Abstract
Non-small cell lung cancer (NSCLC) is a leading cause of cancer-associated mortality worldwide, NCSCLC includes lung adenocarcinoma and lung squamous cell carcinoma. Tumor metastasis is the major cause of mortality of patients with NSCLC. However, the mechanisms underlying NSCLC metastasis remain largely elusive. In present study, the authors focused on exploring the roles of RBAK in NSCLC. The present study demonstrated that RB-associated KRAB zinc finger (RBAK) was upregulated in NSCLC compared with non-tumorous tissues by analyzing Gene Expression Omnibus (GEO) datasets and The Cancer Genome Atlas (TCGA) dataset. High expression of RBAK was associated with poor disease-free survival of patients with NSCLC by analyzing TCGA dataset. Furthermore, an RBAK-mediated protein-protein interaction network was constructed to reveal the potential underlying mechanisms by which RBAK drives NSCLC progression. The authors found that RBAK was involved in regulating a number of transcription factors, including androgen receptor, forkhead box A1, tumor protein 53, and E2F transcription factor 1, 2 and 4, suggesting that RBAK may have a role in regulating gene transcription. GO and KEGG enrichment analyses of the genes co-expressed with RBAK revealed that RBAK is involved in regulating a number of biological functions, including the Wnt signaling pathway, mRNA splicing, protein polyubiquitination, cell-cell adhesion and focal adhesion. Transwell and wound healing assays demonstrated that knockdown of RBAK suppressed NSCLC cell migration and invasion. The present study enhances the current understanding of the important roles of RBAK in NSCLC metastasis and may provide useful information for the development of novel treatment approaches.
Keywords: non-small cell lung cancer, RB associated KRAB zinc finger, prognosis, migration, invasion
Introduction
Non-small cell lung cancer (NSCLC) is a leading cause of cancer-associated mortality worldwide, and the incidence and mortality rates of NSCLC have significantly increased (1,2). NCSCLC includes lung adenocarcinoma (LUAD) and lung squamous cell carcinoma. Tumor metastasis is recognized as the major cause of mortality of patients with NSCLC (3). Previous studies have identified a number of genes that are involved in regulating NSCLC metastasis, including twist family bHLH transcription factor 1 (4,5), patched 1 (PTCH1) (6) and transforming growth factor β-induced long non-coding RNA (7). Elucidation of the molecular mechanisms underlying NSCLC metastasis is urgently required, as this will assist with the identification of novel therapeutic targets for NSCLC.
A previous study has reported that RB-associated KRAB zinc finger (RBAK) interacts with tumor suppressor retinoblastoma 1 (8). However, the roles of RBAK in human diseases remain largely elusive. A number of studies have indicated that RBAK is involved in cancer progression (9). In prostate cancer (PCa), RBAK interacts with androgen receptor (AR) and is involved in the regulation of the cell cycle (10). Wan et al (9) revealed that knockdown of RBAK inhibits PCa growth by inducing cell apoptosis. A high expression level of RBAK has been associated with a shorter survival time for patients with PCa following radical prostatectomy (9). However, the prognostic value and molecular functions of RBAK in NSCLC have remained elusive.
The present study identified that RBAK is upregulated in NSCLC. Furthermore, the potential effects of RBAK on NSCLC cell migration and invasion were investigated. The present study may provide useful information that promotes the understanding of the important roles of RBAK in NSCLC metastasis.
Materials and methods
Analysis of public datasets
In the present study, three Gene Expression Omnibus (GEO) datasets, including GSE19188 (11), GSE19804 (12) and GSE18842 (13), were downloaded from the National Cancer for Biotechnology Information database for analysis of RBAK expression in NSCLC (https://www.ncbi.nlm.nih.gov/geo/). The dataset GSE19188 includes 91 NSCLC samples and 65 normal lung samples. Furthermore, GSE19804 contains 60 paired NSCLC and normal samples, and GSE18842 includes 46 NSCLC samples and 45 control samples. The raw data of the mRNA expression profiles were downloaded and analyzed using R language software. Background correction, quartile data normalization and probe summarization were applied to the original data. The limma tool in Bioconductor software (http://www.bioconductor.org/) was used to identify differentially expressed genes (DEGs) between the NSCLC and normal samples. The significance of each DEG was calculated by a classical t-test and was presented as a P-value. In addition, The Cancer Genome Atlas (TCGA) lung adenocarcinoma (LUAD) dataset, with 59 normal and 517 LUAD samples, which included clinical features such as survival time, were downloaded from cBioPortal (http://www.cbioportal.org/).
Bioinformatics analysis
The Database for Annotation, Visualization and Integrated Discovery (DAVID) bioinformatics tool (https://david.ncifcrf.gov/) was used to perform gene ontology (GO) and Kyoto Encyclopedia of genes and genomes (KEGG) pathway enrichment analyses. The present study used Search Tool for the Retrieval of Interacting Genes (STRING; http://string-db.org/) to construct the PPI network for target genes (minimum required interaction score. >0.4). In addition, Cytoscape software version 3.6.0 (http://www.cytoscape.org/download-platforms.html) was used for visualization of the PPI networks.
