Abstract
Background
Recent studies suggested that miR-17~106 family was involved in the regulation of neural stem/progenitor cells (NPCs). However, distinct function of each family member was reported in regulating stem cells within and without the brain. Hence, to investigate the roles of individual miRNAs in miR-17~106 family and mechanisms underlying their effects on neurogenesis is important to extend our understanding in the CNS development.
Methods
Here, we examined the influence of miR-106a/b on the proliferation, differentiation, and survival of embryonic NPCs using specific mimics and inhibitor. The targets of miR-106a/b were identified from miRNA target prediction database and confirmed by luciferase assay. Specific siRNAs were utilized to erase the effects of miR-106a/b on the expression levels of target genes.
Results
A positive correlation was observed between the temporal reduction of miR-106a/b expression levels and the decline of NPC pools in vivo and in vitro. The perturbation of miR-106’s function approaches revealed that miR-106b, but not miR-106a, facilitated the maintenance of NPCs and repressed the generation of both neuronal and glial cells, without preference to a particular lineage. No effect was observed for miR-106a/b in NPCs’ survival. The influence of miR-106b on NPCs’ proliferation and differentiation is likely achieved by directly inhibiting the expression of Tp53inp1 and Cdkn1a, key components of Tp53inp1-Tp53-Cdkn1a axis.
Conclusion
Our study demonstrated a novel axis, miR-106b-Tp53inp1-Tp53-Cdkn1a, in regulating the proliferation and differentiation of NPCs.
Electronic supplementary material
The online version of this article (10.1186/s13287-019-1387-6) contains supplementary material, which is available to authorized users.
Keywords: miR-106, Neural stem/progenitor cells, Proliferation, Differentiation, Tp53inp1, Cdkn1a
Background
In the development of the vertebrate central nervous system (CNS), multipotent neural stem/progenitor cells (NPCs) generate various types of neurons and glia in a spatially and temporally conserved pattern [1]. Emerging evidences from a variety of approaches show that the maintenance and differentiation of NPCs are regulated in response to the interaction of extracellular signals with the intrinsic properties of NPCs [2, 3]. Although remarkable progress has been made in the identification of cell-intrinsic and cell-extrinsic factors, how these factors fine-tuned gene expression remains largely unknown. Recently, miRNA-mediated gene silencing has been proved as an essential mechanism on the regulation of multiple cellular biological processes.
From the discovery of first microRNA (miRNA) in 1993, miRNAs have emerged as important players in the control of stem cell behavior [4, 5]. miRNAs are evolutionary conserved small non-coding RNAs (22~24 nucleotides), which bind to partially complementary target sites in the 3′ untranslated region (3′UTR) of transcripts and regulate the expression of these transcripts in a post-transcriptional manner [6]. As one of the broadly conserved miRNA polycistron, miR-17~106 family, including miR-17~92 cluster and its two paralogs, miR-106b~25 cluster and miR-106a~363 cluster, is widely associated with multiple cellular processes, including proliferation, differentiation, and apoptosis [7–9]. miR-17~106 family is firstly identified as oncogenic factors in a variety of tumor types by suppressing the expression of anti-tumor genes, such as Cdkn1a (p21), Pten, Timp2, Smad7, and Bim [10–13]. During the CNS development, the functional analyses of miR-17~106 family in the regulation of NPCs majorly focus on the miR-17~92 cluster which plays an important role in maintaining self-renewal and regulating neurogliogenic decision of NPCs [14–17]. Whether or not miR-106b~25 and miR-106a~363 clusters are also involved in the regulation of NPCs during cortical development remains largely unclear. More importantly, Naka-Kaneda et al. observed distinct roles of each miR-17~106 family member in regulating neurogliogenesis in a controlled culture condition, suggesting the necessity to investigate the function of individual miRNAs in this family.
Here, we have examined the effects of miR-106b and miR-106a, key members of miR-106b~25 and miR-106a~363 clusters, on the regulation of embryonic NPCs and identified its target genes. Our observations suggested that miR-106b, but not miR-106a, is essential for the maintenance of NPCs’ proliferation and the repression of NSC’s differentiation. Moreover, we showed that Tp53inp1 and Cdkn1a are direct targets of miR-106b. Finally, the loss of function approach for either Tp53inp1 or Cdkn1a partially compromised the influence of forced miR-106b downregulation on the proliferation and differentiation of NPCs. Hence, our results highlight miR-106b as a key regulator to control the proliferation-differentiation balance of NPCs via Tp53inp1-Tp53-Cdkn1a axis.
Methods
Animal maintenance and use
C57BL/6J mice were housed and maintained in the Comparative Medicine Facility of the Tongji University School of Medicine (Shanghai, China). All procedures were conducted in accordance with the protocols approved by the Institutional Animal Care and Use Committee at the Tongji University School of Medicine.
NPCs’ isolation, enrichment, and differentiation
Cortex from embryonic day 14 (E14) mice were dissected and dissociated as previously described. To generate neurospheres, dissociated E14 cortical cells were cultured for 3–4 days in NSC proliferation medium, containing NeuroCult® NSC Basal Medium (Stem Cell Technologies), NeuroCult® NSC Proliferation Supplements (Stem Cell Technologies), 20 ng/mL bFGF (BioWalkersville), 20 ng/mL EGF (BioWalkersville), 2 μg/mL heparin (Sigma), N2 supplement, 2 mM l-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin. Neurospheres were collected, dissociated, and resuspended into single cells for a second-round neurosphere formation in suspension culture. After 3 rounds of selection and enrichment, neurosphere dissociates were cultured in suspension culture for NPCs’ proliferation experiments. For the NPCs’ differentiation experiments, neurosphere dissociates were cultured on Matrigel-coated 6-well plates or coverslips with NPCs’ differentiation medium, containing DMEM/F12, N2 supplement, 2% Knockout serum, 2 mM l-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin. Both NPCs’ proliferation and differentiation experiments were terminated on 3 days after plating.
miRNA mimics/inhibitor, Tp53inp1/Cdkn1a siRNA, and transfection
The mimics control, miR-106b mimics, inhibitor control, anti-miR-106b inhibitor, scrambled siRNA control, Tp53inp1 siRNA, and Cdkn1a siRNA were purchased from GenePharma (GenePharma Co., Ltd., Shanghai). Transfection of miRNA mimics/inhibitor or siRNAs was performed using the Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer’s instruction.
miRNA antagomir and administration
The antagomir negative control and antagomir-106b were purchased from GenePharma (GenePharma Co., Ltd., Shanghai). Fifty microliters of 100 μM antagomirs (antagomir negative control or antagomir-106b) was administrated intraperitoneally.
Quantitative polymerase chain reaction
The mRNA and miRNA were isolated from cell and tissue samples using miRCURY RNA isolation kit (Exiqon, Woburn, MA). cDNA was synthesized using miScript II RT kit (Qiagen, Valencia, CA). Transcripts were amplified using SYBR green PCR kit (Qiagen, Valencia, CA) with the ABI7500 (Applied Biosystems, Waltham, MA). Sequences of transcript-specific primers are given in Additional file 1: Table S1. All qPCR results measured each sample in triplicate, and no-template blanks were used for negative controls. Amplification curves and gene expressions were normalized to the house-keeping gene Gapdh (for mRNA) and U6 snRNA (for miRNA).
Immunofluorescence analysis
Immunofluorescence analysis for specific proteins was carried out as previously described [18]. Samples were incubated in primary antibody solutions (specifications shown in Additional file 1: Table S2) overnight at 4 °C. After washing with 1× PBS, samples were incubated with secondary antibodies (Cy3 or FITC) for 2 h at room temperature (RT). Samples were mounted using VectaShield (Vector Laboratories, Burlingame, CA), and images were taken using a Zeiss AX10 fluorescence microscope accompanied with ZEN 2.3 (blue edition) software. For quantification of the percentage of specific cell types in each experiment, the numbers of cell type-specific antigen-positive cells were counted in 15 randomly selected fields in 3 coverslips (5 fields each).
TUNEL assay
Terminal deoxynucleotidyltransferase dUTP nick end labeling (TUNEL) was done on cultured cells using the In Situ Cell Death Detection Kit, TMR red (Sigma). Experiments were handled following the manufacturer’s protocol. Briefly, cells cultured on cover slips were fixed in 4% paraformaldehyde (PFA) at RT for 15 min. After two times washing with PBS, the cover slips were treated with a mixture of Label Solution and Enzyme Solution (10:1) for 1 h at 37 °C. Label Solution treatment was used as negative control. DNase I recombinant (1000 U/ml)-treated cover slips (10 min incubation at RT) were used as positive control. The cover slips were then washed twice in PBS. The cover slips were mounted using VectaShield (Vector Laboratories, Burlingame, CA) and processed to microscopy using a Zeiss AX10 fluorescence microscope accompanied with ZEN 2.3 (blue edition) software.
