Abstract
Bacteriophages are the most prevalent biological entities impacting on the ecosystem and are characterized by their extensive diversity. However, there are two aspects of phages that have remained largely unexplored: genetic flux by recombination between phage populations and characterization of specific phages in terms of the pan-genome. Here, we examined the recombination and pan-genome in Helicobacter pylori prophages at both the genome and gene level. In the genome-level analysis, we applied, for the first time, chromosome painting and fineSTRUCTURE algorithms to a phage species, and showed novel trends in inter-population genetic flux. Notably, hpEastAsia is a phage population that imported a higher proportion of DNA fragments from other phages, whereas the hpSWEurope phages showed weaker signatures of inter-population recombination, suggesting genetic isolation. The gene-level analysis showed that, after parameter tuning of the prokaryote pan-genome analysis program, H. pylori phages have a pan-genome consisting of 75 genes and a soft-core genome of 10 genes, which includes genes involved in the lytic and lysogenic life cycles. Quantitative analysis of recombination events of the soft-core genes showed no substantial variation in the intensity of recombination across the genes, but rather equally frequent recombination among housekeeping genes that were previously reported to be less prone to recombination. The signature of frequent recombination appears to reflect the host–phage evolutionary arms race, either by contributing to escape from bacterial immunity or by protecting the host by producing defective phages.
Keywords: homologous recombination, bacteriophage, pan-genome, core genome, Helicobacter pylori, evolution
Data Summary
The genome accession numbers and metadata, including phage population and information from multilocus sequence typing (MLST), are in Table 1.
The protein ID for the phage pan-genome is in Table S2 (available in the online version of this article).
Table 1.
Phage genomes used in the recombination analysis
|
Genome |
Phage population* |
Bacterial population† |
Genome length (bp) |
Accession no. |
References |
|---|---|---|---|---|---|
|
Hac |
na |
na |
28 420 |
[1] |
|
|
KHP30 |
hpEastAsia |
na |
26 215 |
[2] |
|
|
KHP40 |
hpEastAsia |
na |
26 449 |
[2] |
|
|
1961P |
hpEastAsia |
na |
26 836 |
[3] |
|
|
Fr-B58-M |
hpEastAsia |
hpEastAsia |
22 559 |
SRP071277 |
[4] |
|
Pt-B92-G |
hpAfrica1 |
hpEurope |
30 548 |
SRP071282 |
[4] |
|
Pt-212–99 R-U |
hpAfrica1 |
hpEurope |
23 008 |
SRP071292 |
[4] |
|
Pt-1293-U |
hpAfrica1 |
hpEurope |
30 071 |
SRP071280 |
[4] |
|
Pt-5771-G |
hpAfrica1 |
hpEurope |
29 801 |
SRP064707 |
[4] |
|
Pt-B89-G |
hpAfrica1 |
hpEurope |
27 363 |
SRP071278 |
[4] |
|
Pt-5322-G |
hpAfrica1 |
hpEurope |
28 341 |
SRP071284 |
[4] |
|
Fr-ANT170-U |
hpAfrica1 |
hpEurope |
31 200 |
SRP072438 |
[4] |
|
Fr-MEG235-U |
hpAfrica1 |
hpEurope |
31 236 |
SRP072439 |
[4] |
|
Pt-1846-U |
hpAfrica1 |
hpEurope |
27 960 |
SRP071062 |
[4] |
|
Pt-228_99 G |
hpAfrica1 |
hpEurope |
30 078 |
SRP071067 |
[4] |
|
phiHP33 |
hpAfrica1 |
hpEurope |
24 645 |
[5] |
|
|
Pt-4481-G |
hpAfrica1 |
hpEurope |
25 388 |
SRP071279 |
[4] |
|
UK-EN31-U |
hpNEurope |
hspEuropeN† |
30 456 |
SRP071274 |
[4] |
|
UK-EN32-U |
hpNEurope |
hspEuropeN† |
29 882 |
SRP071276 |
[4] |
|
De-M53-M |
hpNEurope |
hspEuropeN† |
28 068 |
SRP064710 |
[4] |
|
India7 |
hpNEurope |
hpAsia2† |
28 310 |
||
|
Cuz20 |
hpNEurope |
hspAmerind† |
28 587 |
||
|
Sw-A626-G |
hpEuropeN |
hspEuropeN† |
30 977 |
SRP071294 |
[4] |
|
Sw-577-G |
hpNEurope |
hspEuropeN† |
26 906 |
SRP071293 |
[4] |
|
Fr-G12-G |
hpSWEurope† |
hpEurope |
28 565 |
SRP064708 |
[4] |
|
Pt-4472-G |
hpSWEurope |
hspEuropeS† |
27 572 |
SRP071271 |
[4] |
|
Fr-GC43-A |
hpSWEurope† |
hpEurope |
32 975 |
SRP072440 |
[4] |
|
Pt-1918-U |
hpSWEurope |
hspEuropeS† |
28 670 |
SRP064706 |
[4] |
|
Pt-4497-U |
hpSWEurope |
hspEuropeS† |
29 393 |
SRP064709 |
[4] |
|
Fr-B41-M |
hpSWEurope |
hspEuropeS* |
29 388 |
SRP072441 |
[4] |
*The phage population was determined by phage sequence typing (PST) using two phage genes [6].
†The bacterial population was determined using seven MLST genes [6]. The asterisks indicate subpopulations determined by fineSTRUCTURE (Fig. S4) [7]. na, not applicable, phage genome only or non-pylori Helicobacter phage.
