Abstract
Exosomes have been demonstrated to be effective in the treatment of a variety of cardiac disorders. However, the effects of mesenchymal stem cell (MSC) exosomes on myocardial infarction is yet to be determined. The current study aimed to investigate the potential therapeutic effects of MSC exosomes on myocardial injuries that are caused by myocardial infarction. MSCs were isolated from rat bone marrow and were used for exosome enrichment using culture medium. Confirmation that MSCs and exosomes had been successfully extracted was performed using flow cytometry, electron microscopy and western blot analysis. A rat myocardial ischemia reperfusion (I/R) model was established by ligation of the left anterior descending coronary artery. Rat myocardial injuries were determined using 2,3,5-triphenyltetrazolium chloride, Masson and TUNEL staining. H9c2 cell proliferation, apoptosis and migration were analyzed using 5-ethynyl-2′-deoxyuridine, Hoechst staining, flow cytometry and Transwell assays. Marker gene expression was evaluated using reverse transcription-quantitative PCR, western blot analysis and immunofluorescence. Rat MSC exosomes were revealed to suppress myocardial injury and the myocardiocyte functions that were induced by I/R. The results also demonstrated decreased apoptotic protease activating factor-1 and increased autophagy-related protein 13 expression. The H9c2 cell proliferation and migration inhibition, as well as cell apoptosis during hypoxia-reoxygenation (H/R), were suppressed by rat MSC exosomes, with an alteration of the expression of apoptotic and autophagic genes also being demonstrated. The application of autophagy inhibitor 3-methyladenine significantly mitigated the effect of exosomes on H9c2 cell proliferation and apoptosis, which were induced by H/R. Rat MSC exosomes inhibited myocardial infarction pathogenesis, possibly by regulating autophagy.
Keywords: exosome, mesenchymal stem cell, myocardial infarction, apoptosis, autophagy
Introduction
Myocardial infarction (MI), also referred to as a heart attack, is characterized by acute heart muscle damage caused by a limited blood supply, which can result in cardiogenic shock and cardiac arrest (1). Globally, >15 million MI cases are reported each year and this incurs an enormous burden on affected patients (2). A number of treatments have been effectively used to treat patients experiencing MI, including aspirin, nitroglycerin, opioids, oxygen supplementation and angioplasty, but long-term medication and treatments are still required (3). Previous epidemiological and pathological studies have demonstrated that the majority of MI cases are induced by coronary artery blockages, which are closely associated with a number of conditions, including high blood pressure, diabetes, high blood cholesterol and smoking (4). Cardiomyocyte apoptosis mediates the development of rapid myocardial damage following MI (5), while autophagy exhibits protection against cardiac damage following MI (6,7). However, the cellular pathogenesis of MI is still poorly understood, and novel cardiomyocyte apoptosis-inhibiting methods require further elucidation.
Mesenchymal stem cells (MSCs) are multipotent cells that can differentiate into a variety of cell types, including myocytes, osteoblasts, chondrocytes and adipocytes (8). MSCs that are derived from bone marrow have been demonstrated to mediate pleiotropic processes, including immunomodulation and tissue damage healing (9). MSCs have also been identified as potential targets for cellular treatments that can be used in a variety of disorders, including autoimmune diseases, graft versus host disease and sepsis (9). MSCs have also been found to be effective in MI treatment. For example, previous studies using experimental animal models demonstrated that the injection of MSCs immediately after MI onset improved myocardial compliance, promoted cardiac nerve sprouting and improved ventricular function (10–12). The effects of autologous MSCs on MI have also been validated in patients exhibiting MI (13), indicating the potential of these cells as a promising therapy for use in MI treatment. Based on the results from previous investigations, the beneficial effects of MSCs on MI have been suggested to be mediated by immuno-inflammatory regulation; functional gene expression modulation, including in the vascular endothelial growth factor (VEGF) gene; and major cellular signaling, including in neuregulin1-erythroblastic leukemia viral oncogene homolog 4 signaling (14–16). It has also been revealed that the VEGF, from bone marrow-derived MSCs, significantly inhibits cardiomyocyte apoptosis during MI via the suppression of microRNA (miRNA)-23a and miRNA-92a expression (17). However, the molecular mechanisms of MSC MI treatment are still to be fully elucidated.
Exosomes are characterized as microRNA-containing vesicles that are derived from a variety of cell types, including stem cells, and are widespread in blood, urine and are often detected in cell culture medium (18,19). Increasing evidence has demonstrated that exosomes exhibit key regulatory functions in a variety of biological processes, including in coagulation and intercellular signaling, and this indicates the potential for their application in clinical diagnosis, treatment and prognosis prediction in numerous human disorders (18–21). It has also been revealed that exosomes are closely associated with cardiac repair and regeneration, due to their anti-apoptotic, anti-fibrotic and pro-angiogenic roles (19). For example, exosomes that are secreted by mouse bone marrow-derived dendritic cells have been indicated to greatly enhance the expression of chemokines and inflammatory cytokines, and improve the cardiac function of mice following MI, via the activation of CD4+ T lymphocytes (22). One recent report demonstrated that exosomes that are derived from embryonic stem cells (ESCs) promoted the endogenous repair system and strengthened cardiac function following MI (23). Furthermore, exosomes that are secreted by MSCs in the bone marrow have also been previously demonstrated to reduce myocardial ischemia/reperfusion (I/R) injury (24). However, further validation of these observations is required, and the underlying mechanisms remain undetermined.
The aim of the current study was to investigate the effects of exosomes derived from bone marrow-derived MSCs on MI, using cellular and animal models, and to determine the underlying mechanisms of this interaction, which may provide novel insights into the therapeutic value of exosomes for the treatment of cardiovascular disease.
