The World Health Organization listed Helicobacter pylori as a high-priority pathogen for antibiotic development. The potential of using peptide transporters in drug design is well recognized. We discovered that the substrate-binding protein of the ABC transporter for peptides, termed dipeptide permease, is an unusual member of its family in that it directly binds peptides of diverse amino acid sequences, ranging between two and eight residues in length. We also provided a structural rationale for the observed broad specificity. Since the ability to import peptides as a source of carbon is critical for H. pylori, our findings will inform drug design strategies based on inhibition or fusion of membrane-impermeant antimicrobials with peptides.
KEYWORDS: binding affinity, crystal structure, mass spectrometry, peptide transport, substrate-binding protein
ABSTRACT
The gastric pathogen Helicobacter pylori has limited ability to use carbohydrates as a carbon source, relying instead on exogenous amino acids and peptides. Uptake of certain peptides by H. pylori requires an ATP binding cassette (ABC) transporter annotated dipeptide permease (Dpp). The transporter specificity is determined by its cognate substrate-binding protein DppA, which captures ligands in the periplasm and delivers them to the permease. Here, we show that, unlike previously characterized DppA proteins, H. pylori DppA binds, with micromolar affinity, peptides of diverse amino acid sequences ranging between two and eight residues in length. We present analysis of the 1.45-Å-resolution crystal structure of its complex with the tetrapeptide STSA, which provides a structural rationale for the observed broad specificity. Analysis of the molecular surface revealed a ligand-binding pocket that is large enough to accommodate peptides of up to nine residues in length. The structure suggests that H. pylori DppA is able to recognize a wide range of peptide sequences by forming interactions primarily with the peptide main chain atoms. The loop that terminates the peptide-binding pocket in DppAs from other bacteria is significantly shorter in the H. pylori protein, providing an explanation for its ability to bind longer peptides. The subsites accommodating the two N-terminal residues of the peptide ligand make the greatest contribution to the protein-ligand binding energy, in agreement with the observation that dipeptides bind with affinity close to that of longer peptides.
IMPORTANCE The World Health Organization listed Helicobacter pylori as a high-priority pathogen for antibiotic development. The potential of using peptide transporters in drug design is well recognized. We discovered that the substrate-binding protein of the ABC transporter for peptides, termed dipeptide permease, is an unusual member of its family in that it directly binds peptides of diverse amino acid sequences, ranging between two and eight residues in length. We also provided a structural rationale for the observed broad specificity. Since the ability to import peptides as a source of carbon is critical for H. pylori, our findings will inform drug design strategies based on inhibition or fusion of membrane-impermeant antimicrobials with peptides.
INTRODUCTION
Extracellular peptides serve as a source of nutrients for bacteria (1–4) and are taken up by either ATP binding cassette (ABC) transporters (5, 6) or ion motive force-driven transporters (7–9). In some bacteria, deletion or inhibition of these transporters results in reduced pathogenicity and virulence (10–14), suggesting that peptide transporters can be considered targets for the development of antimicrobial agents. Furthermore, the peptide transporter systems are used as an entry gate by natural antimicrobial peptides (15). For instance, in Escherichia coli and Erwinia amylovora, the dipeptide and oligopeptide transporter systems Dpp and Opp mediate the import of kasugamycin and blasticidin S (16, 17). In addition, some natural antibiotics that are synthesized with the toxic moiety attached to a small peptide (e.g., bacilysin, bialaphos, and phosphinothricyl) enter target cells via peptide transporters (18). Once inside the cell, the attached peptide is cleaved by a peptidase(s) to release the antibacterial moiety. A similar strategy has been used to deliver otherwise membrane-impermeant synthetic drugs into the bacterial cell by fusing them with peptides, thus creating Trojan horse antibacterials that can be smuggled in via peptide transporters (19–22). Since the potential of using peptide transporters in drug design is well recognized, studies on these systems in pathogenic bacteria can lead to drug discoveries.
The ABC-type peptide transporter systems in bacteria are often encoded by a polycistronic operon and are typically composed of a substrate-binding protein (SBP) referred to as subunit A, two transmembrane permeases (subunits B and C), and two nucleotide-binding proteins (subunits D and F) that couple ATP hydrolysis to substrate translocation (23, 24). The SBPs are the major determinant of the transporter specificity; they capture peptides in the periplasm and deliver them to their cognate permease (25, 26). Some ABC-type peptide transporters take up predominantly dipeptides (dipeptide permease [Dpp] systems) (27). Other ABC transporters have preference for longer peptides (oligopeptide permease [Opp] systems) (28, 29). However, many ABC transporters have a broad specificity and import peptides of various lengths. For example, DppA of E. coli binds tripeptides in addition to dipeptides, albeit with lower affinity (30). OppAs have been reported to bind dipeptides (31), tripeptides (28, 31, 32), tetrapeptides (28, 31), and peptides up to 35 residues long (33, 34).
Analysis of the crystal structures of DppAs from E. coli (35, 36) and a Pseudoalteromonas sp. (27) and OppAs from E. coli (28), Salmonella enterica serovar Typhimurium (31), Thermotoga maritima (37), Lactococcus lactis (34, 38), Yersinia pestis (39), Bacillus subtilis (40), and Burkholderia pseudomallei (41) revealed that they share a common fold consisting of two lobes connected by a hinge. The N- and C-terminal polypeptide segments form a larger lobe consisting of two domains (domains I and II), while the middle segment forms a smaller lobe (domain III). The peptide-binding site is invariably located between domains I and III. In the free form, these proteins adopt an open conformation with two lobes separated from each other. Ligand binding induces a large conformational change in SBP, enclosing the bound peptide in a Venus flytrap manner, and it is in this closed form that the peptide-bound SBP is recognized by the permeases, leading to ATP hydrolysis-driven translocation of the peptide across the membrane into the cytoplasm (24, 42).
Different strains of the gastric carcinogenic pathogen Helicobacter pylori possess seven or eight SBPs, two of which have been annotated as putative ABC peptide transporter substrate-binding proteins DppA and OppA, based on sequence similarity (43–45). The previous mutagenesis study in H. pylori strain 43504 showed that the Dpp and Opp transporter systems are indeed involved in peptide uptake and that the Dpp system in particular is required for H. pylori’s ability to utilize certain dipeptides, hexapeptides, and nonapeptides (46).
Here, we report the crystal structure of DppA from H. pylori strain SS1 (HpDppA, locus tag HPYLSS1_00287), which shares 99% amino acid sequence identity with DppA from H. pylori strain 43504 (46). Based on the analysis of the electron density maps, we hypothesized and proved that the protein copurifies with peptides. We subsequently verified that HpDppA binds peptides of diverse amino acid sequences with micromolar affinity. Bioinformatics and structural analyses allowed us to explore the underlying molecular basis of the broad substrate specificity of HpDppA and to identify residues important for ligand recognition.
RESULTS
Overall structure and comparison with other SBPs.
Crystals of recombinant H. pylori DppA (HpDppA) lacking the N-terminal signal sequence (residues 1 to 22) had P212121 symmetry with one molecule in the asymmetric unit. The structure was determined at a resolution of 1.8 Å using a molecular replacement method. The final model (R/Rfree of 0.146/0.180) (Table 1) contains HpDppA residues 38 to 549 (residues N-terminal to 38 are presumably disordered).
TABLE 1.
Data processing and refinement statistics of HpDppA
| Data set | Value(s)a for: |
|
|---|---|---|
| Native protein | Protein cocrystallized with 1 mM STSA | |
| Data collection | ||
| Wavelength (Å) | 0.9537 | 0.9184 |
| Mosaicity (°) | 0.6 | 0.8 |
| Resolution range (Å) | 46.12–1.80 (1.84–1.80) | 38.92–1.45 (1.47–1.45) |
| No. of observed reflections | 212,403 (12,466) | 239,729 (11,020) |
| No. of unique reflections | 59,276 (3,464) | 95,404 (4,613) |
| Mean I/σ(I) | 7.5 (2.5) | 6.2 (1.9) |
| Completeness (%) | 99 (100) | 86 (85) |
| Multiplicity | 3.6 (3.6) | 2.5 (2.4) |
| Rmerge | 0.102 (0.348) | 0.085 (0.346) |
| CC1/2 (%) | 99 (86) | 99 (75) |
| Refinement statistics | ||
| Resolution range (Å) | 46.12–1.80 | 38.92–1.45 |
| Rwork/Rfree | 0.146/0.180 | 0.147/0.175 |
| No. of residues/atoms/waters | 512/4,911/751 | 512/5,056/875 |
| Bond length deviation from ideality (Å) | 0.011 | 0.012 |
| Bond angle deviation from ideality (°) | 2.0 | 2.2 |
| Average B (Å2) | ||
| Protein | 12.1 | 10.8 |
| Water | 25.4 | 30.3 |
| Ligand | 15.7 | 12.7 |
| Ramachandran plot (%) | ||
| Favored | 98 | 98 |
| Allowed | 2 | 2 |
| Outliers | 0 | 0 |
Values in parentheses are for the highest-resolution shell.
