Abstract
Studies performed previously have indicated that long non-coding RNAs (lncRNAs) may be involved in skeletal muscle regeneration; however, the roles of lncRNAs during the repair of skeletal muscle contusion remain unclear. The present study established a mouse skeletal muscle contusion injury model to identify the roles of lncRNAs that are specifically enriched in the skeletal muscle, namely metastasis-associated lung adenocarcinoma transcript 1 (Malat1), H19, myogenesis-associated lnc (lnc-mg), long intergenic non-protein coding RNAs (linc)-muscle differentiation 1 (linc-MD1), linc-yin yang 1 (linc-YY1) and sirtuin 1-antisense (Sirt1-AS). Morphological analyses revealed that fibrotic scars and regenerating myofibers were formed in the muscle following contusion injury. Gene expression was analyzed by reverse transcription-quantitative polymerase chain reaction. The data revealed that the expression of inflammatory cytokines, myogenic regulatory factors and angiogenic factors increased significantly following skeletal muscle contusion. Additionally, various lncRNAs, including Malat1, H19, lnc-mg, linc-MD1, linc-YY1 and Sirt1-AS were also upregulated. Correlation was also observed between lncRNAs and regulatory factors for skeletal muscle regeneration including transforming growth factor-β1, myogenic differentiation, myogenin, myogenic factor 5 (myf5), myf6, hypoxia-inducible factor-1α and angiopoietin 1. In conclusion, lncRNAs may serve important roles in the regeneration of skeletal muscle following contusion injury, which provides a promising therapy avenue for muscle injury.
Keywords: long non-coding RNAs, skeletal muscle regeneration, inflammatory cytokines, myogenic regulatory factors, angiogenic factors, macrophage
Introduction
Skeletal muscle injury is a common injury in daily life and/or during physical exercise. Skeletal muscle has the remarkable ability to self-regenerate following injury. The mechanism of skeletal muscle repair is one of the major issues surrounding the field of sports medicine. In particular, skeletal muscle contusion is a common form of injury. It is a contact injury caused mainly by an acute, relatively large blunt trauma that is characterized by intact skin and no external damage. The repair of damaged skeletal muscle is a complex process which mainly consists of the inflammatory response, myofiber regeneration, angiogenesis and fibrosis (1). The first phase occurs in the first few days after injury, characterized by muscle fiber rupture, necrosis and infiltration of inflammatory cells. The second phase entails myofiber regeneration, consisting of the phagocytosis of necrotic muscle fibers and formation of new muscle fibers (2). The last phase, namely tissue remodeling, is characterized by the maturation of regenerating myofibers and formation of scar tissue (3–5).
Effective repair of damaged skeletal muscle requires the coordinated action of several cell types and a variety of factors. For example, macrophages serve complex roles in damaged skeletal muscle, and may be involved in all phases of skeletal muscle regeneration mentioned above (6). Inflammatory factors including transforming growth factor (TGF)-β1, interleukin (IL)-10, IL-6, IL-1β, tumor necrosis factor (TNF)-α and interferon (IFN)-γ, produced by macrophages and monocytes, also have the potential to influence muscle repair and regeneration by modulating the proliferation and differentiation of satellite cells in the injured tissue (3). In addition, other physiological processes involved in muscle regeneration, namely myoblast proliferation, migration and subsequent fusion into myotubes, are under the control of a number of regulatory factors including growth factors and myogenic regulatory factors (MRFs), which constitute the key determinants of the progression of satellite cell activation during myogenesis and muscle regeneration (7–9).
In previous years, the roles of long non-coding RNAs (lncRNAs) have become the focus of research. lncRNAs, which can, are non-coding RNAs with a transcript length of >200 nucleotides, which have emerged as an important class of regulators of gene expression, and localize to the nucleus and the cytosol (10,11). lncRNAs participate in various of molecular regulatory processes including transcriptional and post-transcriptional regulation, protein localization, telomere replication and RNA interference (12). Accumulating evidence from myoblast differentiation in vitro, cardiotoxin (CTX)-mediated injury or mdx mice models suggested that certain lncRNAs, including metastasis-associated lung adenocarcinoma transcript 1 (Malat1), H19, long intergenic non-protein coding RNAs (linc)-muscle differentiation 1 (linc-MD1), linc-yin yang 1 (linc-YY1), sirtuin 1-antisense (Sirt1 AS) lncRNA and myogenesis-associated lnc (lnc-mg), can modulate myogenesis and muscle regeneration (13–16). However, few studies have evaluated the roles of lncRNAs in contused muscle (17). In addition, the association between lncRNAs and macrophages, inflammatory factors and angiogenic factors in the regeneration of contused skeletal muscle remains unclear. Therefore, the aim of the present study was to determine whether lncRNAs may be involved in the repair of skeletal muscle following contusion injury.
Materials and methods
Animals
A total of 40, 8 week old C57BL/6 male mice weighing 18.2–22.9 g, purchased from JiesiJie-Lab Animal Research Center (Shanghai JiesiJie Experimental Animal Co., Ltd.), were housed at 21±2°C and 50±5% humidity on a 12 h light/dark cycle, and received water and food ad libitum. Following acclimatization to the local environment for 7 days, the mice were randomly divided into two groups: The uninjured control group (group C) and the muscle contusion group (group M). Mice from group M were used for the induction of contusion injury. All experimental protocols were approved by the Ethics Review Committee for Animal Experimentation of Shanghai University of Sports (approval no. 2016006).
Contusion injury model induction
A simple and reproducible muscle contusion model in mice was applied as previously described with little modification (5). Prior to contusion, mice were anesthetized with 400 mg/kg chloral hydrate administered intraperitoneally. The knee joints of the mice were placed in the extension position at 0° while the ankle joints were placed in the back-extension position at 90°. A 16.8 g stainless steel ball (diameter, 1.59 cm) was dropped from a height of 125 cm through a tube (interior diameter of the tube, 1.60 cm) onto an impactor (surface, 28.26 mm2) resting on the middle of the gastrocnemius muscle (GM) of the mice, resulting in an acute skeletal muscle injury (5). The muscle contusion created by this method was a high-energy blunt trauma that resulted in the formation of a large hematoma, breakage of muscle fibers, exudation of red blood cells and infiltration of inflammatory cells. This was followed by acute inflammatory reactions and extensive muscle regeneration (10), a healing process that is comparable to that observed in humans (18). All mice were sacrificed for GM isolation at days 3, 6, 12 and 24 following the induction of contusion injury.
Hematoxylin and eosin (H&E) staining
At days 3, 6, 12 and 24 following muscle contusion, the right GM was harvested, fixed in 4% paraformaldehyde at 4°C for 24 h and then embedded in paraffin (n=6 mice/group). Cross sections cut at 4 µm were produced from the GM, which were subsequently stained with H&E to evaluate the general morphology using a method described previously (5). Images were captured for each muscle section using a brightfield microscope (magnification, ×200; Labophot-2 microscope; Nikon Corporation).
