lncRNA that promotes EndMT is PVT1. PVT1 associates with tumour cell proliferation, invasion, and metastasis in different cancer models. In prostate cancer, PVT1 promotes cancer invasion and metastasis in part by the induction of EndMT. PVT1 promotes EndMT by acting as a sponge for miRNA-186-5p and positively regulating TWIST1.127
11. lncRNAs inhibiting EndMT
H19 prevents glucose-induced TGF-β1 expression and therefore, EndMT in diabetic retinopathy models. H19 controls TGF-β1 mRNA and protein levels through a SMAD-independent MAPK–ERK1/2 pathway, and this regulation is also independent of the actions of miR-200b, a known miRNA (see above) that interacts with H19.128 Besides the lncRNAs mentioned above, lncRNA n339260, although not directly related to EndMT, controls the expression of VE-Cadherin during the process of vascular mimicry in a human hepatocellular carcinoma.129 Maintenance or reduction of VE-Cadherin in endothelial cells is a critical step in EndMT and therefore, future studies are needed to understand the potential role of lncRNA n339260 in EndMT. Furthermore, this study provided a number of candidate miRNAs that may be regulated by lncRNA n339260 including miR‐31‐3p, miR‐30e‐5p, miR‐519c‐5p, miR‐520c‐5p, miR‐29b‐1‐5p, and miR‐92a‐1‐5p. These will also require further testing to determine their potential role in the EndMT process. A summary of the currently known lncRNAs that modulate EndMT is provided in Figure 6 and Table 2. We expect this list to grow significantly in the coming years.
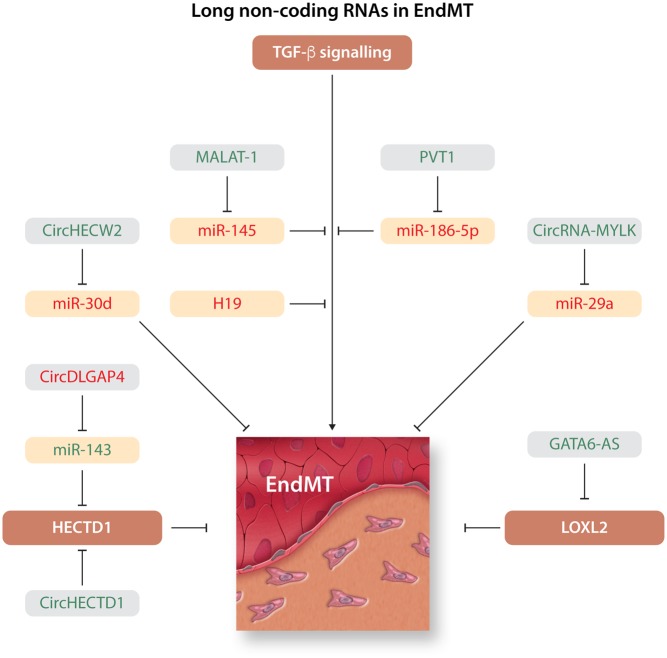
Figure 6.
Long non-coding RNA in EndMT. Overview of long non-coding RNAs (lncRNAs) that induce or inhibit EndMT. lncRNA inducers and inhibitors of EndMT are depicted in green and red, respectively.
Table 2.
Known lncRNAs involved in EndMT
| lncRNAs | Biological context | Targets | Experimental model |
|---|---|---|---|
| Inducing EndMT | |||
| GATA6-AS | Development—Angiogenesis | LOXL2 | In vitro—HUVECs126 |
| MALAT1 | Pathology—Neointimal hyperplasia | miR-145 that targets TGFBR2 and SMAD3 | In vitro—Endothelial progenitor cells125 |
| PVT1 | Pathology—Prostate cancer | miR-186-5p that targets Twist1 | In vitro—Prostate cancer cells127 |
| Inhibiting EndMT | |||
| H19 | Pathology—Diabetic retinopathy | TGF-β1 through control of MAPK-ERK1/2 pathway | In vitro—Human retinal endothelial cells128 |
| In vivo—Diabetic retinopathy mouse model and human samples | |||
List of lncRNAs currently known to regulate EndMT categorized into their inducing or inhibiting role on EndMT.
ERK, extracellular signal-regulated kinase; LOXL2, lysyl oxidase homolog 2; MAPK, mitogen-activated protein kinase; SMAD, SMAD family member; TGF-β1, transforming growth factor beta 1; TGFTGFBR, transforming growth factor beta receptor; Twist1, twist-related protein 1.
12. circRNAs promoting EndMT
circRNAs that are known to promote EndMT include CircHECW2 and CircRNA-MYLK. CircHECW2 promotes EndMT by acting as a sponge for miR-30d and culminating in the increased expression of ATG5, which promotes the activation of the Notch1 pathway to induce pathologic EndMT.130 In the case of CircRNA-MYLK, EndMT is promoted by directly binding and inhibiting miR-29a.131 This promotes EndMT by activating the TGF-β, NF-kB, and β-catenin pathways.132,133 Besides acting as miRNA sponges, circRNAs can control EndMT by directly regulating the expression of genes. In this regard, circHECTD1 regulates the expression of its host gene E6-AP C-terminal domain E3 ubiquitin protein ligase 1 (HECTD1), which is implicated in the maintenance of endothelial fate and controlled by Wnt signalling through APC-Axin interactions.134 Increased expression of circHECTD1 induces EndMT and its derivative fibroblast-like cells contribute to pulmonary fibrosis.135
13. circRNAs inhibiting EndMT
Among the circRNAs, the only currently identified EndMT inhibitor is circRNA DLGAP4. DLGAP4 functions as an endogenous miR-143 sponge to allow for the expression of HECTD1, as mentioned above. circRNA DLGAP4 inhibits EndMT and promotes maintenance of endothelial integrity and in the case of cerebral ischaemia it maintains the integrity of the blood–brain barrier.136 In addition, circRNAs chr5:90817794|90827570, chr8:71336875|71337745, and chr6:22033342|22038870, were found to be increased in a circRNA screening performed on an EndMT assay,137 although their specific functions remain unknown. circRNAs that are currently known to modulate EndMT are summarized in Figure 6 and Table 3.
Table 3.
