Abstract
MicroRNA (miR)-146a levels are reduced in peripheral blood mononuclear cells of patients with systemic lupus erythematosus (SLE); however, its function is not well understood. The present study investigated the role of miR-146a in the regulation of lipopolysaccharide (LPS)-induced inflammation in THP-1 cells. A miR-146a mimic and an inhibitor were used to overexpress and downregulate miR-146a expression, respectively. Reverse transcription-quantitative PCR and western blot analyses were performed to evaluate interleukin (IL)-1 receptor-associated kinase 1 (IRAK1) expression, and western blot analysis was applied to assess nuclear factor-κB activation by analyzing p65 subunit levels in the nucleus. To investigate the effects of miR-146a on LPS-induced inflammation, IL-6 and tumor necrosis factor-α (TNF-α) levels were also measured using ELISA. The results of the present study revealed thatmiR-146a overexpression significantly reduced IRAK1 expression, reduced p65 levels in the nucleus and reduced IL-6 and TNF-α levels in the supernatant of the cell culture medium of THP-1 cells following LPS treatment. Luciferase assays confirmed IRAK1 to be a direct target of miR-146a in THP-1 cells. In conclusion, miR-146a may regulate IRAK1 expression and inhibit the activation of inflammatory signals and secretion of pro-inflammatory cytokines. The present study revealed, at least in part, the mechanisms by which miR-146a regulate the inflammatory response in SLE.
Keywords: microRNA-146a, interleukin-1 receptor-associated kinase 1, nuclear factor-κB, interleukin-6, tumor necrosis factor-α, systemic lupus erythematosus
Introduction
MicroRNAs (miRNAs/miRs) are endogenous non-coding RNA molecules, 19–25 nucleotides in length, that exert post-transcriptional inhibitory effects on gene expression (1). To date, abnormal miRNA expression has been observed in a number of diseases, including cancer, systemic lupus erythematosus (SLE), autoimmune diabetes, atherosclerosis and rheumatoid arthritis (RA) (2–4). In particular, miR-146 is a miRNA that serves an important role in the regulation of the immune response by targeting genes to mediate downstream pathophysiological processes, with the interleukin (IL)-1 receptor-associated kinase l (IRAK1)/tumor necrosis factor (TNF) receptor-associated factor 6 (TRAF6) TRAF6/NF-κB axis as the most important target (5). It has been reported that miR-146a inhibits the activation of donor T cells in acute graft-vs.-host disease by targeting TRAF6, leading to reduced TNF transcription (6). Human miR-146 exists in two forms, miR-146a and miR-146b, the former of which has a seed sequence of 5′-UGAGAACUGAAUUCCAUGGGUU-3′ and is highly expressed in macrophages, mononuclear cells and T lymphocytes (7).
SLE is a complex chronic autoimmune inflammatory disease that is caused by inflammatory responses mediated by the lack of self-tolerance in the immune system. Innate and adaptive immune responses appear to be involved in the development of SLE. A variety of immune cell types, including monocytes, lymphocytes, mediated by a number of cytokines, including type I interferon (IFN) and IL-6, have been previously reported to serve their roles in this process (8). Indeed, previous studies have demonstrated that miR-146a expression is reduced in the peripheral blood mononuclear cells (PBMCs) of patients with SLE (9,10). A pilot study performed by Zhou et al (unpublished data) revealed that miR-146a levels were lower in the serum of patients with SLE. These findings suggested that miR-146a may be associated with the development and progression of SLE.
A previous study revealed that interleukin (IL)-1 receptor-associated kinase l (IRAK1) is a potential target of miR-146a (11). IRAK1 is the critical molecule in the Toll-like receptor (TLR)-4-myeloid differentiation primary response 88 (MyD88) signaling pathway. IRAK1 is phosphorylated after combining with My D88, and forms a complex with TRAF6. This complex then activates inhibitor of nuclear factor (NF)-κB kinase (IΚKβ) and induces the activation and translocation of the NF-κB transcription factor into the nucleus. Activated NF-κB subsequently induces the synthesis and secretion of large quantities of inflammatory cytokines (12).
Immunosuppressive therapy is commonly applied during the clinical management of patients with SLE. Under these conditions, patients are predisposed to infection, including nosocomial and latent infections, which in turn may exacerbate SLE symptoms (13). Pathogen-associated molecular patterns such as lipopolysaccharide (LPS) are the most common etiological agents that induce inflammatory responses by activating the TLR/IRAK1/NF-κB signaling pathway (14). Therefore, the aim of the present study was to investigate the regulatory effects of miR-146a on inflammatory responses by analyzing its effects on the expression of IRAK1 and downstream NF-κB activation. In addition, the current study aimed to investigate the possible infection-associated mechanisms of miR-146a in the pathogenesis of SLE.
Materials and methods
Cell culture and transfection
The human acute monocytic leukemia THP-1 cell line was purchased from the Cell Bank of the Chinese Academy of Sciences. THP-1 cells were cultured using RPMI-1640 culture medium supplemented with 10% fetal bovine serum (both Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100 mg/ml streptomycin at 37°C and 5% CO2. Cells in the logarithmic growth phase were seeded into 6-well plate (1×106 cells/well) and incubated overnight. They were subsequently transfected with 50 nM miR-146a mimic (cat. no. miR 10000449), 50 nM negative control (NC; cat. no. miR 01101) mimic, 100 nM miR-146a inhibitor or (cat. no. miR 20000449) 100 nM inhibitor NC (cat. no. miR 02101). All from Guangzhou RiboBio Co., Ltd.) using Lipofectamine®2000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. For the blank control, corresponding quantities of Opti-MEM (Gibco; Thermo Fisher Scientific, Inc.) solution were used in place of RNA during transfection (15).