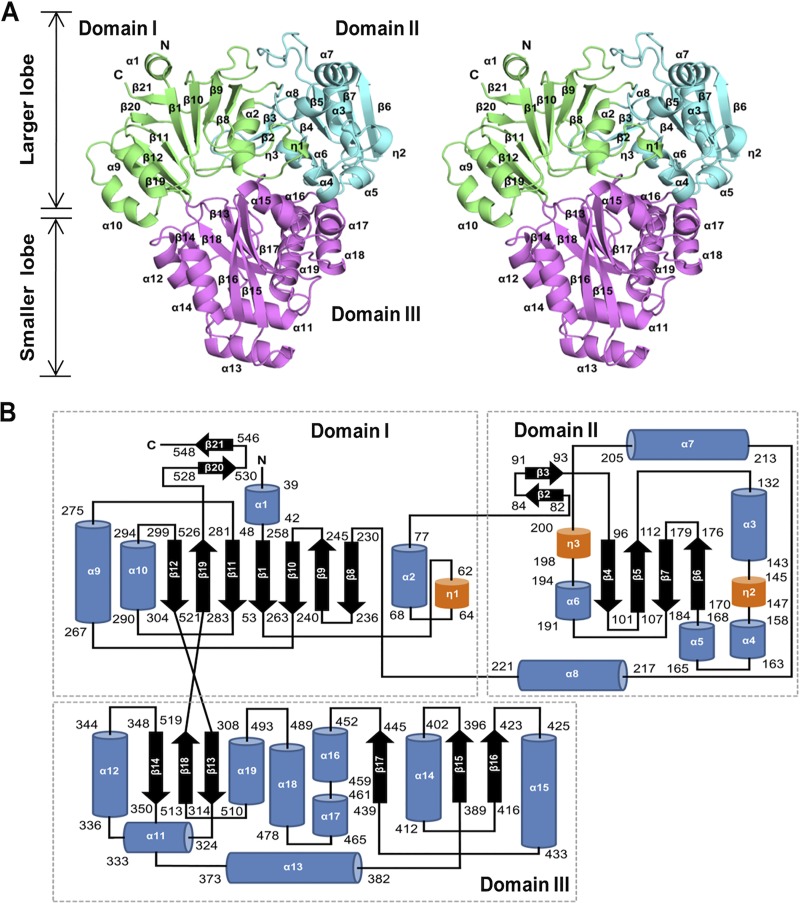
The polypeptide chain of HpDppA comprises a total of 19 α-helices and 21 β-strands and folds into three domains that form two distinct lobes—a larger (domains I and II) and a smaller (domain III) one—separated by a deep cleft, creating the Venus flytrap-like shape that is characteristic for SBPs (Fig. 1). All three domains have a mixed α/β globular fold with a central β-sheet and several flanking helices. Domain I is formed by three separate segments of the polypeptide chain (the protein’s N-terminal segment, residues 38 to 77, midchain segment, residues 227 to 304, and C-terminal segment, residues 521 to 549). Domain II (residues 78 to 226) is inserted between the α-helix α2 and β-strand β8 of domain I. Domain III (smaller lobe, residues 305 to 520) is inserted between β-strands β12 and β19 of domain I. The two crossover segments of the polypeptide chain that connect domain I to the N terminus and C terminus of domain III (residues 305 to 307 and 519 to 521, respectively) form an interlobe hinge. The overall fold and the structure of the hinge region reveal that HpDppA is a type II SBP (47) that belongs to cluster C according to the latest structural classification of SBPs (48).
FIG 1.
Overall structure of H. pylori SS1 DppA. (A) Stereoview representation of the overall fold of HpDppA. The three domains (I, II, and III) are shown in green, cyan, and magenta, respectively, and labeled. (B) The topology of secondary structure elements of HpDppA. The α-helices are represented by blue rods, 310-helices are represented by orange rods, and β-strands are represented by arrows.
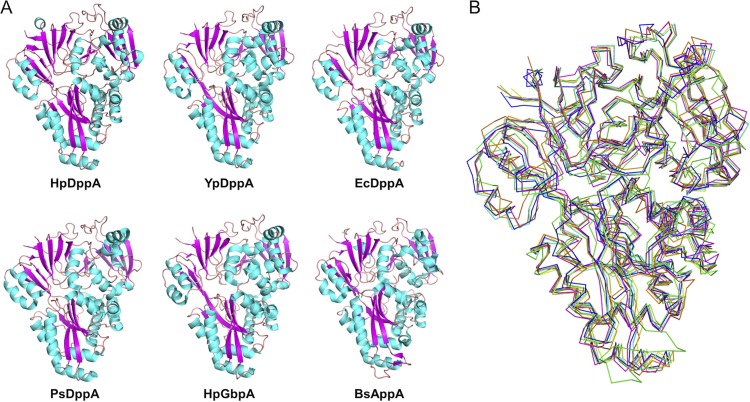
A search for structurally related proteins using PDBeFOLD (49) revealed full-length structural similarities with peptide-binding proteins, such as the putative periplasmic dipeptide-binding protein from Y. pestis (YpDppA) (PDB identification number [ID] 5F1Q), dipeptide-binding protein from E. coli (EcDppA) (PDB ID 1DPP) (35), dipeptide-binding protein from Pseudoalteromonas sp. strain SM9913 (PsDppA) (PDB ID 4QFP) (27), and oligopeptide-binding protein from B. subtilis (BsAppA) (PDB ID 1XOC) (40) (Fig. 2), all of which are also classified as type II/cluster C SBPs. In addition, HpDppA is structurally similar to the glutathione-binding protein from Haemophilus parasuis (HpGbpA) (PDB ID 3M8U) (50, 51).
FIG 2.
Comparison of DppA from H. pylori with other type II/cluster C SBPs. HpDppA, DppA from H. pylori SS1; YpDppA, periplasmic dipeptide transport protein from Y. pestis; EcDppA, dipeptide-binding protein from E. coli; PsDppA, dipeptide-binding protein from Pseudoalteromonas sp. SM9913; HpGbpA, glutathione-binding protein A from H. parasuis SH0165; BsAppA, oligopeptide-binding protein from B. subtilis. (A) A cartoon representation of the structures of different peptide-binding proteins, indicating that the overall fold of HpDppA is largely conserved in other bacterial peptide-binding proteins. (B) Superimpositions of the ribbon diagram of HpDppA (blue) with those of YpDppA (cyan), EcDppA (yellow), PsDppA (pink), HpGbpA (orange), and BsAppA (green).
Location of the putative ligand-binding pocket and identification of ligands copurifying with HpDppA.
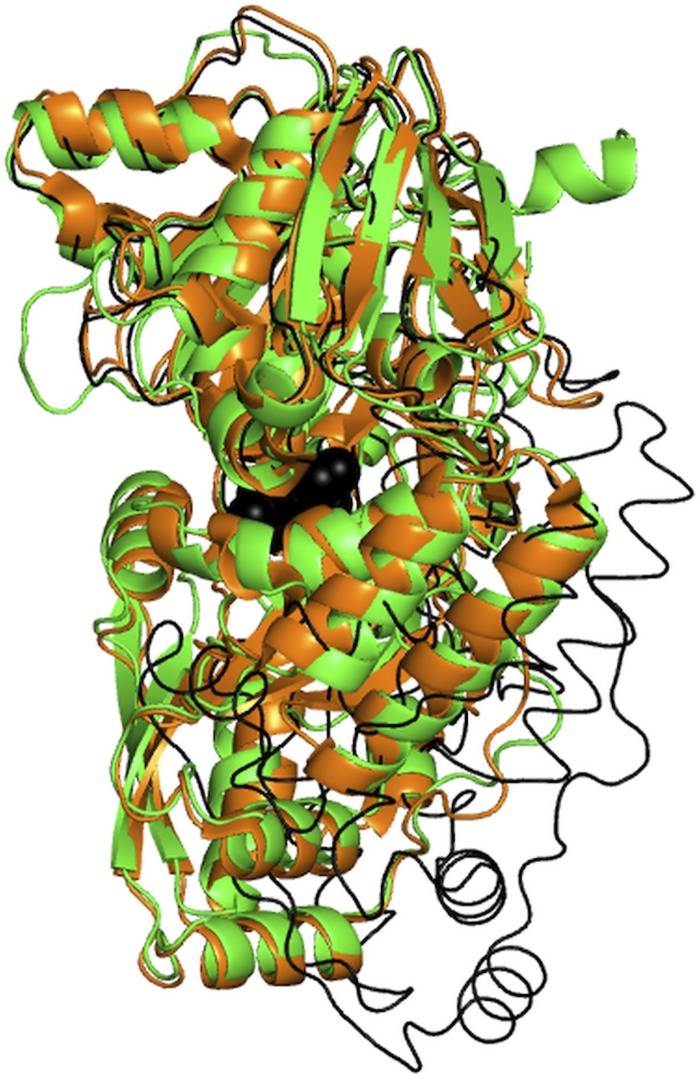
Most of the structurally characterized SBPs exist in an open conformation in the absence of a ligand and adopt a closed conformation upon ligand binding (27, 28, 52). A comparison of the crystal structure of HpDppA with those of the open (free) and closed (peptide-bound) forms of PsDppA (27) (Fig. 3) revealed that HpDppA crystallized in a closed conformation. In agreement with that observation, examination of the interface between the two lobes of HpDppA (a region implicated in ligand binding in other SBPs) revealed an electron density that was unaccounted for by protein atoms. The shape of the density resembled that of a peptide. The electron density for the backbone of four amino acid residues of the peptide was clearly defined. In contrast, the density for the remainder of the peptide’s main chain and for its side chains was ambiguous, indicating that the observed map represented an averaged density for an ensemble of different peptides occupying the same site. No exogenous peptides were added in the protein purification or crystallization steps. Therefore, the ligands trapped in the crystal were likely to be peptides from the culture medium or E. coli cytosol that copurified with the protein. The density at the beta and gamma positions of the side chains was the strongest, and we have modelled the ligand as a peptide with amino acid sequence STSA (see Fig. S1 in the supplemental material).
FIG 3.
Superimposition of the structures of HpDppA (green) and the closed (dipeptide-bound, orange) and open (free, thin black line) forms of PsDppA. The dipeptide ligand of PsDppA is shown as black balls. PDB IDs: 4QFN for the closed form of PsDppA; 4QFK for the open form of PsDppA.