Material and methods
Rat MSC isolation and culture
MSCs were isolated from the bone marrow tissue of 12 3-week-old male Sprague Dawley (SD) rats weighing 80–100 g, which were supplied by the Animal Center of Southern Medical University as previously described (25), housed in cages at 22–24°C in a 12-h light/dark cycle and fed with standard food pellets and tap water ad libitum. All surgical procedures in the current study were performed following The Guidelines for Laboratory Animal Care and Use by Sun Yat-sen University. The present study was approved by the Animal Ethics Committee of The Third Affiliated Hospital of Sun Yat-sen University. Male rats aged three weeks upon purchase were sacrificed for the collection of femora bone marrow tissues, which were flushed with minimal essential medium (Thermo Fisher Scientific, Inc.) supplemented with 15% FBS (Thermo Fisher Scientific, Inc.), penicillin and streptomycin. After filtration and centrifugation, the pellets containing bone marrow-derived MSCs were cultured use in for subsequent assays. MSCs were cultured in dishes for 7 days in a humidified chamber at 37°C, and supplied with 5% CO2. Culture medium was replaced three times per week. The identity of isolated MSCs was validated using flow cytometry as previously described (26) and defined by the positive expression of cell surface antigens CD29 and CD44, without CD31 and CD34 expression. Primary antibodies against CD29 (1:100; cat. no. 11-0291-82), CD44 (1:100; cat. no. MA5-16908), CD31 (1:100; cat. no. MA5-16953) and CD34 (1:100; cat. no. PA5-85917) provided by Thermo Fishier Scientific. Inc. were used for flow cytometry.
Exosome isolation and electron microscopy
The isolation of exosomes from rat MSCs was performed using a previously described method (23). Isolated MSCs were cultured for >40 h and exosomes in culture medium were collected using an ultracentrifugation at 100,000 × g for 70 min at 4°C. Purified exosomes from MSCs were then resuspended and preserved in saline for subsequent experiments. For electron microscopy, exosomes suspensions were first mixed with 4% paraformaldehyde at RT for 15 min, applied onto formvar-coated carbon grids which was pre-coated with 0.01% Poly Lysine at RT for 1 min, stained with 1% phosphotungstic acid at RT for 2 min, the morphological features of isolated exosomes were finally observed using an FEI Tecnai Spirit G2 transmission electron microscope (Thermo Fisher Scientific, Inc.) with 120-kV voltage and ×49,000 magnification.
Rat myocardial I/R model and exosome administration
The establishment of the myocardial I/R model in SD rats was performed using a method previously described (23). A total of 18 male rats aged 7 weeks were randomly divided into the surgery-only, sham and surgery + exosomes groups. Rats in the surgery-only and surgery + exosomes groups were subjected to surgery to ligate the left anterior descending coronary artery. Sham surgery was also performed on 6 rats in the sham group, which were used as negative controls. Subsequently, the exosomes (10 µg) isolated from rat MSCs were intramyocardially injected into the left ventricular wall of 6 rats in the surgery + exosomes group that underwent left anterior descending coronary artery ligation surgery.
Histological analysis of rat myocardial tissues
The hearts of experimental rats were collected, after rats were sacrificed using pentobarbital sodium (135 mg/kg), following the establishment of the myocardial I/R model and the specified treatments. The pathogenic features of rat myocardial tissues were evaluated using histological analysis. The heart tissues were fixed in 4% paraformaldehyde at RT for 24 h, dehydrated with 70% ethonal for 2 min, 90% ethanol for 2 min and 95% ethanol for 2 min, embedded in paraffin and sliced into 4 µm thick serial sections. For hematoxylin-eosin (H&E) staining, tissue slides were stained with hematoxylin at RT for 5 min and stained with eosin at RT for 2 min, finally observed with microscope (TE200, Nikon Corporation) with a ×400 magnification for at least 6 fields. TUNEL staining was performed by incubation with Protease K solution at 37°C for 30 min, incubation with TdT and dUTP mixture (2:29) at 37°C for 2 h, blocking of endogenous peroxidase with 3% hydrogen peroxide solution in methanol in the dark for 15 min and incubation with converter-POD at 37°C for 30 min. mounting with Antifade Mounting Medium (cat. no. P0126; Beyotime Institute of Biotechnology) and observed using an inverted fluorescence microscope (TE2000-E; Nikon Corporation) with a ×400 magnification for at least 6 fields. 2,3,5-triphenyltetrazolium chloride (TTC) staining was performed by immersion in 1% TTC at RT for 15 min. For Masson staining, the tissue slides were stained with Ponceau S acid fuchsin at RT for 5–10 min, followed by incubation with a molybdophosphoric acid solution at RT for 3 min and staining with aniline blue at RT for 5 min, and finally observed with a camera (EOS 80D; Canon, Inc.).
Cellular hypoxia-reoxygenation (H/R) model and treatment
Rat cardiac H9c2 cells were used as the cellular H/R model in the present study. H9c2 cells were obtained from the American Type Culture Collection and were cultured in DMEM (Thermo Fishier Scientific, Inc.) containing high glucose supplemented with 10% FBS (Thermo Fishier Scientific, Inc.), penicillin and streptomycin at 37°C with 5% CO2. As previously described (27), the cellular H/R model was established by culturing H9c2 cells in DMEM containing low glucose with no supplementation of FBS and with 5% CO2 in a hypoxic atmosphere for 4 h, followed by culturing under a normal oxygen supply for 24 h. To analyze the exosome effect, the H9c2 cells were then treated with 2 µg exosomes at 37°C for 48 h. The H9c2 cells were pretreated with 10 mM 3-methyladenine (3-MA) at 37°C for 30 min to inhibit autophagy progression.
