Abstract
Background
SMAD family member 4 (SMAD4) has gained attention as a promising prognostic factor of colorectal cancer (CRC) as well as a key molecule to understand the tumorigenesis and progression of CRC.
Methods
We retrospectively analyzed 1,281 CRC cases immunohistochemically for their expression status of SMAD4, and correlated this status with clinicopathologic and molecular features of CRCs.
Results
A loss of nuclear SMAD4 was significantly associated with frequent lymphovascular and perineural invasion, tumor budding, fewer tumor-infiltrating lymphocytes, higher pT and pN category, and frequent distant metastasis. In contrast, tumors overexpressing SMAD4 showed a significant association with sporadic microsatellite instability. After adjustment for TNM stage, tumor differentiation, adjuvant chemotherapy, and lymphovascular invasion, the loss of SMAD4 was found to be an independent prognostic factor for worse 5-year progression-free survival (hazard ratio [HR], 1.27; 95% confidence interval [CI], 1.01 to 1.60; p=.042) and 7-year cancer-specific survival (HR, 1.45; 95% CI, 1.06 to 1.99; p=.022).
Conclusions
We confirmed the value of determining the loss of SMAD4 immunohistochemically as an independent prognostic factor for CRC in general. In addition, we identified some histologic and molecular features that might be clues to elucidate the role of SMAD4 in colorectal tumorigenesis and progression.
Keywords: Biomarker, SMAD4, Colorectal neoplasms, Prognosis
Ranked third in terms of incidence and second in terms of mortality in the latest global cancer report [1], colorectal cancer (CRC) is a major health burden worldwide. Notably, this report mentioned South Korea as one of the countries where the highest colon cancer incidence rates were observed. Moreover, the report specifically stated that the highest incidence rates of rectal cancer were seen in South Korean males. In this regard, the importance of understanding the biology of Korean CRC cannot be emphasized enough.
SMAD family member 4 (SMAD4) is a transcription factor that acts as the central mediator of the transforming growth factor β (TGF-β) pathway [2]. Also known as deleted pancreatic cancer locus-4, heterozygous or homozygous deletion of SMAD4 was first discovered in pancreatic ductal adenocarcinoma [3], and later detected in other types of cancer, including CRC. Meta-analyses have revealed that the loss of SMAD4 is a negative predictor of overall survival, cancer-specific survival (CSS), and relapse-free survival [4,5]. In CRC, the loss of SMAD4 is associated with poor differentiation [6,7], higher stage [8-12], frequent lymph node metastasis [13-17], loss of immune infiltrates [17], and poor response to 5-fluorouracil [17-19]. Accordingly, numerous studies have reported the value of determining SMAD4 loss as a negative prognostic factor [9-12,17,20-25]. Although some studies failed to identify a significant association [6,8,26-28], a meta-analysis reported pooled hazard ratios over 1 with statistical significance for overall survival, disease-free survival, and CSS [29]. Mechanistically, the loss of SMAD4 has been implicated with activation of the Akt pathway [30] and Wnt pathways [31]. Moreover, germline mutations of SMAD4 and BMPR1A (a gene upstream from SMAD4 in the TGF-β pathway), cause juvenile polyposis syndrome, a genetic cancer predisposition syndrome with increased risk of gastrointestinal cancers [32]. Collectively, these data suggest that SMAD4 is a key molecule to decipher the pathophysiology of CRC while acting as a feasible prognostic marker for optimal surveillance of patients. However, only a handful of studies have explored the expression of SMAD4 and its prognostic significance in a limited number of Korean CRC patients [15,28,33]. Moreover, comprehensive clinicopathologic and molecular characterization of CRCs with respect to SMAD4 expression was rarely performed.
In this study, we analyzed 1,281 CRC cases for their expression status of SMAD4 using immunohistochemistry and demonstrated comprehensive clinicopathologic and molecular characteristics of CRCs including KRAS and BRAF mutation, microsatellite instability (MSI) and CpG island methylator phenotype (CIMP) depending on nuclear SMAD4 expression status. Finally, we evaluated its value as a prognostic factor for CSS and progression-free survival.
MATERIALS AND METHODS
Patients and tissue samples
Under the exclusion criteria described previously [34], 1,370 out of 1,853 CRC cases resected at Seoul National University Hospital, Seoul, Korea, between January 2004 and June 2008 were reviewed. Among them, 1,281 cases with formalin-fixed, paraffin-embedded (FFPE) tumor blocks sufficient for construction of a tissue microarray (TMA) were included in the study. Initial pathologic diagnosis and clinical information, including age, tumor location and radiologic/pathologic evidences of distant metastases were obtained from electronic medical records. Additional histologic parameters including tumor differentiation, tumor budding, representative number of tumor-infiltrating lymphocytes (TILs) per one high power field (400× magnification), were evaluated by two pathologists as described previously.34 FFPE tissues were used for molecular analysis and immunohistochemistry.