Impact Statement.
Despite the large number of bacteriophages (phages) present in the biosphere, little is known about genetic flux by recombination between populations of phages, and the variation in intensity of recombination across phage genes in general. We studied these aspects using genome sequences of phages infecting a highly recombinogenic bacterial species, Helicobacter pylori . Our genome-level analysis showed for the first time the extent and direction of the genetic flux among phage populations, including those that show significantly stronger or weaker signatures of genetic flux. In the subsequent gene-level analysis, we found that parameter tuning of the standard software was necessary to identify well-conserved genes, and it showed equally high recombination among them, including genes that were reported to be less prone to recombination. Our findings suggest that H. pylori phages are among the most recombinogenic phages, reflecting coevolutionary relationships with their hosts.
Introduction
Viruses are the most prevalent biological entities on Earth; they play important roles in ecological balance and are characterized by their extensive diversity [8]. Interactions between bacteria and bacterial viruses, also known as bacteriophages or phages, help to shape the population structure of microbial communities, as well as their ecology and evolution [9]. Phages acquire and donate genes from other phage or bacterial genomes, and thus play an important role in the evolution, physiology and pathogenicity of their bacterial hosts [9]. The high genetic diversity in phages is likely due to their ancient origin (they originated about 3 billion years ago), the huge number of phages present in the biosphere and their frequent infection of permissive hosts. These lead phages to encounter DNA derived from bacteria or prophages (phages integrated into bacterial chromosomes) with which they can recombine [10]. Such recombination is a driving force of evolution, which can occur either through homologous recombination or non-homologous site-specific recombination, generating a mixture of recombinant types. However, as a consequence of such recombination, most recombinants are defective for growth, and natural selection eliminates all but a very small minority in which biological functions are intact, giving rise to a population with non-random recombination sites [10].
Helicobacter pylori is notable among the bacterial host species of phages because it is a long-term colonizer of the human stomach, it has been co-evolving with its human host for more than 100 000 years [11] and it is highly recombinogenic [12, 13]. About 20 % of H. pylori strains contain prophage genes [4, 6, 14, 15], which are present in a phylogeographical pattern that corresponds to that of the host bacterium [6, 14] and provide additional insights into the population structure. For example, the hpEurope population has two distinct subgroups, northern and southwestern, that are segregated by only two prophage genes [6]. Likewise, a comparison of H. pylori whole genomes also pointed to the existence of two European subpopulations [7]. H. pylori phages are highly diverse. They have conserved synteny marked by punctuated gene loss, the introduction of insertion sequences that do not perturb gene synteny and diverse insertion sites with some common traits among H. pylori populations [4]. Mobile elements such as phages may contribute to H. pylori pathogenicity [16–18], which has been suggested by the correlation of prophage-related sequences with the presence of the virulence genes cagA and vacA [19].
Previous studies of recombination events in H. pylori led to a better understanding of genetic flux and showed that subgroups of the Amerind and East Asian origin populations import a significantly smaller number of fragments than they export to other populations [20], and that H. pylori strains from the American continent originated from an admixture between H. pylori strains of European and African origin, without extensive input from pre-Columbian (hspAmerind) strains [7]. In contrast, little is known about the genetic flux between populations of phages in general. In addition, although a recent study showed evidence of recombination or genome mosaicism within some H. pylori prophages [4], little is known about the variation in intensity of recombination across phage genes, as was recently shown in various phages through an analysis of the Earth’s virome [21].
Another aspect of phage biology that is in general poorly understood is the question of how specific phages can be characterized in terms of their pan-genome. The pan-genome corresponds to the entire set of genes in the genomes within a species, while the core genome is the set of genes that are present in all genomes [22]. Notably, phages do not have such a core genome that is conserved among all phages across their phylogeny [23]. The diversity in phage genomes, notable by the presence of genes in many phages with low sequence similarity [24], makes it difficult to study phage core genomes. To overcome this limitation, we conducted analyses based on both genome alignment at the synteny level and gene-by-gene alignment at a finer level. Regarding the latter, we propose parameter tuning of the standard prokaryote pan-genome analysis program, and using the soft-core genome, which is defined here as the genes present in >90 % of the genomes, to verify sequence similarity. We applied this approach to H. pylori prophages, which have preserved genome synteny and a probable modular constitution [4], as little is known about their pan-genome and their genetic diversity.
To explore these two aspects, we conducted a study of recombination in H. pylori prophages at both the genome and gene (soft core) levels using 29 H . pylori genomes and 1 Helicobacter acinonychis genome carrying prophages >20 kb. The genome-level approach was used to elucidate the genetic flux of imported and exported sequences in distinct populations of prophages and their host bacteria. The soft-core gene-level approach was used to examine whether some genes show signatures of increased recombination based on a determination of the soft-core genome and pan-genome.
Methods
Genome sequences, alignment and prediction of prophage activity
We prepared 1 H. acinonychis and 29 Hh. pylori complete prophage sequences (>20 kb) (Table 1), which (i) are present as a single fragment based on Sanger sequencing closing of the genome [1, 3–5, 25] and (ii) have previously been assigned to a population by prophage sequence typing (PST) [6]. The complete prophage genomes were aligned at the synteny level with MAFFT (version 7) [26]. We excluded identical genomes (i.e. UK-EN32-U, which is identical to UK-EN-31-U, and Fr-MEG235-U, which is identical to Fr-ANT170-U) and a genome with a large inversion (Pt-4481-G), resulting in 27 (=29+1–3) complete prophage genome sequences. We excluded the genome with a large inversion because the inverted region did not align well, and the chromosome painting algorithm does not allow missing data in the alignment. The Prophage Hunter tool [27] was used to predict prophage activity, i.e. whether the prophage could be induced into a lytic cycle.
