Abstract
MicroRNAs (mRNAs or miRs) serve an important role in the regulation of gene expression. In the present study, the role of miR-98-5p in bone regeneration was determined. Three osteoblast cell models were established, including primary human stem cells (BMMSC), mouse BMMSC's and MC3T3-E1 cells. miR-98-5p expression was determined using reverse transcription-quantitative (RT-q)PCR. Osteoblast markers, including alkaline phosphatase, runt related transcription factor 2 and transcription factor Sp7, were determined using RT-qPCR and western blot analysis, respectively. Alkaline phosphatase activity was determined in the present study and cell proliferation and apoptosis assays were performed. Furthermore, an association between miR-98-5p and high mobility group AT-Hook 2 (HMGA2) was revealed. This association was determined using TargetScan and a dual luciferase reporter assay. The current study demonstrated that miR-98-5p was downregulated during osteogenic differentiation and further demonstrated that HMGA2 may be a direct target of miR-98-5p. The results also demonstrated that miR-98-5p upregulation significantly inhibited the osteogenic differentiation of MC3T3-E1 cells, an effect that was reversed by an increased HMGA2 expression. Additionally, the results revealed that miR-98-5p upregulation inhibited MC3T3-E1 cell viability and induced cell apoptosis and these effects were eliminated by HMGA2 overexpression. In conclusion, miR-98-5p may prevent bone regeneration through inhibiting osteogenic differentiation and osteoblast growth by targeting HMGA2.
Keywords: bone regeneration, osteogenic differentiation, osteoblast growth, microRNA-98-5p, high mobility group AT-Hook 2
Introduction
MicroRNAs (miRNAs or miRs) are a group of short non-coding RNAs (19–25 nucleotides in length), which are abundant in eukaryotic cells (1). miRNAs regulate post-transcriptional gene expression by binding to the 3′untranslated region (UTR) of target mRNAs and inhibiting their degradation or translation (1,2). It has been demonstrated that miRNAs serve a key role in a variety of biological processes, including cell proliferation, apoptosis and differentiation (2–5). The abnormal expression of miRNA is associated with the development and progression of a variety of diseases including cancer, cardiovascular, metabolic, neurological and bone associated diseases (6–11).
Bone regeneration is critical in many common bone diseases, which include trauma and osteoporosis (12). Although progress has been made in the treatment of these diseases, highly effective therapies are insufficient (13). A study into the molecular mechanisms of bone regeneration will therefore aid the discovery of novel bone disease treatments. In previous studies, it has been demonstrated that miRNAs serve an important role in the regulation of osteoblast differentiation or bone formation (14,15). Tang et al (16) revealed that miR-282 negatively regulated osteoblastic differentiation by targeting special AT-rich-sequence-binding protein 2. A study performed by Arfat et al (17) demonstrated the inhibitory effect of miR-208a-3p on osteoblast differentiation and bone formation by targeting activin A receptor type 1. Furthermore, Zhang et al (18) indicated that miR-335-5p may promote bone formation and regeneration in vivo. miR-342-3p was also demonstrated to increase osteogenic differentiation by inhibiting bone formation or regeneration in vitro (19). These previous studies indicated that miRNAs may serve a role in the promotion or inhibition of bone formation/regeneration. However, the role of miRNAs in these processes remains largely unclear.
miRNA-98-5p has been reported to play important roles in cancer cell proliferation, apoptosis, and metastasis in various types of cancer, including ovarian cancer (20), pancreatic ductal adenocarcinoma (21), non-small-cell lung cancer and hepatocellular carcinoma (22,23). It has also been demonstrated that miR-98-5p serves an important role in the development of Alzheimer's disease (24). It has also been revealed that miR-98-5p serves an important role in the differentiation of human embryonic stem cells (25). The results of the aforementioned studies indicate that miR-98-5p serves a crucial role in the regulation of cell proliferation, apoptosis and differentiation, which are involved in bone formation/regeneration. However, to the best of our knowledge no study has assessed the role of miR-98-5p in bone formation/regeneration.
High mobility group AT-Hook 2 (HMGA2), also known as HMGI-C, is a nuclear-binding protein, which serves a key role in the regulation of cell proliferation and differentiation (26). A previous study revealed that HMGA2 knockdown significantly inhibited the proliferation of bone marrow-derived mesenchymal stem cells (MSCs) (27). However, in recent years, the role of HMGA2 in osteoblast differentiation has been controversial. Wei et al (28) demonstrated that miR let-7 promoted MSC osteogenesis in vitro and in vivo by inhibiting HMGA2 expression (28). It has also been demonstrated that miR-495 prevents new bone regeneration by targeting HMGA2 (29). However, the association between miR-98-5p and HMGA2 has not yet been elucidated.
Therefore, the aim of the current study was to investigate the potential role of miR-98-5p in bone regeneration and to further determine the molecular mechanisms, which govern this.