Immunohistochemistry
For each CRC case, a pair of 2-mm cores of representative tumor areas in the FFPE tissue were extracted to construct the TMA. To evaluate SMAD4 expression in CRCs, TMAs were sectioned at a thickness of 4-μm and stained using a rabbit monoclonal anti-SMAD4 antibody (1:200 dilution, clone EP618Y, Abcam, Cambridge, UK). Stained slides were scanned by an Aperio AT2 slide scanner (Sausalito, CA, USA) at 40× magnification with a resolution of 0.25 μm per pixel. The proportion and the intensity of SMAD4 staining in nuclear compartment were evaluated using TMA Assistant protocol of QuPath [35], open-source software for digital pathology image analysis. In detail, the intensity of SMAD4 staining in nuclear compartment was graded using intensity feature of nuclear diaminobenzidine (DAB) optical densities (OD). Because nuclear DAB ODs of entrapped nonneoplastic epithelium range from 0.2 to 0.6, intensity of nuclear SMAD4 staining of tumor cells were evaluated using following cut-offs: intensity 0, DAB OD<0.2; intensity 1, DAB OD≥0.2 and <0.6; and intensity 2, DAB OD≥0.6. Finally, each case was classified to SMAD4-loss (≥95% of tumor cells showed intensity 0), SMAD4-low (≥5% of tumor cells showed intensity 1, and <30% of tumor cells showed intensity 2), and SMAD4-high (≥30% of tumor cells showed intensity 2). For survival analysis, CRC cases were dichotomized to CRCs with SMAD4 loss and CRCs with retained SMAD4 (SMAD4-low and SMAD4-high). The evaluation method and cut-offs for the expression of cytokeratin 7 (CK7) (clone OV-TL, 12/30, Dako, Carpenteria, CA, USA), cytokeratin 20 (CK20) (clone Ks20.8, Dako), and nuclear protein CDX2 (clone EPR2764Y, ready-to-use, Cell Marque, Rocklin, CA, USA) were described previously [36]. All immunohistochemical procedures in this study were conducted with an automated immunostainer (BenchMark XT, Ventana Medical Systems, Tucson, AZ, USA).
Molecular analyses
Through histological examination, representative tumor portions were marked and then subjected to manual microdissection. The dissected tissues were collected into microtubes containing lysis buffer and proteinase K and were incubated at 55°C for 2 days. DNA from paraffin-embedded tissues was extracted, and the polymerase chain reaction (PCR) was performed. Direct sequencing of KRAS codons 12 and 13 and allele-specific PCR for BRAF codon 600 were performed as described previously [34]. The MSI status of each tumor was determined by evaluating five microsatellite markers (BAT25, BAT26, D2S123, D5S346, and D17S250) as standardized by the National Cancer Institute [37]. A fluorescent label was added to either the forward or reverse primer for each marker, and the PCR products were electrophoresed and analyzed. We classified MSI status as MSI-positive (instability at two or more microsatellite marker), and MSI-negative (no instability or instability at one marker). The CIMP status was evaluated by the MethyLight assay of eight markers (CACNA1G, CDKN2A [p16], CRABP1, IGF2, MLH1, NEUROG1, RUNX3, and SOCS1) [38]. We classified CRCs into CIMPnegative (0–4 methylated markers), CIMP-positive 1 (5–6 methylated markers), and CIMP-positive 2 (CIMP-P2) (7–8 methylated markers), as previously described [34].
Statistical analyses
SAS ver. 9.4 (SAS Institute Inc., Cary, NC, USA) and R software (http://www.r-project.org) were used for statistical analyses. Clinicopathological characteristics were compared between the three SMAD4 expression groups by use of chi-square test or Fisher exact test, as appropriate. Chi-square test for trend was used to compute p for trend. Survival curves after surgery were estimated with the Kaplan-Meier method, and differences in survival curves were tested with the log-rank test. All statistical tests were two-sided, and statistical significance was defined as p<.05.
Ethics statement
This study was approved by the Institutional Review Board which waived the requirement to obtain informed consent (IRB No. C-1502-029-647).
RESULTS
Clinicopathological and molecular correlation of nuclear SMAD4 expression in CRCs
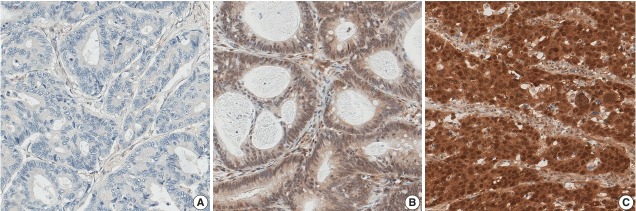
Out of 1,281 CRC cases, 210 (16.4%) showed a loss of nuclear expression of SMAD4 (Fig. 1A). Among the remaining cases with retained SMAD4 expression, 942 cases (73.5%) showed low-level expression (Fig. 1B), while high-level expression was observed in 129 cases (10.1%) (Fig. 1C).
Fig. 1.
Nuclear expression of SMAD4 in colorectal cancers. Representative examples for the loss of expression (A), low-level expression (B), and high-level expression (C).
We then sought to identify clinicopathologic features showing gradual changes according to the loss or overexpression of SMAD4 (Table 1). With decreasing expression of SMAD4, tumors tended to show an infiltrative gross type, more frequent lymphovascular and perineural invasion, tumor budding, and lower TILs (all p<.001). Consequently, CRC cases showed significant associations with higher pT (p<.001) and pN (p<.001) category, and frequent distant metastasis (p=.001) as nuclear SMAD4 expression decreased. Notably, statistically significant increase of SMAD4 loss and concomitant decrease of SMAD4 expression was noted as TNM stage increased (p <.001) (Supplementary Fig. S1).