Stimulation of cells with LPS and cell grouping
At 24 h following transfection, THP-1 cells were washed with Hank's balanced salt solution (Beijing Solarbio Science & Technology Co., Ltd.) before the culture medium was replaced with fresh RPMI-1640 medium. A final concentration of 1 µg/ml LPS (Sigma-Aldrich; Merck KGaA) was subsequently added into the culture medium, followed by incubation for a further 3 h. The cells were then collected for the measurement of IRAK1 and p68 expression, whilst the supernatant of the culture medium was collected for the measurement of cytokine levels, as described previously (15). The cells were divided into the following experimental groups: Blank control, no RNA transfection or LPS treatment; LPS group, treated with LPS only; mimic group, miR-146 amimic+LPS; NC mimic group, NC mimic + LPS; inhibitor group, miR-146a inhibitor + LPS; and inhibitor NC group, inhibitor NC + LPS.
Reverse transcription-quantitative PCR (RT-qPCR)
For the measurement of miRNA expression, small RNA was extracted from cells using the mirVana PARIS kit (Applied Biosystems; Thermo Fisher Scientific, Inc.). cDNA was then reverse transcribed from small RNA using the TaqMan® MicroRNA RT kit (Applied Biosystems; Thermo Fisher Scientific, Inc.) according to manufacturer's protocol. The thermocycling conditions were as follows: 16°C for 30 min, 42°C for 30 min, 85°C for 5 min and 4°C hold. miR-146a expression levels were measured using TaqMan® MicroRNA Assay kit (Assay ID, 000468; Applied Biosystems; Thermo Fisher Scientific, Inc.), the primers, which are patented, were provided by the manufacturer as part of the kit. U6 expression were used for the normalization of miRNA expression.
To determine mRNA expression levels, total RNA was extracted from cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.), and then reverse transcription was performed using High-Capacity cDNA Reverse Transcription Kits (Applied Biosystems; Thermo Fisher Scientific, Inc.) according to manufacturer's protocol. The thermocycling conditions used was as follows: 25°C for 10 min, 37°C for 120 min, 85°C for 5 min and 4°C hold. qPCR was performed to measure IRAK1 mRNA levels in cells using PowerUp™ SYBR™ Green Master Mix (Applied Biosystems; Thermo Fisher Scientific, Inc.). GAPDH was used as an internal reference. The sequences of the primers used were as follows: IRAK1 forward, 5′-GTGCTAGAGACCTTGGCTG-3′ and reverse, 5′-TGTGCTCTGGGTGCTTCTC-3′; and GAPDH forward, 5′-CCATGTTCGTCATGGGTGT-3′ and reverse, 5′-TGAGTCCTTCCACGATACC-3′.
Both qPCR assays aforementioned were performed using ABI Real Time PCR System 7500 (Applied Biosystems; Thermo Fisher Scientific, Inc.) using the following thermocycling protocol: Initiation reaction at 50°C for 2 min and 10 min at 95°C, followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min. The relative expression levels of miR-146a and IRAK1 were determined using the 2−ΔΔCq method (16).
Western blotting
Following treatment with LPS for 3 h, the cells were collected and total protein was extracted using RIPA buffer (Pierce; Thermo Fisher Scientific, Inc.) supplemented with a cocktail of protease and phosphatase inhibitors (Sigma-Aldrich; Merck KGaA). Proteins from the nuclear fraction were extracted using a Nuclear Protein Extraction Kit (cat. R0050; Beijing Solarbio Science & Technology Co., Ltd.), according to the manufacturer's protocols. Protein concentrations were determined using a bicinchoninic acid assay protein quantification kit (Pierce; Thermo Fisher Scientific, Inc.). The protein samples were then heat-denatured, and 5 µg protein/lane was separated on a 10% SDS-PAGE gel, prior to transferral to polyvinylidene difluoride membranes. The membranes were subsequently blocked in 5% bovine serum albumin (Sigma-Aldrich; Merck KGaA), which was prepared in a TBS-T buffer (50 mM Tris, pH 8.0, and 150 mM NaCl, containing 0.05% Tween-20) and incubated overnight at 4°C. Subsequently, the membranes were incubated with primary antibodies against anti-IRAK1 (cat. no. SC-5288; dilution, 1:400), anti-NF-κB p65 (cat. no. SC-8008; dilution, 1:400), anti-β-actin (cat. no. SC-47778; dilution, 1:1,000) (all from Santa Cruz Biotechnology, Inc.) and anti-histone H3 (cat. no. 4499; dilution, 1:2,000; Cell Signaling Technology, Inc.) at 4°C overnight. Following washing with TBS-T for 20 min, the membranes were incubated with the corresponding horseradish peroxidase-conjugated secondary antibodies (cat. nos. 7076 and 7074; dilution 1:2,000; Cell Signaling Technology, Inc.) for 1 h at room temperature. The membranes were visualized using a Millipore Enhanced Chemiluminescence system (EMD Millipore; Merck KGaA) and analyzed using Image Lab (version 3.0; Bio-Rad Laboratories, Inc.). β-actin and histone H3 were used as internal controls. Relative protein expression, normalized to the internal control, is presented as a ratio of the blank control group.
ELISA
Cells in the logarithmic growth phase were seeded into a 24-well plate with a density of 2×105 cells/well. Subsequently, they were transfected with miR-146a mimic, mimic NC, miR-146a inhibitor or and inhibitor NC. For the blank control, the same amount of the Opti-MEM solution was used instead of RNA in the transfection. After 24 h of transfection, the THP-1cells were washed with Hank's balanced salt solution, and the culture medium was replaced with RPMI-1640 medium. LPS (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) was added into culture medium at a final concentration of 1 µg/ml. Following treatment with LPS for 12 h, the culture medium was collected to measure IL-6 and TNF-α levels using ELISA kits (D6050 and DTA00D, R&D Systems, Inc.), according to manufacturer's protocols.
