Abstract
Maintaining salivary gland function is critical for oral health. Loss of saliva is a common side effect of therapeutic irradiation for head and neck cancer or autoimmune diseases such as Sjögren’s syndrome. There is no curative treatment, and current strategies proposed for functional regeneration include gene therapy to reengineer surviving salivary gland tissue, cell-based transplant therapy, use of bioengineered glands, and development of drugs/biologics to stimulate in vivo regeneration or increase secretion. Understanding the genetic and cellular mechanisms required for development and homeostasis of adult glands is essential to the success of these proposed treatments. Recent advances in genetic lineage tracing provide insight into epithelial lineage relationships during murine salivary gland development. During early fetal gland development, epithelial cells expressing keratin 14 (K14) Sox2, Sox9, Sox10, and Trp63 give rise to all adult epithelium, but as development proceeds, lineage restriction occurs, resulting in separate lineages of myoepithelial, ductal, and acinar cells in postnatal glands. Several niche signals have been identified that regulate epithelial development and lineage restriction. Fibroblast growth factor signaling is essential for gland development, and other important factors that influence epithelial patterning and maturation include the Wnt, Hedgehog, retinoic acid, and Hippo signaling pathways. In addition, other cell types in the local microenvironment, such as endothelial and neuronal cells, can influence epithelial development. Emerging evidence also suggests that specific epithelial cells will respond to different types of salivary gland damage, depending on the cause and severity of damage and the resulting damaged microenvironment. Understanding how regeneration occurs and which cell types are affected, as well as which signaling factors drive cell lineage decisions, provides specific targets to manipulate cell fate and improve regeneration. Taken together, these recent advances in understanding cell lineages and the signaling factors that drive cell fate changes provide a guide to develop novel regenerative treatments.
Keywords: lineage tracing, epithelial signaling, salivary gland development, regeneration, progenitor, submandibular gland
Introduction
Developing successful strategies for salivary regeneration requires identification of potent cell sources, signaling pathways involved in communication with their niche, and the cellular interactions essential for tissue homeostasis and regeneration. Major advances in our understanding of salivary gland (SG) progenitors are due to murine genetic lineage tracing, which has revealed lineage relationships during gland development and within postnatal glands. Recently, it was found that several partly overlapping duct populations give rise to and maintain duct cells (Kwak et al. 2016; May et al. 2018; Weng et al. 2018), whereas acinar cells self-duplicate during adult gland homeostasis (Aure, Konieczny, and Ovitt 2015; Emmerson et al. 2018). In addition, normally lineage-restricted cells become bipotent following injury (Weng et al. 2018), highlighting the in vivo plasticity of adult SG cells. This indicates that regenerative potential within cell populations may be influenced by changes in the surrounding microenvironment and secreted factors.
Although some paracrine factors have been identified that might prevent damage or improve regeneration, the signaling cross-talk that stimulates specific cell populations in postnatal glands is not well understood. Much of our knowledge about signaling pathways and progenitor populations derives from studies of murine fetal glands. Here, we review the recent literature and summarize major steps toward understanding epithelial cell lineage relationships and signaling within the developing murine SG that may have implications for functional regeneration of human SGs. We then review recent literature identifying progenitor populations and the signaling pathways involved in postnatal growth and homeostasis and how these respond to injury. Most of our knowledge is from studies on the murine submandibular gland (SMG), and there is much less information on parotid and sublingual glands (SLGs). A challenge for the field is understanding differences among the major glands, in terms of response to the type of damage (e.g., irradiation [IR] vs. immune or chemical/drug damage) and the specific mechanisms of regeneration after a particular type of damage.
Identifying Lineage Relationships during Development
Lineage tracing genetically labels putative progenitors and their progeny with a fluorescent protein or LacZ reporter. These experiments are dependent on the specificity and fidelity of the cell-specific promoters attached to Cre recombinase to permanently mark the cells of interest and their progeny, even if they may no longer express the specific marker (Kretzschmar and Watt 2012). This approach can be used to determine lineage relationships, long-term self-renewal potential, and cell behavior in vivo. The timing of the onset of Cre recombinase induction as well as the temporospatial expression pattern of the specific marker is critical for interpretation of lineage contribution (Fig. 1). Since induction of Cre expression is a permanent and heritable event, interpretation of the results must take into account potentially dynamic gene expression over time. Hence, Cre can be expressed in cells of interest at 1 time point and in other cells at a different time, which may complicate the results of lineage relationship analyses. This issue can be circumvented by using an inducible Cre system, which allows temporal and spatial control of Cre by selective induction in adult tissues via promoters also expressed during embryonic development.
Figure 1.
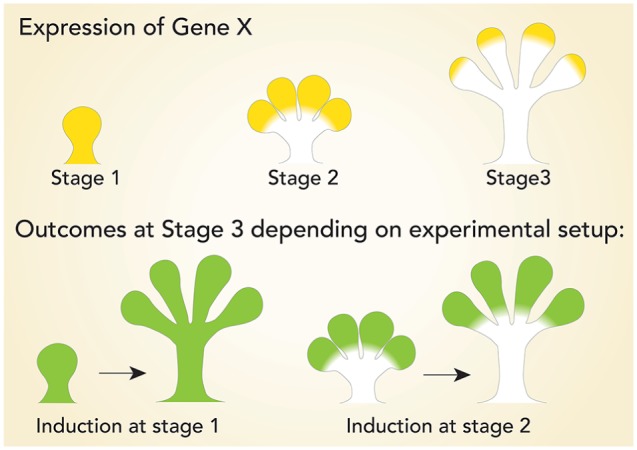
Using genetic models to investigate lineage relationships. Lineage tracing is dependent on knowing the temporospatial expression pattern of the gene of interest driving Cre expression. Expression of Gene-X has a dynamic stage-dependent localization pattern. Outcomes of lineage tracing of Gene-X are dependent on the experimental design. Inducing Gene-X-Cre at stage 1 (green) will lineage trace every cell at stage 3, while induction at stage 2 will lineage trace a subpopulation at stage 3. Both outcomes differ from the expression pattern of Gene-X at stage 3.