Table 1.
Clinicopathologic characteristics of colorectal cancers according to SMAD4 expression
| SMAD4-loss (n = 210, 16.4%) | SMAD4-low (n = 942, 73.5%) | SMAD4-high (n = 129, 10.1%) | p for difference | p for trend | |
|---|---|---|---|---|---|
| Age (yr) | 64 (27–83) | 63 (20–90) | 61 (25–93) | .222 | |
| Sex | .225 | .105 | |||
| Male | 132 (62.9) | 566 (60.1) | 69 (53.5) | ||
| Female | 78 (37.1) | 376 (39.9) | 60 (46.5) | ||
| Location | < .001 | .222 | |||
| Proximal | 61 (29.1) | 216 (22.9) | 50 (38.8) | ||
| Distal/Rectum | 149 (70.9) | 726 (77.1) | 79 (61.2) | ||
| Gross type | .002 | < .001 | |||
| Fungating | 122 (58.1) | 624 (66.2) | 99 (76.7) | ||
| Infiltrative | 88 (41.9) | 318 (33.8) | 30 (23.3) | ||
| pT category | < .001 | < .001 | |||
| pT1-2 | 20 (9.5) | 176 (18.7) | 37 (28.7) | ||
| pT3-4 | 190 (90.5) | 766 (81.3) | 92 (71.3) | ||
| pN category | < .001 | < .001 | |||
| pN0 | 85 (40.5) | 471 (50.0) | 89 (69.0) | ||
| pN1-2 | 125 (59.5) | 471 (50.0) | 40 (31.0) | ||
| Distant metastasis | .001 | .001 | |||
| M0 | 167 (79.5) | 775 (82.3) | 122 (94.6) | ||
| M1 | 43 (20.5) | 167 (17.7) | 7 (5.4) | ||
| TNM stage | < .001 | < .001 | |||
| I, II | 76 (36.2) | 448 (47.6) | 87 (67.4) | ||
| III, IV | 134 (63.8) | 494 (52.4) | 42 (32.6) | ||
| Lymphovascular invasion | < .001 | < .001 | |||
| Absent | 97 (46.2) | 543 (57.6) | 96 (74.4) | ||
| Present | 113 (53.8) | 399 (42.4) | 33 (26.6) | ||
| Perineural invasion | <.001 | < .001 | |||
| Absent | 133 (63.3) | 730 (77.5) | 113 (87.6) | ||
| Present | 77 (36.7) | 212 (22.5) | 16 (12.4) | ||
| Differentiation (grade) | .612 | .923 | |||
| Differentiated (G1/2) | 200 (95.2) | 906 (94.2) | 122 (94.6) | ||
| Undifferentiated (G3/4) | 10 (4.8) | 36 (3.8) | 7 (5.4) | ||
| Tumor budding | < .001 | < .001 | |||
| Absent | 48 (22.9) | 256 (27.2) | 57 (44.2) | ||
| Present | 162 (77.1) | 686 (72.8) | 72 (55.8) | ||
| Tumor-infiltrating lymphocytes (400 × magnification) | < .001 | < .001 | |||
| Low (< 8) | 164 (78.1) | 739 (78.4) | 72 (55.8) | ||
| High (≥ 8) | 46 (21.9) | 203 (21.6) | 57 (44.2) | ||
| Mucin production | .002 | .581 | |||
| Absent | 176 (83.8) | 839 (89.1) | 102 (79.1) | ||
| Present | 34 (16.2) | 103 (10.9) | 27 (20.9) |
Values are presented as median (range) or number (%).
To identify molecular phenotypical correlates of such linear trends, we evaluated MSI, CIMP status, KRAS exon 2, BRAF codon 600, and immunohistochemical expression of CK7, CK20, and CDX2 (Table 2). With increasing expression of SMAD4, tumors tended to be more associated with MSI (p < .001), CIMP-P2 (p=.001), and MLH1 promoter methylation (p< .001). Interestingly, the expression of CK20 and CDX2 showed opposite trends with increasing SMAD2 expression; CDX2 expression was lost as nuclear SMAD4 expression increased (p= .001), while the expression of CK20 increased (p<.001).
Table 2.
