Abstract
The present study applied a circular RNA (circRNA) microarray to examine the circRNA expression profiles in the glomeruli of NZB/W F1 mice with lupus nephritis (LN) during the pathogenesis of the disease. Glomeruli from two groups of female NZB/W F1 mice of the same age with either severe or mild LN were isolated by perfusion with dynabeads. A microarray analysis was then performed to evaluate the differentially expressed circRNAs of the glomeruli in the two groups, which were then confirmed by reverse transcription-quantitative PCR (RT-qPCR) assays. In addition, using a biomathematical strategy, the differentially-expressed circRNAs were identified in severe LN when compared with mild LN, and the commonly expressed circRNA species among these profiles were optimized via competing endogenous RNA (ceRNA) analysis. The predicted microRNAs (miRNAs/miRs) as downstream targets of circRNAs and upstream regulators of mRNAs were verified by RT-qPCR and the final circRNA-miRNA-mRNA network was constructed to identify the circRNA that was a pathogenic link in LN. The present study obtained 116 differentially expressed circRNAs, including 41 up- and 75 downregulated circRNAs, in severe LN when compared with mild LN, and 12 circRNAs were confirmed by RT-qPCR. The most significant difference was in the expression of mmu_circRNA_34428 (P<0.001) when comparing severe and mild LN glomeruli. A network of mmu_circRNA_34428-targeted miRNA-gene interactions was subsequently constructed, including miR-338-3p, miR-670-3p, miR-3066-5p, miR-210-5p and their corresponding mRNA targets. To the best of our knowledge, the present study elucidated, for the first time, circRNA profiling and the circRNA-miRNA interactions in the development of LN in female NZB/W F1 mice. The results revealed that mmu_circRNA_34428 could serve an important role in LN progression; however, the present study did not elucidate the functions of this circRNA or others in LN progression.
Keywords: lupus nephritis, circular RNA, NZB/W F1 mice, biomathematics, competitive endogenous RNA
Introduction
Systemic lupus erythematosus (SLE) is a systemic autoimmune disease characterized by the appearance of the autoantibodies against several nuclear components. The deposition of the formed immune complexes mediates the disease in a wide variety of tissues and organs, including the kidneys, and multiple organ systems. Lupus nephritis (LN) is one of the most common complications in patients with SLE and influences the overall outcome of these patients (1). Estimates of lupus nephritis (LN) incidence among SLE patients during the first 10 years ranges from 35 to 60% (2). It has been reported that two-thirds of patients with SLE have renal disease at some stage, which is the leading cause of mortality in these patients (3). Renal manifestations of lupus vary from asymptomatic urinary abnormalities to rapidly progressive crescentic glomerulonephritis, causing end-stage renal disease (ESRD). However, the pathogenesis of LN remains unclear. Although multiple factors are known to participate in LN development, including genetic susceptibility (4), epigenetic regulation, sex and environmental interactions (5), the present study sought to identify the specific clinical diagnostic markers and therapeutic targets.
Circular RNAs (circRNAs) are RNA molecules with covalently joined 3′ and 5′ ends formed by back-splice events, thereby presenting as covalently closed continuous loops. CircRNAs have been demonstrated to cause loss of microRNA (miRNA/miR) function accompanied by increased levels of endogenous targets, acting as miRNA sponges (6). A number of circRNAs that universally exist in a variety of biological cells are abundant, stable (7) conserved, cell-type specific (8), tissue-specific (9) and potentially function as competing endogenous (ce)-RNAs. Therefore, circRNAs are becoming important biological molecules to understand the molecular mechanisms underlying the disease and for investigating biomarkers for disease diagnosis and targeting treatment. However, the role of circRNAs in glomerular gene expression and their effects on LN remain unknown.
Recent studies have reported and emphasized the importance of circRNAs in regulating immunosenescence- associated immunocytes (10–12). Thus far, little is known about the expression and function of circRNAs in LN. Therefore, the present study, to the best of our knowledge, investigated for the first time the comprehensive expression profile of circRNAs in the glomeruli of LN. In SLE-prone (NZB/W) F1 mice, the systemic bioinformatics analysis was performed to identify circRNAs that are essential for the biological processes of LN, which may provide potential targets for the development of novel diagnostic and therapeutic strategies for LN.
Materials and methods
Mice
Female 12-week-old NZB/W F1 mice (n=30) purchased from the Jackson Laboratory Animal Center (Jackson, MS, USA) were maintained in the Chinese Medical University Animal Laboratory (Liaoning, China). The mice had free access to food and water throughout the experimental period under controlled conditions (humidity, 45±2%; temperature, 22±1°C), and a 12-h light/dark artificial cycle in accordance with the guidelines of the Chinese National Standard (GB 14925–2001). The present study complied with the protocols approved by the Institutional Animal Care and Use Committee at China Medical University. All animals were housed in specific pathogen-free conditions until LN diagnosis, and after 16 weeks, mice were intraperitoneally anesthetized with 1% sodium pentobarbital (40 mg/kg; Beijing Solarbio Science &Technology Co, Ltd.) and sacrificed by exsanguination via the aorta pectoralis.
Mouse groupings
The onset of renal disease was monitored by weekly testing of fresh urine specimens using protein test strips to test for proteinuria (±: 10 mg/dl; +: 30 mg/dl; 2+: 100 mg/dl; 3+, 300 mg/dl; 4+: >1,000 mg/dl). Mice with a proteinuria level ≥300 mg/dl in repeated tests were considered to have ‘severe’ LN, as previously described (13,14). At the age of 28 weeks, the mice were divided into two groups based on their degree of renal disease: Mild LN (pro ±-1+) and severe LN (pro 3+-4+). The body weight of the mice at 28 weeks of age were as follows: Mild LN, 36.433±2.332 g, n=5; and severe LN, 34.933±3.158 g; n=5. Data are expressed as the mean ± standard error of the mean. The remaining 20 animals were used for reverse transcription-quantitative PCR (RT-qPCR) to verify the target circRNA of triple for each group [n=9 (pro±~ +1+); n=9 (pro3+~4+); experiments were repeated three times]. Euthanasia was intended via perfusion; however, 2 mice succumbed to anesthetic due to accidental exsanguination.
Isolation of kidney glomeruli
Glomeruli were isolated through the abdominal aorta as previously described (15), alongside some improvements to achieve the experimental conditions required. Glomeruli were isolated from kidneys perfused via the aorta pectoralis with magnetic Dynabeads (Invitrogen; Thermo Fisher Scientific, Inc.) at 28 weeks of age. First, kidneys were perfused with ice-cold phosphate buffer saline (PBS) to remove any blood and then Dynabeads (diameter, 4.5 µm) (4×106/ml PBS, 25 ml/mice) were perfused into kidneys at a constant rate of 7.4 ml/min/g per kidney. Following the removal, mincing, digestion and filtration of tissue, the cell suspension was obtained and then centrifuged at 200 × g for 5 min in 4°C. Once the supernatant was discarded, the cell pellet was dissolved in 2 ml PBS. Finally, a magnetic particle concentrator (Invitrogen; Thermo Fisher Scientific, Inc.) was used to collect the glomeruli containing Dynabeads; the glomerular RNA was isolated using TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.). Following sacrifice, other samples, such as urine, brain, liver, blood, heart, muscle and lung, were obtained from the animals for future analysis in other studies.
Histological determination of LN
Renal tissue from the severe and mild LN mice, and Dynabead-perfused kidneys were fixed with Formalin-Aceto-Alcohol at room temperature, dehydrated with alcohol and embedded in paraffin for sectioning. Paraformaldehyde-fixed paraffin-embedded histological kidney sections (4-µm-thick) were stained with hematoxylin and eosin (H&E), Masson's trichrome, periodic acid-Schiff and periodic Schiff-methenamine to evaluate disease development by light microscopy. The severe and mild LN groups were diagnosed according to kidney pathology.