Reverse transcription-quantitative (RT-q)PCR assay
Total RNA samples were extracted from rat tissues and cultured cells using TRIzol® solution (cat. no. 12183555; Thermo Fishier Scientific, Inc.) following the manufacturer's protocol. The cDNA synthesis was then performed using the Bestar qPCR RT kit (cat. no. 2220; DBI Bioscience) as described by the manufacturer's protocol, with 3 µg RNA sample used as a template and a setting of 37°C for 15 min and 98°C for 5 min. The relative expression of genes were finally determined by RT-q PCR using the Bestar™ qPCR MasterMix (cat. no. 2043; DBI Bioscience) according to the manufacturer's protocol, on an Agilent Stratagene PCR system (Mx3000P; Agilent Technologies, Inc.). The following conditions were used: Pre-denaturation at 95°C for 2 min followed by 40 cycles of denaturation at 94°C for 20 sec, annealing at 58°C for 20 sec and extension at 72°C for 20 sec. The relative expression of genes was determined using the 2−ΔΔCq method (28). Primers used for quantification were as follows: Apoptotic protease activating factor-1 (Apaf1) forward, 5′-TCAAAAGTCAGGCCACTCAA-3′ and reverse, 5′-TGAAGTGTTTCCACCGTCTG-3′; elevated autophagy-related protein 13 (ATG13) forward, 5′-TCCACCAGGCAATTTGAGAG-3′ and reverse, 5′-TCAAAAGTCAGGCCACTCAA-3′; and GAPDH forward, 5′-CCTCGTCTCATAGACAAGATGGT-3′ and reverse, 5′-GGGTAGAGTCATACTGGAACATG-3′.
Western blot analysis
Total proteins from exosomes, rat tissues and cultured cells were separately extracted using RIPA lysis and extraction buffer (Thermo Fisher Scientific, Inc.) for western blot analysis. The protein concentration was determined using the Thermo Bicinchoninic Acid Protein Assay kit (cat. no. 23227; Thermo Fisher Scientific, Inc.). Total proteins (20 µg) were loaded onto and separated using 10% SDS-PAGE gels and blotted onto a PVDF membrane. After being blocked with 5% lipid-free milk solution for 2 h at room temperature, the PVDF containing targeted proteins were probed with primary (RT; 2 h) and secondary antibodies (RT; 2 h) successively. Blots were developed using enhanced chemiluminescence detection reagents (cat. no. WB KLS0500; EMD Millipore). Band intensity was analyzed using the Image-Pro Plus 6.0 software (Media Cybernetics, Inc.). GAPDH was applied as the internal standard for relative protein abundance determination. The following antibodies were used in the current study: Anti-CD63 (1:1,000; cat. no. ab108950; Abcam), anti-CD81 (1:1,000; cat. no. ab232390; Abcam), anti-ALG-2-interacting protein X (AliX; 1:1,000; cat. no. ab117600; Abcam), anti-ATG13 (1:1,000; cat. no. ab201467; Abcam), anti-Apaf1 (1:2,000; cat. no. ab2001; Abcam), anti-Bcl2 (1:1,000; cat. no. ab196495; Abcam), anti-Beclin-1 (1:1,000; cat. no. ab207612; Abcam), anti-Bax (1:1,000; cat. no. ab32503; Abcam), anti-GAPDH (1:10,000; cat. no. ab8245; Abcam), horseradish peroxidase (HRP) goat anti-Rabbit IgG (1:20,000; cat. no. BA1054; Wuhan Boster Biological Technology) and HRP goat anti-mouse IgG (1:20,000; cat. no. BA1051; Wuhan Boster Biological Technology).
Cell proliferation, apoptosis and migration tests
H9c2 cell proliferation was evaluated using the 5-ethynyl-2′-deoxyuridine (EdU) method. Cultured H9c2 cells (1×105/ml) were stained at 37°C with 1 µM EdU solution (Thermo Fisher Scientific, Inc.) for 30 min, incubated with 1X Hoechst 33342 solution (Beyotime Institute of Biotechnology) for 10 min at room temperature, and observed using a fluorescence microscope (magnification, ×200). The apoptosis of H9c2 cells was analyzed by staining cells with 5 µg/ml Hoechst 33258 for 20 min at room temperature, and observed with a confocal microscope (magnification, ×120). H9c2 cell apoptosis analysis was performed using flow cytometry using the Annexin V-FITC Apoptosis Staining/Detection kit (cat. no. ab14085; Abcam), and cells were incubated with 5 µl Annexin V-FITC and 5 µl propidium iodide at room temperature for 5 min in the dark, and analyzed using WinMDI software (Version 2.8, J.; Trotter, Scripps Research Institute). H9c2 cell migration was evaluated using the Transwell system-based assay. H9c2 cells (5×105/ml) resuspended in serum-free DMEM were seeded and cultured in the upper chamber for 24 h. FBS (10%) in DMEM was used as a chemoattractant. The migrated cells were finally observed using a light microscope (TE200, Nikon Corporation; magnification, ×200) by staining with 0.1% crystal violet at RT for 15 min.
Immunofluorescence
The Apaf1 and autophagy-related protein 13 (ATG13) protein expression in cultured H9c2 cells was performed using an immunofluorescence assay. H9c2 cells seeded on cover slides were fixed using 2% paraformaldehyde at RT for 15 min, blocked with 5% BSA at RT for 70 min and incubated with the Apaf1 or ATG13 antibodies for 30 min at RT and washed three times with PBS for 10 min. After incubation with secondary antibodies at RT for 1 h, the slides were mounted with DAPI-containing SlowFade Antifade Reagents (cat. no. S36939; Thermo Fisher Scientific, Inc.), and finally observed using a confocal microscope (magnification, ×200).
Statistical analysis
Statistical analyses were performed using the SPSS 18.0 software (IBM Corp.), with data presented as the mean ± standard deviation of three replicates. Statistical analyses were performed using an ANOVA statistical test and the Tukey post hoc test. P<0.05 was considered to indicate a statistically significant difference.