Molecular characteristics of colorectal cancers according to SMAD4 expression
| SMAD4-loss (n = 210, 16.4%) | SMAD4-low (n = 942, 73.5%) | SMAD4-high (n = 129, 10.1%) | p for difference | p for trend | |
|---|---|---|---|---|---|
| KRAS mutation | .866 | .606 | |||
| Absent | 143 (68.1) | 657 (69.7) | 91 (70.5) | ||
| Present | 67 (31.9) | 285 (30.3) | 38 (29.5) | ||
| BRAF mutation (n = 1,278) | .094 | .330 | |||
| Absent | 196 (93.3) | 906 (96.5) | 122 (94.6) | ||
| Present | 14 (6.7) | 33 (3.5) | 7 (5.4) | ||
| MSI | < .001 | < .001 | |||
| MSS | 204 (97.1) | 889 (94.4) | 93 (72.1) | ||
| MSI | 6 (2.9) | 53 (5.6) | 36 (27.9) | ||
| CIMP | .229a | .001 | |||
| CIMP-N, P1 | 209 (99.5) | 930 (98.7) | 122 (94.6) | ||
| CIMP-P2 | 1 (0.5) | 12 (1.3) | 7 (5.4) | ||
| MLH1 promoter methylation | < .001 | < .001 | |||
| Unmethylated | 205 (97.6) | 921 (97.8) | 112 (86.8) | ||
| Methylated | 5 (2.4) | 21 (2.2) | 17 (13.2) | ||
| CK7 expression (n = 1,277) | .273 | .120 | |||
| Absent | 186 (89.4) | 868 (92.3) | 121 (93.8) | ||
| Present | 22 (10.6) | 72 (7.7) | 8 (6.2) | ||
| CK20 expression (n = 1,271) | < .001 | < .001 | |||
| Retained | 183 (88.8) | 780 (83.3) | 90 (69.8) | ||
| Loss | 23 (11.2) | 156 (16.7) | 39 (30.2) | ||
| CDX2 expression (n = 1,262) | .001 | .001 | |||
| Retained | 166 (80.6) | 831 (89.4) | 116 (91.3) | ||
| Loss | 40 (19.4) | 98 (10.6) | 11 (8.7) |
Values are presented as median (range) or number (%).
MSI, microsatellite instability; MSS, microsatellite stable; CIMP, CpG island methylator phenotype; CIMP-N, CIMP-negative; CIMP-P1, CIMP-positive 1; CIMPP2, CIMP-positive 2; CK, cytokeratin.
p-value with Fisher exact test.
Prognostic implication of SMAD4 loss in CRCs
A univariate survival analysis revealed that CRCs with a loss of nuclear SMAD4 expression exhibited a significantly worse 5-year progression-free survival (PFS) (p<.001) (Fig. 2A), and 7-year CSS (p=.001) (Fig. 2B). To explore whether there exists a differential prognostic effect of SMAD4 according to TNM stage, we performed the Kaplan-Meier analysis for each TNM stage subgroups (Supplementary Figs. S2, S3). In a stage-specific analysis, SMAD4 loss was associated with worse 5-year PFS (p = .019) in stage II CRCs, worse 5-year PFS (p = .055) and 7-year CSS (p=.011) in stage IV CRCs. In the multivariate Cox proportional hazard analysis, a loss of SMAD4 expression proved to be an independent prognostic factor for 5-year PFS (hazard ratio [HR], 1.27; 95% confidence interval [CI], 1.01 to 1.60; p=.042) (Table 3), and for 7-year CSS (HR, 1.45; 95% CI, 1.06 to 1.99; p=.022), after adjustment for TNM stage, tumor differentiation, adjuvant chemotherapy, and lymphovascular invasion.
Fig. 2.
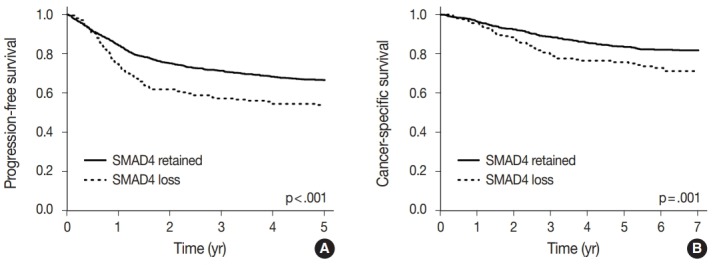
Kaplan-Meier survival curves according to the nuclear SMAD4 expression. (A) 5-Year progression-free survival. (B) 7-Year cancer-specific survival.
Table 3.
Multivariate Cox proportional hazard analysis with respect to 5-year PFS and 7-year CSS
| 5-Year PFS |
7-Year CSS |
|||
|---|---|---|---|---|
| HR (95% CI) | p-value | HR (95% CI) | p-value | |
| Stage (III, IV vs I, II) | 4.57 (3.47–6.00) | < .001 | 4.99 (3.38–7.37) | < .001 |
| Grade (G3/4 vs G1/2) | 1.80 (1.27–2.56) | .001 | 1.62 (0.95–2.75) | .077 |
| Post-operative chemotherapy (yes vs no) | 0.63 (0.49–0.81) | < .001 | 0.34 (0.25–0.47) | < .001 |
| Lymphovascular invasion (yes vs no) | 1.83 (1.48–2.26) | < .001 | 2.22 (1.63–3.03) | < .001 |
| SMAD4 expression (loss vs retained) | 1.27 (1.01–1.60) | .042 | 1.45 (1.06–1.99) | .022 |
PFS, progression-free survival; CSS, cancer-specific survival; HR, hazard ratio; CI, confidence interval.