Cell culture
The human cell lines A549, H1299, 95D and H1975 were purchased from the Cell Bank of the Chinese Academy of Sciences. The NSCLC cell lines were cultured in RPMI 1640 medium containing 10% fetal bovine serum (FBS), 1% sodium pyruvate and 1% penicillin/streptomycin (all Gibco; Thermo Fisher Scientific, Inc.) at 37°C in a humidified atmosphere containing 5% CO2.
Lentiviral constructs and transfection
The RBAK short hairpin (sh)RNA (3′-CCGGGCTGCTTGTATCAATAGCAAACTCGAGTTTGCTATTGATACAAGCAGCTTTTT-5′) was purchased from Shanghai GeneChem Co., Ltd. Recombinant lentiviral plasmids carrying RBAK shRNA were constructed according to the manufacturer's protocol. The empty recombinant lentiviral plasmids were used as negative controls. Reverse transcription-quantitative (RT-q)PCR was performed to detect the knockdown efficiency of RBAK shRNA.
RT-qPCR
Total RNA was extracted from the cells using TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.). Subsequently, 0.5–1.0 µg RNA was reverse-transcribed to complementary (c)DNA using the RevertAid First Strand cDNA Synthesis kit (Takara Biotechnology Co., Ltd.). qPCR was performed with the iQSYBR Green Supermix using the ABI Prism 7900 platform (Applied Biosystems; Thermo Fisher Scientific, Inc.). The PCR conditions were as follows: Initial denaturation at 95°C for 10 sec, followed by 40 cycles of 95°C for 5 sec, 57°C for 15 sec and 72°C for 20 sec. GAPDH was used as an internal control. The relative expression levels of the target genes were calculated using the 2−Δ∆Cq method (14). The following primers were used for qPCR: GAPDH forward, 5′-TGACTTCAACAGCGACACCCA-3′ and reverse, 5′-CACCCTGTTGCTGTAGCCAAA-3′; RBAK forward, 5′-TTACAGGGATGTGATGTTGGA-3′ and reverse, 5′-CTCTTCAGTCAGGGTTTTGCT-3′.
Wound healing assay
A total of 5,000 A549 cells were seeded into a 6-well plate. A circular hole was generated 16 h after serum starvation of the cells using an Oris™ plate. Serum starvation medium was RPMI 1640 containing 1% sodium pyruvate and 1% penicillin/streptomycin (all Gibco; Thermo Fisher Scientific, Inc.). Images were captured after 0, 24 and 48 h using a microscope (CK40-F200; Olympus).
Transwell assay
Transwell assays were performed using 8-mm pore size Transwell® plates (Corning Inc.), according to the manufacturer's protocol. In brief, 50,000 A549 cells were seeded into the upper chamber of the Transwell plate. The upper wells were filled with serum-free RPMI 1640 and the bottom wells were filled with RPMI 1640 containing 10% FBS. After 72 h of incubation, the number of migrated cells was counted and images were captured. Cells were stained with 0.1% crystal violet for 20 min at room temperature. The invasion assay was performed using the above protocol with the Transwell plates pre-coated with Matrigel® basement membrane matrix (BD Biosciences).
Statistical analysis
Statistical analyses were performed using SPSS software (version 16.0.0; SPSS, Inc., Chicago, IL, USA). All experiments were performed three times. Depending on the comparison, a Student's t-test or Mann-Whitney U-test was performed to assess the statistical significance between two groups. Kaplan Meier and Cox regression analyses were applied to measure disease-free survival (DFS), and log-rank test was conducted to determine the survival difference based on RBAK expression. P<0.05 was considered to indicate a statistically significant difference.
Results
RBAK is overexpressed in NSCLC
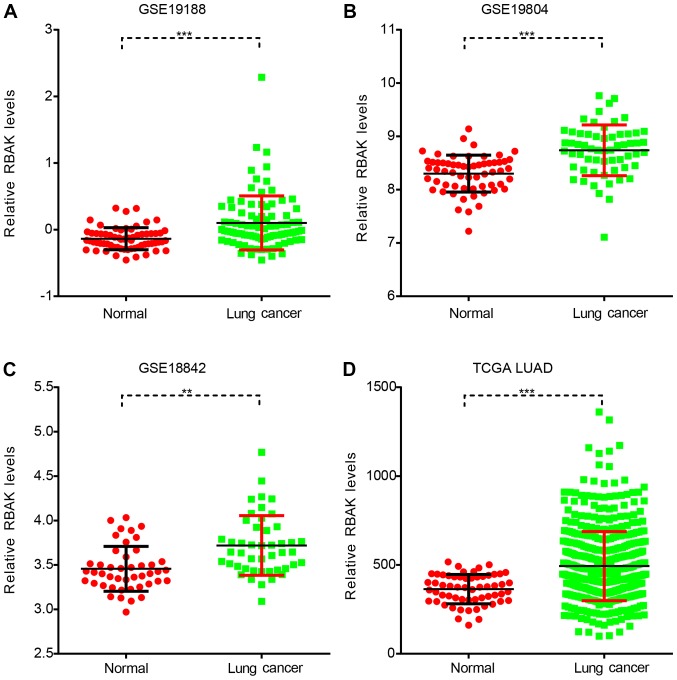
The functional roles and expression pattern of RBAK in human cancer, particularly NSCLC, have remained largely elusive. The present study first analyzed RBAK expression in NSCLC using three GEO datasets, namely GSE19188, GSE19804 and GSE18842. As presented in Fig. 1, it was observed that RBAK was significantly overexpressed in the NSCLC samples compared to normal samples (Fig. 1A-C). Subsequently, TCGA LUAD dataset with 59 normal and 517 LUAD samples was analyzed. It was identified that patients with LUAD had a higher RBAK expression level compared with that in the normal controls (Fig. 1D).