An example of a constitutively expressed Cre model in SGs is the Aquaporin 5-Cre (Aqp5-Cre-IRES-DsRed, called ACID) mouse line (Flodby et al. 2010). In this model, a Cre-IRES-DsRed cassette is knocked into the endogenous Aqp5 gene. Aqp5, a water channel, is a well-established marker for acinar cells in SGs and is detected when acinar differentiation begins at embryonic day 15 (E15; Larsen et al. 2011). To genetically label acinar cells, we crossed the ACID line with the mTmG reporter line (Muzumdar et al. 2007). This led to Cre induction and expression of the GFP reporter in acinar cells, while myoepithelial cells (MECs) and duct cells continue to express the red membrane-bound Tomato reporter (Fig. 2). Furthermore, lineage tracing in combination with injury models allows us to investigate in vivo progenitor behavior in a damaged microenvironment. These approaches are powerful tools to investigate signaling pathways involved in lineage contribution to regeneration after damage.
Figure 2.
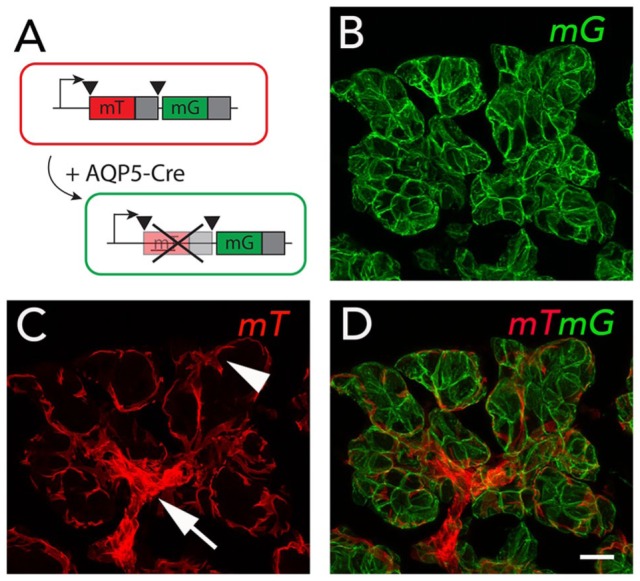
Genetic labeling of AQP5 lineage. (A) Crossing the ACID (Aqp5-Cre-IRES-DsRed) mouse line with the mTmG reporter line leads to constitutive expression of Cre in AQP5+ cells and their progeny. Cre induction mediates recombination by excising the membrane-targeted Tomato (mT) sequence, leading to expression of membrane-targeted GFP (mG). (B) AQP5-Cre is expressed in acinar cells, leading to mG (green) expression in these cells. (C) Cells that are AQP5 negative express mT (red), which includes myoepithelial and duct cells. (D) The overlay shows mG expression in acinar cells (green) and mT (red) expression in other lineages. Arrow indicates intercalated duct; arrowhead indicates myoepithelial cell. Scale bar = 20 µm.
Progenitor Populations during Development
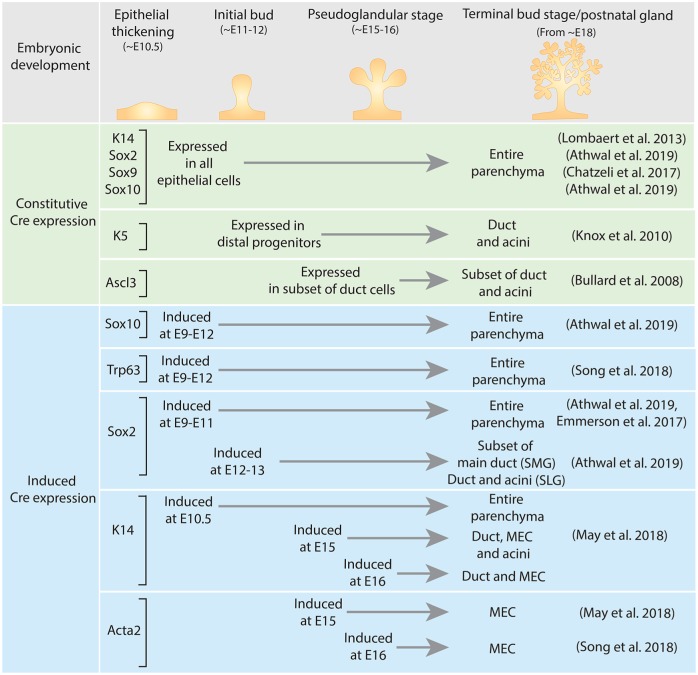
Several studies have investigated lineage contributions in SG epithelium during embryonic development (summarized in Fig. 3). SGs initiate as a thickening of the oral epithelium, called a placode, that proliferates and expands into the underlying mesenchyme to form an initial endbud. The oral epithelium and initial placode express K14, Sox2, Sox9, Sox10, and Trp63. Lineage tracing cell fates from early development for any of these markers confirmed that the entire SG epithelium is derived from the initial placode (Lombaert et al. 2013; Chatzeli et al. 2017; Song et al. 2018; Athwal et al. 2019). Within the initial Sox10 positive (+) endbud, distal and proximal progenitors were identified mainly by position and the cell-specific markers K14+Kit+ and K5+Kit+, respectively (Knox et al. 2010; Lombaert et al. 2013). Lineage tracing of proximal progenitors with a constitutive K5-Cre indicates that this lineage mainly contributes to ducts (Knox et al. 2010).
Figure 3.
Lineage-tracing embryonic glands. Using Cre mouse lines that label the salivary gland anlagen from embryonic day 9.5 (E9.5) to E10.5 lineage traces to the entire gland, confirming that these cells give rise to the entire gland. Inducing lineage tracing from E11.5 to E12.5 gives different results depending on the cells that are traced. MEC, myoepithelial cell; SLG, sublingual gland; SMG, submandibular gland.