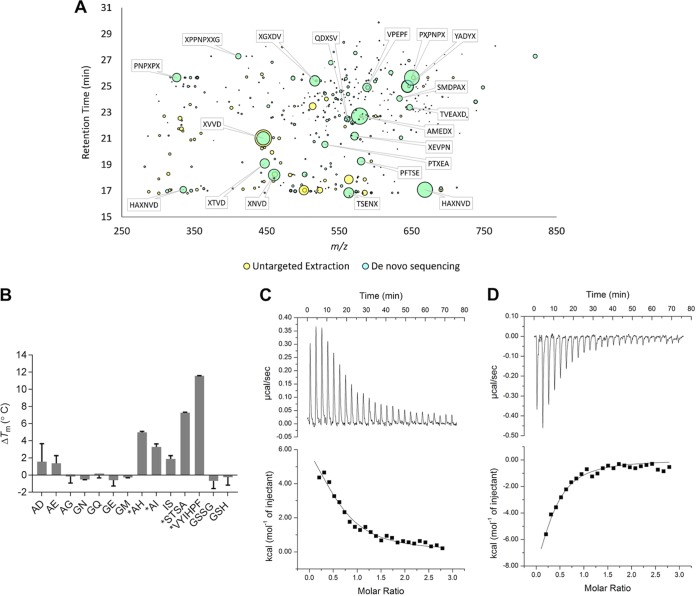
To test the hypothesis that HpDppA copurifies with peptides, the ligands bound to the protein were identified using liquid chromatography-mass spectrometry (LC-MS) following protein denaturation to release the bound small molecules. Untargeted feature extraction identified 180 chromatographic features unique to the HpDppA extract compared with the extract of a negative-control protein (the periplasmic amino acid-binding protein YckK from H. pylori) (Fig. 4A) (53). The molecular masses of all the features were consistent with short peptides, and indeed, subsequent de novo sequencing analysis identified 43% of these unique features as peptides. Critically, this overlap was heavily biased toward the most abundant features, with 9 of the top 10 and 16 of the top 20 untargeted features also identified by de novo sequencing. In contrast, only 34 features of comparatively low abundance were uniquely identified in the YckK extract compared to their abundance in the HpDppA extract, none of which were identified by de novo sequencing (Fig. S2). Taken together, these analyses demonstrate that HpDppA copurifies with a heterologous mix of short peptides, which are predominantly between 4 and 8 amino acids in length.
FIG 4.
Identification of ligands for HpDppA. (A) The chromatographic features uniquely identified in HpDppA by untargeted feature extraction are plotted in m/z and retention time space with the bubble size being proportional to the peak area of the chromatographic curve. The features that were also identified by de novo sequencing are shown in cyan with the most abundant annotated with their putative sequence, where X may be either leucine or isoleucine. (B) Ligand screening using thermal shift assay. Proteins and ligands were assayed at 10 μM and 2 mM concentrations, respectively. *, ΔTm > 2°C. Values shown are the means ± standard deviations (SD) from a triplicate experiment. (C and D) ITC analysis of HpDppA-peptide interactions. Top, raw titration data for VYIHPF (C) and STSA (D). Bottom, normalized integrated heat of the reaction as a function of the ligand/protein molar ratio, with the solid line showing the least-squares fit of the data to a single-site binding model. Thermodynamics parameters derived from the fitting are presented in Table 2.
Quantification of peptide binding to HpDppA.
To measure the affinity of interactions between HpDppA and peptides, we first performed a thermal shift assay with a representative set of 10 different dipeptides, the tetrapeptide STSA, and the hexapeptide VYIHPF. The dipeptides AI and IS and the hexapeptide VYIHPF were chosen because DppA has been previously shown to be involved in their import into H. pylori (46); the remaining eight dipeptides were randomly selected from the ones available in the laboratory. In addition, the reduced and oxidized forms of the modified tripeptide glutathione were included in this analysis, since HpDppA shows a significant degree of sequence and structure similarity with the glutathione-binding protein from H. parasuis (50). Heat-induced protein unfolding in the presence and absence of a peptide was followed by monitoring the fluorescence of the Sypro dye, which increases as the dye binds to the exposed hydrophobic regions of the protein. The melting temperatures (Tm) of the protein in the absence and presence of a candidate ligand were calculated by fitting the data to a sigmoidal denaturation curve. Ligand binding typically increases protein thermal stability, and a more than 2°C increase in the Tm was selected as a threshold for potential binders. Analysis of the thermal unfolding profiles revealed that the hexapeptide VYIHPF, the tetrapeptide STSA, and three of the tested dipeptides (AH, AI, and IS) stabilized the protein (ΔTm > 2°C), while the remaining dipeptides and glutathione did not show a significant stabilizing effect at the concentrations tested (Fig. 4B).
For peptides with stabilizing effects of more than 2°C, ligand binding was confirmed by performing thermal shift assays with increasing concentrations of each potential ligand. Plots for two representative peptides (AH and VYIHPF) (Fig. S3) illustrate that the selected peptides increased the thermal stability of HpDppA in a concentration-dependent manner.
To obtain the thermodynamic parameters of the peptide binding to HpDppA, we performed isothermal titration calorimetry (ITC) measurements. ITC experiments revealed that VYIHPF binds endothermically to HpDppA with an apparent dissociation constant (Kd) of 7.8 ± 0.7 μM (mean ± standard deviation) (Fig. 4C; Table 2). The term “apparent dissociation constant” reflects the fact that the calculations did not factor in the displacement of the bound copurified peptides. In contrast, STSA binds exothermically to HpDppA, albeit with a similar affinity (Kd = 5.9 ± 0.6 μM) (Fig. 4D; Table 2). ITC experiments with dipeptides that stabilized HpDppA in a concentration-dependent manner showed no significant heat change (Fig. S4), indicating that the binding of these dipeptides is entropically driven. The Kd values for dipeptides were therefore calculated using the thermal shift assay data. For validation, we have calculated the Kd values for the tetrapeptide and hexapeptide using both approaches (ITC and thermal shift assay) and demonstrated that they were close (Table 2). Estimation of Kd from the thermal shift assay-derived data gave a value of ∼9 μM for all three dipeptides, AH, AI, and IS. Overall, our results show that HpDppA has a broad specificity both in terms of the amino acid sequence and length of a peptide ligand and that it binds peptides ranging between two and eight amino acids in length with affinity in a low micromolar range.
TABLE 2.
Thermodynamics parameters of peptide binding to HpDppA
| Method | Ligand | Mean value ± SDa |
||
|---|---|---|---|---|
| Kd at 25°C (μM) | Enthalpy (ΔH) (cal/mol) | Entropy (ΔS) (cal/mol/degree) | ||
| Isothermal titration calorimetry | VYIHPF | 7.78 ± 0.65 | 8,474 ± 1,121 | 51.8 ± 3.7 |
| STSA | 5.85 ± 0.56 | −11,800 ± 2,249 | −15.6 ± 7.6 | |
| Thermal shift assay | VYIHPF | 7.15 ± 0.11 | ND | ND |
| STSA | 7.06 ± 0.18 | ND | ND | |
| AH | 9.02 ± 0.01 | ND | ND | |
| AI | 9.04 ± 0.24 | ND | ND | |
| IS | 9.38 ± 0.40 | ND | ND | |
Values shown are from a triplicate experiment. ND, not determined.
Analysis of peptide-binding site.
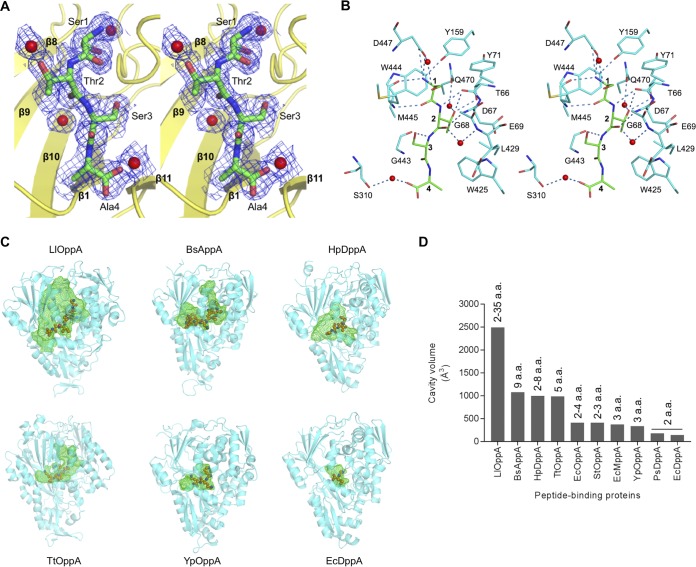
To establish whether peptides bind only at the interlobe cleft, which is a common ligand-binding site for all previously characterized periplasmic peptide-binding proteins, or at multiple sites, the crystal structure of HpDppA cocrystalized with 1 mM STSA was determined to 1.45-Å resolution. The unit cell parameters of the cocrystal with the peptide were very close to those of the crystal grown with no peptide added to the crystallization mixture (Table 1), and the protein backbones of the two crystal structures overlapped very well over their entire lengths (root mean square deviation [RMSD] of 0.2 Å for the aligned Cα atoms), indicating that cocrystallization with STSA also produced a closed form of the protein. Inspection of the electron density maps revealed no peptide-binding sites other than the one in the interlobe cleft. The electron density for both the main and side chains of the STSA molecule was clear and unambiguous (Fig. 5A), indicating that at 1 mM concentration, it outcompeted any peptides that may have copurified with HpDppA.
FIG 5.
Peptide-binding site of HpDppA and comparison to other ABC transporter-associated peptide-binding proteins. (A) Stereoview of the 2mFo-DFc sigma A-weighted electron density for tetrapeptide STSA bound to HpDppA. The map is contoured at 1 σ. (B) A walleye stereodiagram showing the protein side chains that form direct contacts with the bound peptide and hydrogen bonds important for ligand recognition. The ligand is shown with carbon atoms colored green and labeled 1 to 4. (C) Peptide-binding pockets in SBPs from different bacteria, cocrystallized with their native ligands. The protein orientation is similar for all structures, which are sorted according to the size of the cavity, from the largest to the smallest. The protein moieties are shown using a cartoon representation, the bound peptides are drawn using a ball and stick representation, and the ligand-binding cavities are shown using green mesh. LlOppA, oligopeptide-binding protein from L. lactis (PDB ID 3DRF); BsAppA, oligopeptide-binding protein from B. subtilis (PDB ID 1XOC); HpDppA, peptide-binding protein from H. pylori SS1 (this study); TtOppA, peptide-binding protein from T. thermophilus (PDB ID 2D5W); YpOppA, oligopeptide-binding protein from Y. pestis (PDB ID 2Z23); EcDppA, dipeptide-binding protein from E. coli (PDB ID 1DPP). (D) Comparison of the peptide-binding cavity volumes of different bacterial SBPs. The size ranges of their peptide ligands reported in the literature are shown on the top of the bars. a.a., amino acids.