Materials and methods
Clinical samples
Bone marrow (5–7 ml) was obtained from five male healthy voluntary donors (22–32 years old) at Department of Orthopedics, Qinghai Provincial People's Hospital (Quinghai, China) from January 2017 to March 2018. The use of human samples was approved by the Ethical Committee of the Qinghai Provincial People's Hospital and informed consent was obtained from each patient.
Animals
A total of 10 male C57BL6J mice (4–6 weeks old; 18–20 g) were obtained for use in the current study (Charles River Laboratories, Inc.). All mice were housed at 25±5°C with 40–50% humidity and a 12 h dark/light cycle, and access to food and water ad libitum. All animal experiments were performed following the guidelines provided by the National Institutes of Health for the Care and Use of Laboratory Animals. The current study was approved by the Ethics Committee of Qinghai Provincial People's Hospital (Quinghai, China).
Mouse (m)/human (h) bone marrow-derived mesenchymal stem cells (BMMSCs) isolation and cell culture
Primary mBMMSCs were isolated from C57BL/6 mice and hBMMSCs were isolated from human bone marrow as previously described (30). BMMSCs were cultured in DMEM supplemented with 10% fetal bovine serum (FBS) and antibiotics [100 units/ml penicillin G and 100 µg/ml streptomycin (all, Gibco; Thermo Fisher Scientific, Inc.)]. Cells from passages 3–5 were used in the subsequent experiments.
The mouse preosteoblast cell line, MC3T3-E1, was obtained from the American Type Culture Collection (cat. no. CRL-2594). Cells were cultured in α-minimum essential medium (α-MEM; Gibco; Thermo Fisher Scientific, Inc.) containing 10% FBS and 1% penicillin-streptomycin mix. Cells were then incubated at 37°C with 5% CO2.
Osteogenic differentiation
To induce osteogenic differentiation, mBMMSC, hBMMSC and MC3T3-E1 cells (4×104 cells) were seeded into six-well plates. When 80% confluence was achieved, cells were incubated in osteogenic-inducing medium [10% FBS; 5 mM L-glycerophosphate; 100 nM dexamethasone (Sigma-Aldrich; Merck KGaA) and 50 mg/ml ascorbic acid (Sigma-Aldrich; Merck KGaA)] at 37°C for 14 days. Control group cells were cultured in α-MEM supplemented with 10% FBS at 37°C. To assess the osteoblast phenotype, osteoblast marker genes were examined by RT-qPCR and alkaline phosphatase (ALP) activity was determined.
Cell transfection
MC3T3-E1 cells were seeded into six-well plates at a density of 1×106 cells per well and incubated at 37°C for 24 h. Cells were then transfected with 100 nM miR-98-5p mimic (sense 5′-UGAGGUAGUAAGUUGUAUUGUU-3′, antisense 5′-CAAUACAACUUACUACCUCAUU-3′), 100 nM mimic control (sense 5′-UUCUCCGAACGUGUCACGUTT-3′ and antisense, 5′-ACGUGACACGUUCGGAGAATT-3′; both Guangzhou RiboBio Co., Ltd.), 2 µg Control CRISPR activation plasmid (control-plasmid; cat. no. sc-437275), 2 µg HMGI-C CRISPR Activation Plasmid (HMGA2-plasmid; cat. no. sc-420880-ACT; Santa Cruz Biotechnology, Inc.) or 100 nM miR-98-5p mimic with 2 µg HMGA2-plasmid using Lipofectamine 2000® (cat. no. 11668-027; Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Control cells received no treatment. 48 h after treatment, transfection efficiency was assessed using RT-qPCR.
ALP activity measurement
To detect extracellular ALP activity, the spectrophotometric method (31) was performed with an Alkaline Phosphatase Assay kit (cat. no. P0321; Beyotime Institute of Biotechnology) was used according to the manufacturer's protocol. Absorbance was detected at a wavelength of at 405 nm using a microplate reader (BD Biosciences) following the manufacturer's protocol.
Cell viability detection assay
Transfected MC3T3-E1 cells were seeded into 96-well plates (5×103 cells per well). After transfection for 48 h with miR-98-5p mimics, mimic controls, or miR-98-5p mimics in combination with the HMGA2-plasmid, 100 µl α-MEM medium mixed with 10 µl cell counting kit-8 (CCK-8) solution (cat. no. C0041; Beyotime Institute of Biotechnology) was added to each well. Cells were subsequently incubated at 37°C for 30 min. Cell viability was then evaluated by detecting absorbance at 450 nm using a FLUOstar Omega Microplate Reader (BMG Labtech GmbH). Cells without treatment were used as controls. The aforementioned experiment was repeated three times.