Analysis of the population structure and genetic flux in H. pylori phage genomes
We inferred the population structure of H. pylori phages and the genetic flux between populations from phage genome haplotype data (i.e. SNPs without missing data and their positions in the genome alignment) using chromosome painting and fineSTRUCTURE algorithms, according to a procedure used in our previous study of H. pylori genomes [7, 20]. The chromosome painting algorithm is a hidden Markov model (HMM) to ‘paint’ a ‘recipient’ haplotype as a series of recombination-derived DNA fragments (‘chunks’) from a panel of ‘donor’ haplotypes from other individuals in the sample based on sequence similarity between donor and recipient. The interpretation of the painting is that the donor at a given region of the genome has the most recent shared common ancestor with the recipient individual amongst all of the possible donors in the panel. Changes in the identity of the donor along the sequence reflect recombination events that led to different genealogical histories for different parts of the genome [28, 29] . Lawson et al. used the algorithm to summarize information for genome-wide SNPs into chunks based on a ‘co-ancestry matrix’ that tabulated the number of chunks from each donor to each recipient individual. The data reduction from a haplotype matrix to a co-ancestry matrix enables model-based clustering using fineSTRUCTURE. The two-step approach was demonstrated to be effective not only in humans, but also in H. pylori . We used ChromoPainter (version 0.04) to run the chromosome painting algorithm separately for different recipient individuals. Then, fineSTRUCTURE (version 0.02) was used to run 100 000 iterations of both the burn-in and Markov chain Monte Carlo (MCMC) chain to cluster individuals based on the co-ancestry matrix.
Prophage core- and pan-genomes
The core- and pan-genomes of H. pylori prophages were determined using Roary [30]. The H. acinonychis prophage was not included in the pan-genome analysis because H. acinonychis is a non-pylori Helicobacter . Because of the extreme diversity typically found in phages, −i (the minimum percentage identity of the shorter length for blastp) was set at 90, 80, 70, 60, 50 or 40 with or without the −s option, for splitting or not splitting paralogues, respectively. This approach was used to find the most suitable parameters for determining the phage pan-genome by overcoming the limitations of high diversity; the existence of stop codons followed by a second start codon, leading to the annotation of two genes or a smaller gene, which hinders determination of the percentage identity and leads to the misidentification of similar genes; or other annotation limitations, such as the non-identification of overlapping genes, the misidentification of unique genes in a particular genome, or genes that are highly divergent from known homologues [31].
Analysis of recombination at the gene level
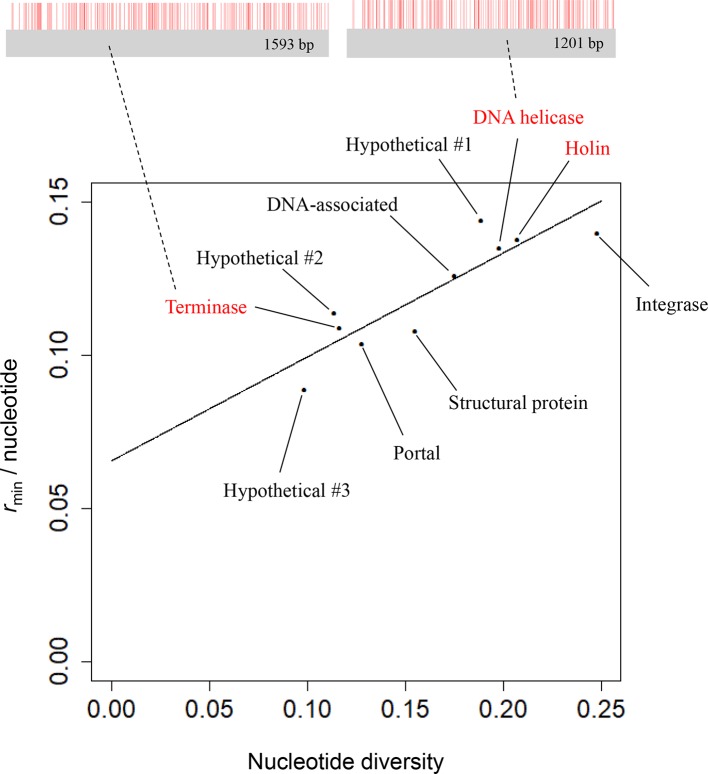
Recombination analysis was carried out at the gene level to determine the genes in the soft-core genome (genes present in at least 90 % of the genomes), which were identified using the Roary pipeline [30] with the −i 70 and −s options. The genes belonging to the soft-core genome that were analysed are integrase, portal protein, structural protein, DNA helicase, terminase, holin, hypothetical protein/DNA-associated and three other hypothetical proteins. For each gene, the minimum number of recombination events (r min) was calculated using the four-gamete test [32], which locates pairs of the closest segregating sites within four haplotypes that are likely to be generated by recombination. We used the method implemented in the PGEToolbox [33], which filters gaps in advance. Basic population genetic statistics (e.g. nucleotide diversity) were also calculated for each gene using DnaSP v5 [34]. Genes with stop codons in the middle of the sequence were excluded from this analysis. We then conducted multiple linear regressions to capture the overall relationship between the r min per nucleotide and nucleotide diversity after controlling for differences in the number of individuals: y i = β 0 + β 1 x 1 , i + β 2 x 2 , i + ε i, where for gene i, yi is the minimum number of recombination events per nucleotide; x1,i is nucleotide diversity; x2,i is the number of aligned sequences; β 0 is the intercept; β 1 and β 2 are regression coefficients; and ε i is error, which is normally distributed. We plotted the regression line in Fig. 1 given the parameter estimates, holding constant x2 as the average number of aligned sequences in a gene.