Residues from all three domains contribute to recognition and binding of the STSA molecule. A total of 10 direct and four water-mediated hydrogen bond interactions are formed between the ligand and the surrounding protein residues. The N-terminal amino group of the ligand forms direct hydrogen bonds with the main chain carbonyl oxygen atom of Met445, side chain phenolic hydroxyl of Tyr159, and side chain Oδ1 and Oδ2 atoms of Asp447 (Fig. 5B). The carbonyl oxygen atom of the Ser1 residue of the ligand accepts a hydrogen bond from the main chain amide of Met445. Ser1 is further stabilized by a water-mediated hydrogen bond between its side chain hydroxyl and the Asp447 Oδ1 atom.
The main chain amide and carbonyl of the Thr2 residue of the ligand form two hydrogen bond interactions with the main chain carbonyl and amide of Thr66 and Gly68, respectively (Fig. 5B). The side chain Oγ1 atom of Thr2 makes direct hydrogen bonds to the main chain carbonyl and side chain Oγ1 of Thr66. The Thr2 residue is further stabilized by two water-mediated hydrogen bonds with protein residues Glu69 and Gln470.
The main chain amide of the third residue of the peptide ligand, Ser3, makes a direct hydrogen bond to the Gly443 O atom. The C-terminal residue of the ligand, Ala4, forms only one water-mediated hydrogen bond to the protein (to residue Ser310).
In addition to the hydrogen bonds, the bound tetrapeptide is stabilized by van der Waals interactions with the protein (Fig. 5B). The side chains of Tyr71 and Met445 form van der Waals contacts with the side chain of Ser1, while the side chain of Trp444 forms van der Waals contacts with its main chain moiety. In addition, the side chain of Trp444 makes van der Waals contacts with the main chain of Thr2. Thr2 is further stabilized by extensive van der Waals interactions of its side chain with the side chains of Asp67, Trp425, and Leu429 and with the main chain of Asp67. Ser3 and Ala4 make van der Waals contacts with the side chains of Met445 and Trp425, respectively.
Insights into structural basis of ligand specificity of HpDppA.
To obtain a structural rationale for the observed ability of HpDppA to bind peptides ranging from two to eight amino acids in length, we first calculated the volume of its ligand-binding pocket and compared it to those of other SBPs specific to peptides (Fig. 5C and D; Table 3). We observed a clear correlation between the size of the cavity in an SBP and the maximum length of a peptide ligand it has been reported to recognize. EcDppA (35) and PsDppA (27), which show a preference for dipeptides, possess the smallest cavities (∼140 Å3 and 179 Å3, respectively). SBPs that recognize peptides of up to three residues in length have ligand-binding pockets with slightly larger volumes, ranging between 333 Å3 (Y. pestis OppA) (39) and 408 Å3 (S. Typhimurium OppA) (32). At the other end of the spectrum, OppA from L. lactis (34), which binds peptides between 2 and 35 residues long, possesses the largest (∼2,494 Å3) cavity among these SBPs. In comparison, the volume of the ligand-binding pocket in HpDppA (∼996 Å3) is closest to those of Thermus thermophilus OppA (∼986 Å3) (PDB ID 2D5W) and BsAppA (∼1,075Å3) (40), which were crystallized in complex with a pentapeptide and a nonapeptide, respectively. Thus, our analysis shows that the ligand-binding pocket in HpDppA is indeed large enough to accommodate peptides of up to ∼9 amino acids in length.
TABLE 3.
Volumes of the peptide-binding cavities in SBPs from different bacteria
| Protein | Microorganism | Reference | Peptide length (amino acids) | Cavity volume (Å3) |
|---|---|---|---|---|
| OppA | L. lactis | 34 | 2–35 | 2,494 |
| AppA | B. subtilis | 40 | 9 | 1,075 |
| DppA | H. pylori | This work | 2–8 | 996 |
| OppA | T. thermophilus | PDB ID 2D5W | 5 | 986 |
| OppA | E. coli | 28 | 2–4 | 413 |
| OppA | S. Typhimurium | 32 | 2 or 3 | 408 |
| MppA | E. coli | 54 | 3 | 373 |
| OppA | Y. pestis | 39 | 3 | 333 |
| DppA | Pseudoalteromonas sp. | 27 | 2 | 179 |
| DppA | E. coli | 35 | 2 | 140 |
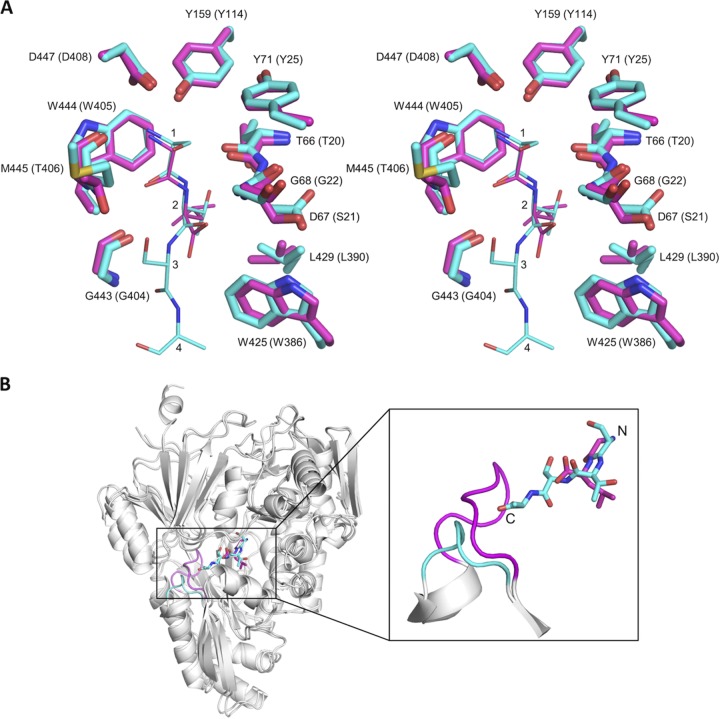
The overall structural similarity between HpDppA and EcDppA (35), which in contrast to the former, shows a strong preference for dipeptides, then allowed us to address the question of the structural factors that define the size of the pocket and, hence, the specificity with regard to the length of the peptide ligand. Figure 6A shows a close-up view of the superposition of the ligand-binding pockets of the two proteins in complex with their respective peptide ligands. The dipeptide bound to EcDppA overlaps remarkably well with residues 1 and 2 of the tetrapeptide ligand bound to HpDppA, and most of the side chains that directly interact with the peptide ligand in HpDppA and EcDppA (in parentheses) are conserved as follows: Tyr71 (Tyr25), Tyr159 (Tyr114), Trp425 (Trp386), Leu429 (Leu390), Trp444 (Trp405), and Asp447 (Asp408), with one conservative amino acid substitution (Asp67 [Ser21]). In both complexes, the α-amino group of the peptide ligand is anchored in a pocket formed by the side chains of Tyr159, Trp444, and Asp447 and the main chain moiety of Met445 (Fig. 6A), forms extensive interactions with the protein, and is completely shielded from the solvent. This conserved pocket defines the N-terminal end of the peptide-binding site in HpDppA and EcDppA, which serves to lock in place the α-amino group of any peptide ligand of these proteins.
FIG 6.
Comparison of the structures of the HpDppA/STSA and EcDppA/GL crystal complexes. (A) Stereoview of the superposition of the ligand-binding sites of HpDppA (carbons colored cyan) and EcDppA (carbons colored magenta). Included are the tetrapeptide STSA bound to HpDppA and the dipeptide GL bound to EcDppA. The residues of the tetrapeptide ligand are labeled 1 to 4. Residue numbering in parentheses is for EcDppA. (B) Ribbon diagram of the HpDppA/STSA complex superimposed on the EcDppA/GL complex (PDB ID 1DPP), highlighting difference in lengths and positions of the loop from residue 397 to 401 in HpDppA (cyan) and the corresponding loop in EcDppA (magenta). The carbons in the tetrapeptide STSA bound to HpDppA and the dipeptide GL bound to EcDppA are also colored cyan and magenta, respectively.
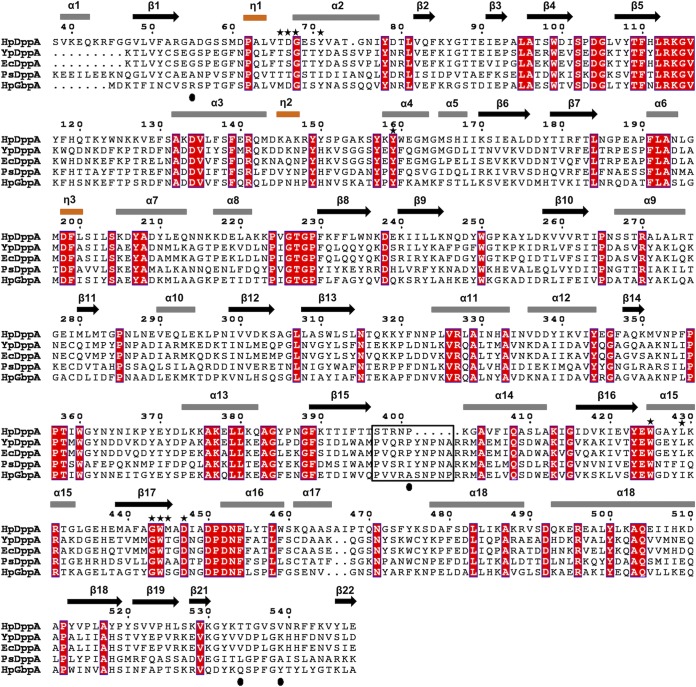
Inspection of the superimposed structures around the C-terminal ends of the peptide ligands revealed that the most noticeable difference between the two proteins concerns the loop from residue 397 to 401 in HpDppA, which is significantly (five residues) shorter than the corresponding loop in EcDppA (Fig. 6B). In the structure of the EcDppA/dipeptide complex, this loop serves as a stereochemical barrier that shortens the peptide-binding pocket at the C-terminal end and restricts the available space to peptides of only 2 or 3 amino acid residues, consistent with EcDppA’s ability to bind dipeptides and, to a lesser degree, tripeptides. In contrast, the absence of this steric barrier in HpDppA creates access to an extended groove that is, in turn, connected to the bulk solvent at the C-terminal end of the STSA peptide (Fig. 6B), which is in agreement with HpDppA’s ability to bind longer peptides, in addition to dipeptides. This finding provides a potential explanation for the strong conservation of the longer loop in the SBPs with specificity toward peptides 2 or 3 residues long (EcDppA, YpDppA, and PsDppA) (Fig. 7) and highlights the important role of HpDppA residues 397 to 401 in defining its broad specificity in terms of the lengths of peptides it can bind.