Masson's trichrome staining
To visualize fibrosis in the muscle injury sites, Total collagen staining was performed to detect fibrosis in injured muscle via Masson's trichrome staining (total collagen staining; Servicebio, Inc.). The procedure was as follows: GM tissue samples were cut into 4-µm-thick sections and stained with hematoxylin for 5 min, 1% hydrochloric acid alcohol for 5 sec, Biebrich scarlet-acid fuchsin for 8 min, Phosphomolybdic acid aqueous solution for 4 min, Aniline blue solution for 5 min, and 1% glacial acetic acid for 1 min. All staining was performed at room temperature. Following Masson's trichrome staining, images were captured for each muscle section viewed under a bright-field microscope (magnification, ×400; Labophot-2; Nikon Corporation). The ratio of the fibrotic area to the total cross-sectional area of the muscle was calculated to estimate the extent of fibrosis formation using Image Pro 6.0 (Media Cybernetics, Inc.). A total of six different fields of view (magnification, ×400) were randomly selected from each section.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis
Total RNA from the GM was extracted using TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.), and the concentration and purity were determined by measuring the absorbance at 260 and 280 nm with a microplate reader (Model 550 Microplate Reader; Bio-Rad Laboratories, Inc.). Total RNA (2 µg) was subsequently reverse transcribed into complementary cDNA (cDNA) using the Revertaid First Strand cDNA Synthesis kit (Thermo Fisher Scientific, Inc.). The temperature protocol for RT was as follows: 25°C for 5 min followed by 42°C for 60 min, termination at 70°C for 5 min and cooling at 4°C. The qPCR reaction system included SYBR Green (Fermentas; Thermo Fisher Scientific, Inc.), nuclease-free water, upstream and downstream primers (designed and synthesized by Shanghai Shenggong Biology Engineering Technology Service, Ltd.; primer sequences presented in Table I) and 1 µl cDNA, made to a total volume of 20 µl/well. An Applied Biosystems 7500 Real-Time PCR System (Thermo Fisher Scientific, Inc.) was used for amplification by applying the following parameters: Denaturation at 95°C for 10 min, 40 cycles of priming at 95°C for 15 sec, and annealing/extension at 60°C for 1 min. Relative expression values were calculated using the comparative quantification cycle (2−ΔΔCq) method and GAPDH was used as the reference gene (19).
Table I.
Primers used for reverse transcription-quantitative PCR.
| Target gene | Forward primer sequence | Reverse primer sequence |
|---|---|---|
| CD68 | 5′-CAAAGCTTCTGCTGTGGAAAT-3′ | 5′-GACTGGTCACGGTTGCAAG-3′ |
| CD163 | 5′-GCAAAAACTGGCAGTGGG-3′ | 5′-GTCAAAATCACAGACGGAGC-3′ |
| CD206 | 5′-GGATTGTGGAGCAGATGGAAG-3′ | 5′-CTTGAATGGAAATGCACAGAC-3′ |
| IFN-γ | 5′-GCTTTGCAGCTCTTCCTCAT-3′ | 5′-GTCACCATCCTTTTGCCAGT-3′ |
| TNF-α | 5′-CTTCTGTCTACTGAACTTCGGG-3′ | 5′-CACTTGGTGGTTTGCTACGAC-3′ |
| IL-1β | 5′-TGACGTTCCCATTAGACAACTG-3′ | 5′-CCGTCTTTCATTACACAGGACA-3′ |
| IL-6 | 5′-GAACAACGATGATGCACTTGC-3′ | 5′CTTCATGTACTCCAGGTAGCTATGGT-3′ |
| TGF-β1 | 5′-TGCGCTTGCAGAGATTAAAA-3′ | 5′-CGTCAAAAGACAGCCACTCA-3′ |
| IL-10 | 5′-CAAGGAGCATTTGAATTCCC-3′ | 5′-GGCCTTGTAGACACCTTGGTC-3′ |
| Myo D | 5′-GAGCGCATCTCCACAGACAG-3′ | 5′-AAATCGCATTGGGGTTTGAG-3′ |
| Myogenin | 5′-CCAGTACATTGAGCGCCTAC-3′ | 5′-ACCGAACTCCAGTGCATTGC-3′ |
| Myf5 | 5′-GGAATGCCATCCGCTACATT-3′ | 5′-CGTCAGAGCAGTTGGAGGTG-3′ |
| Myf6 | 5′-CCTCAGCCTCCAGCAGTCTT-3′ | 5′-TTCTCCACCACCTCCTCCAC-3′ |
| VEGF | 5′-TAACAGTGAAGCGGAGTG-3′ | 5′-TTTGACCCTTTCCCTTTCCTCG-3′ |
| HIF-1α | 5′-GGCGAGAACGAGAAGAAAAAGATGA-3′ | 5′-GCTCACATTGTGGGGAAGTGG-3′ |
| Angpt1 | 5′-AACCGGATTCAACATGGGCA-3′ | 5′-GAGCGTTGGTGTTGTACTGC-3′ |
| Malat1 | 5′-CACTTGTGGGGAGACCTTGT-3′ | 5′-TGTGGCAAGAATCAAGCAAG-3′ |
| H19 | 5′-TGACTTCATCATCTCCCTCCTGTC-3′ | 5′-GGGTAAATGGGGAAACAGAGTCAC-3′ |
| lnc-mg | 5′-CTGCATCACGGAAGGAGATA-3′ | 5′-AACAATCCATCCTCATTGGC-3′ |
| Sirt1 AS | 5′-AATCCAGTCATTAAACGGTCTACAA-3′ | 5′-TAGGACCATTACTGCCAGAGG-3′ |
| linc-MD1 | 5′-GCAAGAAAACCACAGAGGAGG-3′ | 5′-GTGAAGTCCTTGGAGTTTGAGCA-3′ |
| Linc-YY1 | 5′-AGTTACAGGGAAGTTTGGGCTAC-3′ | 5′-AGGCAAAGGACGGCTGTGAG-3′ |
| GAPDH | 5′-ACTCCACTCACGGCAAATTC-3′ | 5′-TCTCCATGGTGGTGAAGACA-3′ |
IL-1β, interleukin-1β; IL-6, interleukin-6; TNF-α, tumor necrosis factor-α; IFN-γ, interferon-γ; IL-10, interleukin-10; TGF-β1, transforming growth factor-β1; MyoD, myogenic differentiation 1; myf5, myogenic factor 5; myf6, myogenic factor 6; HIF-1α, hypoxia-inducible factor-1α; VEGF, vascular endothelial growth factor; Angpt1, angiopoietin 1; Malat1, metastasis associated lung adenocarcinoma transcript 1; lnc-mg, myogenesis-associated long non-coding RNA; Sirt1 AS, sirtuin 1-antisense; linc-MD1, long intergenic non-protein coding RNAs-muscle differentiation 1; linc-YY1, long intergenic non-protein coding RNA-yin yang 1.
Statistical analysis
All data were analyzed using the SPSS 22.0 software (IBM Corp.) and are presented as the mean ± standard deviation of at least three experiments. Statistical analysis was carried out using one-way analysis of variance, and post-hoc multiple comparisons were performed using the Bonferroni test. Image Pro 6.0 software was used to assess fibrosis, which was compared using an independent samples t-test. Correlations were calculated according to Pearson's correlation coefficient. P<0.05 was considered to indicate a statistically significant difference.
