Abstract
Aims:
This study aimed to determine the prevalence and antimicrobial resistance pattern of Salmonella spp., and the genetic relatedness between isolates from broilers and pigs at slaughterhouses in Thailand.
Materials and Methods:
Fecal samples (604 broilers and 562 pigs) were collected from slaughterhouses from April to July 2018. Salmonella spp. were isolated and identified according to the ISO 6579:2002. Salmonella-positive isolates were identified using serotyping and challenged with nine antimicrobial agents: Amoxicillin/clavulanate (AMC, 30 µg), ampicillin (AMP, 10 µg), ceftazidime (30 µg), chloramphenicol (30 µg), ciprofloxacin (CIP, 5 µg), nalidixic acid (NAL, 30 µg), norfloxacin (10 µg), trimethoprim/sulfamethoxazole (SXT, 25 µg), and tetracycline (TET, 30 µg). Isolates of the predominant serovar Salmonella Typhimurium were examined for genetic relatedness using pulsed-field gel electrophoresis (PFGE).
Results:
Salmonella was detected in 18.05% of broiler isolates and 37.54% of pig isolates. The most common serovars were Kentucky, Give, and Typhimurium in broilers and Rissen, Typhimurium, and Weltevreden in pigs. Among broilers, isolates were most commonly resistant to antibiotics, NAL, AMP, TET, AMC, and CIP. Pig isolates most commonly exhibited antimicrobial resistance against AMP, TET, and SXT. Based on PFGE results among 52 S. Typhimurium isolates from broilers and pigs, a high genetic relatedness between broiler and pig isolates (85% similarity) in Cluster A and C from PFGE result was identified.
Conclusion:
The results revealed high cross-contamination between these two animal species across various provinces in Thailand.
Keywords: antimicrobial resistance, broilers, pigs, pulsed-field gel electrophoresis, Salmonella spp
Introduction
Salmonella is a critical zoonotic foodborne pathogen that is a serious public health concern worldwide [1]. The pathogen is internationally recognized as the main cause of diarrheal disease that infects 10% of the population every year [2]. Non-typhoidal Salmonella (NTS) is a major group of Salmonella that causes salmonellosis in humans and animals worldwide. Most cases of human salmonellosis are caused by Salmonella through consumption of food contaminated by the pathogen [3].
Broilers and pigs are important reservoirs of NTS [4]. The most common NTS serovars in Thailand’s broilers are Salmonella Weltevreden, Salmonella Rissen, and Salmonella Corvallis [5-7]. In Thailand’s pigs, the most common serovars are S. Rissen and Salmonella Typhimurium [8-11]. Most NTS serovars are not severe pathogens because of many serovars in pigs and Salmonella Pullorum and Salmonella Gallinarum in broilers are host specific, so they cause disease in animals, but not in humans. Most humans infected with serovars leading to gastroenteritis transmitted by consuming contaminated food or environmental contact can recover without treatment. However, some serovars such as Salmonella Choleraesuis, S. Typhimurium, Salmonella Heidelberg, Salmonella Virchow, Salmonella Infantis, Salmonella Agona, and Salmonella Enteritidis can threaten patient lives, especially infants, elders, and immunosuppressed patients [12].
Antimicrobial-resistant NTS has become a significant problem worldwide. Antimicrobial resistance has led to the failure in the treatment of gastroenteritis patients, prolonged hospitalization, and increased medical costs, leading to massive public health, and economic impacts. Moreover, the presence of multidrug-resistant Salmonella (resistant to more than three antimicrobial agents) exacerbates this problem. Several studies reported that 40-80% of broilers and pigs in Thailand carry multidrug-resistant Salmonella isolates [9,13-16].
Pulsed-field gel electrophoresis (PFGE) is an important tool for controlling and investigating Salmonella outbreaks [17]. The PFGE method is used in many studies to assess the genetic relatedness of Salmonella between human and livestock isolates from animals of the same species in Thailand [11,15,18,19]. However, research into the genetic relatedness between Salmonella isolates across different animal species is still limited.
This study aimed to determine the prevalence of Salmonella serovars, antimicrobial resistance patterns, and genetic relationships between Salmonella isolates from broilers and pigs to provide more data on the dispersal of Salmonella among animals, the environment, and humans.
Materials and Methods
Ethical approval
The study was reviewed and approved by the Institutional Animal Care and Use Committee of Khon Kaen University, Thailand (IACUC-KKU-16/61).
Sample collection
Six hundred and four cloacal swab samples from broilers and 562 rectal swab samples from pigs were randomly collected after stunning the animals at local slaughterhouses in nine provinces of Thailand (n=1,116) (Khon Kaen, Nong Khai, Chiang Rai, Chiang Mai, Lampang, Lamphun, Mae Hong Son, Phayao, and Sa Kaeo) between April and July 2018. All samples were suspended in 5 mL of Cary Blair transport media (Oxoid, England), stored in an icebox (4°C), and transported to the laboratory at the Department of Veterinary Public Health, Khon Kaen University, Khon Kaen, Thailand.
Isolation and identification of Salmonella
Salmonella spp. isolates were purified using the standard method ISO 6579:2002. All samples were enriched in 9 mL buffered peptone water (BPW, Difco, France) for 24 h at 37°C and selected using Modified Semi-solid Rappaport-Vassiliadis medium (MSRV, Difco, France) through incubation for 24 h at 42°C. The plates with white swarming growth were subcultured on xylose-lysine deoxycholate agar (XLD, Difco, France) and incubated at 37°C for 24 h. Positive colonies (red colonies with black centers) were selected and subjected to biochemical tests to confirm Salmonella positive isolates using triple sugar iron agar (TSI, Difco, France) and motility indole lysine (MIL, Difco, France) for 24 h at 37°C [20]. The slide agglutination test used commercial antiserum and followed the Kauffman–White scheme [21]. It was performed for Salmonella spp. serovar identification (ECDC 2012; S&A Reagents Lab, Bangkok, Thailand).