Dual-Luciferase reporter assay
Target Scan bioinformatics software (version 7.2; www.targetscan.org) was used to predict the potential targets of miR-146a by typing the term ‘miR-146-5p’ into the search engine. The software revealed two conserved sites in the 3′-untranslated region (3′-UTR) of IRAK1. A wild-type IRAK1 mRNA 3′-UTR sequence containing the miR-146a target site was cloned into the XbaI site downstream of the firefly luciferase gene in the pGL-3-promoter vector (Promega Corporation) to obtain the wt-IRAK1-3′-UTR vector. In a parallel experiment, the IRAK1 mRNA 3′-UTR sequence was replaced by a mutant sequence (synthesized by ObiO Technology Co., Ltd.) to obtain the mut-IRAK1-3′-UTR vector. The THP-1 cells were seeded into 96-well plates (4×104 cells/well) and cultured for 24 h. Subsequently, the cells were co-transfected with 1 µg plasmids and 100 nM miR-146a mimic using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. An empty pGL-3-promoter vector was used as the blank control, whilst an NC mimic was used as the negative control. The cells were subsequently lysed using Glo Lysis Buffer (Promega Corporation) at 24 h following transfection, and luciferase activity was measured using a Dual-Luciferase Reporter Assay system (Promega Corporation), according to the manufacturer's protocol. All luciferase activities were normalized to that of Renilla luciferase activity.
Statistical analysis
The data are presented as the mean ± standard deviation. One-way analysis of variance followed by the least significance difference post hoc test was performed to determine statistical significance among different groups using the SPSS software package (version 16.0; SPSS Inc.). P<0.05 was considered to indicate a statistically significant difference.
Results
miR-146a regulates IRAK1 expression in THP-1 cells
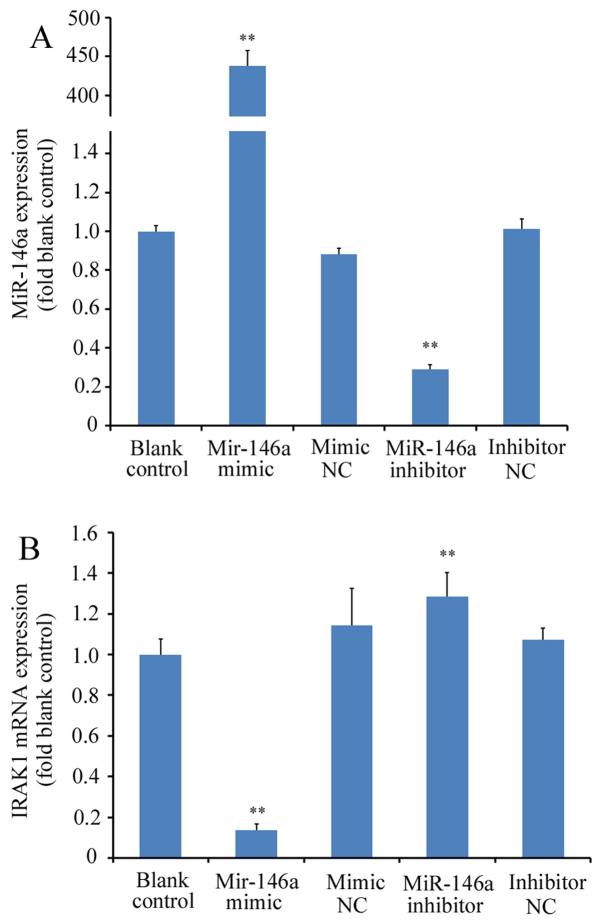
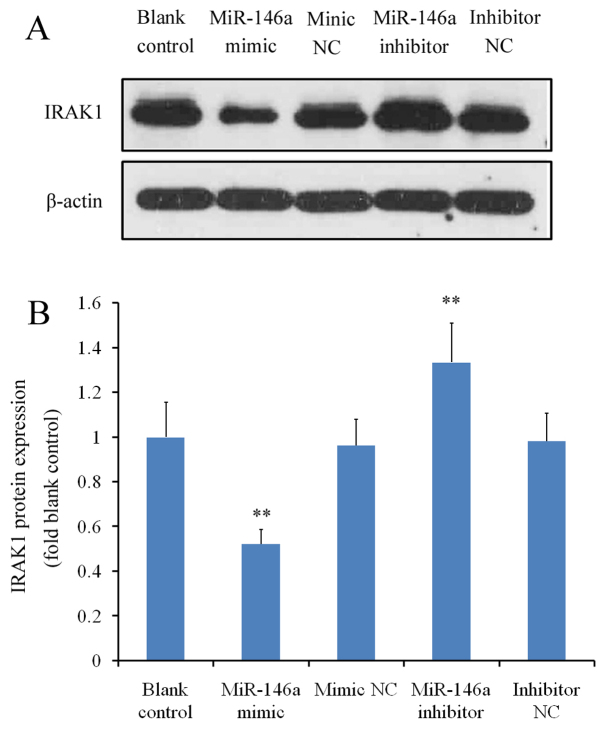
miR-146a mimicor miR-146a inhibitor were first transfected into THP-1 cells prior to RT-qPCR analysis. Transfection with miR-146a mimic significantly increased miR-146a expression (>400-fold) whilst transfection with miR-146a inhibitor significantly reduced miR-146a expression (by ~70%); with differences observed compared with the blank control or corresponding NC groups (P<0.01; Fig. 1A). IRAK1 mRNA expression was subsequently measured in THP-1 cells transfected with miR-146a mimic or inhibitor. miR-146a overexpression using the miR-146a mimic led to a significant reduction in IRAK1 mRNA expression (by ~88% relative to blank control; P<0.01 vs. mimic NC), whereas transfection with the miR-146a inhibitor led to a significant increase in IRAK1 expression (by ~20% relative to blank control; P<0.01 vs. inhibitor NC; Fig. 1B). At the protein level, cells transfected with the miR-146a mimic exhibited a significant reduction in IRAK1 expression compared with the mimic NC, whereas miR-146a inhibitor transfection significantly increased IRAK protein levels (P<0.01 vs. inhibitor NC; Fig. 2A and B). There were no significant differences between the blank control and to two corresponding NC groups (P>0.05 blank control vs. mimic NC and P>0.05 blank control vs. inhibitor NC; Fig. 2A and B).