Fig. 1.
Relationship between the minimum number of recombination events (r min) per gene length and nucleotide diversity among the H. pylori phage soft-core genes. The x and y axes are nucleotide diversity and r min per nucleotide, respectively. The solid line indicates the linear regression between nucleotide diversity and the minimum number of recombination events. Recombination breakpoints in the genes encoding DNA helicase and terminase are shown as red vertical bars at the top.
Results
Recombination and population structure of phage genomes
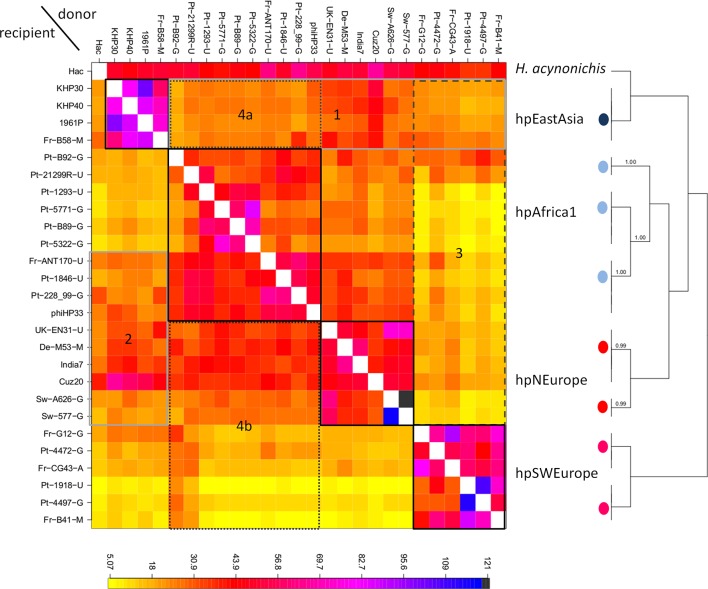
For 27 H . pylori prophages, 19 of which were predicted to be active using Phage Hunter [27] (Table S1), each complete genome was reconstructed using fragments (‘chunks’) of DNA donated by other individual genomes using the chromosome painting algorithm, as summarized and visualized in the co-ancestry matrix (Fig. 2). The algorithm was applied to 7618 core SNPs and their positions in the alignment (53266 bp) of the prophage genome sequences. Based on the matrix, individual phages were assigned to subgroups by the fineSTRUCTURE clustering algorithm. The two-step approach is applicable even when the sample size is small, as shown previously in H. pylori , which is in contrast to STRUCTURE, which is based on allele frequency and thus requires at least 15–20 individuals per hypothesized population [20]. This approach revealed a finer population structure for the genomes classified as hpAfrica1, presenting three subgroups (light blue circles in Fig. 2), and for the genomes in the hpNEurope and hpSWEurope populations (designated in our previous study [6]), each presenting two subgroups (red and pink circles in Fig. 2, respectively). It also revealed notable signatures of inter-population recombination or gene flow, namely, that the hpEastAsia genomes are recipients of DNA chunks from hpAfrica1, hpNEurope and hpSWEurope (box 1 in Fig. 2), while two of the hpAfrica1 subgroups (top and middle light blue circles in Fig. 2) are recipients of DNA chunks from genomes of hpNEurope origin, and the other subgroup received DNA fragments from both hpNEurope and hpEastAsia genomes. In addition, this method revealed that hpNEurope genomes are recipients of DNA from genomes of African and East Asian origin, which is clearer in the genomes of phages from northern continental Europe (top red circle in Fig. 2) than in the genomes of Scandinavian origin (bottom red circle in Fig. 2) (see the Discussion for more quantitative examination and interpretation). Such signatures of inter-population recombination or gene flow were weaker in hpSWEurope genomes, which might be regarded as an outgroup in the H. pylori phages. The H. acinonychis prophage genome, Hac, also seems to show such signatures; however, this is an artifact seen in a sequence that is much more divergent than the others.
Fig. 2.
Co-ancestry matrix of H. pylori phage genomes. Each lane represents the genome of a phage shown on the left. The colour of each square of the matrix represents the expected number of fragments exported from a donor genome (column) to a recipient genome (row). These genomes belong to the phage populations shown on the right. The tree obtained from the fineSTRUCTURE analysis shows the subgroups of each population. The subgroups of each phage population are indicated by coloured circles as follows: hpEastAsia, dark blue; hpAfrica1, light blue; hpNEurope, red ; and hpSWEurope, pink (recipient prefixes: 'De', German; 'Fr', France; 'Pt', Portugal; 'Sw', Sweden; 'UK', United Kingdom).