Antimicrobial susceptibility testing
The Salmonella isolates were tested against nine antimicrobial agents using the agar disk diffusion technique following Clinical and Laboratory Standards Institute M100 27th standard: Ampicillin (AMP, 10 μg), amoxicillin/clavulanate (AMC, 30 μg), chloramphenicol (CHL, 30 μg), ciprofloxacin (CIP, 5 μg), ceftazidime (CAZ, 30 μg), nalidixic acid (NAL, 30 μg), norfloxacin (NOR, 10 μg), trimethoprim/sulfamethoxazole (SXT, 1.25/23.75 μg), and tetracycline (TET, 30 μg) [22]. Escherichia coli ATCC 25922 was used as the control strain.
PFGE
S. Typhimurium isolates were analyzed using PFGE according to the standard operating procedure for PulseNet PFGE of E. coli O157:H7, E. coli non-O157 (STEC), Salmonella serotypes, Shigella sonnei, and Shigella flexneri [23]. Salmonella Braenderup H9812 was used as a reference marker and the isolates were digested using XbaI (New England Biolabs Ipswich, MA, USA). The PFGE profiles were analyzed using BioNumerics software version 7.6 (Applied Maths, Belgium) with the dice coefficient similarity index of 1% optimization and 1% tolerance using the unweighted pair group method with arithmetic means (UPGMA).
Statistical analysis
The prevalence of Salmonella spp. and antimicrobial resistance patterns of broiler and pig samples were analyzed using Pearson’s Chi-square test and significance differences (p<0.05) using SPSS statistical software version 17.0 (IBM, USA).
Results
Salmonella prevalence and serovars
Among the isolates from broilers and pigs, 28.67% (320/1116) were positive for Salmonella spp., 37.54% (211/562) of pig rectal swab samples were positive, and 18.05% (109/604) of isolates from broiler cloacal swab samples were positive for Salmonella spp. The prevalence of Salmonella in pig samples (37.54%) was significantly higher than in broiler samples (18.05%) (p<0.001). Forty-six different serovars were identified in broiler isolates (n=31) and pig isolates (n=19), as shown in Table-1. The most common serovars in broiler isolates were Salmonella Kentucky (22.94%), followed by Salmonella Give (20.18%), S. Typhimurium (7.34%), Salmonella Mbandaka (5.50%), Salmonella Agona (3.67%), Salmonella Derby (3.67%), and Salmonella Singapore (3.67%). The most common serovars in isolates from pigs were S. Rissen (36.97%), followed by S. Typhimurium (21.33%), S. Weltevreden (14.70%), Salmonella Stanley (6.64%), and S. Agona (3.79%). The serovars S. Agona, S. Stanley, S. Typhimurium, and S. Give were found in both broiler and pig samples. The slide agglutination test showed that broiler and pig isolates both had S. Typhimurium as the most common of the previously detected serovars, and we used it as the main isolate for determining genetic relatedness between broiler and pig isolates using PFGE.
Table 1.
Serotypes of Salmonella from pigs and broilers in Thailand.
| Salmonella groups | Salmonella serotypes | Broilers n (%) | Pigs n (%) | Total n (%) |
|---|---|---|---|---|
| B | Salmonella Agona | 4 (3.67) | 8 (3.79) | 12 (3.75) |
| Salmonella Derby | 4 (3.67) | - | 4 (1.25) | |
| Salmonella Haifa | 1 (0.92) | - | 1 (0.31) | |
| Salmonella Heidelberg | - | 1 (0.47) | 1 (0.31) | |
| Salmonella Limete | - | 2 (0.95) | 2 (0.63) | |
| Salmonella Paratyphi B | - | 1 (0.47) | 1 (0.31) | |
| Salmonella Stanley | 1 (0.92) | 14 (6.64) | 15 (4.69) | |
| Salmonella Typhimurium | 8 (7.34) | 45 (21.33) | 53 (16.56) | |
| C | Salmonella Albany | 1 (0.92) | - | 1 (0.31) |
| Salmonella Altona | - | 4 (1.90) | 4 (1.25) | |
| Salmonella Athinai | 2 (1.83) | - | 2 (0.63) | |
| Salmonella Augustenborg | 1 (0.92) | - | 1 (0.31) | |
| Salmonella Bardo | 2 (1.83) | - | 2 (0.63) | |
| Salmonella Bareilly | 2 (1.83) | - | 2 (0.63) | |
| Salmonella Braenderup | 3 (2.75) | - | 3 (0.94) | |
| Salmonella Cayar | 1 (0.92) | - | 1 (0.31) | |
| Salmonella Chomedey | 1 (0.92) | - | 1 (0.31) | |