Figure 1.
Effects of miR-146a modulation on the expression of IRAK1 mRNA levels. THP-1 cells were transfected with a miR-146a mimic or inhibitor. Reverse transcription-quantitative PCR was performed to measure the relative expression levels of (A) miR-146a and (B) IRAK1 mRNA, which were normalized to their respective control genes. Experiments were performed with six replicates for each group. **P<0.01 vs. corresponding NC groups. miR, microRNA; IRAK1, interleukin-1 receptor-associated kinase 1; NC, negative control.
Figure 2.
Effects of miR-146a modulation on IRAK1 protein levels. THP-1 cells were transfected with either a miR-146a mimic or inhibitor. Western blot analysis was performed to measure IRAK1 protein levels relative to the internal reference protein, β-actin. Three independent experiments were performed. (A) Representative image of the western blot analysis. (B) Relative quantification of IRAK1 expression. **P<0.01 vs. corresponding NC groups. miR, microRNA, IRAK1, interleukin-1 receptor-associated kinase 1; NC, negative control.
miR-146a modulates NF-κB activation by regulating IRAK1 expression under inflammatory conditions
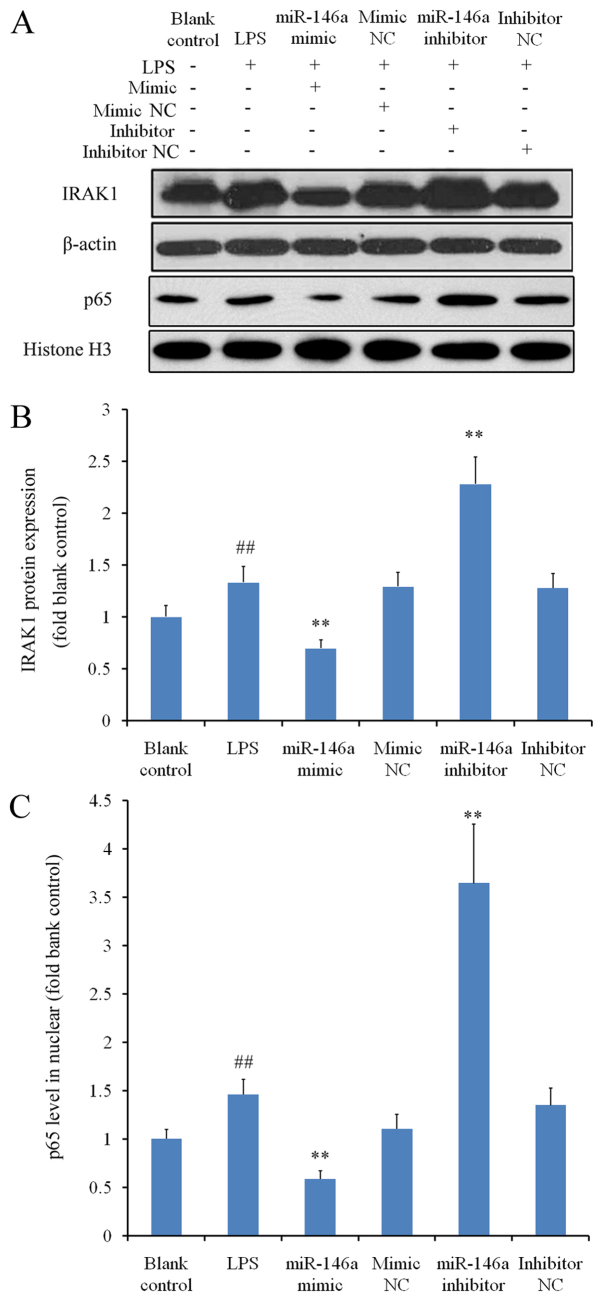
THP-1 cells were treated with LPS to induce inflammation, which significantly augmented IRAK1 expression when compared with the blank control group (P<0.01; Fig. 3A and B). Under inflammatory conditions, transfection with miR-146a mimic significantly reduced IRAK1 expression, whereas transfection with miR-146a inhibitor significantly increased IRAK1 levels compared with the corresponding NC groups (P<0.01; Fig. 3B). As NF-κB is a downstream molecule of IRAK1 (17), western blot analysis was subsequently performed to measure the nuclear protein levels of p65; a subunit of NF-κB. Nuclear p65 levels were significantly higher in the LPS group compared with those in the blank control group (P<0.01; Fig. 3C). The changes in nuclear NF-κB p65 protein levels in cells transfected with either miR-146a mimic or miR-146a inhibitor mirrored those of IRAK1 levels in the corresponding treatment groups (Fig. 3B and C). These results suggested that miR-146a regulates NF-κB activation in the presence of LPS, by modulating the expression of IRAK1.
Figure 3.
