M. smegmatis generates an inadequate amount of energy by substrate-level phosphorylation and mandatorily requires oxidative phosphorylation (OXPHOS) for its growth and metabolism. Here, we explored the influence of suboptimal OXPHOS on M. smegmatis physiology and metabolism. M. smegmatis harbors two identical copies of the atpD gene, which codes for the ATP synthase β subunit. Here, we carried out the deletion of only one copy of atpD in M. smegmatis to understand the bacterial survival response in an energy-deprived state. M. smegmatis ΔatpD shows remarkable phenotypic, metabolic, and physiological differences from the wild type. Our study thus establishes M. smegmatis ΔatpD as an energy-compromised mycobacterial strain, highlights the importance of ATP synthase in mycobacterial physiology, and further paves the way for the identification of novel antimycobacterial drug targets.
KEYWORDS: ATP synthase, Mycobacterium smegmatis, mycobacteria, atpD, bedaquiline, electron transport chain, energy metabolism, oxidative phosphorylation
ABSTRACT
Mycobacterium tuberculosis, a bacterium that causes tuberculosis, poses a serious threat, especially due to the emergence of drug-resistant strains. M. tuberculosis and other mycobacterial species, such as M. smegmatis, are known to generate an inadequate amount of energy by substrate-level phosphorylation and mandatorily require oxidative phosphorylation (OXPHOS) for their growth and metabolism. Hence, antibacterial drugs, such as bedaquiline, targeting the multisubunit ATP synthase complex, which is required for OXPHOS, have been developed with the aim of eliminating pathogenic mycobacteria. Here, we explored the influence of suboptimal OXPHOS on the physiology and metabolism of M. smegmatis. M. smegmatis harbors two identical copies of atpD, which codes for the β subunit of ATP synthase. We show that upon deletion of one copy of atpD (M. smegmatis ΔatpD), M. smegmatis synthesizes smaller amounts of ATP and enters into an energy-compromised state. The mutant displays remarkable phenotypic and physiological differences from the wild type, such as respiratory slowdown, reduced biofilm formation, lesser amounts of cell envelope polar lipids, and increased antibiotic sensitivity compared to the wild type. Additionally, M. smegmatis ΔatpD overexpresses genes belonging to the dormancy operon, the β-oxidation pathway, and the glyoxylate shunt, suggesting that the mutant adapts to a low energy state by switching to alternative pathways to produce energy. Interestingly, M. smegmatis ΔatpD shows significant phenotypic, metabolic, and physiological similarities with bedaquiline-treated wild-type M. smegmatis. We believe that the identification and characterization of key metabolic pathways functioning during an energy-compromised state will enhance our understanding of bacterial adaptation and survival and will open newer avenues in the form of drug targets that may be used in the treatment of mycobacterial infections.
IMPORTANCE M. smegmatis generates an inadequate amount of energy by substrate-level phosphorylation and mandatorily requires oxidative phosphorylation (OXPHOS) for its growth and metabolism. Here, we explored the influence of suboptimal OXPHOS on M. smegmatis physiology and metabolism. M. smegmatis harbors two identical copies of the atpD gene, which codes for the ATP synthase β subunit. Here, we carried out the deletion of only one copy of atpD in M. smegmatis to understand the bacterial survival response in an energy-deprived state. M. smegmatis ΔatpD shows remarkable phenotypic, metabolic, and physiological differences from the wild type. Our study thus establishes M. smegmatis ΔatpD as an energy-compromised mycobacterial strain, highlights the importance of ATP synthase in mycobacterial physiology, and further paves the way for the identification of novel antimycobacterial drug targets.
INTRODUCTION
Tuberculosis is one of the most life-threatening diseases today, leading to very high mortality worldwide (1). Mycobacterium tuberculosis, the etiologic agent of tuberculosis (TB), is therefore considered the pathogen requiring immediate attention. The enhanced metabolic flexibility exhibited by M. tuberculosis makes the eradication of this pathogen difficult (2, 3). M. tuberculosis is known to adapt to its host environment by shifting to alternative carbon sources (4–6). The use of fatty acids as the sole carbon source via β-oxidation is particularly important for M. tuberculosis survival in vivo (5, 7, 8). Upon entering macrophages, M. tuberculosis downregulates its metabolism and primarily utilizes fatty acids and cholesterol rather than the fermentable carbon sugars as its sole carbon source (7–10). This immediately depicts fatty acids as an energy source because of the metabolic dependency of mycobacteria on fatty acids. Additionally, fatty acids also play an important role in mycolic acid and glycopeptidolipid (GPL) biosynthesis (11–13). While mycolic acid is an essential component of the impervious mycobacterial cell wall and is a characteristic feature of the family of mycobacteria (14), GPLs constitute an important class of cell wall-associated lipids which also contribute to mycobacterial pathogenicity (15). These two molecules are the key surface molecules that are found in various pathogenic and nonpathogenic mycobacterial species (13, 16). The presence of these unique lipids in the cell wall confers exceptional hydrophobicity to the cell and is also responsible for maintaining cell envelope integrity. Several antitubercular drugs are known to disrupt cell wall integrity by interfering with the synthesis of mycolic acids (17). However, due to the resistance developed toward these drugs, treatment of mycobacterial infections has become highly challenging.
Recently, oxidative phosphorylation (OXPHOS) has emerged as a novel drug target in mycobacteria (18–20). M. tuberculosis uses its flexible and branched electron transport chain (ETC) to generate the proton motive force (PMF) across the membrane, which is subsequently used by ATP synthase for ATP production (21–23). This key step depicts the importance of OXPHOS in mycobacteria, and, therefore, its inhibition would eventually lead to the cessation of ATP synthesis. Consequently, the bacterium is either eliminated or exhibits phenotypic changes ensuring survival. ATP synthase activity is known to be blocked by one of the diarylquinolone class of drugs called bedaquiline (BDQ), which more effectively targets nonreplicating mycobacteria because of the lower ATP requirements in the dormant state (20, 24–26). Furthermore, upon BDQ treatment, major metabolic alterations occur in the cell and several biosynthetic processes are downregulated (19, 25). Hence, exploring mycobacterial physiology in a low-energy state is required to understand how physiological adaptation occurs in mycobacteria.
Here, we have used Mycobacterium smegmatis mc2155 as our model to understand mycobacterial cellular behavior in an energy-compromised state and to identify changes that occur in its metabolism and physiology. M. smegmatis is known to harbor two copies of the entire ATP synthase operon, due to a large duplicated region in its genome (27). The ATP synthase machinery has been considered essential for mycobacterial survival (27). Indeed, deletion of both copies of atpD, coding for the β subunit of the ATP synthase machinery, hampers the survival of M. smegmatis (27, 28). Here, we carried out the deletion of only one of the two copies of atpD in M. smegmatis to obtain an energy-deprived state in order to understand the bacterial survival response. The M. smegmatis ΔatpD strain generated shows remarkable phenotypic, metabolic, and physiological differences from the wild type (WT) and also mimics to a great extent bedaquiline (BDQ)-treated wild-type M. smegmatis. Our study thus establishes M. smegmatis ΔatpD as an energy-compromised strain of mycobacteria, highlights the importance of ATP synthase in mycobacterial physiology, and further paves the way for the identification of novel antimycobacterial drug targets responsible for cellular adaptation in a low-energy state.
RESULTS
Disruption of one copy of the ATP synthase β-subunit gene results in an energy-compromised state with an altered growth profile.
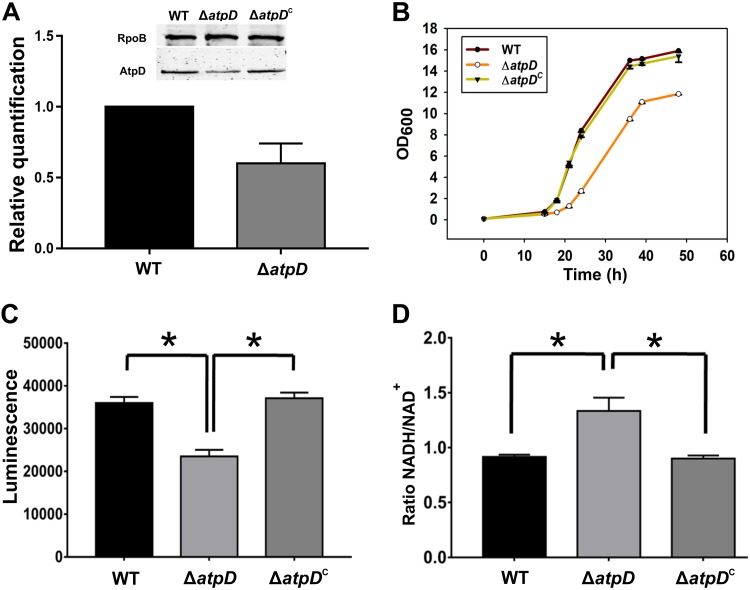
The M. smegmatis atpD gene (MSMEG_4936) belongs to a cluster of 8 genes with the gene order atpBEFHAGDC and codes for the β subunit of FoF1 ATP synthase. To understand the effect of energy depletion on mycobacterial physiology, we disrupted one copy of atpD in M. smegmatis (here referred to as M. smegmatis ΔatpD) by replacing it with a hygromycin resistance marker (Hygr) using an allelic exchange mutagenesis-based strategy (see Fig. S1 in the supplemental material) as described previously (29). Briefly, an allelic exchange substrate (AES) was synthesized by using the oligonucleotides listed in Table S1 and by following the method described previously (30). AES was then used to replace the wild-type gene in the M. smegmatis genome. The generated knockout was further confirmed by PCR and DNA sequencing (Fig. S1B to D). It is important to mention here that only one of the two copies of atpD was disrupted in the cell (Fig. S1E). Additionally, it has been reported earlier that the bacterium carrying the deletion of both copies of atpD does not survive (27). Moreover, we found the β-subunit level in M. smegmatis ΔatpD to be less than that in the wild-type bacterium, as confirmed by the reverse transcription-quantitative PCR (RT-qPCR) and Western blotting data (Fig. 1A), suggesting that, indeed, there are two identical copies of atpD and that only one copy of this gene was disrupted.