| Salmonella Corvallis | 3 (2.75) | - | 3 (0.94) | |
| Salmonella Cremieu | 2 (1.83) | - | 2 (0.63) | |
| Salmonella Dabon | 1 (0.92) | - | 1 (0.31) | |
| Salmonella Cyprus | 1 (0.92) | - | 1 (0.31) | |
| Salmonella Istanbul | 1 (0.92) | - | 1 (0.31) | |
| Salmonella Kentucky | 25 (22.94) | - | 25 (7.81) | |
| Salmonella Litchfield | 1 (0.92) | - | 1 (0.31) | |
| Salmonella Molade | 1 (0.92) | - | 1 (0.31) | |
| Salmonella Newport | 1 (0.92) | - | 1 (0.31) | |
| Salmonella Rissen | - | 78 (36.97) | 78 (36.97) | |
| Salmonella Saint pant | 1 (0.92) | - | - | |
| Salmonella Singapore | 4 (3.67) | - | - | |
| Salmonella Stuttgart | 1 (0.92) | - | - | |
| Salmonella Mbandaka | 6 (5.50) | - | - | |
| Salmonella Wippra | 2 (1.83) | - | - | |
| D | Salmonella Enteritidis | 3 (2.75) | - | 3 (0.94) |
| Salmonella Lome | - | 3 (1.42) | 3 (0.94) | |
| Salmonella Panama | - | 5 (2.37) | 5 (1.56) | |
| Salmonella Powell | - | 1 (0.47) | 1 (0.31) | |
| Salmonella Victoria | - | 1 (0.47) | 1 (0.31) | |
| E | Salmonella Assinie | - | 2 (0.95) | 2 (0.63) |
| Salmonella Biafra | - | 1 (0.47) | 1 (0.31) | |
| Salmonella Give | 22 (20.18) | 1 (0.47) | 23 (7.19) | |
| Salmonella Fulda | - | 6 (2.84) | 6 (1.88) | |
| Salmonella Ugor | 1 (0.92) | - | 1 (0.31) | |
| Salmonella Weltevreden | - | 31 (14.70) | 31 (9.69) | |
| G | Salmonella Kedougou | - | 5 (2.37) | 5 (1.56) |
| I | Salmonella Hvittingfoss | - | 2 (0.95) | 2 (0.63) |
| O | Salmonella Alachua | 2 (1.83) | - | 2 (0.63) |
| Total | 109 (100) | 211 (100) | 320 (100) |
Antimicrobial susceptibility
Among broiler isolates positive for Salmonella, 64.22% were resistant to at least one antimicrobial agent and 28.44% were multidrug-resistant (MDR) (resistant to three or more antimicrobial agents). Antimicrobial resistance of broiler isolates was most commonly observed against NAL (49.54%), followed by AMP (30.28%), TET (27.52%), amoxicillin (26.61%), CIP (23.85%), NOR (19.27%), CHL (4.59%), SXT (4.59%), and CAZ (1.83%). Among pig isolates, 74.88% were resistant to at least one antimicrobial agent and 38.39% were MDR. All isolates were susceptible to NOR. Antimicrobial resistance in pig Salmonella isolates was most commonly observed against AMP (69.05%) and TET (66.19%), as shown in Table-2. There was no significant difference (p=0.077) in the prevalence of MDR in Salmonella isolates from broilers and pigs. The antimicrobial resistance rate for amoxicillin, CIP, NAL, and NOR in Salmonella isolates from broilers was significantly higher than that for Salmonella isolates from pigs (p<0.001), and the resistance of pig isolates to AMP, SXT, and TET was significantly higher than that for broiler isolates (p<0.001). In addition, there was no significant difference in the prevalence of antimicrobial resistance against CHL (p=0.155) and CAZ (p=0.447) between broiler and pig Salmonella isolates.
Table 2.
Salmonella antimicrobial resistance percentages in broiler and pig isolates from Thailand.
| Samples | Amount | Antimicrobial resistance agents (%) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| AMC | AMP | CHL | CAZ | CIP | NAL | NOR | SXT | TET | ||
| Broilers | 109 | 29 (26.60) | 33 (30.27) | 5 (4.58) | 2 (1.83) | 26 (23.85) | 54 (49.54) | 21 (19.26) | 5 (4.58) | 30 (27.52) |
| Pigs | 210 | 2 (0.95) | 145(69.04) | 19 (9.04) | 7 (3.33) | 2 (0.95) | 5 (2.38) | 0 | 75 (35.71) | 139 (66.19) |
*AMP=Ampicillin, AMC=Amoxicillin/clavulanate, CHL=Chloramphenicol, CAZ=Ceftazidime, CIP=Ciprofloxacin, NAL=Nalidixic acid, NOR=Norfloxacin, SXT=Trimethoprim/sulfamethoxazole, TET=Tetracycline
Thirty-one antimicrobial resistance patterns were identified from the 320 Salmonella positive isolates, as shown in Table-3. The most common antimicrobial resistance patterns were NAL (21.10%) and AMC/AMP/CIP/NAL/NOR/TET (16.51%) in broiler isolates and AMP/SXT/TET (28.91%) and AMP/TET (25.12%) in pig isolates.
Table 3.