EdU incorporation assay
DNA synthesis was performed by a Click-iT® EdU Imaging Kits (Thermo Scientific, #C10340) according to the manufacturer’s instructions. For in vivo studies, EdU was administrated intraperitoneally 8 h prior to sacrificing animals. For in vitro studies, NPCs were seeded on 6-well-plates (5 × 105 cells/well) and cultured in NSC proliferation medium for 3 days. EdU was added 6 h prior to fixation. Cryostat tissue sections and cultured cells were fixed with 4% PFA for 15 min at RT, followed by permeabilization step using 0.5% Triton X-100. After permeabilization, the cells were incubated with Click-iT® reaction cocktails for 30 min in dark room. Subsequently, the cells were mounted using VectaShield (Vector Laboratories, Burlingame, CA). Images were taken using Zeiss AX10 fluorescence microscope and AxioVision Rel. 4.8 software.
Dual-Luciferase Reporter Assay
The Cdkn1a 3′UTR and Cdkn1a-mut 3′UTR were synthesized by Genewiz (Genewiz, Suzhou, China) and cloned into the PmeI and SacI site of the pmirGLO vector (Promega, Beijing, China), downstream of the firefly luciferase gene. For the luciferase assay, 3,000 293T cells were cultured in 96-well plates with DMEM, 10% FBS, 100 μg/ml streptomycin, and 100 U/ml penicillin. After the confluency reached ~ 70%, the cells were co-transfected with the miR-106b mimics and either the Cdkn1a 3′UTR or Cdkn1a-mut 3′UTR dual-luciferase reporter vector. Serum-free Opti-MEM was used to prepare the transfection solution, and Lipofectamine 2000 reagent (Invitrogen) was used to facilitate the transfection according to the manufacturer’s instruction. At 24 h post-transfection, Dual-Luciferase® Reporter Assay System (Promega Corporation, Beijing, China) was used to determine the luciferase activities on SpectraMax M5 microplate readers (Molecular Devices). The activity of Renilla luciferase was used to normalize that of firefly luciferase.
Gene Ontology analysis
Mouse miR-106b predicted target genes for Gene Ontology (GO) analyses were extracted from Targetscan.org (http://www.targetscan.org/vert_72/). DAVID bioinformatics platform (david.ncifcrf.gov/home.jsp) and Panther Classification System (http://www.geneontology.org/) were used for GO analyses. Mus musculus genome data was used as annotation background. Biological_Process was selected as Functional_Database for gene function classification. Minimum and maximum numbers of genes in the category were set at 2 and 1000, respectively. Benjamini and Hochberg multiple test adjustment was used to adjust P value of analysis: P value < 0.05 was considered a significantly enriched pathway.
Statistical analysis
Statistical analysis was performed using unpaired two-tail t test for pairwise comparisons (GraphPad Prism Software). Data were shown as mean ± SD, and P values < 0.05 were considered significant.
Results
miR-106b is highly expressed in NPCs
In order to identify candidate miRNAs that may be involved in the regulation of NPCs, we first determined the temporal expression patterns of 44 highly conserved miRNAs during cortical development by qPCR (Fig. 1a). Among these miRNAs, the expression levels of miR-17~106 family members (labeled as red) showed significant reduction in adult stage, compared with developmental stages, except that of miR-19a (Fig. 1b). The sequence alignment suggested that miR-17, miR-106a, miR-106b, miR-20a, and miR-93 share same seed sequence, while other miRNAs in this family were lack of similarity in sequence, implying those 5 miRNAs may be key factors for achieving the function of miR-17~106 family in development (Fig. 1c). Expression analysis further identified two miRNAs (miR-17 and miR-106b) in this family which showed more than 10-fold decrease in their levels during cortical development (Fig. 1d). Although the effects of miR-17 and miR-17~92 cluster in the regulation of NPCs and in CNS development were well investigated [15–17, 19], it remained largely unknown whether or not miR-106b exhibit similar function as miR-17. Our results suggested that the expression of miR-106b could be detected from early developing cortex (E14) (Fig. 1e). Afterwards, their expression decreased steadily, reaching their minimum levels in the adult stage. The temporal expression patterns of miR-106b were positively correlated with the decline of NPC pool during cortical development, confirmed by the expression transcripts corresponding to NPCs and proliferation markers Nestin and Ki67, respectively, implying the relationship of miR-106b expression with the regulation of NPCs (Fig. 1e). To confirm that miR-106b is also abundantly expressed in NPCs, we used embryonic Nestin:EGFP mice [20] and to sorted GFP+ and GFP− cells from the brain (Fig. 1f). qPCR analysis demonstrated that the expression levels of miR-106b are significantly higher in GFP+ cells than GFP− ones, suggesting the enrichment of miR-106b in Nestin+ NPCs.
Fig. 1.
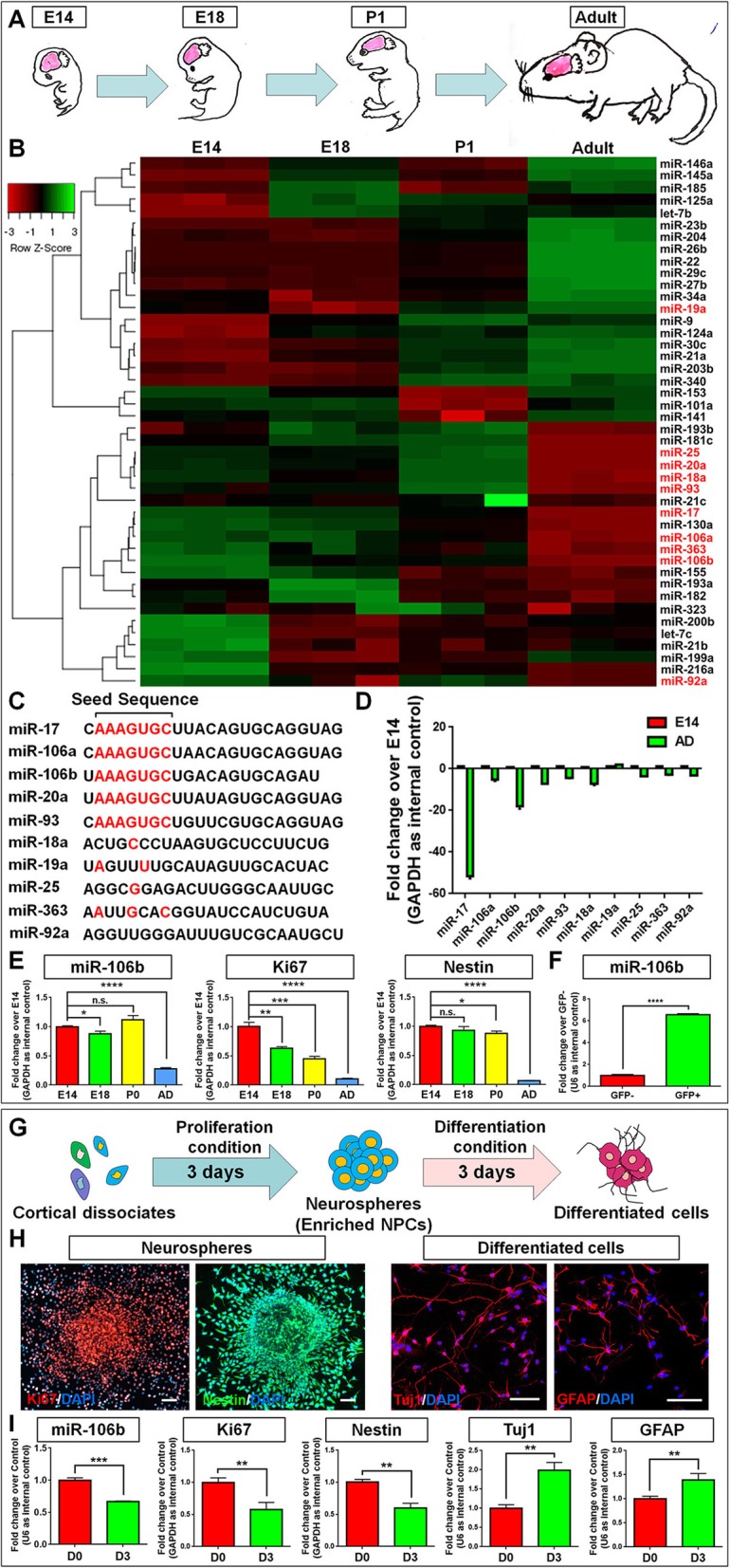
The temporal expression patterns of miR-106b correspond with the decline of proliferating NPC pool. a A schematic representation of the sample collection during brain development. b Hierarchical cluster analysis of 44 miRNAs expressed during cortical development. c Sequence comparison of miRNAs encoded by the miR-17~106 family. d qPCR analysis of each miRNA in miR-17~106 family in E14 and adult mouse cortexes. e The expression levels of the miR-106b decreased with time during brain development, positively correlated with that of transcripts corresponding to proliferating NPC markers, Nestin and Ki67. f qPCR analysis of expression levels of miR-106b in Nestin-GFP+ and Nestin-GFP− cells. g A schematic representation of the enrichment and differentiation of NPCs. h The enrichment and differentiation of NPCs was confirmed by immunoreactivities corresponding to NPCs (Nestin/Ki67) and differentiated cells (Tuj1/GFAP), respectively. i The expression levels of miR-106b decreased during NPCs’ differentiation, positively correlated with that of transcripts corresponding to Nestin and Ki67. Scale bar, 20 μm (g). Data are mean ± SD. ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ∗∗p < 0.01, and ∗p < 0.05
We next determined the expression patterns of miR-106b in NPCs and differentiated cells in vitro (Fig. 1g). E14 cortical dissociates were cultured in the presence of EGF and FGF2, and neurospheres enriched in Ki67+/Nestin + cells were generated 3 days after plating, suggesting the enrichment of NPCs (Fig. 1h). NPCs, cultured in differentiation conditions for 3 days, differentiated into Tuj1+ neuronal and GFAP+ glial cells. qPCR results suggested that miR-106b were abundantly expressed in NPCs but with low expression in differentiated cells, sharing the same expression patterns of transcripts corresponding to Nestin and Ki67 (Fig. 1i). Hence, the in vivo and in vitro studies demonstrated a corresponding positive correlation of miR-106b expression with the maintenance of NPCs, suggesting its functional involvement in the regulation of NPCs.