FIG 7.
A structure-based sequence alignment of HpDppA with DppAs and a glutathione-binding protein. HpDppA, DppA from H. pylori SS1; YpDppA, DppA from Y. pestis; EcDppA, DppA from E. coli; PsDppA, DppA from Pseudoalteromonas sp. SM9913; HpGbpA, glutathione-binding protein GbpA from H. parasuis. The secondary structure elements and the sequence numbering for HpDppA are displayed above the alignment. Conserved residues are boxed in red. The residues of HpDppA that form direct and water-mediated interactions with the bound tetrapeptide are marked with stars. The unique ligand-binding-site residues that are present in HpGbpA but absent in DppAs are indicated by filled circles. Residues that form the loop from position 397 to 401 in HpDppA and the equivalent loop residues in other proteins are highlighted using a black box. The image was generated using ESPript (http://espript.ibcp.fr).
The crystal structure of the HpDppA complex with STSA reveals that the two N-terminal residues of the peptide (Ser1 and Thr2) engage in multiple van der Waals and hydrogen-bonding interactions with HpDppA. Of note, most (9 of 10) of the direct hydrogen bonds made by the ligand involve these two residues. Calculations of the surface area buried upon interaction with the protein give values of 93% and 82% for Ser1 and Thr2, respectively, indicating that, in common with other DppAs, the first two amino acid residues of the ligand are buried deeply in the ligand-binding cleft, illustrating the importance of subsites 1 and 2 for ligand recognition. In contrast, residues Ser3 and Ala4 make relatively few contacts with the protein, with only 69% and 55% of the surface buried. The side chain of Ser3 is fully accessible to the solvent. The side chain of Ala4 is also largely solvent exposed. Thus, the structure of the complex suggests that the two amino acid residue-binding subsites (1 and 2) in HpDppA’s ligand-binding pocket might make an important contribution to the protein-ligand binding energy. This observation is consistent with the results of our binding experiments that show that the binding affinity of HpDppA to dipeptides is only slightly less than the affinity to longer peptides.
To elucidate the structural basis for the observed promiscuous binding of HpDppA to peptides of diverse amino acid sequences, we inspected the interactions stabilizing the bound tetrapeptide STSA, analyzing how substitutions for its residues 1 to 4 might affect the complex formation. Several notable features were observed when examining the mode of peptide binding from this angle. First, 8 of 10 direct hydrogen bonds between the protein and peptide involve the peptide’s main chain, rather than side chain atoms (Fig. 5B). Second, the protein residues forming these bonds are strongly conserved in DppAs from different bacteria (Fig. 7). Third, the protein residues that form van der Waals contacts with the peptide’s main chain atoms (Trp425 and Trp444) are conserved, whereas the few residues that make van der Waals contacts to the peptide’s side chain atoms (Asp67, Tyr71, Leu429, and Met445) are not (Fig. 7). Fourth, the latter group of protein side chains is positioned tangentially to the side chains of the peptide, so that variation of the side chain length at position 1, 2, 3, or 4 would not introduce steric clashes with the protein. These observations suggest that, in common with other DppAs, the H. pylori protein evolved to specifically recognize the invariant moiety of a peptide—its main chain, with the geometry of the ligand-binding pocket imposing few or no restraints on the nature of the peptide’s side chains.
DISCUSSION
The dppA gene is present in all H. pylori strains whose genomic sequences are deposited in the BioCyc database (55), suggesting its important nature. The dpp and opp peptide transporter systems of H. pylori are thought to play a crucial role in fulfilling the carbon requirements of its metabolic pathways, as H. pylori lacks the enzymatic machinery for utilizing complex carbohydrates (56). Due to the proteolytic action of pepsin, peptides are abundant in the stomach. The peptides imported via the dpp and opp systems, including peptidoglycan-recycling products, are hydrolyzed by the cytoplasmic aminopeptidase PepA/M17AP into amino acids (57, 58), which serve as a major source of carbon. Indeed, studies on the growth requirements of different H. pylori strains have shown an absolute need for several amino acids supplied exogenously (59, 60).
The crystal structure of HpDppA revealed that it adopts a type II/cluster C SBP fold. Consistent with its role as an ABC transporter-associated SBP, its gene is located upstream from the genes encoding two ABC transporter permeases (dppBC) and two ATP-binding proteins (dppDF). The results of our study suggest that, to fulfil its role in the utilization of exogenous peptides as nutrients, HpDppA directly binds, with similar affinity, peptides of very diverse amino acid sequences, ranging between two and eight residues in length. Analysis of the crystal structure of the complex of HpDppA with the tetrapeptide STSA provided a structural rationale for the observed broad specificity of this SBP. It revealed a ligand-binding pocket that is large enough to accommodate peptides of up to nine residues in length. The extensive and specific nature of hydrogen bond and van der Waals interactions with the main chain moiety of the peptide, in contrast to a limited number of contacts with the peptide’s side chain atoms, indicates that HpDppA evolved to recognize a wide range of peptides via their common part—the main chain. Furthermore, our analysis revealed that the two subsites accommodating N-terminal residues 1 and 2 of the peptide bound to HpDppA are formed by conserved protein residues and make an important contribution to the protein-ligand binding energy, in agreement with our binding data that showed that dipeptides bind to DppA with affinity that is close to that of longer peptides. The dissociation constants for dipeptide, tetrapeptide, and hexapeptide binding to HpDppA (∼6 to 9 μM) fall within the range of values reported for other SBPs specific to peptides (0.33 nM to 100 μM) (28, 31, 32, 61).
Our finding that, unlike previously characterized DppA proteins, HpDppA binds dipeptides and longer peptides with similar affinity is supported by the previous mutagenesis study that provided evidence that HpDppA is required for uptake of certain dipeptides, hexapeptides, and nonapeptides and contributes to uptake of tri- and tetrapeptides in H. pylori, as the dppA mutants’ ability to use these peptides was drastically reduced (46). It was previously noted that, in this respect, the function of the Dpp transporter in H. pylori is more similar to that of oligopeptide permease (Opp) systems found in other bacteria (46). The annotation was likely based on the fact that the sequences of the H. pylori dpp genes show higher similarity to those of dpp genes than of opp genes in other species. Indeed, we found that DppAs that are structurally similar to HpDppA (YpDppA and EcDppA) share close to 40% amino acid sequence identity with this protein, while the structurally similar substrate-binding protein of the oligopeptide transporter named BsAppA shares only 27% sequence identity with HpDppA.
Intrigued by the observed structural similarity between HpDppA and the glutathione-binding protein GbpA from H. parasuis (HpGbpA) (51), we tested the possibility that HpDppA recognizes glutathione as one of its natural ligands. We did not detect any binding between HpDppA and reduced or oxidized glutathione under conditions where HpDppA interactions with other peptides were significant and measurable. It is well recognized that GbpAs and DppAs are highly similar in their amino acid sequences (50, 51). Vergauwen et al. identified unique conserved residues in the glutathione-binding site of GbpAs that are absent in DppAs and can therefore be used to distinguish between the two (50). Analysis of the structure-guided sequence alignment of HpDppA and HpGbpA (Fig. 7) reveals that HpDppA has an alanine residue (Ala56) at the position equivalent to the conserved arginine that forms an ion pair with the C terminus of the bound glutathione in HpGbpA (50). In addition, in common with many other DppAs, HpDppA has a proline residue (Pro401) at the position where GbpAs use a nonproline residue with its amide group available for hydrogen bonding with the glutathione. Furthermore, HpDppA has a valine residue (Val539) at the position equivalent to the conserved Tyr521, the aromatic ring of which forms favorable interactions with the glutathione molecule in GbpAs. Finally, HpDppA lacks a glutamine residue at position 534, which, in GbpAs, stabilizes the γ-Glu residue of glutathione. These structural differences between HpDppA and GbpAs are consistent with our observation that HpDppA does not bind glutathione.
We also note that HpDppA shares 39% amino acid sequence identity with the dual-specificity protein HbpA from H. influenzae (UniProtKB ID P33950), which is a cognate SBP of the H. influenzae ABC transporter system Dpp. HbpA recognizes both dipeptides and heme (50, 62), but the amino acid residues important for heme recognition have not yet been identified. Although beyond the scope of this study, this similarity raises a question of whether HpDppA recognizes heme in addition to peptides, which may be an interesting area for future research. Furthermore, HpDppA was shown to directly bind and to mediate a negative (repellent) chemotactic response to the quorum signal autoinducer 2 (AI-2) in H. pylori strain G27 (63), which raises an important question of whether AI-2 competes with peptides for the same binding site in HpDppA.
MATERIALS AND METHODS
Gene cloning, overexpression, and protein purification.