RNA extraction
Total RNA was isolated using TRIzol reagent according to the manufacturer's protocol. cDNA for RT-qPCR analysis was synthesized from 1 µg of total RNA. Total RNA was quantified using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Inc.). The RNA integrity and genomic DNA contamination of each sample was assessed by denaturing agarose gel electrophoresis, as previously descried (16,17).
Labeling and hybridization
Sample labeling and array hybridization were performed according to the manufacturers' protocols as described below. To enrich circRNAs, linear RNAs were removed using Rnase R (Epicentre; Illumina, Inc.) to digest total RNAs. Each sample of enriched circRNAs was then amplified and transcribed into fluorescent cRNA using the treating random primers method (Arraystar Super RNA Labeling kit; Arraystar, Inc.). The labeled cRNAs were purified using the RNeasy Mini kit (Qiagen, Inc.). The concentration and specific activity of the labeled cRNAs (pmol Cy3/µg cRNA) were measured using a NanoDrop ND-1000. A total of 1 µg of each labeled cRNA was fragmented by adding 5 µl 10X Blocking Agent (Agilent Technologies, Inc.) and 1 µl of 25X Fragmentation Buffer (Agilent Technologies, Inc.), and the mixture was incubated at 60°C for 30 min. A total of 25 µl 2X Hybridization buffer (Agilent Technologies, Inc.) was added to dilute the labeled cRNA, 50 µl of hybridization solution was added into the gasket slide and assembled with the circRNA expression microarray slide. The slides were then incubated for 17 h at 65°C in an Agilent Hybridization Oven (G2545A; Agilent Technologies, Inc.). Finally, following washing and fixing the slides, the hybridized arrays were scanned using the Agilent Scanner G2505C (Agilent Technologies, Inc.).
Microarray and quality control
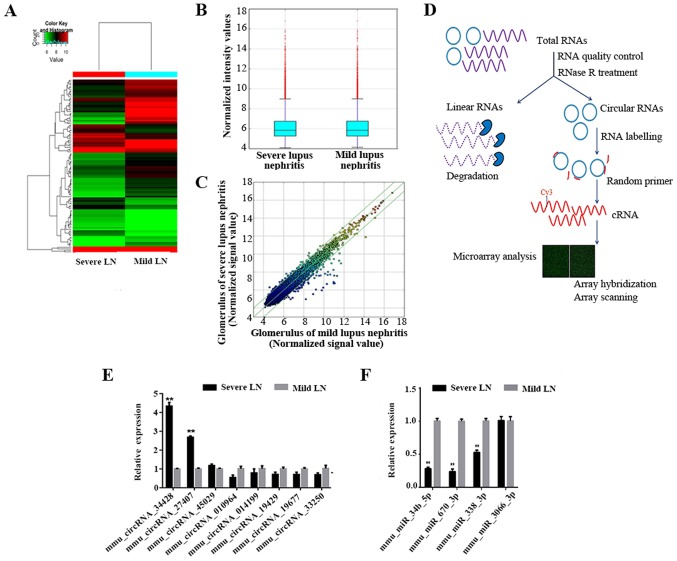
Agilent Feature Extraction software (version 11.0.1.1; Agilent Technologies, Inc.) was used to analyze the acquired array images for raw data extraction. Quantile normalization and subsequent data processing were performed using the R software (version 3.1.2; http://www.r-project.org/) limma package. Following quantile normalization of the raw data, low intensity filtering was performed, and the circRNAs with at least one out of two samples with flags in ‘P’ or ‘M’ (‘All Targets Value’) were retained for further analyses. Differentially expressed circRNAs that were statistically significant between the two groups were identified via Volcano Plot filtering. Differentially expressed circRNAs between two samples were identified via Fold Change filtering. Hierarchical clustering was performed to reveal the distinguishable circRNA expression pattern among samples. circRNAs with fold changes ≥2 were selected as significantly differentially expressed. The experimental workflow is presented in Fig. 3D.
Figure 3.
(A) Analysis of differentially expressed circRNAs in LN. Red indicates higher expression levels, and green indicates lower expression levels. (B) Box plots present fluorescence density distribution following array scanning and revealed that the distribution of circRNAs for the two compared samples were nearly the same following normalization. The Y-axis represents the standardized probe signal value. (C) Scatter plots. The X and Y-axis presents the signal value following normalization of the probe. The red, blue, green and yellow represent the signal intensity of the probe from high to low expression levels. The green lines represent fold change lines. The circRNAs above the top green line and below the bottom green line indicate circRNAs with a fold-change >2.0 between the two compared samples. (D) Experiment workflow of the microarray expression profile of circRNAs. (E) The expression levels of candidate circRNAs for validation in severe and mild LN glomeruli by RT-qPCR. Only mmu_circRNA_34428 and mmu_circRNA_27407 were significantly differentially expressed between the two groups. (F) Expression levels of the predicted four miRNAs by ceRNA analysis of mmu_circRNA_34428 for validation in severe and mild LN glomeruli by RT-qPCR; mmu-miR-34b-5p, mmu-miR-670-3p and mmu-miR-338-3p were significantly differentially expressed between the two groups. **P<0.001. circRNA, circular RNA; LN, lupus nephritis; RT-qPCR, reverse transcription-quantitative PCR; ceRNA, competing endogenous RNA; miRNA/miR, microRNA.
RT-qPCR analysis and statistical analyses
Total RNA was isolated from glomeruli using TRIzol reagent. A total of 1.0 µg of RNA was reverse transcribed to cDNA using the SuperScript™ III Reverse Transcriptase (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. RT-qPCR was performed using the SYBR Green PCR Master Mix (Applied Biosystems; Thermo Fisher Scientific, Inc.) and the fluorescence signal was detected by the ViiA 7 Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.). cDNA samples were prepared from the total RNA of glomeruli by RT-qPCR. In total, 5 upregulated circRNAs and 5 downregulated circRNAs were analyzed by SYBR green I dye-based detection with specific primer sequences (Table I). The following thermocycling conditions were used: 95°C for 10 min; 40 cycles at 95°C for 10 sec and 60°C for 1 min. The relative expression of circRNAs was calculated using the 2−∆∆Cq method (18) with the housekeeping gene-β actin expression to normalize the data. The quantitative data were presented as the mean ± standard error of the mean. The statistical data was calculated using a Student's t-test to analyze expression levels. P<0.05 was considered to indicate a statistically significant difference.
Table I.
Specific circRNAs primers for quantitative polymerase chain reaction analysis.
| Name | Sequence | PS, bp | Name | Sequence | PS, bp |
|---|---|---|---|---|---|
| mouse β-actin | F, 5′-GTACCACCATGTACCCAGGC-3′ | 247 | mmu_circRNA_29625 | F, 5′-TCTGTCCTCGTCATGTTCCAC-3′ | 58 |
| R, 5′-AACGCAGCTCAGTAACAGTCC-3′ | R, 5′CTCTTCTCTCTTCAGGAGTCGTC 3′ | ||||
| mmu_circRNA_010964 | F, 5′-GGCGTGAAACCGTTAAGAGC-3′ | 86 | mmu_circRNA_33250 | F, 5′-AAATGAGTTTGAGACCCTTCG-3′ | 145 |
| R, 5′-CATTCCCAAGCAACCCAACT-3′ | R, 5′-CACCTCGTTGTCCTTAAAATATGT-3′ | ||||
| mmu_circRNA_014199 | F, 5′-ACCTGCAACTTTGAGCAGGACT-3′ | 62 | mmu_circRNA_34414 | F, 5′-TACTCATATCAAAACTTCGCCC-3′ | 103 |
| R, 5′-GAAACCTATGAGAGTGGGTTAGGG-3′ | R, 5′-GTGGGGTTGACTAGGATGATG-3′ | ||||
| mmu_circRNA_19429 | F, 5′-ACAACCCAGAAGAGCCAGGTA-3′ | 158 | mmu_circRNA_34428 | F, 5′-CCAATGATGTGCCTTCTCCATA-3′ | 112 |
| R, 5′-TGAGTTATTCGCCCATACAGC-3′ | R, 5′-GCCTCTTGCAATGTCCACACTT-3′ | ||||
| mmu_circRNA_19677 | F, 5′-CTCTTGACCACGCCACCCTT-3′ | 78 | mmu_circRNA_45029 | F, 5′-AAAAGTGGCTGTATTGGATGG-3′ | 123 |
| R, 5′-AGTGAAGCCAGATGCGAGGAA-3′ | R, 5′-ATGCAAGGACAAGTAACGAATAG-3′ | ||||
| mmu_circRNA_27407 | F, 5′ CTGAGCCTGACGCCATTTCT 3′ | 88 | |||
| R, 5′-′TGTACCTGGCTGCCGTCTC-3′ |
circRNA, circularRNA; PS, product size; bp, base pair; F, forward; R, reverse.