Although the aforementioned markers are expressed in the oral epithelium from the onset of gland initiation, their temporospatial localization changes during embryonic development. To further dissect the fate of embryonic lineages, inducible Cre lines have been used to investigate cell fates. K14 localization becomes restricted during development, and lineage tracing K14 cells showed that this lineage gives rise to duct, myoepithelial, and acinar cells up until E15 (May et al. 2018). However, inducing K14-Cre from E16 highlights the lineage restriction from this point on, where K14 no longer gives rise to acini (May et al. 2018). Lineage restriction was also highlighted through the induction of Sox2-Cre at E9 to E11, which lineage traced the entire gland (Emmerson et al. 2017; Athwal et al. 2019). However, inducing at a later stage (E12 to E13) lineage traced only a subset of the main duct in the SMG (Athwal et al. 2019). In the SLG, Sox2+ cells are major contributors to duct and acini throughout development (Emmerson et al. 2017). Sox10 is specifically expressed within endbud cells, and lineage tracing with both a constitutive and an inducible Cre (E9 to E12) confirmed that these cells give rise to the entire gland (Athwal et al. 2019). Alpha-smooth muscle actin (Acta2) is a marker for MECs within the epithelium after E16 (Gervais et al. 2016), and lineage tracing indicates that MECs are also lineage restricted (May et al. 2018; Song et al. 2018). Last, lineage tracing with a constitutive Cre, a subset of duct cells marked by the transcription factor Ascl3 (expressed from E15.5), gives rise to duct and acinar cells during embryonic development (Bullard et al. 2008). However, little is understood about which signals are instructive for this population or how these cells signal to other lineages. Taken together, these lineage-tracing studies indicate that with cell differentiation comes lineage restriction during embryonic development. These studies also suggest that the major SGs are similar in some respects, but there are specific differences in lineage relationships and intrinsic factors that may be crucial for regeneration. It will also be important to confirm that these similarities and differences occur in human major SGs.
Signaling Pathways Directing Epithelial Patterning
Dynamic expression of signaling factors and cross-talk between signaling pathways and cell types are essential for SG development (for recent reviews covering this topic in more depth, see Liu and Wang 2014; Mattingly et al. 2015; Lombaert 2017; Emmerson and Knox 2018). The critical signals required for the initial thickening of the oral epithelium to form the placode were not well understood (summarized by Som and Miletich 2015; Patel et al. 2006). However, a recent study identified canonical retinoic acid (RA) signaling as the earliest signal instructive for gland initiation (Metzler et al. 2018). RA signaling activity colocalized with mesenchymal expression of retinol metabolic genes Rdh10 and Aldh1a2. Thus, RDH10 and RA are required for initiation of placode formation via canonical signaling through the RA receptor RARα (Metzler et al. 2018).
Once the initial endbud is formed, several Sox transcription factors are expressed in the epithelium. In the placode and initial endbud, Sox9 becomes restricted to the distal endbud cells. Deletion of Sox9 in the epithelium resulted in a smaller initial bud and a reduction in distal progenitors through a loss of Sox10, while proximal progenitors were unaffected (Chatzeli et al. 2017). This indicates that Sox9 is involved in establishing the distal progenitors prior to branching morphogenesis. Although Sox2 is expressed in cells that give rise to duct and acini, genetic ablation of Sox2 in the epithelium resulted in a failure to generate acini but not ducts (Emmerson et al. 2017). It was further shown that Sox2 also is essential for establishing Sox10 and targets acinar specific genes. Genetic deletion of Sox10 in the epithelium resulted in a loss of acinar secretory units, while the ducts remained (Athwal et al. 2019; Fig. 4). Duct cells could not compensate for loss of Sox10 as they lacked the plasticity to form new secretory units. Importantly, overexpression of Sox10 in ductal progenitors enhanced their plasticity, forming Kit+ progenitors that differentiate into secretory units. Thus, Sox10 regulates the plasticity of epithelial progenitors to develop into functional secretory units of the gland during development. Taken together, Sox transcription factors are important players in the initial epithelial patterning and cell type determination, particularly for distal progenitors and acinar cell fate. Understanding the required intrinsic factors driving cell fates is essential for cell-based approaches where one would want direct differentiation toward a specific cell type.
Figure 4.
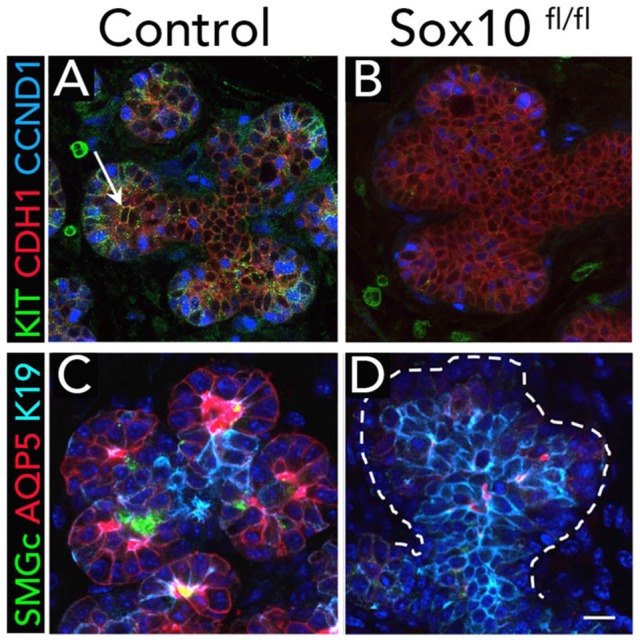
Sox10 is required for secretory cell fate. (A, B) Genetic ablation of Sox10 in epithelial cells leads to loss of Kit+ progenitors (arrow) in embryonic day 16 endbuds. Loss of Sox10 also led to a decrease in proliferation (CCND1) in the epithelium (CDH1). (C, D) Comparing control glands with Sox10fl/fl shows loss of differentiating acinar cells (identified by expression of SMGc and AQP5). AQP5, aquaporin 5; CCND1, cyclin D1; CDH1, cadherin 1; KIT, KIT proto-oncogene receptor tyrosine kinase; SMGc, submandibular gland protein C. Scale bar: 20 µm. From Figure 4 of Athwal et al. (2019).
Fibroblast growth factors (FGFs) are essential for salivary development, and knocking out FGF8, FGF10, or the receptors FGFR2b or FGFR2c leads to gland agenesis (Patel et al. 2006). FGFR signaling differentially interacts with Kit (Lombaert et al. 2013) and Wnt (Patel et al. 2011) signaling to mediate branching. Coordinated FGFR2b/Kit signaling increases distal progenitor proliferation and expansion, which is essential for formation of the endbuds and their subsequent differentiation into proacinar cells. Thus, FGFR and Kit signaling coordinate organ architecture by establishing communication between niches and proximal progenitors (Lombaert et al. 2013).