miR-106b facilitates the proliferation and self-renewal of NPCs
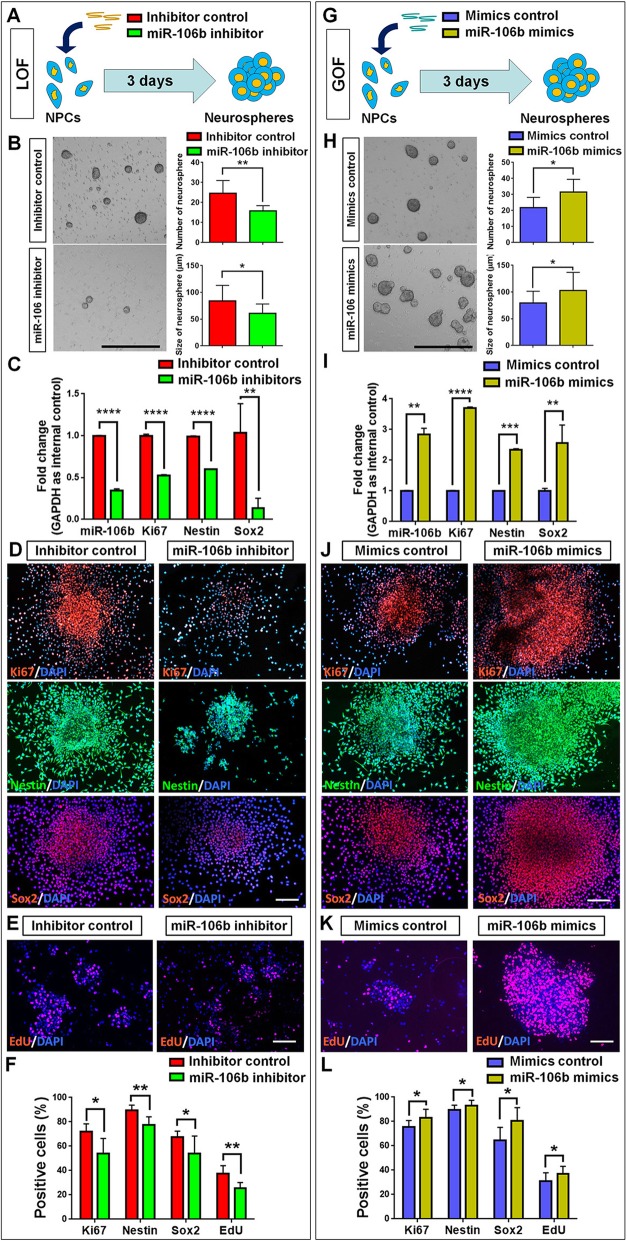
To understand the roles of miR-106 in the regulation of NPCs, we firstly investigated the involvement of miR-106b in the proliferation of NPCs. Both miR-106b loss-of-function (LOF) and gain-of-function (GOF) approaches were carried out using miR-106b-specific inhibitor and mimics, respectively. In the miR-106b LOF approach, NPCs were transfected with either miR-106b inhibitor (=LOF group) or inhibitor control and cultured in proliferation conditions for 3 days in vitro (DIV) (Fig. 2a). The efficiency of transfection is validated by qRT-PCR, where significant downregulation of miR-106 expression levels was observed in miR-106b LOF group, compared to controls (Fig. 2c). We observed a significant decrease in the number and size of neurospheres, demonstrating reduced proliferation and self-renewal of NPCs in the miR-106b LOF group, versus controls (Fig. 2b). Additionally, qRT-PCR analysis revealed a significant decline in the expression levels of transcripts corresponding to proliferation- and NPC-specific markers Ki67 and Nestin/Sox2, respectively (Fig. 2c). A similar effect of miR-106b was observed on the proliferation capacity of NPCs. The proportions of cells expressing immunoreactivities corresponding to Ki67, Nestin, and Sox2 were significantly decreased in the miR-106b LOF group, compared to controls (Fig. 2d, f). The immunocytochemical analysis was corroborated with EdU (5-ethynyl-2-deoxyuridine) assay, that the proportion of EdU+ cells was significantly reduced in the miR-106b LOF group, versus controls, suggesting miR-106b LOF represses the proliferation of NPCs (Fig. 2e, f).
Fig. 2.
miR-106b facilitates the proliferation of NPCs. a A schematic representation of miR-106b LOF approach. b The number and size of neurospheres were quantified in the miR-106b LOF group, compared to controls. c qPCR analysis of expression levels of miR-106b and transcripts corresponding to markers of proliferating cells (Ki67) and NPCs (Nestin/Sox2) in the miR-106b LOF group, compared to controls. d Immunofluorescence analysis of transduced cells displaying proliferating cell (Ki67/EdU)- and NPC (Nestin/Sox2)-specific immunoreactivities. e Immunofluorescence analysis of transduced cells displaying EdU immunoreactivities. f Quantification of cells displaying immunoreactivities corresponding to proliferating cells and NPCs in the miR-106b LOF group, compared to controls. g A schematic representation of miR-106b GOF approach. h The number and size of neurospheres were quantified in the miR-106b GOF group, compared to controls. i qPCR analysis of expression levels of miR-106b and transcripts corresponding to markers of proliferating cells (Ki67) and NPCs (Nestin/Sox2) in the miR-106b GOF group, compared to controls. j Immunofluorescence analysis of transduced cells displaying proliferating cell (Ki67/EdU)- and NPC (Nestin/Sox2)-specific immunoreactivities. k Immunofluorescence analysis of transduced cells displaying EdU immunoreactivities. l Quantification of cells displaying immunoreactivities corresponding to proliferating cells and NPCs in the miR-106b GOF group, compared to controls. Scale bar, 400 μm (b, h) and 50 μm (d, e, j, k). Data are mean ± SD. ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ∗∗p < 0.01, and ∗p < 0.05. Experiments were carried out three times in triplicates for in vitro perturbation
miR-106b GOF was carried out using same strategy of miR-106b LOF, where NPCs were transfected with either miR-106b mimics (=GOF group) or mimics control and cultured in proliferation conditions for 3 DIV (Fig. 2g). qRT-PCR analysis showed a significant increase in the levels of miR-106b, validating the transfection (Fig. 2i). In contrast to miR-106b LOF, ectopic expression of miR-106b accelerated the proliferation and self-renewal of NPCs, which led to more and bigger neurospheres; the upregulation of Ki67, Nestin, and Sox2 transcript levels; and the elevation of proportions of cells with Ki67/Nestin/Sox2/EdU-specific immunoreactivities (Fig. 2h–l). Thus, our results indicated that miR-106b positively regulates the proliferation of NPCs.
In order to examine whether or not the effects of miR-106b were unique, we next examined the involvement of miR-106b paralog, miR-106a, in the proliferation of NPCs. The efficiency for miR-106a LOF and GOF was validated by qRT-PCR (Additional file 1: Figure S1C, I). Unlike miR-106b, the LOF and GOF of miR-106a did not significantly change the numbers and size of neurospheres, generated from NPCs (Additional file 1: Figure S1A, B, G, H). qRT-PCR analysis revealed an inverse correlation in the expression levels of miR-106a and transcripts corresponding to Ki67, Nestin, and Sox2 (Additional file 1: Figure S1C, I). However, the immunofluorescence analysis suggested no significant difference in the proportions of cells with Ki67/Nestin/Sox2/EdU-specific immunoreactivities in both miR-106a LOF and GOF groups versus controls, although a significant decline was observed in the proportions of cells with Ki67- and Sox2-specific immunoreactivities in miR-106a LOF group (Additional file 1: Figure S1D-F, J-L). Thus, our observations suggested that miR-106b, but not miR-106a, may serve as a master regulator to maintain the stemness of NPCs.