Cell apoptosis assay
To assess the effect of miR-98-5p on MC3T3-E1 cell apoptosis, a FITC/propidium iodide (PI) apoptosis detection kit (cat. no. 70-AP101-100; MultiSciences Biotech Co., Ltd.) was utilized. Subsequent to 48 h transfection with miR-98-5p mimics, mimic controls, or miR-98-5p mimics in combination with the HMGA2-plasmid, MC3T3-E1 cells were digested with 0.25% Trypsin, washed with PBS and subsequently stained with 5 µl Annexin V-FITC and 5 µl PI for 30 min at room temperature. Finally, to analyze cell apoptosis, the FACSCanto II flow cytometer (BD Biosciences) was used according to the manufacturer's protocol, and data was analyzed by Cell Quest software (version 5.1; BD Biosciences). Cells without treatment were used as the control.
Western blot analysis
Proteins were extracted from MC3T3-E1 cells using RIPA buffer (cat. no. P0013E; Beyotime Institute of Biotechnology) following the manufacturer's protocol. A Bicinchoninic Acid Assay kit (cat. no. BCA1-1KT; Sigma-Aldrich; Merck KGaA) was used to quantify protein samples. Equal quantities of protein (30 µg per lane) were separated on 10% SDS-PAGE and transferred onto PVDF membranes. Membranes were then blocked with 5% non-fat milk at room temperature for 1 h and incubated with the following primary antibodies (all 1:1,000) overnight at 4°C: HMGA2 (cat. no. 8179; Cell Signaling Technology, Inc.), ALP (cat. no. sc-365765; Santa Cruz Biotechnology), runt related transcription factor 2 (Runx2; cat. no. 12556; Cell Signaling Technology, Inc.), transcription factor sp7 (Osterix; cat. no. sc-393060; Santa Cruz Biotechnology, Inc.) and β-actin (cat. no. 4970; Cell Signaling Technology, Inc.). Samples were then incubated with a horseradish peroxidase-conjugated anti-rabbit immunoglobulin G secondary antibody (cat. no. 7074; 1:1,000; Cell Signaling Technology Inc.) at room temperature for 2 h. Corresponding protein bands were subsequently visualized using Western Blotting Luminol reagent (cat. no. sc-2048; Santa Cruz Biotechnology, Inc.) following the manufacturers' protocol.
Reverse transcription-quantitative (RT-q)PCR
Total RNA was extracted from cells using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) following the manufacturer's protocol. Total RNA was then reverse transcribed into cDNA using the TaqMan MicroRNA Reverse Transcription kit (cat. no. 4366596; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Synthesized cDNA was subsequently analyzed via qPCR using a SYBR Premix Ex TaqTM II (TliRNaseH Plus) kit (cat. no. RR820a; Takara Bio, Inc.). Primer sequences were as following: miR-98-5p forward, 5′-ACACTCCCUAUACAACUUAC-3′ and reverse, 5′-GGGAAAGUAGUGAGGCCTCAGA-3; U6 forward, 5′-GCTTCGGCAGCACATATACTAAAAT-3′ and reverse, 5′-CGCTTCACGAATTTGCGTGTCAT-3; GAPDH forward, 5′-CTTTGGTATCGTGGAAGGACTC-3′ and reverse, 5′-GTAGAGGCAGGGATGATGTTCT-3; RUNX2 forward, 5′-GATGATGACACTGCCACCTCT-3 and reverse, 5′-AGGGCCCAGTTCTGAAGC-3; ALP forward, 5′-CCGAATTCATGTTGGCCTGTTCAACT-3′ and reverse, 5′-ATGTCGACTTAGTTATTTTCATAATACCAAATTCC-3; Osterix forward, 5′-AGAGATCTGAGCTGGGTAG-3; and reverse, 5′-AAGAGAGCCTGGCAAGAGG-3; HMGA2 forward, 5′-TCCCTCTAAAGCAGCTCAAAA-3′ and reverse, 5′-ACTTGTTGTGGCCATTTCCT-3. The following thermocycling conditions were used for the qPCR: Initial denaturation at 95°C for 10 min, and 35 cycles of 95°C for 15 sec and 55°C for 40 sec. U6 or GAPDH was used as an internal control for miRNA and mRNA expression, respectively. The 2−ΔΔCq method was used to calculate relative gene expression (32).
Dual luciferase reporter assay
miR-98-5p targets were predicted using TargetScan bioinformatics software (www.targetscan.org/vert_71) and binding sites between HMGA2 and miR-98-5p were observed. The wild type (WT-HMGA2) and mutant (MUT-HMGA2) 3′-UTR of HMGA2 were cloned into a pmiR-RB-ReportTM dual luciferase reporter gene plasmid vector (Guangzhou RiboBio Co., Ltd.). MC3T3-E1 cells were co-transfected with WT-HMGA2, MUT-HMGA2 and miR-98-5p mimics or mimic control using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Subsequent to 48 h of cell transfection, luciferase activity was measured using the dual-luciferase assay system (Promega Corporation) and normalized to Renilla luciferase activity.
Statistical analysis
All experiments were performed three times. SPSS software version 17.0 was used to analyze data (SPSS, Inc.). Experimental data were expressed as the mean ± standard deviation. Differences between multiple groups were identified using one-way ANOVA followed by a Tukey's post-hoc. A Student's t-test was used to compare the difference between two groups. P<0.05 was considered to indicate a statistically significant difference.
