Abstract
Burkholderia pseudomallei is a Gram-negative saprophytic bacillus and the aetiological agent of melioidosis, a disease of public-health importance throughout Southeast Asia and northern Australia. Infection can occur in humans and a wide array of animal species, though zoonotic transmission and case clusters are rare. Despite its highly plastic genome and extensive strain diversity, fine-scale investigations into the population structure of B. pseudomallei indicate there is limited geographical dispersal amongst sequence types (STs). In the ‘Top End’ of northern Australia, five STs comprise 90 % of the overall abundance, the most prevalent and widespread of which is ST-109. In May 2016, ST-109 was implicated in two fatal cases of melioidosis in juvenile saltwater crocodiles at a wildlife park near Darwin, Australia. To determine the probable source of infection, we sampled the crocodile enclosures and analysed the phylogenetic relatedness of crocodile and culture-positive ST-109 environmental park isolates against an additional 135 ST-109 B. pseudomallei isolates from the Top End. Collectively, our whole-genome sequencing (WGS) and pathology findings confirmed B. pseudomallei detected in the hatchling incubator as the likely source of infection, with zero SNPs identified between clinical and environmental isolates. Our results also demonstrate little variation across the ST-109 genome, with SNPs in recombinogenic regions and one suspected case of ST homoplasy accounting for nearly all observed diversity. Collectively, this study supports the use of WGS for outbreak source attribution in highly recombinogenic pathogens, and confirms the epidemiological and phylogenetic insights that can be gained from high-resolution sequencing platforms.
Keywords: Burkholderia pseudomallei, melioidosis, whole-genome sequencing, source tracing, saltwater crocodile
Data Summary
Raw sequence data from this study are available in the National Center for Biotechnology Information Short Read Archive, under BioProject accession number PRJNA510860 (http://www.ncbi.nlm.nih.gov/bioproject/510860). Accession numbers for these strains and other Burkholderia pseudomallei strains used in the analyses are listed in Table S2 (available with the online version of this article).
Impact Statement.
Melioidosis is a high-mortality tropical disease of humans and animals caused by the saprophytic bacterium Burkholderia pseudomallei . While rare, outbreaks of the infection have been described and are usually attributable to a single point source in the environment. Though traditional molecular fingerprinting methods like multilocus sequence typing have been able to resolve distinct populations of the bacterium, the exceedingly high rate of recombination in B. pseudomallei can significantly confound inferences about infection aetiology and transmission. This has global biosecurity relevance given the bacterium’s recent classification as a tier 1 biological select agent. Here, we used high-resolution whole-genome sequencing (WGS) and comparative phylogenetics to examine the aetiology of a rare cluster of melioidosis infection in two hatchling saltwater crocodiles at a Wildlife Park in northern Australia. Our findings demonstrate the epidemiological insights that can be gained from WGS, and confirm its use to improve knowledge of bacterial genotype diversification and patterns of dispersal in the environment.
Introduction
Melioidosis is a disease of humans and animals endemic to Southeast Asia and northern Australia caused by the saprophytic bacterium Burkholderia pseudomallei [1]. Infection is normally acquired via percutaneous inoculation or ingestion of contaminated soil or water [2], though inhalation, particularly during severe weather events, has been recognized as an important source of infection [3]. While the full extent of melioidosis infection remains poorly understood, recent modelling of the global distribution of melioidosis estimated more than 165,000 human cases and 89, 000 deaths every year [4]. The majority of those who succumb to the infection have at least one identifiable risk factor, including diabetes mellitus, lung or kidney disease, or hazardous alcohol consumption [3]. For healthy individuals, severe disease is uncommon and death is very rare when appropriate antibiotics and intensive care management are available [5].
In endemic areas, melioidosis has also been identified in a wide array of animal species [6–8]. Certain animals are acknowledged to be particularly susceptible to infection and disease, including goats [9], sheep [10], camels [11] and alpacas [12]. Cases have also been reported in domestic pets and native wildlife, with the animals often having prior ill health [12]. Exotic animals imported to zoos in endemic regions appear especially at risk, most notably primates, including iconic species such as gorillas [7, 13]. While rare, outbreaks have also been reported in both endemic and non-endemic settings, including piggeries in Queensland, Australia [14], and several reported in European zoos [7, 15]. Interestingly, B. pseudomallei has been detected in faecal samples from wallabies and chickens [16], as well as from the beaks of healthy native birds [17], suggesting plausible mechanisms for geographical dissemination of the bacterium.
While the global distribution and phylogeny of B. pseudomallei are not fully resolved, whole-genome sequencing (WGS) has been used in combination with traditional typing methods to demonstrate an extensive degree of intra-species diversity [18–22]. More than 1680 multilocus sequence typing (MLST) variants of the bacterium have now been identified (https://pubmlst.org/bpseudomallei/) [19], a consequence of its exceptionally high rate of horizontal gene transfer and recombination [20]. Despite this high degree of diversity, the bacterium is known to spatially cluster and sequence types (STs) have been shown to have limited geographical dispersal [23]. Distinct populations of the bacterium have been identified in Southeast Asia and Australia by WGS, as well as in the Northern Territory, Australia, and Queensland, Australia [23–25]. Interestingly, no variations in diversity have been identified between clinical and environmental STs. The relative abundance and composition of environmental and clinical STs have also been shown to be directly correlated, implying that the majority of culturable environmental B. pseudomallei molecular types have the capability to cause infection [25].
Within the Darwin region of the tropical 'Top End' of northern Australia, five STs have been shown to comprise 90 % of the overall abundance of environmental and clinical isolates [23, 25]. Of the STs uncovered so far in the Northern Territory, ST-109 is the most frequently identified and widely dispersed molecular type, having been detected in environmental samples from Darwin, Northern Territory, as far south as Livingstone, Northern Territory, a linear distance of less than 50 km [23]. It is also one of only a small number of STs to have been isolated from human, animal and environmental samples, including both soil and water [23]. An analysis of 36 ST-109 genomes found the ST to be incredibly diverse, with approximately 3 % variability in the 7.3 Mbp ST-109 genome identified between sequenced isolates [26].