Figure 1.
RBAK is overexpressed in NSCLC samples. (A-C) Analysis of the datasets (A) GSE19188, (B) GSE19804 and (C) GSE18842 revealed that RBAK expression was upregulated in NSCLC samples compared with normal lung tissues. (D) Analysis of the dataset from TCGA demonstrated that RBAK was overexpressed in LUAD compared with normal lung samples. **P<0.01, ***P<0.001. NSCLC, non-small-cell lung cancer; RBAK, RB-associated KRAB zinc finger; TCGA, The Cancer Genome Atlas; LUAD, lung adenocarcinoma.
High RBAK expression is associated with poor survival in patients with NSCLC
Subsequently, Kaplan-Meier analysis was performed to investigate the prognostic value of RBAK in NSCLC. The cut-off finder online system (http://molpath.charite.de/cutoff/) was used to identify the cut-off value to stratify all NSCLC samples into RBAK-high and RBAK-low groups. It was revealed that the DFS rate was lower for patients with LUAD or lung squamous cell carcinoma in the RBAK-high expression group than for those in the RBAK-low expression group (Fig. 2). These results suggest that RBAK may serve as a biomarker for NSCLC.
Figure 2.
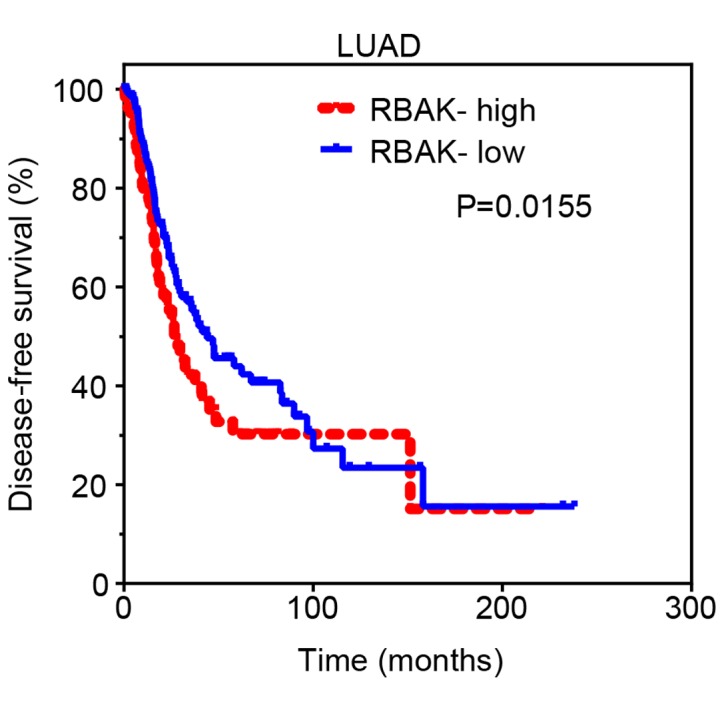
High RBAK expression is associated with shorter disease-free survival for patients with LUAD. High RBAK expression is associated with poor disease-free survival in LUAD. RBAK, RB-associated KRAB zinc finger; TCGA, The Cancer Genome Atlas; LUAD, lung adenocarcinoma.
Bioinformatics analysis of RBAK in NSCLC
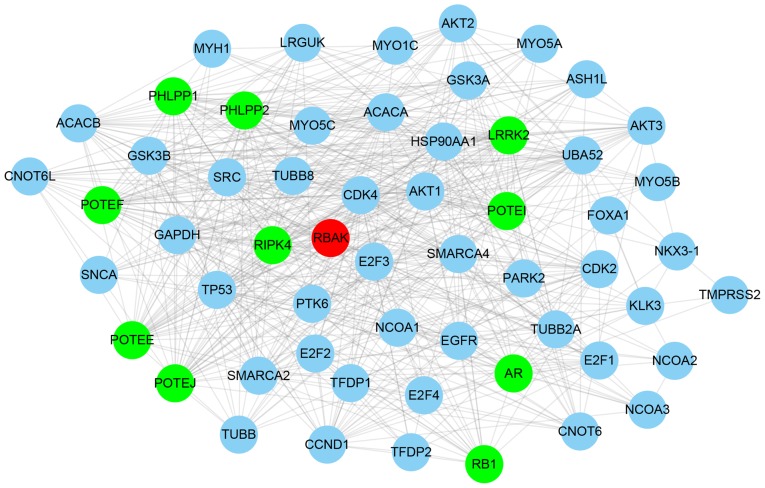
Considering that the function of RBAK in NSCLC has remained elusive, the present study performed a bioinformatics analysis of its roles and interactions. First, an RBAK-mediated protein-protein interaction network was constructed using the STRING online system (https://string-db.org/cgi/network.pl). A total of 57 single nodes and 576 edges were included in the RBAK-mediated protein-protein interaction network (Fig. 3). It was identified that the network included a number of transcription factors, including AR, forkhead box (FOX)A1, tumor protein (TP)53, and E2F transcription factor 1, 2 and 4, suggesting that RBAK may have a role in regulating gene transcription. Therefore, the present study performed a co-expression analysis for RBAK to reveal its potential roles in NSCLC.