Results
Isolation of exosomes secreted by rat MSCs
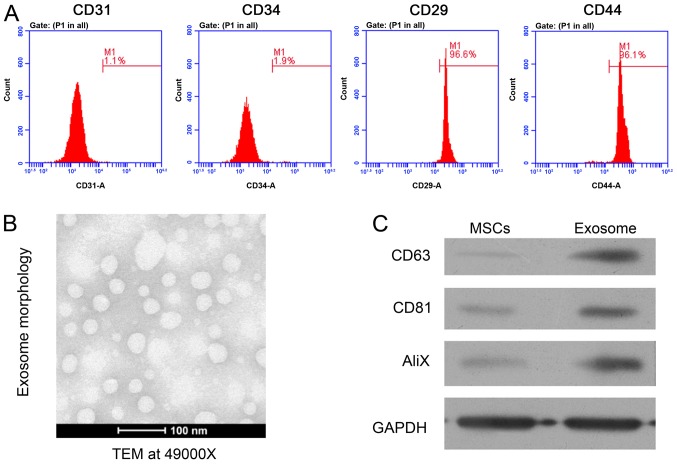
To investigate the potential roles of rat MSC-derived exosomes in MI, primary MSCs were isolated from the bone marrow tissues of SD rats. Flow cytometry was performed to confirm the identity of the collected MSCs. The positive expression of cell surface genes CD29 and CD44, combined with lower expression of CD31 and CD34 were used as biomarkers to confirm the isolation of MSCs (Fig. 1A). Subsequently, exosomes were extracted from the culture medium of isolated rat MSCs. Using a transmission electron microscope, the regular shape of isolated MSC-derived exosomes was observed (Fig. 1B). The expression of three exosome marker genes, CD63, CD81 and AliX, were further analyzed using western blot analysis, which demonstrated the significantly higher expression of these proteins in exosomes compared with MSCs (Fig. 1C). Collectively, these results confirmed the successful isolation of MSCs from rat bone marrow tissues and enriched MSC-derived exosomes, which were used for the subsequent assays.
Figure 1.
Rat mesenchymal stem cell and exosome isolation. (A) Flow cytometry analysis of rat MSCs, confirmed by the positive expression of CD29 and CD44, combined with a lack of expression of CD31 and CD34. (B) TEM (magnification, ×49,000) of the morphological properties of rat mesenchymal stem cell-derived exosomes. (C) Western blot analysis of CD63, CD81 and AliX protein expression in rat MSCs and exosomes. GAPDH was used as an internal control. MSC, mesenchymal stem cell; TEM, transmission electron microscopy; AliX, ALG-2-interacting protein X.
Inhibition of myocardial damage by MSC-derived exosomes
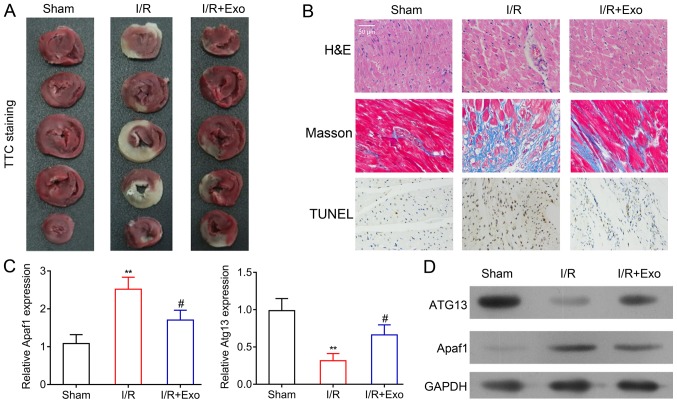
To analyze the protective effects of exosomes isolated from rat MSCs on MI, the myocardial I/R model rats were established using a surgical procedure and treated with exosomes. TTC staining revealed that the I/R model rats showed considerable myocardial damage when compared with the sham surgery group. However, the application of exosomes from the rat MSCs inhibited the progression of myocardial damage that was induced by MI, although this result was not exhibited across all samples (Fig. 2A). The histological features of rat myocardial tissues were further analyzed using H&E and Masson staining, which revealed an irregular alteration of cell shape and lining in the rat myocardium after the induction of MI. The injection of MSC-derived exosomes mitigated the abnormal cellular alterations (Fig. 2B). Furthermore, TUNEL staining was performed to evaluate cell apoptosis in rat myocardial tissues. A marked increase in apoptotic cells was indicated in the I/R rats. Cardiomyocyte apoptosis was suppressed by exosomes treatment in rats that underwent left anterior descending coronary artery ligation (Fig. 2B).
Figure 2.
Inhibition of rat myocardial damages by MSC-derived exosomes. (A) TTC staining and (B) H&E, Masson and TUNEL staining of the Sham, I/R and I/R+Exo groups (magnification, ×400). (C) Reverse transcription-quantitative PCR and (D) western blot analysis of Apaf1 and ATG13 mRNA and protein levels in the Sham, I/R and I/R+Exo groups. GAPDH was applied as the internal standard. **P<0.01 vs. Sham group; #P<0.05 vs. I/R group. MSC, mesenchymal stem cell; TTC, 2,3,5-triphenyltetrazolium chloride; H&E, hematoxylin and eosin; I/R, ischemia reperfusion; Exo, exosomes; Apaf1, apoptotic protease activating factor-1; ATG13, autophagy-related protein 13.
Furthermore, Apaf1 mRNA expression was increased in the myocardial tissues of I/R rats, compared with the sham group, while the expression of ATG13 expression was significantly decreased in the myocardial tissues of I/R rats (Fig. 2C). Consistent with the histological alteration of rat heart tissues, the application of exosomes from rat MSCs recovered Apaf1 and ATG13 mRNA expression in rat myocardial tissues (Fig. 2C). Additionally, the changes in Apaf1 and ATG13 expression in the sham, I/R and exosome treatment groups of rat hearts were further validated using western blot analysis (Fig. 2D). Overall, the histological and molecular findings suggested that exosomes from rat MSCs, could effectively suppress myocardial tissue damage induced by MI.
Exosomes mitigate H/R injuries in H9c2 cells
To further investigate the roles of mesenchymal stem cell-secreted exosomes in MI therapy, a cellar H/R model was applied using rat cardiac H9c2 cells. EdU and Hoechst 33342 staining revealed that MSC-derived exosomes induced no significant changes in the rate of H9c2 cell proliferation compared with normal H9c2 cells (Fig. 3A). However, H9c2 cell proliferation was significantly suppressed by H/R but significantly recovered by exosome treatment (Fig. 3A). The results of Hoechst 33258 staining also demonstrated that H/R induced apoptosis in H9c2 cells, while the mesenchymal stem cell-derived exosome treatment significantly reduced H9c2 apoptosis (Fig. 3B). The inhibitory effects of MSC-derived exosomes on H9c2 cell apoptosis during H/R was further validated using flow cytometry (Fig. 3C). Additionally, the migration capability of H9c2 cells was detected using a Transwell system, and H/R significantly decreased H9c2 migration capacity (Fig. 3D). However, exosome treatment increased H9c2 cell proliferation after H/R (Fig. 3D). Collectively, these cellular biology assays demonstrated that exosomes from rat MSCs could effectively ameliorate the H/R injury induced in rat H9c2 cells. These results further validated the use of exosomes isolated from the rat MSCs in protection against MI.