Known circRNAs involved in EndMT
| circRNAs | Biological context | Targets | Experimental model |
|---|---|---|---|
| Inducing EndMT | |||
| CircHECW2 | Pathology—Neuroinflammatory disorders | miR-30d that targets ATG5 | In vitro—Human brain microvascular endothelial cells130 |
| In vivo—Mouse brain | |||
| CircRNA-MYLK | Pathology—Bladder carcinoma | miR-29a that targets VEGFA | In vitro—Human bladder carcinoma cells131 |
| In vivo—Mouse xenografts | |||
| CircHECTD1 | Pathology—Pulmonary disease | HECTD1 | In vitro—Mouse microvascular lung cells135 |
| In vivo—Lung silicosis in mouse model and human samples | |||
| Inhibiting EndMT | |||
| CircDLGAP4 | Pathology—Ischaemic stroke | miR-143 that targets HECTD1 | In vivo—Mouse brain136 |
List of circRNAs regulating EndMT categorized into their inducing or inhibiting role on EndMT.
ATG5, autophagy related 5; HECTD, HECT domain E3 ubiquitin protein ligase; VEGF, vascular endothelial growth factor.
The identification of the functional relevance of lncRNAs and circRNAs in EndMT is a fast growing field of research. It is expected that our understanding of the regulatory roles of lncRNAs and circRNAs in EndMT will increase accordingly in the coming years and so our understanding of physiologic and pathologic EndMT.
14. Future perspectives: targeting non-coding RNAs to modulate EndMT
As this review has highlighted, non-coding RNAs are key players in the control and modulation of EndMT (Figure 5 and 6). However, our overall knowledge of the role of non-coding RNAs in EndMT is in its infancy. Despite this, taking the data reviewed in this article as a whole, we are already able to draw several important conclusions about this field. Firstly, it is immediately clear that some non-coding RNAs inhibit EndMT such as miR-29, whereas others facilitate or induce EndMT such as miR-21, GATA6-AS, and HECTD1 (Tables 1, 2,and3). Second, most of these non-coding RNAs have only been described in a single context and we do not know if their roles may differ in other contexts. In addition, different markers for EndMT are used in different studies which might not give us the full pictures of how a certain non-coding RNA affects EndMT. The difference in function of these non-coding RNAs between development and various pathological states might also be different and needs to be established. Third, non-coding RNAs also interact with other epigenetic modulators such as DNA methylation and histone modifications to affect EndMT.27 These other epigenetic modulators affecting EndMT are not the focus of this review but are described elsewhere.27 Fourth, it is thought that EMT and EndMT occur via similar mechanisms. Importantly, the miRNA signatures between human corneal endothelium tissues and corneal epithelium tissues did not differ much even though the gene signatures were very different.138 This might suggest that non-coding RNAs function in similar ways in both epithelial and endothelial cells. Of course, we have to establish whether this is similar for other tissues as well. As a further point of consideration, it has been shown that the miRNA signatures of cultured human corneal endothelial cells were very different than those from fresh corneal endothelium tissue.138 The same is true for cultured HUVECs when comparing them to freshly isolate tissue-derived umbilical cord human vascular endothelial cells.139 Furthermore, the overall miRNA expression also decreased significantly in cultured HUVECs when compared to tissue-derived endothelial cells, and miRNA content appears to be lost during passaging of HUVECs.139 The most downregulated miRNA during culturing is miR-126 and the most upregulated miRNA is miR-21.139 Also during culturing of HUVECs, the pro-fibrotic miRNAs miR-21-5p and miR-31-5p were upregulated whereas the anti-fibrotic miRNAs miR-126-3p, Let-7 family, and miR-29 family were downregulated, suggesting a transition towards a mesenchymal cell type in aging cells.139 As a whole this indicates that the miRNA profile is highly adaptable and that cultured cells may not always represent the best model system for studying miRNA function—proper in vivo validation will remain as the gold standard.
It is clear in this emerging field that non-coding RNAs appear to be important players during EndMT. Indeed, as we have reviewed, non-coding RNAs interact with EndMT transcription factors, plus endothelial and mesenchymal genes and signalling pathways, ascribing them a likely pivotal role in regulating this process. As such, they represent promising future clinical targets for modulating EndMT and its accompanied pathologies.140,141 As a whole, the regulation of EndMT via non-coding RNAs represents a challenging but promising field with many potential opportunities for future therapeutic clinical translation.
Conflict of interest: none declared.
Funding
G.K. received support from the Netherlands Organization for Scientific Research/Netherlands Organization for Health Research and Development Innovational Research Incentive #917.16.446. G.d.M.-N.’s research is supported by a Future Leader Fellowship (102036) from the National Heart Foundation of Australia, a Discovery Project (DP190101475) from the Australian Research Council, and start-up funds from Monash University. M.H. received support from a Graduate School of Medical Sciences (GSMS) PhD scholarship, University of Groningen. J.K. acknowledges research support from the National Institutes of Health (R01HL130423 and R01HL135093).