Results
Significant downregulation of miR-98-5p during osteogenic differentiation
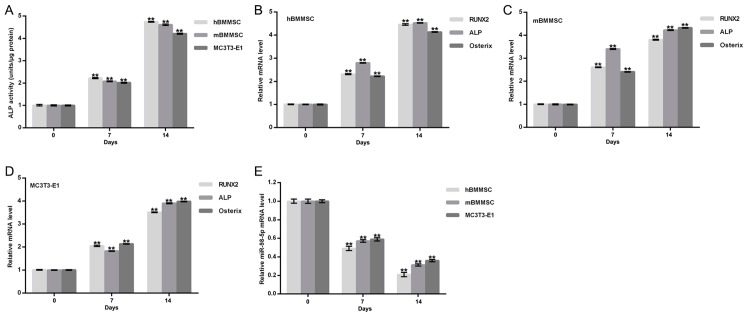
To assess whether miR-98-5p served a role in bone formation, miR-98-5p expression during osteogenic differentiation was detected. Three osteoblast cell models (primary hBMMSC, primary mBMMSC and MC3T3-E1 cells) were used to achieve this. The osteoblast phenotype was confirmed by increased extracellular ALP activity (Fig. 1A) and by the enhanced expression of osteoblast makers including ALP, Runx2 and Osterix (Fig. 1B-D). Compared with day 0, levels of miR-98-5p in hBMMSC, mBMMSC and MC3T3-E1 cells were all significantly reduced at day 7 and day 14 after osteogenic differentiation incubation (Fig. 1E). The results indicate that miR-98-5p may therefore be involved in bone formation.
Figure 1.
miR-98-5p expression during osteogenic differentiation. (A) Extracellular ALP activity of the indicated cell lines. mRNA levels of (B) hBMMSC, (C) mBMMSC and (D) MC3T3-E1 osteoblast makers including Runx2, ALP and Osterix. (E) miR-98-5p expression in hBMMSC, mBMMSC and MC3T3-E1 cells during osteogenic differentiation. Data are presented as the mean ± standard deviation. **P<0.01 vs. day 0. miR, microRNA; ALP, alkaline phosphatase; hBMMSC, human bone marrow-derived mesenchymal stem cells; mBMMSC, mouse bone marrow-derived mesenchymal stem cells; Runx2, runt related transcription factor 2; Osterix, transcription factor sp7.
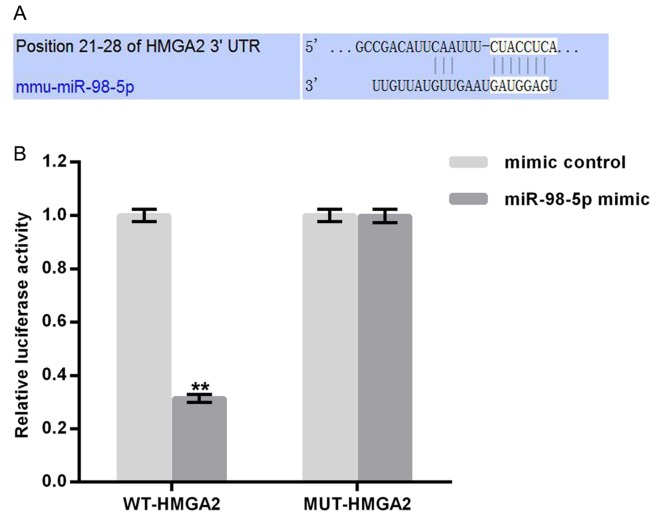
HMGA2 is a target gene of miR-98-5p
TargetScan (http://www.targetscan.org) was utilized to predict potential targets of miR-98-5p. Binding sites between HMGA2 and miR-98-5p were revealed (Fig. 2A). To confirm these, potential binding sites a luciferase reporter assay was performed. As presented in Fig. 2B, compared with MUT-HMGA2 and miR-98-5p mimic transfected cells, luciferase activity was significantly reduced following WT-HMGA2 and miR-98-5p mimic transfection. The results demonstrate that miR-98-5p may directly target HMGA2.
Figure 2.
HMGA2 as a direct target of miR-98-5p. (A) Interaction between miR-98-5p and the 3′UTR of HMGA2 as predicted using TargetScan. (B) Luciferase activity of a reporter containing a WT-HMGA2 3′UTR or a MUT-HMGA2 3′UTR. All data are presented as the mean ± standard deviation of three independent experiments. **P<0.01 vs. mimic control. HMGA2, high mobility group AT-Hook 2; miR, microRNA; UTR, untranslated region; WT, wild-type; MUT, mutant.