Figure 3.
Inhibition of H9c2 cell H/R injuries by MSC exosomes. (A) EdU and Hoechst 33342 staining of the proliferation capability of rat H9c2 cells after H/R and exosome treatment (magnification, ×100). (B) Hoechst 33258 and (C) flow cytometry analysis of apoptosis in rat H9c2 cells after H/R and exosome treatment. (D) Migration capability of H9c2 cells after H/R and MSC exosome treatment (magnification, ×200). **P<0.01, ***P<0.001; #P<0.05, ##P<0.01. EdU, 5-ethynyl-2′-deoxyuridine; MSC, mesenchymal stem cell; H/R, hypoxia-reoxygenation; Exo, exosomes; ns, not significant; PI, propidium iodide.
Apoptosis-related and autophagy-related gene expression in H9c2 cells regulated by exosomes
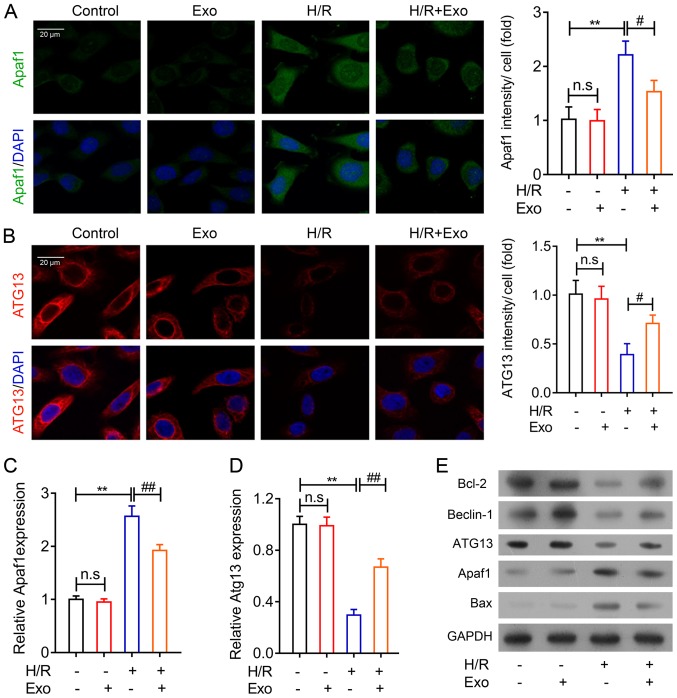
To further confirm the inhibitory effects of rat mesenchymal stem cell-derived exosomes on MI, two major functional genes associated with cell apoptosis and autophagy were analyzed in H9c2 cells following H/R. An immunofluorescence assay was used, and the results demonstrated that Apaf1 expression in H9c2 cells was increased during H/R but was significantly suppressed by exosome treatment (Fig. 4A). The results also indicated that ATG13 expression was repressed by H/R in H9c2 cells, but MSC exosomes enhanced ATG13 expression in H9c2 cells after H/R (Fig. 4B). The alteration of Apaf1 and ATG13 gene and protein expression in H9c2 cells accompanying H/R injury was confirmed using RT-qPCR (Fig. 4C and D) and western blotting (Fig. 4E). Additionally, Bcl-2 and Bax expression were not influenced by exosome treatment in H9c2 cells under normal conditions. However, Bcl-2 gene expression was markedly increased following MSC-derived exosome treatment following H/R, while Bax gene expression was suppressed by MSC exosome treatment in H9c2 cells after H/R treatment (Fig. 4E). Beclin-1 gene expression, an additional autophagy-associated gene, was also suppressed by H/R in H9c2, which was mitigated by exosome treatment (Fig. 4E). These results further indicated that exosomes isolated from rat MSCs could regulate apoptosis and autophagy in rat myocardial cells.
Figure 4.
Apoptosis and autophagy-related gene expression in H9c2 cells regulated by MSC exosomes. Immunofluorescence analysis of (A) Apaf1 and (B) ATG13 protein abundances in rat H9c2 cells after H/R and exosome treatment. Cell nuclei were stained using DAPI (magnification, ×120). Reverse transcription-quantitative PCR analysis of (C) Apaf1 and (D) ATG13 gene expression in rat H9c2 cells after H/R and exosome treatment. (E) Western blot analysis of apoptosis and autophagy-related protein expression in H9c2 cells treated with exosomes. GAPDH was used as the internal standard. **P<0.01; #P<0.05, ##P<0.01. MSC, mesenchymal stem cell; Apaf1, apoptotic protease activating factor-1; ATG13, autophagy-related protein 13; H/R, hypoxia-reoxygenation; Exo, exosomes; ns, not significant.
Role of autophagy in exosome-induced H/R injury inhibition
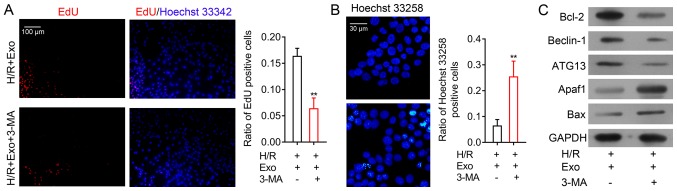
To analyze the role of autophagy in the inhibition of H9c2 cell H/R injury by MSC-derived exosomes, H9c2 cells after H/R and exosome treatment were treated with 3-MA, a known inhibitor of cell autophagy. The results demonstrated that H9c2 cell proliferation after exosome treatment and following H/R was significantly suppressed by a subsequent 3-MA treatment (Fig. 5A). In contrast, H9c2 cell apoptosis after H/R and exosome treatment was greatly increased following 3-MA treatment (Fig. 5B). Beclin-1 and ATG13 expression in H9c2 cells were suppressed by 3-MA (Fig. 5C). Moreover, while the expression of the other apoptosis-associated proteins Bcl-2 was markedly decreased while Apaf1 and Bax were consistently markedly increased by autophagy inhibition in H9c2 cells (Fig. 5C). The cellular and molecular alterations induced by 3-MA in H9c2 cells demonstrated that the inhibition of H/R injury in H9c2 cells by MSC exosomes was mediated by the promotion of autophagy.