References
- 1. Aird WC. Phenotypic heterogeneity of the endothelium: I. Structure, function, and mechanisms. Circ Res 2007;100:158–173. [DOI] [PubMed] [Google Scholar]
- 2. Aird WC. Phenotypic heterogeneity of the endothelium: II. Representative vascular beds. Circ Res 2007;100:174–190. [DOI] [PubMed] [Google Scholar]
- 3. Markwald RR, Fitzharris TP, Manasek FJ.. Structural development of endocardial cushions. Am J Anat 1977;148:85–119. [DOI] [PubMed] [Google Scholar]
- 4. Markwald RR, Fitzharris TP, Smith WN.. Sturctural analysis of endocardial cytodifferentiation. Dev Biol 1975;42:160–180. [DOI] [PubMed] [Google Scholar]
- 5. Ma L, Lu MF, Schwartz RJ, Martin JF.. Bmp2 is essential for cardiac cushion epithelial-mesenchymal transition and myocardial patterning. Development 2005;132:5601–5611. [DOI] [PubMed] [Google Scholar]
- 6. Yamada M, Revelli JP, Eichele G, Barron M, Schwartz RJ.. Expression of chick Tbx-2, Tbx-3, and Tbx-5 genes during early heart development: evidence for BMP2 induction of Tbx2. Dev Biol 2000;228:95–105. [DOI] [PubMed] [Google Scholar]
- 7. Arciniegas E, Neves CY, Carrillo LM, Zambrano EA, Ramirez R.. Endothelial-mesenchymal transition occurs during embryonic pulmonary artery development. Endothelium 2005;12:193–200. [DOI] [PubMed] [Google Scholar]
- 8. DeRuiter MC, Poelmann RE, VanMunsteren JC, Mironov V, Markwald RR, Gittenberger-de Groot AC.. Embryonic endothelial cells transdifferentiate into mesenchymal cells expressing smooth muscle actins in vivo and in vitro. Circ Res 1997;80:444–451. [DOI] [PubMed] [Google Scholar]
- 9. Welch-Reardon KM, Wu N, Hughes CC.. A role for partial endothelial-mesenchymal transitions in angiogenesis? Arterioscler Thromb Vasc Biol 2015;35:303–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Del Monte-Nieto G, Ramialison M, Adam AAS, Wu B, Aharonov A, D'Uva G, Bourke LM, Pitulescu ME, Chen H, de la Pompa JL, Shou W, Adams RH, Harten SK, Tzahor E, Zhou B, Harvey RP.. Control of cardiac jelly dynamics by NOTCH1 and NRG1 defines the building plan for trabeculation. Nature 2018;557:439–445. [DOI] [PubMed] [Google Scholar]
- 11. Ranchoux B, Antigny F, Rucker-Martin C, Hautefort A, Péchoux C, Bogaard HJ, Dorfmüller P, Remy S, Lecerf F, Planté S, Chat S, Fadel E, Houssaini A, Anegon I, Adnot S, Simonneau G, Humbert M, Cohen-Kaminsky S, Perros F.. Endothelial-to-mesenchymal transition in pulmonary hypertension. Circulation 2015;131:1006–1018. [DOI] [PubMed] [Google Scholar]
- 12. Chen P-Y, Qin L, Baeyens N, Li G, Afolabi T, Budatha M, Tellides G, Schwartz MA, Simons M.. Endothelial-to-mesenchymal transition drives atherosclerosis progression. J Clin Invest 2015;125:4514–4528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Souilhol C, Harmsen MC, Evans PC, Krenning G.. Endothelial-mesenchymal transition in atherosclerosis. Cardiovasc Res 2018;114:565–577. [DOI] [PubMed] [Google Scholar]
- 14. Maddaluno L, Rudini N, Cuttano R, Bravi L, Giampietro C, Corada M, Ferrarini L, Orsenigo F, Papa E, Boulday G, Tournier-Lasserve E, Chapon F, Richichi C, Retta SF, Lampugnani MG, Dejana E.. EndMT contributes to the onset and progression of cerebral cavernous malformations. Nature 2013;498:492–496. [DOI] [PubMed] [Google Scholar]
- 15. Hashimoto N, Phan SH, Imaizumi K, Matsuo M, Nakashima H, Kawabe T, Shimokata K, Hasegawa Y.. Endothelial-mesenchymal transition in bleomycin-induced pulmonary fibrosis. Am J Respir Cell Mol Biol 2010;43:161–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zeisberg EM, Potenta SE, Sugimoto H, Zeisberg M, Kalluri R.. Fibroblasts in kidney fibrosis emerge via endothelial-to-mesenchymal transition. J Am Soc Nephrol 2008;19:2282–2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zeisberg EM, Tarnavski O, Zeisberg M, Dorfman AL, McMullen JR, Gustafsson E, Chandraker A, Yuan X, Pu WT, Roberts AB, Neilson EG, Sayegh MH, Izumo S, Kalluri R.. Endothelial-to-mesenchymal transition contributes to cardiac fibrosis. Nat Med 2007;13:952–961. [DOI] [PubMed] [Google Scholar]
- 18. Zeisberg EM, Potenta S, Xie L, Zeisberg M, Kalluri R.. Discovery of endothelial to mesenchymal transition as a source for carcinoma-associated fibroblasts. Cancer Res 2007;67:10123–10128. [DOI] [PubMed] [Google Scholar]
- 19. Hong L, Du X, Li W, Mao Y, Sun L, Li X.. EndMT: a promising and controversial field. Eur J Cell Biol 2018;97:493–500. [DOI] [PubMed] [Google Scholar]
- 20. Moonen J-RAJ, Lee ES, Schmidt M, Maleszewska M, Koerts JA, Brouwer LA, van Kooten TG, van Luyn MJA, Zeebregts CJ, Krenning G, Harmsen MC.. Endothelial-to-mesenchymal transition contributes to fibro-proliferative vascular disease and is modulated by fluid shear stress. Cardiovasc Res 2015;108:377–386. [DOI] [PubMed] [Google Scholar]
- 21. Widyantoro B, Emoto N, Nakayama K, Anggrahini DW, Adiarto S, Iwasa N, Yagi K, Miyagawa K, Rikitake Y, Suzuki T, Kisanuki YY, Yanagisawa M, Hirata K-I.. Endothelial cell-derived endothelin-1 promotes cardiac fibrosis in diabetic hearts through stimulation of endothelial-to-mesenchymal transition. Circulation 2010;121:2407–2418. [DOI] [PubMed] [Google Scholar]
- 22. Maleszewska M, Gjaltema RA, Krenning G, Harmsen MC.. Enhancer of zeste homolog-2 (EZH2) methyltransferase regulates transgelin/smooth muscle-22alpha expression in endothelial cells in response to interleukin-1beta and transforming growth factor-beta2. Cell Signal 2015;27:1589–1596. [DOI] [PubMed] [Google Scholar]
- 23. Xu X, Tan X, Hulshoff MS, Wilhelmi T, Zeisberg M, Zeisberg EM.. Hypoxia-induced endothelial-mesenchymal transition is associated with RASAL1 promoter hypermethylation in human coronary endothelial cells. FEBS Lett 2016;590:1222–1233. [DOI] [PubMed] [Google Scholar]