miR-106b inhibits the differentiation of NPCs
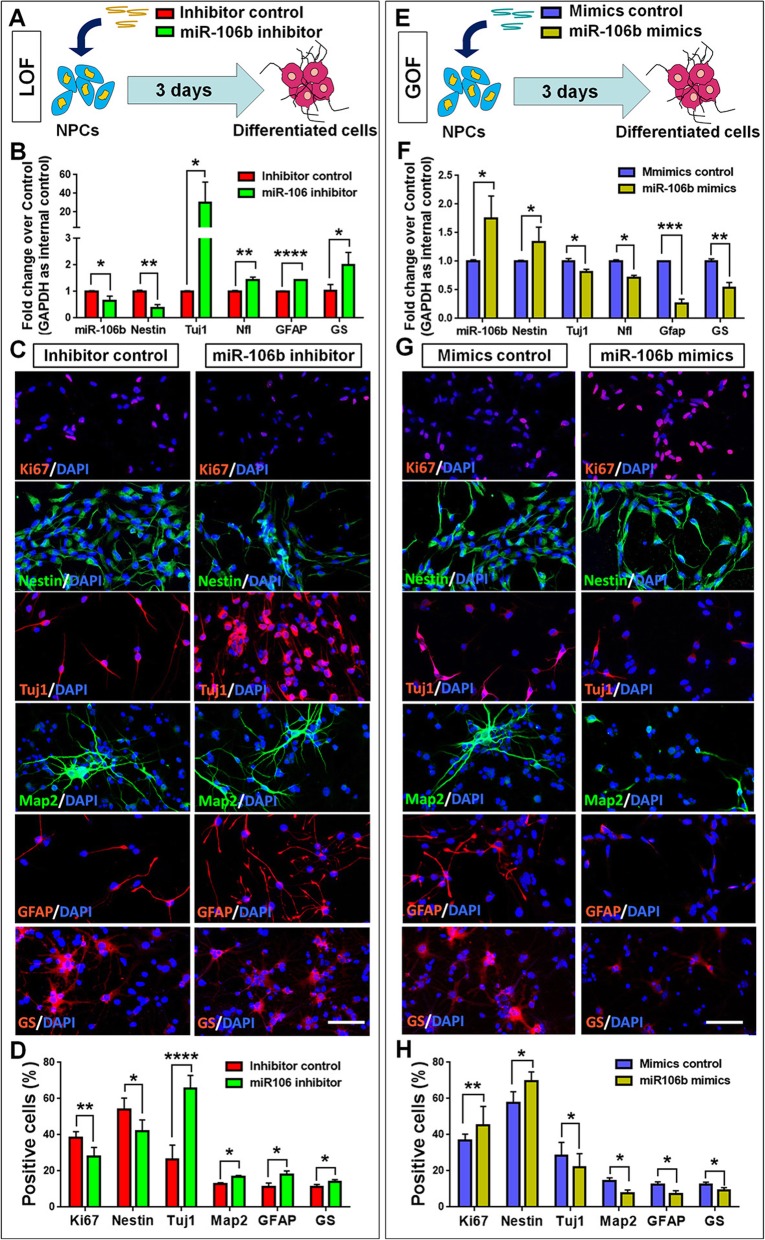
Next, we examined the roles of miR-106b in the differentiation of NPCs. Similar to previous studies, we carried out LOF and GOF approaches to address the involvement of miR-106b in the differentiation conditions. NPCs were firstly transfected with either miR-106b inhibitor (=LOF group) or inhibitor control and cultured in differentiation conditions for 3 DIV (Fig. 3a). qRT-PCR analysis revealed a significant reduction of miR-106b levels, validating the transfection (Fig. 3b). We also observed downregulation of Nestin transcripts expression, together with the increase of expression levels of transcripts corresponding to neuronal- and glial-specific markers Tuj1 and GFAP, respectively (Fig. 3b). qRT-PCR results were corroborated by immunofluorescence analysis, where the proportions of cells immunoreactive for Ki67 and Nestin were reduced significantly while that of cells immunoreactive for Tuj1 and GFAP were significantly increased, confirming that miR-106b LOF promoted the differentiation of NPCs (Fig. 3c, d).
Fig. 3.
miR-106b inhibits the differentiation of NPCs. a A schematic representation of miR-106b LOF approach. b qPCR analysis of expression levels of miR-106b and transcripts corresponding to markers of NPCs (Nestin) and differentiated cells (Tuj1/GFAP) in the miR-106b LOF groups, compared to controls. c Immunofluorescence analysis of transduced cells displaying proliferating NPC (Ki67/Nestin)- and differentiated cell (Tuj1/GFAP)-specific immunoreactivities. d Quantification of cells displaying immunoreactivities corresponding to proliferating NPCs and differentiated cells in the miR-106b LOF group, compared to controls. e A schematic representation of miR-106b GOF approach. f qPCR analysis of expression levels of miR-106b and transcripts corresponding to markers of NPCs (Nestin) and differentiated cells (Tuj1/GFAP) in the miR-106b GOF group, compared to controls. g Immunofluorescence analysis of transduced cells displaying proliferating NPC (Ki67/Nestin)- and differentiated cell (Tuj1/GFAP)-specific immunoreactivities. h Quantification of cells displaying immunoreactivities corresponding to proliferating NPCs and differentiated cells in the miR-106b GOF group, compared to controls. Scale bar, 50 μm (c, g). Data are mean ± SD. ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ∗∗p < 0.01, and ∗p < 0.05. Experiments were carried out three times in triplicates for in vitro perturbation
In the miR-106b GOF approach, NPCs were transfected with either miR-106b mimics or inhibitor control and cultured in differentiation conditions for 3 DIV (Fig. 3e). In contrast to the results obtained by the LOF approach, the ectopic expression of miR-106b significantly inhibited the differentiation of both neuronal and glial cells, as ascertained by significant decreases in levels of transcripts corresponding to cell type-specific markers and the number of cells displaying cell-type-specific immunoreactivities (Fig. 3f–h). Therefore, both LOF and GOF studies demonstrated that miR-106b negatively regulated the differentiation of NPCs, regardless of neuronal and glial lineages.
Our results revealed that the function of miR-106b is different from that of miR-17 which regulates the neurogliogenic decision. To examine whether miR-106b paralog, miR-106a, has similar function, we tested the effects of miR-106a in NPCs’ differentiation using the same LOF and GOF approaches in differentiation conditions. The upregulation or downregulation of miR-106a levels in NPCs did not significantly regulate the differentiation of NPCs, confirmed by qRT-PCR and immunofluorescence analyses (Additional file 1: Figure S2). Hence, our observations indicated that miR-106b, but not miR-106a, significantly regulates the proliferation and differentiation of NPCs, suggesting that miR-106b may play a central role in the regulation of NPCs, compared with its paralogs.
miR-106b does not regulate the neuronal subtype specification of NPCs
To further examine the influence of miR-106b on the cell fate determination of NPCs, especially on the neuronal subtype specification, we carried out long-term differentiation (14 DIV) of NPCs for sufficient cell fate commitment and maturation of differentiated cells under both miR-106b LOF and GOF conditions. Similar to our observations in short-term differentiation study, extended culture did not affect the commitment of neuronal and glial lineages. The generation of neurons and astrocytes was equally enhanced and inhibited in miR-106b LOF and GOF conditions, respectively (Additional file 1: Figure S3). The immunofluorescence analysis using neuronal subtype-specific antibodies corresponding to glutamatergic neurons (vGlut), GABAergic neurons (Gaba), and cholinergic neurons (ChAT) revealed that miR-106b had no preference in regulating the differentiation of neuronal subtypes, which was validated by the coincident patterns of the proportions of each cell type in both miR-106b LOF and GOF conditions (Additional file 1: Figure S3).
miR-106b has no effects on the survival of NPCs
To examine the contribution of apoptosis in the effects of miR-106 in NPCs’ regulation, we carried out terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay, detecting DNA fragmentation by labeling the terminal end of nucleic acids. The cell counting results revealed similar proportions of TUNEL+ cells either in miR-106b mimics- or in miR-106b inhibitor-treated groups, compared with controls, when NPCs were cultured in proliferation medium (Additional file 1: Figure S4A, B). Same results were observed during NPCs’ differentiation, that the modification of miR-106b expression levels did not affect the survival of differentiated cells (Additional file 1: Figure S4C, D). Similar results were obtained that there was no significant difference in the proportions of TUNEL+ cells either in miR-106a mimics- or in miR-106a inhibitor-treated groups versus controls in both proliferation and differentiation conditions (Additional file 1: Figure S5). Thus, our results suggested that both miR-106b and miR-106a had no significant effects on the survival of NPCs in both the proliferation and differentiation conditions.