Classifying the H. pylori prophage soft core- and pan-genome genes
For the pan-genome analysis of the prophage genomes, given the high diversity, we tested several combinations of parameters related to the percentage sequence identity (sequence identity from 40 to 90 %), with and without paralogue disabling (Fig. S1). The default parameters for the prokaryote pan-genome analysis program, i.e. 90 % percentage identity and paralogue enabled, yielded no core genes, which contradicted previous results, since it was previously known that these genomes shared genes (see below). Indeed, at sequence percentage identity values greater than 70, the size of the pan-genome increased exponentially. While no genes were classified as core or soft core, genes such as integrase were present in every single prophage genome (see Fig. S2, which illustrates the genome alignment, location and alignment of the integrase gene). A closer inspection of the results with or without paralogue split, revealed that genes considered to be paralogues were in fact genes with a stop codon and an initiation codon in the middle of the gene, and they did not correspond to true gene duplications. Using a minimum sequence identity of 70 % with paralogue split disabled, the pan-genome has 75 genes; 13 % (10/75) of these genes belong to the soft-core genome, and 21 % (16/75) of the genes are singletons present in a single genome. A DNA primase gene was present in all phage genomes, but in two different clusters of orthologous genes, and thus was not considered to be a soft-core gene. Most (75 %, 12/16) singletons had unassigned functions, while 30 % of the soft-core genes had no functional assignment (Table S2).
Recombination among phage soft-core genes
For each soft-core gene, we calculated the minimum number of recombination events (r min) using the four-gamete test, a conservative method for locating pairs of the closest segregating sites within four haplotypes that were likely to have been generated by recombination. The r min per nucleotide was at least 0.089, which was higher than the highest value (0.053) among the 211 viral (phage species) groups using the Earth’s virome data [21]. Although the value itself depends on nucleotide diversity, it suggests frequent recombination among the H. pylori phages, which is similar to the host bacterium. To account for the dependence of the r min per nucleotide on nucleotide diversity, we took an approach similar to that used in previous studies [21, 35]. We plotted the r min per nucleotide versus the nucleotide diversity of a gene and calculated the linear regression that captured the overall relationship between nucleotide diversity and the minimum number of recombination events per nucleotide (Fig. 1). The extent of deviation from the regression line is a measure of the intensity of recombination after accounting for nucleotide diversity. Overall, the soft-core genes did not show a clear deviation from the regression line, suggesting the absence of substantial variation in the intensity of recombination across phage genes. In contrast, housekeeping genes encoding holin, DNA helicase and terminase (red in Fig. 1), which were reported to be less prone to recombination [36, 37], were similarly close to the regression line, suggesting equally frequent recombination among genes.
Discussion
This is the first study to apply chromosome painting and fineSTRUCTURE to prophage genomes. This approach allowed us to elucidate the extent and direction of the genetic flux among distinct phage populations. The phage populations we identified were consistent with those determined by phage typing using integrase and holin genes [6]. However, we uniquely identified subgroups within these populations and genetic flux among these subgroups (Figs 1 and S3).
Notably, the hpEastAsia phage population imported nearly the same percentages of DNA fragments from all other populations (median: 24.4 % per individual recipient), which differed from the percentages of imports that other populations received from different populations (median: 13.2 %, P value=0.017, Wilcoxon’s rank sum test; box 1 in Fig. 2). The hpEastAsia phage population exported more DNA fragments to the hpNEurope population and one of the hpAfrica1 subgroups (median exports of hpEastAsia phages to hpNEurope and subgroup of hpAfrica1 : 12.8 %; box 2 in Fig. 2) than to hpSWEurope and the other subgroups of hpAfrica1 (median: 10.0 % per donor population; P value=1.3e-11, Wilcoxon’s rank sum test). This pattern was observed among phages in the hpEastAsia population isolated in both East Asian countries (KHP30, KHP40 and P1961) and Europe (Fr-B58-M). This result seems to contradict the findings of a previous report showing that hpEastAsia H. pylori genomes imported a smaller number of fragments from other populations than they exported [20]. One hypothesis is that hpEastAsia phages have been exposed to more phages from other populations than the phages in other populations have been exposed to hpEastAsia phages.
The hpSWEurope phage population exported fewer DNA fragments to other phage populations (median exports of hpSWEurope to other populations: 11.9 %; box 3 in Fig. 2) than other populations did (median: 21.3 % per donor population; P value<2.2e-16, Wilcoxon’s rank sum test). The two observable groups of hpSWEurope phages show signs of genetic isolation, which was previously suggested by the long branches in the phylogenetic trees [4], especially for the subgroup including phages Fr-G12-G, Pt-4472-G, Fr-GC43-A, Pt-1918-U, Pt-4497-G and Fr-B41-M, in which the host belongs to the hpEurope population.
There is a bidirectional genetic flux between the hpNEurope and hpAfrica1 phage populations; however, it should be noted that the hpAfrica1 strains were isolated in Europe, and thus may be more exposed to other European phages, which could influence the results. Although isolated in Europe, these strains were originally defined as hpAfrica1 based on the typing of the integrase and holin genes, and also because they were previously shown to cluster with other H. pylori hpAfrica1 prophages isolated from African countries [6, 14]. It has been suggested that directional gene flow may indicate that a group of genomes act as donors during recombination events, representing a reservoir for diversity [38], a role that appears to be played by hpAfrica1 phages, acting as substantial donors to other populations. Indeed, hpAfrica1 phages exported significantly more DNA fragments (median exports of hpAfrica1 to other populations: 30.2 %; boxes 4a and 4b in Fig. 2) than other populations did (median: 12.6 % per donor population; P value=0.0001, Wilcoxon’s rank sum test).