- 24. Mahler GJ, Farrar EJ, Butcher JT.. Inflammatory cytokines promote mesenchymal transformation in embryonic and adult valve endothelial cells. Arterioscler Thromb Vasc Biol 2013;33:121–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. van Meeteren LA, ten Dijke P.. Regulation of endothelial cell plasticity by TGF-beta. Cell Tissue Res 2012;347:177–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Xu S, Kamato D, Little PJ, Nakagawa S, Pelisek J, Jin ZG.. Targeting epigenetics and non-coding RNAs in atherosclerosis: from mechanisms to therapeutics. Pharmacol Ther 2019;196:15–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hulshoff MS, Xu X, Krenning G, Zeisberg EM.. Epigenetic regulation of endothelial-to-mesenchymal transition in chronic heart disease. Arterioscler Thromb Vasc Biol 2018;38:1986–1996. [DOI] [PubMed] [Google Scholar]
- 28. Kim J. MicroRNAs as critical regulators of the endothelial to mesenchymal transition in vascular biology. BMB Rep 2018;51:65–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kovacic JC, Dimmeler S, Harvey RP, Finkel T, Aikawa E, Krenning G, Baker AH.. Endothelial to mesenchymal transition in cardiovascular disease: JACC State-of-the-Art Review. J Am Coll Cardiol 2019;73:190–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Garside VC, Chang AC, Karsan A, Hoodless PA.. Co-ordinating Notch, BMP, and TGF-beta signaling during heart valve development. Cell Mol Life Sci 2013;70:2899–2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yamagishi T, Ando K, Nakamura H.. Roles of TGFbeta and BMP during valvulo-septal endocardial cushion formation. Anat Sci Int 2009;84:77–87. [DOI] [PubMed] [Google Scholar]
- 32. Derynck R, Zhang YE.. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature 2003;425:577–584. [DOI] [PubMed] [Google Scholar]
- 33. Zhang YE. Non-Smad pathways in TGF-beta signaling. Cell Res 2009;19:128–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hata A, Chen YG.. TGF-beta signaling from receptors to Smads. Cold Spring Harb Perspect Biol 2016;8:a022061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Peinado H, Olmeda D, Cano A.. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer 2007;7:415–428. [DOI] [PubMed] [Google Scholar]
- 36. Lander R, Nasr T, Ochoa SD, Nordin K, Prasad MS, Labonne C.. Interactions between Twist and other core epithelial-mesenchymal transition factors are controlled by GSK3-mediated phosphorylation. Nat Commun 2013;4:1542.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Peiro S, Escriva M, Puig I, Barbera MJ, Dave N, Herranz N.. Snail1 transcriptional repressor binds to its own promoter and controls its expression. Nucleic Acids Res 2006;34:2077–2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Takkunen M, Grenman R, Hukkanen M, Korhonen M, Garcia de Herreros A, Virtanen I.. Snail-dependent and -independent epithelial-mesenchymal transition in oral squamous carcinoma cells. J Histochem Cytochem 2006;54:1263–1275. [DOI] [PubMed] [Google Scholar]
- 39. Wels C, Joshi S, Koefinger P, Bergler H, Schaider H.. Transcriptional activation of ZEB1 by Slug leads to cooperative regulation of the epithelial-mesenchymal transition-like phenotype in melanoma. J Invest Dermatol 2011;131:1877–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dufton NP, Peghaire CR, Osuna-Almagro L, Raimondi C, Kalna V, Chuahan A, Webb G, Yang Y, Birdsey GM, Lalor P, Mason JC, Adams DH, Randi AM.. Dynamic regulation of canonical TGFbeta signalling by endothelial transcription factor ERG protects from liver fibrogenesis. Nat Commun 2017;8:895.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rivera-Feliciano J, Lee K-H, Kong SW, Rajagopal S, Ma Q, Springer Z, Izumo S, Tabin CJ, Pu WT.. Development of heart valves requires Gata4 expression in endothelial-derived cells. Development 2006;133:3607–3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rivera-Feliciano J, Tabin CJ.. Bmp2 instructs cardiac progenitors to form the heart-valve-inducing field. Dev Biol 2006;295:580–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. McCulley DJ, Kang JO, Martin JF, Black BL.. BMP4 is required in the anterior heart field and its derivatives for endocardial cushion remodeling, outflow tract septation, and semilunar valve development. Dev Dyn 2008;237:3200–3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Xu X, Tan X, Tampe B, Nyamsuren G, Liu X, Maier LS, Sossalla S, Kalluri R, Zeisberg M, Hasenfuss G, Zeisberg EM.. Epigenetic balance of aberrant Rasal1 promoter methylation and hydroxymethylation regulates cardiac fibrosis. Cardiovasc Res 2015;105:279–291. [DOI] [PubMed] [Google Scholar]
- 45. Timmerman LA, Grego-Bessa J, Raya A, Bertran E, Perez-Pomares JM, Diez J, et al. Notch promotes epithelial-mesenchymal transition during cardiac development and oncogenic transformation. Genes Dev 2004;18:99–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Niessen K, Fu Y, Chang L, Hoodless PA, McFadden D, Karsan A.. Slug is a direct Notch target required for initiation of cardiac cushion cellularization. J Cell Biol 2008;182:315–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Luna-Zurita L, Prados B, Grego-Bessa J, Luxán G, del Monte G, Benguría A, Adams RH, Pérez-Pomares JM, de la Pompa JL.. Integration of a Notch-dependent mesenchymal gene program and Bmp2-driven cell invasiveness regulates murine cardiac valve formation. J Clin Invest 2010;120:3493–3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hurlstone AFL, Haramis A-PG, Wienholds E, Begthel H, Korving J, Van Eeden F, Cuppen E, Zivkovic D, Plasterk RHA, Clevers H.. The Wnt/beta-catenin pathway regulates cardiac valve formation. Nature 2003;425:633–637. [DOI] [PubMed] [Google Scholar]
- 49. Liebner S, Cattelino A, Gallini R, Rudini N, Iurlaro M, Piccolo S, Dejana E.. Beta-catenin is required for endothelial-mesenchymal transformation during heart cushion development in the mouse. J Cell Biol 2004;166:359–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gessert S, Kuhl M.. The multiple phases and faces of wnt signaling during cardiac differentiation and development. Circ Res 2010;107:186–199. [DOI] [PubMed] [Google Scholar]