Multiple Wnts are expressed in the embryonic gland epithelium (Wnt4, Wnt5b, Wnt7b, Wnt10a) and parasympathetic ganglion (Wnt2, Wnt5b, Wnt9a; Knosp et al. 2015). Inhibiting downstream Wnt signaling genetically or pharmacologically decreased branching morphogenesis, suggesting that mesenchymal Wnts influence epithelial patterning. Embryonic SMGs cultured ex vivo with Wnt pathway inhibitors displayed decreased branching morphogenesis (Knosp et al. 2015). Just prior to a major wave of ductal differentiation at E14.5, Wnt signaling decreases in the mesenchyme and increases in the ductal epithelium (Patel et al. 2011). Wnt5b promotes duct maturation by positively regulating Tcfcp2l1, a duct marker and gene essential for duct differentiation, and the absence of FGF signaling leads to premature ductal lumen formation (Patel et al. 2011).
Hippo pathway signaling is also involved in gland growth, and its inhibition perturbs branching morphogenesis ex vivo (Enger et al. 2013). Ablation of the Hippo pathway effector Yap in salivary epithelium led to a reduced expression of epiregulin and severely affected the epithelial patterning with a loss of ductal specification. Furthermore, restricting nuclear Yap by ablating the Lats1/2 genes resulted in an expansion of ductal/proximal progenitors and loss of luminal duct cells. This indicates that the Hippo pathway promotes the identity of proximal progenitors and proper expression of Yap is critical for duct maturation (Szymaniak et al. 2017).
RA signaling also has an important function during branching morphogenesis. Chemical inhibition of RA signaling in ex vivo cultures of E13 SGs reduces epithelial growth and branching morphogenesis in a dose-dependent manner (Wright et al. 2015). Reduced RA signaling decreases proliferation and upregulates K5 expression, whereas Kit expression is decreased (Abashev et al. 2017). Specifically, the ductal progenitor cell population is reciprocally regulated by the 2 isoforms RARα and RARγ (DeSantis et al. 2017). Taken together, these data indicate that RA signaling directly affects duct progenitors and ductal patterning in the developing SG.
Epithelial morphogenesis also involves the cross-talk between the epithelial progenitors and other cell types, such as endothelial and neuronal cells. SGs are highly vascularized, and the vascular network develops with the branching epithelium. By E13.5, endothelial precursors form a continuous network surrounding the branched epithelia. The vasculature regulates epithelial patterning through VEGFR2-dependent angiocrine factors that promote branching and expansion of the distal progenitors while suppressing premature ductal differentiation (Kwon et al. 2017).
Parasympathetic innervation is also essential for SG morphogenesis (Knox et al. 2010). Loss of innervation reduces the proximal K5+ progenitor pool, which differentiates and is not maintained. In turn, K5+ ductal progenitors recruit the developing parasympathetic ganglion through secretion of multiple Wnt ligands (Knosp et al. 2015). Furthermore, the distal K14+Sox10+ progenitors secrete neurturin (Nrtn), a neurotrophic factor that supports survival and axonal growth (Knox et al. 2013; Lombaert et al. 2013). Thus, distal and proximal progenitors are involved in promoting gland innervation, which is essential to growth and function.
Mapping lineage relationships during embryonic development has been instructive to understand how specific signals and cell interactions are affecting them. Taken together, recent advances highlight how formation, differentiation, and expansion of ducts differ from endbuds. Uncovering factors and signaling pathways important for specific lineage expansion is crucial for development of novel cell-based therapies.
Progenitor Populations during Postnatal Growth and Maintenance
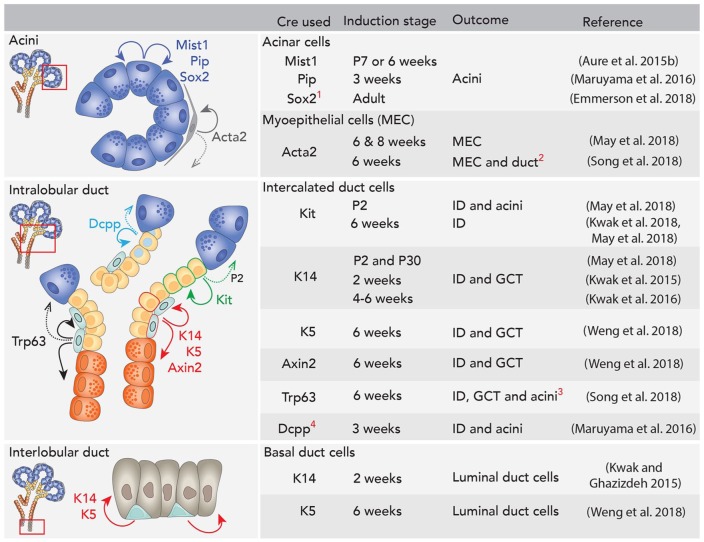
SGs are not fully mature at birth and undergo postnatal growth and differentiation. Recent reports have identified some lineage relationships in vivo in postnatal glands (summarized in Fig. 5). Historically, acinar cells were viewed as postmitotic and continually replenished by progenitors residing within ducts (Aure, Arany, and Ovitt 2015). However, lineage tracing of differentiated acinar cells with an inducible Mist1-Cre or a Pip-Cre indicated that the acinar cells are self-duplicating and thereby maintain themselves during postnatal growth and homeostasis (Aure, Konieczny, and Ovitt 2015; Maruyama et al. 2016). Interestingly, the acinar lineage in the SLG is different from the SMG. In the SLG, a subpopulation within Mist1+ acinar cells expressing Sox2 is important for acinar cell maintenance, as shown by Sox2 lineage tracing (Emmerson et al. 2018).
Figure 5.
Lineage tracing in adult glands. Duct, myoepithelial, and acinar cells are mainly maintained by separate lineages. 1Sox2 lineage tracing in adult mice is specific to sublingual gland. 2A modest number of duct cells were lineage traced from Acta2+ cells in this study. 3A modest number of acinar cells were lineage traced from Trp63 following long-term chase in the submandibular gland (SMG). 4In parotid gland (PG) specifically, Dcpp+ intercalated ducts (IDs) infrequently also give rise to acinar cells. Dcpp, demilune cell and parotid protein 1; GCT, granular convoluted tubules; P, postnatal day. Dotted arrows indicate age-specific or rare events reported.