Figure 5.
Autophagy inhibitor treatment ameliorates exosome H/R injury-inhibiting effects. (A) EdU assay analysis of proliferation rates of H9c2 cells treated with 3-MA following H/R and exosome treatment (magnification, ×100). (B) Hoechst 33258 staining of apoptosis of H9c2 cells treated with 3-MA following H/R and exosome treatment (magnification, ×200). (C) Western blot analysis of apoptosis and autophagy-related protein expression in H9c2 cells treated with 3-MA after H/R and exosome treatment. GAPDH was used as the internal standard. **P<0.01 vs. control. H/R, hypoxia-reoxygenation; EdU, 5-ethynyl-2′-deoxyuridine; 3-MA, 3-Methyladenine; Apaf1, apoptotic protease activating factor-1; ATG13, autophagy-related protein 13; Bax, BCL2-associated X protein; H/R, hypoxia-reoxygenation.
Discussion
Exosomes were first characterized in mammalian reticulocytes as being important in plasma membrane protein removal during erythrocyte formation (29). The major constituents of exosomes are proteins, numerous mRNAs and miRNAs, and exosomes are widely associated with multiple biological processes, including immune response regulation, due to their capability for membrane vesicle trafficking (19,20,29). Exosomes have been recognized for their therapeutic potential in injury repair and a variety of human disorders, especially exosomes derived from multipotent stem cells (19,20). Exosomes from MSCs have been reported to promote would healing by activating numerous cellular signaling pathways (30). However, the effects of MSC-derived exosomes on injuries associated with MI have not been fully determined, despite reports demonstrating the significant roles of exosomes isolated from stem and dendritic cells in this process (22–24). In the present study, MSCs were isolated from rat bone marrow tissues using a widely applied method. The identity of MSCs was validated using flow cytometry to detect CD29 and CD44 expression on the cell surface, which are previously identified specific biomarkers for MSC-derived exosomes (31). CD31 and CD34 were also detected as the negative control. The identification of additional biomarkers, including CD90 and CD45, would further increase the characterization specificity. Exosomes isolated from the culture medium of rat MSCs were further verified using TEM and the confirmed expression of several exosome-specific proteins. The isolation and validation of rat MSCs and secreted exosomes provided a basis for the subsequent analyses.
The establishment of an I/R model by left anterior descending coronary artery ligation has been successfully applied in pathogenesis and pharmaceutical studies of MI, due to its ease of use, low cost and availability of adequate tissue samples for mechanistic investigation (23). To explore the effects of rat mesenchymal stem cell-derived exosomes on MI, rat myocardial I/R was performed and cells were then treated with exosomes that were isolated from rat MSCs. Using multiple histological and cellular methods, the results demonstrated that exosomes from MSCs suppressed the myocardial injury and cell apoptosis induced by I/R. Apaf1 is a key regulator of the mitochondrial pathways in cell apoptosis (32), and in the current study it was indicated that its expression was significantly repressed by MSC-derived exosomes. Furthermore, ATG13 expression, which promotes autophagy machinery through the mechanistic target of the mTOR signaling pathway (33), was increased by MSC exosome treatment in rats undergoing I/R. These in vivo assays using I/R rats demonstrated that exosomes secreted by MSCs could inhibit the myocardial injuries caused by MI, possibly by regulating autophagy.
The rat myoblast cell line H9c2 is a beneficial model used to study myocardial physiology and pathogenic processes, including MI (27,34). To provide evidence of MSC exosome inhibition of MI-related cell damage, H9c2 cells were treated with H/R. Using this cellular model, it was demonstrated that MSC exosomes inhibited the suppression of cell proliferation and migration during H/R, and also suppressed the myocardiocyte apoptosis that was induced by H/R. Furthermore Apaf1 and ATG13 gene expression, as well as a number of apoptosis-associated and autophagy-associated functional proteins in H9c2 cells treated with MSC exosomes following H/R, indicated alterations that were consistent with the rat I/R models. These results further validated the significant regulatory effects of MSC exosomes on myocardial injuries that are associated with infarctions. Together with other evidence obtained using exosomes from embryonic stem cells and dendritic cells (22,23), it is reasonable to conclude that exosomes secreted by certain cell types might be useful as therapeutic agents for use in MI treatment.
The autophagy process is associated with the pathogenic progression during MI (35,36). The results of the current study revealed that the inhibition of autophagy using 3-MA mitigated the injury-suppressing effects of MSC exosomes on H9c2 cells, demonstrating that the autophagy process serves a critical role in the pathogenesis of MI, as well as in exosome-mediated infarction-induced tissue damage. It was also suggested that modulation of autophagy progression might be an effective strategy for the treatment of MI. The evidence of the current study, of autophagy mediating infarction-induced tissue damage, is limited by a lack of in vivo investigation. The roles of autophagy, in MI inhibition, by MSC-derived exosomes need further determination using animal models treated with a combination of exosomes and autophagy inhibitors.
In summary, animal and cellular models were used to validate exosome secretion by rat MSCs and if these exosomes could exert inhibitory effects on myocardiocyte apoptosis associated with MI, which is mediated by the regulation of autophagy machinery. Conclusions of the present study provided a basis for the development of a novel exosome-based therapy for MI.
Acknowledgements
Not applicable.