- 51. Aisagbonhi O, Rai M, Ryzhov S, Atria N, Feoktistov I, Hatzopoulos AK.. Experimental myocardial infarction triggers canonical Wnt signaling and endothelial-to-mesenchymal transition. Dis Models Mech 2011;4:469–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Thornton TM, Pedraza-Alva G, Deng B, Wood CD, Aronshtam A, Clements JL, Sabio G, Davis RJ, Matthews DE, Doble B, Rincon M.. Phosphorylation by p38 MAPK as an alternative pathway for GSK3beta inactivation. Science 2008;320:667–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cheng SL, Shao JS, Behrmann A, Krchma K, Towler DA.. Dkk1 and MSX2-Wnt7b signaling reciprocally regulate the endothelial-mesenchymal transition in aortic endothelial cells. Arterioscler Thromb Vasc Biol 2013;33:1679–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Maleszewska M, Moonen JR, Huijkman N, van de Sluis B, Krenning G, Harmsen MC.. IL-1beta and TGFbeta2 synergistically induce endothelial to mesenchymal transition in an NFkappaB-dependent manner. Immunobiology 2013;218:443–454. [DOI] [PubMed] [Google Scholar]
- 55. Wu Y, Deng J, Rychahou PG, Qiu S, Evers BM, Zhou BP.. Stabilization of snail by NF-kappaB is required for inflammation-induced cell migration and invasion. Cancer Cell 2009;15:416–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Julien S, Puig I, Caretti E, Bonaventure J, Nelles L, van Roy F, Dargemont C, de Herreros AG, Bellacosa A, Larue L.. Activation of NF-kappa B by Akt upregulates Snail expression and induces epithelium mesenchyme transition. Oncogene 2007;26:7445–7456. [DOI] [PubMed] [Google Scholar]
- 57. Johnson DE, Williams LT.. Structural and functional diversity in the FGF receptor multigene family. Adv Cancer Res 1993;60:1–41. [DOI] [PubMed] [Google Scholar]
- 58. Chen PY, Qin L, Tellides G, Simons M.. Fibroblast growth factor receptor 1 is a key inhibitor of TGFbeta signaling in the endothelium. Sci Signal 2014;7:ra90.. [DOI] [PubMed] [Google Scholar]
- 59. Mahmoud MM, Kim HR, Xing R, Hsiao S, Mammoto A, Chen J, Serbanovic-Canic J, Feng S, Bowden NP, Maguire R, Ariaans M, Francis SE, Weinberg PD, van der Heiden K, Jones EA, Chico TJA, Ridger V, Evans PC.. TWIST1 integrates endothelial responses to flow in vascular dysfunction and atherosclerosis. Circ Res 2016;119:450–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mahmoud MM, Serbanovic-Canic J, Feng S, Souilhol C, Xing R, Hsiao S, Mammoto A, Chen J, Ariaans M, Francis SE, Van der Heiden K, Ridger V, Evans PC.. Shear stress induces endothelial-to-mesenchymal transition via the transcription factor Snail. Sci Rep 2017;7:3375.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Semenza GL. HIF-1: mediator of physiological and pathophysiological responses to hypoxia. J Appl Physiol 2000;88:1474–1480. [DOI] [PubMed] [Google Scholar]
- 62. Xu X, Tan X, Tampe B, Sanchez E, Zeisberg M, Zeisberg EM.. Snail is a direct target of Hypoxia-inducible Factor 1alpha (HIF1alpha) in hypoxia-induced endothelial to mesenchymal transition of human coronary endothelial cells. J Biol Chem 2015;290:16653–16664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yang M-H, Wu M-Z, Chiou S-H, Chen P-M, Chang S-Y, Liu C-J, Teng S-C, Wu K-J.. Direct regulation of TWIST by HIF-1alpha promotes metastasis. Nat Cell Biol 2008;10:295–305. [DOI] [PubMed] [Google Scholar]
- 64. Evrard SM, Lecce L, Michelis KC, Nomura-Kitabayashi A, Pandey G, Purushothaman K-R, d’Escamard V, Li JR, Hadri L, Fujitani K, Moreno PR, Benard L, Rimmele P, Cohain A, Mecham B, Randolph GJ, Nabel EG, Hajjar R, Fuster V, Boehm M, Kovacic JC.. Endothelial to mesenchymal transition is common in atherosclerotic lesions and is associated with plaque instability. Nat Commun 2016;7:11853.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Dong W-Q, Chao M, Lu Q-h, Chai W-L, Zhang W, Chen X-y, Liang E-S, Wang L-B, Tian H-L, Chen Y-G, Zhang M-X.. Prohibitin overexpression improves myocardial function in diabetic cardiomyopathy. Oncotarget 2016;7:66–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Gu S, Kay MA.. How do miRNAs mediate translational repression? Silence 2010;1:11.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kung JT, Colognori D, Lee JT.. Long noncoding RNAs: past, present, and future. Genetics 2013;193:651–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 2015;22:256–264. [DOI] [PubMed] [Google Scholar]
- 69. Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J.. Natural RNA circles function as efficient microRNA sponges. Nature 2013;495:384–388. [DOI] [PubMed] [Google Scholar]
- 70. Zhang Y, Zhang X-O, Chen T, Xiang J-F, Yin Q-F, Xing Y-H, Zhu S, Yang L, Chen L-L.. Circular intronic long noncoding RNAs. Mol Cell 2013;51:792–806. [DOI] [PubMed] [Google Scholar]
- 71. Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S.. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell 2014;56:55–66. [DOI] [PubMed] [Google Scholar]
- 72. Hentze MW, Preiss T.. Circular RNAs: splicing’s enigma variations. EMBO J 2013;32:923–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wilusz JE, Sharp PA.. Molecular biology. A circuitous route to noncoding RNA. Science 2013;340:440–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ghosh AK, Nagpal V, Covington JW, Michaels MA, Vaughan DE.. Molecular basis of cardiac endothelial-to-mesenchymal transition (EndMT): differential expression of microRNAs during EndMT. Cell Signal 2012;24:1031–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Suzuki HI, Katsura A, Mihira H, Horie M, Saito A, Miyazono K.. Regulation of TGF-beta-mediated endothelial-mesenchymal transition by microRNA-27. J Biochem 2017;161:417–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Guo Y, Li P, Bledsoe G, Yang ZR, Chao L, Chao J.. Kallistatin inhibits TGF-beta-induced endothelial-mesenchymal transition by differential regulation of microRNA-21 and eNOS expression. Exp Cell Res 2015;337:103–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Kumarswamy R, Volkmann I, Jazbutyte V, Dangwal S, Park DH, Thum T.. Transforming growth factor-beta-induced endothelial-to-mesenchymal transition is partly mediated by microRNA-21. Arterioscler Thromb Vasc Biol 2012;32:361–369. [DOI] [PubMed] [Google Scholar]