DISCUSSION
To the best of our knowledge, this is the second largest study focusing on the clinicopathologic and prognostic implications of SMAD4 expression in CRC, and the largest in an Asian population. While the largest previous study was performed on 1,381 stage II or III CRC patients enrolled at a pan-European clinical trial for adjuvant chemotherapy [12], our cohort consisted of 1,281 retrospectively collected patients ranging from stage I to IV. By combining CIMP analysis with additional histologic features such as tumor budding and TILs, we identified the general expression status of SMAD4 and its association with clinicopathologic and molecular features of CRC.
Most previous studies evaluated SMAD4 expression based on two-tier classification: loss versus no loss, or low versus high expression [7,8,10,11,13-15,17,18,20]. Because of ambiguities in the definition of low expression, the reported prevalence of low-level SMAD4 expression varied significantly from 2.34% to 75.2%. On the other hand, studies that adopted a multi-tier classification reported relatively homogeneous results, with the prevalence of low-level SMAD4 expression ranging from 9.3% to 37.7% [6,9,12,16]. Considering the presence of an apparent loss, overexpression, and intermediate expression of SMAD4, we felt it more appropriate to use a three-tier classification (loss, low-level, and high-level) of SMAD4 expression. As a result, 16.4% of our cases were identified as SMAD4 loss, which falls within the range of previous reports.
Consistent with the previous studies [8-17], we observed a significant association between SMAD4 loss and higher pT and pN category, and frequent distant metastasis. Consequently, we confirmed the value of SMAD4 loss as a prognostic factor for poor CSS and PFS of CRC. In colonic epithelial cells, TGF-β signaling reduces proliferation and promotes apoptosis and differentiation [39]. Because SMAD4 translocates to the nucleus by forming a heterodimeric complex with SMAD2/3 that is phosphorylated by the activated TGF-β receptor [2], it is intuitive that the immunohistochemical loss of nuclear SMAD4 expression suggests a concomitant loss of TGF-β signaling, which leads to uncontrolled proliferative behavior.
Interestingly, we observed that some tumors overexpressed SMAD4 and those tumors tend to be MSI, CIMP-P2, and have their MLH1 promoter methylated. Along with the association between fungating gross type, these data collectively suggested an association between the sporadic MSI-high (MSI-H)/CIMP-high phenotype and overexpression of nuclear SMAD4. Although the meaning of SMAD4 overexpression is not straightforward, such a trend has been reported previously [9,12,40]. One possibility is that overexpression of SMAD4 in MSI-H tumors might be the consequence of a compensatory mechanism for mutational inactivation of the TGF-β signaling pathway; i.e., some machineries in the pathway, such as transforming growth factor β receptor II (TGFBR2) or activin type II receptor (ACVR2), are prone to mutations when mismatch repair is impaired [39]. At the same time, there are reports suggesting various bypass mechanisms to overcome such mutations [41-44]. Further mechanistic studies are needed to confirm this hypothesis.
A novel finding in our study was the gradual increase of CDX2 expression as nuclear SMAD4 increased, while CK20 showed the opposite trend. Although this is in contrast with our previous report that expression of both CK20 and CDX2 got lost according to CpG island methylation [34], the proportional relationship of CDX2 and SMAD4 is consistent with some previous reports. In stomach, it has been reported that SMAD4 can activate the promoter of CDX2, and knockdown of SMAD4 led to the decreased expression of CDX2 [45]. Concomitant loss of SMAD4 and CDX2 was also observed in colorectal juvenile polyps obtained from juvenile polyposis syndrome patients [46]. The loss of CK20 expression could be explained as a consequence of epithelial mesenchymal transition (EMT). It is known that switch of intermediate filaments from cytokeratin to vimentin occurs during EMT [47]. It has been reported that the immunohistochemical expression of SMAD4 was positively correlated with that of EMT-related transcription factors such as Snail-1 and Twist-1 [48], and silencing of SMAD4 inhibited EMT [49]. Consequently, an intriguing hypothesis emerges that overexpression of SMAD4 accompanied loss of CK20 by promoting EMT.
Consistent with a previous study [17], we observed a significant association between low TIL infiltration and SMAD4 loss. Although this could be a secondary effect of the MSI status, there are a series of reports on the correlation between SMAD4 loss and CCL15 expression [50-52]. These researchers demonstrated that the loss of SMAD4 upregulated CCL15, which resulted in recruitment of CCR1+ cells at the invasive front. CCR1+ cells are phenotypically myeloid-derived suppressor cells (MDSC) and express immunosuppressive molecules such as indoleamine 2,3- dioxygenase. Using mouse models, these researchers demonstrated that the loss of SMAD4 promoted pulmonary and hepatic metastasis through the CCL15-CCR1 axis. It is plausible that the MDSCs are also responsible for the lower TIL infiltration we observed for tumors with SMAD4 loss.
In conclusion, we confirmed the value of determining expression of SMAD4 immunohistochemically as an independent prognostic factor for CRC in general. Furthermore, we identified some histologic and molecular features that might be clues to elucidate the role of SMAD4 in colorectal tumorigenesis and progression. Further studies are needed to validate these findings with an independent large-scale series of CRC cases.
Footnotes
Author contributions
Conceptualization: JMB, GHK.
Data curation: SYY, JMB.
Formal analysis: SYY, JMB.
Funding acquisition: JMB, GHK.
Investigation: SYY, JAL, JMB.