Construction of the competing endogenous RNA (ceRNA) network for candidate circRNAs
It was hypothesized that the RNA transcripts can crosstalk by competing for common miRNAs, with miRNA response elements (MREs) as the foundation of this interaction (19). These RNA transcripts have been termed as ceRNAs (20). Any RNA transcript with MREs that may function as ceRNAs, and ceRNAs containing pseudogene transcripts, long non-coding RNAs, circRNAs and mRNAs, can compete for the same MREs. To identify the potential target of miRNAs, the target miRNAs were predicted with miRNA target prediction software TargetScan 7.2 (http://www.targetscan.or) and miRanda (http://www.microrna.org/microrna/home.do) (21–28). Validated candidate circRNAs were used as seeds to enrich the circRNA/miRNA/gene network according to the analysis.
Statistical analysis
Statistical analyses were performed using GraphPad software (version 6.01; GraphPad Software, Inc.). The Student's t-test was applied for comparison of two groups. P<0.05 were considered to indicate a statistically significant difference. Gene Ontology (GO; www.geneontology.org) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway (www.genome.jp/kegg) analysis was utilized based on the predicted gene from the online TargetScan and miRanda analysis. A network map of circRNA-miRNA-mRNA interactions was constructed with Cytoscape 3.5.0 (www.cytoscape.org).
Results
Pathology of kidneys and efficiency of glomeruli isolation
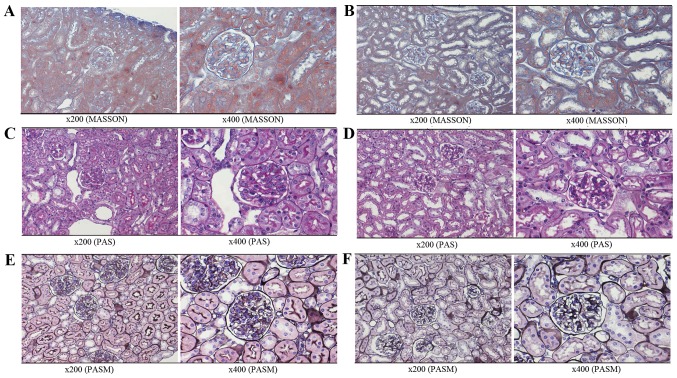
With the development of renal disease, NZB/W F1 mice present with severe proteinuria at only a few weeks of age. According to the different groups (as presented in Fig. 1A), the predominant pathological manifestations observed in severe LN included glomerular diffuse mesangial cell proliferation, matrix expansion and segmental-spherical endothelial cell proliferation (Figs. 1B and 2A, C and E). The pathology of the kidneys in the mild LN group was decreased when compared with the severe group (Figs. 1C and 2B, D and F).
Figure 1.
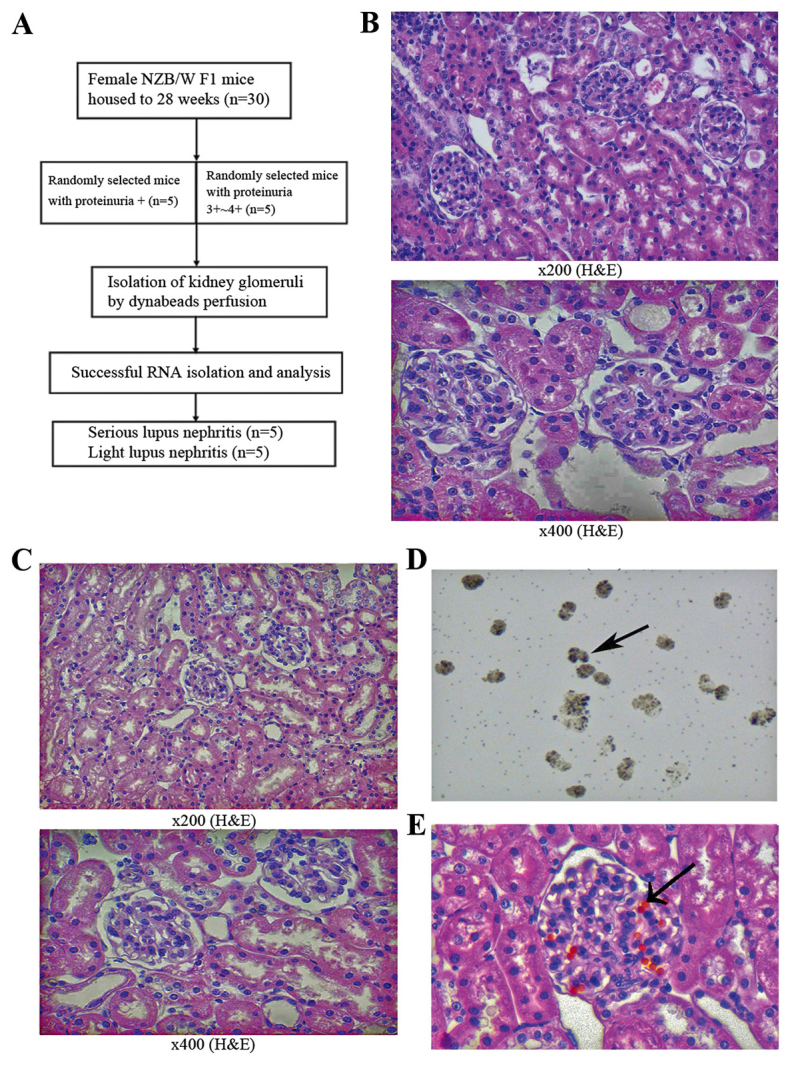
(A) Flow diagram of the groupings, detailing the selection of NZB/W F1 mice. (B) NZB/W F1 pro=4+ (28 weeks) and (C) NZB/WF1 pro=± (28 weeks). Pathological examination of mouse kidneys following Dynabeads perfusion. (D) Completely isolated mice glomeruli with Dynabeads (28 weeks; magnivication, ×100) and (E) pathological examination (magnification, ×400; H&E staining) of mouse glomeruli with magnetic bead perfusion. Arrowheads indicate Dynabeads. H&E, hematoxylin and eosin.
Figure 2.
Pathological analysis of the kidney in NZB/W F1 mice. Masson staining of (A) severe LN group and (B) mild LN group. PAS staining of (C) severe LN group and (D) mild LN group. PASM staining of (E) severe LN group and (F) mild LN group. LN, lupus nephritis.
The mouse glomeruli were isolated by perfusion with Dynabeads (diameter, 4.5 µm) through the aorta pectoralis. The glomerular structure was only slightly affected by the collagenase digestion of the kidney (15). Dynabeads accumulated in the glomerular vessels, making the glomeruli easy to isolate using a magnet with a low degree of contaminating tissues. The isolated glomeruli with Dynabeads were distributed across the slide when observed under a light microscope (Fig. 1D). H& E staining of the kidneys from mice perfused by Dynabeads revealed that the beads were primarily distributed in the glomeruli, and that only a few beads could be detected in the surrounding renal tissues and were mainly concentrated in the afferent and efferent arterioles (Fig. 1E).