- 78. Kwon O-S, Kim K-T, Lee E, Kim M, Choi S-H, Li H, Fornace AJ, Cho J-H, Lee Y-S, Lee J-S, Lee Y-J, Cha H-J.. Induction of MiR-21 by stereotactic body radiotherapy contributes to the pulmonary fibrotic response. PLoS One 2016;11:e0154942.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Katsura A, Suzuki HI, Ueno T, Mihira H, Yamazaki T, Yasuda T, Watabe T, Mano H, Yamada Y, Miyazono K.. MicroRNA-31 is a positive modulator of endothelial-mesenchymal transition and associated secretory phenotype induced by TGF-beta. Genes Cells 2016;21:99–116. [DOI] [PubMed] [Google Scholar]
- 80. Colangelo T, Fucci A, Votino C, Sabatino L, Pancione M, Laudanna C, Binaschi M, Bigioni M, Alberto Maggi C, Parente D, Forte N, Colantuoni V.. MicroRNA-130b promotes tumor development and is associated with poor prognosis in colorectal cancer. Neoplasia 2013;15:1086–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Li L, Kim IK, Chiasson V, Chatterjee P, Gupta S.. NF-kappaB mediated miR-130a modulation in lung microvascular cell remodeling: implication in pulmonary hypertension. Exp Cell Res 2017;359:235–242. [DOI] [PubMed] [Google Scholar]
- 82. Jiang L, Hao C, Li Z, Zhang P, Wang S, Yang S, Wei F, Zhang J.. miR-449a induces EndMT, promotes the development of atherosclerosis by targeting the interaction between AdipoR2 and E-cadherin in Lipid Rafts. Biomed Pharmacother 2019;109:2293–2304. [DOI] [PubMed] [Google Scholar]
- 83. Vanchin B, Offringa E, Friedrich J, Brinker MG, Kiers B, Pereira AC, et al. MicroRNA-374b induces endothelial-to-mesenchymal transition and early lesion formation through the inhibition of MAPK7 signaling. J Pathol 2019;247:456–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Chen Y, Yang Q, Zhan Y, Ke J, Lv P, Huang J.. The role of miR-328 in high glucose-induced endothelial-to-mesenchymal transition in human umbilical vein endothelial cells. Life Sci 2018;207:110–116. [DOI] [PubMed] [Google Scholar]
- 85. Chakraborty S, Zawieja DC, Davis MJ, Muthuchamy M.. MicroRNA signature of inflamed lymphatic endothelium and role of miR-9 in lymphangiogenesis and inflammation. Am J Physiol, Cell Physiol 2015;309:C680–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Yan X‐C, Cao J, Liang L, Wang L, Gao F, Yang Z‐Y, Duan J‐L, Chang T‐F, Deng S‐M, Liu Y, Dou G‐R, Zhang J, Zheng Q‐J, Zhang P, Han H.. miR-342-5p is a notch downstream molecule and regulates multiple angiogenic pathways including notch, vascular endothelial growth factor and transforming growth factor beta signaling. J Am Heart Assoc 2016;5: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Bonet F, Dueñas Á, López-Sánchez C, García-Martínez V, Aránega AE, Franco D.. MiR-23b and miR-199a impair epithelial-to-mesenchymal transition during atrioventricular endocardial cushion formation. Dev Dyn 2015;244:1259–1275. [DOI] [PubMed] [Google Scholar]
- 88. Lagendijk AK, Goumans MJ, Burkhard SB, Bakkers J.. MicroRNA-23 restricts cardiac valve formation by inhibiting Has2 and extracellular hyaluronic acid production. Circ Res 2011;109:649–657. [DOI] [PubMed] [Google Scholar]
- 89. Yi M, Liu B, Tang Y, Li F, Qin W, Yuan X.. Irradiated human umbilical vein endothelial cells undergo endothelial-mesenchymal transition via the Snail/miR-199a-5p axis to promote the differentiation of fibroblasts into myofibroblasts. Biomed Res Int 2018;2018:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Esmerats JF, Villa-Roel N, Kumar S, Gu L, Salim MT, Ohh M.. Disturbed flow increases UBE2C (Ubiquitin E2 Ligase C) via loss of miR-483-3p, inducing aortic valve calcification by the HIF-1alpha (Hypoxia-Inducible Factor-1alpha) pathway in endothelial cells. Arterioscler Thromb Vasc Biol 2019;ATVBAHA118312233.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. He M, Chen Z, Martin M, Zhang J, Sangwung P, Woo B, Tremoulet AH, Shimizu C, Jain MK, Burns JC, Shyy JY-J.. miR-483 targeting of CTGF suppresses endothelial-to-mesenchymal transition: therapeutic implications in Kawasaki disease. Circ Res 2017;120:354–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Song H, Wang Q, Wen J, Liu S, Gao X, Cheng J, Zhang D.. ACVR1, a therapeutic target of fibrodysplasia ossificans progressiva, is negatively regulated by miR-148a. Int J Mol Sci 2012;13:2063–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Miscianinov V, Martello A, Rose L, Parish E, Cathcart B, Mitić T, Gray GA, Meloni M, Al Haj Zen A, Caporali A.. MicroRNA-148b targets the TGF-beta pathway to regulate angiogenesis and endothelial-to-mesenchymal transition during skin wound healing. Mol Ther 2018;26:1996–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Chen P-Y, Qin L, Barnes C, Charisse K, Yi T, Zhang X, Ali R, Medina PP, Yu J, Slack FJ, Anderson DG, Kotelianski V, Wang F, Tellides G, Simons M.. FGF regulates TGF-beta signaling and endothelial-to-mesenchymal transition via control of let-7 miRNA expression. Cell Rep 2012;2:1684–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Terzuoli E, Nannelli G, Giachetti A, Morbidelli L, Ziche M, Donnini S.. Targeting endothelial-to-mesenchymal transition: the protective role of hydroxytyrosol sulfate metabolite. Eur J Nutr 2019; in press. Epub doi: https://doi.org/10.1007/s00394-019-01920-x. [DOI] [PubMed] [Google Scholar]
- 96. Takagi K, Yamakuchi M, Matsuyama T, Kondo K, Uchida A, Misono S, Hashiguchi T, Inoue H.. IL-13 enhances mesenchymal transition of pulmonary artery endothelial cells via down-regulation of miR-424/503 in vitro. Cell Signal 2018;42:270–280. [DOI] [PubMed] [Google Scholar]
- 97. Geng H, Guan J.. MiR-18a-5p inhibits endothelial-mesenchymal transition and cardiac fibrosis through the Notch2 pathway. Biochem Biophys Res Commun 2017;491:329–336. [DOI] [PubMed] [Google Scholar]