In May 2016, B. pseudomallei was isolated from two deceased saltwater crocodile (Crocodylus porosus) hatchings at a Wildlife Park just outside of Darwin, Northern Territory. ST-109 was identified as the strain responsible for infection in both animals. Though case reports of melioidosis in crocodiles have been documented in Thailand, Malaysia and Australia, crocodiles are thought to be highly resistant to infection [2, 8, 12, 27]. In an overview of bacterial isolates obtained from diagnostic samples of 159 sick or dead farmed hatchling and juvenile saltwater crocodiles over 5 years in the Northern Territory, B. pseudomallei was not identified [28]. Moreover, melioidosis is not usually considered transmissible between animals and case clusters are rare [3, 12]. This prompted us to further investigate whether the two cases were caused by a single point source in the park environment. We undertook three rounds of environmental sampling at the park, and performed WGS and MLST on cultured isolates. Using WGS data already available through Menzies School of Health Research, Australia, we analysed the genetic relatedness and population structure of clinical and environmental ST-109 isolates (n=140) collected and sequenced over the last 29 years from the Northern Territory. Used in conjunction with the crocodile pathology findings, these data allowed us to infer the most likely source of infection in the hatchlings.
Methods
Animals and animal sampling
The deceased saltwater crocodiles were from a Wildlife Park and crocodile breeding facility in Darwin, Australia. The index case was a hatchling less than 1-week-old, found dead in the hatchling pen in early May 2016. The second case was a 3-week-old hatchling, found dead in the same pen 5 days later. Both crocodiles were presumed to originate from the same hatchery incubator batch. Complete gross necropsies were performed on both crocodiles. For histology, samples of liver, lung, heart, spleen, kidney, adrenal gland, thymus, brain, stomach and intestine from both crocodiles, plus eye from the second crocodile, were fixed in 10 % phosphate-buffered formalin, processed in a standard fashion and stained with haematoxylin and eosin. For general aerobic bacterial culture, using aseptic technique, samples of the internalized yolk sacs and liver were taken from both crocodiles, plus lung from the second crocodile. These samples were homogenized with 0.85% physiological saline, plated onto sheep blood and MacConkey agars and incubated at 35 °C for 48 h. Bacterial growth was speciated using the API 20 NE identification system (bioMérieux).
Environmental sample collection
Environmental sampling took place in May 2016 at a Wildlife Park located approximately 10 km outside of Darwin, Northern Territory, Australia (latitude 12°S). Three rounds of sampling were completed over an 18 day period, starting 3 days after the death of the second crocodile. Surveying efforts were concentrated on the areas surrounding the juvenile holding ponds and crocodile hatchling incubator. A total of 44 samples were collected from the park, including 1 soil sample, 18 water samples, 14 environmental swabs and 1 air sample (Table S1).
Environmental sample processing and confirmation
Culture of B. pseudomallei from water, soil, air and swab specimens was carried out using methods previously developed in our laboratory [29–31]. Briefly, samples were enriched in Ashdown’s broth containing colistin (50 mg l−1) and incubated at 37 °C aerobically for 2 and 7 days. Enriched broth was plated onto Ashdown's agar with gentamicin (8 mg l−1) and incubated for 48 h, and all colonies resembling B. pseudomallei were sub-cultured onto Ashdown’s agar. DNA from suspected colonies was extracted using 10 % Chelex-100 resin [32] and confirmation of B. pseudomallei was carried out using a real-time PCR assay targeting a 115 bp segment within the type three secretion system 1 (TTS1) gene cluster [33] specific to B. pseudomallei .
ST-109 isolates used for analysis
One hundred and forty ST-109 B. pseudomallei isolates from the Northern Territory, Australia (132 clinical isolates, including 126 human and 6 animal isolates, plus 8 environmental isolates), collected between January 1990 and June 2017 were analysed in this study (Table S2).
Human isolates
Genomes from 126 human ST-109 isolates were obtained through the ongoing Darwin Prospective Melioidosis Study (DPMS). The study began in October 1989 and comprises all known human cases of culture-confirmed melioidosis in the Top End of the Northern Territory over the past 29 years [3]. All clinical isolates were cultured and stored at −80 °C at Menzies School of Health Research, Australia. Between October 1 1989 and June 1 2018, the study enrolled 1052 patients. B. pseudomallei isolates were not available for 36 (3.4 %) of these cases. All remaining clinical cases had WGS and in silico MSLT data available for at least one isolate. B. pseudomallei isolates from 126 of these human cases (12.4 %) had been assigned as ST-109 and were consequently selected for the study. Of these, 30 genomes were already publicly available [26, 34] (Table S2).
Animal isolates
A single B. pseudomallei yolk sac isolate was selected for analysis from each of the two deceased crocodiles. Four additional ST-109 animal isolates belonging to a rhesus macaque (Macaca mulatta), common marmoset (Callithrix jacchus), black-capped capuchin (Sapajus apella) and green iguana (Iguana iguana) were also included in the study. All four of these melioidosis cases in exotic species occurred at the same Wildlife Park where the two deceased crocodiles were discovered. Isolates were collected between the years 2004 and 2016, and were cultured at Berrimah Veterinary Laboratory, Australia, and transferred to Menzies School of Health Research, Australia, for confirmation and strain typing.
Environmental isolates
Three ST-109 environmental isolates collected as part of the crocodile case cluster investigation (two holding tank water isolates and one incubator wall swab isolate) were included in the analysis. Five additional ST-109 environmental isolates (MSHR4462, MSHR4483, MSHR7797, MSHR8251, MSHR8316) with available WGS and MLST data were also included in the study. Two of these isolates, MSHR4462 and MSHR4483, had publicly available genomes [26, 34] (Table S2).
MLST assignment
MLST types for human clinical isolates were assigned in silico prior to the study commencing based on WGS data. This was carried out using BIGSdb [35], an integrated tool available on the B. pseudomallei MLST website (http://pubmlst.org/bpseudomallei/). MLST strain typing of animal isolates (n=5) and environmental park isolates collected as part of the investigation (n=3) was performed using a set of in-house PCR primers designed to rapidly identify the common Darwin ST types 36, 109, 132 and 562 [26]. Only isolates designated as ST-109 were included in the study. The five environmental isolates not originating from the park had MLST types assigned from WGS data in silico (MSH4462, MSHR8251, MSHR8316) or via in-house PCR primers (MSHR4483, MSHR7797).
WGS of clinical and environmental Wildlife Park isolates
Genomic DNA was extracted using the Qiagen DNeasy blood and tissue kit (Qiagen), as previously described [21]. Samples were sequenced at Macrogen, Inc. (Gasan-dong, Seoul, Republic of Korea), Australian Genome Research Facility Ltd. (Melbourne, Australia) or the Centre for Microbial Genetics and Genomics and Translational Genomics Research Institute (Flagstaff, AZ, USA) using the Illumina HiSeq2000 and Illumina HiSeq2500 platforms.