Chromosome painting and fineSTRUCTURE algorithms were applied to the 7618 core SNPs and their positions in the alignment of the prophage genome sequences. If the number of SNPs were too small, the co-ancestry matrix (heatmap) would become uniform because each recipient genome would have similar, and small, numbers of fragments (chunks) of DNA inferred to be donated by other donor individuals. Clearly, that is not the case, as observed in the co-ancestry matrix in Fig. 2, which shows variation in the number of fragments (chunks) of DNA for each recipient genome and population structure. Therefore, even though the prophage genomes are much smaller than those of the host, the current dataset has sufficient information for the algorithms to work.
Although no genomic study has yet quantified recombination, phages have generally been considered as actively engaged with horizontal genetic exchange (recombination between different individuals), and their genomes are pervasively mosaic in their architectures [39]. In this context, our study contributes in general to phage biology by (1) showing the extent and direction of the genetic flux among phage populations based on the first application of chromosome painting and fineSTRUCTURE algorithms and (2) presenting a quantitative analysis of recombination events of soft-core genes that revealed frequent recombination among housekeeping genes previously reported to be less prone to recombination.
It is important to keep in mind that sampling bias and sample size may limit our ability to interpret results related to population structure and recombination [20]. If this is the case for hpSWEurope phages, further studies are needed with increased sample size and more geographically diverse isolates.
Within a bacterial genome, prophages are considered to be part of the accessory genome and are not considered to be a part of the bacterial species’ core genome [40]. However, the phages themselves have their own pan-genome, although an accurate determination is challenging due to their high diversity. Our sample of H. pylori phages had a pan-genome composed of 75 genes, and a small soft-core genome comprising 13 % of the pan-genome genes, which is in agreement with the small-core genomes of mycobacteriophages [10], marine roseophages [41] and the phages that infect Bacillus cereus [42], Escherichia coli [43] and Clostridium difficile [44]. The percentage identity used for pan-genome determination is of paramount importance, and for H. pylori phages, we realized that it should not be higher than 70 % to avoid missing similar genes due to diversity. At one end of the pan-genome spectrum, there are 16 singleton genes in 11 genomes, which may have no selective benefit to the phage that carries them, or they could serve as a ‘gene nursery’, where novel genetic functions could develop [39], while at the other end, there are 10 soft-core genes, which should have essential roles in the phage life cycle. Indeed, these genes function in lysogeny (integrase); cell lysis (holin); DNA packaging (terminase, portal protein) and ejection (terminase); DNA replication, recombination and nucleotide excision repair; RNA transcription and splicing (helicase); and as structural components (structural protein). Furthermore, soft-core genome sequences are useful for clarifying the evolutionary relationships among prophages [45].
Analysis of recombination at the gene level revealed that the H. pylori phage soft-core genes were equally highly prone to recombination, indicating a continuous shuffle of sequences shaping phage genomes. Thus, the previous proposal that genes evolve at different speeds, with integrase evolving very quickly and holin nearly standing still [37], does not appear to be true for H. pylori phages [21]. A comparison to the analysis of the Earth’s virome suggested that H. pylori phages are among the most recombinogenic phages on Earth. The high recombination rate in H. pylori phages suggests the existence of frequent interactions between episomal phages and prophages, or the existence of recombination between multiple infected strains carrying prophages. Such frequent recombination may aid in the escape from bacterial immunity, especially given that restriction and modification systems are extremely abundant in H. pylori [46, 47]. Although 70 % of the H. pylori prophages were predicted to be active, it might also indicate that some of them are in decay due to frequent recombination, making the difference in the recombination intensity of genes undetectable. In other words, recombination may also protect the bacterial host by inactivating prophages, suggesting a new aspect of recombination that should be further examined in subsequent studies.
Data bibliography
1. Vale FF. Sequence Read Archive (SRA), SRP064706 to SRP064710, SRP071062,SRP071067, SRP071271, SRP071274, SRP071276 to SRP071280, SRP071282, SRP071284, SRP071289 to SRP071296 and SRP072438 to SRP072441 (2016).
2. Vale FF. Genbank, KX119174 to KX119206 (2016).
Supplementary Data
Funding information
F.F.V. is the recipient of a postdoctoral fellowship (SFRH/BPD/95125/2013) and a project grant (PTDC/BTM-SAL/28978/2017) from the Fundação para a Ciência e a Tecnologia (FCT), which supported this work. This research was supported by a Grant-in-Aid for Scientific Research on Innovative Areas from the Ministry of Education, Culture, Science, Sports, and Technology (MEXT) of Japan (nos 16H06429, 16K21723 and 19H04846).
Acknowledgements
The authors thank Andrew Page for discussion and instruction. Computational calculations were performed at the Human Genome Center at the Institute of Medical Science (the University of Tokyo) and at the National Institute of Genetics.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
No human or animal experimentation is reported.
Footnotes
Abbreviations: MLST, multilocus sequence typing; PST, prophage sequence typing.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Four supplementary figures and two supplementary tables are available with the online version of this article.