- 98. Bayoumi AS, Teoh J-P, Aonuma T, Yuan Z, Ruan X, Tang Y, Su H, Weintraub NL, Kim I-M.. MicroRNA-532 protects the heart in acute myocardial infarction, and represses prss23, a positive regulator of endothelial-to-mesenchymal transition. Cardiovasc Res 2017;113:1603–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Zhu K, Pan Q, Jia L-Q, Dai Z, Ke A-W, Zeng H-y, Tang Z-y, Fan J, Zhou J.. MiR-302c inhibits tumor growth of hepatocellular carcinoma by suppressing the endothelial-mesenchymal transition of endothelial cells. Sci Rep 2015;4:5524.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Zhu Y-T, Li F, Han B, Tighe S, Zhang S, Chen S-Y, Liu X, Tseng SCG.. Activation of RhoA-ROCK-BMP signaling reprograms adult human corneal endothelial cells. J Cell Biol 2014;206:799–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Lun W, Wu X, Deng Q, Zhi F.. MiR-218 regulates epithelial-mesenchymal transition and angiogenesis in colorectal cancer via targeting CTGF. Cancer Cell Int 2018;18:83.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Chen Y, Banda M, Speyer CL, Smith JS, Rabson AB, Gorski DH.. Regulation of the expression and activity of the antiangiogenic homeobox gene GAX/MEOX2 by ZEB2 and microRNA-221. Mol Cell Biol 2010;30:3902–3913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Chen S, Zhao G, Miao H, Tang R, Song Y, Hu Y, Wang Z, Hou Y.. MicroRNA-494 inhibits the growth and angiogenesis-regulating potential of mesenchymal stem cells. FEBS Lett 2015;589:710–717. [DOI] [PubMed] [Google Scholar]
- 104. Kim DY, Woo YM, Lee S, Oh S, Shin Y, Shin J-O, Park EY, Ko JY, Lee EJ, Bok J, Yoo KH, Park JH.. Impact of miR-192 and miR-194 on cyst enlargement through EMT in autosomal dominant polycystic kidney disease. FASEB J 2019;33:2870–2884. [DOI] [PubMed] [Google Scholar]
- 105. Liu F, Zhang S, Xu R, Gao S, Yin J.. Melatonin attenuates endothelial-to-mesenchymal transition of glomerular endothelial cells via regulating miR-497/ROCK in diabetic nephropathy. Kidney Blood Press Res 2018;43:1425–1436. [DOI] [PubMed] [Google Scholar]
- 106. Kanasaki K, Shi S, Kanasaki M, He J, Nagai T, Nakamura Y, Ishigaki Y, Kitada M, Srivastava SP, Koya D.. Linagliptin-mediated DPP-4 inhibition ameliorates kidney fibrosis in streptozotocin-induced diabetic mice by inhibiting endothelial-to-mesenchymal transition in a therapeutic regimen. Diabetes 2014;63:2120–2131. [DOI] [PubMed] [Google Scholar]
- 107. Shi S, Kanasaki K, Koya D.. Linagliptin but not Sitagliptin inhibited transforming growth factor-beta2-induced endothelial DPP-4 activity and the endothelial-mesenchymal transition. Biochem Biophys Res Commun 2016;471:184–190. [DOI] [PubMed] [Google Scholar]
- 108. Srivastava SP, Shi S, Kanasaki M, Nagai T, Kitada M, He J, Nakamura Y, Ishigaki Y, Kanasaki K, Koya D.. Effect of antifibrotic microRNAs crosstalk on the action of N-acetyl-seryl-aspartyl-lysyl-proline in diabetes-related kidney fibrosis. Sci Rep 2016;6:29884.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Nagai T, Kanasaki M, Srivastava SP, Nakamura Y, Ishigaki Y, Kitada M, Shi S, Kanasaki K, Koya D.. N-acetyl-seryl-aspartyl-lysyl-proline inhibits diabetes-associated kidney fibrosis and endothelial-mesenchymal transition. Biomed Res Int 2014;2014:1.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Zhang H, Hu J, Liu L.. MiR-200a modulates TGF-beta1-induced endothelial-to-mesenchymal shift via suppression of GRB2 in HAECs. Biomed Pharmacother 2017;95:215–222. [DOI] [PubMed] [Google Scholar]
- 111. Cao Y, Feng B, Chen S, Chu Y, Chakrabarti S.. Mechanisms of endothelial to mesenchymal transition in the retina in diabetes. Invest Ophthalmol Vis Sci 2014;55:7321–7331. [DOI] [PubMed] [Google Scholar]
- 112. Feng B, Cao Y, Chen S, Chu X, Chu Y, Chakrabarti S.. miR-200b mediates endothelial-to-mesenchymal transition in diabetic cardiomyopathy. Diabetes 2016;65:768–779. [DOI] [PubMed] [Google Scholar]
- 113. Chan YC, Khanna S, Roy S, Sen CK.. miR-200b targets Ets-1 and is down-regulated by hypoxia to induce angiogenic response of endothelial cells. J Biol Chem 2011;286:2047–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Maleki S, Cottrill KA, Poujade F-A, Bhattachariya A, Bergman O, Gådin JR, Simon N, Lundströmer K, Franco-Cereceda A, Björck HM, Chan SY, Eriksson P.. The mir-200 family regulates key pathogenic events in ascending aortas of individuals with bicuspid aortic valves. J Intern Med 2019;285:102–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Yang B, Zhou W, Jiao J, Nielsen JB, Mathis MR, Heydarpour M, Lettre G, Folkersen L, Prakash S, Schurmann C, Fritsche L, Farnum GA, Lin M, Othman M, Hornsby W, Driscoll A, Levasseur A, Thomas M, Farhat L, Dubé M-P, Isselbacher EM, Franco-Cereceda A, Guo D-C, Bottinger EP, Deeb GM, Booher A, Kheterpal S, Chen YE, Kang HM, Kitzman J, Cordell HJ, Keavney BD, Goodship JA, Ganesh SK, Abecasis G, Eagle KA, Boyle AP, Loos RJF, Eriksson P, Tardif J-C, Brummett CM, Milewicz DM, Body SC, Willer CJ.. Protein-altering and regulatory genetic variants near GATA4 implicated in bicuspid aortic valve. Nat Commun 2017;8:15481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Gill JG, Langer EM, Lindsley RC, Cai M, Murphy TL, Murphy KM.. Snail promotes the cell-autonomous generation of Flk1(+) endothelial cells through the repression of the microRNA-200 family. Stem Cells Dev 2012;21:167–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Kim Y, Kim N, Park SW, Kim H, Park HJ, Han YM.. Lineage-specific expression of miR-200 family in human embryonic stem cells during in vitro differentiation. Int J Stem Cells 2017;10:28–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Nagai N, Ohguchi H, Nakaki R, Matsumura Y, Kanki Y, Sakai J, Aburatani H, Minami T.. Downregulation of ERG and FLI1 expression in endothelial cells triggers endothelial-to-mesenchymal transition. PLoS Genet 2018;14:e1007826.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Zhang J, Zhang Z, Zhang DY, Zhu J, Zhang T, Wang C.. microRNA 126 inhibits the transition of endothelial progenitor cells to mesenchymal cells via the PIK3R2-PI3K/Akt signalling pathway. PLoS One 2013;8:e83294.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Xu Y-P, He Q, Shen Z, Shu X-L, Wang C-h, Zhu J-J, Shi L-P, Du L-Z.. MiR-126a-5p is involved in the hypoxia-induced endothelial-to-mesenchymal transition of neonatal pulmonary hypertension. Hypertens Res 2017;40:552–561. [DOI] [PubMed] [Google Scholar]