miR-98-5p expression prevents the osteogenic differentiation of MC3T3-E1 cells
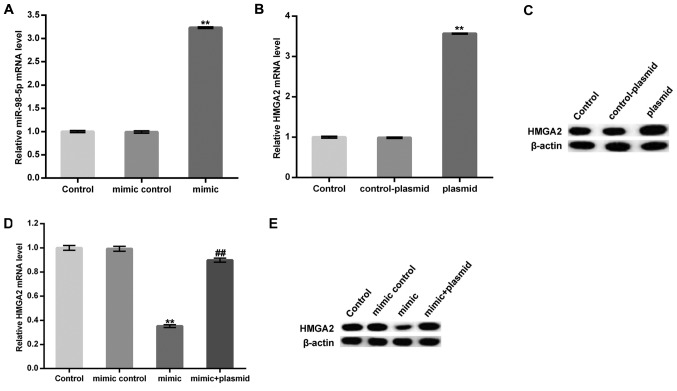
MC3T3-E1 cells were transfected with a miR-98-5p mimic, a mimic control, a control-plasmid, a HMGA2-plasmid, or a miR-98-5p mimic in combination with a HMGA2-plasmid for 48 h and subsequently subjected to osteogenic differentiation induction. Transfection efficiency was measured using RT-qPCR. As presented in Fig. 3A-C, the miR-98-5p mimic significantly upregulated miR-98-5p in MC3T3-E1 cells. Furthermore, protein and mRNA levels of HMGA2 in MC3T3-E1 cells were enhanced following treatment with the HMGA2-plasmid. The data demonstrated that the miR-98-5p mimic significantly reduced the mRNA level of HMGA2. The protein level of HMGA2 was markedly decreased by miR-98-5p mimic. However, these decreases were reversed by HMGA2-plasmid co-transfection (Fig. 3D and E).
Figure 3.
miR-98-5p mimic treatment inhibited HMGA2 expression in MC3T3-E1 cells. (A) Relative miR-98-5p expression in MC3T3-E1 cells of different transfection treatments. (B) Relative HMGA2 mRNA and (C) protein expression in MC3T3-E1 control, control-plasmid and plasmid cell treatments. (D) Relative HMGA2 mRNA and (E) protein expression in MC3T3-E1 control, mimic control, mimic and mimic + plasmid treatments. Data are presented as the mean ± standard deviation. **P<0.01 vs. control group; ##P<0.01 vs. mimic group. miR, microRNA; HMGA2, high mobility group AT-Hook 2.
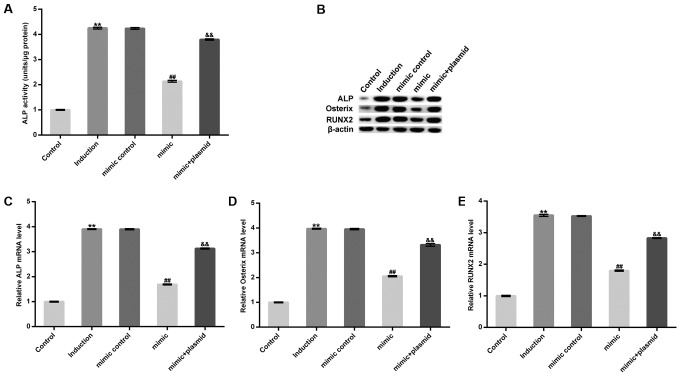
On day 14 after induction, extracellular ALP activity increased and was subsequently inhibited by treatment with the miR-98-5p mimic. However, this inhibition was eliminated following HMGA2-plasmid treatment (Fig. 4A). Furthermore, treatment with the miR-98-5p mimic markedly decreased the protein levels, and significantly decreased the mRNA levels of osteoblast makers including ALP, Runx2 and Osterix. However, these decreases were reversed following HMGA2-plasmid transfection (Fig. 4B-E). The results demonstrate that miR-98-5p may prevent osteogenic differentiation.
Figure 4.
Effect of the miR-98-5p mimic on MC3T3-E1 cell osteogenic differentiation. Osteoblast phenotypes were assessed by determining the expression of osteoblast marker genes and extracellular ALP activity. (A) Extracellular ALP activity in MC3T3-E1 cells. (B) Protein level of ALP, Osterix and Runx2 and mRNA levels of (C) ALP, (D) Osterix and (E) Runx2. Data are presented as the mean ± standard deviation. **P<0.01 vs. Control; ##P<0.01 vs. Induction; &&P<0.01 vs. mimic. miR, microRNA; ALP, alkaline phosphatase; Osterix, transcription factor sp7; Runx2, runt related transcription factor 2.