K14 and Kit mark distal progenitors during embryonic development but are expressed in distinct populations in the intercalated duct (ID) postnatally (May et al. 2018). K14 labels basal duct cells, while Kit is also expressed in a subset of striated ducts. K14 lineage tracing shows that they contribute to granular ducts (Kwak et al. 2016; May et al. 2018) and luminal cells in the excretory duct (Kwak and Ghazizadeh 2015), while Kit cells gave rise to IDs (Kwak et al. 2018; May et al. 2018). K14 and K5 are coexpressed in basal duct cells and MECs of postnatal glands (Yamamoto et al. 2016). Lineage tracing with an inducible K5-Cre showed, similar to K14, that these cells give rise to granular ducts while K5+ basal cells in the excretory ducts give rise to luminal cells (Weng et al. 2018). MECs also coexpress K5 and K14, and lineage tracing indicates that they are self-renewing (May et al. 2018; Weng et al. 2018). To further support this, experiments with an inducible Acta2-Cre confirmed that MECs are self-renewing (May et al. 2018; Song et al. 2018). Taken together, this implies that ducts, MECs, and acini are maintained by separate lineages. However, some data suggest multipotency of some lineages between ducts and acini during homeostasis. Although Kit+ cells are lineage restricted in adult glands, inducing Kit-Cre at postnatal day 2 gave rise to duct and acini (May et al. 2018), suggesting that they are multipotent in early postnatal development. In parotid glands, Dcpp labels ID cells, and lineage tracing indicates that these cells occasionally give rise to acinar cells (Maruyama et al. 2016). Similarly, lineage tracing Trp63 in adult SGs suggests that these cells give rise to some acinar cells in addition to ducts (Song et al. 2018).
These recent advances indicate that there are several partly overlapping cell populations involved in postnatal growth and homeostasis. A future challenge will be to identify signaling pathways that promote plasticity and multipotency. The recent development of single-cell genomic analysis will help identify cell populations producing signaling factors and those cells receiving them. Single-cell analyses of development, postnatal homeostasis, and regeneration are likely to provide novel information to challenge the current paradigms.
Signaling Pathways during Postnatal Homeostasis
Signals governing homeostasis of the adult gland are not well defined or understood. The signals involve proliferation, self-renewal, morphogenesis, and differentiation of the epithelial lineages. Studies with genetic reporters to investigate Wnt signaling show that it is active within the ID in postnatal glands (Hai et al. 2010; Weng et al. 2018). Lineage tracing of Wnt-responsive cells with an Axin2-Cre showed that these cells are restricted to the duct lineage during homeostasis (Weng et al. 2018). The Axin2-Cre expressing cells were partly overlapping with the K5 population in the ID. This could be interpreted as either a subpopulation of K5+ cells being Wnt responsive or that there is dynamic Wnt signaling activity in ID cells. While specific inhibition of Wnt signaling in the K5 lineage led to a significantly reduced granular convoluted tubule area, forced activation of Wnt or Hedgehog (Hh) pathways promoted expansion of ductal structures (Hai et al. 2010; Fiaschi et al. 2011). This implies that Wnt and Hh signaling is important for proliferation of the K5 lineage and, subsequently, duct homeostasis. It was also reported that manipulating Wnt and Hh pathways in the K5 lineage led to less differentiation or loss of acinar cells. However, it is not clear whether altering signaling pathways in the K5 lineage is directly or indirectly affecting acinar cells, and this warrants further study.
As we learn more about lineage relationships and how the gland epithelium is maintained, we can ask more defined questions to elucidate how to direct cell lineages during gland regeneration.
Response to Injury
Combining genetic tools with injury models increases our understanding of injury response and regenerative potential. By ligating the main duct, SGs undergo dramatic but reversible acinar atrophy, and this is a widely used model to investigate regeneration. Upon removal of the ligation, if innervation to the gland is maintained, cell proliferation is induced, and the gland completely regenerates within 2 wk.
Genetic labeling of acinar cells prior to duct ligation showed that not all acinar cells are lost, and surviving atrophied acinar cells are likely involved in replenishing the acinar compartment following deligation (Aure, Konieczny, and Ovitt 2015). It is not clear whether the surviving acinar cells undergo transdifferentiation during the ligation period, and the signaling pathways important for their regeneration have not been investigated directly with genetic models. However, epiregulin, HB-EGF, and EGFR increase in ducts following deligation in parallel with acinar regeneration, indicating that the EGFR signaling pathway may be involved in repair after injury (Nagai et al. 2014).
In ducts, increased Wnt/β-catenin and Hh activity were detected after ligation (Hai et al. 2010). Accordingly, lineage tracing of K5+ and Axin2+ cells showed that these populations are involved in lineage-restricted duct regeneration following injury (Weng et al. 2018). The role of MECs during the ligation and regeneration process has not been specifically investigated, but labeling prior to ligation showed that preexisting MECs are present in the regenerated glands (Weng et al. 2018). Following partial gland surgical resection, another injury model in SGs, MECs were found to undergo morphologic changes in size and length and number of cell processes during the time of increased epithelial proliferation (Kawabe et al. 2016). Although the significance of these changes is unclear, it could indicate that increased interaction between MECs and surrounding epithelium is important for regeneration. Little is known about the function of MECs during gland homeostasis, let alone during response to injury and regeneration, and further investigation is warranted.
SGs are sensitive to IR damage, which leads to irreversible gland hypofunction and is widely used as a severe injury model. In mouse models, IR induces rapid hypofunction of the gland in the absence of obvious cell death and histologic changes. However, with time, chronic IR damage results in changes in cell proliferation and senescence, a relative increase in innervation, and eventually fibrosis. Although a major outcome of IR damage is loss of functional acinar cells, they may survive and proliferate months after IR (Emmerson et al. 2018; Weng et al. 2018). Lineage tracing of duct populations after IR showed that although they initially remain lineage restricted, their plasticity increases over time (May et al. 2018; Weng et al. 2018; Fig. 6). It may be that after damage, cells eventually become less lineage restricted as a result of a change in niche signals. The possibility that cells gain plasticity in vivo and could aid in regeneration may have implications for new therapeutic approaches.
Figure 6.