Identification of orthologous core biallelic SNP variants was performed on WGS data using SPANDx [36]. MSHR0605, a genome obtained from a Darwin clinical isolate in 1995, was used as reference for read mapping, being a high quality ST-109 assembly (N50- 348,030; total length- 7,136,846 bp; contigs- 35). Two maximum parsimony (MP) trees were generated using paup v4.0a162 from 31, 688 SNPs identified amongst 140 ST-109 genomes and 479 SNPs identified amongst 129 ST-109 genomes, respectively [37]. Incorporation of small insertions or deletions (InDels) in our analysis did not improve the resolution in this study and, thus, were excluded from the analysis [25]. Bootstrapping using 1000 replicates was carried out to establish the robustness of branches. Phylogenetic trees were visualized and manipulated in FigTree v1.4.3 (http://tree.bio.ed.ac.uk/software/figtree/) and iTOL v3: Interactive Tree of Life [38]. Recombinogenic SNPs were identified with Gubbins (Genealogies Unbiased by Recombination In Nucleotide Sequences, v.2.3.1) using default parameters [39].
Pan-genome analysis
ST-109 assemblies (n=139) were annotated using Prokka (v1.13). A pan-genome was calculated for the 139 annotated assemblies using Roary (v3.12.0) with a default protein Basic Local Alignment Search Tool (blastp) minimum percentage identity threshold of 95 % [40]. Based on results from Gubbins and phylogenetic trees, MSHR2174 was not included in the pan-genome analysis.
Virulence factor assignment
The presence of four known genetic virulence markers was investigated to further elucidate strain relatedness. Variably present filamentous haemagglutinin 3 (fhaB3) and the mutually exclusive virulence factors Burkholderia thailandensis-like flagella and chemotaxis (BTFC) cluster and Yersinia-like fimbriae (YLF) cluster loci, lipopolysaccharide (LPS) types A/B/B2 and Burkholderia intracellular motility factor A (bimA)Bm/Bp were examined using blast, as previously described [41–43]. In brief, the genes were blast searched against the B. pseudomallei ST-109 database using the nucleotide blast (blastn) parameter. Each genome was assigned as fhaB3 (encoded by BPSS2053 in B. pseudomallei K96243) positive or negative; as a carrier of BTFC or YLF [lafU in B. pseudomallei MSHR668 (GenBank accession no. NC_006350) and BPSS0124 in B. pseudomallei K96243 (GenBank accession no. CP009545.1), respectively]; as either LPS type A, B or B2 [LPS A, wbil to apaH in K96243 (GenBank accession no. NC_006350); LPS B, BUC_3392 to apaH in B. pseudomallei 579 (GenBank accession no. NZ_ACCE01000003); LPS B2, BURP840_LPSb01 to BURP840_LPSb21 in B. pseudomallei MSHR840 (GenBank accession no. 146 GU574442)]; and finally as carriers of bimA Bm [BURPS668_A2118 in B. pseudomallei MSHR668 (GenBank accession no. 147 NZ_CP009545)] or bimA Bp (BPSS1492 in B. pseudomallei K96243).
Results
Pathology and bacterial culture
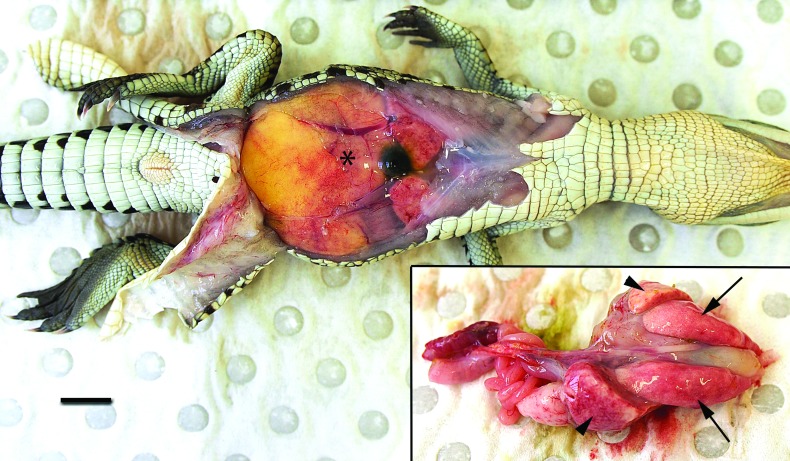
On a gross scale, both crocodiles had large, unabsorbed internal yolk sacs with overlying reddened serosae. In the first crocodile, the yolk appeared of normal consistency (pale yellow, liquid and homogeneous), while in the second case, the yolk was of uneven liquid/firm consistency and discoloured grey–yellow. Both crocodiles exhibited patchy subcutaneous and serosal oedema, and mild enlargement and abnormal pale pink-red mottling of the liver (Fig. 1). The lungs in the first crocodile were pale and wet, while those of the second crocodile exhibited random multifocal pinpoint white foci. Histologically, both cases were similar, with severe, random multifocal lytic necrosis with associated fibrin and marked heterophil and moderate macrophage infiltration involving the liver, spleen, lungs and heart most extensively, but with scattered foci also present in intestinal lamina propria, thymus and adrenal gland (Fig. S1). In both crocodiles, Gram staining revealed scattered Gram-negative bacilli within necrotic foci. Bacterial culture yielded pure growth of B. pseudomallei from all samples tested (yolk sac and liver from both crocodiles, plus lung from the second crocodile).
Fig. 1.
Gross necropsy image of the first crocodile showing a large internal yolk sac with overlying congested serosa (asterisk). Bar, 1 cm. Inset: dorsal view of viscera dissected out and yolk sac removed. The liver (arrowheads) is mottled pale pink and red, and the lungs (arrows) are pale pink and moist.
Environmental sampling
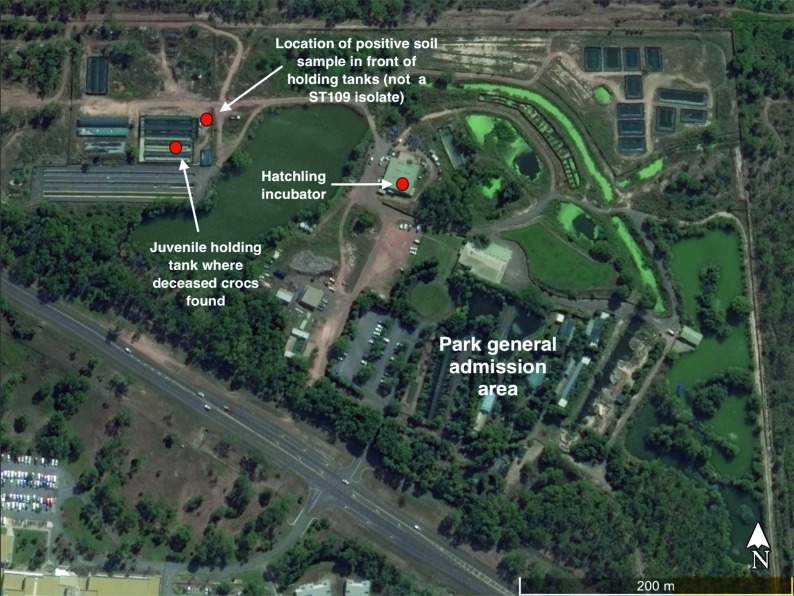
Of the 33 environmental samples collected from the park as part of the investigation, 4 (4/33, 12.1%) tested positive for B. pseudomallei (Table 1). This included one soil sample (1/1) from the grass lawn next to the juvenile holding pens, two water samples (2/18, 11.1%) both taken from the same holding tank in which the crocodiles were discovered deceased and one environmental swab (1/13, 7.7%) taken from a slime-like biofilm on the back wall of the crocodile egg incubator (Figs 2 and 3). The two positive water samples were taken 18 days apart from one another. No other replicate samples tested positive.