Inhibitory effects of miR-146a on IRAK1 expression and NF-κB activation under inflammatory conditions. THP-1 cells were treated with LPS following transfection with either a miR-146a mimic or inhibitor to activate the inflammatory signaling response. Western blot analysis was used to measure relative protein expression levels of IRAK1 and nuclear NF-κB p65 subunit, normalized to their respective internal references. Three independent experiments were performed. (A) Representative images of the western blot analysis. Relative protein expression levels of (B) IRAK1 and (C) the nuclear p65 subunit. ##P<0.01 vs. blank control and **P<0.01 vs. corresponding NC groups. miR, microRNA; IRAK1, interleukin-1 receptor-associated kinase 1; NF-κB, nuclear factor κB; LPS, lipopolysaccharide; NC, negative control.
miR-146a inhibits IL-6 and TNF-α secretion
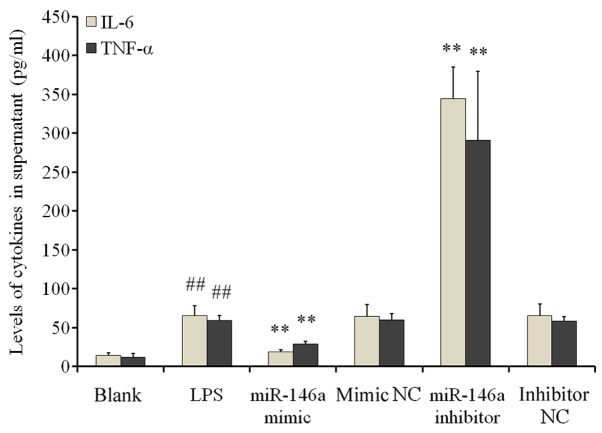
ELISA results demonstrated significantly increased IL-6 and TNF-α levels in the supernatant of cells in the LPS group when compared with cells in the blank control group (P<0.01; Fig. 4). miR-146a overexpression significantly reduced IL-6 and TNF-α secretion compared with the NC mimic groups (P<0.01). By contrast, transfection with the miR-146a inhibitor significantly increased IL-6 and TNF-α secretion when compared with the inhibitor NC group (P<0.01; Fig. 4). These findings suggested that LPS stimulation induced inflammatory responses in THP-1 cells by increasing the secretion of inflammatory cytokines; a process that is negatively regulated by miR-146a.
Figure 4.
Regulatory effects of miR-146a on IL-6 and TNF-α expression under inflammatory conditions. THP-1 cells were treated with LPS following transfection with either a miR-146a mimic or inhibitor to activate the inflammatory signaling response via Toll-like receptor 4. ELISA was performed to measure the levels of IL-6 and TNF-α in the supernatant of the culture medium. Experiments were performed with 6 replicates for each group. ##P<0.01 vs. blank control and **P<0.01 vs. corresponding NC groups. miR, microRNA; IL-6, interleukin-6; TNF-α, tumor necrosis factor-α; LPS, lipopolysaccharide; NC, negative control.
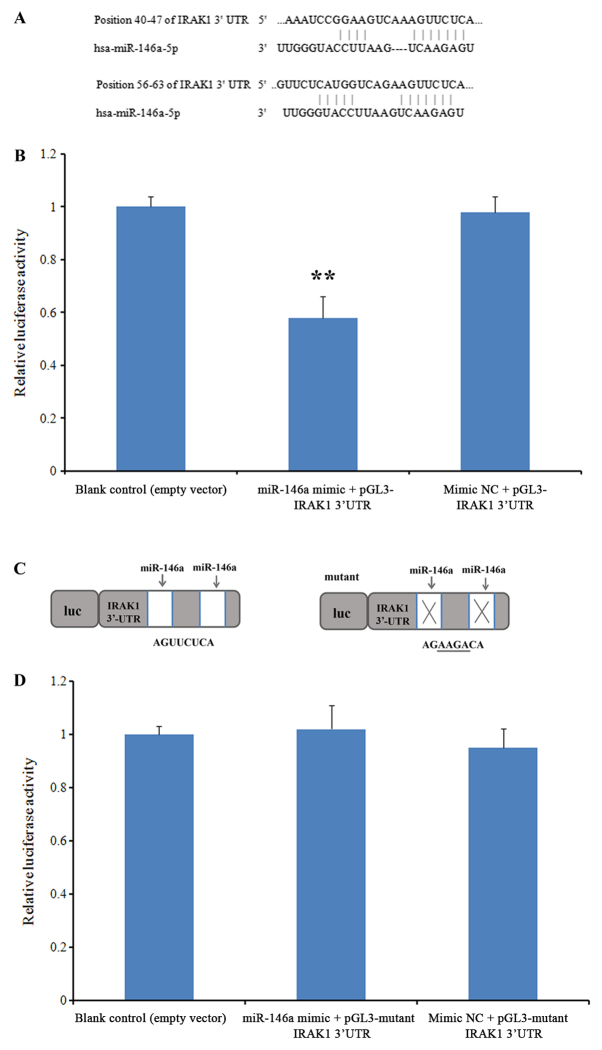
IRAK1 is a direct target of miR-146a
To investigate whether miR-146a can interact with the IRAK1 mRNA 3′-UTR, a luciferase reporter assay was performed. Prior to this assay, the predicted targets of this miRNA were revealed using TargetScan (version 7.2). Two evolutionarily conserved potential miR-146a target sites were discovered in the 3′-UTR of IRAK1 mRNA (positions 40–47 and 56–63; Fig. 5A). In the group co-transfected with miR-146a mimics and the wild-type 3′-UTR IRAK1 sequence, luciferase activity was significantly repressed when compared with the blank control group, while no significant difference was observed in cells co-transfected with the NC mimic (P<0.01; Fig. 5B). When the IRAK1 3′-UTR sequence was replaced with a mutant sequence (Fig. 5C), no significant differences were observed in the luciferase activity of cells co-transfected with the miR-146a mimic when compared with the blank control group (Fig. 5D). These results suggested that miR-146a directly targets the IRAK1 3′-UTR.