FIG 1.
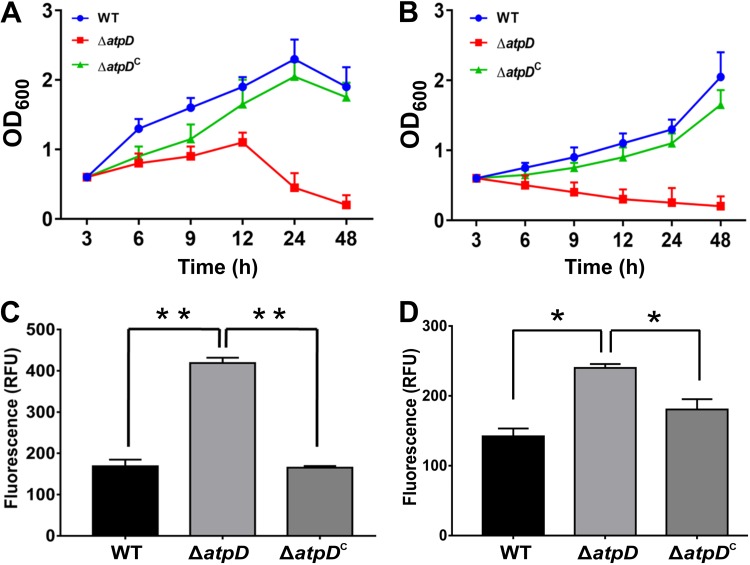
Monitoring of the expression of atpD, growth, and energy level in the M. smegmatis strains. (A) The plot shows the RT-qPCR-based atpD expression data obtained from the wild-type and the atpD-knockout strains of M. smegmatis. (Inset) Western blot of the AtpD protein in the wild-type, ΔatpD, and ΔatpDC strains. Western blotting was performed with anti-AtpD antibody, and the RNA polymerase anti-RpoB (β-subunit) antibody was used as a loading control. The experiment was repeated at least thrice; only one representative image is shown here. (B) Growth analysis of the WT, ΔatpD, and ΔatpDC strains in MB7H9 broth supplemented with 2% glucose as a carbon source at 37°C. The knockout grew slower than the other two bacteria. (C) Intracellular ATP estimation in the WT, ΔatpD, and ΔatpDC strains, plotted as luminescence. A reduced ATP level, in the form of lower luminescence in the ΔatpD strain than in the WT strain and the complemented strain, was observed. (D) Cellular NADH/NAD+ ratio in all three strains. In all panels, the data represent averages from at least three independent experiments, with standard deviations shown as error bars. *, a P value of ≤0.05 was considered significant.
ATP synthase is essential for mycobacterial survival in both the replicating and dormant states (18, 19, 25). Thus, inhibition of this essential pathway via any means is, in all likelihood, bound to affect the key metabolic pathways in the cell. We therefore asked how the inhibition of ATP synthase by disrupting only one copy of atpD affects the growth, energy profile, and physiology of the bacterium. We first recorded the M. smegmatis ΔatpD growth profile and compared it with that of the wild-type (WT) and the atpD-complemented (ΔatpDC) M. smegmatis strains. Complementation was achieved by introducing the wild-type atpD gene on a plasmid, pMVAcet (31), and expressing it from an acetamide-inducible promoter. M. smegmatis ΔatpD showed a slow growth phenotype, in contrast to the WT and the ΔatpDC strains (Fig. 1B), when grown in the presence of 2% glucose in Middlebrook 7H9 (MB7H9) medium. Additionally, we monitored the growth of the atpD-knockout strain in LB broth containing 0.05% Tween 80 and 20 mM glucose (LBT medium) at three different temperatures, 28, 37, and 42°C, as has been reported earlier (27). We found no difference in the growth of the wild-type and knockout strains in this medium at any of the mentioned temperatures (Fig. S2A to C). This suggests that the effect of the atpD gene deletion is largely confined to defined medium, such as MB7H9.
We next determined the amount of ATP in M. smegmatis ΔatpD and compared it with that in the WT and the complemented strains. We observed lower intracellular ATP levels in M. smegmatis ΔatpD strain than in the WT and ΔatpDC strains (Fig. 1C), which suggests that the mutant is unable to form sufficient amounts of ATP. This clearly indicates that an energy-compromised state prevails in the mycobacterial cell upon deletion of one copy of the atpD gene. Interestingly, such a difference in the ATP level was not observed when the cells were grown in LBT medium (Fig. S2D), further indicating that the effect of atpD deletion is indeed confined to the defined medium. We next determined the intracellular levels of NADH, a central redox cofactor of the bacterial cell, and its oxidized form, NAD+. We observe an elevated NADH/NAD+ ratio in M. smegmatis ΔatpD compared to the WT and ΔatpDC strains (Fig. 1D; Table S2). This indicates an altered redox state of the cell, which can be attributed to the partial loss of atpD, which likely exerts a stress on the mycobacterial ETC, thereby hindering the flow of electrons through the respiratory chain and, in turn, reducing NADH oxidation. Taken together, our results show that the M. smegmatis ΔatpD mutant has low ATP levels and an elevated NADH/NAD+ ratio and grows poorly compared to the wild-type bacterium.
The antibiotic BDQ targets the ATP synthase machinery and, therefore, disrupts normal ATP synthesis in the cell (20). We therefore asked if BDQ-treated wild-type M. smegmatis phenocopies the atpD-deleted strain. We first examined the production of AtpD in the wild-type strain as well as the atpD-deleted strain of M. smegmatis by Western blotting. Interestingly, in both the wild-type and the knockout strains, BDQ treatment resulted in a lowering of the AtpD level (Fig. S3A). Furthermore, growth of both the wild-type and the ΔatpD-knockout strains in the presence of a sublethal concentration of BDQ was reduced (Fig. S3B) compared to that in the untreated cultures (Fig. 1B). Very interestingly, the ATP levels in the wild-type cell upon BDQ treatment were comparable to those present in atpD-knockout cells without BDQ treatment (Fig. S3C). Taken together, the data suggest that, upon BDQ treatment, the wild-type bacterium shows phenotypic similarities to the knockout strain.
M. smegmatis ΔatpD shows altered cell surface properties.
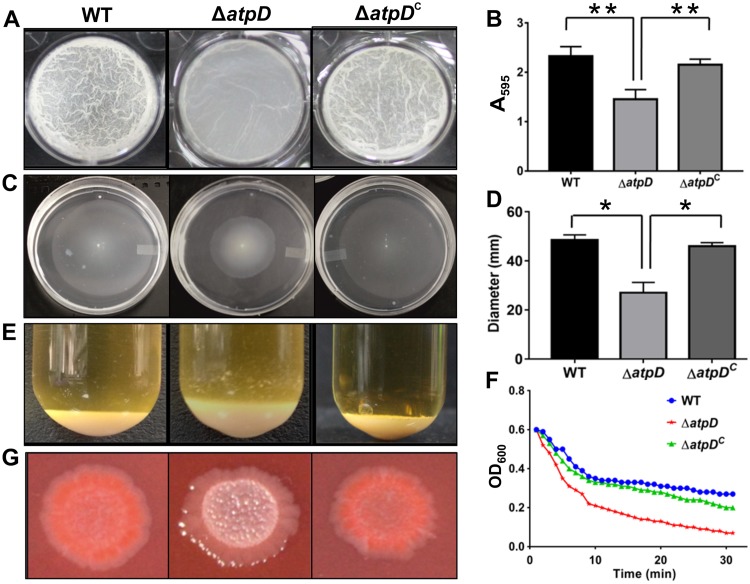
We next assessed the biofilm-forming ability of the M. smegmatis strain devoid of the atpD gene. A biofilm allows bacteria to survive in a stressful environment. Our data show that M. smegmatis ΔatpD was unable to form a mature biofilm (Fig. 2A), as its biofilm lacked the characteristic reticulation that was clearly seen in the biofilm formed by the WT and the ΔatpDC strains. A quantitative assay for the biofilm carried out with the crystal violet dye also showed that the mutant formed significantly less biofilm than the wild-type and the complemented strains (Fig. 2B). Further, an assessment of the level of expression of the genes involved in biofilm formation demonstrated that all the genes in question showed lower levels of expression than they did in the wild-type bacterium (Fig. S4). We also examined the sliding motility of the atpD-knockout strain. Mycobacteria are known to exhibit sliding motility, despite being nonflagellated, by means of the production of glycopeptidolipids (GPLs) (32). M. smegmatis ΔatpD showed a marked reduction in sliding motility compared to the wild-type and the complemented strains (Fig. 2C and D). Additionally, the cellular aggregation profile, as monitored by the change in the values of the optical density at 600 nm (OD600) with time, revealed that ΔatpD cells tended to aggregate more than the cells of the WT and the complemented strains (Fig. 2E and F). Here, a change in the aggregation profile of ΔatpD cells can be attributed to the enhanced hydrophobicity of the cell surface. To further confirm this observation, we examined the cell surface hydrophobicity of the atpD-knockout strain by estimating the degree of adherence of bacteria to hydrophobic molecules, such as n-hexadecane and xylene (33). Upon challenging the atpD-knockout strain with hydrophobic solvents, we observed a significant reduction in the bacterial density present in the aqueous phase, which suggests that, due to increased hydrophobicity, ΔatpD cells adhere more to the hydrophobic solvent than cells of the WT and the ΔatpDC strains do (Fig. S5). Additionally, we performed a Congo red (CR) dye-binding assay. CR dye is known to bind to lipoproteins present on the mycobacterial surface and is frequently used to identify cell wall modifications (34). Our ΔatpD mutant exhibited an altered colony morphology on a CR agar plate. Whereas the WT bacterium exhibited a reddish and shiny colony morphotype, the mutant formed smooth and whitish colonies (Fig. 2G); the atpD-complemented strain showed the WT phenotype. Taken together, our data suggest that distinct changes occur on the cell surface upon deletion of atpD.