Funding
The current study was supported by the National Natural Science Foundation of China (grant no. 81370214).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
LZ, XM and CP conceived and supervised the study. LZ, XM and SL designed and performed experiments. SL, BW and YC analyzed the data. LZ, BW, YC and CP drafted the manuscript and made revisions. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Animal Ethics Committee of The Third Affiliated Hospital of Sun Yat-sen University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Thygesen K, Alpert JS, White HD, Joint ESC/ACCF/AHA/WHF Task Force for the Redefinition of Myocardial Infarction Universal definition of myocardial infarction. Eur Heart J. 2008;28:2525. doi: 10.1093/eurheartj/ehm355. [DOI] [PubMed] [Google Scholar]
- 2.GBD 2016 Disease, Injury Incidence, Prevalence Collaborators. Global, regional, and national incidence, prevalence, and years lived with disability for 328 diseases and injuries for 195 countries, 1990–2016: A systematic analysis for the Global Burden of Disease Study 2016. Lancet. 2017;390:1211–1259. doi: 10.1016/S0140-6736(17)32154-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levine GN, Dai X, Henry TD, Calfon Press M, Denktas AE, Garberich RF, Jacobs AK, Jaski BE, Kaul P, Kontos MC, et al. In-hospital ST-segment elevation myocardial infarction: Improving diagnosis, triage, and treatment. JAMA Cardiol. 2018;3:527–531. doi: 10.1001/jamacardio.2017.5356. [DOI] [PubMed] [Google Scholar]
- 4.Varghese T, Hayek SS, Shekiladze N, Schultz WM, Wenger NK. Psychosocial risk factors related to ischemic heart disease in women. Curr Pharm Des. 2016;22:3853–3870. doi: 10.2174/1381612822666160519113605. [DOI] [PubMed] [Google Scholar]
- 5.Abbate A, Salloum FN, Vecile E, Das A, Hoke NN, Straino S, Biondi-Zoccai GG, Houser JE, Qureshi IZ, Ownby ED, et al. Anakinra, a recombinant human interleukin-1 receptor antagonist, inhibits apoptosis in experimental acute myocardial infarction. Circulation. 2008;117:2670–2683. doi: 10.1161/CIRCULATIONAHA.107.740233. [DOI] [PubMed] [Google Scholar]
- 6.Riquelme JA, Chavez MN, Mondaca-Ruff D, Bustamante M, Vicencio JM, Quest AF, Lavandero S. Therapeutic targeting of autophagy in myocardial infarction and heart failure. Expert Rev Cardiovasc Ther. 2016;14:1007–1019. doi: 10.1080/14779072.2016.1202760. [DOI] [PubMed] [Google Scholar]
- 7.Bravo-San Pedro JM, Kroemer G, Galluzzi L. Autophagy and mitophagy in cardiovascular disease. Circ Res. 2017;120:1812–1824. doi: 10.1161/CIRCRESAHA.117.311082. [DOI] [PubMed] [Google Scholar]
- 8.Ankrum JA, Ong JF, Karp JM. Mesenchymal stem cells: Immune evasive, not immune privileged. Nat Biotechnol. 2014;32:252–260. doi: 10.1038/nbt.2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heirani-Tabasi A, Hassanzadeh M, Hemmati-Sadeghi S, Shahriyari M, Raeesolmohaddeseen M. Mesenchymal stem cells; Defining the future of regenerative medicine. J Genes Cells. 2015;1:34–39. doi: 10.15562/gnc.15. [DOI] [Google Scholar]
- 10.Berry MF, Engler AJ, Woo YJ, Pirolli TJ, Bish LT, Jayasankar V, Morine KJ, Gardner TJ, Discher DE, Sweeney HL. Mesenchymal stem cell injection after myocardial infarction improves myocardial compliance. Am J Physiol Heart Circ Physiol. 2006;290:2196–2203. doi: 10.1152/ajpheart.01017.2005. [DOI] [PubMed] [Google Scholar]
- 11.Pak HN, Qayyum M, Kim DT, Hamabe A, Miyauchi Y, Lill MC, Frantzen M, Takizawa K, Chen LS, Fishbein MC, et al. Mesenchymal stem cell injection induces cardiac nerve sprouting and increased tenascin expression in a Swine model of myocardial infarction. J Cardiovasc Electrophysiol. 2010;14:841–848. doi: 10.1046/j.1540-8167.2003.03124.x. [DOI] [PubMed] [Google Scholar]
- 12.Price MJ, Chou CC, Frantzen M, Miyamoto T, Kar S, Lee S, Shah PK, Martin BJ, Lill M, Forrester JS, et al. Intravenous mesenchymal stem cell therapy early after reperfused acute myocardial infarction improves left ventricular function and alters electrophysiologic properties. Int J Cardiol. 2004;111:231–239. doi: 10.1016/j.ijcard.2005.07.036. [DOI] [PubMed] [Google Scholar]
- 13.Mohyeddin-Bonab M, Mohamad-Hassani MR, Alimoghaddam K, Sanatkar M, Gasemi M, Mirkhani H, Radmehr H, Salehi M, Eslami M, Farhig-Parsa A, et al. Autologous in vitro expanded mesenchymal stem cell therapy for human old myocardial infarction. Arch Iran Med. 2007;10:467–473. [PubMed] [Google Scholar]
- 14.Du YY, Zhou SH, Zhou T, Su H, Pan HW, Du WH, Liu B, Liu QM. Immuno-inflammatory regulation effect of mesenchymal stem cell transplantation in a rat model of myocardial infarction. Cytotherapy. 2008;10:469–478. doi: 10.1080/14653240802129893. [DOI] [PubMed] [Google Scholar]
- 15.Deuse T, Peter C, Fedak PW, Doyle T, Reichenspurner H, Zimmermann WH, Eschenhagen T, Stein W, Wu JC, Robbins RC, Schrepfer S. Hepatocyte growth factor or vascular endothelial growth factor gene transfer maximizes mesenchymal stem cell-based myocardial salvage after acute myocardial infarction. Circulation 120 (11 Suppl) 2009:S247–S254. doi: 10.1161/CIRCULATIONAHA.108.843680. [DOI] [PubMed] [Google Scholar]