Figure 5.
miR-146a interacts directly with the IRAK1 3′-UTR. A luciferase reporter assay was performed to investigate the potential interaction between miR-146a and the IRAK1 3′-UTR. (A) Potential miR-146a target sites in the 3′-UTR of the IRAK1 sequence, as revealed using TargetScan analysis. (B) Luciferase activity in cells co-transfected with miR-146a mimics or mimics NC and the wild-type 3′-UTR sequence, relative to the blank control. (C) The mutant sequence of the IRAK1 3′-UTR. (D) Luciferase activity in cells co-transfected with miR-146a mimics or mimics NC and the mutant 3′-UTR sequence, relative to the blank control. Experiments were performed with 3 replicates for each group. **P<0.01 vs. blank control. miR, microRNA; IRAK1, interleukin-1 receptor-associated kinase 1; 3′-UTR, 3′-untranslated region; NC, negative control.
Discussion
SLE is a chronic diffuse connective tissue disease that is caused by inflammatory responses associated with autoimmunity (18). Although the mechanisms underlying SLE pathogenesis are unclear, the current consensus is that SLE may be associated with disturbances in the immune response, predisposing genetic factors, sexual hormone levels and exogenous stimulation (19). Dai et al (19) reported significant differences in the expression profiles of miRNAs between PBMCs derived from patients with SLE and healthy controls. Specifically, seven miRNAs were discovered to be downregulated whilst nine miRNAs were found to be upregulated in patients with SLE. Tang et al (20) discovered that the expression of miR-146a in PBMCs was significantly lower in patients with SLE compared with the control group, further suggesting that miR-146a negatively regulates the IFN-1 signaling pathway. It has been established that type I IFN serves a key role in SLE (20,21). Immune complexes of autoantibodies against self-nucleoproteins and self-DNA from lupus patients are known to induce IFN-α production in plasmacytoid dendritic cells (22). Once induced, type I IFN binds to its receptors to initiate downstream signaling by activation of STAT proteins, ultimately leading to the transcription of target genes (20). Reduced expression of miR-146a in patients with SLE and correlation between miR-146a levels and disease activity have been previously reported (20,23). miR-146a has been demonstrated to inhibit the type I IFN pathway by downregulating IFN regulatory factor 5 (IRF-5) and STAT-1 (20).
IRAK1, one of the regulatory targets of miR-146a, is a critical signaling molecule involved in the TLR4 signal transduction pathway. When the TLR is activated by pathogens or antigen-antibody complexes, it activates the associated adaptor protein IRAKI by the downstream My D88-dependent signaling pathway, leading to the activation of TRAF6, IΚKβ and eventually NF-κB (24,25). The findings of the present study demonstrated that miR-146a overexpression may downregulate the expression of IRAK1, reduce NF-κB activation and inhibit the secretion of pro-inflammatory cytokines, a signaling cascade that may serve as one of the mechanisms underlying miR-146a-mediated regulation of inflammatory responses in SLE.
Patients with SLE are predisposed to infection due to alterations in the immune system as a result of immunosuppressive therapy. As a result, infections may exacerbate SLE by promoting inflammatory responses. The development and progression of SLE is associated with enhanced inflammatory responses and dysregulated pro-inflammatory cytokines secretion (26). IL-6 and TNF-α expression are generally increased in patients with SLE compared with healthy individuals (27–29). The levels of IL-6 and TNF-α are positively associated with disease progression and serum levels of anti-double-stranded (ds) DNA antibody (29,30). IL-6 is an important type II T helper cell cytokine that is secreted by a variety of cell types, including T cells and monocytes, and stimulates the production of anti-ds DNA autoimmune antibodies in patients with SLE. This subsequently leads to the production of IL-6, and the cycle begins again (30,31). TNF-α activates a number of inflammatory cells, which infiltrate multiple organs and induce inflammation-associated damage (30–32). The activation of NF-κB promotes the expression of pro-inflammatory cytokines including IL-6 and TNF-α (33). The results of the present study indicated that miR-146a may inhibit the activation of NF-κB, thereby decreasing the expression of IL-6 and TNF-α. Similarly, Du et al (34) demonstrated that miR-146 a suppresses the β-glucan-induced production of IL-6 and TNF-α by inhibiting the dectin-1/tyrosine-protein kinase SYK/NF-κB signaling pathway. These observations suggest that the downregulation of miR-146a may eliminate its regulatory effects on the secretion of pro-inflammatory cytokines, leading to an increase in IL-6 and TNF-α levels. This may promote the development of SLE, particularly under conditions of infection. Ultimately, miR-146a may regulate IRAK1 expression and inhibit the activation of inflammatory signals, leading to the secretion of pro-inflammatory cytokines that may be involved in SLE pathogenesis.
Although an association between miR-146a and inflammation was observed in vitro in the present study, the clinical significance of miR-146a would need to be addressed in future studies. Previous studies have observed aberrant miR-146a expression in blood cells, including PBMCs, T and B cells (35–37). Procedures involving cell isolation and extraction are complex, and therefore not suitable for clinical detection. Alternatively, it has been clearly demonstrated that miRNA remains stable in the serum and other bodily fluids (38). Therefore, the serum levels of miR-146a between patients with SLE and healthy individuals, and the potential application of serum miRNA as a tool for monitoring SLE in a clinical setting, remain potential avenues for further investigation. The in vitro experiments performed in the present study revealed that miR-146a may regulate the expression of IL-6 and TNF-α in THP-1 cells; therefore, in vivo miR-146a levels in patients with SLE may be associated with the levels of IL-6 and TNF-α. A clinical trial is currently in progress involving >120 patients with SLE, in which the patients will be divided into active and inactive groups according to the Systemic Lupus Erythematosus Disease Activity Index. In addition, the role of the miR-146a/IRAK1/NF-κB axis in vivo will need to be investigated. Finally, the association between miR-146a and other autoimmune diseases, including RA and Sjogren's syndrome, are also interesting topics of future study.
Acknowledgements
Not applicable.