Figure 3.
Construction of an RBAK-mediated protein-protein interaction network. An RBAK-mediated protein-protein interaction network in non-small-cell lung cancer was generated, which included 57 single nodes and 576 edges. RBAK is shown in red, genes that were positively co-expressed with RBAK are denoted as green nodes and genes that were negatively co-expressed with RBAK are denoted as blue nodes. RBAK, RB-associated KRAB zinc finger.
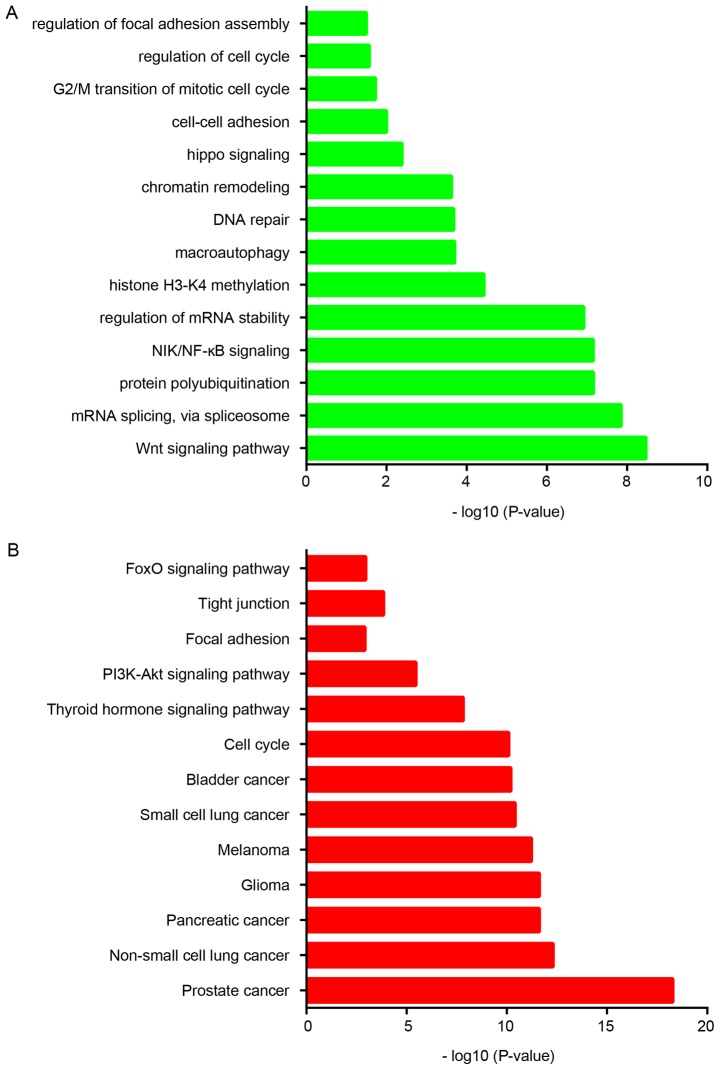
Subsequently, a functional analysis of RBAK in NSCLC was performed based on its co-expressed genes. Gene ontology enrichment analysis demonstrated that RBAK is involved in regulating the Wnt signaling pathway, mRNA splicing, protein polyubiquitination, NF-κB-inducing kinase (NIK)/NF-κB signaling, regulation of mRNA stability, histone H3-K4 methylation, macroautophagy, DNA repair, chromatin remodeling, Hippo signaling, cell-cell adhesion, G2/M transition of mitotic cell cycle and focal adhesion assembly (Fig. 4A). Furthermore, Kyoto Encyclopedia of Genes and Genomes analysis revealed that RBAK is significantly associated with various types of human cancer, including NSCLC, in addition to the cell cycle, thyroid hormone signaling pathway, tight junction and FOXO signaling pathways (Fig. 4B). Metastasis is a key cause of NSCLC-associated mortality. According to the aforementioned analysis, RBAK is involved in regulating cell-cell adhesion and focal adhesion, suggesting that RBAK may affect the progression of metastasis in NSCLC.
Figure 4.
Bioinformatics analysis of RBAK in NSCLC. (A) Gene Ontology and (B) Kyoto Encyclopedia of Genes and Genomes analyses were performed to predict potential functions of RBAK in NSCLC using its co-expressing genes. NSCLC, non-small-cell lung cancer; RBAK, RB-associated KRAB zinc finger. NIK, NF-κB-inducing kinase.