Results
Evaluation of skeletal muscle repair following contusion injury by H&E staining
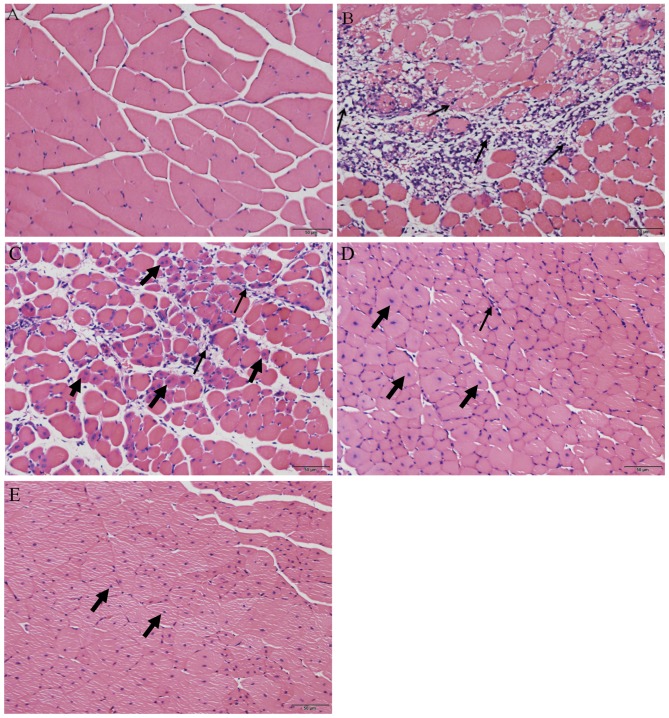
Following H&E staining, the histological appearance of the skeletal muscle was compared between the uninjured control group and the muscle contusion group. Skeletal muscles that were not injured exhibited cells that were arranged regularly with the nuclei, stained blue-black, located primarily in the cell periphery (Fig. 1A). On day 3 following injury induction, a greater number of inflammatory cells and necrotic muscle fibers were observed (Fig. 1B). However, 6 days after contusion injury, the necrotic muscle fibers had been replaced mostly by muscle fibers containing centrally localized nuclei or polynucleated myoblasts/myotubes in the injured areas (Fig. 1C). In addition, inflammatory cells gradually disappeared from the injury site from day 6 to day 24 (Fig. 1C-E). On day 12 following injury induction, a small number of developing myofibers with centrally localized nuclei could be observed in the injured area (Fig. 1D). Finally, on day 24 post-injury, the regenerated fibers appeared to have matured with their nuclei having migrated from the center of the cell to the periphery (20) (Fig. 1E). This indicates that muscle regeneration was on the verge of completion on day 24 following injury.
Figure 1.
Representative images from the hematoxylin and eosin staining of gastrocnemius muscle tissues from the (A) uninjured control group, and (B) 3, (C) 6, (D) 12 and (E) 24 days after injury induction. Thin arrows indicate inflammatory cells, bold arrows indicate central nucleation. Scale bars, 50 µm.
Fibrosis during damaged skeletal muscle repair
Following Masson's trichrome staining, the tissue in the injured area of the GM was assessed. Fibrotic scar tissues, in the form of collagen, were stained in blue, whereas skeletal muscle cells were stained in red (Fig. 2). Little or no blue collagen fibers were observed in the uninjured muscle (Fig. 2A), whereas intense deposition of blue collagen fibers were noted surrounding the regenerating myofibers 12 days after the induction of contusion injury (Fig. 2B). The fibrotic area in the muscle contusion group on day 24 post-injury was slightly smaller compared with that in day 12 post-injury (Fig. 2C), but the difference was not statistically significant (P>0.05; Fig. 2D).
Figure 2.
Histological evaluation of scar tissue formation in the injured and uninjured GM by Masson's trichrome staining. Scar tissues are indicated in blue and muscle tissue in red. Representative images of the (A) uninjured control group, (B) muscle contusion group (12 days post-injury) and (C) muscle contusion group (24 days post-injury). (D) Quantification of the scar tissue area following GM injury. Data are presented as the mean ± standard deviation. Scale bars, 100 µm. d, days; GM, gastrocnemius muscle.
Expression of macrophage-specific markers following skeletal muscle injury
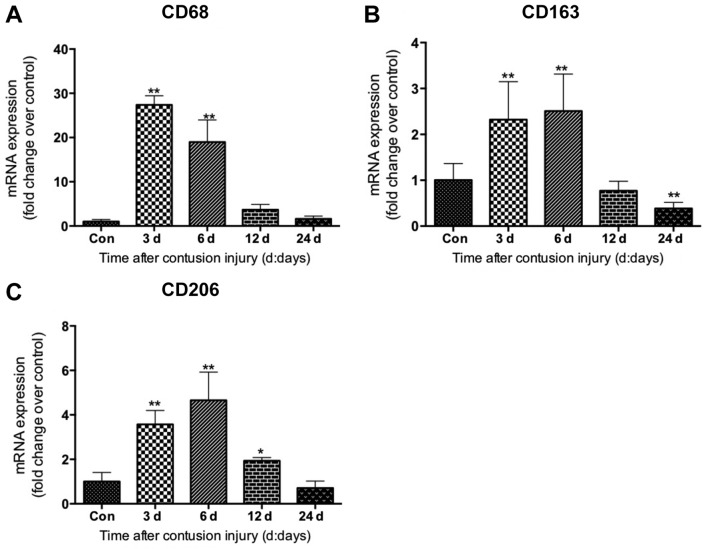
The mRNA levels of specific markers of macrophages in muscle were evaluated. Compared with the uninjured control group, the mRNA levels of CD68, which is a specific marker of M1 macrophages (21), increased significantly in the muscle samples on days 3 and 6 following injury induction (both P<0.01), peaking at 3 days post-injury (Fig. 3A). The data also revealed that the mRNA levels of CD163, a molecular marker of M2 macrophages (M2c) (21), increased significantly on days 3 and 6 (both P<0.01) after injury, peaking on day 6 post-injury (Fig. 3B). Similarly, the mRNA levels of CD206, another marker of the M2 macrophage (M2a and M2c) subset (22), increased significantly on days 3, 6 and 12 (all P<0.05) after injury induction compared with the uninjured control group (Fig. 3C).
Figure 3.
Expression of macrophage-specific markers in gastrocnemius muscle samples after muscle contusion. (A) mRNA expression of CD68, (B) CD163 (marker of M2 macrophages) and (C) CD206. Data are presented as the mean ± standard deviation (n=8). *P<0.05, **P<0.01 vs. Con. Con, control; d, days; CD, cluster of differentiation.
Expression of inflammatory cytokines following skeletal muscle injury
The present study evaluated the expression of inflammatory cytokines (IL-1β, IL-6, TNF-α, INF-γ, IL-10 and TGF-β1) in isolated GM samples. The mRNA levels of proinflammatory cytokines IL-1β, TNF-α, and IFN-γ increased significantly in skeletal muscle samples on days 3 and 6 following contusion compared with control (all P<0.01; Fig. 4A, C and D). The mRNA levels of IL-6 also increased significantly on days 3 and 6 after injury compared with the control (both P<0.01; Fig. 4B), and were higher compared with those in the uninjured control group at 24 days post-injury (P<0.05; Fig. 4B). The levels of TGF-β1 mRNA were significantly higher in the GM muscle samples at 3, 6 and 12 days after injury compared with those in the uninjured control group (all P<0.01; Fig. 4F). Lastly, the mRNA levels of the anti-inflammatory factor IL-10 increased significantly at 3 and 6 days after injury compared with the uninjured control group, and returned to a level comparable to that of the control by 24 days (all P<0.01; Fig. 4E).