miR-98-5p overexpression inhibits osteoblast viability and induces apoptosis
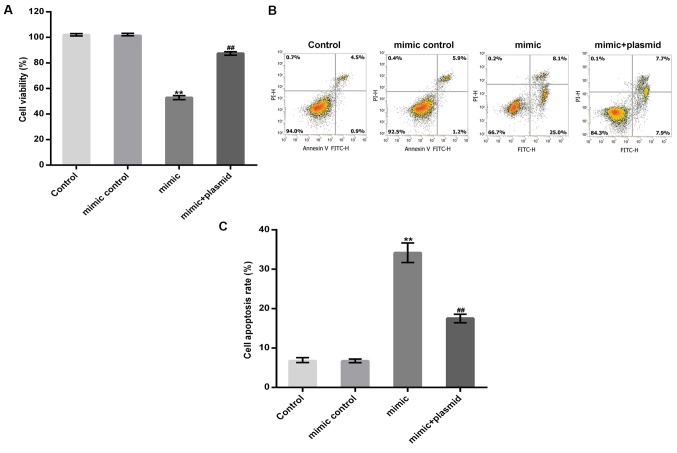
MC3T3-E1 cells were transfected with a miR-98-5p mimic, a mimic control, or a miR-98-5p mimic in combination with a HMGA2-plasmid for 48 h. A CCK-8 assay and flow cytometry were subsequently performed to detect MC3T3-E1 cell viability and apoptosis, respectively. The results revealed that the miR-98-5p mimic significantly reduced MC3T3-E1 cell viability and induced MC3T3-E1 cell apoptosis and these changes were reversed by HMGA2 upregulation (Fig. 5A-C).
Figure 5.
Effect of the miR-98-5p mimic on MC3T3-E1 cell viability and apoptosis. (A) A cell counting kit-8 assay and (B) flow cytometry were performed to detect MC3T3-E1 cell viability and apoptosis, respectively. (C) Cell apoptosis rate was subsequently calculated and presented. Data are presented as the mean ± standard deviation. **P<0.01 vs. Control; ##P<0.01 vs. mimic. miR, microRNA.
Discussion
The results of the present study demonstrated that miR-98-5p was significantly downregulated during osteogenic differentiation in vitro and that HMGA2 may be a direct target of miR-98-5p. miR-98-5p upregulation inhibited ALP activity and the expression of osteoblast markers, including ALP, Runx2 and Osterix. This inhibition may have been due to miR-98-5p targeting HMGA2, demonstrating the inhibitory effect of miR-98-5p on osteogenic differentiation. In addition, the results of the current study demonstrated that miR-98-5p upregulation inhibited cell viability and induced cell apoptosis in the mouse preosteoblast cell line, MC3T3-E1. miR-98-5p may have also inhibited bone regeneration by targeting HMGA2 and therefore may be an encouraging therapeutic target for the enhancement of bone regeneration.
New bone regeneration is essential in many common bone diseases, including trauma and osteoporosis (12). Although previous studies have identified a variety of therapies to treat these diseases, the efficacy of these treatments remains unsatisfactory (13). Evidence has revealed that miRNAs serve a regulatory role in cell proliferation, differentiation and apoptosis (2–5). Increasing evidence has also revealed that miRNAs are involved in the regulation of osteoblasts differentiation, bone metabolism and bone formation (14–19). miR-467g has been demonstrated to prevent new bone regeneration by regulating the indian hedgehog protein/Runx2 signaling pathway (33). miR-221 was revealed to suppress osteoblast differentiation and bone formation by regulating Runx2 expression (34). miR-214 has also been identified to inhibit osteogenesis by targeting baculoviral IAP repeat-containing protein 7 (35). Therefore, miRNAs may serve as novel therapeutic targets for the treatment of bone degenerative disorders and other abnormal bone formation disorders.
In the present study, three osteoblast cell models (primary hBMMSC, primary mBMMSC and MC3T3-E1 cells) were used to investigate the role of miR-98-5p in bone regeneration. miR-98-5p significantly decreased during osteogenic differentiation in vitro. Furthermore, HMGA2 was revealed to be a direct target of miR-98-5p. HMGA2 is a nuclear-binding protein that serves an important role in the regulation of cell proliferation and differentiation (26). A previous study has demonstrated that HMGA2 knockdown results in the inhibition of MSC proliferation (27). The effect of miR-98-5p on the osteogenic differentiation of the mouse preosteoblast cell line, MC3T3-E1, was subsequently investigated in the current study. Consistent with previous studies (27,29), the results of the present study demonstrated that miR-98-5p significantly inhibited MC3T3-E1 cell osteogenic differentiation by repressing the expression of HMGA2. In addition, miR-98-5p upregulation significantly inhibited cell viability and induced apoptosis in MC3T3-E1 cells and these effects were eliminated by HMGA2 overexpression.
In summary, the results obtained in the present study indicated that miR-98-5p was downregulated during osteogenic differentiation in vitro and its overexpression could inhibit osteogenic differentiation and suppress the growth of osteoblast cells by targeting HMGA2. Therefore, miR-98-5p may be a promising therapeutic target for the enhancement of new bone regeneration.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
FZ was responsible for experimental design and manuscript preparation. FW performed most of the experiments and ZX analyzed the data.