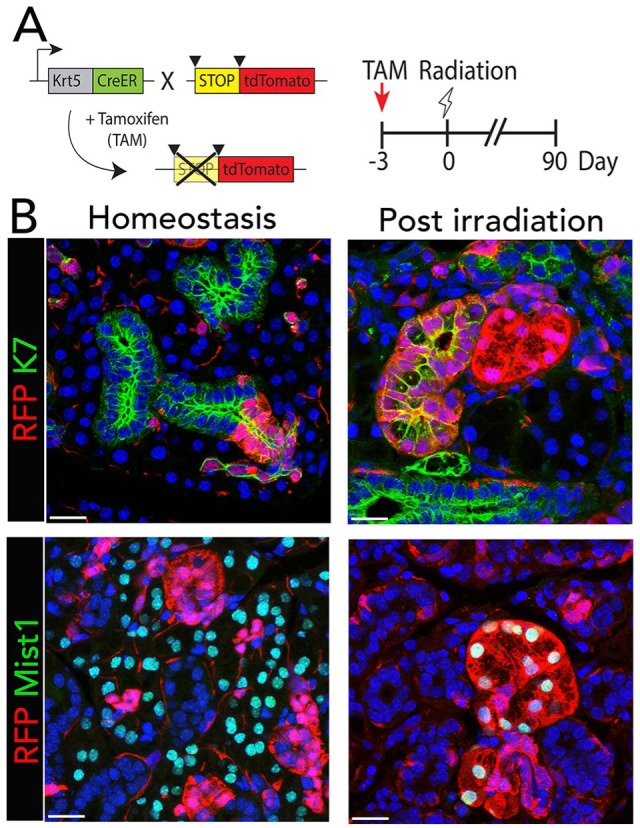
Lineage plasticity after irradiation. (A) Tamoxifen (TAM)–inducible K5CreER mice were crossed with the R26tdTomato reporter strain to follow K5 lineage in postnatal glands. Tamoxifen treatment induces Cre-mediated recombination in K5+ cells, which excises the stop codon allowing tdTomato (RFP) expression in the K5+ cells and their descendants. Experimental timeline for tamoxifen-dependent Cre induction, radiation, and tissue harvest. Mice were irradiated 3 d after tamoxifen treatment, and irradiated glands were compared with nonirradiated glands following a 90-d chase period. (B) After 90 d, the expanded number of RFP-labeled cells in female submandibular glands remain colocalized with K7, a specific duct marker, but not with Mist1, an acinar cell marker. However, 90 d after irradiation, RFP labels clusters of cells that colocalize with duct and acinar cells, indicating that K5 cells have given rise to duct and acini following irradiation. Nuclei are stained with DAPI (blue). Scale bars: 25 µm. From Figures 1 and 4 of Weng et al. (2018).
Changes in signaling pathways following IR are not well understood. Several studies have shown that modifying the environment by introducing “signaling factors” can prevent cell loss and have a positive effect on saliva secretion following IR. For example, injecting IGF1 immediately before or after IR had a protective effect through an improvement of DNA repair (Meyer et al. 2017; Chibly et al. 2018). Neurotrophic factors have also been implicated as potential therapeutic candidates in tissue regeneration (Xiao and Le 2016). The expression of glial cell–derived neurotropic factor (GDNF) increases following IR, and GDNF promotes epithelial proliferation (Peng et al. 2017). Nrtn can also attenuate IR damage. Gene therapy with adenovirus-expressing Nrtn increased the expression of Nrtn in SGs and prevented a reduction in secretion following IR. There was increased parasympathetic innervation and saliva secretion following IR (Ferreira et al. 2018).
In addition, adenoviral vectors expressing sonic hedgehog (Shh) have been delivered to SGs after IR in mice and minipigs and significantly increase salivary function (Hai et al. 2018; Hu et al. 2018). Although IR damage itself does not activate Hh signaling, forced transient activation following IR increases proliferation and glandular function (Hai et al. 2014). Shh reduced senescence by promoting DNA repair and reducing oxidative stress. Gland innervation was maintained and increased autophagy occurred with reduced fibrosis. The direct effect and mechanisms of Shh on specific populations remain to be explored.
FGF signaling is important for regeneration of various tissues where it promotes proliferation and/or differentiation (Maddaluno et al. 2017). Treating glands with FGF7 after IR reduced apoptosis and prevented salivary hypofunction in rat SGs (Choi et al. 2017). In mice with IR-induced salivary hypofunction, transplanting mesenchymal stem cells led to an increase in FGF10 and restoration of saliva flow, indicating that FGF10 signaling has regenerative potential (Shin et al. 2018). Mesenchymal/stromal cells are in all major human SGs, and they express genes encoding members of the FGF and platelet-derived growth factor signaling pathways (Togarrati et al. 2017). The role of endogenous FGF signaling and stromal cells in adult SGs in homeostasis or during regeneration is not well understood but could be investigated with available genetic tools. Improving functional regeneration after IR remains an important clinical problem to resolve.
Concluding Remarks
The knowledge gained from studying progenitors and signaling pathways in embryonic development of SGs has been instructive in designing putative regenerative strategies for adult glands. In vivo plasticity following IR within normally lineage-restricted cell populations indicates that niche signals, including those from a damaged niche, may direct cell fates in adult glands. This supports the concept that “stemness” is a dynamic state mainly directed by the cell’s niche (Chacon-Martinez et al. 2018). The SG field is poised to use the information about lineage relationships and signaling pathways to target specific cells/pathways to prevent or reduce SG damage or regenerate SGs. An emerging concept is that cells involved in regeneration will vary depending on the type/severity of damage and the resulting microenvironment that emerges from this damage. This underscores the unique challenges that we face when restoring gland function caused by specific types of damage, such as IR, immune damage, or chemical/drug-induced damage. Understanding how signaling pathways regulate cell-specific expansion and fate determination in the context of a specific damaged niche will be essential for developing regenerative strategies.
Author Contributions
M.H. Aure, J.M. Symonds, contributed to conception and design, drafted and critically revised the manuscript; J.W. Mays, M.P. Hoffman, contributed to conception and design, critically revised the manuscript. All authors gave final approval and agree to be accountable for all aspects of the work ensuring integrity and accuracy.
Acknowledgments
The authors thank Dr. A. Chibly and Dr. V. Patel for critical reading of the manuscript.
Footnotes
This work was funded by the Intramural Research Program of the National Institute of Dental and Craniofacial Research at the National Institutes of Health.