Analysis of differentially expressed circRNAs
In total, 13,538 circRNAs were detected using the Arraystar mouse circRNA Microarray. Through the circRNA array profiles, differentially expressed circRNAs were identified in each pairwise comparison. Hierarchical clustering was performed to present the circRNAs based on the expression levels of two samples (Fig. 3A). Box plots revealed that the distribution of the circRNAs for the compared samples were nearly the same following normalization (Fig. 3B). The present study set a threshold of fold-change ≥2.0.
Among the differentially-expressed circRNAs, 41 were significantly upregulated in severe LN when compared with mild LN according to the pre-defined fold-change of ≥2.0 (the top 25 are presented in Table II). In addition, 75 circRNAs that were significantly downregulated in severe LN when compared with mild LN were also identified (the top 25 are presented in Table III).
Table II.
the top up-regulated 25 circRNAs between serious LN and light LN.
| circRNA | FC | circRNA_type | Gene symbol | MRE1 | MRE2 | MRE3 | MRE4 | MRE5 |
|---|---|---|---|---|---|---|---|---|
| mmu_circRNA_30815 | 3.25 | Exonic | L3mbtl4 | mmu-miR-6986-3p | mmu-miR-216c-3p | mmu-miR-9768-5p | mmu-miR-873b | mmu-miR-219b-5p |
| mmu_circRNA_36259 | 3.02 | Exonic | Lmo4 | mmu-miR-6337 | mmu-miR-143-3p | mmu-miR-5621-3p | mmu-miR-547-5p | mmu-miR-433-3p |
| mmu_circRNA_30673 | 2.93 | Exonic | Adgre1 | mmu-miR-207 | mmu-miR-7007-5p | mmu-miR-7054-5p | mmu-miR-6925-5p | mmu-miR-6965-5p |
| mmu_circRNA_41054 | 2.61 | Exonic | Bcat1 | mmu-miR-298-5p | mmu-miR-7080-5p | mmu-miR-544-5p | mmu-miR-6931-5p | mmu-miR-5113 |
| mmu_circRNA_45030 | 2.6 | Exonic | Wdr82 | mmu-miR-3471 | mmu-miR-493-3p | mmu-miR-7230-3p | mmu-miR-3069-5p | mmu-miR-6354 |
| mmu_circRNA_35594 | 2.46 | Intronic | mmu-miR-7092-3p | mmu-miR-1187 | mmu-miR-185-5p | mmu-miR-297c-5p | mmu-miR-6925-5p | |
| mmu_circRNA_45029 | 2.38 | Exonic | Wdr82 | mmu-miR-7232-5p | mmu-miR-345-3p | mmu-miR-3097-5p | mmu-miR-466c-5p | mmu-miR-221-5p |
| mmu_circRNA_39054 | 2.37 | Exonic | Wdfy3 | mmu-miR-6919-3p | mmu-miR-3544-3p | mmu-miR-138-5p | mmu-miR-1902 | mmu-miR-7072-3p |
| mmu_circRNA_27407 | 2.36 | Exonic | Sh3bp5 | mmu-miR-22-5p | mmu-miR-207 | mmu-miR-6938-3p | mmu-miR-877-3p | mmu-miR-484 |
| mmu_circRNA_27357 | 2.32 | Exonic | Tkt | mmu-miR-6403 | mmu-miR-7074-3p | mmu-miR-1668 | mmu-miR-182-5p | mmu-miR-5134-3p |
| mmu_circRNA_42341 | 2.28 | Exonic | Itgam | mmu-miR-136-5p | mmu-miR-3084-5p | mmu-miR-29b-1-5p | mmu-miR-1198-5p | mmu-miR-107-3p |
| mmu_circRNA_31618 | 2.28 | Exonic | Hars2 | mmu-miR-361-3p | mmu-miR-6370 | mmu-miR-1968-3p | mmu-miR-3087-3p | mmu-miR-6926-3p |
| mmu_circRNA_19193 | 2.25 | Senseoverlapping | Adgre1 | mmu-miR-6957-5p | mmu-miR-207 | mmu-miR-7007-5p | mmu-miR-6925-5p | mmu-miR-877-3p |
| mmu_circRNA_38229 | 2.24 | Exonic | Orc5 | mmu-miR-34c-5p | mmu-miR-34b-5p | mmu-miR-195a-3p | mmu-miR-299a-3p | mmu-miR-6916-5p |
| mmu_circRNA_43151 | 2.24 | Exonic | Smad1 | mmu-miR-7085-5p | mmu-miR-667-5p | mmu-miR-92a-2-5p | mmu-miR-3094-5p | mmu-miR-3100-5p |
| mmu_circRNA_42194 | 2.22 | Exonic | Arl6ip1 | mmu-miR-3100-5p | mmu-miR-761 | mmu-miR-214-3p | mmu-miR-6901-5p | mmu-miR-6370 |
| mmu_circRNA_38710 | 2.21 | Exonic | Limch1 | mmu-miR-7034-3p | mmu-miR-1903 | mmu-miR-103-1-5p | mmu-miR-103-2-5p | mmu-miR-107-5p |
| mmu_circRNA_24654 | 2.2 | Senseoverlapping | AK135963 | mmu-miR-5110 | mmu-miR-7665-5p | mmu-miR-6976-5p | mmu-miR-7012-5p | mmu-miR-5113 |
| mmu_circRNA_24113 | 2.2 | Exonic | Kat7 | mmu-miR-1904 | mmu-miR-6344 | mmu-miR-137-5p | mmu-miR-7671-3p | mmu-miR-8120 |
| mmu_circRNA_32604 | 2.19 | Exonic | Tm9sf3 | mmu-miR-7011-3p | mmu-miR-421-5p | mmu-miR-7016-5p | mmu-miR-6384 | mmu-miR-107-5p |
| mmu_circRNA_30668 | 2.18 | Exonic | Adgre1 | mmu-miR-181d-5p | mmu-miR-191-3p | mmu-miR-181b-5p | mmu-miR-7668-3p | mmu-miR-6360 |
| mmu_circRNA_41078 | 2.18 | Exonic | Tm7sf3 | mmu-miR-7012-5p | mmu-miR-7057-5p | mmu-miR-8113 | mmu-miR-504-3p | mmu-miR-7686-5p |
| mmu_circRNA_34428 | 2.15 | Exonic | Pdia3 | mmu-miR-1903 | mmu-miR-7092-3p | mmu-miR-495-5p | mmu-miR-7649-3p | mmu-miR-338-3p |
| mmu_circRNA_29625 | 2.14 | Exonic | L3mbtl4 | mmu-miR-6986-3p | mmu-miR-216c-3p | mmu-miR-9768-5p | mmu-miR-873b | mmu-miR-219b-5p |
| mmu_circRNA_19115 | 2.12 | Senseoverlapping | Tkt | mmu-miR-6973a-5p | mmu-miR-880-3p | mmu-miR-298-5p | mmu-miR-6974-3p | mmu-miR-3099-5p |
circRNA, circularRNA; LN, lupus nephritis; FC, fold change; MRE, miRNA response element; miR, microRNA.
Table III.