Salmonella antimicrobial resistance pattern in broiler and pig isolates from Thailand.
| Antimicrobial resistance pattern | Number of isolates (%) | |
|---|---|---|
| Broilers (n=109) | Pigs (n=211) | |
| AMP | 14 (6.63) | |
| CHL | 4 (3.67) | |
| CAZ | 2 (0.95) | |
| CIP | 1 (0.48) | |
| NAL | 23 (21.10) | 1 (0.48) |
| SXT | 1 (0.92) | |
| TET | 2 (1.84) | 5 (2.37) |
| AMP- NAL | 4 (3.67) | |
| AMP-TET | 53 (25.12) | |
| CHL-SXT | 1 (0.92) | |
| CIP-NAL | 4 (3.67) | |
| SXT-TET | 1 (0.48) | |
| AMC-AMP-TET | 5 (4.59) | |
| AMP-CHL-TET | 2 (0.95) | |
| AMP-SXT-TET | 61 (28.91) | |
| CHL-NAL-TET | 1 (0.48) | |
| CHL-SXT-TET | 1 (0.48) | |
| AMC-AMP-CAZ-CIP | 1 (0.92) | |
| AMC-AMP-CIP-NAL | 1 (0.92) | |
| AMC-AMP-SXT-TET | 2 (1.84) | 1 (0.48) |
| AMP-CHL-CAZ-TET | 2 (0.95) | |
| AMP-CHL-SXT-TET | 9 (4.27) | |
| AMP-CIP-NAL-TET | 1 (0.48) | |
| CAZ-CIP-NAL-NOR | 1 (0.92) | |
| AMC-CIP-NAL-NOR-TET | 1 (0.92) | |
| AMP-CHL-CAZ-NAL-TET | 1 (0.48) | |
| AMP-CHL-CAZ-SXT-TET | 1 (0.48) | |
| AMP-CIP-NAL-NOR-TET | 1 (0.92) | |
| AMC-AMP-NAL-SXT-TET | 1 (0.92) | |
| AMC-AMP-CHL-NAL-SXT-TET | 1 (0.48) | |
| AMC-AMP-CIP-NAL-NOR-TET | 18 (16.51) | |
| Susceptible to all | 39 (35.78) | 53 (25.12) |
AMP=Ampicillin, CHL=Chloramphenicol, CAZ=Ceftazidime, CIP=Ciprofloxacin, NAL=Nalidixic acid, SXT=Trimethoprim/sulfamethoxazole, TET=Tetracycline, AMC=Amoxicillin/clavulanate, NOR=Norfloxacin
PFGE profiles
The PFGE dendrogram of 52 S. Typhimurium isolates from broilers (n=7) and pigs (n=45) with 85% similarity valuation is shown in Figure-1. Nine clusters (A-I) with 21 PFGE patterns were created. Cluster A was the predominant group, containing PFGE patterns with three broiler isolates collected from Chiang Mai and 17 pig isolates from Khon Kaen, Chiang Mai, Chiang Rai, Phayao, and Mae Hong Son. Cluster B consisted of three PFGE patterns from 12 pig isolates collected from Chiang Rai. Cluster C had four PFGE patterns from two broiler isolates from Chiang Mai and Chiang Rai and nine pig isolates from Chiang Mai and Chiang Rai. Cluster D contained one PFGE pattern from two broiler isolates (Sa Kaeo), and Clusters E, F, and H each had one PFGE pattern from one pig isolate (Chiang Mai). Cluster G had one PFGE pattern from three pig isolates from Chiang Mai. Cluster I had one PFGE pattern from a pig isolate from Chiang Rai.
Figure-1.
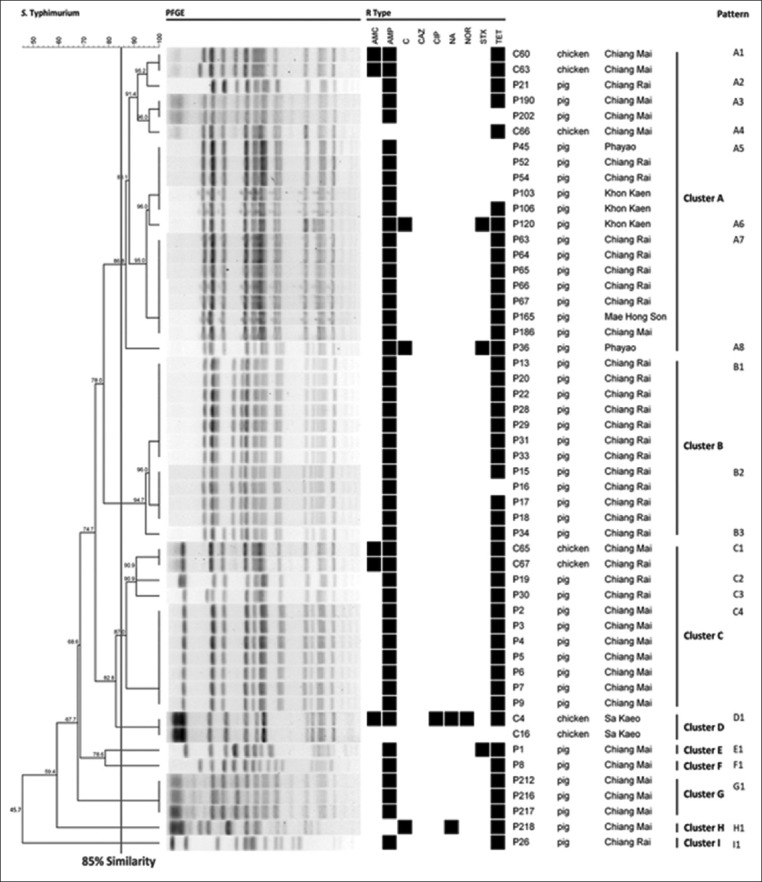
Dendrogram of 52 PFGE-Xbal profiles of Salmonella Typhimurium isolated from broilers and pigs at slaughterhouses in Thailand (n=Resistance, o=Susceptibility).