Table 1.
Environmental samples collected from the park over three rounds of sampling
|
Sample type |
Total no. of samples collected |
No. of B. pseudomallei -positive samples |
|---|---|---|
|
Soil |
1 |
1 |
|
Water |
18 |
2 |
|
Air |
1 |
0 |
|
Environmental swab |
13 |
1 |
|
Total |
33 |
4 |
Fig. 2.
- Map of the Wildlife Park showing sites where B. pseudomallei was isolated. Two water samples taken from the same holding tank tested positive, as did a swab taken from the back wall of the hatchling incubator and a soil sample from in front of the holding tanks.
Fig. 3.
Locations of the ST-109 positive samples. (a) The crocodile egg incubator and (b) the location of the swab sample obtained from a biofilm growth on a strip of black paint on the back wall of the incubator. (c) The outside of the juvenile holding ponds where the deceased crocodiles were found and (d) the location where the two ST-109 positive water samples were retrieved.
ST-109 was the strain responsible for the infection and was present in the park environment
Isolates from both crocodile specimens were designated as ST-109 using the described SNP ST PCR assays [26]. ST-109 was also assigned to isolates from three of four positive environmental samples, including the swab taken from the back wall of the incubator and both water samples taken from the crocodile holding pond (Fig. 3). The fourth sample, soil collected from the area immediately adjacent to the holding tanks, did not test positive for any of the four common STs and, thus, was not included in further analysis.
WGS analysis of ST-109 shows there is little intra-ST diversity
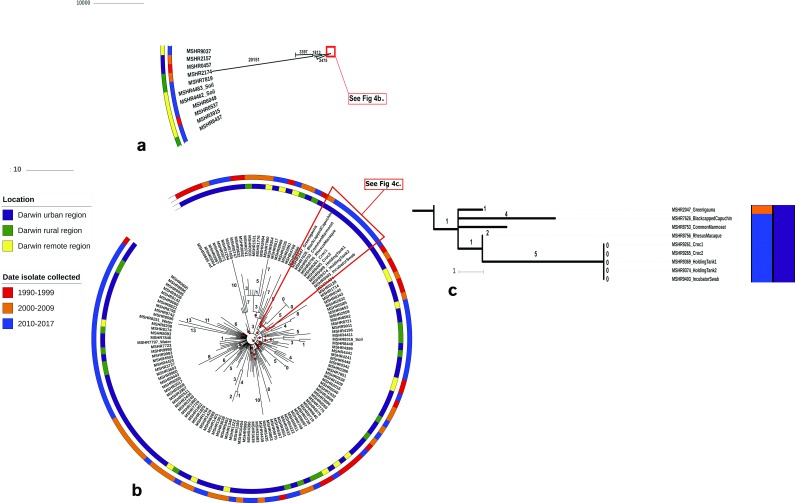
To further investigate the potential source of infection in the crocodiles, we examined five ST-109 clinical and environmental Wildlife Park isolates together with 135 ST-109 isolates from the Top End of the Northern Territory. These additional genomes served as close references for our crocodile case cluster findings. The total genomic size of the 140 ST-109 B. pseudomallei isolates ranged from 6,979,682 to 7,311,795 bp with a mean length of 7,135,942 bp. The two crocodile isolates were slightly larger than the mean total length. MP phylogenetic reconstruction of orthologous SNPs from the 140 ST-109 isolates showed only minimal differences across the genome for the majority of isolates (Fig. 4a, b, c). Nearly all of the variation observed across ST-109 stemmed from 11 outliers (MSHR0457, MSHR0537, MSHR2157, MSHR2174, MSHR3915, MSHR4462, MSHR4483, MSHR6049, MSHR6437, MSHR7819, MSHR9037) (Fig. 4a). While two of these isolates (MSHR3915, MSHR6437) contained 77 and 88 SNPs compared to the ST-109 reference genome, respectively, the remaining nine variants contained between 2,760 to 24, 564 orthologous SNPs, causing a large reduction in the overall resolution of the phylogeny. Of note, 5 of the 11 outliers were isolated from clinical or environmental samples originating from Adelaide River, Northern Territory, a town 90 linear km south of Darwin. Four of these Adelaide River isolates (two soil isolates, MSHR4462 and MSHR4483, and two human, MSHR0537 and MSHR6049) clustered closely together on the whole-genome level (<8 SNPs differentiating them) (Fig. 4a). Despite the limited number of SNPs differentiating the isolates, the earliest of these four isolates (MSHR0537) was collected 14 years earlier than the latest (MSHR6049).
Fig. 4.
(a, b) MP phylogeny of 140 B. pseudomallei ST-109 genomes reconstructed using core-genome orthologous SNPs [overall consistency index (CI)=0.913] (a) and high-resolution ST-109 reconstructed SNP phylogeny with 11 outliers removed (n=129) (CI=0.9937) (b). MSHR0605 was used as the reference for both phylogenies. Environmental and Wildlife Park isolates are labelled, while human samples are denoted by respective isolate (MSHR) IDs. Red circles on branches denote bootstrap values<80. (c) The inset shows a high-resolution view of the Wildlife Park samples from (b). Bars indicate number of SNP’s.
To check for large-scale recombination in the ST-109 genomes, we reconstructed our phylogeny using Gubbins (v.2.3.1). Recombinogenic SNPs were detected in all ST-109 strains, including the outlying strains. SNP density filtering was applied to remove recombined regions and 3,632 (12.7 %) SNPs were identified in recombinogenic regions, consequently changing the topology of our phylogeny for all but one of the outliers, MSHR2174. For this isolate, 24,041 SNPs were still present in non-recombinogenic regions after filtering, suggesting this could be a case of MLST homoplasy as has been described before for B. pseudomallei [18].