Top 25 downregulated circRNAs between severe LN and light LN.
| circRNA | FC | circRNA_type | Gene symbol | MRE1 | MRE2 | MRE3 | MRE4 | MRE5 |
|---|---|---|---|---|---|---|---|---|
| mmu_circRNA_26644 | −20.03 | Exonic | Edil3 | mmu-miR-215-3p | mmu-miR-3075-3p | mmu-miR-8118 | mmu-miR-129-5p | mmu-miR-8092 |
| mmu_circRNA_35752 | −18.88 | Exonic | Dap3 | mmu-miR-3104-3p | mmu-miR-7116-3p | mmu-miR-7051-5p | mmu-miR-337-3p | mmu-miR-181d-3p |
| mmu_circRNA_003795 | −16.94 | Senseoverlapping | Cep350 | mmu-miR-1249-5p | mmu-miR-504-3p | mmu-miR-6399 | mmu-miR-7054-5p | mmu-miR-667-5p |
| mmu_circRNA_013216 | −14.06 | Senseoverlapping | Sulf1 | mmu-miR-7012-3p | mmu-miR-486b-5p | mmu-miR-486a-5p | mmu-miR-181a-1-3p | |
| mmu_circRNA_34193 | −13.39 | Exonic | Lgr4 | mmu-miR-146a-3p | mmu-miR-450a-2-3p | mmu-miR-6359 | mmu-miR-7087-5p | mmu-miR-7647-3p |
| mmu_circRNA_26765 | −12.68 | Senseoverlapping | Wdr41 | mmu-miR-3082-5p | mmu-miR-466f | mmu-miR-466i-5p | mmu-miR-1187 | mmu-miR-466a-5p |
| mmu_circRNA_23904 | −12.36 | Exonic | Acaca | mmu-miR-320-5p | mmu-miR-1903 | mmu-miR-677-3p | mmu-miR-7090-5p | mmu-miR-20b-3p |
| mmu_circRNA_006620 | −10.69 | Exonic | Mllt10 | mmu-miR-5098 | mmu-miR-7059-3p | mmu-miR-7092-3p | mmu-miR-7210-5p | mmu-miR-136-5p |
| mmu_circRNA_25774 | −10.41 | Exonic | Ppp4r4 | mmu-miR-3097-5p | mmu-miR-7235-3p | mmu-miR-7033-5p | mmu-miR-7661-5p | mmu-miR-7214-5p |
| mmu_circRNA_45713 | −9.33 | Senseoverlapping | Tex16 | mmu-miR-7092-3p | mmu-miR-297a-5p | mmu-miR-297c-5p | mmu-miR-1187 | mmu-miR-466c-5p |
| mmu_circRNA_41925 | −8.9 | Exonic | Ints4 | mmu-miR-708-3p | mmu-miR-6342 | mmu-miR-670-3p | mmu-miR-6938-5p | mmu-miR-3075-3p |
| mmu_circRNA_38586 | −7.49 | Exonic | Adgra2 | mmu-miR-3058-5p | mmu-miR-3059-5p | mmu-miR-677-3p | mmu-miR-1946a | mmu-miR-7219-3p |
| mmu_circRNA_014199 | −6.64 | Senseoverlapping | Mamdc4 | mmu-miR-125a-3p | mmu-miR-6930-5p | mmu-miR-6932-5p | mmu-miR-7068-5p | mmu-miR-5710 |
| mmu_circRNA_33250 | −6.55 | Exonic | Tbc1d13 | mmu-miR-7089-3p | mmu-miR-7018-5p | mmu-miR-3113-5p | mmu-miR-6899-3p | mmu-miR-1956 |
| mmu_circRNA_20247 | −6.44 | Intronic | Ino80d | mmu-miR-7661-5p | mmu-miR-3089-5p | mmu-miR-339-5p | mmu-miR-7222-3p | mmu-miR-7222-5p |
| mmu_circRNA_010498 | −5.25 | Antisense | Dstn | mmu-miR-7661-5p | mmu-miR-152-3p | mmu-miR-93-5p | mmu-miR-350-5p | mmu-miR-20a-5p |
| mmu_circRNA_29589 | −4.93 | Exonic | Heg1 | mmu-miR-7075-5p | mmu-miR-7029-3p | mmu-miR-7081-5p | mmu-miR-149-3p | mmu-miR-6935-5p |
| mmu_circRNA_010964 | −4.85 | Intergenic | mmu-miR-20a-3p | mmu-miR-7019-5p | mmu-miR-1969 | mmu-miR-7092-3p | mmu-miR-7684-5p | |
| mmu_circRNA_19429 | −4.77 | Senseoverlapping | Plxnd1 | mmu-miR-6953-5p | mmu-miR-7054-5p | mmu-miR-6925-5p | mmu-miR-7047-5p | mmu-miR-665-5p |
| mmu_circRNA_016016 | −4.15 | Exonic | Nhlrc2 | mmu-miR-320-5p | mmu-miR-7233-3p | mmu-miR-7668-5p | mmu-miR-6976-3p | mmu-miR-130c |
| mmu_circRNA_24437 | 3.77 | Exonic | Rnf157 | mmu-miR-3087-5p | mmu-miR-5107-5p | mmu-miR-7663-5p | mmu-miR-877-3p | mmu-miR-1946b |
| mmu_circRNA_45841 | 3.44 | Exonic | Kdm5c | mmu-miR-148b-5p | mmu-miR-7667-5p | mmu-miR-7659-3p | mmu-miR-320-5p | mmu-miR-6959-5p |
| mmu_circRNA_20692 | 3.4 | Exonic | Epb4.1l5 | mmu-miR-7078-3p | mmu-miR-206-5p | mmu-miR-129b-5p | mmu-miR-1187 | mmu-miR-7079-5p |
| mmu_circRNA_35336 | 3.35 | Exonic | Slc7a11 | mmu-miR-509-5p | mmu-miR-541-5p | mmu-miR-7226-5p | mmu-miR-6539 | mmu-miR-1946b |
| mmu_circRNA_19322 | 3.32 | Senseoverlapping | Reck | mmu-miR-298-5p | mmu-miR-7012-5p | mmu-miR-7067-5p | mmu-miR-1966-5p | mmu-miR-6953-5p |
circRNA, circularRNA; LN, lupus nephritis; FC, fold change; MRE, miRNA response element; miR, microRNA.
RT-qPCR validation of differential circRNAs
Based on the raw intensity, the gene length and fold change of the circRNA profile, the present study selected 10 dysregulated circRNAs, including five upregulated (34414, 29625, 27407, 34428 and 45029) and five downregulated circRNAs (010964, 014199, 19429, 19677 and 33250) for RT-qPCR validation in the samples. The RT-qPCR results revealed that, except for two upregulated circRNAs (34414 and 29625), three upregulated circRNAs and all five downregulated circRNAs were expressed consistently with microarray results (Fig. 3E). The relative fold changes in circRNA expression were calculated using the ΔΔCq method, and the values were expressed as 2−ΔΔCq are presented as the expression level relative to the control group with the standard deviation of the mean of triplicate measures for each group. mmu_circRNA_34428 was the most significantly differentially expressed when compared between the two groups (P<0.001).
Construction of the circRNA-miRNA-mRNA interaction network and predicting the circRNAs that act as ceRNAs
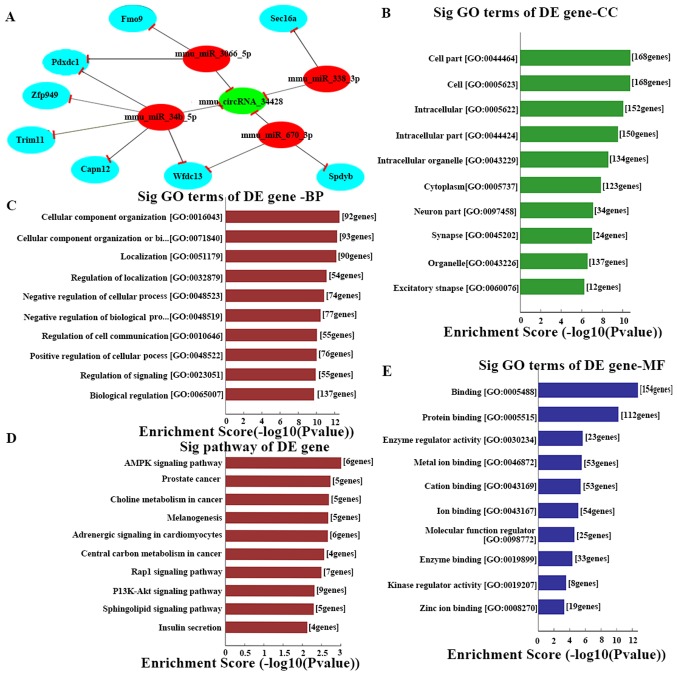
In order to identify more targeted miRNAs, the present study constructed a ceRNA network. Based on the results of the RT-qPCR, the key circRNAs (27407, 34428, 45029, 010964, 014199 and 19429) were selected to predict ceRNAs, and a network map of circRNA-miRNA-mRNA interactions was constructed with Cytoscape (Fig. 4A). Through specific base pairing, the genetic crosstalk between the selected circRNAs and the predicted miRNA targets were detected using an miRNA target prediction software created based on TargetScan & miRanda. The target miRNA of mmu_circRNA_34428 was investigated further. Based on the ceRNA analysis, four miRNAs were predicted to have an interaction with mmu_circRNA_34428, which was consistent with the results of RT-qPCR of these four miRNAs (Fig. 3F). A total of four miRNAs and eight mRNAs were demonstrated to interact with mmu_circRNA_34428, as presented in Fig. 4A. The ceRNA analysis of the interaction network of mmu_circRNA_34428 indicated that miR-34b-5p exhibited the greatest number of interactions, followed by mmu-miR-670-3p and mmu-miR-338-3p (Fig. 4A).