References
- 1.Eppinger M, Baar C, Linz B, Raddatz G, Lanz C, et al. Who ate whom? adaptive Helicobacter genomic changes that accompanied a host jump from early humans to large felines. PLoS Genet. 2006;2:e120. doi: 10.1371/journal.pgen.0020120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Uchiyama J, Takeuchi H, Kato S-ichiro, Takemura-Uchiyama I, Ujihara T, et al. Complete genome sequences of two Helicobacter pylori bacteriophages isolated from Japanese patients. J Virol. 2012;86:11400–11401. doi: 10.1128/JVI.01767-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Luo C-H, Chiou P-Y, Yang C-Y, Lin N-T. Genome, integration, and transduction of a novel temperate phage of Helicobacter pylori . J Virol. 2012;86:8781–8792. doi: 10.1128/JVI.00446-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vale FF, Nunes A, Oleastro M, Gomes JP, Sampaio DA, et al. Genomic structure and insertion sites of Helicobacter pylori prophages from various geographical origins. Sci Rep. 2017;7:42471. doi: 10.1038/srep42471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lehours P, Vale FF, Bjursell MK, Melefors O, Advani R, et al. Genome sequencing reveals a phage in Helicobacter pylori . mBio. 2011;2:pii: e00239–11. doi: 10.1128/mBio.00239-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vale FF, Vadivelu J, Oleastro M, Breurec S, Engstrand L, et al. Dormant phages of Helicobacter pylori reveal distinct populations in Europe. Sci Rep. 2015;5:14333. doi: 10.1038/srep14333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thorell K, Yahara K, Berthenet E, Lawson DJ, Mikhail J, et al. Rapid evolution of distinct Helicobacter pylori subpopulations in the Americas. PLoS Genet. 2017;13:e1006546. doi: 10.1371/journal.pgen.1006546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paez-Espino D, Eloe-Fadrosh EA, Pavlopoulos GA, Thomas AD, Huntemann M, et al. Uncovering earth's virome. Nature. 2016;536:425–430. doi: 10.1038/nature19094. [DOI] [PubMed] [Google Scholar]
- 9.Paez-Espino D, Sharon I, Morovic W, Stahl B, Thomas BC, et al. CRISPR immunity drives rapid phage genome evolution in Streptococcus thermophilus . mBio. 2015;6:e00262-15. doi: 10.1128/mBio.00262-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pedulla ML, Ford ME, Houtz JM, Karthikeyan T, Wadsworth C, et al. Origins of highly mosaic mycobacteriophage genomes. Cell. 2003;113:171–182. doi: 10.1016/S0092-8674(03)00233-2. [DOI] [PubMed] [Google Scholar]
- 11.Moodley Y, Linz B, Bond RP, Nieuwoudt M, Soodyall H, et al. Age of the association between Helicobacter pylori and man. PLoS Pathog. 2012;8:e1002693. doi: 10.1371/journal.ppat.1002693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thorell K, Lehours P, Vale FF. Genomics of Helicobacter pylori . Helicobacter. 2017;22:hel.12409. doi: 10.1111/hel.12409. [DOI] [PubMed] [Google Scholar]
- 13.Berthenet E, Sheppard S, Vale FF. Recent "omics" advances in Helicobacter pylori . Helicobacter. 2016;21:14–18. doi: 10.1111/hel.12334. [DOI] [PubMed] [Google Scholar]
- 14.Secka O, Vale FF, Buissonnière A, Thomas JE, Mégraud F, et al. Phylogeographic agreement between prophage and bacterial housekeeping genes in Helicobacter pylori strains from The Gambia. Helicobacter. 2017;22:hel.12394. doi: 10.1111/hel.12394. [DOI] [PubMed] [Google Scholar]
- 15.Vale FF, Matos APA, Carvalho P, Vítor JMB. Helicobacter pylori Phage Screening. Microsc Microanal. 2008;14:150–151. doi: 10.1017/S1431927608089721. [DOI] [Google Scholar]
- 16.Vitoriano I, Vítor JMB, Oleastro M, Roxo-Rosa M, Vale FF. Proteome variability among Helicobacter pylori isolates clustered according to genomic methylation. J Appl Microbiol. 2013;114:1817–1832. doi: 10.1111/jam.12187. [DOI] [PubMed] [Google Scholar]
- 17.Silva B, Nunes A, Vale FF, Rocha R, Gomes JP, et al. The expression of Helicobacter pylori tfs plasticity zone cluster is regulated by pH and adherence, and its composition is associated with differential gastric IL-8 secretion. Helicobacter. 2017;22:hel.12390. doi: 10.1111/hel.12390. [DOI] [PubMed] [Google Scholar]
- 18.Delahay RM, Croxall NJ, Stephens AD. Phylogeographic diversity and mosaicism of the Helicobacter pylori tfs integrative and conjugative elements. Mob DNA. 2018;9:5. doi: 10.1186/s13100-018-0109-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kyrillos A, Arora G, Murray B, Rosenwald AG. The presence of phage orthologous genes in Helicobacter pylori correlates with the presence of the virulence factors CagA and VacA . Helicobacter. 2016;21:226–233. doi: 10.1111/hel.12282. [DOI] [PubMed] [Google Scholar]
- 20.Yahara K, Furuta Y, Oshima K, Yoshida M, Azuma T, et al. Chromosome painting in silico in a bacterial species reveals fine population structure. Mol Biol Evol. 2013;30:1454–1464. doi: 10.1093/molbev/mst055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meier-Kolthoff JP, Uchiyama J, Yahara H, Paez-Espino D, Yahara K. Investigation of recombination-intense viral groups and their genes in the earth's virome. Sci Rep. 2018;8:11496. doi: 10.1038/s41598-018-29272-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu B, Xie G, Lo C-C, Starkenburg SR, Chain PSG. Pathogen comparative genomics in the next-generation sequencing era: genome alignments, pangenomics and metagenomics. Brief Funct Genomics. 2011;10:322–333. doi: 10.1093/bfgp/elr042. [DOI] [PubMed] [Google Scholar]