miR-106b regulates the maintenance of NPC pool in vivo
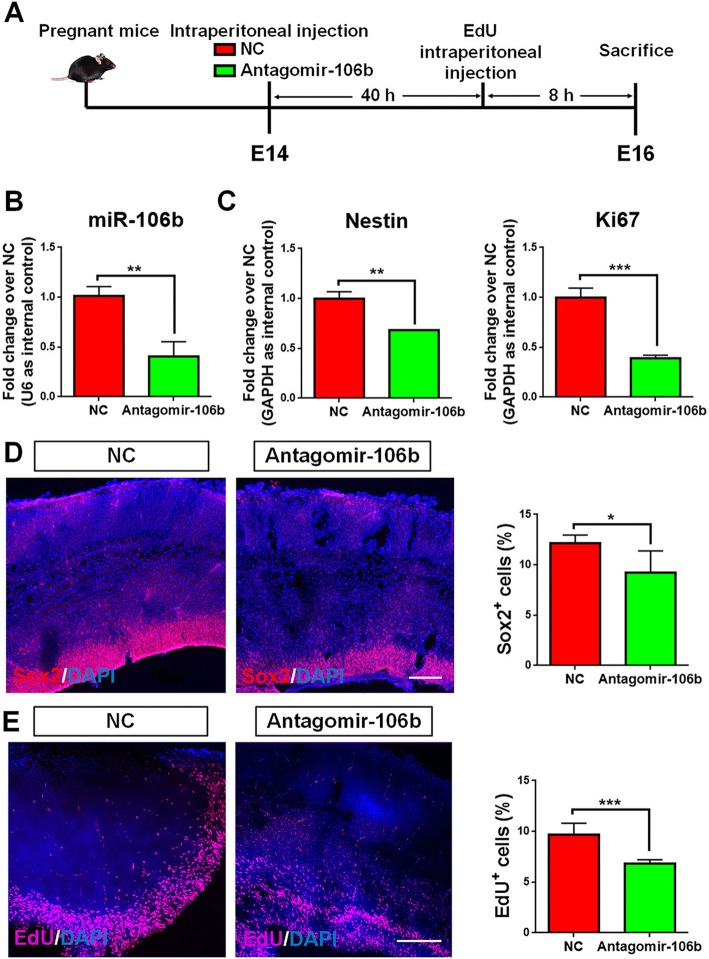
Our in vitro studies suggested the importance of miR-106b in the maintenance of NPCs’ proliferation and stemness. To further validate our observations, we carried out miR-106b knockdown in E14 C57BL/6J mice (Fig. 4a). Either negative control (NC) or antagomir-106b (=LOF group) was administrated into pregnant mice intraperitoneally, and animals were sacrificed after 48 h. The expression of miR-106b in the cortex of E14 mice was repressed in the miR-106b LOF group, versus controls, indicating the successful knockdown of miR-106b in vivo (Fig. 4b). qPCR results revealed a significant reduction for the expression of transcripts corresponding to Nestin and Ki67, in the miR-106b LOF group, compared to controls (Fig. 4c). The miR-106b LOF group also exhibited a significant decrease in the proportions of cells displaying Sox2 and EdU immunoreactivities, when compared to NC group (Fig. 4d, e). Besides, the miR-106b LOF enhanced the differentiation of NPCs, ascertained by the increase of Tuj1 and GS transcripts in the miR-106b LOF group, versus controls (Additional file 1: Figure S6). Thus, our results suggested that miR-106b plays an important role in the maintenance of NPC pool in vivo.
Fig. 4.
miR-106b regulates the maintenance of NPCs in vivo. a A schematic representation of miR-106b LOF approach in vivo. b qPCR analysis of expression levels of miR-106b in the miR-106b LOF and control groups, compared to their respective controls. c qPCR analysis of transcripts corresponding to markers of NPCs (Nestin) and proliferating cells (Ki67) in the miR-106b LOF and control groups. (d, e) Immunofluorescence analysis and quantification of transduced cells displaying Sox2- (d) and EdU (e)-specific immunoreactivities. Scale bar, 10 μm (d, e). Data are mean ± SD. ∗∗∗p < 0.001, ∗∗p < 0.01, and ∗p < 0.05. Experiments were carried out three times in triplicates with 5–7 E14 embryos per group three times in triplicates for in vivo perturbation
Tp53inp1-Tp53-Cdkn1a axis is a target of miR-106b
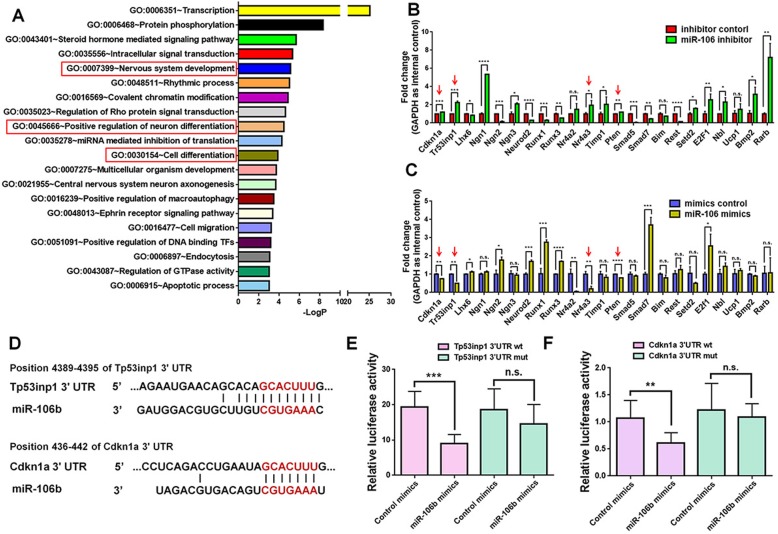
To investigate the underlying mechanisms of miR-106b-mediated regulation of NPCs, we firstly extracted the predicted targets of miR-106b from Targetscan.org, a miRNA target prediction database. Over 900 transcripts exhibited conserved miR-106b target sites on their 3′UTR. In order to identify the putative candidates of miR-106b, Gene Ontology (GO) analysis of the predicted targets was carried out. The highest enriched GO terms, sorted out by the DAVID bioinformatics platform and Panther Classification System based on the biological function of genes, demonstrated the predicted targets of miR-106b were abundantly clustered into the categories of Nervous system development (GO: 0007399), Positive regulation of neuron differentiation (GO: 0045666), and Cell differentiation (GO: 0030154) (Fig. 5a).
Fig. 5.
miR-106b targets Tp53inp1-Tp53-Cdkn1a axis in NPCs. a The GO analysis of top 20 enriched biological processes in the predicted miR-106b target genes. b, c qPCR analysis of candidate miR-106b target genes in the miR-106b LOF (b) and GOF (c) groups, compared to their respective controls. d The predicted consequential pairing of 3′UTR of candidate genes (top) and miR-106b (bottom) on the TargetScan website. e, f Repression of luciferase activities by the Tp53inp1 (e) and Cdkn1a (f) 3′UTR were dependent on miR-106b. Firefly luciferase activities were normalized to the internal control, Renilla luciferase activities. Data are mean ± SD. ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ∗∗p < 0.01, and ∗p < 0.05. Experiments were carried out three times in triplicates for in vitro perturbation
To select putative targets of miR-106b, we filtered out 23 genes, which were reported to regulate the proliferation or differentiation of stem cells, in above three GO terms. The qPCR analysis of these genes demonstrated that the expression levels of the transcripts corresponding to Tp53inp1, Cdkn1a, Nr4a3, and Pten were inversely correlated with that of miR-106b in both miR-106b LOF and GOF approaches in the proliferation conditions (Fig. 5b, c).
Recently, the Tp53inp1-Tp53-Cdkn1a axis was shown to regulate multiple cellular activities including proliferation and survival of tumor cells [21, 22]. Interestingly, two factors in the axis have miR-106b target sites, and their expression was negatively regulated by miR-106b, revealing this axis as potential key downstream pathways of miR-106b-mediated effects on NPCs. To confirm the interaction between miR-106b and Tp53inp1/Cdkn1a, Luciferase assay was carried out (Fig. 5d–f). Co-transfection of miR-106b mimics and Dual-Luciferase reporter constructs containing the wild-type 3′UTR of Tp53inp1 and Cdkn1a, but not miR-106b target site mutated 3′UTR of Tp53inp1 and Cdkn1a, significantly decreased the firefly activity in HEK293A cells, normalized by the Rellina activity, indicating miR-106b directly targets Tp53inp1 (Fig. 5e) and Cdkn1a (Fig. 5f).
miR-106b regulates the proliferation and differentiation of NPCs through Tp53inp1-Tp53-Cdkn1a axis
Since miR-106b directly targeted and regulated the expression of Tp53inp1 and Cdkn1a, we hypothesized that miR-106b might influence the proliferation and differentiation of NPCs through Tp53inp1-Tp53-Cdkn1a axis. To test our premise, we knocked down the expression of Tp53inp1 and Cdkn1a using siRNAs after miR-106b inhibitor treatment, where both Tp53inp1 and Cdkn1a expression was significantly upregulated. Four siRNAs targeting different sites of transcripts corresponding to Tp53inp1 and Cdkn1a were transfected into NPCs, and the siRNA silencing efficiency was examined by qPCR analysis 72 h post-transfection (Additional file 1: Figure S7). siRNAs showing the highest silencing efficiency were selected for the following studies.
First, we examined the roles of Tp53inp1 and Cdkn1a in the proliferation and differentiation of NPCs. The knockdown of either Tp53inp1 or Cdkn1a by siRNA significantly increased the numbers and sizes of neurospheres generated by NPCs, compared with controls (Additional file 1: Figure S8A-C, H-J). qRT-PCR analysis and immunocytochemical analyses revealed that either Tp53inp1 or Cdkn1a siRNA treatment significantly elevated the expression of Ki67, Nestin, and Sox2 in both transcripts and protein levels (Additional file 1: Figure S8D-G, K-N). Moreover, the Tp53inp1 and Cdkn1a siRNA treatment preserved the stemness of NPCs and significantly downregulated the differentiation capacity of NPCs into both neuronal and glial lineages, by examining the transcript and protein expression of markers for NPCs and differentiated cells through qRT-PCR and immunofluorescence analyses (Additional file 1: Figure S9). No significant effects were observed for Tp53inp1 and Cdkn1a on the survival of NPCs in both siRNA-treated groups versus controls (Additional file 1: Figure S10). Thus, in contrast with the effects of miR-106b, both Tp53inp1 and Cdkn1a inhibit the proliferation and accelerate the differentiation of NPCs.