The gene encoding DppA from H. pylori strain SS1 (GenBank accession number AQM65333.1) minus the signal peptide (residues 1 to 22) was codon optimized for expression in E. coli, synthesized, and ligated into the pet151/D-TOPO expression vector (Invitrogen, Waltham, MA) using GenScript. The construct contained an N-terminal His6 tag followed by the V5 epitope and a tobacco etch virus (TEV) protease cleavage site. The expression vector was transformed into E. coli BL21 (Novagen), and cells were cultured to exponential phase at 37°C in Luria Bertani (LB) medium containing 50 μg/ml ampicillin. Protein expression was induced by adding 1 mM isopropyl β-d-1-thiogalactopyranoside (Thermo Scientific) at an optical density at 600 nm (OD600) of 0.6 to 0.8. The cells were further grown for 4 h at 37°C and then harvested by centrifugation at 6,000 × g for 15 min at 4°C. The cell pellet was resuspended in buffer A (20 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1 mM phenylmethanesulfonyl fluoride) and lysed using an Avestin EmulsiFlex-C5 high-pressure homogenizer (Avestin, Ottawa, Canada). The cell lysate was centrifuged at 10,000 × g for 15 min at 4°C to pellet debris and inclusion bodies. Analysis of the pellet and supernatant using SDS-PAGE showed that the recombinant protein was mostly in the soluble fraction. The supernatant was passed through a 0.45-μm Whatman membrane filter, and NaCl and imidazole were added to final concentrations of 500 mM and 20 mM, respectively. The sample was loaded onto a 5-ml HiTrap chelating HP column (GE Healthcare, Chicago, IL) preequilibrated with buffer B (20 mM Tris-HCl [pH 8.0], 500 mM NaCl, 20 mM imidazole). The column was washed with 30 column volumes of buffer B, and the protein was eluted with buffer C (20 mM Tris-HCl, pH 8.0, 500 mM NaCl, 500 mM imidazole). For all other experiments, the His6-V5 tag was cleaved using His-tagged TEV protease (1:10 [wt/wt] TEV/protein ratio; Invitrogen, Waltham, MA, USA) while dialyzing the sample at 4°C against buffer D (20 mM Tris-HCl [pH 8.0], 150 mM NaCl, 2 mM dithiothreitol [DTT], 1% [vol/vol] glycerol) overnight. NaCl and imidazole were then added to the sample to final concentrations of 500 mM and 20 mM, respectively. The uncleaved protein, cleaved tag, and TEV protease were removed on a HiTrap chelating HP column. The protein was further purified by passing through a Superdex 200 HiLoad 26/60 size exclusion column (GE Healthcare, Chicago, IL) equilibrated with buffer E (20 mM Tris [pH 8.0], 150 mM NaCl). The protein concentration was determined by Bradford assay, and purity was estimated by SDS-PAGE to be >95%.
LC-MS analysis.
Small molecules that copurified with HpDppA were extracted, subjected to liquid chromatography-mass spectrometry (LC-MS) analysis, and identified based on their masses. The substrate-binding protein YckK from H. pylori, prepared as previously described (53), was used as a negative control. Prior to extraction, samples were dialyzed against 30 mM ammonium bicarbonate, pH 8.5. Five hundred microliters of protein solution at a concentration of 2 mg/ml was denatured by boiling at 100°C for 10 min, and supernatant was collected after centrifugation. Samples were analyzed using a Dionex ultimate 3000 RSLCnano nanoscale-LC system coupled to a Q Exactive classic mass spectrometer (Thermo Scientific) through a nanoscale electrospray ionization (ESI) source. The LC was plumbed to allow 1 μl of sample to be analyzed on an Acclaim PepMap rapid separation LC (RSLC) analytical column (75-μm by 15-cm, 2-μm, 100-Å, C18 nanoViper column; Thermo Scientific) with an Acclaim PepMap 100 (100-μm by 2-cm, 5-μm, 100-Å, C18 nanoViper column; Thermo Scientific) acting as a guard column. The mass spectrometer was operated in positive ion and data-dependent acquisition mode, selecting the top 12 peaks from each cycle for fragmentation with a dynamic exclusion of 12.0 s. The full MS resolution was set to 140,000, with an automatic gain control (AGC) target of 3 × 106, maximum injection time (IT) of 120 ms, and scan range of 150 to 1,500 m/z. The tandem MS (MS/MS) resolution was set to 35,000, with an AGC target of 105, maximum IT of 120 ms, loop count of 12, isolation window of 1.0 m/z, and fixed first mass of “none.”
The LC-MS data were processed using two distinct workflows: untargeted feature extraction and de novo peptide sequencing. Untargeted feature extraction was performed by the unbiased extraction of chromatographic features based on the MS1 data using MZmine 2.26 (64) (a complete list of the parameters used for this processing is included in the supplemental material). Untargeted extraction yielded several hundred features even in blank controls due to the small-molecule background. In order to identify features uniquely associated with a specific sample, only those features which were at least 10 times more abundant than a corresponding control (YckK) were retained.
De novo peptide sequencing was performed by converting all files to .mzXML format using MSConvert (65) and processing the resulting .mzXML files using Peaks Studio 64-bit version 7.5 (Bioinformatics Solutions, Inc.) with the parameters detailed in the supplemental material. Chromatographic features corresponding to peptides with an average local confidence (ALC) of >50% were then extracted using MZmine 2.26. In order to identify features unique to the HpDppA protein extract, only those features which were at least 10 times more intense than in the corresponding YckK control were retained.
Thermal shift assays.
Thermal shift assays were carried out using a Rotor-Gene Q real-time PCR instrument (Qiagen, Hilden, Germany). Each 25-μl reaction mixture contained 10 μM HpDppA, 10× Sypro orange reagent (5,000× stock, catalogue number S5692; Sigma-Aldrich), and ligand at the required concentration in buffer E. The experiment with reduced glutathione was performed in the presence of 0.5 mM DTT to prevent conversion to oxidized glutathione. Samples were thermally denatured by heating from 35°C to 90°C at a ramp rate of 0.5°C/min. Protein unfolding was monitored by following the Sypro orange fluorescence emission (excitation/emission wavelengths, λex 530 nm/λem 555 nm). The denaturation data were fit to a derivation of the Boltzmann equation for the two-state unfolding model, using GraphPad Prism, in order to obtain the midpoint of denaturation (the melting temperature [Tm]) (66). All experiments were performed in triplicate. Screening for potential binders was performed at a saturating concentration of ligands (2 mM, 200:1 ligand/protein ratio). For estimation of Kd, a 20:1 ligand/protein ratio was used (10 μM protein, 200 μM ligand). Calculations of the Kd values based on the thermal shift data at 25°C were performed as described previously (67).
Isothermal titration calorimetry.
The protein sample was dialyzed against buffer F (100 mM Tris [pH 8.0], NaCl 150 mM). Ligand solutions at a concentration of 300 μM were prepared in the same dialysis buffer. Measurements of heat change upon ligand binding to HpDppA were performed using a VP-ITC calorimeter (Malvern Instruments, Malvern, UK) at 25°C. An amount of 1.45 ml of 20 μM protein solution in a reaction cell was injected with 25 successive 10-μl aliquots of the ligand solution. Binding isotherms were generated by plotting the heat change per injection versus molar ratio of ligands to HpDppA. The data were fitted to the one-site binding model using nonlinear least-squares regression (Origin 7; OriginLab, USA), yielding the binding enthalpy ΔH, dissociation constant Kd, and binding entropy ΔS.
Crystallization, X-ray diffraction data collection, and processing.
Prior to crystallization, HpDppA was concentrated to 15 mg/ml using an Amicon Ultracel 10-kDa-cutoff concentrator (Merck Millipore, Darmstadt, Germany). Initial screening of crystallization conditions was performed by the hanging-drop vapor diffusion method using a Phoenix crystallization robot (Art Robbins Instruments, Sunnyvale, CA) and the JCSG+ suite (Qiagen). Crystals appeared after 10 days under conditions of 0.18 M triammonium citrate and 20% (wt/vol) polyethylene glycol (PEG) 3350. To improve the crystal quality, the initial crystallization conditions were optimized by increasing the concentrations of PEG 3350 and triammonium citrate to 23% and 0.25 M, respectively. Crystals formed after 3 days in trays set up manually using conditions of a 2-μl hanging drop over 500 μl reservoir solution. The crystals had symmetry of space group P212121 with unit cell dimensions of a = 65.1 Å, b = 76.0 Å, c = 127.9 Å, and had one subunit in the asymmetric unit. Cocrystals of HpDppA with 1 mM tetrapeptide Ser-Thr-Ser-Ala (STSA) were obtained under similar conditions. They had the same symmetry and similar unit cell parameters (a = 64.1 Å, b = 75.7 Å, c = 128.3 Å).
X-ray diffraction data were collected for the cryocooled crystal of HpDppA to 1.8 Å and for the HpDppA cocrystal with STSA to 1.45 Å at –173°C using the MX1 beamline of the Australian Synchrotron. Data were processed and scaled using XDS (68) and AIMLESS (69, 70).
Structure determination and analysis.
The phase problem was solved by molecular replacement with Phaser (71) using the coordinates of the putative periplasmic dipeptide-binding protein from Y. pestis (PDB ID 5F1Q) as a search model. Automated building of the preliminary model was performed with ARP/wARP (72). Model building was completed using alternating rounds of manual adjustment in Coot (73) and refinement using Phenix (74) and Refmac5 (75). The quality of the final models was evaluated using the MolProbity server (76). The volumes of the ligand-binding pockets were calculated using the online tool POCASA 1.1 with a probe radius of 2 Å (77).
Accession numbers.
The coordinates and structure factors of the HpDppA/STSA complex obtained by cocrystallization with 1 mM peptide have been deposited in the RCSB PDB under PDB ID 6OFQ. The coordinates and structure factors of HpDppA that was crystallized with no exogenous peptide added have been deposited under PDB ID 6PU3. The de novo peptide sequencing data are publicly available at https://store.erc.monash.edu/experiment/view/10585/.
Supplementary Material
ACKNOWLEDGMENTS
We thank Danuta Maksel and Geoffrey Kwai Wai Kong at the Monash Macromolecular Crystallization Facility for their assistance in setting up robotic crystallization screens. Part of this research was undertaken on the MX1 beamline of the Australian Synchrotron (AS), Victoria, Australia. We also thank the AS staff for their help with data collection.
M.M.R. acknowledges the support provided by the Australian Government Research Training Program Scholarship. M.A.M. is indebted to the Departamento Administrativo de Ciencia, Tecnología e Innovación COLCIENCIAS for a doctoral scholarship.
M. M. Rahman, M. A. Machuca, M. F. Khan, C. K. Barlow, and A. Roujeinikova designed the research; M. M. Rahman and C. K. Barlow performed the research; all authors analyzed data and wrote the paper.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00400-19.