To further examine the intra-strain relatedness, a pan-genome was calculated for 139 of the 140 ST-109 isolates based on WGS data. MSHR2174 was removed from the analysis due to suspicions of ST homoplasy. We identified a total of 10,203 predicted coding sequences, with 4,926 and 5,277 genes assigned to the core (present in 99 % of isolates) and accessory (variably present) genome, respectively (Fig. S2). While the number of genes assigned to the core was similar to that reported elsewhere, we identified fewer accessory genes than has previously been observed in pan-genome studies of B. pseudomallei [22, 44]. Moreover, there was only limited isolate-specific gene clustering observed, which occurred within the accessory genomes of nine of the ten ST-109 outlier strains included in the pan-genome analysis (MSHR0457, MSHR0537, MSHR2157, MSHR3915, MSHR4462, MSHR4483, MSHR6049, MSHR6437, MSHR9037) (Fig S2a).
We also investigated the variably present virulence gene markers fhaB3, BTFC/YLF, LPS types A/B/B2 and bimA Bm/Bp, which are frequently used as determinants of geographical origin, strain relatedness and pathogenicity in B. pseudomallei [41–43]. All isolates were carriers of LPS A, bimA Bp and BTFC, consistent with their close phylogenomic relatedness. Additionally, all were positive for fhaB3 except for a single isolate, MSHR9632, a clinical patient strain from urban Darwin.
ST-109 crocodile and Wildlife Park environmental isolates are identical by WGS SNP analysis
Lastly, the 11 outliers (MSHR0457, MSHR0537, MSHR2157, MSHR2174, MSHR3915, MSHR4462, MSHR4483, MSHR6049, MSHR6437, MSHR7819, MSHR9037) were removed to improve the resolution of our SNP phylogeny. Upon removal, we were able to differentiate the remaining 129 ST-109 isolates. Within-ST diversity was minor; all 129 genomes differed by fewer than 476 total orthologous SNPs (Fig. 4). Isolates did not appear to cluster by geographical location or date of isolate collection.
Clustering was observed in isolates originating from the Wildlife Park. Nine animal and environmental isolates (MSHR2047, MSHR7626, MSHR8753, MSHR8756, MSHR9261, MSHR9265, MSHR9369, MSHR9374, MSHR9403) grouped on one branch of the phylogenetic tree, differing by fewer than 13 total orthologous SNPs (Fig. 4c). Moreover, we observed no SNP differences between the two clinical crocodile isolates (MSHR9261 and MSHR9265) and the three ST-109 environmental samples (MSHR9369, MSHR9374, MSHR9403) collected as part of the case cluster investigation (Fig. 4c). These SNP results suggest that melioidosis infection in both crocodiles was most likely the result of one of these point sources.
Discussion
Source attribution and population dynamics can now be examined on an increasingly small and well-defined scale thanks to high-resolution sequencing platforms [20, 45]. We investigated the aetiology of a ST-109 melioidosis case cluster in two juvenile crocodiles by comparing WGS data from 140 Northern Territory, Australia, ST-109 isolates. All genomes isolated from the wildlife park clustered together on one branch and no orthologous SNPs were detected amongst the three environmental and two clinical crocodile ST-109 isolates collected as part of the investigation. While phylogenetic reconstruction of 140 ST-109 genomes showed limited intra-ST diversity overall, almost no other ST-109 genome clusters were identified. This further supports that the environmental and clinical isolates were epidemiologically related to one another.
The pathology exhibited by both crocodiles is consistent with death due to severe, acute septicaemia with haematogenous spread to numerous filtering organs. Both deceased crocodiles had unabsorbed yolk sacs. This may have been normal for the first crocodile, given that it was less than 1 week of age, but the yolk sac should have largely been resorbed in the second crocodile by 3 weeks of age. Retention of the yolk sac can transpire when the yolk becomes infected prior to hatching, or if the open umbilicus becomes infected immediately after hatching [46]. Since B. pseudomallei was isolated from the yolk in both crocodiles, a nidus of infection in the yolk sac and subsequent systemic spread is a possible pathogenesis. Alternatively, the bacterium may have been harboured within the intestinal tract, eventually spreading systemically from there. The Wildlife Park reportedly kept the newborn crocodiles inside the incubator for several days before transferring them to the holding ponds, suggesting the infections were acquired from within the incubator rather than the holding-tank water. It is thought that once in the holding ponds, the infected crocodiles most likely shed bacteria into the water, as B. pseudomallei has been detected in faecal samples from other species including wallabies and chickens [16]. Since both water samples were collected within a month of the crocodile deaths, this could explain why we observed no SNP diversity between these isolates and those from the incubator. It is conceivable that water vapour generated by the incubator’s internal humidifying system came in contact with the B. pseudomallei -containing biofilm on the back wall and that B. pseudomallei -containing aerosols within the incubator subsequently made contact with the eggs or yolk sac prior to or immediately after hatching [30]. Nevertheless, the single air sample taken from inside the incubator tested negative for the bacterium by both routine culture and direct molecular PCR detection, which has been demonstrated to be the most sensitive technique for the detection of B. pseudomallei in the environment [47, 48]. Therefore, the exact mechanism of transmission remains speculative.
Previous work on the phylogenetic relatedness of ST-109 isolates identified a considerable amount of intra-ST diversity (~9,500 SNPs) across this ST when compared to other common Northern Territory STs [26]. Results from this study, however, revealed significant levels of recombination in a limited number of isolates accounting for nearly all observed intra-ST-109 variation. The B. pseudomallei genome is highly plastic, with frequent horizontal gene transfer events and a recombination-to-mutation rate more than double that of Streptococcus pneumoniae [20]. Substantial genetic variation that confounds the phylogenetic evolution of B. pseudomallei is, thus, common [26, 49]. In addition to the Wildlife Park cluster, we observed a cluster of four environmental and clinical ST-109 outlying genomes from Adelaide River (90 km south of Darwin). While we identified 2,760 SNPs separating these four isolates and the reference ST-109 genome, they varied from one another by fewer than 8 SNPs. This suggests a large-scale recombination event took place at some point in the evolutionary history of ST-109, followed by subsequent dispersal in the environment in Adelaide River. This recombinant Adelaide River ST-109 was first identified in an isolate from a patient with melioidosis in 1998, but when the recombinant ST-109 became established there and the mechanisms behind its dispersal remain undetermined.