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
References
- Abashev TM, Metzler MA, Wright DM, Sandell LL. 2017. Retinoic acid signaling regulates Krt5 and Krt14 independently of stem cell markers in submandibular salivary gland epithelium. Dev Dyn. 246(2):135–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athwal HK, Murphy G, Tibbs E, Cornett A, Hill E, Yeoh K, Berenstein E, Hoffman MP, Lombaert IMA. 2019. SOX10 regulates plasticity of epithelial progenitors toward secretory units of exocrine glands. Stem Cell Rep. 12(2):366–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aure MH, Arany S, Ovitt CE. 2015. Salivary glands: stem cells, self-duplication, or both? J Dent Res. 94(11):1502–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aure MH, Konieczny SF, Ovitt CE. 2015. Salivary gland homeostasis is maintained through acinar cell self-duplication. Dev Cell. 33(2):231–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullard T, Koek L, Roztocil E, Kingsley PD, Mirels L, Ovitt CE. 2008. Ascl3 expression marks a progenitor population of both acinar and ductal cells in mouse salivary glands. Dev Biol. 320(1):72–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacon-Martinez CA, Koester J, Wickstrom SA. 2018. Signaling in the stem cell niche: regulating cell fate, function and plasticity. Development. 145(15):dev165399. [DOI] [PubMed] [Google Scholar]
- Chatzeli L, Gaete M, Tucker AS. 2017. Fgf10 and Sox9 are essential for the establishment of distal progenitor cells during mouse salivary gland development. Development. 144(12):2294–2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chibly AM, Wong WY, Pier M, Cheng H, Mu Y, Chen J, Ghosh S, Limesand KH. 2018. aPKCζ-dependent repression of Yap is necessary for functional restoration of irradiated salivary glands with IGF-1. Sci Rep. 8(1):6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JS, Shin HS, An HY, Kim YM, Lim JY. 2017. Radioprotective effects of keratinocyte growth factor-1 against irradiation-induced salivary gland hypofunction. Oncotarget. 8(8):13496–13508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis KA, Stabell AR, Spitzer DC, O’Keefe KJ, Nelson DA, Larsen M. 2017. RARα and RARγ reciprocally control K5(+) progenitor cell expansion in developing salivary glands. Organogenesis. 13(4):125–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmerson E, Knox SM. 2018. Salivary gland stem cells: a review of development, regeneration and cancer. Genesis. 56(5):e23211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmerson E, May AJ, Berthoin L, Cruz-Pacheco N, Nathan S, Mattingly AJ, Chang JL, Ryan WR, Tward AD, Knox SM. 2018. Salivary glands regenerate after radiation injury through SOX2-mediated secretory cell replacement. EMBO Mol Med. 10(3):e8051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmerson E, May AJ, Nathan S, Cruz-Pacheco N, Lizama CO, Maliskova L, Zovein AC, Shen Y, Muench MO, Knox SM. 2017. Sox2 regulates acinar cell development in the salivary gland. Elife. 6:e26620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enger TB, Samad-Zadeh A, Bouchie MP, Skarstein K, Galtung HK, Mera T, Walker J, Menko AS, Varelas X, Faustman DL, et al. 2013. The hippo signaling pathway is required for salivary gland development and its dysregulation is associated with Sjogren’s syndrome. Lab Invest. 93(11):1203–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira JNA, Zheng C, Lombaert IMA, Goldsmith CM, Cotrim AP, Symonds JM, Patel VN, Hoffman MP. 2018. Neurturin gene therapy protects parasympathetic function to prevent irradiation-induced murine salivary gland hypofunction. Mol Ther Methods Clin Dev. 9:172–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiaschi M, Kolterud A, Nilsson M, Toftgard R, Rozell B. 2011. Targeted expression of GLI1 in the salivary glands results in an altered differentiation program and hyperplasia. Am J Pathol. 179(5):2569–2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flodby P, Borok Z, Banfalvi A, Zhou B, Gao D, Minoo P, Ann DK, Morrisey EE, Crandall ED. 2010. Directed expression of Cre in alveolar epithelial type 1 cells. Am J Respir Cell Mol Biol. 43(2):173–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gervais EM, Sequeira SJ, Wang W, Abraham S, Kim JH, Leonard D, DeSantis KA, Larsen M. 2016. Par-1b is required for morphogenesis and differentiation of myoepithelial cells during salivary gland development. Organogenesis. 12(4):194–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hai B, Qin L, Yang Z, Zhao Q, Shangguan L, Ti X, Zhao Y, Kim S, Rangaraj D, Liu F. 2014. Transient activation of hedgehog pathway rescued irradiation-induced hyposalivation by preserving salivary stem/progenitor cells and parasympathetic innervation. Clin Cancer Res. 20(1):140–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hai B, Yang Z, Millar SE, Choi YS, Taketo MM, Nagy A, Liu F. 2010. Wnt/β-catenin signaling regulates postnatal development and regeneration of the salivary gland. Stem Cells Dev. 19(11):1793–1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hai B, Zhao Q, Deveau MA, Liu F. 2018. Delivery of sonic hedgehog gene repressed irradiation-induced cellular senescence in salivary glands by promoting DNA repair and reducing oxidative stress. Theranostics. 8(4):1159–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L, Zhu Z, Hai B, Chang S, Ma L, Xu Y, Li X, Feng X, Wu X, Zhao Q, et al. 2018. Intragland Shh gene delivery mitigated irradiation-induced hyposalivation in a miniature pig model. Theranostics. 8(16):4321–4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabe Y, Mizobe K, Bando Y, Sakiyama K, Taira F, Tomomura A, Araki H, Amano O. 2016. Morphological changes of myoepithelial cells in the rat submandibular gland following the application of surgical stimuli. Acta Histochem Cytochem. 49(6):159–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knosp WM, Knox SM, Lombaert IM, Haddox CL, Patel VN, Hoffman MP. 2015. Submandibular parasympathetic gangliogenesis requires sprouty-dependent wnt signals from epithelial progenitors. Dev Cell. 32(6):667–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox SM, Lombaert IM, Haddox CL, Abrams SR, Cotrim A, Wilson AJ, Hoffman MP. 2013. Parasympathetic stimulation improves epithelial organ regeneration. Nat Commun. 4:1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox SM, Lombaert IM, Reed X, Vitale-Cross L, Gutkind JS, Hoffman MP. 2010. Parasympathetic innervation maintains epithelial progenitor cells during salivary organogenesis. Science. 329(5999):1645–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretzschmar K, Watt FM. 2012. Lineage tracing. Cell. 148(1–2):33–45. [DOI] [PubMed] [Google Scholar]