Knockdown of RBAK inhibits cell migration in NSCLC
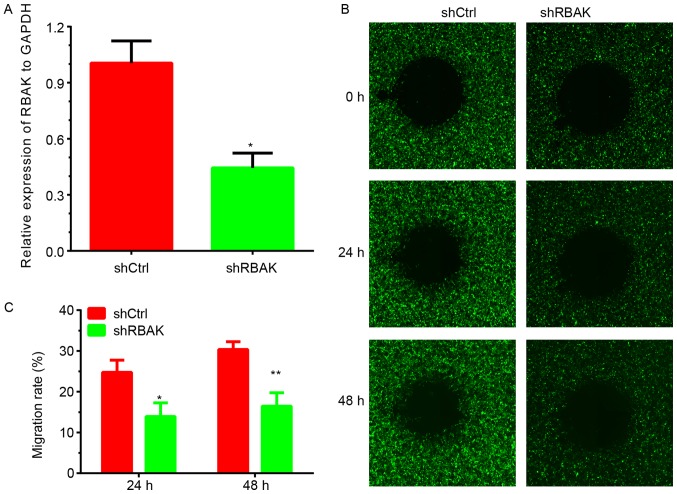
By detecting the endogenous expression of RBAK in the human A549, H1299, 95D and H1975 cell lines, it was identified that RBAK expression is higher in A549 cells. To further validate the effect of RBAK on cell metastasis, the present study performed a loss-of-function assay with NSCLC A549 cells. As presented in Fig. 5, a wound-healing assay was used to investigate the role of RBAK in the regulation of NSCLC cell migration. Images of the area void of cells were obtained at 0, 24 and 48 h. After 24 h, ~25% of the wound area was repaired by migrating cells in the control group, while only 11% of the wound area was repaired by cells in the RBAK-knockdown group (Fig. 5B and C). After 48 h, ~30% of the wound area was repaired by migrating cells in the control group, while only 12% of the wound area was repaired in the RBAK-knockdown group (Fig. 5B and C). The transfection efficiency is presented in Fig. 5A. The knockdown efficiency is presented in Fig. 5A and the transfection efficiency is presented in Fig. S1.
Figure 5.
Wound-healing assay revealing that RBAK knockdown inhibits NSCLC cell migration. (A) The knockdown efficiency was evaluated. (B) A wound-healing assay demonstrated that RBAK knockdown significantly inhibits the migration of NSCLC cells. Magnification, ×400. (C) Quantified results of the wound-healing assay. Values are expressed as the mean ± standard deviation (n=3). *P<0.05, **P<0.01 vs. shCtrl. NSCLC, non-small-cell lung cancer; RBAK, RB-associated KRAB zinc finger; shRNA, short hairpin RNA; shCtrl, control shRNA; shRBAK, shRNA targeting RBAK.
Furthermore, the present study investigated the roles of RBAK in the regulation of NSCLC cell migration by using a Transwell assay. As illustrated in Fig. 6, it was revealed that RBAK knockdown significantly decreased the number of migrating A549 cells compared with that in the negative control group (Fig. 6A and B).
Figure 6.
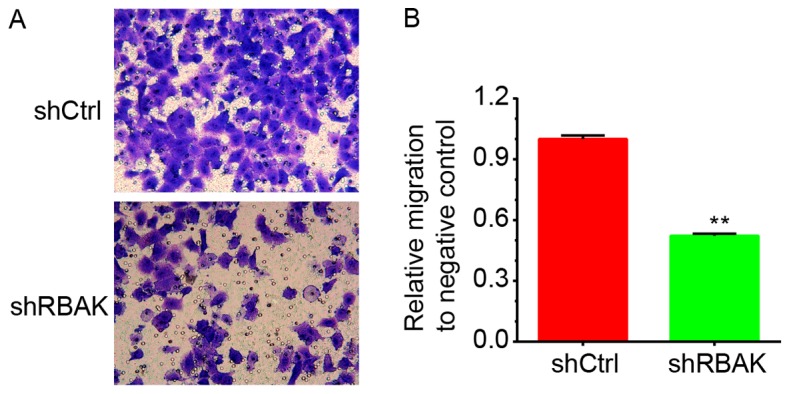
Transwell assay revealed that RBAK knockdown inhibits NSCLC cell migration. (A) Representative images of cells migrated through the Transwell filter. Cells were stained with 0.1% crystal violet for 20 min at room temperature. Magnification, ×400. (B) Quantified results of the Transwell migration assay. Values are expressed as the mean ± standard deviation (n=3). **P<0.01 vs. shCtrl. NSCLC, non-small-cell lung cancer; RBAK, RB-associated KRAB zinc finger; shCtrl, control shRNA; shRBAK, shRNA targeting RBAK; shRNA, short hairpin RNA.
Knockdown of RBAK suppresses the invasive ability of NSCLC cells
In addition, the roles of RBAK in the regulation of NSCLC cell invasion were investigated. At 72 h post-transfection, the ability of A549 cells to invade through Matrigel was assessed. It was identified that knockdown of RBAK significantly inhibited A549 cell invasion. RBAK knockdown decreased the number of invading A549 cells by 88% compared with that in the negative control group (Fig. 7A and B).
Figure 7.