- 23.Clokie MR, Millard AD, Letarov AV, Heaphy S. Phages in nature. Bacteriophage. 2011;1:31–45. doi: 10.4161/bact.1.1.14942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pope WH, Bowman CA, Russell DA, Jacobs-Sera D, Asai DJ, et al. Whole genome comparison of a large collection of mycobacteriophages reveals a continuum of phage genetic diversity. Elife. 2015;4:e06416. doi: 10.7554/eLife.06416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uchiyama J, Takeuchi H, Kato S-ichiro, Gamoh K, Takemura-Uchiyama I, et al. Characterization of Helicobacter pylori bacteriophage KHP30. Appl Environ Microbiol. 2013;79:3176–3184. doi: 10.1128/AEM.03530-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song W, Sun H-X, Zhang C, Cheng L, Peng Y, et al. Prophage Hunter: an integrative hunting tool for active prophages. Nucleic Acids Res. 2019;47:W74–W80. doi: 10.1093/nar/gkz380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lawson DJ, Hellenthal G, Myers S, Falush D. Inference of population structure using dense haplotype data. PLoS Genet. 2012;8:e1002453. doi: 10.1371/journal.pgen.1002453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yahara K, Didelot X, Ansari MA, Sheppard SK, Falush D. Efficient inference of recombination hot regions in bacterial genomes. Mol Biol Evol. 2014;31:1593–1605. doi: 10.1093/molbev/msu082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mills R, Rozanov M, Lomsadze A, Tatusova T, Borodovsky M. Improving gene annotation of complete viral genomes. Nucleic Acids Res. 2003;31:7041–7055. doi: 10.1093/nar/gkg878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hudson RR, Kaplan NL. Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics. 1985;111:147–164. doi: 10.1093/genetics/111.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cai JJ. PGEToolbox: a Matlab toolbox for population genetics and evolution. J Hered. 2008;99:438–440. doi: 10.1093/jhered/esm127. [DOI] [PubMed] [Google Scholar]
- 34.Librado P, Rozas J. DnaSP V5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 35.Yahara K, Furuta Y, Morimoto S, Kikutake C, Komukai S, et al. Genome-wide survey of codons under diversifying selection in a highly recombining bacterial species, Helicobacter pylori . DNA Res. 2016;23:135–143. doi: 10.1093/dnares/dsw003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goerke C, Pantucek R, Holtfreter S, Schulte B, Zink M, et al. Diversity of prophages in dominant Staphylococcus aureus clonal lineages. J Bacteriol. 2009;191:3462–3468. doi: 10.1128/JB.01804-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ackermann HW, Elzanowski A, Fobo G, Stewart G. Relationships of tailed phages: a survey of protein sequence identity. Arch Virol. 1995;140:1871–1884. doi: 10.1007/BF01384350. [DOI] [PubMed] [Google Scholar]
- 38.Reuter S, Corander J, de Been M, Harris S, Cheng L, et al. Directional gene flow and ecological separation in Yersinia enterocolitica . Microb Genom. 2015;1:e000030. doi: 10.1099/mgen.0.000030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hatfull GF, Hendrix RW. Bacteriophages and their genomes. Curr Opin Virol. 2011;1:298–303. doi: 10.1016/j.coviro.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ali A, Naz A, Soares SC, Bakhtiar M, Tiwari S, et al. Pan-genome analysis of human gastric pathogen H. pylori: comparative genomics and pathogenomics approaches to identify regions associated with pathogenicity and prediction of potential core therapeutic targets. Biomed Res Int. 2015;2015:139580. doi: 10.1155/2015/139580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chan JZ-M, Millard AD, Mann NH, Schäfer H. Comparative genomics defines the core genome of the growing N4-like phage genus and identifies N4-like Roseophage specific genes. Front Microbiol. 2014;5:506. doi: 10.3389/fmicb.2014.00506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Geng P, Tian S, Yuan Z, Hu X. Identification and genomic comparison of temperate bacteriophages derived from emetic Bacillus cereus . PLoS One. 2017;12:e0184572. doi: 10.1371/journal.pone.0184572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Petrov VM, Ratnayaka S, Nolan JM, Miller ES, Karam JD. Genomes of the T4-related bacteriophages as windows on microbial genome evolution. Virol J. 2010;7:292. doi: 10.1186/1743-422X-7-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ramírez-Vargas G, Goh S, Rodríguez C. The novel phages phicd5763 and phicd2955 represent two groups of big plasmidial siphoviridae phages of Clostridium difficile . Front Microbiol. 2018;9:26. doi: 10.3389/fmicb.2018.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Uchiyama I, Albritton J, Fukuyo M, Kojima KK, Yahara K, et al. A novel approach to Helicobacter pylori pan-genome analysis for identification of genomic islands. PLoS One. 2016;11:e0159419. doi: 10.1371/journal.pone.0159419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vale FF, Mégraud F, Vítor JMB. Geographic distribution of methyltransferases of Helicobacter pylori: evidence of human host population isolation and migration. BMC Microbiol. 2009;9:193. doi: 10.1186/1471-2180-9-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vale FF, Vítor JMB. Genomic methylation: a tool for typing Helicobacter pylori isolates. Appl Environ Microbiol. 2007;73:4243–4249. doi: 10.1128/AEM.00199-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.