Figure 4.
Expression of inflammatory factors in gastrocnemius muscle samples following muscle contusion. mRNA expression levels of (A) IL-1β, (B) IL-6, (C) TNF-α, (D) IFN-γ, (E) IL-10 and (F) TGF-β1. Data are presented as the mean ± standard deviation (n=8). *P<0.05, **P<0.01 vs. con. Con, control; IL-1β, interleukin-1β; IL-6, interleukin-6; TNF-α, tumor necrosis factor-α; IFN-γ, interferon-γ; IL-10, interleukin-10; TGF-β1, transforming growth factor-β1.
Expression of myogenic regulatory factors following skeletal muscle injury
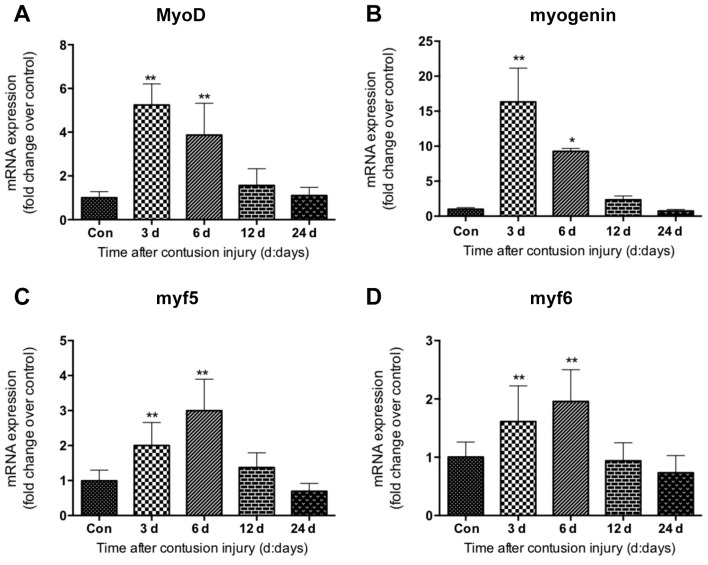
The expression of myogenic regulatory factors including MyoD, myogenin, myf5 and myf6 was investigated in GM samples following contusion injury. The data revealed that MyoD, myogenin, myf5 and myf6 displayed similar gene expression patterns. Their mRNA levels were elevated significantly at 3 and 6 days after injury compared with uninjured control, which returned to normal 24 days after injury (Fig. 5). The expression of MyoD and myogenin peaked at 3 days following injury induction (Fig. 5A and B); whereas the levels of myf5 and myf6 peaked on day 6 following injury (Fig. 5C and D).
Figure 5.
Expression of myogenic regulatory factors in gastrocnemius muscle samples following muscle contusion. mRNA expression levels of (A) MyoD, (B) myogenin, (C) myf5 and (D) myf6. Data are presented as the mean ± standard deviation (n=8). *P<0.05, **P<0.01 vs. Con. Con, control; MyoD, myogenic differentiation 1; myf5, myogenic factor 5; myf6, myogenic factor 6.
Expression of angiogenic factors following skeletal muscle injury
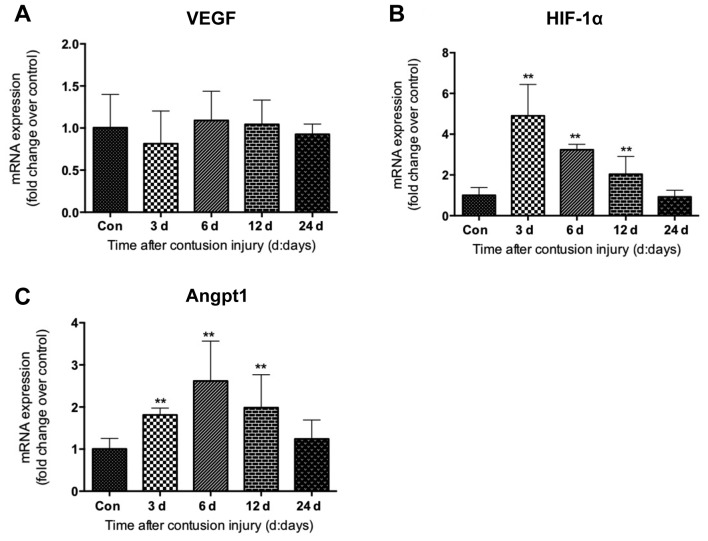
Next, the expression of angiogenic factors was evaluated in the skeletal muscle tissues isolated following muscle contusion injury. Vascular endothelial growth factor (VEGF), hypoxia-inducible factor-1α (HIF-1α) and angiopoietin-1 (Angpt-1) exhibited differential expression patterns. The mRNA levels of VEGF did not appear to be significantly altered during the healing process following muscle injury (Fig. 6A). However, compared with the uninjured control group, HIF-1α mRNA levels were revealed to be significantly increased at 3, 6 and 12 days after injury induction (all P<0.01), peaking on day 3 (Fig. 6B). Increased expression was observed for Angpt-1 at days 3, 6 and 12 after injury, which was significantly higher compared with that in the uninjured control group (all P<0.01; Fig. 6C).
Figure 6.
Expression of angiogenic factors in gastrocnemius muscle samples following muscle contusion. mRNA expression levels of (A) VEGF, (B) HIF-1α and (C) Angpt1. Data are presented as the mean ± standard deviation (n=8). **P<0.01 vs. Con. Con, control; HIF-1α, hypoxia-inducible factor-1α; VEGF, vascular endothelial growth factor; Angpt1, angiopoietin 1.