Ethics approval and consent to participate
The use of human samples was approved by the Ethical Committee of the Qinghai Provincial People's Hospital (Quinghai, China) and informed consent was obtained from each patient.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Hammond SM. An overview of microRNAs. Adv Drug Deliv Rev. 2015;87:3–14. doi: 10.1016/j.addr.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Soifer HS, Rossi JJ, Saetrom P. MicroRNAs in disease and potential therapeutic applications. Mol Ther. 2017;15:2070–2079. doi: 10.1038/sj.mt.6300311. [DOI] [PubMed] [Google Scholar]
- 4.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 5.O'Connell RM, Rao DS, Chaudhuri AA, Baltimore D. Physiological and pathological roles for microRNAs in the immune system. Nat Rev Immunol. 2010;10:111–122. doi: 10.1038/nri2708. [DOI] [PubMed] [Google Scholar]
- 6.Tutar Y. miRNA and cancer; computational and experimental approaches. Curr Pharm Biotechnol. 2014;15:429. doi: 10.2174/138920101505140828161335. [DOI] [PubMed] [Google Scholar]
- 7.Zhong Z, Hou J, Zhang Q, Zhong W, Li B, Li C, Liu Z, Yang M, Zhao P. Circulating microRNA expression profiling and bioinformatics analysis of dysregulated microRNAs of patients with coronary artery disease. Medicine (Baltimore) 2018;97:e11428. doi: 10.1097/MD.0000000000011428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang S, Zhang R, Wu F, Li X. MicroRNA-208a regulates H9c2 cells simulated ischemia-reperfusion myocardial injury via targeting CHD9 through Notch/NF-kappa B signal pathways. Int Heart J. 2018;59:580–588. doi: 10.1536/ihj.17-147. [DOI] [PubMed] [Google Scholar]
- 9.Price NL, Ramírez CM, Fernández-Hernando C. Relevance of microRNA in metabolic diseases. Crit Rev Clin Lab Sci. 2014;51:305–320. doi: 10.3109/10408363.2014.937522. [DOI] [PubMed] [Google Scholar]
- 10.Qiu L, Tan EK, Zeng L. microRNAs and neurodegenerative diseases. Adv Exp Med Biol. 2015;888:85–105. doi: 10.1007/978-3-319-22671-2_6. [DOI] [PubMed] [Google Scholar]
- 11.Rossi M, Tagliaferri P, Tassone P. MicroRNAs in multiple myeloma and related bone disease. Ann Transl Med. 2015;3:334. doi: 10.3978/j.issn.2305-5839.2015.12.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Majidinia M, Sadeghpour A, Yousefi B. The roles of signaling pathways in bone repair and regeneration. J Cell Physiol. 2018;233:2937–2948. doi: 10.1002/jcp.26042. [DOI] [PubMed] [Google Scholar]
- 13.Hu C, Zhang T, Ren B, Deng Z, Cai L, Lei J, Ping A. Effect of vacuum-assisted closure combined with open bone grafting to promote rabbit bone graft vascularization. Med Sci Monit. 2015;21:1200–1206. doi: 10.12659/MSM.892939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lian JB, Stein GS, van Wijnen AJ, Stein JL, Hassan MQ, Gaur T, Zhang Y. MicroRNA control of bone formation and homeostasis. Nat Rev Endocrinol. 2012;8:212–227. doi: 10.1038/nrendo.2011.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Valenti MT, Dalle Carbonare L, Mottes M. Role of microRNAs in progenitor cell commitment and osteogenic differentiation in health and disease (Review) Int J Mol Med. 2018;41:2441–2449. doi: 10.3892/ijmm.2018.3452. [DOI] [PubMed] [Google Scholar]
- 16.Tang J, Zhang Z, Jin X, Shi H. miR-383 negatively regulates osteoblastic differentiation of bone marrow mesenchymal stem cells in rats by targeting Satb2. Bone. 2018;114:137–143. doi: 10.1016/j.bone.2018.06.010. [DOI] [PubMed] [Google Scholar]
- 17.Arfat Y, Basra MAR, Shahzad M, Majeed K, Mahmood N, Munir H. miR-208a-3p suppresses osteoblast differentiation and inhibits bone formation by targeting ACVR1. Mol Ther Nucleic Acids. 2018;11:323–336. doi: 10.1016/j.omtn.2017.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang L, Tang Y, Zhu X, Tu T, Sui L, Han Q, Yu L, Meng S, Zheng L, Valverde P, et al. Overexpression of MiR-335-5p promotes bone formation and regeneration in mice. J Bone Miner Res. 2017;32:2466–2475. doi: 10.1002/jbmr.3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang M, Qing Y, Shi Q, Cao Y, Song K. miR-342-3p elevates osteogenic differentiation of umbilical cord mesenchymal stem cells via inhibiting Sufu in vitro. Biochem Biophys Res Commun. 2017;491:571–577. doi: 10.1016/j.bbrc.2017.07.163. [DOI] [PubMed] [Google Scholar]