FIG 2.
Phenotypic characterization of the atpD-knockout strain of M. smegmatis. A comparison of the phenotypes obtained for the WT, ΔatpD, and ΔatpDC strains under different assay conditions is shown. (A) The 7-day-old biofilm formed at the liquid-air interface in the presence of glucose as the carbon source by the three bacterial strains. (B) The graph shows the quantification of the biofilms performed using the crystal violet dye assay. The absorbance measured at 595 nm in each case is plotted. The data show that the mutant forms less biofilm than the WT and the complemented strains. (C) Sliding motility, monitored after 3 days of incubation at 37°C, is shown for all three strains. (D) The zone occupied by each bacterium on the agar plate was measured in millimeters and plotted. The mutant showed less sliding motility than the wild-type and the complemented strains. (E) The cellular aggregation profiles of the three strains. The mutant bacterium clearly showed a larger amount of cellular aggregation than the other two strains. (F) Aggregation of the indicated strains, as measured in the form of the change in OD600 values over a period of time after suspension of the cells in detergent-free medium, is plotted. The knockout strain showed higher aggregation with time than the other two strains. (G) The colony morphology of all three bacteria on a Congo red agar plate is shown. The mutant did not retain the red color in this assay. For all panels, experiments were carried out at least thrice; only one representative image is shown in each of panels A, C, E, F, and G. In panels B and D, the data are representative of those from three independent experiments, with error bars representing standard deviations. *, P ≤ 0.05; **, P ≤ 0.01.
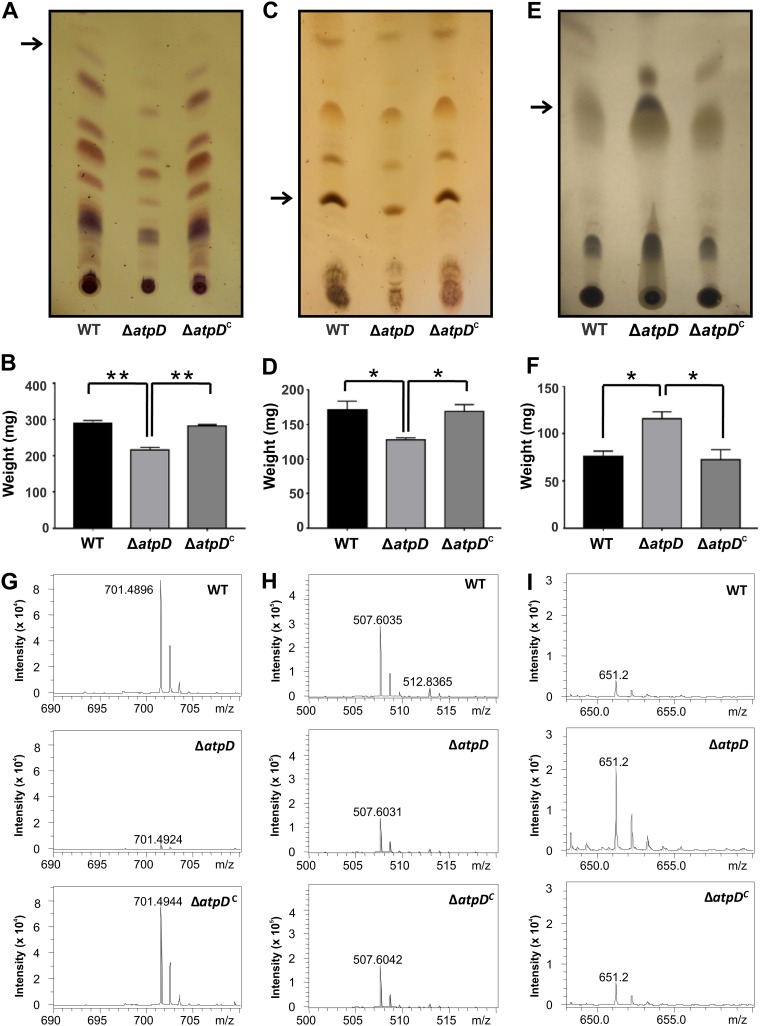
Thus, in the M. smegmatis ΔatpD mutant, we observed a reduction in sliding motility, enhanced cellular aggregation and hydrophobicity, a change in colony morphology, and reduced biofilm formation, which strongly indicate significant adjustments in the cell envelope lipid composition. Previous studies have also shown that an alteration in GPL content is known to alter the cell surface profile (33, 35). Furthermore, GPLs are known to regulate a wide variety of phenotypes, such as sliding motility, cellular aggregation, hydrophobicity, and colony morphology (13, 15, 32, 36). Conceivably, therefore, the mutant should show differences in the lipid profile. Hence, we next examined the effect of atpD disruption on bacterial lipids using thin-layer chromatography (TLC) and electron spray ionization mass spectrometry (ESI-MS) and compared the effect with that on the WT and the complemented strains. We observed that the ΔatpD mutant showed significantly lower levels of GPLs than the other strains (Fig. 3A and B).
FIG 3.
Comparison of the GPL, polar lipid, and apolar lipid profiles. Thin-layer chromatograms, quantification, and mass spectrometry data are shown for GPLs (A, B, and G), polar lipids (C, D, and H), and apolar lipids (E, F, and I). The lipids were extracted from the M. smegmatis wild-type (WT), atpD-knockout (ΔatpD), and complemented (ΔatpDC) strains. (A) TLC of the GPLs shows remarkable differences for the atpD-knockout strain. (B) Quantification of the GPLs shows lower values for the knockout strain. (G) Further, the ESI-MS data correspond to those for the band extracted from the TLC (marked with an arrow in panel A). Only one representative peak is shown; complete mass spectrometric data are available in Fig. S6 in the supplemental material. (C) Similarly, TLC of the polar lipids shows remarkable differences for the atpD-knockout strain. (D) Quantification of the polar lipids shows lower values for the knockout strain. (H) Further, the ESI-MS data correspond to those for the band extracted from the TLC (marked with an arrow in panel D). Only one representative peak is shown; complete mass spectrometric data are available in Fig. S7. (E) TLC of the apolar lipids shows remarkable differences for the atpD-knockout strain. (F) Quantification of the apolar lipids shows higher values for the knockout strain. (I) Further, the ESI-MS data correspond to those for the band extracted from the TLC (marked with an arrow in panel E). Only one representative peak is shown; complete mass spectrometric data are available in Fig. S8. For the experiments whose results are presented in panels A, C, and E, the experiments were repeated at least thrice, and only one representative image is shown in each case. In panels B, D, and F, the error bars represent standard deviations. *, P ≤ 0.05; **, P ≤ 0.01.
Additionally, TLC analysis of both polar and apolar lipids showed that while the amount of polar lipids was significantly less in the atpD-knockout strain than in the others (Fig. 3C and D), more apolar lipids were present (Fig. 3E and F). This is also in concurrence with our hydrophobicity data, where we observed an increase in the hydrophobicity in the cell envelope of the atpD-knockout strain (Fig. S5). Additionally, we performed ESI-MS analysis of the TLC-separated lipids. The mass spectrum of one representative peak in each of the lipid profiles for GPL and the polar and apolar lipids is presented (Fig. 3G to I). The mass spectrum shows that in the case of GPL, one specific peak of m/z 701.48 (H+) was present in the case of the wild-type and the complemented strains, whereas the same peak appeared with an extremely reduced intensity in the knockout. This mass corresponds to phosphatidylglycerol (37). Similarly, the m/z 507.60 (H+) peak, corresponding to diglycosyl diacylglycerol, is one representation of polar lipids (38). Further, the m/z 651.2 (H+) peak, corresponding to diacylglycerol (37), was largely present in the knockout strain. All of these data are summarized in Table S3. Further, complete mass spectrometric profiles of the extracted lipids are given in the supplemental material (Fig. S6, S7, and S8). Here, while the WT and complemented strains showed largely similar m/z profiles, the m/z profiles of the knockout strain showed significant differences from those of the WT and complemented strains. Thus, the ESI-MS data strongly suggest that atpD knockout has a huge effect on the cell envelope.
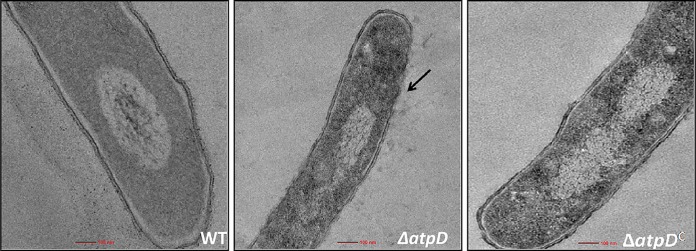
We imagined that such differences in the lipid profile would result in an altered cell envelope. Therefore, we analyzed the ultrastructure of the cell by transmission electron microscopy (TEM). Our studies demonstrated that the cell boundary in the case of the WT was intact but was altered in M. smegmatis ΔatpD (Fig. 4; Fig. S9); here, the complemented strain showed a wild-type-like structure. Taken together, these data strongly suggest that deletion of the atpD gene results in an altered cell envelope, lipid composition, and phenotype.
FIG 4.
Transmission electron microscopy-based ultrastructural analysis of M. smegmatis devoid of atpD. TEM micrographs of the WT, ΔatpD, and ΔatpDC strains (as indicated in each panel) are shown. The ultrastructure of the ΔatpD bacterium shows remarkable differences (marked with an arrow pointing to the altered cell envelope) from the other two. The experiments were performed at least thrice. Only one representative image is shown here. For more images, see Fig. S9 in the supplemental material.
An altered cell envelope leads to increased susceptibility to stresses and antibiotics.