Methodology: SYY, JAL, YS, NYC, JMB.
Project administration: JMB, GHK.
Resources: JMB, GHK.
Supervision: JMB, GHK.
Validation: JMB, GHK.
Visualization: SYY, JMB.
Writing—original draft: SYY, JMB.
Writing—review & editing: JMB, GHK.
Conflicts of Interest
The authors declare that they have no potential conflicts of interest.
Funding
This study was supported by grants from the National Research Foundation (NRF) funded by the Korean Ministry of Science, ICT and Future Planning (2011-0030049, 2016M3 A9B6026921), Korean Health Technology R&D Project, Ministry of Health and Welfare (HI14C1277), and a grant from the SNUH Research Fund (04-2018-0620).
Electronic Supplementary Material
Supplementary materials are available at Journal of Pathology and Translational Medicine (http://jpatholtm.org).
REFERENCES
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Zhao M, Mishra L, Deng CX. The role of TGF-beta/SMAD4 signaling in cancer. Int J Biol Sci. 2018;14:111–23. doi: 10.7150/ijbs.23230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schutte M. DPC4/SMAD4 gene alterations in human cancer, and their functional implications. Ann Oncol. 1999;10 Suppl 4:56–9. [PubMed] [Google Scholar]
- 4.Du Y, Zhou X, Huang Z, et al. Meta-analysis of the prognostic value of smad4 immunohistochemistry in various cancers. PLoS One. 2014;9:e110182. doi: 10.1371/journal.pone.0110182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang JD, Jin K, Chen XY, Lv JQ, Ji KW. Clinicopathological significance of SMAD4 loss in pancreatic ductal adenocarcinomas: a systematic review and meta-analysis. Oncotarget. 2017;8:16704–11. doi: 10.18632/oncotarget.14335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kouvidou C, Latoufis C, Lianou E, et al. Expression of Smad4 and TGF-beta2 in colorectal carcinoma. Anticancer Res. 2006;26:2901–7. [PubMed] [Google Scholar]
- 7.Xu WQ, Jiang XC, Zheng L, Yu YY, Tang JM. Expression of TGF-beta1, TbetaRII and Smad4 in colorectal carcinoma. Exp Mol Pathol. 2007;82:284–91. doi: 10.1016/j.yexmp.2006.10.011. [DOI] [PubMed] [Google Scholar]
- 8.Bacman D, Merkel S, Croner R, Papadopoulos T, Brueckl W, Dimmler A. TGF-beta receptor 2 downregulation in tumour-associated stroma worsens prognosis and high-grade tumours show more tumour-associated macrophages and lower TGF-beta1 expression in colon carcinoma: a retrospective study. BMC Cancer. 2007;7:156. doi: 10.1186/1471-2407-7-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Isaksson-Mettaväinio M, Palmqvist R, Dahlin AM, et al. High SMAD4 levels appear in microsatellite instability and hypermethylated colon cancers, and indicate a better prognosis. Int J Cancer. 2012;131:779–88. doi: 10.1002/ijc.26473. [DOI] [PubMed] [Google Scholar]
- 10.Voorneveld PW, Jacobs RJ, De Miranda NF, et al. Evaluation of the prognostic value of pSMAD immunohistochemistry in colorectal cancer. Eur J Cancer Prev. 2013;22:420–4. doi: 10.1097/CEJ.0b013e32835e88e2. [DOI] [PubMed] [Google Scholar]
- 11.Kozak MM, von Eyben R, Pai J, et al. Smad4 inactivation predicts for worse prognosis and response to fluorouracil-based treatment in colorectal cancer. J Clin Pathol. 2015;68:341–5. doi: 10.1136/jclinpath-2014-202660. [DOI] [PubMed] [Google Scholar]
- 12.Yan P, Klingbiel D, Saridaki Z, et al. Reduced expression of SMAD4 is associated with poor survival in colon cancer. Clin Cancer Res. 2016;22:3037–47. doi: 10.1158/1078-0432.CCR-15-0939. [DOI] [PubMed] [Google Scholar]
- 13.Jain S, Singhal S, Francis F, et al. Association of overexpression of TIF1gamma with colorectal carcinogenesis and advanced colorectal adenocarcinoma. World J Gastroenterol. 2011;17:3994–4000. doi: 10.3748/wjg.v17.i35.3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Handra-Luca A, Olschwang S, Fléjou JF. SMAD4 protein expression and cell proliferation in colorectal adenocarcinomas. Virchows Arch. 2011;459:511–9. doi: 10.1007/s00428-011-1152-4. [DOI] [PubMed] [Google Scholar]
- 15.Chung Y, Wi YC, Kim Y, et al. The Smad4/PTEN expression pattern predicts clinical outcomes in colorectal adenocarcinoma. J Pathol Transl Med. 2018;52:37–44. doi: 10.4132/jptm.2017.10.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oyanagi H, Shimada Y, Nagahashi M, et al. SMAD4 alteration associates with invasive-front pathological markers and poor prognosis in colorectal cancer. Histopathology. 2019;74:873–82. doi: 10.1111/his.13805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wasserman I, Lee LH, Ogino S, et al. SMAD4 loss in colorectal cancer patients correlates with recurrence, loss of immune infiltrate, and chemoresistance. Clin Cancer Res. 2019;25:1948–56. doi: 10.1158/1078-0432.CCR-18-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alhopuro P, Alazzouzi H, Sammalkorpi H, et al. SMAD4 levels and response to 5-fluorouracil in colorectal cancer. Clin Cancer Res. 2005;11:6311–6. doi: 10.1158/1078-0432.CCR-05-0244. [DOI] [PubMed] [Google Scholar]