The group of indistinguishable isolates from different provinces revealed that 80% of the isolates shared a similar antimicrobial resistance pattern (A5 pattern) composed of pig isolates from the northeastern region (Khon Kaen) and the north region (Chiang Rai and Phayao). Similar antimicrobial resistance patterns for the A7 pattern composed of pig isolates from Chiang Rai, Chiang Mai, and Mae Hong Son, and the C1 pattern of broiler isolates from Chiang Mai and Chiang Rai were also observed.
Discussion
The prevalence of Salmonella spp. in pig isolates (37.54%) was significantly higher (p<0.001) than in broiler isolates (18.05%). The prevalence of Salmonella in broilers was lower than that reported by previous studies for Northern Thailand [24], Southern Thailand [9], Bangkok, and Central Thailand [25], but it was higher than that reported for Chiang Mai [5] and Sa Kaeo [26]. The prevalence of Salmonella spp. in pig isolates in this study was lower than that reported for Chiang Mai, Chiang Mai’s surrounding areas [11], and Southern Thailand [9]. However, the prevalence of Salmonella in pig isolates was higher than that reported for Sa Kaeo [15,26] and Central Thailand [25]. In this study, the prevalence of Salmonella spp. in broilers and pigs was higher than that reported for broilers and pigs from European countries: 2.35% in Belgium, 1.56% in Italy, 0% in Poland, and 11.72% in Spain from pig carcasses, and 0.26% in Belgium, 0.01% in Italy, 0.08% in Poland, and 0.06% in Spain for broiler flocks reported by food business operator [27]. As the Salmonella prevalence control and reduction program have not been established for broilers and pigs in Thailand, these results indicate a noticeable difference between Thailand and Europe with respect to Salmonella infection in food animals. This is a significant issue that Thailand should address as soon as possible. The most common serovars in broiler isolates were S. Kentucky, S. Give, and S. Typhimurium. This finding was inconsistent with the findings of Padungtod and Kaneene [7], who reported that the most common serovars in Thailand’s chickens were S. Weltevreden and S. Rissen. Other studies reported that S. Corvallis and S. Rissen were the most common serovars in chicken [5]. Several studies in European countries also reported that S. Enteritidis and S. Typhimurium were the most common serovars in broilers [28-30], while S. Rissen, S. Typhimurium, and S. Weltevreden were the most common in Salmonella isolates from pigs. These findings agree with several Thai studies which reported that S. Rissen was the most common serovar in pigs [8,9,11,19]. They are also consistent with reports from European countries that the most common serovar in pig was S. Typhimurium [31-33]. Another study in Thailand indicated that S. Weltevreden and S. Dumfries were the most common serovars in swine samples from Sa Kaeo [15]. These results revealed the variation of Salmonella spp. serovars and prevalence based on location and animal species [7].
The prevalence of MDR between broilers (28.44%) and pigs (38.39%) was not significantly different (p=0.077) in this study. However, the prevalence of MDR was high compared to other European studies, 19.70% in broiler flocks from Spain [34] and 19.30% in pigs from Denmark [33]. To deal with this issue, Thailand should adopt a more restrictive policy toward antimicrobial use in food animal production. The most common MDR pattern in the two species was different: AMC/AMP/CIP/NAL/NOR/TET for broilers and AMP/SXT/TET for pigs. Furthermore, the resistance prevalence of some antimicrobial agents differed between broiler and pig isolates. Resistance to antimicrobial agents (amoxicillin, CIP, NAL, and NOR) was higher in broiler isolates than pig isolates, but resistance to antimicrobial agents (AMP, SXT, and TET) was higher in pig isolates than broiler isolates. These differences indicated a potential difference in the use and frequency of antimicrobial agents in the broiler industry compared to the pig industry. There are few reports on antimicrobial use in food animal production in Thailand, which report the common use of amoxicillin, colistin, doxycycline, oxytetracycline, and tilmicosin in Thai poultry farms [35]. The top ten most commonly used veterinary antimicrobials in food producing animals (pigs, poultry, cattle, and aquatic animals) were found to be amoxicillin, halquinol, chlortetracycline, tiamulin, doxycycline, sulfadimidine, colistin, tilmicosin, gentamicin, and tylosin [36]. These results are consistent with the findings of our study regarding the most common antimicrobial resistance in broiler (NAL, AMP, TET, and amoxicillin) and pig isolates (AMP, TET, and SXT). The inconsistent results can be explained by the fact that the use of antimicrobial agents in Thai food animal production varies depending on the level and farm type [37]. The fluoroquinolone group is the recommended antimicrobial agent for treating severe and multidrug-resistant salmonellosis both in humans and animals [38,39]. The previous studies in Thailand reported that Salmonella isolates from poultry were still susceptible or resistant at a lower proportion to CIP and NOR [3,5,7,9,24,26]. However, the prevalence of broiler isolates resistant to CIP and NOR was higher here than in previous studies. The higher prevalence of resistance indicated a possible decrease in fluoroquinolone susceptibility, which may reflect an association between the high proportion of resistance to NAL (49.50%), CIP, and NOR in broiler isolates [40,41]. The NAL and fluoroquinolone-resistant Salmonella spp. can lead to severe Salmonella infections, treatment failure, or extended hospital stays for patients with multidrug-resistant salmonellosis compared to patients with susceptible Salmonella spp. [42].