During host cell infection, M. tuberculosis encounters various stresses, such as acidic stress, oxidative stress, and chemical stress (39). These factors play an important role in determining mycobacterial growth and survival within the host cell. The thick rigid cell envelope enables mycobacteria to withstand a variety of unfavorable conditions, and therefore, an altered cell envelope, as examined earlier, may lower the tolerance of mycobacteria to stresses and also affect cell permeability.
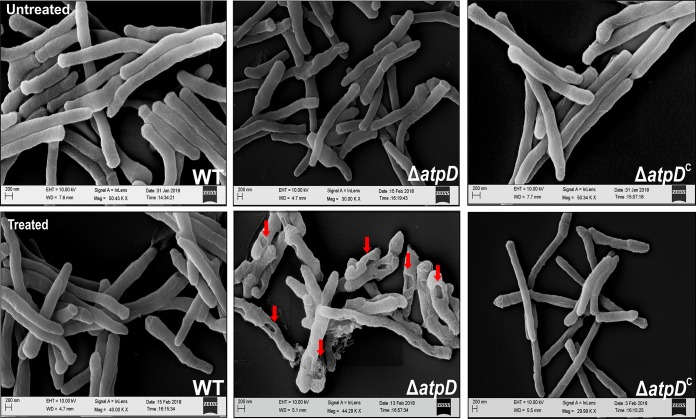
In order to assess the effect of external stress agents on the mycobacterial cell, we subjected the WT, ΔatpD, and ΔatpDC strains to various treatments, such as SDS, lysozyme, and acidic pH. We observed that the ΔatpD strain was more susceptible to 0.1% SDS treatment than the wild-type and the complemented strains (Fig. S10A). Additionally, examination of the cell surface architecture of both the wild-type and the mutant bacteria by scanning electron microscope (SEM) upon treatment with SDS revealed interesting characteristics (Fig. 5). While the cells generally appeared to be cylindrical in the case of the WT and ΔatpDC strains, distinct differences were apparent when the ΔatpD cells were treated with SDS. Whereas untreated cells of all three strains showed a normal morphology, treatment with 0.1% SDS altered the overall surface of only the cells of the mutant bacterium; in this case, the cell surface showed local pits that were absent in both the WT and the complemented strains.
FIG 5.
Scanning electron microscopy-based assessment of the effect of SDS on the cell surface of the atpD-knockout strain. SEM images of the WT, ΔatpD, and ΔatpDC strains with or without treatment (as indicated in the images) with 0.1% SDS are shown. The pits formed in the case of the atpD-knockout due to SDS treatment are marked with arrows. The experiments were performed at least thrice; only one representative image is shown in each case. EHT, extra-high tension; WD, working distance.
Next, upon treatment with different concentrations of lysozyme (25 μg/ml, 250 μg/ml), we found effective killing of the ΔatpD strain compared to that of the WT and the complemented strains (Fig. S10B). Similarly, bacterial survival assays carried out after subjecting the cells to acidic stress at pH 5.5 for 24 h showed the ΔatpD strain to be less tolerant than the others (Fig. S10C). Additionally, treatment of an M. smegmatis lawn with 5 mM H2O2, which is a known natural oxidant and which causes oxidative damage to the cell, on the MB7H9 agar plate resulted in a larger zone of clearance in the case of the atpD mutant than in the case of the wild-type and the complemented strains (Fig. S10D).
We next examined the sensitivity of the atpD-knockout strain to various antibiotics, such as tetracycline, chloramphenicol, rifampin, streptomycin, and vancomycin, at a concentration that is not lethal to wild-type M. smegmatis and found the mutant to be highly sensitive to all the antibiotics tested (Fig. S11). We also tested the sensitivity of M. smegmatis ΔatpD to isoniazid (Fig. 6A and Fig. S12A and B), which inhibits mycolic acid synthesis (39), and triclosan (Fig. 6B and Fig. S12C and D), which is a known fatty acid synthesis inhibitor in mycobacteria (40, 41), and compared the cultures with the untreated cultures (Fig. S12E). Our results clearly showed that the ΔatpD strain was rapidly killed by both the antibiotics tested at concentrations at which the wild-type and the complemented strains remained viable. Here, we hypothesized that the enhanced sensitivity to drugs in the case of the atpD mutant either was due to an increased uptake because of altered cellular permeability or was because the generally poor fitness of the mutant bacterium makes it more susceptible to antibiotics. In order to examine the former, we determined the intracellular accumulation of Nile red dye and ethidium bromide, which are known to be indicative of the uptake of hydrophobic and hydrophilic compounds, respectively (34). We observed an enhanced uptake of both the Nile red and ethidium bromide dyes by the atpD mutant compared to the wild-type and the complemented strains (Fig. 6C and D). Although these experiments strongly suggest that an increased killing of the mutant bacterium by the antibiotics may be due to increased uptake, we do not rule out the possibility that the poor metabolic profile of the mutant is the reason for the antibiotic-mediated enhanced cell death.
FIG 6.
Susceptibility to antibiotics and altered cellular permeability in the ΔatpD strain. (A and B) Growth profiles of the WT, ΔatpD, and ΔatpDC strains in the presence of 20 μg/ml of isoniazid (INH) and 30 μg/ml of triclosan, respectively. The data were recorded every 3 h after antibiotic addition at time zero. Reduced growth was observed for the ΔatpD strain compared to the other two. (C and D) Uptake of Nile red dye and ethidium bromide, respectively. In both cases, the knockout strain showed higher fluorescence due to increased uptake. In each panel, data are representative of those from three independent experiments, with error bars denoting standard deviations. *, P ≤ 0.05; **, P ≤ 0.01. RFU, relative fluorescence units.
In our experiments, we observed that the atpD-knockout strain was more sensitive to antibiotic treatment than the other strains. This made us postulate whether BDQ treatment of M. smegmatis makes it more sensitive to other antibiotics, in order to develop a combination therapy. We therefore tested the effect of BDQ and its combination with different antibiotics (isoniazid, rifampin, and tetracycline) on the M. smegmatis WT and ΔatpD strains. Very interestingly, we observed that addition of a sublethal concentration of BDQ (0.0001 μg/ml) along with a sublethal concentration of the other antibiotics, as specified below, enhanced the killing compared to the effect of the respective antibiotic alone in the culture medium. Our data thus clearly show that the treatment of the M. smegmatis WT and atpD-knockout strains with BDQ sensitizes the bacterium to other antibiotics (Fig. S13A and B), leading to enhanced killing of the bacterium.
Taken together, our results suggest that the disruption of a single copy of atpD alters the cell surface and the cell envelope architecture, which results in increased permeability to antibiotics as well as enhanced susceptibility to external stress agents. These results also tempt us to suggest that mycobacterial infections can be treated rapidly by the use of a combinatorial therapy, wherein an ATP synthase inhibitor, such as bedaquiline, is used along with another antibiotic(s).
The ΔatpD mutant shows respiratory slowdown with a higher level of ROS coupled with the upregulation of antioxidant genes.
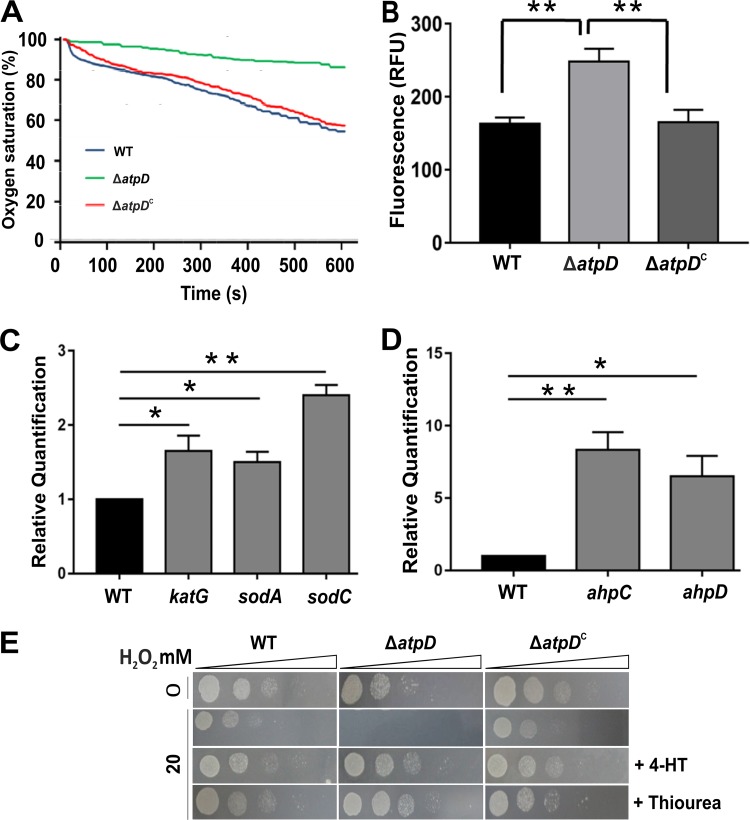
Mycobacterium is an obligate aerobe which depends on its ETC for energy production. Upon being targeted with an ETC inhibitor, M. tuberculosis is known to increase the total respiration rate in order to maintain cellular ATP levels (22). We therefore asked if the disruption of one copy of atpD results in a change in the respiration rate of the cell so as to maintain ATP levels and measured the oxygen consumption by the bacterium on a Clark-type oxygen electrode. We observed that the ΔatpD mutant showed a decreased respiration rate (i.e., decreased O2 consumption) compared with the wild-type and the complemented strains (Fig. 7A). Here, the control experiments were performed by using the known ETC inhibitors rotenone and antimycin (Fig. S14A to C). The data suggest that the mutant undergoes respiratory slowdown, most likely because of the lower availability of functional ATP synthase.
FIG 7.