- 20.Wang Y, Bao W, Liu Y, Wang S, Xu S, Li X, Li Y, Wu S. miR-98-5p contributes to cisplatin resistance in epithelial ovarian cancer by suppressing miR-152 biogenesis via targeting Dicer1. Cell Death Dis. 2018;9:447. doi: 10.1038/s41419-018-0390-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fu Y, Liu X, Chen Q, Liu T, Lu C, Yu J, Miao Y, Wei J. Downregulated miR-98-5p promotes PDAC proliferation and metastasis by reversely regulating MAP4K4. J Exp Clin Cancer Res. 2018;37:130. doi: 10.1186/s13046-018-0807-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu F, Mo Q, Wan X, Dan J, Hu H. NEAT1/hsa-mir-98- 5p/MAPK6 axis is involved in non-small-cell lung cancer development. J Cell Biochem. 2019;120:2836–2846. doi: 10.1002/jcb.26442. [DOI] [PubMed] [Google Scholar]
- 23.Jiang T, Li M, Li Q, Guo Z, Sun X, Zhang X, Liu Y, Yao W, Xiao P. MicroRNA-98-5p inhibits cell proliferation and induces cell apoptosis in hepatocellular carcinoma via targeting IGF2BP1. Oncol Res. 2017;25:1117–1127. doi: 10.3727/096504016X14821952695683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Q, Li X, Wang L, Zhang Y, Chen L. miR-98-5p acts as a target for Alzheimer's disease by regulating Aβ production through modulating SNX6 expression. J Mol Neurosci. 2016;60:413–420. doi: 10.1007/s12031-016-0815-7. [DOI] [PubMed] [Google Scholar]
- 25.Sahu M, Mallick B. Deciphering synergistic regulatory networks of microRNAs in hESCs and fibroblasts. Int J Biol Macromol. 2018;113:1279–1286. doi: 10.1016/j.ijbiomac.2018.03.061. [DOI] [PubMed] [Google Scholar]
- 26.Park SM, Shell S, Radjabi AR, Schickel R, Feig C, Boyerinas B, Dinulescu DM, Lengyel E, Peter ME. Let-7 prevents early cancer progression by suppressing expression of the embryonic gene HMGA2. Cell Cycle. 2007;6:2585–2590. doi: 10.4161/cc.6.21.4845. [DOI] [PubMed] [Google Scholar]
- 27.Kalomoiris S, Cicchetto AC, Lakatos K, Nolta JA, Fierro FA. Fibroblast growth factor 2 regulates high mobility group A2 expression in human bone marrow-derived mesenchymal stem cells. J Cell Biochem. 2016;117:2128–2137. doi: 10.1002/jcb.25519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wei J, Li H, Wang S, Li T, Fan J, Liang X, Li J, Han Q, Zhu L, Fan L, Zhao RC. let-7 enhances osteogenesis and bone formation while repressing adipogenesis of human stromal/mesenchymal stem cells by regulating HMGA2. Stem Cells Dev. 2014;23:1452–1463. doi: 10.1089/scd.2013.0600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tian Z, Zhou H, Xu Y, Bai J. MicroRNA-495 inhibits new bone regeneration via targeting high mobility group AT-Hook 2 (HMGA2) Med Sci Monit. 2017;23:4689–4698. doi: 10.12659/MSM.904404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsubara T, Suardita K, Ishii M, Sugiyama M, Igarashi A, Oda R, Nishimura M, Saito M, Nakagawa K, Yamanaka K, et al. Alveolar bone marrow as a cell source for regenerative medicine: Differences between alveolar and iliac bone marrow stromal cells. J Bone Miner Res. 2005;20:399–409. doi: 10.1359/JBMR.041117. [DOI] [PubMed] [Google Scholar]
- 31.Ni P, Xie J, Chen C, Jiang Y, Zhao Z, Zhang Y, Lu Y, Yu J. Spectrophotometric determination of the activity of alkaline phosphatase and detection of its inhibitors by exploiting the pyrophosphate-accelerated oxidase-like activity of nanoceria. Mikrochim Acta. 2019;186:320. doi: 10.1007/s00604-019-3423-8. [DOI] [PubMed] [Google Scholar]
- 32.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 33.Kureel J, John AA, Dixit M, Singh D. MicroRNA-467g inhibits new bone regeneration by targeting Ihh/Runx-2 signaling. Int J Biochem Cell Biol. 2017;85:35–43. doi: 10.1016/j.biocel.2017.01.018. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Y, Gao Y, Cai L, Li F, Lou Y, Xu N, Kang Y, Yang H. MicroRNA-221 is involved in the regulation of osteoporosis through regulates RUNX2 protein expression and osteoblast differentiation. Am J Transl Res. 2017;9:126–135. [PMC free article] [PubMed] [Google Scholar]
- 35.Liu J, Li Y, Luo M, Yuan Z, Liu J. MicroRNA-214 inhibits the osteogenic differentiation of human osteoblasts through the direct regulation of baculoviral IAP repeat-containing 7. Exp Cell Res. 2017;351:157–162. doi: 10.1016/j.yexcr.2017.01.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.