REFERENCES
- 1.Juillard V, Le Bars D, Kunji ER, Konings WN, Gripon JC, Richard J. 1995. Oligopeptides are the main source of nitrogen for Lactococcus lactis during growth in milk. Appl Environ Microbiol 61:3024–3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matthews DM. 1975. Intestinal absorption of peptides. Physiol Rev 55:537–608. doi: 10.1152/physrev.1975.55.4.537. [DOI] [PubMed] [Google Scholar]
- 3.Smid EJ, Plapp R, Konings WN. 1989. Peptide uptake is essential for growth of Lactococcus lactis on the milk protein casein. J Bacteriol 171:6135–6140. doi: 10.1128/jb.171.11.6135-6140.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zheng F, Shao ZQ, Hao X, Wu Q, Li C, Hou H, Hu D, Wang C, Pan X. 2018. Identification of oligopeptide-binding protein (OppA) and its role in the virulence of Streptococcus suis serotype 2. Microb Pathog 118:322–329. doi: 10.1016/j.micpath.2018.03.061. [DOI] [PubMed] [Google Scholar]
- 5.Tynkkynen S, Buist G, Kunji E, Kok J, Poolman B, Venema G, Haandrikman A. 1993. Genetic and biochemical characterization of the oligopeptide transport system of Lactococcus lactis. J Bacteriol 175:7523–7532. doi: 10.1128/jb.175.23.7523-7532.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pletzer D, Lafon C, Braun Y, Kohler T, Page MG, Mourez M, Weingart H. 2014. High-throughput screening of dipeptide utilization mediated by the ABC transporter DppBCDF and its substrate-binding proteins DppA1-A5 in Pseudomonas aeruginosa. PLoS One 9:e111311. doi: 10.1371/journal.pone.0111311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Steiner HY, Naider F, Becker JM. 1995. The PTR family: a new group of peptide transporters. Mol Microbiol 16:825–834. doi: 10.1111/j.1365-2958.1995.tb02310.x. [DOI] [PubMed] [Google Scholar]
- 8.Hagting A, Kunji ER, Leenhouts KJ, Poolman B, Konings WN. 1994. The di- and tripeptide transport protein of Lactococcus lactis. A new type of bacterial peptide transporter. J Biol Chem 269:11391–11399. [PubMed] [Google Scholar]
- 9.Gomolplitinant KM, Saier MH Jr.. 2011. Evolution of the oligopeptide transporter family. J Membrane Biol 240:89–110. doi: 10.1007/s00232-011-9347-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Detmers FJ, Lanfermeijer FC, Poolman B. 2001. Peptides and ATP binding cassette peptide transporters. Res Microbiol 152:245–258. doi: 10.1016/S0923-2508(01)01196-2. [DOI] [PubMed] [Google Scholar]
- 11.Moraes PM, Seyffert N, Silva WM, Castro TL, Silva RF, Lima DD, Hirata R Jr, Silva A, Miyoshi A, Azevedo V. 2014. Characterization of the Opp peptide transporter of Corynebacterium pseudotuberculosis and its role in virulence and pathogenicity. Biomed Res Int 2014:1. doi: 10.1155/2014/489782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Samen U, Gottschalk B, Eikmanns BJ, Reinscheid DJ. 2004. Relevance of peptide uptake systems to the physiology and virulence of Streptococcus agalactiae. J Bacteriol 186:1398–1408. doi: 10.1128/jb.186.5.1398-1408.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garai P, Chandra K, Chakravortty D. 2017. Bacterial peptide transporters: messengers of nutrition to virulence. Virulence 8:297–309. doi: 10.1080/21505594.2016.1221025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang C-H, Lin C-Y, Luo Y-H, Tsai P-J, Lin Y-S, Lin MT, Chuang W-J, Liu C-C, Wu J-J. 2005. Effects of oligopeptide permease in group A streptococcal infection. Infect Immun 73:2881–2890. doi: 10.1128/IAI.73.5.2881-2890.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parra-Lopez C, Baer MT, Groisman EA. 1993. Molecular genetic analysis of a locus required for resistance to antimicrobial peptides in Salmonella typhimurium. EMBO J 12:4053–4062. doi: 10.1002/j.1460-2075.1993.tb06089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shiver AL, Osadnik H, Kritikos G, Li B, Krogan N, Typas A, Gross CA. 2016. A chemical-genomic screen of neglected antibiotics reveals illicit transport of kasugamycin and blasticidin S. PLoS Genet 12:e1006124. doi: 10.1371/journal.pgen.1006124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ge Y, Lee JH, Hu B, Zhao Y. 2018. Loss-of-function mutations in the Dpp and Opp permeases render Erwinia amylovora resistant to kasugamycin and blasticidin S. Mol Plant Microbe Interact 31:823–832. doi: 10.1094/MPMI-01-18-0007-R. [DOI] [PubMed] [Google Scholar]
- 18.Higgins C. 1987. Microbiology. Synthesizing designer drugs. Nature 327:655–656. doi: 10.1038/327655a0. [DOI] [PubMed] [Google Scholar]
- 19.Pletzer D, Braun Y, Weingart H. 2016. Swarming motility is modulated by expression of the putative xenosiderophore transporter SppR-SppABCD in Pseudomonas aeruginosa PA14. Antonie Van Leeuwenhoek 109:737–753. doi: 10.1007/s10482-016-0675-8. [DOI] [PubMed] [Google Scholar]
- 20.St Georgiev V. 2000. Membrane transporters and antifungal drug resistance. Curr Drug Targets 1:261–284. doi: 10.2174/1389450003349209. [DOI] [PubMed] [Google Scholar]
- 21.Garmory HS, Titball RW. 2004. ATP-binding cassette transporters are targets for the development of antibacterial vaccines and therapies. Infect Immun 72:6757–6763. doi: 10.1128/IAI.72.12.6757-6763.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hammond SM, Claesson A, Jansson AM, Larsson LG, Pring BG, Town CM, Ekstrom B. 1987. A new class of synthetic antibacterials acting on lipopolysaccharide biosynthesis. Nature 327:730–732. doi: 10.1038/327730a0. [DOI] [PubMed] [Google Scholar]
- 23.Davidson AL, Dassa E, Orelle C, Chen J. 2008. Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol Mol Biol Rev 72:317–364. doi: 10.1128/MMBR.00031-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.ter Beek J, Guskov A, Slotboom DJ. 2014. Structural diversity of ABC transporters. J Gen Physiol 143:419–435. doi: 10.1085/jgp.201411164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guyer CA, Morgan DG, Osheroff N, Staros JV. 1985. Purification and characterization of a periplasmic oligopeptide binding protein from Escherichia coli. J Biol Chem 260:10812–10818. [PubMed] [Google Scholar]
- 26.Olson ER, Dunyak DS, Jurss LM, Poorman RA. 1991. Identification and characterization of dppA, an Escherichia coli gene encoding a periplasmic dipeptide transport protein. J Bacteriol 173:234–244. doi: 10.1128/jb.173.1.234-244.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li CY, Chen XL, Qin QL, Wang P, Zhang WX, Xie BB, Su HN, Zhang XY, Zhou BC, Zhang YZ. 2015. Structural insights into the multispecific recognition of dipeptides of deep-sea gram-negative bacterium Pseudoalteromonas sp. strain SM9913. J Bacteriol 197:1125–1134. doi: 10.1128/JB.02600-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klepsch MM, Kovermann M, Low C, Balbach J, Permentier HP, Fusetti F, de Gier JW, Slotboom DJ, Berntsson RP. 2011. Escherichia coli peptide binding protein OppA has a preference for positively charged peptides. J Mol Biol 414:75–85. doi: 10.1016/j.jmb.2011.09.043. [DOI] [PubMed] [Google Scholar]
- 29.Doeven MK, Abele R, Tampe R, Poolman B. 2004. The binding specificity of OppA determines the selectivity of the oligopeptide ATP-binding cassette transporter. J Biol Chem 279:32301–323017. doi: 10.1074/jbc.M404343200. [DOI] [PubMed] [Google Scholar]
- 30.Smith MW, Tyreman DR, Payne GM, Marshall NJ, Payne JW. 1999. Substrate specificity of the periplasmic dipeptide-binding protein from Escherichia coli: experimental basis for the design of peptide prodrugs. Microbiology 145:2891–2901. doi: 10.1099/00221287-145-10-2891. [DOI] [PubMed] [Google Scholar]
- 31.Tame JR, Dodson EJ, Murshudov G, Higgins CF, Wilkinson AJ. 1995. The crystal structures of the oligopeptide-binding protein OppA complexed with tripeptide and tetrapeptide ligands. Structure 3:1395–1406. doi: 10.1016/S0969-2126(01)00276-3. [DOI] [PubMed] [Google Scholar]
- 32.Sleigh SH, Seavers PR, Wilkinson AJ, Ladbury JE, Tame JR. 1999. Crystallographic and calorimetric analysis of peptide binding to OppA protein. J Mol Biol 291:393–415. doi: 10.1006/jmbi.1999.2929. [DOI] [PubMed] [Google Scholar]
- 33.Detmers FJ, Lanfermeijer FC, Abele R, Jack RW, Tampe R, Konings WN, Poolman B. 2000. Combinatorial peptide libraries reveal the ligand-binding mechanism of the oligopeptide receptor OppA of Lactococcus lactis. Proc Natl Acad Sci U S A 97:12487–12492. doi: 10.1073/pnas.220308797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berntsson RP, Doeven MK, Fusetti F, Duurkens RH, Sengupta D, Marrink SJ, Thunnissen AM, Poolman B, Slotboom DJ. 2009. The structural basis for peptide selection by the transport receptor OppA. EMBO J 28:1332–1340. doi: 10.1038/emboj.2009.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dunten P, Mowbray SL. 1995. Crystal structure of the dipeptide binding protein from Escherichia coli involved in active transport and chemotaxis. Protein Sci 4:2327–2334. doi: 10.1002/pro.5560041110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nickitenko AV, Trakhanov S, Quiocho FA. 1995. 2 A resolution structure of DppA, a periplasmic dipeptide transport/chemosensory receptor. Biochemistry 34:16585–16595. doi: 10.1021/bi00051a006. [DOI] [PubMed] [Google Scholar]
- 37.Yoon HJ, Kim HJ, Mikami B, Yu YG, Lee HH. 2016. Crystal structure of a putative oligopeptide-binding periplasmic protein from a hyperthermophile. Extremophiles 20:723–731. doi: 10.1007/s00792-016-0861-7. [DOI] [PubMed] [Google Scholar]