The ΔatpD strain shows respiratory slowdown and is hypersensitive to peroxide and oxidative stresses. (A) Change in dissolved oxygen (i.e., percent oxygen saturation) that occurs in each case with time. At the start of the experiment, this value was considered 100%. Oxygen consumption in all three bacteria, viz., the WT, ΔatpD, and ΔatpDC strains, is shown as the reduction in oxygen saturation, which is observed as a downward slope. The mutant bacterium showed reduced oxygen consumption compared to the wild-type and the complemented strains. (B) Cellular ROS levels, as determined by use of the DCFH-DA dye. Higher fluorescence is suggestive of higher ROS levels, which was observed in the case of the knockout strain. (C and D) Gene expression analyses of several genes involved in the antioxidant defense system, carried out by RT-qPCR. The expression levels of katG, sodA, and sodC (C) and ahpC and ahpD (D) in the atpD-knockout strain with respect to those in the wild-type (control), which was considered 1, are shown. (E) MB7H9 agar plate images of the results of the spot assays carried out with the ΔatpD strain in the presence of H2O2 are shown. The knockout strain was found to be hypersensitive to peroxide stress. Bacteria were treated for 6 h with either 0 mM (top row) or 20 mM (subsequent three rows) of H2O2 and in the presence of 4-HT or thiourea (bottom two rows). Dilutions of the spotted cultures are marked as triangles above the panels. The ΔatpD strain was found to be susceptible to 20 mM H2O2 and could be rescued by antioxidants, such as 4-HT and thiourea. For the experiments whose results are presented in both panels A and E, the experiments were repeated at least thrice, and only representative data are shown. In panels B, C, and D, the data represent averages from at least three independent experiments with standard deviations. *, P ≤ 0.05; **, P ≤ 0.01.
We next asked whether this respiratory slowdown triggers an oxidative stress response within the cell. A previous report has suggested that inhibition of ATP synthase via bedaquiline results in increased reactive oxygen species (ROS) production (42). Furthermore, a higher cellular NADH/NAD+ ratio is thought to increase the reducing state of respiratory complexes, leading to an oxidative stress response in the cell (42). We therefore examined the ROS levels in the ΔatpD strain using an ROS-sensitive fluorescent dye, dichlorodihydrofluorescein diacetate (DCFH-DA). A significant increase in ROS production was observed in the ΔatpD strain compared with the WT and the complemented strains (Fig. 7B). This suggests that the deletion of a copy of atpD interferes with the functioning of ETC, thus creating oxidative stress and leading to cellular ROS production. In addition, we also measured the expression levels of several antioxidant genes, such as those for superoxide dismutase (sod), catalase (katG), and alkyl hydroperoxidase (ahpC and ahpD), and found them to be upregulated in the ΔatpD strain compared with the wild-type and the complemented strains (Fig. 7C and D).
Hydrogen peroxide is a known natural oxidant and causes oxidative damage to the cell, leading to a variety of cellular perturbations involving DNA, proteins, and lipids. We observed that the ΔatpD strain was more sensitive to H2O2 treatment than the other strains (Fig. S10D). To further test the effect of peroxide stress on the ΔatpD strain, we performed spot assays with different concentrations of H2O2. The ΔatpD cells were found to be sensitive to 20 mM H2O2, in contrast to the WT (Fig. 7E). This growth defect of the ΔatpD strain due to H2O2 could be completely rescued in the presence of 4-hydroxyl-2,2,6,6-tetramethylpiperidyl-1-oxy (4-hydroxy-TEMPO or 4-HT), which is a known chelator of peroxides (Fig. 7E). Similar results were obtained when thiourea was used as a reducing agent (Fig. 7E). These results strongly indicate that the deletion of one copy of atpD results in the disruption of cellular redox homeostasis in M. smegmatis. Additionally, BDQ-treated M. smegmatis wild-type cells, very interestingly, showed an enhanced upregulation of oxidative stress response genes (Fig. S15A), further suggesting that the effect of treatment of M. smegmatis with a sublethal concentration of BDQ mimics, to a great extent, that in the atpD gene-knockout strain.
atpD deletion causes modulation and rerouting of major metabolic pathways.
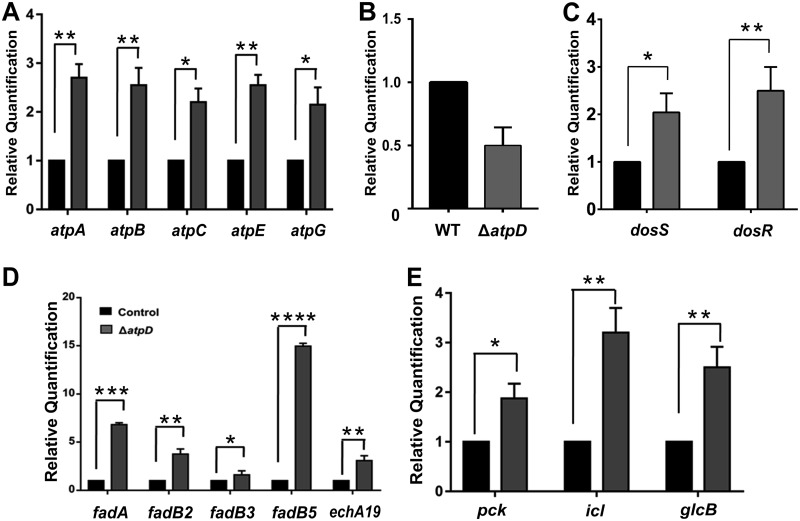
Reduced energy levels alter bacterial energy metabolism, which is essential for cell survival in an energy-compromised state. When ATP levels are low, bacteria modulate their major metabolic pathways in order to maintain their viability in a low-energy state (20). Therefore, to explore the bacterial survival pathways functional under such conditions, we examined the differential expression of the genes involved in various metabolic pathways in M. smegmatis ΔatpD. We observed the overexpression of genes belonging to the ATP synthase operon (Fig. 8A), which demonstrates the bacterium’s attempt to restore the ATP synthase machinery for the generation of ATP. We wish to add here that the overexpression of the ATP synthase operon in such an energy-compromised state appears to be a general response by the bacterium to survive, since a similar behavior was observed in wild-type M. smegmatis upon its treatment with a sublethal concentration of BDQ (Fig. S15B).
FIG 8.
The atpD-knockout strain shows modulation of major metabolic pathways. (A to C) The relative mRNA transcript levels of ATP synthase operon genes atpA, atpB, atpC, atpE, and atpG (A), MSMEG_3630 (B), dormancy operon genes dosS and dosR (C), (D and E) β-oxidation pathway genes fadA, fadB2, fadB3, fadB5, and echA19 (D), and central metabolic pathway enzyme genes pck, icl, and glcB (E), as assessed by RT-qPCR, are shown. In each panel, the transcript level in the atpD-knockout strain is compared with that in the wild type, which was used as a control. The experiments in each case were performed at least thrice. The error bars represent standard deviations. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.005; ****, P ≤ 0.001.
The transcriptional repressor for the β-lactam antibiotic resistance operon, BlaI, has also been shown to be involved in the regulation of ATP synthase operon expression in M. tuberculosis (43). A bioinformatics search for the M. tuberculosis BlaI homolog in M. smegmatis using the BLAST tool revealed MSMEG_3630, which shares 83% identity with the protein in question (data not shown). Therefore, we checked the expression level of the MSMEG_3630 gene and found it to be downregulated in the ΔatpD mutant (Fig. 8B), which further concurs with the finding of ATP synthase operon overexpression. Our data thus suggest that this is a general response of the cell to counteract the depletion of cellular ATP levels.
In addition, genes belonging to the dormancy operon, dosS and dosR (20), were also found to be overexpressed (Fig. 8C). It is likely that the perturbed NADH/NAD+ redox balance enhances the reducing state of respiratory complexes, thus activating dosS, which encodes a known sensor kinase and which is known to regulate the dormancy operon (20). We also monitored the expression of genes belonging to the lipid catabolism pathway. β oxidation serves as an essential pathway for fatty acid catabolism to generate energy (4). We found an upregulation of the genes involved in the β-oxidation pathway, such as fadA, fadB2, fadB3, fadB5, and echA19 (Fig. 8D). Additionally, a similar observation was made when wild-type M. smegmatis was treated with BDQ (Fig. S15C). We thus conclude that in an energy-compromised state with low cellular ATP levels, the β-oxidation pathway serves as a major catabolic process to generate the ATP that is required to maintain cellular viability. Here, interestingly, we saw an upregulation of the genes involved in the β-oxidation pathway even in the presence of glucose in the culture medium.
Furthermore, the upregulation of genes for central metabolic enzymes, viz., isocitrate lyase (icl), malate synthase (glcB), and phosphoenolpyruvate carboxykinase (pck), was also noticed in the M. smegmatis ΔatpD strain (Fig. 8E). The genes for these enzymes belong to the glyoxylate shunt, and their upregulation strongly indicates that M. smegmatis lacking atpD diverts the central metabolic pathway to reduce the burden on the ETC while carrying out the key metabolic processes.
DISCUSSION
All organisms use ATP as the energy currency for their growth and survival. Mycobacteria, including M. tuberculosis and M. smegmatis, are obligated to use the oxidative phosphorylation pathway to meet their ATP requirement, using the ATP synthase enzyme. In mycobacteria, FoF1 ATP synthase has been shown to meet the high energy demands (44, 45). Not surprisingly, M. tuberculosis bacteria grown on nonfermentable carbon sources are killed more rapidly than M. tuberculosis bacteria grown on fermentable carbon sources (20).
The essentiality of FoF1 ATP synthase has also been reported in other organisms. For example, the knockdown of ATP synthase expression in Staphylococcus aureus has been shown to strongly suppress the growth of the bacterium, which further confirms the crucial role of ATP synthase in bacterial growth and metabolism (46). A recent study in Candida albicans showed the significance of the β subunit of FoF1 ATP synthase, where it plays a crucial role during carbon metabolism and helps in Candida pathogenicity (47). Furthermore, in Saccharomyces cerevisiae, deletion of the F1 proteins of ATP synthase leads to a defect in oxidative phosphorylation, which does not allow the organism to grow on nonfermentable carbon sources (48).