The high genetic relatedness (85% similarity) in Clusters A and C between these two animal species and the results of PFGE of 52 S. Typhimurium isolates from broilers and pigs indicated high genetic relatedness between broiler and pig isolates. The identical PFGE patterns (A5 and A7) of pig isolates were collected from various provinces. Most of the isolates in each identical PFGE pattern had the same antimicrobial resistance pattern, except for P106 in A5, which revealed cross-contamination in pig isolates from Khon Kaen, Chiang Rai, and Phayao (A5), and Chiang Rai, Chiang Mai, and Mae Hong Son (A7). The cross-contamination could be explained by the following: Worker’s hands that contaminated by Salmonella, contaminated transport cages, personnel moving among the provinces, or flies as a transmission vector [43]. However, the high genetic relatedness between broiler and pig isolates should be further investigated to develop a deeper understanding of the epidemiological characteristics and to prevent further cross-contamination between these two animal species.
Conclusion
This study revealed the prevalence of Salmonella spp. in pigs and broilers from multiple provinces of Thailand. Moreover, the problem of MDR Salmonella spp. and the increasing resistance to CIP and NOR compared to previous studies should be noted as a concerning public health issue. We also revealed high genetic relatedness between Salmonella isolates from broilers and pigs and cross-contamination in pig isolates across different provinces. These findings should be investigated further to gain a better understanding of the epidemiological characteristics and to prevent cross-contamination in food animal productions. We also obtained noticeably different prevalence and MDR resistance results in our study compared to those from European countries, which already has established control and reduction programs for Salmonella in poultry and pig production. We also conclude that Thailand’s government should implement new policies such as control and reduction programs for Salmonella to control and reduce the prevalence of Salmonella and MDR resistance issues in Thailand.
Authors’ Contributions
SA conceived and designed the experiments, provided materials and reagent. SA, DP, and SK collected samples. DP performed the experiments, analyzed data and wrote the manuscript. All authors read and approved the final manuscript.
Acknowledgments
The authors are thankful to Dr. Rungtip Chuanchuen and staff of the Center for Antimicrobial Resistance in Foodborne Pathogens, Faculty of Veterinary Medicine, Chulalongkorn University, for their assistance in Salmonella serotyping; Dr. Prapas Patchanee and staff from Faculty of Veterinary Medicine, Chiangmai University, for assistance in the analysis of PFGE profiles; the staff from the Veterinary Diagnostic Laboratory Research and Academic Service Unit, KKU Animal Hospital, for laboratory assistance; and Dr. Royce Tsukayama for manuscript editing and Editage (www.editage.com) for English language editing. This study was supported by graduate school Khon Kaen University (Grant no. 591JH105), Thailand.
Competing Interests
The authors declare that they have no competing interests.
Publisher’s Note
Veterinary World remains neutral with regard to jurisdictional claims in published institutional affiliation.
References
- 1.Uyttendaele M.R, Debevere J.M, Lips R.M, Neyts K.D. Prevalence of Salmonella in poultry carcasses and their products in Belgium. Int. J. Food Microbiol. 1998;40(1-2):1–8. doi: 10.1016/s0168-1605(98)00012-9. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization. Salmonella (non-typhoidal) 2018. [Last accessed on 10-01-2019]. Available from: https://www.who.int/news-room/fact-sheets/detail/salmonella-(non-typhoidal)
- 3.Angkititrakul S, Chomvarin C, Chaita T, Kanistanon K, Waethewutajarn S. Epidemiology of antimicrobial resistance in Salmonella isolated from pork, chicken meat and humans in Thailand. Southeast Asian J. Trop. Med. Public Health. 2005;36(6):1510–1515. [PubMed] [Google Scholar]
- 4.Vadhanasin S, Bangtrakulnonth A, Chidkrau T. Critical control points for monitoring Salmonella reduction in Thai commercial frozen broiler processing. J. Food Prot. 2004;67(7):1480–1483. doi: 10.4315/0362-028x-67.7.1480. [DOI] [PubMed] [Google Scholar]
- 5.Chotinun S, Rojanasthien S, Unger F, Tadee P, Patchanee P. Prevalence and antimicrobial resistance of Salmonella isolated from carcasses, processing facilities and the environment surrounding small scale poultry slaughterhouses in Thailand. Southeast Asian J. Trop. Med. Public Health. 2014;45(6):1392–1400. [PubMed] [Google Scholar]
- 6.Na Lampang K, Chailangkarn S, Padundtod P. Prevalence and antimicrobial resistance of Salmonella serovars in chicken farm, Chiang Mai and Lamphun province, Northern of Thailand. Chiang Mai Vet. J. 2013;12(2):85–93. [Google Scholar]
- 7.Padungtod P, Kaneene J.B. Salmonella in food animals and humans in northern Thailand. Int. J. Food Microbiol. 2006;108(3):346–354. doi: 10.1016/j.ijfoodmicro.2005.11.020. [DOI] [PubMed] [Google Scholar]
- 8.Hendriksen R.S, Bangtrakulnonth A, Pulsrikarn C, Pornreongwong S, Hasman H, Song S.W, Aarestrup F.M. Antimicrobial resistance and molecular epidemiology of Salmonella Rissen from animals, food products, and patients in Thailand and Denmark. Foodborne Pathog. Dis. 2008;5(5):605–619. doi: 10.1089/fpd.2007.0075. [DOI] [PubMed] [Google Scholar]
- 9.Lertworapreecha M, Sutthimusik S, Tontikapong K. Antimicrobial resistance in Salmonella enterica isolated from pork, chicken, and vegetables in Southern Thailand. Jundishapur J. Microbiol. 2012;6(1):36–41. [Google Scholar]
- 10.Tadee P, Boonkhot P, Patchanee P. Quantification of contamination levels and particular risk of Salmonella spp. in pigs in slaughterhouses in Chiang Mai and Lamphun provinces, Thailand. Jpn. J. Vet. Res. 2014;62(4):171–179. [PubMed] [Google Scholar]
- 11.Patchanee P, Tansiricharoenkul K, Buawiratlert T, Wiratsudakul A, Angchokchatchawal K, Yamsakul P, Yano T, Boonkhot P, Rojanasatien S, Tadee P. Salmonella in pork retail outlets and dissemination of its pulsotypes through pig production chain in Chiang Mai and surrounding areas, Thailand. Prev. Vet. Med. 2016;130:99–105. doi: 10.1016/j.prevetmed.2016.06.013. [DOI] [PubMed] [Google Scholar]
- 12.Bush L.M, Perez M.T. Overview of Salmonella Infections. 2018. [Last accessed on 22-02-2019]. Available from: https://www.msdmanuals.com/professional/infectious-disease/gram-negative-bacilli/overview-of-salmonella-infections .