- 19.Boulay JL, Mild G, Lowy A, et al. SMAD4 is a predictive marker for 5-fluorouracil-based chemotherapy in patients with colorectal cancer. Br J Cancer. 2002;87:630–4. doi: 10.1038/sj.bjc.6600511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alazzouzi H, Alhopuro P, Salovaara R, et al. SMAD4 as a prognostic marker in colorectal cancer. Clin Cancer Res. 2005;11:2606–11. doi: 10.1158/1078-0432.CCR-04-1458. [DOI] [PubMed] [Google Scholar]
- 21.Isaksson-Mettavainio M, Palmqvist R, Forssell J, Stenling R, Oberg A. SMAD4/DPC4 expression and prognosis in human colorectal cancer. Anticancer Res. 2006;26:507–10. [PubMed] [Google Scholar]
- 22.Mesker WE, Liefers GJ, Junggeburt JM, et al. Presence of a high amount of stroma and downregulation of SMAD4 predict for worse survival for stage I-II colon cancer patients. Cell Oncol. 2009;31:169–78. doi: 10.3233/CLO-2009-0478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baraniskin A, Munding J, Schulmann K, et al. Prognostic value of reduced SMAD4 expression in patients with metastatic colorectal cancer under oxaliplatin-containing chemotherapy: a translational study of the AIO colorectal study group. Clin Colorectal Cancer. 2011;10:24–9. doi: 10.3816/CCC.2011.n.003. [DOI] [PubMed] [Google Scholar]
- 24.Wang C, Zhou Y, Ruan R, Zheng M, Han W, Liao L. High expression of COUP-TF II cooperated with negative Smad4 expression predicts poor prognosis in patients with colorectal cancer. Int J Clin Exp Pathol. 2015;8:7112–21. [PMC free article] [PubMed] [Google Scholar]
- 25.Coates RF, Gardner JA, Gao Y, et al. Significance of positive and inhibitory regulators in the TGF-beta signaling pathway in colorectal cancers. Hum Pathol. 2017;66:34–9. doi: 10.1016/j.humpath.2017.05.021. [DOI] [PubMed] [Google Scholar]
- 26.Xie W, Rimm DL, Lin Y, Shih WJ, Reiss M. Loss of Smad signaling in human colorectal cancer is associated with advanced disease and poor prognosis. Cancer J. 2003;9:302–12. doi: 10.1097/00130404-200307000-00013. [DOI] [PubMed] [Google Scholar]
- 27.Gulubova M, Manolova I, Ananiev J, Julianov A, Yovchev Y, Peeva K. Role of TGF-beta1, its receptor TGFbetaRII, and Smad proteins in the progression of colorectal cancer. Int J Colorectal Dis. 2010;25:591–9. doi: 10.1007/s00384-010-0906-9. [DOI] [PubMed] [Google Scholar]
- 28.Chun HK, Jung KU, Choi YL, et al. Low expression of transforming growth factor beta-1 in cancer tissue predicts a poor prognosis for patients with stage III rectal cancers. Oncology. 2014;86:159–69. doi: 10.1159/000358064. [DOI] [PubMed] [Google Scholar]
- 29.Voorneveld PW, Jacobs RJ, Kodach LL, Hardwick JC. A Meta-analysis of SMAD4 immunohistochemistry as a prognostic marker in colorectal cancer. Transl Oncol. 2015;8:18–24. doi: 10.1016/j.tranon.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang B, Zhang B, Chen X, et al. Loss of Smad4 in colorectal cancer induces resistance to 5-fluorouracil through activating Akt pathway. Br J Cancer. 2014;110:946–57. doi: 10.1038/bjc.2013.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Voorneveld PW, Kodach LL, Jacobs RJ, et al. The BMP pathway either enhances or inhibits the Wnt pathway depending on the SMAD4 and p53 status in CRC. Br J Cancer. 2015;112:122–30. doi: 10.1038/bjc.2014.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chow E, Macrae F. A review of juvenile polyposis syndrome. J Gastroenterol Hepatol. 2005;20:1634–40. doi: 10.1111/j.1440-1746.2005.03865.x. [DOI] [PubMed] [Google Scholar]
- 33.Ahn BK, Jang SH, Paik SS, Lee KH. Smad4 may help to identify a subset of colorectal cancer patients with early recurrence after curative therapy. Hepatogastroenterology. 2011;58:1933–6. doi: 10.5754/hge11186. [DOI] [PubMed] [Google Scholar]