Figure 4.
(A) The predicted mmu_circRNA_34428 targeted gene network (circRNA-miRNA-mRNA) according to sequence-pairing prediction. The interactions predicted by ceRNA. Nodes colored red are miRNAs, light blue nodes are protein coding RNAs and green nodes are circRNAs. Edges with T-shape arrows represent the direction of associations and edges without arrows represent the general association. GO and Kyoto Encyclopedia of Genes and Genomes pathway analysis of mRNAs based on mmu_circRNA_34428. The top 10 enrichment scores of (B) CC, (C) BP, (D) signaling pathway and (E) MF. BP, biological process; CC, cellular component; MF, molecular function; circRNA, circular RNA; miRNA, microRNA; ceRNA, competing endogenous RNA; GO, Gene Ontology; DE, differentially expressed.
However, these eight mRNAs were not enough to predict the function of the four miRNAs as well as mmu_circRNA_34428. A total of four miRNAs were conserved between the online TargetScan and miRanda prediction software, including, mmu-miR-34b-5p, mmu-miR-670-3p, mmu-miR-338-3p and-mmu-miR-3066-3p. These four miRNAs were validated by RT-qPCR, and mmu-miR-34b-5p, mmu-miR-670-3p and mmu-miR-338-3p were significantly differentially expressed between the two groups (P<0.001; Fig. 3F).
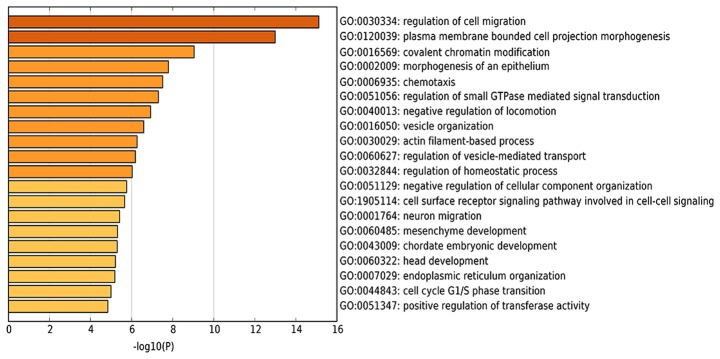
To expand the current understanding of the genetic functions of mmu_circRNA_34428, GO and KEGG pathway analysis was utilized based on the predicted results from the online TargetScan and miRanda analysis (Figs. 4A and S1). mmu_circRNA_34428 had the greatest number of interactions with mmu_miR_34b_5p, mmu_miR_670_3p and mmu_mirR_338_3p within the biological process of cell migration (Figs. 4B-E and 5).
Figure 5.
Heatmap of enriched terms across the input gene list. GO analysis based on the mmu_circRNA_34428-miRNA-mRNAs network. The top 20 significantly enriched biological processes and their scores (negative logarithm of P-value) are presented and listed as the X-axis and the Y-axis, respectively. circRNA, circularRNA; miRNA, microRNA; GO, Gene Ontology.
Discussion
With the growing amount of attention being paid to circRNAs in the field, increasing evidence has demonstrated their roles and mechanisms underlying the pathogenesis of many diseases. Xia et al (29) reported that the overexpressed circRNA cia-cGAS is the effective inhibitor of cGAS-mediated autoimmune disease. The most predominant function of circRNAs is acting as miRNA sponges, and therefore, they could be ideal biomarkers for diagnosing disease. However, little is known about the role of circRNAs in LN, particularly in the glomeruli. In the present study, high-throughput circRNA microarray was utilized to detect differentially expressed circRNAs that were in the glomeruli of those with serious LN when compared with corresponding mild LN glomeruli. Following validation of the expression of 10 circRNAs among the expressed profile, mmu_circRNA_34428 and mmu_circRNA_27407 were significantly upregulated in severe LN (P<0.001). Therefore, the most significantly upregulated circRNA (mmu_circRNA_34428) was selected for investigating the multiple biological processes of LN development.
An increasing body of evidence has reported that circRNAs may act as ceRNAs and serve roles in biological functions, including acting as miRNA sponges to regulate mRNA transcription and protein production (6,30). In addition, a previous study reported that circRNAs have more miRNA binding sites and may be more effective in silencing miRNAs when compared with liner RNAs (31–33). Therefore, the immune-inflammatory response mechanisms of circRNAs may occur in LN by miRNA-mediated effects. The present study demonstrated that mmu_circRNA_34428 was the most likely to interact (i.e., contain complementary base-pair sites) with the following four miRNAs: mmu-miR-34b-5p, mmu-miR-670-3p, mmu-miR-338-3p and mmu-miR-3066-3p. In addition, the present study validated via RT-qPCR that three miRNAs (mmu-miR-34b-5p, mmu-miR-670-3p and mmu-miR-338-3p) had significantly different expression levels between the two groups (P<0.001).
A previous study demonstrated that miR-34b and miR-34c are targets of p53 and cooperate in the control of cell proliferation and adhesion-independent growth (34); miR-34 serves a redundant function in the p53 pathway, suggesting that there may be additional p53-independent functions for this family of miRNAs, which was identified in miR-34-deficient mice (35). In addition, miR-449 and miR-34b/c function was redundant in murine testes by targeting the E2F transcription factor-retinoblastoma protein (E2F-pRb) pathway (36). Research on the p53 pathway in SLE has discovered the pathway participating in the activation of inflammatory factors and B-cells (37,38). In the present ceRNA analysis, mmu-miR-34b-5p was revealed to be one of the binding miRNAs that strongly associates with mmu_circRNA_34428. Finally, in line with all the aforementioned studies that focus on mmu-miR-34b-5p in immune disease, it was hypothesized that mmu_circRNA_34428 acts as an miRNA sponge to inhibit the expression and function of mmu-miR-34b-5p.
Similarly, a previous study demonstrated that miR-338-3p expression inhibits cell proliferation following expression profile analysis in LO2/HBx-d382 cells (39), which indicates that downregulation of miR-338-3p may promote cell proliferation. Furthermore, miR-338-3p was reportedly downregulated in patients with celiac disease with more severe histological lesions when compared with the controls, and this affected the expression of innate and adaptive immunity proteins (40). Zhang et al (41) demonstrated that miR-338-3p serves a role as a novel tumor suppressor to prevent the invasion of renal cell carcinoma by affecting ALK5 (activin receptor-like kinase 5, ALK5) expression. Thus, the low expression level and inhibited function of miR-338-3p and miR-670-3p in cell proliferation as well as in the immune system support the hypothesis that mmu_circRNA_34428 acts as a miRNA sponge to accelerate LN development.