Although ST-109 is commonly isolated from patients and the environment in the urban Darwin area [23], our results also demonstrate medium-range dispersal of ST-109. While approximately 500 MLST strain types of B. pseudomallei have now been identified in the Northern Territory (https://pubmlst.org/bpseudomallei/), the majority of isolates comprising each ST are found over a maximum linear distance of less than 45 km [23]. Though prior investigations into ST-109 in the Top End indicated a greater than normal distribution diameter for this ST [23], the results from this study suggest even further dispersal of ST-109, such that isolates have been retrieved from both Darwin and Adelaide River. B. pseudomallei is characteristically spatially clustered in the environment and the distribution of MLST STs remains surprisingly restricted despite animal migration, severe weather events and other anthropogenic influences [25]. Still, instances of long-range ST dispersal have been reported for the bacterium [26, 50, 51]. Most notably, an Asian strain of B. pseudomallei, ST-562, was recently identified in Darwin, Australia [26]. The strain has become well established in the region and is now one of the more frequently isolated STs in urban Darwin patients [26]. The mechanism and timing of the proposed introduction of ST-562 into Darwin remains unknown, but potential long-range spread by severe weather events was postulated to have occurred when a strain previously implicated in an outbreak in West Kimberley, Western Australia, between 1997 and 1999 was detected approximately 500 km inland after a severe weather event in 2005 [51]. In the Northern Territory, distant dispersal of B. pseudomallei ST-149 was recently implicated in linking isolates from a remote island off the northern coast and an inland isolate from Katherine, Northern Territory (460 linear km) [50]. In this instance, WGS was utilized to rule out MLST homoplasy. The exact mechanisms driving the dispersion of ST-109 in the Top End environment will require continued investigation.
Results from this study additionally suggest a MLST homoplasy event may have taken place in a clinical isolate from rural Darwin. Phylogenomic analysis identified>24, 000 non-recombinogenic SNPs in this isolate, a genetic separation typically seen in strains belonging to different STs [50]. B. pseudomallei isolates commonly share the same ST despite being considerably different on the whole-genome level, a result of the bacterium’s exceedingly high rate of recombination [50]. Though MLST has been able to resolve distinct populations on both the intra and intercontinental level [25, 45, 52], shared STs due to homoplasy are an inevitable certainty in highly recombinogenic pathogens like B. pseudomallei . This can significantly confound inferences about infection aetiology and transmission. Other instances of ST homoplasy have been reported in isolates originating from Cambodia and Australia [18], and most recently in two separate long-range intracontinental cases from Australia [50]. For all three of these suspected cases of ST homoplasy, WGS was necessary to determine the genuine relatedness of isolates. Similarly, our study supports the use of higher-resolution methods like WGS to investigate instances of homoplasy and recombination events in isolates with matching STs.
While cases of melioidosis in animals are frequently described throughout the Top End [12], there is only a single report from native saltwater crocodiles, which was a limb wound infection occurring over 30 years ago [53]. In the current investigation, we combined epidemiological findings with high-resolution comparative genomics to determine the source of the case cluster in juvenile crocodiles. Collectively, these data enabled us to identify the probable source of infection as the hatchling incubator. Environmental remediation at the Wildlife Park consisted of thorough bleach-based disinfection of the crocodile incubator and hatchling ponds. No additional melioidosis cases have since been reported in the park crocodiles, although since it is not routine procedure to submit deceased newly born hatchling crocodiles for pathology, it is possible that subsequent cases have gone undiagnosed. Furthermore, in addition to the limited amount of SNP diversity observed across the isolates, we also identified a smaller number of accessory genes in our pan-genome analysis than that previously described [22, 44] and all ST-109 strains contained nearly identical virulence marker profiles. This collectively suggests that despite being the Top End’s most prevalent and widely dispersed genotype, variation in ST-109 is generally low. Exceptions, like those identified in the cluster of isolates from Adelaide River, were due to extensive rates of recombination. Ongoing work examining the phylogenetic relationships between other prevalent ST types in the Top End will ultimately improve our knowledge of genotype diversification and patterns of dispersal in the environment for this highly pathogenic bacterium. This has global biosecurity relevance given recent prediction mapping of melioidosis that suggests substantial numbers of undetected cases and deaths are occurring in many countries [4], and genotyping studies suggesting dispersal from Asia to Madagascar [52] and from West Africa to the Americas [22].
Data bibliography
1. Rachlin A et al. NCBI BioProject PRJNA510860 (2018).
2. Johnson SL et al. NCBI BioProject PRJNA230723 (2015).
3. Price EP et al. NCBI BioProject PRJNA300580 (2015).
Supplementary Data
Funding information
This study was supported by grants from the Australian National Health and Medical Research Council: grant numbers 1046812, 1098337 and 1131932 (The HOT NORTH initiative) and A. R. is supported by a Charles Darwin University International PhD scholarship.
Acknowledgements
We thank staff at Crocodylus Park, Northern Territory, Australia, and Berrimah Veterinary Laboratory, Australia, for their support in undertaking this investigation. We also thank Grahame J. W. Webb and S. Charlie Manolis for their expert knowledge of crocodile ecology and giving us access to sample the wildlife park. Additionally, we thank Glenda Harrington and Barbara Machunter for laboratory support, Erin Price and Derek Sarovich for genomics expertise, and the microbiology staff at the Royal Darwin Hospital, Australia, for their expertise in identifying B. pseudomallei and providing clinical isolates for this study.
Author contributions
The study was conceptualized by B. J. C., M. M. and A. R. Funding was acquired by B. J. C. Data curation and investigation were undertaken by A. R., M. M., V. R., C. S., S. B. and K. D. Formal analysis was done by A. R., M. KI., M. Ka. and J. R. W. The original draft was written by A. R., and B. J. C., M. M., M. Ka., and C. S. revised and edited the manuscript.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
Ethical approval for patient data was acquired through the Human Research Ethics Committee of the Northern Territory Department of Health and Families, approval number HREC 02/38, and Menzies School of Health Research. All patient information was made anonymous prior to analysis. Animal ethics approval was not required for veterinary diagnostic investigation of the deceased crocodiles, since they were kept under normal farm conditions and died naturally (they were not experimental subjects). However, approval for work with animal samples was obtained by Menzies School of Health Research and Berrimah Veterinary Laboratory before commencement of the study. Bacterial culture work was carried out at Berrimah Veterinary Laboratory. Berrimah Veterinary Laboratory supplied Menzies School of Health Research with bacterial B. pseudomallei isolates.
Footnotes
Abbreviations: MLST, multilocus sequence typing; MP, maximum parsimony; ST, sequence type; WGS, whole-genome sequencing.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Two supplementary figures and two supplementary tables are available with the online version of this article.