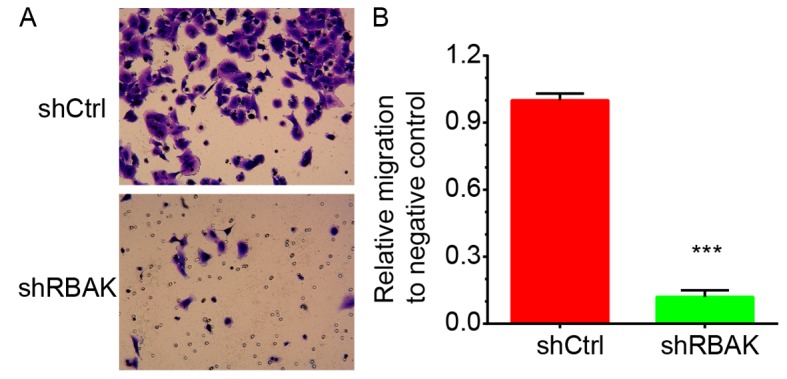
Transwell assay revealed that RBAK knockdown inhibits NSCLC cell invasion. (A) Representative images of cells migrated through the Transwell filter membrane. Cells were stained with 0.1% crystal violet for 20 min at room temperature. Magnification, ×400. (B) Quantified results of the Transwell invasion assay. Values are expressed as the mean ± standard deviation (n=3). ***P<0.001 vs. shCtrl. NSCLC, non-small-cell lung cancer; RBAK, RB-associated KRAB zinc finger; shRNA, short hairpin RNA; shCtrl, control shRNA; shRBAK, shRNA targeting RBAK.
Discussion
Tumor metastasis is a major cause of mortality in patients with NSCLC (3). The mechanisms underlying NSCLC metastasis remain to be further investigated. The present study was the first, to the best of our knowledge, to identify RBAK as a novel regulator of metastasis in NSCLC. RBAK was observed to be overexpressed in NSCLC and high expression was associated with shorter DFS times in patients with NSCLC. Bioinformatics analysis demonstrated that RBAK is involved in regulating various metastasis-associated biological functions. These roles were experimentally validated, and in vitro assays confirmed that knockdown of RBAK suppressed NSCLC cell migration and invasion.
A limited number of studies have indicated that RBAK is involved in regulating cancer progression. Higher expression levels of RBAK have been associated with a shorter survival time for patients with PCa (9). However, to the best of our knowledge, the expression pattern of RBAK in NSCLC has remained largely elusive. In the present study, analysis of public datasets revealed that the expression levels of RBAK in NSCLC were higher compared with those in normal samples. Furthermore, a high RBAK expression level was associated with poorer DFS of NSCLC patients. These results suggest that RBAK may serve as a novel prognostic marker for NSCLC.
RBAK has been identified to interact with AR and promote cell proliferation and the cell cycle in PCa (15). In the present study, a Bioinformatics analysis was performed to investigate the potential roles of RBAK in NSCLC. RBAK-mediated protein-protein interaction networks were constructed, which revealed that RBAK interacts with various key regulators in NSCLC, including AKT1 (16), AKT2 (16), AKT3 (17), E2F1 (18), E2F2 (19), E2F3 (20), RB1 (21), cyclin D 1 (21), cyclin-dependent kinase 4 (22), epidermal growth factor receptor (23) and TP53 (24). The present study indicated that RBAK may be involved in regulating NSCLC progression and metastasis by influencing cell-cell adhesion and focal adhesion. Tumor metastasis is the major cause of mortality for patients with NSCLC. In previous studies, a number of metastasis regulators have been identified in NSCLC. For instance, Wan et al (6) reported that the 3′-untranslated region of PTCH1 may promote NSCLC metastasis by activating the micro (mi)RNA-101-3p/solute carrier family 39 member 6 axis. Ras association domain-containing protein 1 suppresses the invasion of NSCLC by inhibiting Yes-associated protein 1 (25). miR-483-5p promotes LUAD metastasis by targeting Rho GDP dissociation inhibitor 1 and activated leukocyte cell adhesion molecule (26). The present study reported that silencing of RBAK suppresses NSCLC migration and invasion, suggesting that RBAK promotes metastasis-associated processes in NSCLC.
Several limitations of the present study should be noted. First, Bioinformatics analysis demonstrated that RBAK is involved in regulating a number of biological functions and pathways, including the Wnt signaling pathway and mRNA splicing. These pathways have crucial roles in NSCLC progression. Wnt signaling regulators have widely been observed to de dysregulated in NSCLC. Wnt signaling may regulate NSCLC cell proliferation (27), apoptosis (28) and metastasis (29). Furthermore, the Wnt pathway may enhance resistance to chemotherapy and radiotherapy (30). However, the effect of RBAK on these pathways requires further validation. In addition, in vivo experimental validation of the functions of RBAK remains to be performed in future studies. Furthermore, the upstream regulators of RBAK still require to be investigated. Future studies focusing on these points may promote the understanding regarding the important roles of RBAK in NSCLC progression
In conclusion, the present study demonstrated that RBAK is upregulated in NSCLC vs. non-tumorous tissues samples and may serve as a novel prognostic marker. Furthermore, the present study was the first, to the best of our knowledge, to reveal that RBAK is a novel regulator of cell metastasis. The present study may serve as a basis for the development of novel potential therapeutic approaches for NSCLC.
Supplementary Material
Acknowledgements
Not applicable.
Glossary
Abbreviations
- NSCLC
non-small-cell lung cancer
- RBAK
RB-associated KRAB zinc finger
- PCa
prostate cancer
- GO
Gene Ontology
- TCGA
The Cancer Genome Atlas
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- RT-qPCR
reverse transcription-quantitative PCR
- LUAD
lung adenocarcinoma
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
Conception and design: BH and GY. Development of methodology and in vitro assays: BH, BW and HW. Analysis and interpretation of data: BH, CZ, YW and LF. Writing, review and/or revision of the manuscript: BH and GY. All authors read and approved the final manuscript.