- 34.Bae JM, Kim JH, Kwak Y, et al. Distinct clinical outcomes of two CIMP-positive colorectal cancer subtypes based on a revised CIMP classification system. Br J Cancer. 2017;116:1012–20. doi: 10.1038/bjc.2017.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bankhead P, Loughrey MB, Fernández JA, et al. QuPath: open source software for digital pathology image analysis. Sci Rep. 2017;7:16878. doi: 10.1038/s41598-017-17204-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bae JM, Lee TH, Cho NY, Kim TY, Kang GH. Loss of CDX2 expression is associated with poor prognosis in colorectal cancer patients. World J Gastroenterol. 2015;21:1457–67. doi: 10.3748/wjg.v21.i5.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boland CR, Thibodeau SN, Hamilton SR, et al. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248–57. [PubMed] [Google Scholar]
- 38.Weisenberger DJ, Siegmund KD, Campan M, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38:787–93. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- 39.Jung B, Staudacher JJ, Beauchamp D. Transforming growth factor beta superfamily signaling in development of colorectal cancer. Gastroenterology. 2017;152:36–52. doi: 10.1053/j.gastro.2016.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Salovaara R, Roth S, Loukola A, et al. Frequent loss of SMAD4/DPC4 protein in colorectal cancers. Gut. 2002;51:56–9. doi: 10.1136/gut.51.1.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ilyas M, Efstathiou JA, Straub J, Kim HC, Bodmer WF. Transforming growth factor beta stimulation of colorectal cancer cell lines: type II receptor bypass and changes in adhesion molecule expression. Proc Natl Acad Sci U S A. 1999;96:3087–91. doi: 10.1073/pnas.96.6.3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deacu E, Mori Y, Sato F, et al. Activin type II receptor restoration in ACVR2-deficient colon cancer cells induces transforming growth factor-beta response pathway genes. Cancer Res. 2004;64:7690–6. doi: 10.1158/0008-5472.CAN-04-2082. [DOI] [PubMed] [Google Scholar]
- 43.Baker K, Raut P, Jass JR. Microsatellite unstable colorectal cancer cell lines with truncating TGFbetaRII mutations remain sensitive to endogenous TGFbeta. J Pathol. 2007;213:257–65. doi: 10.1002/path.2235. [DOI] [PubMed] [Google Scholar]
- 44.de Miranda NF, van Dinther M, van den Akker BE, van Wezel T, ten Dijke P, Morreau H. Transforming growth factor beta signaling in colorectal cancer cells with microsatellite instability despite biallelic mutations in TGFBR2. Gastroenterology. 2015;148:1427–37.:e8. doi: 10.1053/j.gastro.2015.02.052. [DOI] [PubMed] [Google Scholar]
- 45.Barros R, Pereira B, Duluc I, et al. Key elements of the BMP/SMAD pathway co-localize with CDX2 in intestinal metaplasia and regulate CDX2 expression in human gastric cell lines. J Pathol. 2008;215:411–20. doi: 10.1002/path.2369. [DOI] [PubMed] [Google Scholar]
- 46.Barros R, Mendes N, Howe JR, et al. Juvenile polyps have gastric differentiation with MUC5AC expression and downregulation of CDX2 and SMAD4. Histochem Cell Biol. 2009;131:765–72. doi: 10.1007/s00418-009-0579-z. [DOI] [PubMed] [Google Scholar]
- 47.Savagner P. The epithelial-mesenchymal transition (EMT) phenomenon. Ann Oncol. 2010;21 Suppl 7:vii89–92. doi: 10.1093/annonc/mdq292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ioannou M, Kouvaras E, Papamichali R, Samara M, Chiotoglou I, Koukoulis G. Smad4 and epithelial-mesenchymal transition proteins in colorectal carcinoma: an immunohistochemical study. J Mol Histol. 2018;49:235–44. doi: 10.1007/s10735-018-9763-6. [DOI] [PubMed] [Google Scholar]
- 49.Deckers M, van Dinther M, Buijs J, et al. The tumor suppressor Smad4 is required for transforming growth factor beta-induced epithelial to mesenchymal transition and bone metastasis of breast cancer cells. Cancer Res. 2006;66:2202–9. doi: 10.1158/0008-5472.CAN-05-3560. [DOI] [PubMed] [Google Scholar]
- 50.Itatani Y, Kawada K, Fujishita T, et al. Loss of SMAD4 from colorectal cancer cells promotes CCL15 expression to recruit CCR1+ myeloid cells and facilitate liver metastasis. Gastroenterology. 2013;145:1064–75.:e11. doi: 10.1053/j.gastro.2013.07.033. [DOI] [PubMed] [Google Scholar]
- 51.Inamoto S, Itatani Y, Yamamoto T, et al. Loss of SMAD4 promotes colorectal cancer progression by accumulation of myeloid-derived suppressor cells through the CCL15-CCR1 chemokine axis. Clin Cancer Res. 2016;22:492–501. doi: 10.1158/1078-0432.CCR-15-0726. [DOI] [PubMed] [Google Scholar]
- 52.Yamamoto T, Kawada K, Itatani Y, et al. Loss of SMAD4 promotes lung metastasis of colorectal cancer by accumulation of CCR1+ tumor-associated neutrophils through CCL15-CCR1 axis. Clin Cancer Res. 2017;23:833–44. doi: 10.1158/1078-0432.CCR-16-0520. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.