Funding
This study was supported by the National High Technology Research and Development Program (grant no. 2015AA021107), the Natural Science Foundation of Tianjin (grant nos. 17JCYBJC27500 and 17JCYBJC26200), the National Natural Science Foundation of China (grant no. 81602403) and the Clinical Research Foundation of Tianjin Fist Central Hospital (grant no. CF201812).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
HM and CZ designed the study, analyzed the data, and contributed significantly to the preparation of the manuscript. LZ and KW performed the reverse transcription-quantitative PCR and western blot experiments. QQ and MW performed the cell culture experiments. PS and LY performed the ELISA experiments.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Ranganathan K, Sivasankar V. MicroRNAs-biology and clinical applications. J Oral Maxillofac Pathol. 2014;18:229–234. doi: 10.4103/0973-029X.140762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pileczki V, Cojocneanu-Petric R, Maralani M, Neagoe IB, Sandulescu R. MicroRNAs as regulators of apoptosis mechanisms in cancer. Clujul Med. 2016;89:50–55. doi: 10.15386/cjmed-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miao CG, Yang YY, He X, Huang C, Huang Y, Zhang L, Lv XW, Jin Y, Li J. The emerging role of microRNAs in the pathogenesis of systemic lupus erythematosus. Cell Signal. 2013;25:1828–1836. doi: 10.1016/j.cellsig.2013.05.006. [DOI] [PubMed] [Google Scholar]
- 4.Sharma AR, Sharma G, Lee SS, Chakraborty C. MiRNA-regulated key components of cytokine signaling pathways and inflammation in rheumatoid arthritis. Med Res Rev. 2016;36:425–439. doi: 10.1002/med.21384. [DOI] [PubMed] [Google Scholar]
- 5.Habibi F, Ghadiri Soufi F, Ghiasi R, Khamaneh AM, Alipour MR. Alteration in inflammation-related miR-146a expression in NF-κB signaling pathway in diabetic rat hippocampus. Adv Pharm Bull. 2016;6:99–103. doi: 10.15171/apb.2016.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stickel N, Prinz G, Pfeifer D, Hasselblatt P, Schmitt-Graeff A, Follo M, Thimme R, Finke J, Duyster J, Salzer U, Zeiser R. MiR-146a regulates the TRAF6/TNF-axis in donor T cells during GVHD. Blood. 2014;124:2586–2595. doi: 10.1182/blood-2014-04-569046. [DOI] [PubMed] [Google Scholar]
- 7.Taganov KD, Boldin MP, Baltimore D. MicroRNAs and immunity: Tiny players in a big field. Immunity. 2007;26:133–137. doi: 10.1016/j.immuni.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 8.Perl A. Pathogenic mechanisms in systemic lupus erythematosus. Autoimmunity. 2010;43:1–6. doi: 10.3109/08916930903374741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang H, Peng W, Ouyang X, Li W, Dai Y. Circulating microRNAs as candidate biomarkers in patients with systemic lupus erythematosus. Transl Res. 2012;160:198–206. doi: 10.1016/j.trsl.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 10.Ji JD, Cha ES, Lee WJ. Association of miR-146a polymorphisms with systemic lupus erythematosus: A meta-analysis. Lupus. 2014;23:1023–1030. doi: 10.1177/0961203314534512. [DOI] [PubMed] [Google Scholar]
- 11.Gao M, Wang X, Zhang X, Ha T, Ma H, Liu L, Kalbfleisch JH, Gao X, Kao RL, Williams DL, Li C. Attenuation of cardiac dysfunction in polymicrobial sepsis by MicroRNA-146a is mediated via targeting of IRAK1 and TRAF6 expression. J Immunol. 2015;195:672–682. doi: 10.4049/jimmunol.1403155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaisho T, Aldra S. Toll-like receptor function and signaling. J Allergy Clin Immunol. 2006;117:979–988. doi: 10.1016/j.jaci.2006.02.023. [DOI] [PubMed] [Google Scholar]
- 13.Rodriguez-Pla A, Patel P, Maecker HT, Rossello-Urgell J, Baldwin N, Bennett L, Cantrell V, Baisch J, Punaro M, Gotte A, et al. IFN priming is necessary but not sufficient to turn on a migratory dendritic cell program in lupus monocytes. J Immunol. 2014;192:5586–5598. doi: 10.4049/jimmunol.1301319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang P, Han X, Mo B, Huang G, Wang C. LPS enhances TLR4 expression and IFN-γ production via the TLR4/IRAK/NF-κB signaling pathway in rat pulmonary arterial smooth muscle cells. Mol Med Rep. 2017;16:3111–3116. doi: 10.3892/mmr.2017.6983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nahid MA, Pauley KM, Satoh M, Chan EK. MiR-146a is critical for endotoxin-induced tolerance: Implication in innate immunity. J Biol Chem. 2009;284:34590–34599. doi: 10.1074/jbc.M109.056317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 17.Chen Y, Zeng Z, Shen X, Wu Z, Dong Y, Cheng JC. MicroRNA-146a-5p negatively regulates pro-inflammatory cytokine secretion and cell activation in lipopolysaccharide stimulated human hepatic stellate cells through inhibition of Toll-like receptor 4 signaling pathways. Int J Mol Sci1. 2016;17(pii):E1076. doi: 10.3390/ijms17071076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muñoz LE, Janko C, Schulze C, Schorn C, Sarter K, Schett G, Herrmann M. Autoimmunity and chronic inflammation-two clearance-related steps in the etiopathogenesis of SLE. Autoimmun Rev. 2010;10:38–42. doi: 10.1016/j.autrev.2010.08.015. [DOI] [PubMed] [Google Scholar]