Here, we attempted to understand the M. smegmatis metabolic profile by partially suppressing the functioning of ATP synthase. M. smegmatis is known to have two identical copies of the atpD gene, which codes for the β subunit, and a deletion of both copies has been shown to cause lethality in M. smegmatis (27). Here, we report that the genetic disruption of only one copy of atpD in M. smegmatis renders the bacterium in an energy-compromised state, wherein it exhibits phenotypic defects along with cell surface alterations. We observed that in such an energy-compromised state there was an overall decrease in ATP levels, higher NADH levels, enhanced ROS production, a decrease in oxygen consumption, and increased sensitivity toward oxidative stress and antibiotics. The ΔatpD mutant also exhibited growth defects, an atypical colony morphotype, and altered sliding motility and biofilm formation. Moreover, the glycolipid content in the cell envelope of the mutant strain was also less than that in the cell envelope of the WT and the complemented strains. These phenotypes have been shown to be associated with GPL-deficient strains (49).
Lowered intracellular energy levels result in bacterial metabolic pathway remodeling, which is essential for bacterial survival in an energy-compromised state. Indeed, we observed a higher expression of genes belonging to the ATP synthase operon, dormancy operon, and lipid catabolism and glyoxylate pathways in the M. smegmatis ΔatpD strain. Upregulation of the ATP synthase operon suggests the existence of a feedback mechanism that is triggered by the depletion of cellular ATP. Furthermore, the dormancy operon has been shown to be related to metabolic conditions that require a reduced amount of ATP and is thought to play a major role in maintaining bacterial ATP levels (20). It may also act as a defense mechanism to counteract the depletion of ATP levels within the cell. β oxidation acts as an essential pathway of fatty acid oxidation when energy supplies are low within the cell (4). The overexpression of β-oxidation pathway genes suggests that bacteria sense low ATP levels and switch to fatty acid oxidation for the supply of energy, even in the presence of glucose in the culture medium. Isocitrate lyase is the key enzyme in the glyoxylate pathway that is upregulated in an energy-compromised state (50). Here, the induction of isocitrate lyase indicates that the tricarboxylic acid (TCA) cycle is being bypassed most likely by means of the feedback inhibition of the TCA cycle because of the poor functioning of the ETC; such a bypass will therefore help in the maintenance of the energy pool. Overall, the metabolic changes observed in the ΔatpD strain of M. smegmatis reflect a general strategy to refashion its metabolic pathways to maintain the energy pool and ensure cell survival.
BDQ is the first-in-class ATP synthase inhibitor which displays high selectivity toward mycobacterial ATP synthase and therefore renders the bacterium low on energy. Furthermore, it has been reported that, upon BDQ treatment, major metabolic alterations occur in the cell and several biosynthetic processes are downregulated (19, 25). Hence, we were tempted to explore if the atpD-knockout strain mimics BDQ-treated wild-type M. smegmatis. Upon exposure to sublethal concentrations of BDQ, M. smegmatis exhibits reduced growth along with depletion of cellular ATP levels. This is found to be in accordance with a previous report which suggested that, upon treatment with BDQ, M. tuberculosis shows decreased growth along with a 10-fold decrease in ATP levels (20). Furthermore, our atpD-knockout strains largely mimicked the BDQ-treated wild-type M. smegmatis strain in its basic physiology, such as upregulation of the ATP operon, β-oxidation pathway, and oxidative stress response genes. We believe that such a comparison of the mycobacterial physiology observed in a low-energy state with that observed in the presence of an ATP synthase inhibitor will shed light on how physiological adaptation by employing alternative pathways occurs in mycobacteria. Furthermore, inhibition of such alternative metabolic pathways while targeting the ATP synthase in the form of a combinatorial therapy may be employed as a novel strategy to overcome mycobacterial infection.
Our understanding of M. smegmatis adaptation in an energy-compromised state will allow us to examine bacterial adaptation during stress. One such adaptive response greatly studied in mycobacteria is the stringent response, wherein the bacterium synthesizes unusual guanosine nucleotides, ppGpp and pppGpp (51, 52). During carbon starvation, M. smegmatis has been shown to accumulate these nucleotides and displays altered cellular phenotypes (53–55). It did not escape our notice that most of the phenotypic changes that have been reported during carbon starvation were also observed during the energy-compromised state that occurred upon atpD deletion. It remains to be seen, however, how the stringent response is related to the suboptimal functioning of FoF1 ATP synthase. The identification of such a novel mediator(s) involved in M. smegmatis in an altered physiological state will generate a more in-depth molecular understanding of the pathways mediating adaptation in M. smegmatis.
MATERIALS AND METHODS
The experimental methods regarding growth, protein expression analysis, colony morphology, ROS estimation, oxygen consumption, RNA extraction, and cDNA synthesis, etc., are detailed in the supplemental material. The oligonucleotides used for the generation of the gene knockout and its confirmation and in the RT-qPCR experiments are given in Table S1 in the supplemental material.
Bacterial strains, plasmids, reagents, media, and growth conditions.
Wild-type M. smegmatis mc2155 or its transformed versions were grown in Middlebrook 7H9 (MB7H9) broth (Difco) in the presence of 2% glucose and 0.05% Tween 80, which was supplemented with 50 μg/ml kanamycin (Sigma) or 100 μg/ml hygromycin (MP Biomedical) wherever required. Cultures were induced with 2% acetamide (wherever required) and further incubated at 37°C with constant shaking at 200 rpm. To monitor the growth of the different strains, the optical density of the culture at 600 nm (OD600) was measured every 3 h and plotted. Additionally, for spot assays, the required culture at each specified time point was placed on MB7H9 medium supplemented with 1.5% agar and 2% glucose, and the plate was further incubated at 37°C.
Genetic knockout construction and complementation.
One copy of the atpD gene in M. smegmatis mc2155 was replaced with a hygromycin resistance cassette (Hygr) by following the method described elsewhere (30, 56), which is detailed in the supplemental material. Knockout was confirmed by PCR using oligonucleotides specific for the gene sequence, as depicted in Fig. S1, followed by DNA sequencing of the amplified fragments. For complementation, the PCR-amplified atpD gene was cloned in an Escherichia coli-mycobacterium shuttle vector, pMVAcet, under the control of an acetamide-inducible promoter (31) and was introduced in both the wild-type and the atpD-deleted bacteria, as detailed in the supplemental material.
Measurement of intracellular ATP level and NADH/NAD+ ratio.
Cellular ATP levels were determined by using a BacTiter-Glo microbial cell viability assay kit (Promega) following the manufacturer’s instructions. The luminescence was measured on a SpectraMax M5 plate reader (Molecular Devices, Sunnyvale, CA). NADH/NAD+ ratios were measured using an NADH/NAD+ assay kit (catalog number MAK037; Sigma-Aldrich) following the manufacturer’s instructions. Total NAD (NAD and NADH) or NADH levels were quantified in a colorimetric assay at 450 nm on SpectraMax M5 plate reader.
Biofilm quantification and sliding motility assay.
For biofilm experiments, all the strains were first grown in MB7H9 medium supplemented with 2% glucose and 0.05% Tween 80 at 37°C until they reached stationary phase. Secondary cultures were then inoculated in MB7H9 medium, and the log-phase cultures were then used for biofilm formation. For biofilm formation, all bacterial strains were grown undisturbed in Sauton’s medium (HiMedia) in a 24-well plate supplemented with 2% glycerol and 2% acetamide, as described elsewhere (57, 58). The plates were wrapped with Parafilm and incubated at 37°C in a humidified chamber for 7 days. For biofilm quantification, the protocol was adapted as described previously (58), with some modifications. Briefly, the wells containing the biofilm were emptied, dried, and stained with 1% crystal violet. The wells were then rinsed with water and dried. The bound dye was solubilized in acetic acid, and the eluted stain was quantified by recording the absorbance at 595 nm. For the sliding motility assay, the protocol was adapted essentially as described previously (59). The degree of spread was determined by measuring the diameter of the growth zone.
Cellular aggregation and hydrophobicity index or bacterial adherence to hydrocarbons (BATH) assay.
The method for cellular aggregation was adapted as described previously (33). Briefly, the bacterial strains were grown in tryptone soy broth without Tween 80 and centrifuged to separate the single-cell mycobacteria from the aggregates. Further, for determining the cellular aggregation index, the bacterial strains were grown without Tween 80 in tryptone soy broth until saturation. One milliliter of the log-phase cell suspension (OD600, ∼0.8) was subjected to spectrophotometric analysis, and the aggregation was recorded at regular time intervals. Relative hydrophobicity was assessed by using the hexadecane partition procedure (33, 60), wherein the adherence of bacteria to hydrocarbons n-hexadecane and xylene is measured (33). The hydrophobicity was further assessed by calculating the reduction in the OD650 values. The hydrophobicity index was calculated as follows: [(A − B) × 100]/(A + B), where A represents the number of cells before the addition of hydrocarbons and B represents the number of cells in the aqueous phase after the addition of hydrocarbons.
Lipid extraction and analysis by TLC.
Glycopeptidolipids (GPLs) were extracted as described previously (49), with some modifications, as detailed in the supplemental material. Briefly, mycobacterial cells were grown in MB7H9 medium supplemented with 2% glucose and 0.05% Tween 80. GPLs were extracted by treating the cells with CHCl3, CH3OH, and 0.9% NaCl. The organic phase containing GPLs was recovered, and equal amounts were subjected to thin-layer chromatography (TLC), in which the plates were developed in CHCl3-CH3OH. The plates were allowed to dry, sprayed with orcinol-sulfuric acid, and charred until the color developed.
Nile red and ethidium bromide uptake assay.