References
- 1.Wiersinga WJ, Currie BJ, Peacock SJ. Melioidosis. N Engl J Med Overseas Ed. 2012;367:1035–1044. doi: 10.1056/NEJMra1204699. [DOI] [PubMed] [Google Scholar]
- 2.Cheng AC, Currie BJ. Melioidosis: epidemiology, pathophysiology, and management. Clin Microbiol Rev. 2005;18:383–416. doi: 10.1128/CMR.18.2.383-416.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Currie BJ, Ward L, Cheng AC. The epidemiology and clinical spectrum of melioidosis: 540 cases from the 20 year Darwin prospective study. PLoS Negl Trop Dis. 2010;4:e900. doi: 10.1371/journal.pntd.0000900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Limmathurotsakul D, Golding N, Dance DAB, Messina JP, Pigott DM, et al. Predicted global distribution of Burkholderia pseudomallei and burden of melioidosis. Nat Microbiol. 2016;1:15008. doi: 10.1038/nmicrobiol.2015.8. [DOI] [PubMed] [Google Scholar]
- 5.Stewart JD, Smith S, Binotto E, McBride WJ, Currie BJ, et al. The epidemiology and clinical features of melioidosis in far North Queensland: implications for patient management. PLoS Negl Trop Dis. 2017;11:e0005411. doi: 10.1371/journal.pntd.0005411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sprague LD, Neubauer H. Melioidosis in animals: a review on epizootiology, diagnosis and clinical presentation. J Vet Med B Infect Dis Vet Public Health. 2004;51:305–320. doi: 10.1111/j.1439-0450.2004.00797.x. [DOI] [PubMed] [Google Scholar]
- 7.Dance DA, King C, Aucken H, Knott CD, West PG, et al. An outbreak of melioidosis in imported primates in Britain. Vet Rec. 1992;130:525–529. doi: 10.1136/vr.130.24.525. [DOI] [PubMed] [Google Scholar]
- 8.Limmathurotsakul D, Thammasart S, Warrasuth N, Thapanagulsak P, Jatapai A, et al. Melioidosis in animals, Thailand, 2006-2010. Emerg Infect Dis. 2012;18:325–327. doi: 10.3201/eid1802.111347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thomas AD, Spinks GA, D'Arcy TL, Norton JH, Trueman KF. Evaluation of four serological tests for the diagnosis of caprine melioidosis. Aust Vet J. 1988;65:261–264. doi: 10.1111/j.1751-0813.1988.tb16138.x. [DOI] [PubMed] [Google Scholar]
- 10.Cottew GS. Melioidosis in sheep in queens land; a description of the causal organism. Aust J Exp Biol Med Sci. 1950;28:677–683. [PubMed] [Google Scholar]
- 11.Forbes-Faulkner JC, Townsend WL, Thomas AD. Pseudomonas pseudomallei infection in camels. Aust Vet J. 1992;69:148. doi: 10.1111/j.1751-0813.1992.tb07492.x. [DOI] [PubMed] [Google Scholar]
- 12.Choy JL, Mayo M, Janmaat A, Currie BJ. Animal melioidosis in Australia. Acta Trop. 2000;74:153–158. doi: 10.1016/S0001-706X(99)00065-0. [DOI] [PubMed] [Google Scholar]
- 13.Sim S, Ong C, Gan Y, Wang D, Koh V, et al. Melioidosis in Singapore: clinical, veterinary, and environmental perspectives. Trop Med Infect Dis. 2018;3:31. doi: 10.3390/tropicalmed3010031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ketterer PJ, Webster WR, Shield J, Arthur RJ, Blackall PJ, et al. Melioidosis in intensive piggeries in South eastern Queensland. Aust Vet J. 1986;63:146–149. doi: 10.1111/j.1751-0813.1986.tb02953.x. [DOI] [PubMed] [Google Scholar]
- 15.Dodin A, Galimand M. [Origin, course and recession of an infectious disease, melioidosis, in temperate countries] Arch Inst Pasteur Tunis. 1986;63:69–73 (in French). [PubMed] [Google Scholar]
- 16.Höger ACR, Mayo M, Price EP, Theobald V, Harrington G, et al. The melioidosis agent Burkholderia pseudomallei and related opportunistic pathogens detected in faecal matter of wildlife and livestock in northern Australia. Epidemiol Infect. 2016;144:1924–1932. doi: 10.1017/S0950268816000285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hampton V, Kaestli M, Mayo M, Choy JL, Harrington G, et al. Melioidosis in birds and Burkholderia pseudomallei dispersal, Australia. Emerg Infect Dis. 2011;17:1310–1312. doi: 10.3201/eid1707.100707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Smet B, Sarovich DS, Price EP, Mayo M, Theobald V, et al. Whole-genome sequencing confirms that Burkholderia pseudomallei multilocus sequence types common to both Cambodia and Australia are due to homoplasy. J Clin Microbiol. 2015;53:323–326. doi: 10.1128/JCM.02574-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Godoy D, Randle G, Simpson AJ, Aanensen DM, Pitt TL, et al. Multilocus sequence typing and evolutionary relationships among the causative agents of melioidosis and glanders, Burkholderia pseudomallei and Burkholderia mallei . J Clin Microbiol. 2003;41:2068–2079. doi: 10.1128/JCM.41.5.2068-2079.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pearson T, Giffard P, Beckstrom-Sternberg S, Auerbach R, Hornstra H, et al. Phylogeographic reconstruction of a bacterial species with high levels of lateral gene transfer. BMC Biol. 2009;7:78. doi: 10.1186/1741-7007-7-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Currie BJ, Gal D, Mayo M, Ward L, Godoy D, et al. Using BOX-PCR to exclude a clonal outbreak of melioidosis. BMC Infect Dis. 2007;7:68. doi: 10.1186/1471-2334-7-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chewapreecha C, Holden MTG, Vehkala M, Välimäki N, Yang Z, et al. Global and regional dissemination and evolution of Burkholderia pseudomallei . Nat Microbiol. 2017;2:16263. doi: 10.1038/nmicrobiol.2016.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chapple SNJ, Price EP, Mayo M, Currie BJ, Kaestli M, et al. Burkholderia pseudomallei genotype distribution in the Northern Territory, Australia. Am J Trop Med Hyg. 2016;94:68–72. doi: 10.4269/ajtmh.15-0627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng AC, Ward L, Godoy D, Norton R, Mayo M, et al. Genetic diversity of Burkholderia pseudomallei isolates in Australia. J Clin Microbiol. 2008;46:249–254. doi: 10.1128/JCM.01725-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McRobb E, Kaestli M, Price EP, Sarovich DS, Mayo M, et al. Distribution of Burkholderia pseudomallei in northern Australia, a land of diversity. Appl Environ Microbiol. 2014;80:3463–3468. doi: 10.1128/AEM.00128-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Price EP, Sarovich DS, Smith EJ, MacHunter B, Harrington G, et al. Unprecedented melioidosis cases in northern Australia caused by an Asian Burkholderia pseudomallei strain identified by using large-scale comparative genomics. Appl Environ Microbiol. 2016;82:954–963. doi: 10.1128/AEM.03013-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kommanee J, Preecharram S, Daduang S, Temsiripong Y, Dhiravisit A, et al. Antibacterial activity of plasma from crocodile (Crocodylus siamensis) against pathogenic bacteria. Ann Clin Microbiol Antimicrob. 2012;11:22. doi: 10.1186/1476-0711-11-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benedict S, Shilton CM. Providencia rettgeri septicaemia in farmed crocodiles. Microbiol Aust. 2016;37:114–117. doi: 10.1071/MA16039. [DOI] [Google Scholar]