- 16.Liang X, Ding Y, Zhang Y, Chai YH, He J, Chiu SM, Gao F, Tse HF, Lian Q. Activation of NRG1-ERBB4 signaling potentiates mesenchymal stem cell-mediated myocardial repairs following myocardial infarction. Cell Death Dis. 2015;6:e1765. doi: 10.1038/cddis.2015.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song YS, Joo HW, Park IH, Shen GY, Lee Y, Shin JH, Kim H, Kim KS. Bone marrow mesenchymal stem cell-derived vascular endothelial growth factor attenuates cardiac apoptosis via regulation of cardiac miRNA-23a and miRNA-92a in a rat model of myocardial infarction. PLoS One. 2017;12:e0179972. doi: 10.1371/journal.pone.0179972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Min PK, Chan SY. The biology of circulating microRNAs in cardiovascular disease. Eur J Clin Invest. 2015;45:860–874. doi: 10.1111/eci.12475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singla DK. Stem cells and exosomes in cardiac repair. Curr Opin Pharmacol. 2016;27:19–23. doi: 10.1016/j.coph.2016.01.003. [DOI] [PubMed] [Google Scholar]
- 20.Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, Chen D, Gu J, He X, Huang S. Circular RNA is enriched and stable in exosomes: A promising biomarker for cancer diagnosis. Cell Res. 2015;25:981–984. doi: 10.1038/cr.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taylor DD, Gerceltaylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008;110:13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 22.Liu H, Gao W, Yuan J, Wu C, Yao K, Zhang L, Ma L, Zhu J, Zou Y, Ge J. Exosomes derived from dendritic cells improve cardiac function via activation of CD4(+) T lymphocytes after myocardial infarction. J Mol Cell Cardiol. 2016;91:123–133. doi: 10.1016/j.yjmcc.2015.12.028. [DOI] [PubMed] [Google Scholar]
- 23.Khan M, Nickoloff E, Abramova T, Johnson J, Verma SK, Krishnamurthy P, Mackie AR, Vaughan E, Garikipati VN, Benedict C, et al. Embryonic stem cell-derived exosomes promote endogenous repair mechanisms and enhance cardiac function following myocardial infarction. Circ Res. 2015;117:52–64. doi: 10.1161/CIRCRESAHA.117.305990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lai RC, Arslan F, Lee MM, Sze NS, Choo A, Chen TS, Salto-Tellez M, Timmers L, Lee CN, El Oakley RM, et al. Exosome secreted by MSC reduces myocardial ischemia/reperfusion injury. Stem Cell Res. 2010;4:214–222. doi: 10.1016/j.scr.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 25.Zhang L, Chan C. Isolation and enrichment of rat mesenchymal stem cells (MSCs) and separation of single-colony derived MSCs. J Vis Exp. 2010;37:e185. doi: 10.3791/1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boxall S, Jones E. The use of multiparameter flow cytometry and cell sorting to characterize native human bone marrow mesenchymal stem cells (MSC) Methods Mol Biol 1235. 2015:121–130. doi: 10.1007/978-1-4939-1785-3_11. [DOI] [PubMed] [Google Scholar]
- 27.Jia X, Zhang L, Mao X. S-propranolol protected H9C2 cells from ischemia/reperfusion-induced apoptosis via downregultion of RACK1 Gene. Int J Clin Exp Pathol. 2015;8:10335–10344. [PMC free article] [PubMed] [Google Scholar]
- 28.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 29.Van NG, Porto-Carreiro I, Simoes S, Raposo G. Exosomes: A common pathway for a specialized function. J Biochem. 2006;140:13–21. doi: 10.1093/jb/mvj128. [DOI] [PubMed] [Google Scholar]
- 30.Silva AM, Teixeira JH, Almeida MI, Gonçalves RM, Barbosa MA, Santos SG. Extracellular Vesicles: Immunomodulatory messengers in the context of tissue repair/regeneration. Eur J Pharm Sci. 2017;98:86–95. doi: 10.1016/j.ejps.2016.09.017. [DOI] [PubMed] [Google Scholar]
- 31.Liao W, Yang S, Song C, Li X, Li Y, Xiong Y. Construction of ureteral grafts by seeding bone marrow mesenchymal stem cells and smooth muscle cells into bladder acellular matrix. Transplantat Proc. 2013;45:730–734. doi: 10.1016/j.transproceed.2012.08.023. [DOI] [PubMed] [Google Scholar]
- 32.Yoshida H, Kong YY, Yoshida R, Elia AJ, Hakem A, Hakem R, Penninger JM, Mak TW. Apaf1 is required for mitochondrial pathways of apoptosis and brain development. Cell. 1998;94:739–750. doi: 10.1016/S0092-8674(00)81733-X. [DOI] [PubMed] [Google Scholar]
- 33.Jung CH, Jun CB, Ro SH, Kim YM, Otto NM, Cao J, Kundu M, Kim DH. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20:1992–2003. doi: 10.1091/mbc.e08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jana S, Patel D, Patel S, Upadhyay K, Thadani J, Mandal R, Das S, Devkar R. Anthocyanin rich extract of Brassica oleracea L. alleviates experimentally induced myocardial infarction. PLoS One. 2017;12:e0182137. doi: 10.1371/journal.pone.0182137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liao LZ, Chen YL, Lu LH, Zhao YH, Guo HL, Wu WK. Polysaccharide from Fuzi likely protects against starvation-induced cytotoxicity in H9c2 cells by increasing autophagy through activation of the AMPK/mTOR pathway. Am J Chin Med. 2013;41:353–367. doi: 10.1142/S0192415X13500262. [DOI] [PubMed] [Google Scholar]
- 36.Lu L, Wu W, Yan J, Li X, Yu H, Yu X. Adriamycin-induced autophagic cardiomyocyte death plays a pathogenic role in a rat model of heart failure. Int J Cardiol. 2009;134:9. doi: 10.1016/j.ijcard.2008.01.043. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.