- 38.Berntsson RP, Thunnissen AM, Poolman B, Slotboom DJ. 2011. Importance of a hydrophobic pocket for peptide binding in lactococcal OppA. J Bacteriol 193:4254–4256. doi: 10.1128/JB.00447-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tanabe M, Mirza O, Bertrand T, Atkins HS, Titball RW, Iwata S, Brown KA, Byrne B. 2007. Structures of OppA and PstS from Yersinia pestis indicate variability of interactions with transmembrane domains. Acta Crystallogr D Biol Crystallogr 63:1185–1193. doi: 10.1107/S0907444907048299. [DOI] [PubMed] [Google Scholar]
- 40.Levdikov VM, Blagova EV, Brannigan JA, Wright L, Vagin AA, Wilkinson AJ. 2005. The structure of the oligopeptide-binding protein, AppA, from Bacillus subtilis in complex with a nonapeptide. J Mol Biol 345:879–892. doi: 10.1016/j.jmb.2004.10.089. [DOI] [PubMed] [Google Scholar]
- 41.Lassaux P, Peri C, Ferrer-Navarro M, Gourlay LJ, Gori A, Conchillo-Sole O, Rinchai D, Lertmemongkolchai G, Longhi R, Daura X, Colombo G, Bolognesi M. 2013. A structure-based strategy for epitope discovery in Burkholderia pseudomallei OppA antigen. Structure 21:167–175. doi: 10.1016/j.str.2012.10.005. [DOI] [PubMed] [Google Scholar]
- 42.Locher KP. 2016. Mechanistic diversity in ATP-binding cassette (ABC) transporters. Nat Struct Mol Biol 23:487–493. doi: 10.1038/nsmb.3216. [DOI] [PubMed] [Google Scholar]
- 43.Draper JL, Hansen LM, Bernick DL, Abedrabbo S, Underwood JG, Kong N, Huang BC, Weis AM, Weimer BC, van Vliet AHM, Pourmand N, Solnick JV, Karplus K, Ottemann KM. 2017. Fallacy of the unique genome: sequence diversity within single Helicobacter pylori strains. mBio 8:e02321-16. doi: 10.1128/mBio.02321-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baltrus DA, Amieva MR, Covacci A, Lowe TM, Merrell DS, Ottemann KM, Stein M, Salama NR, Guillemin K. 2009. The complete genome sequence of Helicobacter pylori strain G27. J Bacteriol 191:447–448. doi: 10.1128/JB.01416-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, Ketchum KA, Klenk HP, Gill S, Dougherty BA, Nelson K, Quackenbush J, Zhou L, Kirkness EF, Peterson S, Loftus B, Richardson D, Dodson R, Khalak HG, Glodek A, McKenney K, Fitzegerald LM, Lee N, Adams MD, Hickey EK, Berg DE, Gocayne JD, Utterback TR, Peterson JD, Kelley JM, Cotton MD, Weidman JM, Fujii C, Bowman C, Watthey L, Wallin E, Hayes WS, Borodovsky M, Karp PD, Smith HO, Fraser CM, Venter JC. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 46.Weinberg MV, Maier RJ. 2007. Peptide transport in Helicobacter pylori: roles of Dpp and Opp systems and evidence for additional peptide transporters. J Bacteriol 189:3392–3402. doi: 10.1128/JB.01636-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fukami-Kobayashi K, Tateno Y, Nishikawa K. 1999. Domain dislocation: a change of core structure in periplasmic binding proteins in their evolutionary history. J Mol Biol 286:279–290. doi: 10.1006/jmbi.1998.2454. [DOI] [PubMed] [Google Scholar]
- 48.Scheepers GH, Lycklama A, Poolman B. 2016. An updated structural classification of substrate-binding proteins. FEBS Lett 590:4393–4401. doi: 10.1002/1873-3468.12445. [DOI] [PubMed] [Google Scholar]
- 49.Krissinel E, Henrick K. 2004. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr 60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 50.Vergauwen B, Elegheert J, Dansercoer A, Devreese B, Savvides SN. 2010. Glutathione import in Haemophilus influenzae Rd is primed by the periplasmic heme-binding protein HbpA. Proc Natl Acad Sci U S A 107:13270–13275. doi: 10.1073/pnas.1005198107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vergauwen B, Van der Meeren R, Dansercoer A, Savvides SN. 2011. Delineation of the Pasteurellaceae-specific GbpA-family of glutathione-binding proteins. BMC Biochem 12:59. doi: 10.1186/1471-2091-12-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sleigh SH, Tame JR, Dodson EJ, Wilkinson AJ. 1997. Peptide binding in OppA, the crystal structures of the periplasmic oligopeptide binding protein in the unliganded form and in complex with lysyllysine. Biochemistry 36:9747–9758. doi: 10.1021/bi970457u. [DOI] [PubMed] [Google Scholar]
- 53.Rahman M, Germantsis D, Machuca M, Ud-Din A, Roujeinikova A. 2017. Crystallisation and preliminary crystallographic analysis of Helicobacter pylori periplasmic binding protein YckK. Crystals 7:330. doi: 10.3390/cryst7110330. [DOI] [Google Scholar]
- 54.Maqbool A, Levdikov VM, Blagova EV, Herve M, Horler RS, Wilkinson AJ, Thomas GH. 2011. Compensating stereochemical changes allow murein tripeptide to be accommodated in a conventional peptide-binding protein. J Biol Chem 286:31512–31521. doi: 10.1074/jbc.M111.267179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Caspi R, Billington R, Ferrer L, Foerster H, Fulcher CA, Keseler IM, Kothari A, Krummenacker M, Latendresse M, Mueller LA, Ong Q, Paley S, Subhraveti P, Weaver DS, Karp PD. 2016. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res 44:D471–D480. doi: 10.1093/nar/gkv1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Doig P, de Jonge BL, Alm RA, Brown ED, Uria-Nickelsen M, Noonan B, Mills SD, Tummino P, Carmel G, Guild BC, Moir DT, Vovis GF, Trust TJ. 1999. Helicobacter pylori physiology predicted from genomic comparison of two strains. Microbiol Mol Biol Rev 63:675–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ki MR, Lee JH, Yun SK, Choi KM, Hwang SY. 2015. Roles of the peptide transport systems and aminopeptidase PepA in peptide assimilation by Helicobacter pylori. J Microbiol Biotechnol 25:1629–1633. doi: 10.4014/jmb.1505.05099. [DOI] [PubMed] [Google Scholar]
- 58.Modak JK, Rut W, Wijeyewickrema LC, Pike RN, Drag M, Roujeinikova A. 2016. Structural basis for substrate specificity of Helicobacter pylori M17 aminopeptidase. Biochimie 121:60–71. doi: 10.1016/j.biochi.2015.11.021. [DOI] [PubMed] [Google Scholar]
- 59.Reynolds DJ, Penn CW. 1994. Characteristics of Helicobacter pylori growth in a defined medium and determination of its amino acid requirements. Microbiology 140:2649–2656. doi: 10.1099/00221287-140-10-2649. [DOI] [PubMed] [Google Scholar]
- 60.Nedenskov P. 1994. Nutritional requirements for growth of Helicobacter pylori. Appl Environ Microbiol 60:3450–3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guyer CA, Morgan DG, Staros JV. 1986. Binding specificity of the periplasmic oligopeptide-binding protein from Escherichia coli. J Bacteriol 168:775–779. doi: 10.1128/jb.168.2.775-779.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Letoffe S, Delepelaire P, Wandersman C. 2006. The housekeeping dipeptide permease is the Escherichia coli heme transporter and functions with two optional peptide binding proteins. Proc Natl Acad Sci U S A 103:12891–12896. doi: 10.1073/pnas.0605440103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Anderson JK, Huang JY, Wreden C, Sweeney EG, Goers J, Remington SJ, Guillemin K. 2015. Chemorepulsion from the quorum signal autoinducer-2 promotes Helicobacter pylori biofilm dispersal. mBio 6:e00379. doi: 10.1128/mBio.00379-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pluskal T, Castillo S, Villar-Briones A, Oresic M. 2010. MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics 11:395. doi: 10.1186/1471-2105-11-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chambers MC, Maclean B, Burke R, Amodei D, Ruderman DL, Neumann S, Gatto L, Fischer B, Pratt B, Egertson J, Hoff K, Kessner D, Tasman N, Shulman N, Frewen B, Baker TA, Brusniak M-Y, Paulse C, Creasy D, Flashner L, Kani K, Moulding C, Seymour SL, Nuwaysir LM, Lefebvre B, Kuhlmann F, Roark J, Rainer P, Detlev S, Hemenway T, Huhmer A, Langridge J, Connolly B, Chadick T, Holly K, Eckels J, Deutsch EW, Moritz RL, Katz JE, Agus DB, MacCoss M, Tabb DL, Mallick P. 2012. A cross-platform toolkit for mass spectrometry and proteomics. Nat Biotechnol 30:918. doi: 10.1038/nbt.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Orwig SD, Lieberman RL. 2011. Biophysical characterization of the olfactomedin domain of myocilin, an extracellular matrix protein implicated in inherited forms of glaucoma. PLoS One 6:e16347. doi: 10.1371/journal.pone.0016347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pantoliano MW, Petrella EC, Kwasnoski JD, Lobanov VS, Myslik J, Graf E, Carver T, Asel E, Springer BA, Lane P, Salemme FR. 2001. High-density miniaturized thermal shift assays as a general strategy for drug discovery. J Biomol Screen 6:429–440. doi: 10.1177/108705710100600609. [DOI] [PubMed] [Google Scholar]
- 68.Kabsch W. 2010. XDS. Acta Crystallogr D Biol Crystallogr 66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Evans PR, Murshudov GN. 2013. How good are my data and what is the resolution? Acta Crystallogr D Biol Crystallogr 69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. 2011. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr 67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J Appl Crystallogr 40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Langer G, Cohen SX, Lamzin VS, Perrakis A. 2008. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat Protoc 3:1171–1179. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. 2011. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr 67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB III, Snoeyink J, Richardson JS, Richardson DC. 2007. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res 35:W375–383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yu J, Zhou Y, Tanaka I, Yao M. 2010. Roll: a new algorithm for the detection of protein pockets and cavities with a rolling probe sphere. Bioinformatics 26:46–52. doi: 10.1093/bioinformatics/btp599. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.