- 121. Wang J, He W, Xu X, Guo L, Zhang Y, Han S, et al. The mechanism of TGF-beta/miR-155/c-Ski regulates endothelial-mesenchymal transition in human coronary artery endothelial cells. Biosci Rep 2017;37, article identifier BSR20160603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Bijkerk R, G. de Bruin R, van Solingen C, M.G.J. Duijs J, Kobayashi K, P. van der Veer E, ten Dijke P, J. Rabelink T, J. Goumans M, J. van Zonneveld A.. MicroRNA-155 functions as a negative regulator of RhoA signaling in TGF-beta-induced endothelial to mesenchymal transition. MicroRNA 2012;1:2–10. [DOI] [PubMed] [Google Scholar]
- 123. Bai Y, Wang J, Morikawa Y, Bonilla-Claudio M, Klysik E, Martin JF.. Bmp signaling represses Vegfa to promote outflow tract cushion development. Development 2013;140:3395–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Correia AC, Moonen JR, Brinker MG, Krenning G.. FGF2 inhibits endothelial-mesenchymal transition through microRNA-20a-mediated repression of canonical TGF-beta signaling. J Cell Sci 2016;129:569–579. [DOI] [PubMed] [Google Scholar]
- 125. Xiang Y, Zhang Y, Tang Y, Li Q.. MALAT1 modulates TGF-beta1-Induced endothelial-to-mesenchymal transition through downregulation of miR-145. Cell Physiol Biochem 2017;42:357–372. [DOI] [PubMed] [Google Scholar]
- 126. Neumann P, Jaé N, Knau A, Glaser SF, Fouani Y, Rossbach O, Krüger M, John D, Bindereif A, Grote P, Boon RA, Dimmeler S.. The lncRNA GATA6-AS epigenetically regulates endothelial gene expression via interaction with LOXL2. Nat Commun 2018;9:237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Chang Z, Cui J, Song Y.. Long noncoding RNA PVT1 promotes EMT via mediating microRNA-186 targeting of Twist1 in prostate cancer. Gene 2018;654:36–42. [DOI] [PubMed] [Google Scholar]
- 128. Thomas AA, Biswas S, Feng B, Chen S, Gonder J, Chakrabarti S.. lncRNA H19 prevents endothelial-mesenchymal transition in diabetic retinopathy. Diabetologia 2019;62:517.. [DOI] [PubMed] [Google Scholar]
- 129. Zhao X, Sun B, Liu T, Shao B, Sun R, Zhu D, Zhang Y, Gu Q, Dong X, Liu F, Zhao N, Zhang D, Li Y, Meng J, Gong W, Zheng Y, Zheng X.. Long noncoding RNA n339260 promotes vasculogenic mimicry and cancer stem cell development in hepatocellular carcinoma. Cancer Sci 2018;109:3197–3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Yang L, Han B, Zhang Y, Bai Y, Chao J, Hu G, Yao H.. Engagement of circular RNA HECW2 in the nonautophagic role of ATG5 implicated in the endothelial-mesenchymal transition. Autophagy 2018;14:404–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Zhong Z, Huang M, Lv M, He Y, Duan C, Zhang L, Chen J.. Circular RNA MYLK as a competing endogenous RNA promotes bladder cancer progression through modulating VEGFA/VEGFR2 signaling pathway. Cancer Lett 2017;403:305–317. [DOI] [PubMed] [Google Scholar]
- 132. Luo M, Hou L, Li J, Shao S, Huang S, Meng D, Liu L, Feng L, Xia P, Qin T, Zhao X.. VEGF/NRP-1axis promotes progression of breast cancer via enhancement of epithelial-mesenchymal transition and activation of NF-kappaB and beta-catenin. Cancer Lett 2016;373:1–11. [DOI] [PubMed] [Google Scholar]
- 133. Park HY, Kim JH, Park CK.. VEGF induces TGF-beta1 expression and myofibroblast transformation after glaucoma surgery. Am J Pathol 2013;182:2147–2154. [DOI] [PubMed] [Google Scholar]
- 134. Tran H, Bustos D, Yeh R, Rubinfeld B, Lam C, Shriver S, Zilberleyb I, Lee MW, Phu L, Sarkar AA, Zohn IE, Wertz IE, Kirkpatrick DS, Polakis P.. HectD1 E3 ligase modifies adenomatous polyposis coli (APC) with polyubiquitin to promote the APC-axin interaction. J Biol Chem 2013;288:3753–3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Fang S, Guo H, Cheng Y, Zhou Z, Zhang W, Han B, Luo W, Wang J, Xie W, Chao J.. circHECTD1 promotes the silica-induced pulmonary endothelial-mesenchymal transition via HECTD1. Cell Death Dis 2018;9:396.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Bai Y, Zhang Y, Han B, Yang L, Chen X, Huang R, Wu F, Chao J, Liu P, Hu G, Zhang JH, Yao H.. Circular RNA DLGAP4 ameliorates ischemic stroke outcomes by targeting miR-143 to regulate endothelial-mesenchymal transition associated with blood-brain barrier integrity. J Neurosci 2018;38:32–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Huang X, Chen Y, Xiao J, Huang Z, He L, Xu D, et al. Identification of differentially expressed circular RNAs during TGF-ss1-induced endothelial-to-mesenchymal transition in rat coronary artery endothelial cells. Anatol J Cardiol 2018;19:192–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Ueno M, Asada K, Toda M, Schlötzer-Schrehardt U, Nagata K, Montoya M, Sotozono C, Kinoshita S, Hamuro J.. Gene signature-based development of ELISA assays for reproducible qualification of cultured human corneal endothelial cells. Invest Ophthalmol Vis Sci 2016;57:4295–4305. [DOI] [PubMed] [Google Scholar]
- 139. Kuosmanen SM, Kansanen E, Sihvola V, Levonen AL.. MicroRNA profiling reveals distinct profiles for tissue-derived and cultured endothelial cells. Sci Rep 2017;7:10943.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Jackson AO, Zhang J, Jiang Z, Yin K.. Endothelial-to-mesenchymal transition: a novel therapeutic target for cardiovascular diseases. Trends Cardiovasc Med 2017;27:383–393. [DOI] [PubMed] [Google Scholar]
- 141. Chen PY, Simons M.. Future targets in endothelial biology: endothelial cell to mesenchymal transition. Curr Drug Targets 2016;17:1707–1713. [DOI] [PubMed] [Google Scholar]