In addition, the results from the GO and KEGG pathway analysis support and identify the important mRNAs in mmu-circRNA-34428 including biological progresses, cellular components, molecular function and meaningful biological signaling pathways. In the KEGG pathway analysis, the AMP kinase signaling pathway serves a key role in controlling cell growth, cell proliferation and stability; it also participates in SLE (42,43). The phosphoinositide-3 kinase/protein kinase B signaling pathway is also closely associated with SLE (44,45). mmu_circRNA_34428 was predicted to serve an important role in cell migration and plasma membrane bounded cell projection morphogenesis, which is involved in immune cell proliferation and immune inflammatory responses in SLE (46–48). It was also assumed that mmu_circRNA_34428 functions as an miRNA sequestering factor for its predicted miRNA binding partners, as aforementioned. It has also been reported that cell migration and morphogenesis were involved in immune pathogenesis and disease activities in SLE and LN (47). It is therefore possible to hypothesize that mmu_circRNA_34428 functions in the regulation of chemokine-mediated cell migration, causing immune factors and cell proliferation to affect LN development; however, further studies are required in order to elucidate its mechanism.
There is evidence to suggest that mesangial cell proliferation is involved in the pathogenesis of LN (49), which can eventually lead to renal failure. Notably, the present study identified that mmu_circRNA_34428 was positively associated with LN disease deterioration in mice depending on the degree of mouse glomerular lesion, which indicates that a high expression of mmu_circRNA_34428 in LN glomeruli may promote mesangial cell and matrix proliferation, and glomerular sclerosis progression. Therefore, the results of the present study indicate that mmu_circRNA_34428 may be an ideal potential diagnostic biomarker for LN with a high degree of accuracy, specificity and sensitivity. In the future, further studies focusing on the circRNA function as miRNA sequestering factors in the regulation of LN occurrence and development will be completed. The small sample size in the present study is one limitation and thus, future experiments using a bigger sample size are underway. To identify the candidate circRNA for the development of albuminuria in LN in vivo and in vitro, future studies will be pursued, including gene sequencing, molecular biological function analysis, intervention experiment and an investigation of functional linkage and crosstalk between these differentially expressed circRNAs and relative proteins of LN.
In conclusion, the present study provided a unique circRNA profile of LN in NZB/W F1 mice, based on which a substantial circRNA signature was demonstrated to indicate possible involvement in LN development. Furthermore, the present study characterized and functionally evaluated one abundant circRNA, mmu_circRNA_34428, thereby offering a potential and credible pathogenicity link and treatment target for LN in the future.
Supplementary Material
Acknowledgements
The authors would like to thank Professors XIAOLI LI, ZILONG LI and LINING WANG affiliated with the Department of Nephrology, The First Hospital of China Medical University (Shenyang, China) for acquisition of funding.
Funding
The present study was supported by the National Natural Science Foundation of China (grant no. 81273297), the Science and Technology Plan of Liaoning Provincial Technology Department (grant no. 2012225021), the National Science and Technology Support Program during the 12th five-year plant period (grant no. 2011BAI10B04) and the Natural Science Foundation of Liaoning Province (grant no. 201202254).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
ST and YL designed the study, curated the data and performed the analysis. YL and LY acquired the funding. ST and XL performed the experiments. YL performed the project administration and supervised the experiments. ST, YL, QF, JM and LY assisted in analysing, integrating and checking the data. ST assisted with the application of the software and wrote the manuscript. YL wrote and reviewed the manuscript.
Ethics approval and consent to participate
The present study complied with the protocols approved by the Institutional Animal Care and Use Committee at China Medical University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Plantinga L, Lim SS, Patzer R, McClellan W, Kramer M, Klein M, Pastan S, Gordon C, Helmick C, Drenkard C. Incidence of end-stage renal disease among newly diagnosed systemic lupus erythematosus patients: The georgia lupus registry. Arthritis Care Res (Hoboken) 2016;68:357–365. doi: 10.1002/acr.22685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hahn BH, McMahon MA, Wilkinson A, Wallace WD, Daikh DI, Fitzgerald JD, Karpouzas GA, Merrill JT, Wallace DJ, Yazdany J, et al. American College of Rheumatology guidelines for screening, treatment, and management of lupus nephritis. Arthritis Care Res (Hoboken) 2012;64:797–808. doi: 10.1002/acr.21664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cameron JS. Lupus nephritis. J Am Soc Nephrol. 1999;10:413–424. doi: 10.1681/ASN.V102413. [DOI] [PubMed] [Google Scholar]
- 4.Tsao BP. Genetic susceptibility to lupus nephritis. Lupus. 1998;7:585–590. doi: 10.1191/096120398678920695. [DOI] [PubMed] [Google Scholar]
- 5.Frangou EA, Bertsias GK, Boumpas DT. Gene expression and regulation in systemic lupus erythematosus. Eur J Clin Invest. 2013;43:1084–1096. doi: 10.1111/eci.12130. [DOI] [PubMed] [Google Scholar]
- 6.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 7.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9:e1003777. doi: 10.1371/annotation/f782282b-eefa-4c8d-985c-b1484e845855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Szabo L, Morey R, Palpant NJ, Wang PL, Afari N, Jiang C, Parast MM, Murry CE, Laurent LC, Salzman J. Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development. Genome Biol. 2015;16:126. doi: 10.1186/s13059-015-0690-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang YH, Yu XH, Luo SS, Han H. Comprehensive circular RNA profiling reveals that circular RNA100783 is involved in chronic CD28-associated CD8(+)T cell ageing. Immun Ageing. 2015;12:17. doi: 10.1186/s12979-015-0042-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lal N, White BS, Goussous G, Pickles O, Mason MJ, Beggs AD, Taniere P, Willcox BE, Guinney J, Middleton GW. KRAS mutation and consensus molecular subtypes 2 and 3 are independently associated with reduced immune infiltration and reactivity in colorectal cancer. Clin Cancer Res. 2018;24:224–233. doi: 10.1158/1078-0432.CCR-17-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ouyang Q, Wu J, Jiang Z, Zhao J, Wang R, Lou A, Zhu D, Shi GP, Yang M. Microarray expression profile of circular RNAs in peripheral blood mononuclear cells from rheumatoid arthritis patients. Cell Physiol Biochem. 2017;42:651–659. doi: 10.1159/000477883. [DOI] [PubMed] [Google Scholar]