Ethics approval and informed consent
Not applicable.
Patients' consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Kumar MS, Hancock DC, Molina-Arcas M, Steckel M, East P, Diefenbacher M, Armenteros-Monterroso E, Lassailly F, Matthews N, Nye E, et al. The GATA2 transcriptional network is requisite for RAS oncogene-driven non-small cell lung cancer. Cell. 2012;149:642–655. doi: 10.1016/j.cell.2012.02.059. [DOI] [PubMed] [Google Scholar]
- 2.Ji P, Diederichs S, Wang W, Böing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 3.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 4.Millien G, Cao Y, O'Hara CJ, Tagne JB, Hinds A, Williams MC, Ramirez MI, Kathuria H. ETS1 regulates Twist1 transcription in a Kras (G12D) /Lkb1(−/-) metastatic lung tumor model of non-small cell lung cancer. Clin Exp Metastasis. 2018;35:149–165. doi: 10.1007/s10585-018-9912-z. [DOI] [PubMed] [Google Scholar]
- 5.Seo SK, Kim JH, Choi HN, Choe TB, Hong SI, Yi JY, Hwang SG, Lee HG, Lee YH, Park IC. Knockdown of TWIST1 enhances arsenic trioxide- and ionizing radiation-induced cell death in lung cancer cells by promoting mitochondrial dysfunction. Biochem Biophys Res Commun. 2014;449:490–495. doi: 10.1016/j.bbrc.2014.05.030. [DOI] [PubMed] [Google Scholar]
- 6.Wan X, Kong Z, Chu K, Yi C, Hu J, Qin R, Zhao C, Fu F, Wu H, Li Y, Huang Y. Co-expression analysis revealed PTCH1-3′UTR promoted cell migration and invasion by activating miR-101-3p/SLC39A6 axis in non-small cell lung cancer: Implicating the novel function of PTCH1. Oncotarget. 2018;9:4798–4813. doi: 10.18632/oncotarget.23219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu Z, Li Y, Che Y, Huang J, Sun S, Mao S, Lei Y, Li N, Sun N, He J. The TGFβ-induced lncRNA TBILA promotes non-small cell lung cancer progression in vitro and in vivo via cis-regulating HGAL and activating S100A7/JAB1 signaling. Cancer Lett. 2018;432:156–168. doi: 10.1016/j.canlet.2018.06.013. [DOI] [PubMed] [Google Scholar]
- 8.So A, Jeske YW, Gordon RD, Duffy D, Kelemen L, Stowasser M. No evidence for coding region mutations in the retinoblastoma-associated Kruppel-associated box protein gene (RBaK) causing familial hyperaldosteronism type II. Clin Endocrinol (Oxf) 2006;65:829–831. doi: 10.1111/j.1365-2265.2006.02674.x. [DOI] [PubMed] [Google Scholar]
- 9.Wan X, Pu H, Huang W, Yang S, Zhang Y, Kong Z, Yang Z, Zhao P, Li A, Li T, Li Y. Androgen-induced miR-135a acts as a tumor suppressor through downregulating RBAK and MMP11, and mediates resistance to androgen deprivation therapy. Oncotarget. 2016;7:51284–51300. doi: 10.18632/oncotarget.9992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao Y, Zhou L, Liu B, Deng Y, Wang Y, Wang Y, Huang W, Yuan W, Wang Z, Zhu C, et al. ZNF325, a novel human zinc finger protein with a RBaK-like RB-binding domain, inhibits AP-1- and SRE-mediated transcriptional activity. Biochem Biophys Res Commun. 2006;346:1191–1199. doi: 10.1016/j.bbrc.2006.06.031. [DOI] [PubMed] [Google Scholar]
- 11.Hou J, Aerts J, den Hamer B, van Ijcken W, den Bakker M, Riegman P, van der Leest C, van der Spek P, Foekens JA, Hoogsteden HC, et al. Gene expression-based classification of non-small cell lung carcinomas and survival prediction. PLoS One. 2010;5:e10312. doi: 10.1371/journal.pone.0010312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu TP, Tsai MH, Lee JM, Hsu CP, Chen PC, Lin CW, Shih JY, Yang PC, Hsiao CK, Lai LC, Chuang EY. Identification of a novel biomarker, SEMA5A, for non-small cell lung carcinoma in nonsmoking women. Cancer Epidemiol Biomarkers Prev. 2010;19:2590–2597. doi: 10.1158/1055-9965.EPI-10-0332. [DOI] [PubMed] [Google Scholar]
- 13.Sanchez-Palencia A, Gomez-Morales M, Gomez-Capilla JA, Pedraza V, Boyero L, Rosell R, Fárez-Vidal ME. Gene expression profiling reveals novel biomarkers in nonsmall cell lung cancer. Int J Cancer. 2011;129:355–364. doi: 10.1002/ijc.25704. [DOI] [PubMed] [Google Scholar]
- 14.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 15.Hofman K, Swinnen JV, Claessens F, Verhoeven G, Heyns W. The retinoblastoma protein-associated transcription repressor RBaK interacts with the androgen receptor and enhances its transcriptional activity. J Mol Endocrinol. 2003;31:583–596. doi: 10.1677/jme.0.0310583. [DOI] [PubMed] [Google Scholar]