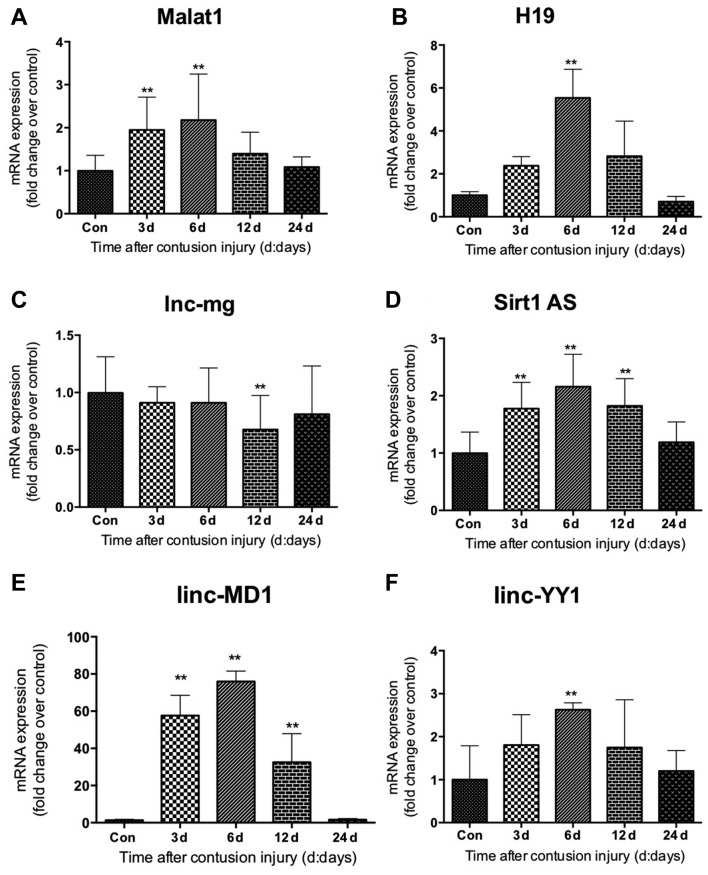
Expression of lncRNAs following skeletal muscle injury
The expression levels of lncRNAs (Malat1, H19, lnc-mg, Sirt1 AS, linc-MD1 and linc-YY1) during GM regeneration were subsequently determined using RT-qPCR. The expression levels of linc-MD1 and Sirt1 AS were significantly increased compared with the uninjured control group at 3, 6 and 12 days following injury (all P<0.01), and returned to normal levels 24 days after injury (Fig. 7D and E). Compared with the uninjured control group, Malat1 expression in the skeletal muscle of the muscle contusion group also increased on day 3 following injury induction (P<0.01), and remained elevated on day 6 (P<0.01; Fig. 7A). linc-YY1 and H19 exhibited similar gene expression patterns, as both were elevated significantly 6 days post-injury compared with the control (both P<0.01; Fig. 7B and F). However, their gene expression levels did not change significantly 3, 12 or 24 days post-injury in the muscle contusion group compared with the uninjured control group. It was additionally demonstrated that lnc-mg mRNA levels did not appear to be significantly altered during the process of regeneration following muscle contusion injury, although the levels were observed to be decreased 12 days following injury compared with the control group (P<0.01; Fig. 7C).
Figure 7.
Expression of long non-coding RNAs in gastrocnemius muscle samples following muscle contusion. mRNA expression levels of (A) Malat1, (B) H19, (C) lnc-mg, (D) Sirt1 AS, (E) linc-MD1 and (F) linc-YY1. Data are presented as the mean ± standard deviation (n=8). **P<0.01 vs. Con. Con, control; Malat1, metastasis associated lung adenocarcinoma transcript 1; lncRNA, long non-coding RNA; lnc-mg, myogenesis-associated long non-coding RNA; Sirt1 AS, sirtuin 1-antisense; linc-MD1, long intergenic non-protein coding RNAs-muscle differentiation 1; linc-YY1, long intergenic non-protein coding RNA-yin yang 1.
Correlation between the lncRNAs and the specific markers of macrophages, inflammatory cytokines, myogenic regulatory factors and angiogenic factors
To assess the association between lncRNAs and specific markers of macrophages, inflammatory cytokines, myogenic regulatory factors and angiogenic factors, Pearson's correlations analysis was performed. The results of this analysis are summarized in Table II. Positive correlations were revealed between Malat1 and inflammatory cytokines (TGF-β1, IL-10, IL-6 and TNF-α), myogenic regulatory factors (MyoD and myogenin) and angiogenic factors (HIF-1α and Angpt1) (Table II). Although both H19 and Sirt1 AS were demonstrated to correlate positively with myogenic regulatory factors (myogenin, myf5 and myf6), only H19 correlated with angiogenic factors (HIF-1α and Angpt1), whereas only Sirt1 AS was correlated with MyoD (Table II). Significant positive correlations were revealed between linc-MD1 and macrophage markers (CD163 and CD206), inflammatory cytokines (TGF-β1, IL-10, IL-6, TNF-α and IFN-γ) and myogenic regulatory factors (MyoD, myogenin, myf5 and myf6). A moderate correlation was observed between linc-YY1 and the myogenic regulatory factors (MyoD and myogenin). No correlation was found between lnc-mg and any of the genes tested (Table II).
Table II.
Correlation between the lncRNAs and the specific markers of macrophages, inflammatory cytokines, myogenic regulatory factors and angiogenic factors.
| lncRNA | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Malat1 | H19 | lnc-mg | Sirt1 AS | linc-MD1 | linc-YY1 | |||||||
| Gene | r | P-value | r | P-value | r | P-value | r | P-value | r | P-value | r | P-value |
| CD68 | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. |
| CD163 | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. | 0.793 | 0.015 | N.S. | N.S. |
| CD206 | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. | 0.862 | 0.002 | N.S. | N.S. |
| TGF-β1 | 0.916 | 0.029 | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. | 0.912 | <0.001 | N.S. | N.S. |
| IL-10 | 0.986 | <0.001 | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. | 0.896 | <0.001 | N.S. | N.S. |
| IL-6 | 0.598 | 0.005 | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. | 0.850 | 0.024 | N.S. | N.S. |
| IL-1β | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. |
| TNF-α | 0.886 | 0.046 | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. | 0.906 | 0.034 | N.S. | N.S. |
| IFN-γ | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. | 0.884 | 0.037 | N.S. | N.S. |
| MyoD | 0.558 | 0.003 | N.S. | N.S. | N.S. | N.S. | 0.563 | <0.001 | 0.825 | <0.001 | 0.474 | 0.003 |
| myogenin | 0.600 | 0.012 | 0.470 | 0.003 | N.S. | N.S. | 0.535 | <0.001 | 0.773 | <0.001 | 0.423 | 0.007 |
| Myf5 | N.S. | N.S. | 0.797 | 0.001 | N.S. | N.S. | 0.703 | <0.001 | 0.782 | <0.001 | N.S. | N.S. |
| Myf6 | N.S. | N.S. | 0.674 | 0.007 | N.S. | N.S. | 0.620 | 0.001 | 0.897 | 0.039 | N.S. | N.S. |
| VEGF | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. |
| HIF-1α | 0.785 | 0.016 | 0.504 | 0.001 | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. |
| Angpt1 | 0.653 | 0.040 | 0.593 | <0.001 | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. | N.S. |
N.S., not significant; IL-1β, interleukin-1β; IL-6, interleukin-6; TNF-α, tumor necrosis factor-α; IFN-γ, interferon-γ; IL-10, interleukin-10; TGF-β1, transforming growth factor-β1; MyoD, myogenic differentiation 1; myf5, myogenic factor 5; myf6, myogenic factor 6; HIF-1α, hypoxia-inducible factor-1α; VEGF, vascular endothelial growth factor; Angpt1, angiopoietin 1; Malat1, metastasis associated lung adenocarcinoma transcript 1; lnc-mg, myogenesis-associated long non-coding RNA; Sirt1 AS, sirtuin 1-antisense; linc-MD1, long intergenic non-protein coding RNAs-muscle differentiation 1; linc-YY1, long intergenic non-protein coding RNA-yin yang 1.
Discussion
Skeletal muscle retains the ability to regenerate following damage. The present study employed a mouse skeletal muscle contusion injury model which can induce inflammatory responses with macrophage infiltration as one of the signatures, followed by regeneration. Histologically, a large number of inflammatory cells and factors infiltrated the injured area in the early stages of skeletal muscle injury. The infiltration patterns of inflammatory cells were consistent with a previous study (23). In addition, the necrotic muscle fibers were replaced mostly by centrally-nucleated developing muscle fibers or myotubes at day 6 following injury. This indicated that satellite cells were committed into the myoblast pathway to differentiate into myotubes following contusion injury.