- 13.Inatsu Y, Hosotani Y, Kawasaki S, Ananchaipattana C. Detection and Characterization of Salmonella spp. in Various Kinds of Food Sold in Bangkok, Thailand. Proceeding of International Symposium on Agri-Foods for Health and Wealth (AFHW) 2013:321–327. [Google Scholar]
- 14.Khemtong S, Chuanchuen R. Class 1 integrons and Salmonella genomic island 1 among Salmonella enterica isolated from poultry and swine. Microb. Drug Resist. 2008;14(1):65–70. doi: 10.1089/mdr.2008.0807. [DOI] [PubMed] [Google Scholar]
- 15.Pulsrikarn C, Chaichana P, Pornruangwong S, Morita Y, Yamamoto S, Boonmar S. Serotype, antimicrobial susceptibility, and genotype of Salmonella isolates from swine and pork in Sa Kaew province, Thailand. Thai Vet. Med. 2012;42(1):21–27. [Google Scholar]
- 16.Wannaprasat W, Padungtod P, Chuanchuen R. Class 1 integrons and virulence genes in Salmonella enterica isolates from pork and humans. Int. J. Antimicrob. Agents. 2011;37(5):457–461. doi: 10.1016/j.ijantimicag.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 17.Torpdahl M, Skov M.N, Sandvang D, Baggesen D.L. Genotypic characterization of Salmonella by multilocus sequence typing, pulsed-field gel electrophoresis and amplified fragment length polymorphism. J. Microbiol. Methods. 2005;63(2):173–184. doi: 10.1016/j.mimet.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 18.Pornruangwong S, Sriyapai T, Pulsrikarn C, Sawanpanyalert P, Boonmar S, Bangtrakulnonth A. The epidemiological relationship between Salmonella enterica serovar Typhimurium and Salmonella enterica serovar 4,[5],12:i:- isolates from humans and swine in Thailand. Southeast Asian J. Trop. Med. Public Health. 2008;39(2):288–296. [PubMed] [Google Scholar]
- 19.Tadee P, Boonkhot P, Pornruangwong S, Patchanee P. Comparative phenotypic and genotypic characterization of Salmonella spp. in pig farms and slaughterhouses in two provinces in Northern Thailand. PLoS One. 2015;10(2):e0116581. doi: 10.1371/journal.pone.0116581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.International Organization for Standardization. Microbiology of the Food Chain —Horizontal Method for the Detection, Enumeration and Serotyping of Salmonella. Part 1:Detection of Salmonella spp. 2017. [Last accessed on 08-07-2019]. Available from: https://www.iso.org/standard/56712.html .
- 21.Popoff M.Y, Le Minor L. Antigenic Formulas of the Salmonella Serovars. 8th ed. France: WHO Collaborating Centre for Reference and Research on Salmonella Pasteur Institute; 2007. [Google Scholar]
- 22.Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing. 27th edition. Wayne, PA, USA: Information Supplement. Clinical and Laboratory Standards Institute; 2017. [Google Scholar]
- 23.Centers for Disease Control and Prevention. Standard Operating Procedure for PulseNet PFGE of Escherichia coli O157:H7Escherichia coli non-O157 (STEC)Salmonella serotypes Shigella sonnei and Shigella flexneri. 2017. [Last accessed on 19-10-2018]. Available from: https://www.cdc.gov/pulsenet/pdf/ecoli-shigella-salmonella-pfge-protocol-508c.pdf .