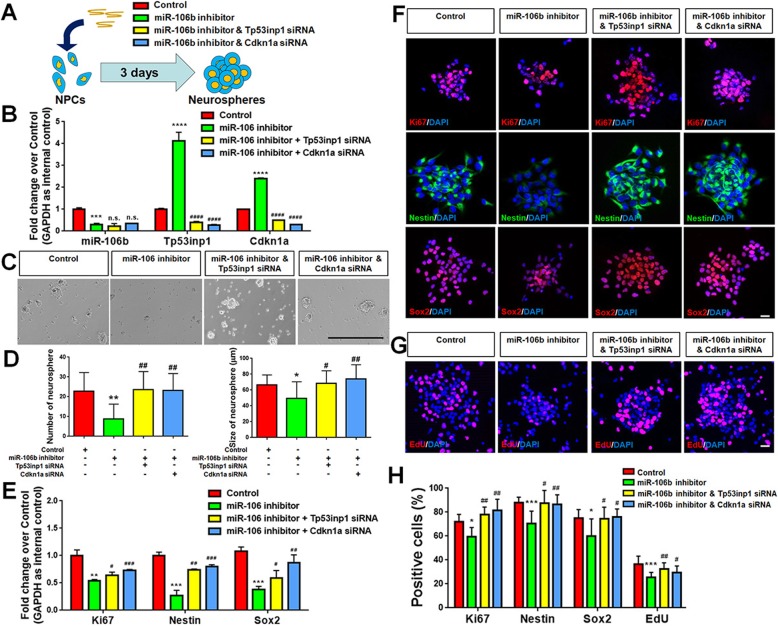
Second, we examined the roles of Tp53inp1 and Cdkn1a in the miR-106b-mediated regulation of NPCs’ proliferation (Fig. 6a). NPCs were divided into 4 groups based on the inhibitor and siRNA transfection: control group (inhibitor control + siRNA control), miR-106b LOF group (miR-106b inhibitor + siRNA control), miR-106b and Tp53inp1 LOF group (miR-106b inhibitor + Tp53inp1 siRNA), and miR-106b and Cdkn1a LOF group (miR-106b inhibitor + Cdkn1a siRNA). The transfected cells were cultured in proliferation condition for 3 DIV to generate neurospheres. The LOF of Tp53inp1 and Cdkn1a was validated by qPCR analysis (Fig. 6b). The negative influence of miR-106b knockdown on the number and size of neurospheres was abrogated in both Tp53inp1 and Cdkn1a LOF groups (Fig. 6c, d). We also observed a significant increase in the expression of transcripts corresponding to Ki67, Nestin, and Sox2 in Tp53inp1 and Cdkn1a LOF groups, versus miR-106b LOF group, suggesting the restorative effects of Tp53inp1 and Cdkn1a LOF on the proliferative capacity and stem cells properties of NPCs in miR-106b knockdown conditions (Fig. 6e). Our observations were corroborated with immunocytochemical analysis that the significant reduction of proportion of cells expressing immunoreactivities corresponding to Ki67, Nestin, Sox2, and EdU in miR-106b LOF groups was compromised by the silencing of Tp53inp1 and Cdkn1a expression (Fig. 6f–h). Thus, our results suggested Tp53inp1 and Cdkn1a as key downstream effectors of miR-106b in the regulation of NPCs’ proliferation.
Fig. 6.
a–h miR-106b promotes the proliferation of NPCs through Tp53inp1-Tp53-Cdkn1a axis. a A schematic representation of the experimental approach. b qPCR analysis of expression levels of miR-106b and transcripts corresponding to Tp53inp1 and Cdkn1a in all experimental groups versus controls. c, d The number and size of neurospheres were quantified in all experimental groups versus controls. e qPCR analysis of levels of miR-106b and transcripts corresponding to Ki67, Nestin, Sox2, Tp53inp1, and Cdkn1a in all experimental groups versus controls. f Immunofluorescence analysis of transduced cells displaying proliferating cells (Ki67)- and NPCs (Nestin/Sox2)-specific immunoreactivities in all experimental groups versus controls. g Immunofluorescence analysis of transduced cells displaying EdU immunoreactivities in all experimental groups versus controls. h Quantification of cells expressing Ki67/Nestin/Sox2/EdU immunoreactivities in all experimental groups versus controls. Scale bar, 400 μm (b) and 20 μm (e, f). Data are mean ± SD. ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ∗∗p < 0.01, and ∗p < 0.05. ####p < 0.0001, ###p < 0.001, ##p < 0.01, and #p < 0.05 versus the miR-106b LOF groups. Experiments were carried out three times in triplicates for in vitro perturbation
Third, we tested the roles of Tp53inp1 and Cdkn1a in the miR-106b-mediated regulation of NPCs’ differentiation. NPCs were divided into four groups using the same group setting in the proliferation study and cultured in differentiation conditions for 3 DIV (Fig. 7a). The transcript levels of Tp53inp1 and Cdkn1a were significantly reduced in siRNA treatment groups, versus controls, validating the transfection efficiency (Fig. 7b). Moreover, the inhibitory effects of miR-106b knockdown in the expression of transcripts corresponding to Nestin, Tuj1, and GFAP were compromised by the Tp53inp1 and Cdkn1a LOF (Fig. 7c). The restoration of NPCs’ differentiation was confirmed by the immunocytochemical analysis by examining the proportion of cells displaying immunoreactivities corresponding to NPCs (Ki67/Nestin), neuronal (Tuj1/Map 2), and glial (GFAP/GS) markers (Fig. 7d, e). We observed that the positive effects of miR-106b LOF on the differentiation of both neuronal cell and glia were abrogated by either Tp53inp1 or Cdkn1a LOF. Besides, the reduction of Ki67+/Nestin+ cells in the miR-106b LOF group was restored by either Tp53inp1 or Cdkn1a LOF. Hence, our observation suggested that miR-106b regulates the proliferation and differentiation of NPCs via Tp53inp1 and Cdkn1a.
Fig. 7.
miR-106b inhibits the differentiation of NPCs through Tp53inp1-Tp53-Cdkn1a axis. a A schematic representation of the experimental approach. b qPCR analysis of levels of miR-106b and transcripts corresponding to Tp53inp1 and Cdkn1a in all experimental groups versus controls. c qPCR analysis of levels of transcripts corresponding to Ki67, Tuj1, and GFAP in all experimental groups versus controls. d Immunofluorescence analysis of transduced cells displaying Ki67/Nestin and Tuj1/GFAP immunoreactivities in all experimental groups versus controls. e Quantification of cells expressing Ki67/Nestin and Tuj1/GFAP immunoreactivities in all experimental groups versus controls. f A schematic representation of miR-106b-mediated regulation of NPCs to facilitate proliferation: Tp53inp1-Tp53-Cdkn1a axis is a key inducer for cell cycle exit and differentiation of NPCs. The high levels of miR-106b inhibit Tp53inp1 and Cdkn1a expression, which maintains the proliferation of NPCs. The downregulation of miR-106b expression activates Tp53inp1-Tp53-Cdkn1a axis, leading to the decline of NPCs pool and the generation of both neurons and glia. Scale bar, 20 μm (d). Data are mean ± SD. ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ∗∗p < 0.01, and ∗p < 0.05. ####p < 0.0001, ###p < 0.001, ##p < 0.01, and #p < 0.05 versus the miR-106b LOF groups. Experiments were carried out three times in triplicates for in vitro perturbation
Discussion
The development of the vertebrate CNS is a highly conserved dynamic process that involves progression through distinct stages, starting from the formation of neural tube [23, 24]. Neurogenesis, especially the maintenance and differentiation of NPCs, plays a central role in all these stages [25]. The impairment of neurogenesis during CNS development can cause severe developmental defects, such as malformations and amentia [24]. Identification of key factors in the regulation of neurogenesis is essential to fully understand the CNS development, which will benefit from preventing and treating developmental defects. Recently, miR-17~106 family was reported to regulate neurogenesis of mouse developing/adult NPCs or ESC-derived NPCs [8, 15, 17]. Publications from multiple independent groups have shown that miR-17~106 family serves as a cell cycle facilitator, mostly in cancer cells [10, 26, 27]. However, conflicting observations were also reported by Zhi et al. that miR-106a might negatively regulate proliferation through targeting FASTK in astrocytoma cells [28]. Besides the controversy for the effects of miR-17~106 family on the proliferation of tumor cells, conflicting results were reported for that of miR-17~106 family on the regulation of NPCs [8, 15]. In 2011, Brett et al. showed that miR-106b~25 cluster can promote the neuronal differentiation of adult NPCs. However, in 2013, Bian et al. demonstrated the miR-17~106 family inhibits the transition of NPCs to intermediate progenitors, which may block the generation of neuronal cells. Thus, these contradictory findings suggest two propositions: (1) the function of miR-17~106 family, including miR-106, could be tissue- and cell type-specific in regulating proliferation and differentiation and (2) the roles of individual miRNAs in the family could be diverse although they may share similar seed sequence.