Fibrosis may occur when skeletal muscles experience severe injury, which is characterized by the accumulation of fibroblasts and myofibroblasts, and high levels of extracellular matrix deposition (24). According to the collagen staining performed in the present study, scar tissues could be observed in the contused muscle 12 and 24 days after injury; with more fibrotic scars recorded on day 12 than on day 24. Total collagen staining was performed to detect fibrosis in the injured muscle as previously described (25). These observations are concordant with those reported by Ghaly et al (26). Fibrosis of skeletal muscle is a characteristic feature of skeletal muscle repair, usually beginning between the 2nd and 3rd week after injury. Resultant scar tissue continues to develop and mature over time (27). The results of the present study revealed that fibrosis occurred in the late stages of skeletal muscle repair (12 and 24 days after injury). This indicates that the skeletal muscle contusion injury was successfully induced in the present study, which was followed by muscle repair.
Despite the rapidly increasing number of studies investigating the functions of lncRNAs, their specific roles in myogenesis remain poorly defined. The findings presented in this study provided a comprehensive analysis of lncRNA (Malat1, H19, lnc-mg, Sirt1 AS, linc-MD1 and linc-YY1) expression during skeletal muscle regeneration following contusion injury. In addition, their association with the expression levels of lincRNAs and macrophage markers, inflammatory cytokines, myogenic factors and angiogenic factors was elucidated. Malat1 and Sirt1 AS are lincRNAs that are expressed in high abundance in proliferating and differentiating myoblasts (15,28). The present study revealed that the expression of Malat1 and Sirt1 AS were significantly upregulated throughout the skeletal muscle regeneration process. It has been suggested previously that Malat1 and Sirt1 AS can promote myoblast proliferation and inhibit myoblast differentiation (15,16,29). Therefore, it would be feasible that Malat1 and Sirt1 AS may serve important functions in the regeneration of contused skeletal muscle. In addition, Malat1 and Sirt1 AS were revealed to positively correlate with myogenic transcription factors myogenin and MyoD. Wang et al (15) demonstrated that Sirt1 AS lncRNA overexpression downregulated the expression of MyoD and myogenin; furthermore, Malat1 has been previously postulated to modulate the expression of myogenin and the activity of MyoD (16). Consequently, it may be hypothesized that Malat1 and Sirt1 AS lncRNA serve key roles in the regeneration of contused muscle, possibly by regulating the expression of MyoD and myogenin.
Additionally, lnc-mg has also been suggested to be a skeletal muscle-enriched lncRNA (30). A main purpose of the present study was to investigate the role of lnc-mg in the regeneration of skeletal muscle following contusion. The expression of lnc-mg was significantly decreased on day 12 following contusion. This suggests that lnc-mg may act as a negative regulator in the regeneration of skeletal muscle following contusion. This result is consistent with an earlier finding that the knockdown of lnc-mg resulted in marked inhibition of muscle satellite cell differentiation by downregulating MyoD and myogenin expression (30). This observation supported the hypothesis that lnc-mg can regulate the expression of MyoD and myogenin in the present study. However, no correlation was found between lnc-mg and MyoD or myogenin expression. This discrepancy could be due to differences in the animal models used in the two studies. Therefore, further research is necessary to elucidate whether lnc-mg is involved in skeletal muscle regeneration following contusion.
Although several lncRNAs have been demonstrated to serve a number of roles in skeletal muscle cell differentiation and myogenesis in vitro (13,28,29), little is known about their function during the regeneration of skeletal muscle following contusion. Therefore, the present study also investigated the roles of H19, linc-MD1 and lncYY1 during skeletal muscle regeneration. The present study revealed that the expression of linc-MD1 was significantly increased throughout the skeletal muscle regeneration process, whereas the upregulation of H19 and lncYY1 was only observed on day 6 after muscle contusion. This suggests that H19 and lncYY1 may perform important functions during the early repair phase of contused skeletal muscle. Indeed, a number of studies have demonstrated that skeletal muscle deficient in lncRNAs H19, linc-MD1 and/or lncYY1 displays aberrant skeletal muscle regeneration post-injury due to the downregulation of MyoD and myogenin (14,31,32). Data from the present study revealed that H19, linc-MD1 and lncYY1 all correlated positively with MyoD and/or myogenin. Therefore, it may be hypothesized that H19, linc-MD1 and lnc-YY1 promote the regeneration of contused skeletal muscle, possibly by modulating MyoD and myogenin expression.
Myogenic factors Myf5 and Myf6 are essential for muscle regeneration and can promote myoblast differentiation (33). However, investigations into the role of lncRNAs in regulating myf5 and myf6 expression during contused muscle regeneration are lacking. In the present study, a positive correlation was revealed between the expression of lncRNAs (linc-MD1, Sirt AS and H19) and myf5/myf6; however, it remains unclear how this relationship can impact contused muscle regeneration. Further studies are required to investigate this underlying mechanism.
During the skeletal muscle repair process, lncRNAs have also been reported to be involved in the regulation of the skeletal muscle inflammatory response, angiogenesis and fibrosis (34–36). The inflammatory response is an integral part of the reaction to muscle injury and serves a pivotal role in subsequent muscle regeneration (37). Macrophages either induce inflammation or repair damaged tissues by secreting a large quantity of inflammatory cytokines. M1 macrophages produce TNF-α, IL-1β and IL-6, while M2 macrophages produce IL-10 and TGF-β1 (21). A number of studies performed previously have illustrated that Malat1 can regulate the inflammatory response in skeletal muscle, the knockdown of which increases the lipopolysaccharide-induced expression of TNF-α and IL-6 (34,35,38). These findings suggest that Malat1 may function as a regulator of the inflammatory response in this organ. The present study revealed that Malat1 correlated positively with the expression of a number of inflammatory cytokines (TGF-β1, TNF-α, IL-6 and IL-10) in contused muscle. As a result, this suggests that Malat1 can modulate the inflammatory response during the regeneration of contused skeletal muscle.
Vascular regeneration is part of the complete regeneration of damaged skeletal muscle. In the present study, the expression of HIF-1α and Angpt1 was markedly increased following muscle contusion, which correlated positively with Malat1. Michalik et al (39) observed that Malat1-deficient mice displayed a reduction in blood vessel density, suggesting that Malat1 may be involved in angiogenesis. Therefore, Malat1 may contribute to angiogenesis in regeneration after skeletal muscle injury, possibly by modulating the expression of HIF-1α and Angpt1. However, further research is necessary to investigate the underlying mechanism.
Recently, a growing body of evidence suggested that lncRNAs are also involved in tissue fibrosis in several organs, including the lungs, liver and heart (36,40). However, the role of lncRNAs in injury-induced skeletal muscle fibrosis remains unclear. Results from the present study revealed that linc-MD1 and Malat1 significantly correlated with the profibrotic factor TGF-β1. Therefore, lncRNAs may be involved in the fibrosis of contused skeletal muscle by interacting with TGF-β1. However, further research is necessary to investigate the underlying mechanism.
lncRNAs such as Malat1 serve important roles in the inflammatory response and angiogenesis of injured skeletal muscle. To the best of our knowledge, only a small number of studies have evaluated the role of lncRNAs in the inflammatory response and angiogenesis following skeletal muscle injury (34,35). The present study revealed that a number of lncRNAs are associated with inflammatory and angiogenic factors. This suggests that the inflammatory response and angiogenesis during skeletal muscle regeneration are regulated by lncRNAs.