Log-phase cultures (OD600, ∼0.6) of all the strains were harvested and washed with 1× phosphate-buffered saline–Tween 80. Two hundred microliters of the cell suspension was added to a 96-well plate, to which Nile red or ethidium bromide was added to final concentrations of 2 μM and 2 μg/ml, respectively (34, 61). Intracellular accumulation of these dyes was recorded by measuring the fluorescence on a SpectraMax M5 plate reader, with excitation and emission wavelengths of 540 and 630 nm, respectively, being used for Nile Red and 545 and 600 nm, respectively, being used for ethidium bromide.
SEM and TEM.
For scanning electron microscopy (SEM), the procedure was adapted as described previously (34, 53), with some modifications, as detailed in the supplemental material. Briefly, the bacterial cell pellet obtained from a log-phase culture was fixed in the required buffer, attached to stubs, sputter coated with gold, and imaged on an UltraPlus scanning electron microscope (Zeiss, Germany) at 10.0 kV. Transmission electron microscopy (TEM) was carried out as described previously (58), with some modifications, as detailed in the supplemental material. M. smegmatis cells fixed in sodium cacodylate buffer containing tannic acid were treated with osmium tetroxide, followed by embedding in molten agarose. After ultrasectioning, samples were stained with uranyl acetate and phosphotungstic acid and imaged on an FEI Talos 200S system equipped with a 200-kV field emission gun.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Abhishek A. Dubey for initial experiments with an atpD transposon mutant in M. smegmatis that nucleated the study. We also thank Lakshmi Dwivedi and Gayatri S. Nimbalkar for their help with the experiments. The pVV16 plasmid was obtained through BEI Resources, NIAID, NIH. pJV53 was a kind gift from Graham Hatfull, University of Pittsburgh. We also thank the DST-FIST facility at IISER, Bhopal, India, for the TEM imaging.
V.P. acknowledges the receipt of an INSPIRE senior research fellowship from the Department of Science and Technology (DST), Government of India. This work is supported in part by a grant [grant 27(0325)/17/EMR-II] from the Council of Scientific and Industrial Research, Government of India, and by intramural funds from IISER, Bhopal, India, to V.J.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00210-19.
REFERENCES
- 1.World Health Organization. 2018. Global tuberculosis report 2018. http://www.who.int/tb/publications/global_report/en/.
- 2.Barry CE III, Boshoff HI, Dartois V, Dick T, Ehrt S, Flynn J, Schnappinger D, Wilkinson RJ, Young D. 2009. The spectrum of latent tuberculosis: rethinking the biology and intervention strategies. Nat Rev Microbiol 7:845–855. doi: 10.1038/nrmicro2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boshoff HI, Barry CE III.. 2005. Tuberculosis—metabolism and respiration in the absence of growth. Nat Rev Microbiol 3:70–80. doi: 10.1038/nrmicro1065. [DOI] [PubMed] [Google Scholar]
- 4.Lee W, VanderVen BC, Fahey RJ, Russell DG. 2013. Intracellular Mycobacterium tuberculosis exploits host-derived fatty acids to limit metabolic stress. J Biol Chem 288:6788–6800. doi: 10.1074/jbc.M112.445056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Niederweis M. 2008. Nutrient acquisition by mycobacteria. Microbiology 154:679–692. doi: 10.1099/mic.0.2007/012872-0. [DOI] [PubMed] [Google Scholar]
- 6.Russell DG, VanderVen BC, Lee W, Abramovitch RB, Kim MJ, Homolka S, Niemann S, Rohde KH. 2010. Mycobacterium tuberculosis wears what it eats. Cell Host Microbe 8:68–76. doi: 10.1016/j.chom.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fujita Y, Matsuoka H, Hirooka K. 2007. Regulation of fatty acid metabolism in bacteria. Mol Microbiol 66:829–839. doi: 10.1111/j.1365-2958.2007.05947.x. [DOI] [PubMed] [Google Scholar]
- 8.Williams KJ, Boshoff HI, Krishnan N, Gonzales J, Schnappinger D, Robertson BD. 2011. The Mycobacterium tuberculosis beta-oxidation genes echA5 and fadB3 are dispensable for growth in vitro and in vivo. Tuberculosis (Edinb) 91:549–555. doi: 10.1016/j.tube.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jensen PR, Michelsen O. 1992. Carbon and energy metabolism of atp mutants of Escherichia coli. J Bacteriol 174:7635–7641. doi: 10.1128/jb.174.23.7635-7641.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pieters J. 2008. Mycobacterium tuberculosis and the macrophage: maintaining a balance. Cell Host Microbe 3:399–407. doi: 10.1016/j.chom.2008.05.006. [DOI] [PubMed] [Google Scholar]
- 11.Fujiwara N, Ohara N, Ogawa M, Maeda S, Naka T, Taniguchi H, Yamamoto S, Ayata M. 2015. Glycopeptidolipid of Mycobacterium smegmatis J15cs affects morphology and survival in host cells. PLoS One 10:e0126813. doi: 10.1371/journal.pone.0126813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Russell DG, Mwandumba HC, Rhoades EE. 2002. Mycobacterium and the coat of many lipids. J Cell Biol 158:421–426. doi: 10.1083/jcb.200205034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schorey JS, Sweet L. 2008. The mycobacterial glycopeptidolipids: structure, function, and their role in pathogenesis. Glycobiology 18:832–841. doi: 10.1093/glycob/cwn076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang L, Slayden RA, Barry CE III, Liu J. 2000. Cell wall structure of a mutant of Mycobacterium smegmatis defective in the biosynthesis of mycolic acids. J Biol Chem 275:7224–7229. doi: 10.1074/jbc.275.10.7224. [DOI] [PubMed] [Google Scholar]
- 15.Freeman R, Geier H, Weigel KM, Do J, Ford TE, Cangelosi GA. 2006. Roles for cell wall glycopeptidolipid in surface adherence and planktonic dispersal of Mycobacterium avium. Appl Environ Microbiol 72:7554–7558. doi: 10.1128/AEM.01633-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ortalo-Magne A, Lemassu A, Laneelle MA, Bardou F, Silve G, Gounon P, Marchal G, Daffe M. 1996. Identification of the surface-exposed lipids on the cell envelopes of Mycobacterium tuberculosis and other mycobacterial species. J Bacteriol 178:456–461. doi: 10.1128/jb.178.2.456-461.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Favrot L, Ronning DR. 2012. Targeting the mycobacterial envelope for tuberculosis drug development. Expert Rev Anti Infect Ther 10:1023–1036. doi: 10.1586/eri.12.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bald D, Koul A. 2010. Respiratory ATP synthesis: the new generation of mycobacterial drug targets? FEMS Microbiol Lett 308:1–7. doi: 10.1111/j.1574-6968.2010.01959.x. [DOI] [PubMed] [Google Scholar]
- 19.Bald D, Villellas C, Lu P, Koul A. 2017. Targeting energy metabolism in Mycobacterium tuberculosis, a new paradigm in antimycobacterial drug discovery. mBio 8:e00272-17. doi: 10.1128/mBio.00272-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koul A, Vranckx L, Dhar N, Gohlmann HW, Ozdemir E, Neefs JM, Schulz M, Lu P, Mortz E, McKinney JD, Andries K, Bald D. 2014. Delayed bactericidal response of Mycobacterium tuberculosis to bedaquiline involves remodelling of bacterial metabolism. Nat Commun 5:3369. doi: 10.1038/ncomms4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kalia NP, Hasenoehrl EJ, Ab Rahman NB, Koh VH, Ang MLT, Sajorda DR, Hards K, Gruber G, Alonso S, Cook GM, Berney M, Pethe K. 2017. Exploiting the synthetic lethality between terminal respiratory oxidases to kill Mycobacterium tuberculosis and clear host infection. Proc Natl Acad Sci U S A 114:7426–7431. doi: 10.1073/pnas.1706139114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lamprecht DA, Finin PM, Rahman MA, Cumming BM, Russell SL, Jonnala SR, Adamson JH, Steyn AJ. 2016. Turning the respiratory flexibility of Mycobacterium tuberculosis against itself. Nat Commun 7:12393. doi: 10.1038/ncomms12393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rao SP, Alonso S, Rand L, Dick T, Pethe K. 2008. The protonmotive force is required for maintaining ATP homeostasis and viability of hypoxic, nonreplicating Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 105:11945–11950. doi: 10.1073/pnas.0711697105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hards K, Robson JR, Berney M, Shaw L, Bald D, Koul A, Andries K, Cook GM. 2015. Bactericidal mode of action of bedaquiline. J Antimicrob Chemother 70:2028–2037. doi: 10.1093/jac/dkv054. [DOI] [PubMed] [Google Scholar]
- 25.Iqbal IK, Bajeli S, Akela AK, Kumar A. 2018. Bioenergetics of Mycobacterium: an emerging landscape for drug discovery. Pathogens 7:E24. doi: 10.3390/pathogens7010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koul A, Dendouga N, Vergauwen K, Molenberghs B, Vranckx L, Willebrords R, Ristic Z, Lill H, Dorange I, Guillemont J, Bald D, Andries K. 2007. Diarylquinolines target subunit c of mycobacterial ATP synthase. Nat Chem Biol 3:323–324. doi: 10.1038/nchembio884. [DOI] [PubMed] [Google Scholar]
- 27.Tran SL, Cook GM. 2005. The F1Fo-ATP synthase of Mycobacterium smegmatis is essential for growth. J Bacteriol 187:5023–5028. doi: 10.1128/JB.187.14.5023-5028.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu P, Lill H, Bald D. 2014. ATP synthase in mycobacteria: special features and implications for a function as drug target. Biochim Biophys Acta 1837:1208–1218. doi: 10.1016/j.bbabio.2014.01.022. [DOI] [PubMed] [Google Scholar]