Here, we examined the involvement of miR-106a/b in the regulation of NPCs. We observed that, during brain development, the expression of miR-106b decreased with time, coincided with that of Nestin and Ki67. Similar expression patterns were also observed during NPCs’ differentiation in vitro. It is most likely that under the decrease of miR-106b expression, the maintenance of NPCs was attenuated, as demonstrated by a significant reduction of levels of transcripts corresponding to NPCs. The perturbation of function approaches also revealed that miR-106b, but not miR-106a, is important in the regulation of proliferation and differentiation of embryonic NPCs. It has been reported that individual component of miRNA families could exhibit distinct functions in the regulation of cell fate [17, 29]. Thus, our observations match with these reports that the effects of miR-106a and miR-106b on the regulation NPCs are different, even though they belong to the same family and share the same seed sequence.
In the in vitro studies, we demonstrated for the first time that, miR-106b positively and negatively regulated embryonic NPC’s proliferation and differentiation, respectively. When subjected to proliferation, NPCs’ phenotype was maintained by high levels of miR-106b and significant reduction of NPC pool was observed once miR-106b levels were downregulated. This result was matched with others’ findings that miR-106b, together with other miR-17~106 family members, was tightly associated with the maintenance of NPCs [8, 15, 16]. When induced to differentiate, NPCs downregulated miR-106b and generated both neurons and glia. However, when the expression of miR-106b was perturbed, NPCs’ differentiation along neuronal and glial lineages was similarly compromised or enhanced. Our results suggested that miR-106b may serve as a proliferation “accelerator” and differentiation “break” and have no instructive effect in the commitment of certain lineages. It is highly possible that miR-106b may also be a key factor in regulating the balance of embryonic NPCs between proliferation and differentiation during brain development, due to the same positive correlation of the expression levels of miR-106b and the proportion of NPCs in vitro and in vivo, matching with Bian et al.’s observations.
The role of miR-106b in regulating proliferation and differentiation in extra-neural tissues is well known. As firstly investigated in cancer cells, miR-106b, together with all other miR-17~106 family members, is considered as an oncogenic miRNA [10–13]. miR-106b is reported to promote the tumor growth through targeting tumor suppressor genes, such as Pten, Cdkn1a, E2F1, Setd2, Runx3, Smad7, Cdkn1a, and Bim [10, 13, 26, 30–33]. Besides, miR-106b negatively regulates the differentiation of various types of non-neural cells. For instance, miR-106b suppresses the differentiation of brown adipocytes and osteoblast through targeting Ucp1 and Smad5, respectively [34, 35]. Thus, our observations matched with the situation in extra-neural tissues, suggesting a coincident function of miR-106b on diverse types of stem cells. Furthermore, our study identified Tp53inp1-Tp53-Cdkn1a axis as a key downstream regulatory network of miR-106b-mediated regulation of NPCs. Previously identified in cancer cells and mouse embryonic fibroblasts, Tp53inp1-Tp53-Cdkn1a axis is considered as a tumor suppressor pathway [21, 36, 37]. Tp53inp1, as a Tp53-induced nuclear protein, inhibits cell-cycle progression and promotes of apoptosis in a Tp53-dependent manner [21, 38]. Tp53inp1 also physically interacts with Tp53 and enhances its activity by Ser 46 phosphorylation, forming a positive feedback loop with Tp53 [21, 39]. The activation of Tp53inp1 and Tp53 leads to the elevation of Cdkn1a expression, resulting in cell-cycle arrest and proliferation reduction [40, 41]. In CNS, Tp53inp and Cdkn1a were also reported to regulate the expansion of postnatal and adult NPCs by regulating key genes for NPCs’ phenotype, such as Sox2 [16, 40, 42]. Hence, miR-106b-Tp53inp1-Tp53-Cdkn1a axis may act as an upstream pathway of Sox2-Lin28-let-7 axis, an essential molecular mechanism for NPC proliferation and neurogenic potential, which forms a complete and comprehensive network on the regulation of NPCs [43].
Conclusions
In summary, we demonstrated that miR-106b is highly expressed in embryonic NPCs. The decrease in miR-106b expression progressively shifts the balance toward NPCs’ differentiation. The mechanism involved is likely via the loss of miR-106b-mediated repression on the cell-cycle inhibitory network, Tp53inp-Tp53-Cdkn1a axis (Fig. 7f). Our findings demonstrate a unique master pathway on the regulation of NPCs, suggesting the necessity in exploring the exact roles and mechanisms underlying the effects of other miRNAs in miR-17~106 family, which is currently under investigation.
Additional file
Figure S1. miR-106a does not promote the proliferation of NPCs. Figure S2. miR-106a does not regulate the differentiation of NPCs. Figure S3. miR-106b has no effect on neuronal subtype specification. Figure S4. miR-106b has no effect on NPCs’ survival. Figure S5. miR-106a has no effect on NPCs’ survival. Figure S6. miR-106b regulates the differentiation of NPCs in vivo. Figure S7. The validation of Tp53inp1 and Cdkn1a siRNAs. Figure S8. Tp53inp1 and Cdkn1a siRNAs promote the proliferation of NPCs. Figure S9. Tp53inp1 and Cdkn1a siRNAs inhibit the differentiation of NPCs. Figure S10. Tp53inp1 and Cdkn1a have no effect on NPCs’ survival. Table S1. List of specific primers. Table S2. List of primary antibodies. (DOCX 4511 kb)
Acknowledgements
We kindly acknowledge Dr. Xinrui Qi, Ms. Wenping Cai, and Ms. Jie Zhu, who provided valuable comments and suggestions about the manuscript.
Abbreviations
- CNS
Central nervous system
- DIV
Days in vitro
- GOF
Gain-of-function
- LOF
Loss-of-function
- miRNA
MicroRNA
- NPCs
Neural stem/progenitor cells
- PFA
Paraformaldehyde
- RT
Room temperature
Authors’ contributions
JCZ and XX conceived and designed the experiments. XX, HL, CL, and XY performed the experiments. XX, HL, CL, YH, and YW analyzed the data. XX, HL, CL, and XY contributed reagents/materials/analysis tools. XX, YH, YW, and JCZ wrote the paper. All authors read and approved the final manuscript.
Funding
This work was funded by research grants from the State Key Program of the National Natural Science Foundation of China (No. 81830037 to JCZ); the National Basic Research Program of China (973 Program Grant No. 2014CB965001 to JCZ); Innovative Research Groups of the National Natural Science Foundation of China (No. 81221001 to JCZ); Joint Research Fund for Overseas Chinese, Hong Kong and Macao Young Scientists of the National Natural Science Foundation of China (No. 81329002 to JCZ); the National Institutes of Health (No. 1R01NS097195-01 to JCZ); Shanghai Sailing Program (No. 19YF1451700 to XX); and China Postdoctoral Science Foundation Grant (No. 2018 M642087 to XX).
Availability of data and materials
Data could be accessed through emails with the corresponding authors.
Ethics approval and consent to participate
All work involving animals was approved by the Institutional Animal Care and Use Committee of Tongji University.
Consent for publication
No human sample or data is used in the study.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Xiaohuan Xia, Email: xiaohuan_xia1@163.com.
Hongfang Lu, Email: lu_ashin@163.com.
Chunhong Li, Email: chlige@foxmail.com.
Yunlong Huang, Email: yhuan1@unmc.edu.
Yi Wang, Email: windyyiwang@foxmail.com.
Xiaoyu Yang, Email: yxy15316098979@163.com.