This present investigation was the first to demonstrate that lncRNAs are associated with the regeneration of contused skeletal muscle. The changes in the expression of a number of candidate lncRNAs at multiple timepoints following skeletal muscle contusion, as well as their association with other physiological factors, were assessed. Results illustrated in the present study support the hypothesis that lncRNAs may play important roles in the regeneration of contused skeletal muscle, but further research is needed to elucidate the underlying mechanism. However, there are several limitations to the study; for example, knockdown or overexpression experiments on the lncRNAs were not performed. Although Pearson's correlation analysis indicated correlations between lncRNAs and macrophage infiltration, inflammation and angiogenesis, this did not reveal the mechanism underlying the role of lncRNAs in contused muscle regeneration. Nevertheless, this present investigation do lay the foundation for further research into the functional role of lncRNAs in skeletal muscle regeneration.
In conclusion, the expression of inflammatory cytokines, myogenic regulatory factors and angiogenic factors were demonstrated to be significantly increased following the induction of skeletal muscle contusion, along with lncRNAs including Malat1, H19, lnc-mg, linc-MD1, linc-YY1 and Sirt1 AS. There was a correlation between lncRNAs and a variety of established regulatory factors (TGF-β1, MyoD, myogenin, myf5, myf6, HIF-1α and Angpt1) during the skeletal muscle regeneration process. These results suggest that lncRNAs may serve important roles in the regeneration of damaged skeletal muscle. Effective muscle regeneration is essential for the treatment of muscle diseases including muscle atrophy, muscular dystrophy and sporting injuries. Therefore, these findings serve as a basis for the effective treatment of muscle atrophy and muscular dystrophy.
Acknowledgements
The present study was previously presented at a conference (https://ojs.uclouvain.be/index.php/EBR/article/view/8693).
Funding
The present study was supported by the National Natural Science Foundation of China (grant nos. 31271273 and 31300975), Shanghai Natural Science Fund Project (grant no. 18ZR1437100) and Shanghai Key Laboratory of Human Movement Development and Protection (Shanghai University of Sport; grant no. 11DZ2261100).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
LZ analyzed the results and drafted the manuscript. LZ performed histological staining and PCR. XL assisted with PCR. PC and WX designed the current study and provided funds. All authors reviewed and critiqued the manuscript and agreed to the final submission of the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Review Committee for Animal Experimentation of Shanghai University of Sports (approval no. 2016006).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Lu H, Huang D, Saederup N, Charo IF, Ransohoff RM, Zhou L. Macrophages recruited via CCR2 produce insulin-like growth factor-1 to repair acute skeletal muscle injury. FASEB J. 2011;25:358–369. doi: 10.1096/fj.10-171579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carlson BM, Faulkner JA. The regeneration of skeletal muscle fibers following injury: A review. Medicine Sci Sports Exerc. 1983;15:187–198. doi: 10.1249/00005768-198315030-00003. [DOI] [PubMed] [Google Scholar]
- 3.Tidball JG. Mechanisms of muscle injury, repair and regeneration. Compr Physiol. 2011;1:2029–2062. doi: 10.1002/cphy.c100092. [DOI] [PubMed] [Google Scholar]
- 4.Baggiolini M. Chemokines in pathology and medicine. J Intern Med. 2001;250:91–104. doi: 10.1046/j.1365-2796.2001.00867.x. [DOI] [PubMed] [Google Scholar]
- 5.Xiao W, Liu Y, Chen P. Macrophage depletion impairs skeletal muscle regeneration: The roles of pro-fibrotic factors, inflammation and oxidative stress. Inflammation. 2016;39:2016–2028. doi: 10.1007/s10753-016-0438-8. [DOI] [PubMed] [Google Scholar]
- 6.Angelini G, Salinari S, Bertuzzi A, Iaconelli A, Mingrone G. Metabolic surgery improves insulin resistance through the reduction of gut-secreted heat shock proteins. Commun Biol. 2018;1:69. doi: 10.1038/s42003-018-0069-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zammit PS. Function of the myogenic regulatory factors Myf5, MyoD, Myogenin and MRF4 in skeletal muscle, satellite cells and regenerative myogenesis. Semin Cell Dev Biol. 2017;72:19–32. doi: 10.1016/j.semcdb.2017.11.011. [DOI] [PubMed] [Google Scholar]
- 8.Kassar-Duchossoy L, Gayraud-Morel B, Gomès D, Rocancourt D, Buckingham M, Shinin V, Tajbakhsh S. Mrf4 determines skeletal muscle identity in Myf5: Myod double-mutant mice. Nature. 2004;431:466–471. doi: 10.1038/nature02876. [DOI] [PubMed] [Google Scholar]
- 9.Zanou N, Gailly P. Skeletal muscle hypertrophy and regeneration: Interplay between the myogenic regulatory factors (MRFs) and insulin-like growth factors (IGFs) pathways. Cell Mol Life Sci. 2013;70:4117–4130. doi: 10.1007/s00018-013-1330-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Heesch S, van Iterson M, Jacobi J, Boymans S, Essers PB, de Bruijn E, Hao W, MacInnes AW, Cuppen E, Simonis M. Extensive localization of long noncoding RNAs to the cytosol and mono- and polyribosomal complexes. Genome Biol. 2014;15:R6. doi: 10.1186/gb-2014-15-1-r6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Batista PJ, Chang HY. Long noncoding RNAs: Cellular address codes in development and disease. Cell. 2013;152:1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang J, Shao N, Ding X, Tan B, Song Q, Wang N, Jia Y, Ling H, Cheng Y. Crosstalk between transforming growth factor-beta signaling pathway and long non-coding RNAs in cancer. Cancer Lett. 2016;370:296–301. doi: 10.1016/j.canlet.2015.11.007. [DOI] [PubMed] [Google Scholar]