- 24.Hanson R, Kaneene J.B, Padungtod P, Hirokawa K, Zeno C. Prevalence of Salmonella and E. coli and their resistance to antimicrobial agents, in farming communities in Northern Thailand. Southeast Asian J. Trop. Med Public Health. 2002;33(3):120–126. [PubMed] [Google Scholar]
- 25.Niyomdecha N, Mungkornkaew N, Samornsuk W. Serotypes and antimicrobial resistance of Salmonella enterica isolated from pork, chicken meat and lettuce, Bangkok and central Thailand. Southeast Asian J. Trop. Med. Public Health. 2016;47(1):31–39. [PubMed] [Google Scholar]
- 26.Trongjit S, Angkititrakul S, Tuttle R.E, Poungseree J, Padungtod P, Chuanchuen R. Prevalence and antimicrobial resistance in Salmonella enterica isolated from broiler chickens, pigs and meat products in Thailand-Cambodia border provinces. Microbiol. Immunol. 2017;61(1):23–33. doi: 10.1111/1348-0421.12462. [DOI] [PubMed] [Google Scholar]
- 27.EFSA and ECDC (European Food Safety Authority and European Centre for Disease Prevention and Control) The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2017. EFSA J. 2018;16(12):5500. doi: 10.2903/j.efsa.2018.5500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Witkowska D, Kuncewicz M, Żebrowska J.P, Sobczak J, Sowińska J. Prevalence of Salmonella spp. in broiler chicken flocks in Northern Poland in 2014-2016. Ann. Agric. Environ. Med. 2018;25(4):693–697. doi: 10.26444/aaem/99528. [DOI] [PubMed] [Google Scholar]
- 29.Marin C, Balasch S, Vega S, Lainez M. Sources of Salmonella contamination during broiler production in Eastern Spain. Prev. Vet. Med. 2011;98(1):39–45. doi: 10.1016/j.prevetmed.2010.09.006. [DOI] [PubMed] [Google Scholar]
- 30.Pieskus J, Maria F, Patrizia P, Felix R, Kazeniauskas E, Ambrozeviciene C.B, Mykolas M, Bolder N. Preliminary investigations on Salmonella spp. incidence in meat chicken farms in Italy, Germany, Lithuania and the Netherlands. Int. J. Poult. Sci. 2008;7(8):813–817. [Google Scholar]
- 31.Arguello H, Carvajal A, Collazos J.A, García-Feliz C, Rubio P. Prevalence and serovars of Salmonella enterica on pig carcasses, slaughtered pigs and the environment of four Spanish slaughterhouses. Food Res. Int. 2012;45(2):905–912. [Google Scholar]
- 32.Rowe T.A, Leonard F.C, Kelly G, Lynch P.B, Egan J, Quirke A.M, Quinn P.J. Salmonella serotypes present on a sample of Irish pig farms. Vet. Rec. 2003;153(15):453–456. doi: 10.1136/vr.153.15.453. [DOI] [PubMed] [Google Scholar]
- 33.Arguello H, Sorensen G, Carvajal A, Baggesen D.L, Rubio P, Pedersen K. Prevalence, serotypes and resistance patterns of Salmonella in Danish pig production. Res. Vet. Sci. 2013;95(2):334–342. doi: 10.1016/j.rvsc.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 34.Lamas A, Fernandez-No I.C, Miranda J.M, Vázquez B, Cepeda A, Franco C.M. Prevalence, molecular characterization and antimicrobial resistance of Salmonella serovars isolated from Northwestern Spanish broiler flocks (2011-2015) Poult. Sci. 2016;95(9):2097–2105. doi: 10.3382/ps/pew150. [DOI] [PubMed] [Google Scholar]
- 35.Wongsuvan G, Wuthiekanun V, Hinjoy S, Day N.P, Limmathurotsakul D. Antibiotic use in poultry:A survey of eight farms in Thailand. Bull. World Health Organ. 2018;96(2):94–100. doi: 10.2471/BLT.17.195834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thai Working Group on Health Policy and Systems Research on Antimicrobial Resistance (HPSR-AMR) Consumption of antimicrobial agents in Thailand in 2017. First Report. Thai Working Group on Health Policy and Systems Research on Antimicrobial Resistance (HPSR-AMR) 2018 [Google Scholar]
- 37.Archawakulathep A, Kim C.T.T, Meunsene D, Handijatno D, Hassim H, Rovira H, Myint K, Baldrias L, Sothy M, Aung M, Wahyu N, Chea R, Boonmasawai S, Vannamahaxay S, Angkititrakul S, Collantes T, Van T.N, Punyapornwithaya V, Zakaria Z, Chuanchuen R. Perspectives on antimicrobial resistance in livestock and livestock products in ASEAN countries. Thai J. Vet. Med. 2014;44(1):5–13. [Google Scholar]
- 38.Archambault M, Petrov P, Hendriksen R.S, Asseva G, Bangtrakulnonth A, Hasman H, Aarestrup F.M. Molecular characterization and occurrence of extended-spectrum beta-lactamase resistance genes among Salmonella enterica serovar Corvallis from Thailand, Bulgaria, and Denmark. Microb. Drug Resist. 2006;12(3):192–198. doi: 10.1089/mdr.2006.12.192. [DOI] [PubMed] [Google Scholar]
- 39.Dalhoff A. Global fluoroquinolone resistance epidemiology and implictions for clinical use. Interdiscip Perspect Infect. Dis. 2012;2012:976273. doi: 10.1155/2012/976273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kownhar H, Shankar E.M, Rajan R, Rao U.A. Emergence of nalidixic acid-resistant Salmonella enterica serovar Typhi resistant to ciprofloxacin in India. J. Med. Microbiol. 2007;56(1):136–137. doi: 10.1099/jmm.0.46763-0. [DOI] [PubMed] [Google Scholar]
- 41.Michael P.R, Dillon C, Catherine C.A. Nalidixic acid-resistant strains of Salmonella showing decreased susceptibility to fluoroquinolones in the Midwestern region of the Republic of Ireland due to mutations in the gyrA Gene. J. Clin. Microbiol. 2011;49(5):2077–2079. doi: 10.1128/JCM.02574-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Helms M, Simonsen J, Molbak K. Quinolone resistance is associated with increased risk of invasive illness or death during infection with Salmonella serotype Typhimurium. J. Infect. Dis. 2004;190(9):1652–1654. doi: 10.1086/424570. [DOI] [PubMed] [Google Scholar]
- 43.Boonprasert N, Nuanualsuwan S, Pulsrikarn C, Pornaem S, Chokesajjawatee N. Sources and dissemination of Salmonella spp. in an integrated broiler meat production. Thai J. Vet. Med. 2014;44(1):117–124. [Google Scholar]