- 16.Lee MW, Kim DS, Lee JH, Lee BS, Lee SH, Jung HL, Sung KW, Kim HT, Yoo KH, Koo HH. Roles of AKT1 and AKT2 in non-small cell lung cancer cell survival, growth, and migration. Cancer Sci. 2011;102:1822–1828. doi: 10.1111/j.1349-7006.2011.02025.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sung JS, Park KH, Kim ST, Kim YH. Discovery and evaluation of polymorphisms in the AKT2 and AKT3 promoter regions for risk of Korean lung cancer. Genomics Inform. 2012;10:167–174. doi: 10.5808/GI.2012.10.3.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang T, Chen X, Qiao W, Kong L, Sun D, Li Z. Transcription factor E2F1 promotes EMT by regulating ZEB2 in small cell lung cancer. BMC Cancer. 2017;17:719. doi: 10.1186/s12885-017-3701-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feliciano A, Garcia-Mayea Y, Jubierre L, Mir C, Hummel M, Castellvi J, Hernández-Losa J, Paciucci R, Sansano I, Sun Y, et al. miR-99a reveals two novel oncogenic proteins E2F2 and EMR2 and represses stemness in lung cancer. Cell Death Dis. 2017;8:e3141. doi: 10.1038/cddis.2017.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cooper CS, Nicholson AG, Foster C, Dodson A, Edwards S, Fletcher A, Roe T, Clark J, Joshi A, Norman A, et al. Nuclear overexpression of the E2F3 transcription factor in human lung cancer. Lung Cancer. 2006;54:155–162. doi: 10.1016/j.lungcan.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 21.Betticher DC, Heighway J, Thatcher N, Hasleton PS. Abnormal expression of CCND1 and RB1 in resection margin epithelia of lung cancer patients. Br J Cancer. 1997;75:1761–1768. doi: 10.1038/bjc.1997.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu A, Wu B, Guo J, Luo W, Wu D, Yang H, Zhen Y, Yu X, Wang H, Zhou Y, et al. Elevated expression of CDK4 in lung cancer. J Transl Med. 2011;9:38. doi: 10.1186/1479-5876-9-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yun CH, Boggon TJ, Li Y, Woo MS, Greulich H, Meyerson M, Eck MJ. Structures of lung cancer-derived EGFR mutants and inhibitor complexes: Mechanism of activation and insights into differential inhibitor sensitivity. Cancer Cell. 2007;11:217–227. doi: 10.1016/j.ccr.2006.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schwaederlé M, Lazar V, Validire P, Hansson J, Lacroix L, Soria JC, Pawitan Y, Kurzrock R. VEGF-A expression correlates with TP53 mutations in non-small cell lung cancer: Implications for antiangiogenesis therapy. Cancer Res. 2015;75:1187–1190. doi: 10.1158/0008-5472.CAN-14-2305. [DOI] [PubMed] [Google Scholar]
- 25.Dubois F, Keller M, Calvayrac O, Soncin F, Hoa L, Hergovich A, Parrini MC, Mazières J, Vaisse-Lesteven M, Camonis J, et al. RASSF1A suppresses the invasion and metastatic potential of human non-small cell lung cancer cells by inhibiting YAP activation through the GEF-H1/RhoB pathway. Cancer Res. 2016;76:1627–1640. doi: 10.1158/0008-5472.CAN-15-1008. [DOI] [PubMed] [Google Scholar]
- 26.Song Q, Xu Y, Yang C, Chen Z, Jia C, Chen J, Zhang Y, Lai P, Fan X, Zhou X, et al. miR-483-5p promotes invasion and metastasis of lung adenocarcinoma by targeting RhoGDI1 and ALCAM. Cancer Res. 2014;74:3031–3042. doi: 10.1158/0008-5472.CAN-13-2193. [DOI] [PubMed] [Google Scholar]
- 27.Wang XH, Cui YX, Wang ZM, Liu J. Down-regulation of FOXR2 inhibits non-small cell lung cancer cell proliferation and invasion through the Wnt/beta-catenin signaling pathway. Biochem Biophys Res Commun. 2018;500:229–235. doi: 10.1016/j.bbrc.2018.04.046. [DOI] [PubMed] [Google Scholar]
- 28.Wu C, Zhuang Y, Jiang S, Tian F, Teng Y, Chen X, Zheng P, Liu S, Zhou J, Wu J, Wang R, Zou X. Cinnamaldehyde induces apoptosis and reverses epithelial-mesenchymal transition through inhibition of Wnt/β-catenin pathway in non-small cell lung cancer. Int J Biochem Cell Biol. 2017;84:58–74. doi: 10.1016/j.biocel.2017.01.005. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Q, Li YD, Zhang SX, Shi YY. Centromere protein U promotes cell proliferation, migration and invasion involving Wnt/β-catenin signaling pathway in non-small cell lung cancer. Eur Rev Med Pharmacol Sci. 2018;22:7768–7777. doi: 10.26355/eurrev_201811_16400. [DOI] [PubMed] [Google Scholar]
- 30.Stewart DJ. Wnt signaling pathway in non-small cell lung cancer. J Natl Cancer Inst. 2014;106:djt356. doi: 10.1093/jnci/djt356. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.