- 13.Xu X, Ji S, Li W, Yi B, Li H, Zhang H, Ma W. LncRNA H19 promotes the differentiation of bovine skeletal muscle satellite cells by suppressing Sirt1/FoxO1. Cell Mol Biol Lett. 2017;22:10. doi: 10.1186/s11658-017-0040-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dey BK, Pfeifer K, Dutta A. The H19 long noncoding RNA gives rise to microRNAs miR-675-3p and miR-675-5p to promote skeletal muscle differentiation and regeneration. Genes Dev. 2014;28:491–501. doi: 10.1101/gad.234419.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang GQ, Wang Y, Xiong Y, Chen XC, Ma ML, Cai R, Gao Y, Sun YM, Yang GS, Pang WJ. Sirt1 AS lncRNA interacts with its mRNA to inhibit muscle formation by attenuating function of miR-34a. Sci Rep. 2016;6:21865. doi: 10.1038/srep21865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen X, He L, Zhao Y, Li Y, Zhang S, Sun K, So K, Chen F, Zhou L, Lu L, et al. Malat1 regulates myogenic differentiation and muscle regeneration through modulating MyoD transcriptional activity. Cell Discov. 2017;3:17002. doi: 10.1038/celldisc.2017.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun Y, Sun X, Liu S, Liu L, Chen J. The overlap between regeneration and fibrosis in injured skeletal muscle is regulated by phosphatidylinositol 3-kinase/Akt signaling pathway-A bioinformatic analysis based on lncRNA microarray. Gene. 2018;672:79–87. doi: 10.1016/j.gene.2018.06.001. [DOI] [PubMed] [Google Scholar]
- 18.Diaz JA, Fischer DA, Rettig AC, Davis TJ, Shelbourne KD. Severe quadriceps muscle contusions in athletes. A report of three cases. Am J Sports Med. 2003;31:289–293. doi: 10.1177/03635465030310022201. [DOI] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Ota S, Uehara K, Nozaki M, Kobayashi T, Terada S, Tobita K, Fu FH, Huard J. Intramuscular transplantation of muscle-derived stem cells accelerates skeletal muscle healing after contusion injury via enhancement of angiogenesis. Am J Sports Med. 2011;39:1912–1922. doi: 10.1177/0363546511415239. [DOI] [PubMed] [Google Scholar]
- 21.Manferdini C, Paolella F, Gabusi E, Gambari L, Piacentini A, Filardo G, Fleury-Cappellesso S, Barbero A, Murphy M, Lisignoli G. Adipose stromal cells mediated switching of the pro-inflammatory profile of M1-like macrophages is facilitated by PGE2: In vitro evaluation. Osteoarthritis Cartilage. 2017;25:1161–1171. doi: 10.1016/j.joca.2017.01.011. [DOI] [PubMed] [Google Scholar]
- 22.Tidball JG, Villalta SA. Regulatory interactions between muscle and the immune system during muscle regeneration. Am J Physiol Regul Integr Comp Physiol. 2010;298:R1173–R1187. doi: 10.1152/ajpregu.00735.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tonkin J, Temmerman L, Sampson RD, Gallego-Colon E, Barberi L, Bilbao D, Schneider MD, Musarò A, Rosenthal N. Monocyte/macrophage-derived IGF-1 orchestrates murine skeletal muscle regeneration and modulates autocrine polarization. Mol Ther. 2015;23:1189–1200. doi: 10.1038/mt.2015.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wynn TA, Ramalingam TR. Mechanisms of fibrosis: Therapeutic translation for fibrotic disease. Nat Med. 2012;18:1028–1040. doi: 10.1038/nm.2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen W, Li Y, Zhu J, Schwendener R, Huard J. Interaction between macrophages, TGF-beta1 and the COX-2 pathway during the inflammatory phase of skeletal muscle healing after injury. J Cell Physiol. 2008;214:405–412. doi: 10.1002/jcp.21212. [DOI] [PubMed] [Google Scholar]
- 26.Ghaly A, Marsh DR. Ischaemia-reperfusion modulates inflammation and fibrosis of skeletal muscle after contusion injury. Int J Exp Pathol. 2010;91:244–255. doi: 10.1111/j.1365-2613.2010.00708.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Y, Cummins J, Huard J. Muscle injury and repair. Curr Opin Orthop. 2001;12:409–415. doi: 10.1097/00001433-200110000-00008. [DOI] [Google Scholar]
- 28.Watts R, Johnsen VL, Shearer J, Hittel DS. Myostatin-induced inhibition of the long noncoding RNA Malat1 is associated with decreased myogenesis. Am J Physiol Cell Physiol. 2013;304:C995–C1001. doi: 10.1152/ajpcell.00392.2012. [DOI] [PubMed] [Google Scholar]
- 29.Han X, Yang F, Cao H, Liang Z. Malat1 regulates serum response factor through miR-133 as a competing endogenous RNA in myogenesis. FASEB J. 2015;29:3054–3064. doi: 10.1096/fj.14-259952. [DOI] [PubMed] [Google Scholar]
- 30.Zhu M, Liu J, Xiao J, Yang L, Cai M, Shen H, Chen X, Ma Y, Hu S, Wang Z, et al. Lnc-mg is a long non-coding RNA that promotes myogenesis. Nat Commun. 2017;8:14718. doi: 10.1038/ncomms14718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou L, Sun K, Zhao Y, Zhang S, Wang X, Li Y, Lu L, Chen X, Chen F, Bao X, et al. Linc-YY1 promotes myogenic differentiation and muscle regeneration through an interaction with the transcription factor YY1. Nat Commun. 2015;6:10026. doi: 10.1038/ncomms10026. [DOI] [PubMed] [Google Scholar]
- 33.Petersson SJ, Jørgensen LH, Andersen DC, Nørgaard RC, Jensen CH, Schrøder HD. SPARC is up-regulated during skeletal muscle regeneration and inhibits myoblast differentiation. Histol Histopathol. 2013;28:1451–1460. doi: 10.14670/HH-28.1451. [DOI] [PubMed] [Google Scholar]
- 34.Huang JK, Ma L, Song WH, Lu BY, Huang YB, Dong HM, Ma XK, Zhu ZZ, Zhou R. LncRNA-MALAT1 promotes angiogenesis of thyroid cancer by modulating tumor-associated macrophage FGF2 protein secretion. J Cell Biochem. 2017;118:4821–4830. doi: 10.1002/jcb.26153. [DOI] [PubMed] [Google Scholar]
- 35.Marques-Rocha JL, Samblas M, Milagro FI, Bressan J, Martinez JA, Marti A. Noncoding RNAs, cytokines and inflammation-related diseases. FASEB J. 2015;29:3595–3611. doi: 10.1096/fj.14-260323. [DOI] [PubMed] [Google Scholar]
- 36.Duru N, Wolfson B, Zhou Q. Mechanisms of the alternative activation of macrophages and non-coding RNAs in the development of radiation-induced lung fibrosis. World J Biol Chem. 2016;7:231–239. doi: 10.4331/wjbc.v7.i4.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang C, Li Y, Wu Y, Wang L, Wang X, Du J. Interleukin-6/signal transducer and activator of transcription 3 (STAT3) pathway is essential for macrophage infiltration and myoblast proliferation during muscle regeneration. J Biol Chem. 2013;288:1489–1499. doi: 10.1074/jbc.M112.419788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao G, Su Z, Song D, Mao Y, Mao X. The long noncoding RNA MALAT1 regulates the lipopolysaccharide-induced inflammatory response through its interaction with NF-kappaB. FEBS Lett. 2016;590:2884–2895. doi: 10.1002/1873-3468.12315. [DOI] [PubMed] [Google Scholar]
- 39.Michalik KM, You X, Manavski Y, Doddaballapur A, Zörnig M, Braun T, John D, Ponomareva Y, Chen W, Uchida S, et al. Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth. Circ Res. 2014;114:1389–1397. doi: 10.1161/CIRCRESAHA.114.303265. [DOI] [PubMed] [Google Scholar]
- 40.Piccoli MT, Gupta SK, Viereck J, Foinquinos A, Samolovac S, Kramer FL, Garg A, Remke J, Zimmer K, Batkai S, Thum T. Inhibition of the cardiac fibroblast-enriched lncRNA Meg3 prevents cardiac fibrosis and diastolic dysfunction. Circ Res. 2017;121:575–583. doi: 10.1161/CIRCRESAHA.117.310624. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.