- 29.van Kessel JC, Hatfull GF. 2007. Recombineering in Mycobacterium tuberculosis. Nat Methods 4:147–152. doi: 10.1038/nmeth996. [DOI] [PubMed] [Google Scholar]
- 30.Dubey AA, Wani SR, Jain V. 2018. Methylotrophy in mycobacteria: dissection of the methanol metabolism pathway in Mycobacterium smegmatis. J Bacteriol 200:e00288-18. doi: 10.1128/JB.00288-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pohane AA, Joshi H, Jain V. 2014. Molecular dissection of phage endolysin: an interdomain interaction confers host specificity in lysin A of Mycobacterium phage D29. J Biol Chem 289:12085–12095. doi: 10.1074/jbc.M113.529594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Recht J, Martinez A, Torello S, Kolter R. 2000. Genetic analysis of sliding motility in Mycobacterium smegmatis. J Bacteriol 182:4348–4351. doi: 10.1128/JB.182.15.4348-4351.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Etienne G, Astarie-Dequeker C, Dupont M-A, Daffé M, Villeneuve C, Billman-Jacobe H. 2002. The impact of the absence of glycopeptidolipids on the ultrastructure, cell surface and cell wall properties, and phagocytosis of Mycobacterium smegmatis. Microbiology 148:3089–3100. doi: 10.1099/00221287-148-10-3089. [DOI] [PubMed] [Google Scholar]
- 34.Li QM, Zhou ML, Fan XY, Yan JL, Li WM, Xie JP. 2016. Mycobacteriophage SWU1 gp39 can potentiate multiple antibiotics against Mycobacterium via altering the cell wall permeability. Sci Rep 6:28701. doi: 10.1038/srep28701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miyamoto Y, Mukai T, Takeshita F, Nakata N, Maeda Y, Kai M, Makino M. 2004. Aggregation of mycobacteria caused by disruption of fibronectin-attachment protein-encoding gene. FEMS Microbiol Lett 236:227–234. doi: 10.1111/j.1574-6968.2004.tb09651.x. [DOI] [PubMed] [Google Scholar]
- 36.Recht J, Kolter R. 2001. Glycopeptidolipid acetylation affects sliding motility and biofilm formation in Mycobacterium smegmatis. J Bacteriol 183:5718–5724. doi: 10.1128/JB.183.19.5718-5724.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hsu FF, Turk J. 1999. Structural characterization of triacylglycerols as lithiated adducts by electrospray ionization mass spectrometry using low-energy collisionally activated dissociation on a triple stage quadrupole instrument. J Am Soc Mass Spectrom 10:587–599. doi: 10.1016/S1044-0305(99)00035-5. [DOI] [PubMed] [Google Scholar]
- 38.Tatituri RV, Brenner MB, Turk J, Hsu FF. 2012. Structural elucidation of diglycosyl diacylglycerol and monoglycosyl diacylglycerol from Streptococcus pneumoniae by multiple-stage linear ion-trap mass spectrometry with electrospray ionization. J Mass Spectrom 47:115–123. doi: 10.1002/jms.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Singh P, Rao RN, Reddy JR, Prasad RB, Kotturu SK, Ghosh S, Mukhopadhyay S. 2016. PE11, a PE/PPE family protein of Mycobacterium tuberculosis is involved in cell wall remodeling and virulence. Sci Rep 6:21624. doi: 10.1038/srep21624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gomez A, Andreu N, Ferrer-Navarro M, Yero D, Gibert I. 2016. Triclosan-induced genes Rv1686c-Rv1687c and Rv3161c are not involved in triclosan resistance in Mycobacterium tuberculosis. Sci Rep 6:26221. doi: 10.1038/srep26221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McMurry LM, McDermott PF, Levy SB. 1999. Genetic evidence that InhA of Mycobacterium smegmatis is a target for triclosan. Antimicrob Agents Chemother 43:711–713. doi: 10.1128/AAC.43.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu P, Heineke MH, Koul A, Andries K, Cook GM, Lill H, van Spanning R, Bald D. 2015. The cytochrome bd-type quinol oxidase is important for survival of Mycobacterium smegmatis under peroxide and antibiotic-induced stress. Sci Rep 5:10333. doi: 10.1038/srep10333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sala C, Haouz A, Saul FA, Miras I, Rosenkrands I, Alzari PM, Cole ST. 2009. Genome-wide regulon and crystal structure of BlaI (Rv1846c) from Mycobacterium tuberculosis. Mol Microbiol 71:1102–1116. doi: 10.1111/j.1365-2958.2008.06583.x. [DOI] [PubMed] [Google Scholar]
- 44.Cox RA, Cook GM. 2007. Growth regulation in the mycobacterial cell. Curr Mol Med 7:231–245. doi: 10.2174/156652407780598584. [DOI] [PubMed] [Google Scholar]
- 45.Haagsma AC, Driessen NN, Hahn MM, Lill H, Bald D. 2010. ATP synthase in slow- and fast-growing mycobacteria is active in ATP synthesis and blocked in ATP hydrolysis direction. FEMS Microbiol Lett 313:68–74. doi: 10.1111/j.1574-6968.2010.02123.x. [DOI] [PubMed] [Google Scholar]
- 46.Balemans W, Vranckx L, Lounis N, Pop O, Guillemont J, Vergauwen K, Mol S, Gilissen R, Motte M, Lancois D, De Bolle M, Bonroy K, Lill H, Andries K, Bald D, Koul A. 2012. Novel antibiotics targeting respiratory ATP synthesis in Gram-positive pathogenic bacteria. Antimicrob Agents Chemother 56:4131–4139. doi: 10.1128/AAC.00273-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li SX, Wu HT, Liu YT, Jiang YY, Zhang YS, Liu WD, Zhu KJ, Li DM, Zhang H. 2018. The F1Fo-ATP synthase beta subunit is required for Candida albicans pathogenicity due to its role in carbon flexibility. Front Microbiol 9:1025. doi: 10.3389/fmicb.2018.01025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lai-Zhang J, Xiao Y, Mueller DM. 1999. Epistatic interactions of deletion mutants in the genes encoding the F1-ATPase in yeast Saccharomyces cerevisiae. EMBO J 18:58–64. doi: 10.1093/emboj/18.1.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tatham E, Sundaram Chavadi S, Mohandas P, Edupuganti UR, Angala SK, Chatterjee D, Quadri LE. 2012. Production of mycobacterial cell wall glycopeptidolipids requires a member of the MbtH-like protein family. BMC Microbiol 12:118. doi: 10.1186/1471-2180-12-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nandakumar M, Nathan C, Rhee KY. 2014. Isocitrate lyase mediates broad antibiotic tolerance in Mycobacterium tuberculosis. Nat Commun 5:4306. doi: 10.1038/ncomms5306. [DOI] [PubMed] [Google Scholar]
- 51.Chatterji D, Ojha AK. 2001. Revisiting the stringent response, ppGpp and starvation signaling. Curr Opin Microbiol 4:160–165. doi: 10.1016/S1369-5274(00)00182-X. [DOI] [PubMed] [Google Scholar]
- 52.Jain V, Kumar M, Chatterji D. 2006. ppGpp: stringent response and survival. J Microbiol 44:1–10. [PubMed] [Google Scholar]
- 53.Gupta KR, Baloni P, Indi SS, Chatterji D. 2016. Regulation of growth, cell shape, cell division, and gene expression by second messengers (p)ppGpp and cyclic di-GMP in Mycobacterium smegmatis. J Bacteriol 198:1414–1422. doi: 10.1128/JB.00126-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ojha AK, Mukherjee TK, Chatterji D. 2000. High intracellular level of guanosine tetraphosphate in Mycobacterium smegmatis changes the morphology of the bacterium. Infect Immun 68:4084–4091. doi: 10.1128/IAI.68.7.4084-4091.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ojha AK, Varma S, Chatterji D. 2002. Synthesis of an unusual polar glycopeptidolipid in glucose-limited culture of Mycobacterium smegmatis. Microbiology 148:3039–3048. doi: 10.1099/00221287-148-10-3039. [DOI] [PubMed] [Google Scholar]
- 56.van Kessel JC, Marinelli LJ, Hatfull GF. 2008. Recombineering mycobacteria and their phages. Nat Rev Microbiol 6:851–857. doi: 10.1038/nrmicro2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Anand A, Verma P, Singh AK, Kaushik S, Pandey R, Shi C, Kaur H, Chawla M, Elechalawar CK, Kumar D, Yang Y, Bhavesh NS, Banerjee R, Dash D, Singh A, Natarajan VT, Ojha AK, Aldrich CC, Gokhale RS. 2015. Polyketide quinones are alternate intermediate electron carriers during mycobacterial respiration in oxygen-deficient niches. Mol Cell 60:637–650. doi: 10.1016/j.molcel.2015.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gupta KR, Kasetty S, Chatterji D. 2015. Novel functions of (p)ppGpp and cyclic di-GMP in mycobacterial physiology revealed by phenotype microarray analysis of wild-type and isogenic strains of Mycobacterium smegmatis. Appl Environ Microbiol 81:2571–2578. doi: 10.1128/AEM.03999-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zanfardino A, Migliardi A, D’Alonzo D, Lombardi A, Varcamonti M, Cordone A. 2016. Inactivation of MSMEG_0412 gene drastically affects surface related properties of Mycobacterium smegmatis. BMC Microbiol 16:267. doi: 10.1186/s12866-016-0888-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rosenberg M, Gutnick D, Rosenberg E. 1980. Adherence of bacteria to hydrocarbons—a simple method for measuring cell-surface hydrophobicity. FEMS Microbiol Lett 9:29–33. doi: 10.1111/j.1574-6968.1980.tb05599.x. [DOI] [Google Scholar]
- 61.Yu J, Tran V, Li M, Huang X, Niu C, Wang D, Zhu J, Wang J, Gao Q, Liu J. 2012. Both phthiocerol dimycocerosates and phenolic glycolipids are required for virulence of Mycobacterium marinum. Infect Immun 80:1381–1389. doi: 10.1128/IAI.06370-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.