- Kwak M, Alston N, Ghazizadeh S. 2016. Identification of stem cells in the secretory complex of salivary glands. J Dent Res. 95(7):776–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak M, Ghazizadeh S. 2015. Analysis of histone H2BGFP retention in mouse submandibular gland reveals actively dividing stem cell populations. Stem Cells Dev. 24(5):565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak M, Ninche N, Klein S, Saur D, Ghazizadeh S. 2018. C-kit(+) cells in adult salivary glands do not function as tissue stem cells. Sci Rep. 8(1):14193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon HR, Nelson DA, DeSantis KA, Morrissey JM, Larsen M. 2017. Endothelial cell regulation of salivary gland epithelial patterning. Development. 144(2):211–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen HS, Aure MH, Peters SB, Larsen M, Messelt EB, Kanli Galtung H. 2011. Localization of AQP5 during development of the mouse submandibular salivary gland. J Mol Histol. 42(1):71–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Wang S. 2014. Molecular cues for development and regeneration of salivary glands. Histol Histopathol. 29(3):305–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombaert I. 2017. Implications of salivary gland developmental mechanisms for the regeneration of adult damaged tissues. In: Cha S, editor. Salivary gland development and regeneration. Berlin (Germany): Springer; p. 3–22. [Google Scholar]
- Lombaert IM, Abrams SR, Li L, Eswarakumar VP, Sethi AJ, Witt RL, Hoffman MP. 2013. Combined KIT and FGFR2B signaling regulates epithelial progenitor expansion during organogenesis. Stem Cell Rep. 1(6):604–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddaluno L, Urwyler C, Werner S. 2017. Fibroblast growth factors: key players in regeneration and tissue repair. Development. 144(22):4047–4060. [DOI] [PubMed] [Google Scholar]
- Maruyama EO, Aure MH, Xie X, Myal Y, Gan L, Ovitt CE. 2016. Cell-specific Cre strains for genetic manipulation in salivary glands. PLoS One. 11(1):e0146711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattingly A, Finley JK, Knox SM. 2015. Salivary gland development and disease. Wiley Interdiscip Rev Dev Biol. 4(6):573–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May AJ, Cruz-Pacheco N, Emmerson E, Gaylord EA, Seidel K, Nathan S, Muench MO, Klein OD, Knox SM. 2018. Diverse progenitor cells preserve salivary gland ductal architecture after radiation-induced damage. Development. 145(21):12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzler MA, Raja S, Elliott KH, Friedl RM, Quang N, Brugmann SA, Larsen M, Sandell LL. 2018. RDH10-mediated retinol metabolism and RARα-mediated retinoic acid signaling are required for submandibular salivary gland initiation. Development. 145(15):dev164822. Erratum in: Development; 2018;145(17):dev170795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer S, Chibly AM, Burd R, Limesand KH. 2017. Insulin-like growth factor-1-mediated DNA repair in irradiated salivary glands is sirtuin-1 dependent. J Dent Res. 96(2):225–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. 2007. A global double-fluorescent cre reporter mouse. Genesis. 45(9):593–605. [DOI] [PubMed] [Google Scholar]
- Nagai K, Arai H, Okudera M, Yamamura T, Oki H, Komiyama K. 2014. Epiregulin is critical for the acinar cell regeneration of the submandibular gland in a mouse duct ligation model. J Oral Pathol Med. 43(5):378–387. [DOI] [PubMed] [Google Scholar]
- Patel N, Sharpe PT, Miletich I. 2011. Coordination of epithelial branching and salivary gland lumen formation by Wnt and FGF signals. Dev Biol. 358(1):156–167. [DOI] [PubMed] [Google Scholar]
- Patel VN, Rebustini IT, Hoffman MP. 2006. Salivary gland branching morphogenesis. Differentiation. 74(7):349–364. [DOI] [PubMed] [Google Scholar]
- Peng X, Varendi K, Maimets M, Andressoo JO, Coppes RP. 2017. Role of glial-cell-derived neurotrophic factor in salivary gland stem cell response to irradiation. Radiother Oncol. 124(3):448–454. [DOI] [PubMed] [Google Scholar]
- Shin HS, Lee S, Kim YM, Lim JY. 2018. Hypoxia-activated adipose mesenchymal stem cells prevents irradiation-induced salivary hypofunction by enhanced paracrine effect through fibroblast growth factor 10. Stem Cells. 36(7):1020–1032. [DOI] [PubMed] [Google Scholar]
- Som PM, Miletich I. 2015. The embryology of the salivary glands: an update. Neurographics. 5(4):167–177(11). [Google Scholar]
- Song EAC, Min S, Oyelakin A, Smalley K, Bard JE, Liao L, Xu JM, Romano RA. 2018. Genetic and scRNA-seq analysis reveals distinct cell populations that contribute to salivary gland development and maintenance. Sci Rep. 8(1):14043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymaniak AD, Mi R, McCarthy SE, Gower AC, Reynolds TL, Mingueneau M, Kukuruzinska M, Varelas X. 2017. The hippo pathway effector YAP is an essential regulator of ductal progenitor patterning in the mouse submandibular gland. Elife. 6:e23499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Togarrati PP, Sasaki RT, Abdel-Mohsen M, Dinglasan N, Deng X, Desai S, Emmerson E, Yee E, Ryan WR, da Silva MCP, et al. 2017. Identification and characterization of a rich population of CD34(+) mesenchymal stem/stromal cells in human parotid, sublingual and submandibular glands. Sci Rep. 7(1):3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng PL, Aure MH, Maruyama T, Ovitt CE. 2018. Limited regeneration of adult salivary glands after severe injury involves cellular plasticity. Cell Rep. 24(6):1464–1470e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright DM, Buenger DE, Abashev TM, Lindeman RP, Ding J, Sandell LL. 2015. Retinoic acid regulates embryonic development of mammalian submandibular salivary glands. Dev Biol. 407(1):57–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao N, Le QT. 2016. Neurotrophic factors and their potential applications in tissue regeneration. Arch Immunol Ther Exp (Warsz). 64(2):89–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M, Nakata H, Kumchantuek T, Sakulsak N, Iseki S. 2016. Immunohistochemical localization of keratin 5 in the submandibular gland in adult and postnatal developing mice. Histochem Cell Biol. 145(3):327–339. [DOI] [PubMed] [Google Scholar]