- 13.Zhang D, Fujio K, Jiang Y, Zhao J, Tada N, Sudo K, Tsurui H, Nakamura K, Yamamoto K, Nishimura H, et al. Dissection of the role of MHC class II A and E genes in autoimmune susceptibility in murine lupus models with intragenic recombination. Proc Natl Acad Sci USA. 2004;101:13838–13843. doi: 10.1073/pnas.0405807101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sitrin J, Suto E, Wuster A, Eastham-Anderson J, Kim JM, Austin CD, Lee WP, Behrens TW. The Ox40/Ox40 ligand pathway promotes pathogenic Th cell responses, plasmablast accumulation, and lupus nephritis in NZB/W F1 mice. J Immunol. 2017;199:1238–1249. doi: 10.4049/jimmunol.1700608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takemoto M, Asker N, Gerhardt H, Lundkvist A, Johansson BR, Saito Y, Betsholtz C. A new method for large scale isolation of kidney glomeruli from mice. Am J Pathol. 2002;161:799–805. doi: 10.1016/S0002-9440(10)64239-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adachi H, Yu YT. Purification of radiolabeled RNA products using denaturing gel electrophoresis. Curr Protoc Mol Biol. 2014;105 doi: 10.1002/0471142727.mb0420s105. Unit 4.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aranda PS, LaJoie DM, Jorcyk CL. Bleach gel: A simple agarose gel for analyzing RNA quality. Electrophoresis. 2012;33:366–369. doi: 10.1002/elps.201100335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Phelps M, Coss C, Wang H, Cook M, Reproducibility Project Cancer Biology; Reproducibility Project Cancer Biology: Registered report: Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Elife. 2016;5 doi: 10.7554/eLife.12470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jin X, Feng CY, Xiang Z, Chen YP, Li YM. CircRNA expression pattern and circRNA-miRNA-mRNA network in the pathogenesis of nonalcoholic steatohepatitis. Oncotarget. 2016;7:66455–66467. doi: 10.18632/oncotarget.12186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen L, Zhang S, Wu J, Cui J, Zhong L, Zeng L, Ge S. circRNA_100290 plays a role in oral cancer by functioning as a sponge of the miR-29 family. Oncogene. 2017;36:4551–4561. doi: 10.1038/onc.2017.89. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Huang M, Zhong Z, Lv M, Shu J, Tian Q, Chen J. Comprehensive analysis of differentially expressed profiles of lncRNAs and circRNAs with associated co-expression and ceRNA networks in bladder carcinoma. Oncotarget. 2016;7:47186–47200. doi: 10.18632/oncotarget.9706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garcia DM, Baek D, Shin C, Bell GW, Grimson A, Bartel DP. Weak seed-pairing stability and high target-site abundance decrease the proficiency of lsy-6 and other microRNAs. Nat Struct Mol Biol. 2011;18:1139–1146. doi: 10.1038/nsmb.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in Drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–D97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xia P, Wang S, Ye B, Du Y, Li C, Xiong Z, Qu Y, Fan Z. A circular RNA protects dormant hematopoietic stem cells from DNA Sensor cGAS-mediated exhaustion. Immunity. 2018;48:688.e7–701.e7. doi: 10.1016/j.immuni.2018.03.016. [DOI] [PubMed] [Google Scholar]
- 30.Chen I, Chen CY, Chuang TJ. Biogenesis, identification, and function of exonic circular RNAs. Wiley Interdiscip Rev RNA. 2015;6:563–579. doi: 10.1002/wrna.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilusz JE, Sharp PA. Molecular biology. A circuitous route to noncoding RNA. Science. 2013;340:440–441. doi: 10.1126/science.1238522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thomas LF, Sætrom P. Circular RNAs are depleted of polymorphisms at microRNA binding sites. Bioinformatics. 2014;30:2243–2246. doi: 10.1093/bioinformatics/btu257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dudekula DB, Panda AC, Grammatikakis I, De S, Abdelmohsen K, Gorospe M. CircInteractome: A web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 2016;13:34–42. doi: 10.1080/15476286.2015.1128065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Concepcion CP, Han YC, Mu P, Bonetti C, Yao E, D'Andrea A, Vidigal JA, Maughan WP, Ogrodowski P, Ventura A. Intact p53-dependent responses in miR-34-deficient mice. PLoS Genet. 2012;8:e1002797. doi: 10.1371/journal.pgen.1002797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bao J, Li D, Wang L, Wu J, Hu Y, Wang Z, Chen Y, Cao X, Jiang C, Yan W, Xu C. MicroRNA-449 and microRNA-34b/c function redundantly in murine testes by targeting E2F transcription factor-retinoblastoma protein (E2F-pRb) pathway. J Biol Chem. 2012;287:21686–21698. doi: 10.1074/jbc.M111.328054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miret C, Molina R, Filella X, García-Carrasco M, Claver G, Ingelmo M, Ballesta A, Font J. Relationship of p53 with other oncogenes, cytokines and systemic lupus erythematosus activity. Tumour Biol. 2003;24:185–188. doi: 10.1159/000074428. [DOI] [PubMed] [Google Scholar]
- 38.Luo S, Liu Y, Liang G, Zhao M, Wu H, Liang Y, Qiu X, Tan Y, Dai Y, Yung S, et al. The role of microRNA-1246 in the regulation of B cell activation and the pathogenesis of systemic lupus erythematosus. Clin Epigenetics. 2015;7:24. doi: 10.1186/s13148-015-0063-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fu X, Tan D, Hou Z, Hu Z, Liu G, Ouyang Y, Liu F. The effect of miR-338-3p on HBx deletion-mutant (HBx-d382) mediated liver-cell proliferation through CyclinD1 regulation. PLoS One. 2012;7:e43204. doi: 10.1371/journal.pone.0043204. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 40.Magni S, Buoli Comani G, Elli L, Vanessi S, Ballarini E, Nicolini G, Rusconi M, Castoldi M, Meneveri R, Muckenthaler MU, et al. miRNAs affect the expression of innate and adaptive immunity proteins in celiac disease. Am J Gastroenterol. 2014;109:1662–1674. doi: 10.1038/ajg.2014.203. [DOI] [PubMed] [Google Scholar]
- 41.Zhang X, Wang C, Li H, Niu X, Liu X, Pei D, Guo X, Xu X, Li Y. miR-338-3p inhibits the invasion of renal cell carcinoma by downregulation of ALK5. Oncotarget. 2017;8:64106–64113. doi: 10.18632/oncotarget.19329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Benatti FB, Miyake CNH, Dantas WS, Zambelli VO, Shinjo SK, Pereira RMR, Silva MER, Sá-Pinto AL, Borba E, Bonfá E, Gualano B. Exercise increases insulin sensitivity and skeletal muscle AMPK expression in systemic lupus erythematosus: A randomized controlled trial. Front Immunol. 2018;9:906. doi: 10.3389/fimmu.2018.00906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schuiveling M, Vazirpanah N, Radstake TRDJ, Zimmermann M, Broen JCA. Metformin, a new era for an old drug in the treatment of immune mediated disease? Curr Drug Targets. 2018;19:945–959. doi: 10.2174/1389450118666170613081730. [DOI] [PubMed] [Google Scholar]
- 44.Badr G, Sayed A, Abdel-Maksoud MA, Mohamed AO, El-Amir A, Abdel-Ghaffar FA, Al-Quraishy S, Mahmoud MH. Infection of female BWF1 lupus mice with malaria parasite attenuates B cell autoreactivity by modulating the CXCL12/CXCR4 axis and its downstream signals PI3K/AKT, NFκB and ERK. PLoS One. 2015;10:e0125340. doi: 10.1371/journal.pone.0125340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Beşliu AN, Pistol G, Marica CM, Bănică LM, Chiţonu C, Ionescu R, Tănăseanu C, Tamsulea I, Matache C, Stefănescu M. PI3K/Akt signaling in peripheral T lymphocytes from systemic lupus erythematosus patients. Roum Arch Microbiol Immunol. 2009;68:69–79. [PubMed] [Google Scholar]
- 46.Qingjuan L, Xiaojuan F, Wei Z, Chao W, Pengpeng K, Hongbo L, Sanbing Z, Jun H, Min Y, Shuxia L. miR-148a-3p overexpression contributes to glomerular cell proliferation by targeting PTEN in lupus nephritis. Am J Physiol Cell Physiol. 2016;310:C470–C478. doi: 10.1152/ajpcell.00129.2015. [DOI] [PubMed] [Google Scholar]
- 47.Adalid-Peralta L, Mathian A, Tran T, Delbos L, Durand-Gasselin I, Berrebi D, Peuchmaur M, Couderc J, Emilie D, Koutouzov S. Leukocytes and the kidney contribute to interstitial inflammation in lupus nephritis. Kidney Int. 2008;73:172–180. doi: 10.1038/sj.ki.5002625. [DOI] [PubMed] [Google Scholar]
- 48.Schiffer L, Bethunaickan R, Ramanujam M, Huang W, Schiffer M, Tao H, Madaio MP, Bottinger EP, Davidson A. Activated renal macrophages are markers of disease onset and disease remission in lupus nephritis. J Immunol. 2008;180:1938–1947. doi: 10.4049/jimmunol.180.3.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ichinose K, Rauen T, Juang YT, Kis-Toth K, Mizui M, Koga T, Tsokos GC. Cutting edge: Calcium/Calmodulin-dependent protein kinase type IV is essential for mesangial cell proliferation and lupus nephritis. J Immunol. 2011;187:5500–5504. doi: 10.4049/jimmunol.1102357. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.