Jialin C. Zheng, Email: jialinzheng@tongji.edu.cn
References
- 1.Okano H, Temple S. Cell types to order: temporal specification of CNS stem cells. Curr Opin Neurobiol. 2009;19(2):112–119. doi: 10.1016/j.conb.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 2.Mori N, et al. Contributions of cell-extrinsic and cell-intrinsic factors to the differentiation of a neural-crest-derived neuroendocrine progenitor cell. Cold Spring Harb Symp Quant Biol. 1990;55:255–264. doi: 10.1101/SQB.1990.055.01.028. [DOI] [PubMed] [Google Scholar]
- 3.Gil-Perotin S, Casaccia-Bonnefil P. Extrinsic and intrinsic factors modulating proliferation and self-renewal of multipotential CNS progenitors and adult neural stem cells of the subventricular zone. In: Levison SW, editor. Mammalian subventricular zones: their roles in brain development, cell replacement and disease. Boston: Springer US; 2006. pp. 30–83. [Google Scholar]
- 4.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- 5.Blakaj A, Lin H. Piecing together the mosaic of early mammalian development through microRNAs. J Biol Chem. 2008;283(15):9505–9508. doi: 10.1074/jbc.R800002200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136(4):642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Trompeter HI, et al. MicroRNAs MiR-17, MiR-20a, and MiR-106b act in concert to modulate E2F activity on cell cycle arrest during neuronal lineage differentiation of USSC. PLoS One. 2011;6(1):e16138. doi: 10.1371/journal.pone.0016138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brett JO, et al. The microRNA cluster miR-106b~25 regulates adult neural stem/progenitor cell proliferation and neuronal differentiation. Aging (Albany NY) 2011;3(2):108–124. doi: 10.18632/aging.100285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mendell JT. miRiad roles for the miR-17-92 cluster in development and disease. Cell. 2008;133(2):217–222. doi: 10.1016/j.cell.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kan T, et al. The miR-106b-25 polycistron, activated by genomic amplification, functions as an oncogene by suppressing p21 and Bim. Gastroenterology. 2009;136(5):1689–1700. doi: 10.1053/j.gastro.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luo B, et al. Oncogene miR-106a promotes proliferation and metastasis of prostate cancer cells by directly targeting PTEN in vivo and in vitro. Minerva Med. 2018;109(1):24–30. doi: 10.23736/S0026-4806.17.05342-3. [DOI] [PubMed] [Google Scholar]
- 12.Wang Z, et al. Oncogenic miR-20a and miR-106a enhance the invasiveness of human glioma stem cells by directly targeting TIMP-2. Oncogene. 2015;34(11):1407–1419. doi: 10.1038/onc.2014.75. [DOI] [PubMed] [Google Scholar]
- 13.Smith AL, et al. The miR-106b-25 cluster targets Smad7, activates TGF-beta signaling, and induces EMT and tumor initiating cell characteristics downstream of Six1 in human breast cancer. Oncogene. 2012;31(50):5162–5171. doi: 10.1038/onc.2012.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Y, et al. The silencing effect of microRNA miR-17 on p21 maintains the neural progenitor pool in the developing cerebral cortex. Front Neurol. 2014;5:132. doi: 10.3389/fneur.2014.00132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bian S, et al. MicroRNA cluster miR-17-92 regulates neural stem cell expansion and transition to intermediate progenitors in the developing mouse neocortex. Cell Rep. 2013;3(5):1398–1406. doi: 10.1016/j.celrep.2013.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garg N, et al. microRNA-17-92 cluster is a direct Nanog target and controls neural stem cell through Trp53inp1. EMBO J. 2013;32(21):2819–2832. doi: 10.1038/emboj.2013.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Naka-Kaneda H, et al. The miR-17/106-p38 axis is a key regulator of the neurogenic-to-gliogenic transition in developing neural stem/progenitor cells. Proc Natl Acad Sci U S A. 2014;111(4):1604–1609. doi: 10.1073/pnas.1315567111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peng H, et al. Stromal cell-derived factor 1-mediated CXCR4 signaling in rat and human cortical neural progenitor cells. J Neurosci Res. 2004;76(1):35–50. doi: 10.1002/jnr.20045. [DOI] [PubMed] [Google Scholar]
- 19.Liu XS, et al. MicroRNA-17-92 cluster mediates the proliferation and survival of neural progenitor cells after stroke. J Biol Chem. 2013;288(18):12478–12488. doi: 10.1074/jbc.M112.449025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma K, et al. Direct conversion of mouse astrocytes into neural progenitor cells and specific lineages of neurons. Transl Neurodegener. 2018;7:29. doi: 10.1186/s40035-018-0132-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tomasini R, et al. TP53INP1s and homeodomain-interacting protein kinase-2 (HIPK2) are partners in regulating p53 activity. J Biol Chem. 2003;278(39):37722–37729. doi: 10.1074/jbc.M301979200. [DOI] [PubMed] [Google Scholar]
- 22.Jiang F, et al. MiR-125b promotes proliferation and migration of type II endometrial carcinoma cells through targeting TP53INP1 tumor suppressor in vitro and in vivo. BMC Cancer. 2011;11:425. doi: 10.1186/1471-2407-11-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rakic P. Evolution of the neocortex: a perspective from developmental biology. Nat Rev Neurosci. 2009;10(10):724–735. doi: 10.1038/nrn2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jessell TM, Sanes JR. Development. The decade of the developing brain. Curr Opin Neurobiol. 2000;10(5):599–611. doi: 10.1016/S0959-4388(00)00136-7. [DOI] [PubMed] [Google Scholar]
- 25.Bergstrom T, Forsberg-Nilsson K. Neural stem cells: brain building blocks and beyond. Ups J Med Sci. 2012;117(2):132–142. doi: 10.3109/03009734.2012.665096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ivanovska I, et al. MicroRNAs in the miR-106b family regulate p21/CDKN1A and promote cell cycle progression. Mol Cell Biol. 2008;28(7):2167–2174. doi: 10.1128/MCB.01977-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poliseno L, et al. Identification of the miR-106b~25 microRNA cluster as a proto-oncogenic PTEN-targeting intron that cooperates with its host gene MCM7 in transformation. Sci Signal. 2010;3(117):ra29. doi: 10.1126/scisignal.2000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhi F, et al. miR-106a-5p inhibits the proliferation and migration of astrocytoma cells and promotes apoptosis by targeting FASTK. PLoS One. 2013;8(8):e72390. doi: 10.1371/journal.pone.0072390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parsi S, et al. Preclinical evaluation of miR-15/107 family members as multifactorial drug targets for Alzheimer’s disease. Mol Ther Nucleic Acids. 2015;4:e256. doi: 10.1038/mtna.2015.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu Y, et al. MicroRNA-106b regulates the tumor suppressor RUNX3 in laryngeal carcinoma cells. FEBS Lett. 2013;587(19):3166–3174. doi: 10.1016/j.febslet.2013.08.024. [DOI] [PubMed] [Google Scholar]
- 31.Xiang W, et al. miR-106b-5p targets tumor suppressor gene SETD2 to inactive its function in clear cell renal cell carcinoma. Oncotarget. 2015;6(6):4066–4079. doi: 10.18632/oncotarget.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y, et al. Role of the miR-106b-25 microRNA cluster in hepatocellular carcinoma. Cancer Sci. 2009;100(7):1234–1242. doi: 10.1111/j.1349-7006.2009.01164.x. [DOI] [PubMed] [Google Scholar]
- 33.Zheng L, et al. MiR-106b induces cell radioresistance via the PTEN/PI3K/AKT pathways and p21 in colorectal cancer. J Transl Med. 2015;13:252. doi: 10.1186/s12967-015-0592-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fang T, et al. miR-106b-5p and miR-17-5p suppress osteogenic differentiation by targeting Smad5 and inhibit bone formation. Exp Cell Res. 2016;347(1):74–82. doi: 10.1016/j.yexcr.2016.07.010. [DOI] [PubMed] [Google Scholar]
- 35.Wu Y, et al. Identification of miR-106b-93 as a negative regulator of brown adipocyte differentiation. Biochem Biophys Res Commun. 2013;438(4):575–580. doi: 10.1016/j.bbrc.2013.08.016. [DOI] [PubMed] [Google Scholar]
- 36.Cano CE, et al. Tumor protein 53-induced nuclear protein 1 is a major mediator of p53 antioxidant function. Cancer Res. 2009;69(1):219–226. doi: 10.1158/0008-5472.CAN-08-2320. [DOI] [PubMed] [Google Scholar]
- 37.Narayanan BA, et al. p53/p21(WAF1/CIP1) expression and its possible role in G1 arrest and apoptosis in ellagic acid treated cancer cells. Cancer Lett. 1999;136(2):215–221. doi: 10.1016/S0304-3835(98)00323-1. [DOI] [PubMed] [Google Scholar]
- 38.Okamura S, et al. p53DINP1, a p53-inducible gene, regulates p53-dependent apoptosis. Mol Cell. 2001;8(1):85–94. doi: 10.1016/S1097-2765(01)00284-2. [DOI] [PubMed] [Google Scholar]
- 39.D'Orazi G, et al. Homeodomain-interacting protein kinase-2 phosphorylates p53 at Ser 46 and mediates apoptosis. Nat Cell Biol. 2002;4(1):11–19. doi: 10.1038/ncb714. [DOI] [PubMed] [Google Scholar]
- 40.Chen M, Pereira-Smith OM, Tominaga K. Loss of the chromatin regulator MRG15 limits neural stem/progenitor cell proliferation via increased expression of the p21 Cdk inhibitor. Stem Cell Res. 2011;7(1):75–88. doi: 10.1016/j.scr.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cayrol C, Knibiehler M, Ducommun B. p21 binding to PCNA causes G1 and G2 cell cycle arrest in p53-deficient cells. Oncogene. 1998;16(3):311–320. doi: 10.1038/sj.onc.1201543. [DOI] [PubMed] [Google Scholar]
- 42.Marques-Torrejon MA, et al. Cyclin-dependent kinase inhibitor p21 controls adult neural stem cell expansion by regulating Sox2 gene expression. Cell Stem Cell. 2013;12(1):88–100. doi: 10.1016/j.stem.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cimadamore F, et al. SOX2-LIN28/let-7 pathway regulates proliferation and neurogenesis in neural precursors. Proc Natl Acad Sci U S A. 2013;110(32):E3017–E3026. doi: 10.1073/pnas.1220176110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. miR-106a does not promote the proliferation of NPCs. Figure S2. miR-106a does not regulate the differentiation of NPCs. Figure S3. miR-106b has no effect on neuronal subtype specification. Figure S4. miR-106b has no effect on NPCs’ survival. Figure S5. miR-106a has no effect on NPCs’ survival. Figure S6. miR-106b regulates the differentiation of NPCs in vivo. Figure S7. The validation of Tp53inp1 and Cdkn1a siRNAs. Figure S8. Tp53inp1 and Cdkn1a siRNAs promote the proliferation of NPCs. Figure S9. Tp53inp1 and Cdkn1a siRNAs inhibit the differentiation of NPCs. Figure S10. Tp53inp1 and Cdkn1a have no effect on NPCs’ survival. Table S1. List of specific primers. Table S2. List of primary antibodies. (DOCX 4511 kb)
Data Availability Statement
Data could be accessed through emails with the corresponding authors.