- 19.Dai Y, Huang YS, Tang M, Lv TY, Hu CX, Tan YH, Xu ZM, Yin YB. Microarray analysis of microRNA expression in peripheral blood cells of systemic lupus erythematosus patients. Lupus. 2007;16:939–946. doi: 10.1177/0961203307084158. [DOI] [PubMed] [Google Scholar]
- 20.Tang Y, Luo X, Cui H, Ni X, Yuan M, Guo Y, Huang X, Zhou H, de Vries N, Tak PP, et al. MicroRNA-146A contributes to abnormal activation of the type I interferon pathway in human lupus by targeting the key signaling proteins. Arthritis Rheum. 2009;60:1065–1075. doi: 10.1002/art.24436. [DOI] [PubMed] [Google Scholar]
- 21.Wigren M, Nilsson J, Kaplan MJ. Pathogenic immunity in systemic lupus erythematosus and atherosclerosis: Common mechanisms and possible targets for intervention. J Intern Med. 2015;278:494–506. doi: 10.1111/joim.12357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Means TK, Latz E, Hayashi F, Murali MR, Golenbock DT, Luster AD. Human lupus autoantibody-DNA complexes activate DCs through cooperation of CD32 and TLR9. J Clin Invest. 2005;115:407–417. doi: 10.1172/JCI23025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Husakova M. MicroRNAs in the key events of systemic lupus erythematosus pathogenesis. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2016;160:327–342. doi: 10.5507/bp.2016.004. [DOI] [PubMed] [Google Scholar]
- 24.Fang H, Wang PF, Zhou Y, Wang YC, Yang QW. Toll-like receptor 4 signaling in intracerebral hemorrhage-induced inflammation and injury. J Neuroinflammation. 2013;10:27. doi: 10.1186/1742-2094-10-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He X, Zheng Y, Liu S, Shi S, Liu Y, He Y, Zhang C, Zhou X. MiR-146a protects small intestine against ischemia/reperfusion injury by down-regulatingTLR4/TRAF6/NF-κB pathway. J Cell Physiol. 2018;233:2476–2488. doi: 10.1002/jcp.26124. [DOI] [PubMed] [Google Scholar]
- 26.Nahid MA, Satoh M, Chan EK. MicroRNA in TLR signaling and endotoxin tolerance. Cell Mol Immunol. 2011;8:388–403. doi: 10.1038/cmi.2011.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Willis R, Seif AM, McGwin G, Jr, Martinez-Martinez LA, González EB, Dang N, Papalardo E, Liu J, Vilá LM, Reveille JD, et al. Effect of hydroxychloroquine treatment on pro-inflammatory cytokines and disease activity in SLE patients: Data from LUMINA (LXXV), a multiethnic US cohort. Lupus. 2012;21:830–835. doi: 10.1177/0961203312437270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ripley BJ, Goncalves B, Isenberg DA, Latchman DS, Rahman A. Raised levels of interleukin 6 in systemic lupus erythematosus correlate with anaemia. Ann Rheum Dis. 2005;64:849–853. doi: 10.1136/ard.2004.022681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sabry A, Sheashaa H, EI-Husseini A, Mahmoud K, Eldahshan KF, George SK, Abdel-Khalek E, El-Shafey EM, Abo-Zenah H. Proinflammatory cytokines (TNF-alpha and IL-6) in Egyptian patients with SLE: Its correlation with disease activity. Cytokine. 2006;35:148–153. doi: 10.1016/j.cyto.2006.07.023. [DOI] [PubMed] [Google Scholar]
- 30.Arora V, Verma J, Marwah V, Kumar A, Anand D, Das N. Cytokine imbalance in systemic lupus erythematosus: A study on northern Indian subjects. Lupus. 2012;21:596–603. doi: 10.1177/0961203311434937. [DOI] [PubMed] [Google Scholar]
- 31.Gigante A, Gasperini ML, Afeltra A, Barbano B, Margiotta D, Cianci R, De Francesco I, Amoroso A. Cytokines expression in SLE nephritis. Eur Rev Med Pharmacol Sci. 2011;15:15–24. [PubMed] [Google Scholar]
- 32.Robinson ES, Werth VP. The role of cytokines in the pathogenesis of cutaneous lupus erythematosus. Cytokine. 2015;73:326–334. doi: 10.1016/j.cyto.2015.01.031. [DOI] [PubMed] [Google Scholar]
- 33.Nahid MA, Satoh M, Chan EK. Mechanistic role of microRNA-146a in endotoxin-induced differential cross-regulation of TLR signaling. J Immunol. 2011;186:1723–1734. doi: 10.4049/jimmunol.1002311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Du L, Chen X, Duan Z, Liu C, Zeng R, Chen Q, Li M. MiR-146a negatively regulatesdectin-1-induced inflammatory responses. Oncotarget. 2017;8:37355–37366. doi: 10.18632/oncotarget.16958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mookherjee N, El-Gabalawy HS. High degree of correlation between whole blood and PBMC expression levels of miR-155 and miR-146a in healthy controls and rheumatoid arthritis patients. J Immunol Methods. 2013;400-401:106–110. doi: 10.1016/j.jim.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 36.Sheppard HM, Verdon D, Brooks AE, Feisst V, Ho YY, Lorenz N, Fan V, Birch NP, Didsbury A, Dunbar PR. MicroRNA regulation in human CD8+ T cell subsets-cytokine exposure alone drives miR-146a expression. J Transl Med. 2014;12:292. doi: 10.1186/s12967-014-0292-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cho S, Lee HM, Yu IS, Choi YS, Huang HY, Hashemifar SS, Lin LL, Chen MC, Afanasiev ND, Khan AA, et al. Differential cell-intrinsic regulations of germinal center B and T cells by miR-146a and miR-146b. Nat Commun. 2018;9:2757. doi: 10.1038/s41467-018-05196-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu AM, Wang W, Luk JM. miRNAs: New tools for molecular classification, diagnosis and prognosis of hepatocellular carcinoma. Hepat Oncol. 2014;1:323–329. doi: 10.2217/hep.14.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.