- 29.Mayo M, Kaesti M, Harrington G, Cheng AC, Ward L, et al. Burkholderia pseudomallei in unchlorinated domestic bore water, tropical northern Australia. Emerg Infect Dis. 2011;17:1283–1285. doi: 10.3201/eid1707.100614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Currie BJ, Price EP, Mayo M, Kaestli M, Theobald V, et al. Use of whole-genome sequencing to link Burkholderia pseudomallei from air sampling to mediastinal melioidosis, Australia. Emerg Infect Dis. 2015;21:2052–2054. doi: 10.3201/eid2111.141802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Limmathurotsakul D, Dance DAB, Wuthiekanun V, Kaestli M, Mayo M, et al. Systematic review and consensus guidelines for environmental sampling of Burkholderia pseudomallei . PLoS Negl Trop Dis. 2013;7:e2105. doi: 10.1371/journal.pntd.0002105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Lamballerie X, Zandotti C, Vignoli C, Bollet C, de Micco P. A one-step microbial DNA extraction method using "Chelex 100" suitable for gene amplification. Res Microbiol. 1992;143:785–790. doi: 10.1016/0923-2508(92)90107-Y. [DOI] [PubMed] [Google Scholar]
- 33.Novak RT, Glass MB, Gee JE, Gal D, Mayo MJ, et al. Development and evaluation of a real-time PCR assay targeting the type III secretion system of Burkholderia pseudomallei . J Clin Microbiol. 2006;44:85–90. doi: 10.1128/JCM.44.1.85-90.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson SL, Baker AL, Chain PS, Currie BJ, Daligault HE, et al. Whole-Genome sequences of 80 environmental and clinical isolates of Burkholderia pseudomallei . Genome Announc. 2015;3:e01282-14. doi: 10.1128/genomeA.01282-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jolley KA, Maiden MCJ. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics. 2010;11:595. doi: 10.1186/1471-2105-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sarovich DS, Price EP. SPANDx: a genomics pipeline for comparative analysis of large haploid whole genome re-sequencing datasets. BMC Res Notes. 2014;7:618. doi: 10.1186/1756-0500-7-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Swofford DL. Sunderland, MA: Sinauer Associates; 2001. PAUP*: phylogenetic analysis using parsimony (*and other methods) 4.0.b5. [Google Scholar]
- 38.Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 2015;43:e15. doi: 10.1093/nar/gku1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sarovich DS, Price EP, Webb JR, Ward LM, Voutsinos MY, et al. Variable virulence factors in Burkholderia pseudomallei (melioidosis) associated with human disease. PLoS One. 2014;9:e91682. doi: 10.1371/journal.pone.0091682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tuanyok A, Auerbach RK, Brettin TS, Bruce DC, Munk AC, et al. A horizontal gene transfer event defines two distinct groups within Burkholderia pseudomallei that have dissimilar geographic distributions. J Bacteriol. 2007;189:9044–9049. doi: 10.1128/JB.01264-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Webb JR, Sarovich DS, Price EP, Ward LM, Mayo M, et al. Burkholderia pseudomallei lipopolysaccharide genotype does not correlate with severity or outcome in melioidosis: host risk factors remain the critical determinant. Open Forum Infect Dis. 2019;6:ofz091. doi: 10.1093/ofid/ofz091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spring-Pearson SM, Stone JK, Doyle A, Allender CJ, Okinaka RT, et al. Pangenome analysis of Burkholderia pseudomallei: genome evolution preserves gene order despite high recombination rates. PLoS One. 2015;10:e0140274. doi: 10.1371/journal.pone.0140274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dale J, Price EP, Hornstra H, Busch JD, Mayo M, et al. Epidemiological tracking and population assignment of the non-clonal bacterium, Burkholderia pseudomallei . PLoS Negl Trop Dis. 2011;5:e1381. doi: 10.1371/journal.pntd.0001381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huchzermeyer F. Crocodiles: Biology, Husbandry and Diseases. Wallingford: CABI Publishing; 2003. [Google Scholar]
- 47.Kaestli M, Mayo M, Harrington G, Watt F, Hill J, et al. Sensitive and specific molecular detection of Burkholderia pseudomallei, the causative agent of melioidosis, in the soil of tropical northern Australia. Appl Environ Microbiol. 2007;73:6891–6897. doi: 10.1128/AEM.01038-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Knappik M, Dance DAB, Rattanavong S, Pierret A, Ribolzi O, et al. Evaluation of molecular methods to improve the detection of Burkholderia pseudomallei in soil and water samples from Laos. Appl Environ Microbiol. 2015;81:3722–3727. doi: 10.1128/AEM.04204-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Price EP, Sarovich DS, Viberg L, Mayo M, Kaestli M, et al. Whole-genome sequencing of Burkholderia pseudomallei isolates from an unusual melioidosis case identifies a polyclonal infection with the same multilocus sequence type. J Clin Microbiol. 2015;53:282–286. doi: 10.1128/JCM.02560-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aziz A, Sarovich DS, Harris TM, Kaestli M, McRobb E, et al. Suspected cases of intracontinental Burkholderia pseudomallei sequence type homoplasy resolved using whole-genome sequencing. Microb Genom. 2017;3:e000139. doi: 10.1099/mgen.0.000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Inglis TJJ, Levy A, Merritt AJ, Hodge M, McDonald R, et al. Melioidosis risk in a tropical industrial environment. Am J Trop Med Hyg. 2009;80:78–84. doi: 10.4269/ajtmh.2009.80.78. [DOI] [PubMed] [Google Scholar]
- 52.Sarovich DS, Garin B, De Smet B, Kaestli M, Mayo M, et al. Phylogenomic analysis reveals an Asian origin for African Burkholderia pseudomallei and further supports melioidosis endemicity in Africa. mSphere. 2016;1:e00089-15. doi: 10.1128/mSphere.00089-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Asche V. Melioidosis - a disease for all organs. Today's Life Sci. 1991;3:34–40. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.